Abstract
During the second half of embryogenesis, the ellipsoidal Caenorhabditis elegans embryo elongates into a long, thin worm. This elongation requires a highly organized cytoskeleton composed of actin microfilaments, microtubules and intermediate filaments throughout the epidermis of the embryo. This architecture allows the embryonic epidermal cells to undergo a smooth muscle-like actin/myosin-based contraction that is redundantly controlled by LET- 502/Rho kinase and MEL-11/myosin phosphatase in one pathway and FEM-2/PP2c phosphatase and PAK-1/p21-activated kinase in a parallel pathway(s). Although actin microfilaments surround the embryo, the force for contraction is generated mainly in the lateral (seam) epidermal cells whose actin microfilaments appear qualitatively different from those in their dorsal/ventral neighbors. We have identified FHOD-1, a formin family actin nucleator, which acts in the lateral epidermis. fhod-1 mutants show microfilament defects in the embryonic lateral epidermal cells and FHOD-1 protein is detected only in those cells. fhod-1 genetic interactions with let-502, mel-11, fem-2 and pak-1 indicate that fhod-1 preferentially regulates those microfilaments acting with let-502 and mel-11, and in parallel to fem-2 and pak-1. Thus, FHOD-1 may contribute to the qualitative differences in microfilaments found in the contractile lateral epidermal cells and their non-contractile dorsal and ventral neighbors. Different microfilament populations may be involved in the different contractile pathways.
Keywords: C. elegans, morphogenesis, actin, formin, cytoskeleton, embryo, genetics
Introduction
Morphogenesis is the process by which the organism attains its three dimensional body form. In multicellular organisms, this process can involve organization of many different cell types and concerted changes in cell-cell adhesion, cellular migration and cell size and shape. For proper morphogenesis, an organism must coordinate changes in multiple cell types (or tissues), each of which must be at the right location at the right time and must also be receptive to morphogenetic signals.
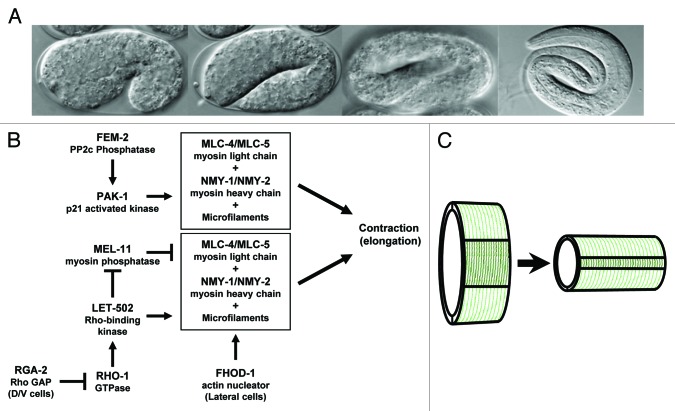
During C. elegans embryonic morphogenesis, an ellipsoidal ball of cells is converted to the vermiform shape characteristic of nematodes. This morphogenetic event, which results in a 4-fold lengthening of the embryo (Fig. 1A), takes place without a change in cell number or cell size and is initially driven by contraction of a single cell layer that surrounds the embryo, the epidermis (also known as hypodermis in nematodes).1-3 Early elongation is driven by an actin/myosin-based contraction of a network of circumferential epidermal microfilaments that is homologous to vertebrate smooth muscle contraction.4,5 Phosphorylation of MLC-4/regulatory myosin light chain triggers its assembly with the non-muscle myosins NMY-1/2, resulting in contraction6-9 (Fig. 1B). Prior to elongation, contraction is blocked by myosin light chain dephosphorylation by myosin phosphatase (of which MEL-11 is the C. elegans MYPT targeting subunit9,10). At the appropriate time, RHO-1 GTPase activates Rho kinase/LET-502 to phosphorylate both the regulatory light chain (MLC-4) to activate myosin, and MEL-11 to inhibit myosin phosphatase, which would otherwise continue to dephosphorylate MLC-4 and block contraction.4,6,11 After this initial phase of elongation to the 2-fold stage (twice the length of the eggshell), muscle function mediates further lengthening of the embryo.3,12,13
Figure 1. Diagram of C. elegans embryonic elongation. (A) DIC photographs illustrate the 1.2-fold (fold embryo length relative to the egg shell), 2-fold, 3-fold and 4-fold stages of elongation. (B) The proposed regulatory interactions during elongation are based on C. elegans genetic interactions and analogy to vertebrate smooth muscle contraction. Phosphorylation of MLC-4 results in contraction in conjunction with NMY-1/NMY-2 and actin microfilaments. MEL-11 dephosphorylates MLC-4, blocking contraction, until LET-502 is activated by RHO-1 to both inhibit MEL-11 and to phosphorylate MLC-4. RGA-2 blocks RHO-1 activation in the dorsal and ventral cells. FHOD-1 regulates microfilaments used in this pathway in lateral cells. PAK-1 and FEM-2 activate a parallel contractile actin-myosin contractile cassette that does not depend on FHOD-1. (C) The cartoon illustrates the contraction of the microfilaments (dark green) in the lateral epidermal cells, which lengthens the embryo. Microfilaments in the dorsal and ventral epidermis (light green) remain passive.
The let-502 mutant phenotype (as well as the phenotype upon simultaneous loss of nmy-1 and nmy-2) is failure to elongate, with arrest at the 2-fold stage or sooner, depending on the severity of the allele.6,14 Conversely, loss of mel-11, the inhibitor of contraction, has the opposite phenotype: mel-11 mutants result in a hypercontraction and rupture of the embryo. When let-502 and mel-11 are both lost, a redundant pathway mediates contraction.14 In vertebrate smooth muscle, this redundant pathway is activated by Calmodulin and myosin light chain kinase.5 However, in C. elegans, there is no obvious myosin light chain kinase homolog.15 The C. elegans redundant pathway includes FEM-2 (PP2c phosphatase), mutations of which genetically enhance and suppress mutations of let-502 and mel-11, respectively. Although fem-2 mutations alone show only weak elongation defects, no elongation occurs in let-502; mel-11; fem-2 triple mutants because both redundant pathways are inactive.14 PAK-1/p21 kinase and MRCK-1/myotonic dystrophy kinase-related Cdc42-binding kinase both show elongation defects and enhance let-502,7 although unlike fem-2 and let-502, they do not show genetic interactions with mel-11.
Although circumferential actin is found throughout the epidermis during early elongation, the contractile force that drives elongation occurs primarily in the lateral (seam) cells with actin cables in the dorsal/ventral cells acting as passive struts.2,7,13 Part of this difference between the contractile lateral cells and their neighbors is explained by differential activity of the inhibitory Rho GTPase-activating protein RGA-2 in the non-contractile dorsal and ventral cells.13 In addition, zygotic LET-502 and MEL-11 are differentially expressed, with LET-502 expression being relatively higher in the lateral vs. dorsal/ventral cells and MEL-11 showing the reciprocal pattern.16 The actin microfilament network in the contractile lateral cells is qualitatively different from that in their dorsal/ventral neighbors. Unlike the dorsal and ventral cells, circumferential actin filaments in the lateral cells are not associated with the apical membrane. Lateral cells form parallel microfilament bundles later than their neighbors, with microfilaments initially organized as a meshwork before coalescing into a circumferential orientation as contraction proceeds.2,7 Here we describe a new player, the formin family member FHOD-1, which may be responsible for nucleating the differing actin network in the contractile lateral cells.
Nucleation of actin is the rate-limiting step of actin filament assembly.4,17 Members of the formin family nucleate unbranched microfilaments. Formins stabilize the initial actin dimer, recruit profilin, an actin monomer binding protein and then remain bound to the growing (+) end of the filament and prevent capping proteins from terminating filament extension. The resultant long, unbranched actin filaments can be bundled into stress fibers. Formins are involved in multiple cellular processes,17,18 including cytokinesis, cell movement and changes in cell shape. The mammalian genome encodes 15 formins while the C. elegans genome encodes seven.
Formin homology domain-containing proteins (FHODs) constitute one of the formin sub-families. FHOD proteins have characteristic proline-rich FH1 domains that bind to profilin.19 The FH2 domain is involved in formin homodimerization, forming a ring that is involved in the nucleation/processive capping of actin. Like many formins, mammalian FHOD1 is initially inactive due to an autoinhibitory interaction between the formin’s C-terminal diaphanous autoregulatory domain (DAD) with the N-terminal diaphanous inhibitory domain (DID, sometimes referred to as FH3). For FHOD1, autoinhibition can be relieved by the phosphorylation of the C terminus by Rho-kinase.20-23 FHOD1 also contains an N-terminal GTPase-binding domain (GBD), which binds Rac in a GTP-independent manner. This Rac binding may regulate localization rather than activity.20,24 Depending upon the cell line, transfection of constitutively active FHOD1 constructs that lack the autoinhibitory domains (N- or C-terminal) produces actin stress fibers that can either elongate the cells,20,25 form filopodia or enhance cell migration.26 Constitutively active FHOD1 can localize along the length of microfilaments and microtubules to organize these two cytoskeletal components into parallel fibers, but again, only in certain cell lines.25 In addition, mammalian FHOD3 acts to regulate myofilament organization in cardiomyocytes.23,27-29
fhod-1 encodes the only C. elegans FHOD homolog, and FHOD-1 is highly similar to both human FHOD1 and FHOD330 (BLAST E value scores of 10−173). FHOD-1 has recently been shown to regulate contractile lattice growth and maintenance in C. elegans striated muscle.30 Here we show that FHOD-1 is specifically expressed in the lateral epidermal cells that drive embryonic elongation and fhod-1 loss is associated with a defect in formation of stress fibers in those cells. Genetic interactions indicate that fhod-1 acts downstream of let-502 and mel-11 while acting in parallel to contraction driven by pak-1 and fem-2. This implies that the parallel let-502/mel-11 and pak-1/fem-2 pathways could act on different sets of microfilaments. Previous work showed that fem-2 and pak-1 each acts in parallel to let-502 and mel-11; however, while fem-2 suppresses mel-11,14pak-1 does not.7 Here we show that fem-2 and pak-1 act in the same elongation pathway, but fem-2 suppression of mel-11 requires pak-1(+).
Results
RNAi screen for genes affecting elongation
A targeted RNAi screen to find mediators of elongation was performed on worms bearing let-502(sb118) (Rho kinase, which activates contraction) or mel-11(it26) (myosin phosphatase, which inhibits contraction). Both mutations are ts, and at appropriate temperatures, the strains are sensitized to relatively small changes in other pathway components and so genetic interactions can be detected by changes in hatching rates. By looking for genes that act reciprocally with let-502 and mel-11, that is, that enhance (exacerbate) the phenotype of one mutation and suppress (alleviate) that of the other, we can have more confidence that the genetic interactions are specific for the elongation pathway. Knockdown of a gene potentiating contraction should enhance the let-502 phenotype while suppressing mel-11, while knockdown of genes inhibiting elongation would show the opposite pattern. The ts allele let-502(sb118) behaves as a strong hypomorph for let-502’s role in elongation at the restrictive temperature of 25°C (Materials and Methods). The mutation in allele sb118 appears to uncouple the elongation functions of let-502 from its other roles in embryonic cytokinesis and in the uterine contractions necessary for egg laying. mel-11 mutants have a phenotype opposite that of let-502, with the lateral epidermal cells hypercontracting during elongation and embryos rupturing before the 2-fold stage due to the strain of contraction on the adherens junctions. A ts allele mel-11(it26) is 100% embryonic lethal at 25°C (behaving as a genetic null at this temperature for this phenotype), has very few escapers (1–2%) at 20°C (strongly hypomorphic) and only 18% survival at 15°C.16,31
We tested the role of formin protein family members in nucleating the unbranched microfilaments that are involved in elongation. There are seven formin-encoding genes in C. elegans. Whereas daam-1, inft-1, cyk-1 and fozi-1 all enhanced lethality of both let-502 and mel-11 using RNAi feeding clones (data not shown), only fhod-1 showed reciprocal interactions with let-502 and mel-11. fhod-1(RNAi) feeding enhanced let-502(ts), decreasing the number of progeny reaching adulthood from 10% to 3% at 25°C. In contrast, fhod-1(RNAi) feeding suppressed mel-11(ts), increasing hatching at 20°C from 3% to 10% (injection of dsRNA increased hatching to 20%). fhod-1(RNAi) by feeding or injection had no effect on wild-type worms.
We also tested RNAi or mutations of genes involved in planar cell polarity and Wnt signaling, which interact with Drosophila Rho kinase,32 and the Rac pathway, which we have previously implicated in elongation.16 However, we found none that resulted in reciprocal enhancement/suppression of mel-11 and let-502 (genes tested included dsh-1, dsh-2, bar-1, hmp-2, mom-5, rac-2 and ced-12).
fhod-1 elongation phenotypes
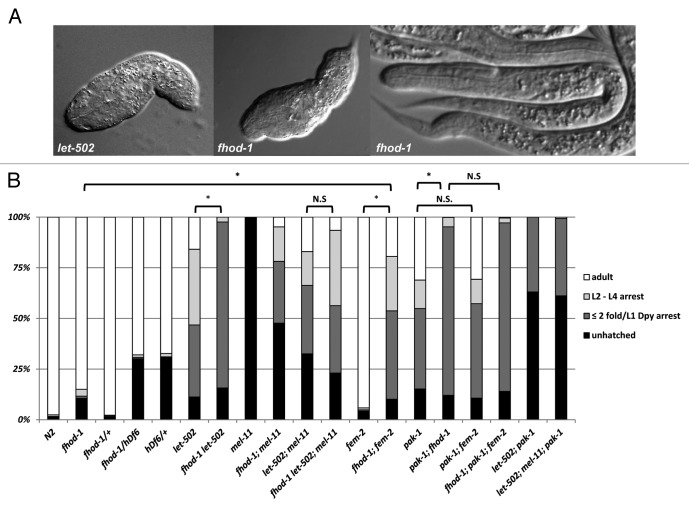
To avoid the caveats associated with incomplete knockdown by RNAi, we obtained fhod-1(tm2363) from the National Bioresources Project.33 This allele is predicted to delete part of the FH2 domain, leading to a frameshift and a premature stop, which likely leads to a null mutation (Fig. 2). fhod-1(tm2363) is homozygous viable with 85% of embryos surviving to adulthood at 25°C. Of the small number (~1%) of animals that had arrested development after hatching, all exhibited an unelongated phenotype similar to strong let-502 alleles (Fig. 3), whereas the majority of arrested development occurred prior to hatching. When developing fhod-1 embryos were examined microscopically to determine if there were additional early phenotypes, we found that only 2/73 arrested before the onset morphogenesis (0/61 wild-type embryos arrested before elongation). Of 44 embryos examined by time-lapse microscopy, two arrested pre-morphogenesis, one ceased elongation at the 1.2-fold stage, three arrested at the 1.5-fold stage and eight arrested at the 2-fold stage. Arrested embryos had differentiated tissue (e.g., pharynx and gut, Fig. 3A) indicating that embryos had not simply died upon reaching this stage of elongation but rather continued other aspects of development. The embryonic and larval arrest phenotypes were recessive. When fhod-1(tm2363) was in trans to a deficiency (hDf6), embryonic lethality did not increase beyond that observed for the deficiency alone (Fig. 3B). Furthermore, the proportion of animals hatching with elongation defects was not increased, in contrast to what was seen when fhod-1 was combined with other elongation pathway mutants (see below). These data suggest that this allele behaves as a strong loss of function or null allele.
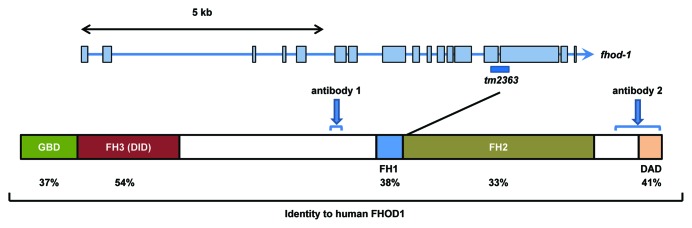
Figure 2. Diagram of the fhod-1 locus. Exons are shown as blue boxes and amino acid similarities to corresponding domains of human FHOD1 are indicated. Domain assignments are based on those of Shulte et al.24 for human FHOD1. tm2363 results in a C-terminal truncation beginning at the position indicated. Blue arrows denote regions to which antisera were raised.
Figure 3.fhod-1 phenotypes and genetic interactions. (A) A low percentage of fhod-1 embryos arrest with limited elongation similar to that seen with the strong dominant-negative allele of let-502(ca201) but most fhod-1 embryos hatch into normal L1 larvae. Note that these elongation-arrested larvae have differentiated gut and pharynx and so did not simply die during elongation. The let-502(sb118) allele used in part (B) is hypomorphic (see Materials and Methods) and arrests slightly later (≥ 2-fold). (B) Distribution of elongation phenotypes seen for indicated mutant backgrounds. See text for interpretations and Materials and Methods for alleles. Animals with ≤ 2-fold arrest are combined with the ≥ 2-fold L1 arrest that were also Dpy and lumpy (“≤ 2 fold/L1 Dpy”). n > 400 for each genotype and all experiments were done at 25°C. Statistical tests were performed by pooling those categories corresponding to the strongest elongation defects, namely unhatched embryos (most of which arrested during morphogenesis) plus hatched and arrested at ≤ 2-fold/L1 Dpy. For the differences indicated as significant (*), p < 10−5, while for those indicated as not significant (n.s.), p > 0.05 (see Materials and Methods).
Genetic interactions of fhod-1 with mutants of other elongation pathway genes
Like fhod-1(RNAi), fhod-1(tm2363) (hereafter referred to as simply “fhod-1”) showed reciprocal genetic interactions with let-502 and mel-11. fhod-1 enhanced the elongation phenotype of the hypomorphic let-502(ts) allele, dramatically increasing the proportion of hatched larvae with severe elongation defects from 36% to 82% at the restrictive temperature of 25° (Fig. 3B). These included those arrested at ≤ 2-fold and arrested L1 stage animals with a lumpy and Dpy appearance (most escapers that arrested at later larval stages were also lumpy Dpy). None of the double mutants grew to adulthood. This range of phenotypes is typical of genotypes where elongation is partially compromised.6,14 The increase in unhatched plus hatched with ≤ 2-fold/L1 Dpy arrest was highly significant (p < 10−5) (see Materials and Methods) when comparing let-502(ts) and fhod-1 let-502(ts). Conversely, fhod-1 suppressed mel-11(ts), increasing hatching from 0% to 52%. Most hatchlings arrested as larva that had undergone little elongation, but 5% did survive to adulthood (Fig. 3B).
To determine how fhod-1 acts genetically during morphogenesis, we combined fhod-1 with mutations that eliminate the let-502/mel-11 branch of the elongation pathway (as in let-502; mel-11 double mutants). fhod-1 should not enhance other mutants in the same pathway. That is, once a pathway is shut down, adding other lesions in the same pathway is irrelevant. Indeed, we found that addition of fhod-1 to the let-502; mel-11 background had at best modest effects as differences in progeny total arrested by 2-fold were not statistically different between the double and triple mutants. In contrast, 100% progeny arrested before the 2-fold stage when either fem-214 or pak-17 are added to the let-502; mel-11.
The mild effect of fhod-1(null) on let-502; mel-11 suggests that fhod-1 functions primarily in the let-502/mel-11 branch of the elongation pathway. In agreement with this conclusion, the fhod-1 mutant did have strong effects on fem-2, increasing the number of hatched larva arresting with ≤ 2-fold elongation/L1 Dpy arrest from ~1% in each of the parents to 44% in the double, resulting in a highly significant (p < 10−5) enhancement of the total percentage of progeny arrested prior to 2-fold (Fig. 3B). This strong interaction between fhod-1 and fem-2 again suggests fhod-1 acts primarily in the let-502/mel-11 branch of the elongation pathway, in parallel to fem-2. Finally, enhancement of fhod-1 by let-502 or fem-2 was largely expressed as an increase in animals with elongation defects rather than pre-elongation defects: both fhod-1 let-502 and fhod-1; fem-2 double mutants each resulted in a low proportion of embryos that arrested prior to elongation (2/47 and 4/68, respectively) that was comparable to the levels seen in the single mutants. Together, these argue against a mechanism, whereby these mutant combinations induce novel phenotypes unrelated to elongation.
pak-1, like fem-2, also acts in parallel with let-502/mel-11, suggesting pak-1 should act in parallel to fhod-1. Hatching animals from the pak-1(ok448) null are slightly shorter than wild-type and substantial numbers arrest as L1’s.7,34 All arrested animals, as well as many of those that grew to later stages, had the characteristic lumpy and Dpy appearance commonly seen when elongation is partially compromised.6,14 Unhatched plus ≤ 2-fold elongation/L1 Dpy arrested larvae increased from 55% to 95% when pak-1 was combined with fhod-1 (significant at p < 10−5) (Fig. 3B). Unlike the parental strains, no animals reached adulthood. The action of both pak-1 and fem-2 in parallel to both let-502/mel-11 and fhod-1 suggests that pak-1 and fem-2 act in the same branch of the elongation pathway. Indeed, fem-2 did not enhance pak-1 (Fig. 3B), nor did addition of fem-2 further enhance fhod-1; pak-1, reinforcing the interpretation that fem-2 and pak-1 act together and in parallel to fhod-1.
The genetic interactions of fem-2 and pak-1 during elongation are similar, but there are notable differences. As previously reported,7 and as shown in Figure 3B, pak-1 and fem-2 each enhance let-502 and block elongation of let-502; mel-11.14 However, as expected for the loss of an activator of elongation, fem-2 suppresses mel-11 hypercontraction14 but pak-1 does not.7 We examined this further at different temperatures, under conditions where suppression by fem-2 was readily apparent, but were unable to detect any suppression of mel-11 by pak-1 (Table 1). The fact that fem-2, but not pak-1, can suppress mel-11 hypercontraction is unexpected since elongation defects of pak-1 single mutants are much more penetrant than those of fem-2 (Fig. 3B). Finally, we noted that no suppression of mel-11 was present in mel-11; fem-2; pak-1 triple mutants, indicating that pak-1(+) is required for fem-2-mediated suppression of mel-11 (Table 1, see Discussion).
Table 1.pak-1 blocks suppression of mel-11 lethality by fem-2.
| Percent hatching | ||
|---|---|---|
| |
20°C |
25°C |
|
mel-11 |
1 |
0 |
|
mel-11; fem-2 |
32 |
12 |
|
mel-11; pak-1 |
0.6 |
0 |
| mel-11; fem-2; pak-1 | 0.5 | 0 |
n > 400 for each genotype. Alleles are indicated in the Materials and Methods.
fhod-1 is associated with actin defects in the lateral epidermal cells
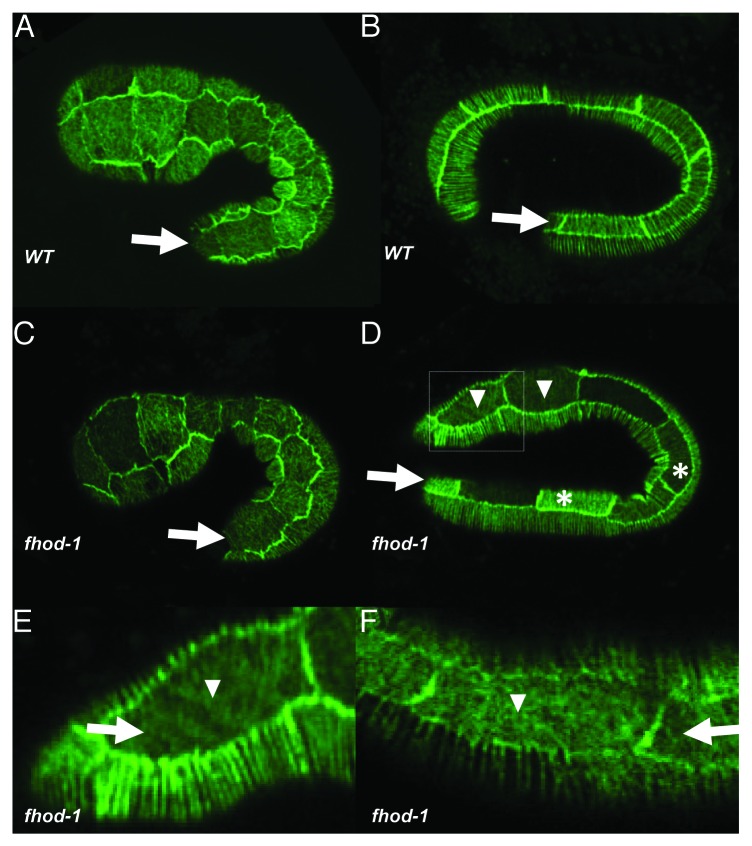
Because human FHOD1 stimulates the assembly of actin filaments in actin stress fibers,20,21,27,35 we examined actin in the fhod-1 mutant to determine if defects in the cytoskeletal network cause fhod-1’s elongation phenotypes. As visualized with a reporter consisting of the actin-binding domain of VAB-10 fused to GFP,7 34% (n = 41) of fhod-1 mutant embryos showed lateral cell actin defects apparent by the 2-fold stage, wherein microfilaments failed to coalesce into parallel bundles, compared with only 3% (n = 35) of wild-type embryos (Fig. 4). Similar results were seen with phalloidin staining, as 30% (n = 20) of fhod-1 embryos displayed actin defects in the lateral epidermal cells, lacked circumferential filaments or showed actin puncta in place of the stress fibers. These values are higher than the number of fhod-1 animals with elongation defects, perhaps indicating that the weaker actin defects do not always lead to elongation arrest.
Figure 4.fhod-1 microfilament defects. Actin is marked in living embryos using an integrated version of the lin-26p::ABDvab-10::GFP described by Gally et al.7 Note that expression levels of the transgene sometimes differ between cells. In all panels, the arrow denotes the lateral row of epidermal cells and anterior is to the left and dorsal to the top. (A) Microfilaments are found throughout the epidermis of a 1.5-fold wild-type embryo. Note that at this stage, the microfilament bundles form a meshwork in the lateral cells while microfilaments form parallel rows in the dorsal and ventral epidermal cells. (B) A 2- to 2.5-fold wild-type embryo, where the microfilaments in the lateral row are now aligned in parallel. (C) A fhod-1 embryo at the 1.5-fold stage appears like wild type. (D) A 2- to 2.5-fold fhod-1 embryo showing a mixture of lateral cells with normal (i.e., parallel) microfilament bundles (asterisks) and cells with abnormal patterns (arrowheads). The boxed region is magnified in (E). (F) A fhod-1 embryo showing similar defects to (E).
FHOD-1 epidermal expression is limited to the lateral cells
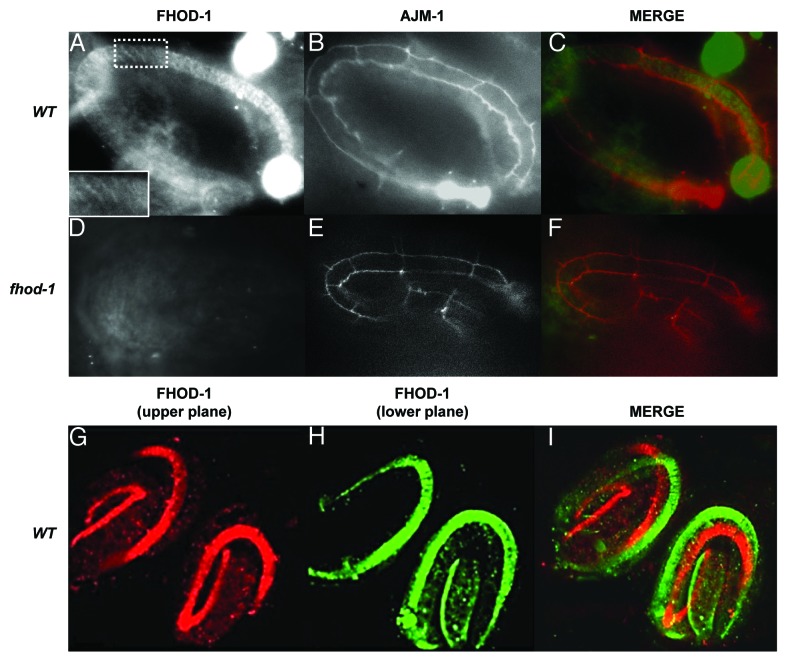
fhod-1 lateral-cell defects could be related to previous results showing that the actin cytoskeleton forms differently in the lateral vs. dorsal/ventral epidermal cell populations.2,7 Two polyclonal antibodies, raised to either a peptide in the central region or at the C terminus of FHOD-1 (Fig. 2), revealed that the protein is expressed in the lateral epidermis in a circumferential pattern, similar to the actin cytoskeleton in these cells (Fig. 5). Co-staining with the adherens junction marker, anti-AJM-1, which highlights epithelial boundaries,36 demonstrated that FHOD-1 was present in the lateral epidermal cells (Fig. 5A‒C). An actin capping protein might be expected to localize to the ends of filaments once assembly is completed, but anti-FHOD-1 staining showed a filamentous pattern (Fig. 5A). This pattern was similar to human anti-FHOD1 staining in cultured cells where staining is seen along the entire length of stress fibers rather than just their distal ends.35,37,38 No staining was seen in fhod-1(tm2363) (Fig. 5D and E), consistent with the interpretation that this is a null allele. We did not detect the embryonic muscle expression that Mi-Mi et al.30 found using a fhod-1::gfp, perhaps because FHOD-1 muscle expression was incompatible with our fixation conditions. Likewise, we did not detect epidermal expression using the fhod-1::gfp, possibly because the transgene lacked epidermal regulatory regions.
Figure 5. FHOD-1 is expressed in lateral epidermal cells. Anti-FHOD-1 staining (A) using antibody 1 (Fig. 2) is seen in wild-type lateral epidermal cells. Staining appeared striated (inset shows magnification of the boxed area). Anti-FHOD-1 was within the lateral epidermal cells, which are outlined by anti-AJM-1 staining (B). Merged image is shown in (C) with FHOD-1 (green) and AJM-2 (red). (D) No anti-FHOD-1 labeling is apparent in fhod-1 mutants, with lateral cells marked with anti-AJM-1 (E). (F) Merged image. A similar pattern is seen using antibody 2 (Fig. 2). FHOD-1 is false colored red in the upper focal plane (G) and green in the lower focal plane (H). Merged image (I) demonstrates FHOD-1 expression in the lateral cells on both sides of the animals.
FHOD-1 expression was apparent only relatively late in elongation, from the 2.5-fold stage. Although actin forms circumferential bundles from the outset of elongation in the dorsal and ventral cells, microfilament bundles are initially in a mesh-like pattern in the lateral cells, coalescing into circumferential filaments by the 2-fold stage.2,7 The relatively late appearance of FHOD-1 may imply that the protein is primarily involved in organizing the latter, circumferential microfilaments. However, as noted earlier, some fhod-1 embryos arrest as early as the 1.2-fold stage (although most arrest later), perhaps indicating that low (undetectable) amounts of FHOD-1 are required before parallel filaments form.
Discussion
C. elegans elongation is a precisely regulated process, and failure to elongate results in eventual death.2,3 Although the cytoskeletal elements throughout the epidermis are superficially similar, contraction only occurs in a subset of the epidermis: lateral (seam) cells contract while their dorsal and ventral neighbors play a more passive role.2,13 Some of these differences occur at the level of regulation of contraction. The Rho GAP RGA-2, an inhibitor of Rho (and therefore LET-502/Rho kinase), is required in the non-contractile dorsal and ventral cells.13 In addition, zygotic expression of LET-502/Rho kinase is at higher levels in the lateral than in the dorsal/ventral cells. In contrast, MEL-11/myosin phosphatase inhibits contraction and shows the opposite pattern.16 Here we describe a new player, FHOD-1, a member of the family of actin nucleators that form unbranched microfilaments.17,18 We show that fhod-1 is specifically involved in organizing actin filaments in the contractile lateral cells.
The actin cytoskeleton differs between the lateral and dorsal/ventral epidermal cells.2,7 Actin filaments organize later in the lateral cells, initially forming a meshwork before coalescing into a circumferential pattern after elongation has initiated. In addition, the microfilaments are found further from the apical surface in the lateral cells. FHOD-1 is only expressed in the lateral epidermis and appears to act specifically in organizing the lateral actin network (Fig. 4). Mutants show impenetrant elongation defects (Fig. 3) associated with disorganization of the microfilament network in the lateral cells (Fig. 4). However, FHOD-1 is only apparent at the 2.5-fold stage, perhaps indicating it is required (or that its requirements are greatest) only after the microfilaments organize into a banded pattern. By weakening the microfilament network in the lateral epidermis, fhod-1 mutants enhance the hypocontractile defects of let-502 mutants while suppressing the hypercontraction phenotype of mel-11. However, let-502 cannot act exclusively through fhod-1 as the let-502 elongation defects are much stronger than those of fhod-1.
In addition to the let-502/mell-11 pathway, elongation of the C. elegans embryo is mediated by a redundant path involving fem-2.14 fhod-1 shows much stronger genetic interactions with fem-2 than it does with let-502; mel-11 (Fig. 3), suggesting that fhod-1 may function mainly in the let-502/mel-11 branch of the elongation pathway. pak-1 also functions in parallel to the let-502/mel-11,7 but its relationship to fem-2 was unknown. We found that pak-1 was not enhanced by fem-2, but both fem-2 and pak-1 were enhanced by fhod-1 (Fig. 3). These results suggest that pak-1 and fem-2 are in the same pathway and that fhod-1 microfilaments preferentially function in the let-502/mel-11 pathway. Thus, the let-502/mel-11 and fem-2/pak-1 pathways may mediate elongation using different (but perhaps overlapping) subsets of microfilaments. Parallel pathways may also function at slightly different times relative to the reorganization of the lateral cell actin network, with fhod-1 acting differently before and after the reorganization. Finally, later stages of elongation (after the 2-fold stage) are driven by muscle function.3,12,13 pak-1 functions in hemidesmosomes that attach muscle to epidermis,34 and so perhaps the parallel contractile pathways represent elongation mediated by epidermis vs. muscle. Although FHOD-1 is also present in embryonic muscle,30 with respect to elongation, our genetic analysis shows that FHOD-1 acts with let-502 and, therefore, in the epidermal cells.
While pak-1 appears to act with fem-2 to mediate contraction, pak-1 genetic interactions differ from those of mutations of fem-2. As expected for an activator of contraction acting in parallel to let-502, mutations in either gene enhance let-502 and let-502; mel-11. However, pak-1, unlike fem-2, cannot suppress mel-11 hypercontraction7,14 (Table 1). Suppression of mel-11 by fem-2 but not pak-1 was perhaps unexpected, given that pak-1 single mutants have stronger elongation defects than do fem-2 (Fig. 3),14,34 i.e., the mutant with a greater degree of hypocontraction, pak-1, might be expected to more readily suppress mel-11 hypercontraction. One scenario to explain these results would suppose that FEM-2 (either directly or indirectly) activates PAK-1. Inactive PAK-1(+), present in fem-2 mutants, may have different effects on elongation than does the complete absence of PAK-1 in pak-1 mutants. Indeed, we observed that suppression of mel-11 by fem-2 requires pak-1(+) (Table 1). It should be noted that mammalian PP2c homologs of FEM-2 act as inhibitors rather than activators of PAK1,39,40 and so the regulatory relationship between C. elegans fem-2 and pak-1 could be more complex.
In mammalian systems, autoinhibition of FHOD1 is relieved by Rho kinase, after which the FHOD1-nucleated actin filaments, in turn, respond to Rho kinase-mediated contraction.20-23 However, since fhod-1-like actin defects are absent in let-502/Rho kinase mutants,6,7,41 C. elegans FHOD-1 appears not to require Rho kinase activation during elongation. Whether C. elegans FHOD-1 requires a different form of regulation is unknown.
fhod-1 functions in other C. elegans contractile cells, including in anchoring actin filaments in striated muscle.30 Like mammalian FHODs,26C. elegans fhod-1 functions in cell migration, during distal tip cell migration of the somatic gonad (C.A. Vanneste and P.E. Mains, unpublished). fhod-1 defines a new player in C. elegans morphogenesis, highlighting how differential organization of the actin cytoskeleton may result in differing contractile properties of cells.
Materials and Methods
Strains and alleles
The C. elegans wild-type strain N2 (var. Bristol) was maintained under standard conditions.42 The following alleles were used: mel-11(it26), fem-2(b245), let-502(sb118), fhod-1(tm2363), pak-1(ok448) and dpy-5(e61). Descriptions of genes and alleles used in this work are found in WormBase (www.wormbase.org). Strains were constructed using standard procedures. cis-linked morphological markers often were used to follow alleles of interest through crosses, in combination with PCR to identify deletion alleles. Homozygous lethal or sterile mutations were maintained as heterozygous stocks balanced either with appropriate crossover suppressors or normal chromosomes with flanking morphological markers.
The actin reporter strain mcIs50(lin-26p::ABDvab-10::GFP) was kindly provided by M. Labouesse (IGBMC). This is an integrated derivative of the transgene described by Gally et al.7 The insertion site was closely-linked to fhod-1, and so we built fhod-1 dpy-5 mcIs50 by picking green Dpy worms from fhod-1 dpy-5/mcIs50. dpy-5 mcIs50 was similarly constructed as the control.
We identified let-502(sb118) as a temperature-sensitive (ts) lethal linked to src-1(cj293) in strain TW416.43 sb118 was removed by recombination and outcrossed 11 times. This let-502 allele behaves as a hypomorph at the restrictive temperature of 25°, arresting at 2-fold or later, compared with ~1.2-fold for dominant-negative alleles.14,16 let-502(RNAi) enhanced sb118, demonstrating that this allele is a hypomorph. sb118 encodes an R168H substitution in a conserved region of the catalytic domain where most other Rho kinases have R or K. We eliminated the possibility that the 1.2-fold arrest of let-502 dominant-negative alleles is a neomorphic property as let-502(RNAi) in combination with let-502(ca201) did not alter the 1.2-fold arrest point. fhod-1(tm2363) was provided by the National Bioresources Project (NBP), Japan,33 but our original isolate of tm2363 contained a 367 bp deletion in the FH2 domain and a closely linked duplication of fhod-1(+). The strain reacted with an anti-FHOD-1 antibody 2, (Fig. 2) which was directed against a region predicted to be eliminated by the deletion allele. Although this allele showed lateral epidermal cell actin defects, it differed from fhod-1(RNAi) by enhancing both let-502 and mel-11 rather than showing reciprocal genetic interactions. We recovered a deletion without the associated duplication in a second shipment of this strain. This latter strain did not react with anti-FHOD-1 and all data we report was generated using this isolate.
To determine hatching rates of different genetic combinations, four or more L4 hermaphrodites were brooded at the appropriate temperatures until they ceased to lay fertilized embryos, as described in reference 44; a minimum of 400 progeny were scored. ts strains were shifted to the restrictive temperature at least 16 h prior to fixing and staining, or were shifted to the restrictive temperature for at least 16 h after RNAi injection. Injected animals were transferred to new plates at this time if their progeny were to be scored.
The percentages of animals for each genotype that arrested prior to the 2-fold stage were subjected to one-factor analysis of variance. As unhatched animals mainly arrest during elongation, we combined unhatched and hatched and arrested at ≤ 2-fold/L1 Dpy into one category for each genotype. Fisher's least significant difference posthoc testing was used to identify statistically significant differences between genotypes. As we brooded multiple animals per plate, this does not measure variance between individual mothers.
RNAi
RNA interference (RNAi) was performed as previously described45,46 using a clone from the Ahringer RNAi library (www.lifesciences.sourcebioscience.com) that targets the 3′ end of the gene. For feeding RNAi, L3 larvae were placed on the RNAi plate and raised at specified temperatures until they laid eggs. The worms where then transferred to a fresh RNAi plate every day until they stopped laying eggs and scored for hatching as described above. This first brood was not scored to allow time for RNAi to become fully effective.
For RNAi by injection, double-stranded RNA was generated making use of the T7 promoters found in the L4440-based plasmids in the Ahringer RNAi library. Plasmid-specific L4440 primers were used to amplify the appropriate DNA segment, the DNA was purified and in vitro RNA transcription was performed using the T7 polymerase (Megascript T7 Transcription Kit, Ambion Systems). dsRNA was injected into gravid hermaphrodites at 100 ng/ml.
PCR and reverse transcription
PCR was performed under standard conditions using Expand Long Template Kit (Roche). The primers GCCAATTGCATATCCATAAGGGG and GCCTAAACCATTCCTCAATCC were used to confirm the presence of the deletion allele tm2363 during strain construction. Products were gel purified, cloned and sequenced to confirm identity.
Microscopy and immunofluorescence
A polyclonal rabbit antibody (antibody 1, Fig. 2) was made and affinity purified by AnaSpec (www.anaspec.com) directed against the peptide PEKVAPPPPRAKIED in the central region of FHOD-1. Antibody 2 (Fig. 2) was made using a GST-tagged version of the FHOD-1 C-terminal domain using primers AACCGAATTCTCATGGTCCAGGAACTGTAGCG and AACCGTCGACTGTTTCGAGCTGTCCATCAACC. The product was cloned into pGEX-4T3 (Pharmacia) using EcoRI and SalI sites included in the primers. The protein was expressed, purified on a GST column and used to immunize rabbits.
Embryos were collected for staining by placing gravid hermaphrodites on unseeded NMG plates for up to 4 h (absence of food causes worms to retain older, elongation-stage embryos). These hermaphrodites were placed on polylysine-coated slides and gentle pressure was applied to the coverslip until the embryos were extruded. After freeze cracking,47 embryos were fixed in MeOH at -20°C and rehydrated through an ethanol/PBS series. Slides were blocked in 10 mg/ml bovine serum albumin (BSA) or 20% (v/v) normal donkey and goat serum (Jackson ImmunoResearch) in PBS with 0.1% Triton X100 or with 0.1% Tween-20 for at least 1 h at 37°. The slides were incubated with primary anti-FHOD-1 diluted (1:10) and with the adherens junction marker anti-AJM-1 (1:200)36 in blocking buffer (10 mg/ml BSA or 5% donkey and goat serums in PBS) for 1 h at 37° or overnight at 4°. Slides were washed three times with PBS with 0.1% Triton X100 in PBS and then incubated with anti-rabbit Alexa Fluor 568 and anti-mouse Alexa Fluor 488 fluorescent secondary antibodies (Invitrogen) diluted 1:500 in blocking buffer for 1 h at 37° or 4° overnight. Slides were mounted in a drop of Antifade (Invitrogen). Embryos were examined with a Zeiss Axioplan 2 imaging microscope and images where collected with Hamatsu Orca ER digital camera using Axiovision 4 software (Zeiss).
Actin was visualized using an integrated version of the lin-26p::ABDvab-10::GFP that expresses the actin-binding domain of VAB-10 fused to GFP in embryonic epidermal cells.7 Phalloidin staining of actin in formaldehyde fixed embryos was done as previously described.48 Actin filaments were visualized on a Zeiss LSM microscope.
For Differential Interference Contrast (DIC) microscopy, embryos were mounted on agar pads49 and were examined with a Zeiss Axioplan 2 microscope and photographed with a Hamatsu Orca ER digital camera. For time-lapse video recordings, a Z-stack of at least four focal planes was taken every 10 min for 5 h. The microscope stage temperature was typically between 22°C and 25°C, but was not controlled.
Acknowledgments
We would like to thank J.D. McGhee, J. Gaudet and D. Hansen and members of their labs for their input during the course of this project. We thank Michel Labouesse for stains and H. Yee, E. Raharjo and B. Chan for technical assistance. S. Davis and J.D. McGhee are thanked for assistance with microscopy. Some of the strains were obtained from the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440) or the National Bioresource Project. This work was supported by grants from the Canadian Institute of Health Research to P.E.M. and the American Heart Association to D.P.
Submitted
03/14/13
Revised
05/10/13
Accepted
05/14/13
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/25040
References
- 1.Quintin S, Gally C, Labouesse M. Epithelial morphogenesis in embryos: asymmetries, motors and brakes. Trends Genet. 2008;24:221–30. doi: 10.1016/j.tig.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Priess JR, Hirsh DI. Caenorhabditis elegans morphogenesis: the role of the cytoskeleton in elongation of the embryo. Dev Biol. 1986;117:156–73. doi: 10.1016/0012-1606(86)90358-1. [DOI] [PubMed] [Google Scholar]
- 3.Chisholm AD, Hardin J. Epidermal morphogenesis. In: The_C._elegans_Research_Community, ed. WormBook: www.wormbook.org), 2005:1-22. [DOI] [PMC free article] [PubMed]
- 4.Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124:679–83. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 5.Somlyo AP, Somlyo AV. Signal transduction by G-proteins, rho-kinase and protein phosphatase to smooth muscle and non-muscle myosin II. J Physiol. 2000;522:177–85. doi: 10.1111/j.1469-7793.2000.t01-2-00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piekny AJ, Johnson JL, Cham GD, Mains PE. The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 function as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogenesis. Development. 2003;130:5695–704. doi: 10.1242/dev.00807. [DOI] [PubMed] [Google Scholar]
- 7.Gally C, Wissler F, Zahreddine H, Quintin S, Landmann F, Labouesse M. Myosin II regulation during C. elegans embryonic elongation: LET-502/ROCK, MRCK-1 and PAK-1, three kinases with different roles. Development. 2009;136:3109–19. doi: 10.1242/dev.039412. [DOI] [PubMed] [Google Scholar]
- 8.Shelton CA, Carter JC, Ellis GC, Bowerman B. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J Cell Biol. 1999;146:439–51. doi: 10.1083/jcb.146.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wissmann A, Ingles J, McGhee JD, Mains PE. Caenorhabditis elegans LET-502 is related to Rho-binding kinases and human myotonic dystrophy kinase and interacts genetically with a homolog of the regulatory subunit of smooth muscle myosin phosphatase to affect cell shape. Genes Dev. 1997;11:409–22. doi: 10.1101/gad.11.4.409. [DOI] [PubMed] [Google Scholar]
- 10.Hartshorne DJ, Ito M, Erdödi F. Myosin light chain phosphatase: subunit composition, interactions and regulation. J Muscle Res Cell Motil. 1998;19:325–41. doi: 10.1023/A:1005385302064. [DOI] [PubMed] [Google Scholar]
- 11.Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, et al. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase) J Biol Chem. 1996;271:20246–9. doi: 10.1074/jbc.271.34.20246. [DOI] [PubMed] [Google Scholar]
- 12.Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–90. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diogon M, Wissler F, Quintin S, Nagamatsu Y, Sookhareea S, Landmann F, et al. The RhoGAP RGA-2 and LET-502/ROCK achieve a balance of actomyosin-dependent forces in C. elegans epidermis to control morphogenesis. Development. 2007;134:2469–79. doi: 10.1242/dev.005074. [DOI] [PubMed] [Google Scholar]
- 14.Piekny AJ, Wissmann A, Mains PE. Embryonic morphogenesis in Caenorhabditis elegans integrates the activity of LET-502 Rho-binding kinase, MEL-11 myosin phosphatase, DAF-2 insulin receptor and FEM-2 PP2c phosphatase. Genetics. 2000;156:1671–89. doi: 10.1093/genetics/156.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batchelder EL, Thomas-Virnig CL, Hardin JD, White JG. Cytokinesis is not controlled by calmodulin or myosin light chain kinase in the Caenorhabditis elegans early embryo. FEBS Lett. 2007;581:4337–41. doi: 10.1016/j.febslet.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wissmann A, Ingles J, Mains PE. The Caenorhabditis elegans mel-11 myosin phosphatase regulatory subunit affects tissue contraction in the somatic gonad and the embryonic epidermis and genetically interacts with the Rac signaling pathway. Dev Biol. 1999;209:111–27. doi: 10.1006/dbio.1999.9242. [DOI] [PubMed] [Google Scholar]
- 17.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 18.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 19.Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasteier JE, Madrid R, Krautkrämer E, Schröder S, Muranyi W, Benichou S, et al. Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J Biol Chem. 2003;278:38902–12. doi: 10.1074/jbc.M306229200. [DOI] [PubMed] [Google Scholar]
- 21.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 2008;27:618–28. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannemann S, Madrid R, Stastna J, Kitzing T, Gasteier J, Schönichen A, et al. The Diaphanous-related Formin FHOD1 associates with ROCK1 and promotes Src-dependent plasma membrane blebbing. J Biol Chem. 2008;283:27891–903. doi: 10.1074/jbc.M801800200. [DOI] [PubMed] [Google Scholar]
- 23.Iskratsch T, Reijntjes S, Dwyer J, Toselli P, Dégano IR, Dominguez I, et al. Two distinct phosphorylation events govern the function of muscle FHOD3. Cell Mol Life Sci. 2013;70:893–908. doi: 10.1007/s00018-012-1154-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulte A, Stolp B, Schönichen A, Pylypenko O, Rak A, Fackler OT, et al. The human formin FHOD1 contains a bipartite structure of FH3 and GTPase-binding domains required for activation. Structure. 2008;16:1313–23. doi: 10.1016/j.str.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Gasteier JE, Schroeder S, Muranyi W, Madrid R, Benichou S, Fackler OT. FHOD1 coordinates actin filament and microtubule alignment to mediate cell elongation. Exp Cell Res. 2005;306:192–202. doi: 10.1016/j.yexcr.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Koka S, Neudauer CL, Li X, Lewis RE, McCarthy JB, Westendorf JJ. The formin-homology-domain-containing protein FHOD1 enhances cell migration. J Cell Sci. 2003;116:1745–55. doi: 10.1242/jcs.00386. [DOI] [PubMed] [Google Scholar]
- 27.Taniguchi K, Takeya R, Suetsugu S, Kan-O M, Narusawa M, Shiose A, et al. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J Biol Chem. 2009;284:29873–81. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iskratsch T, Lange S, Dwyer J, Kho AL, dos Remedios C, Ehler E. Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J Cell Biol. 2010;191:1159–72. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kan-o M, Takeya R, Taniguchi K, Tanoue Y, Tominaga R, Sumimoto H. Expression and subcellular localization of mammalian formin Fhod3 in the embryonic and adult heart. PLoS One. 2012;7:e34765. doi: 10.1371/journal.pone.0034765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mi-Mi L, Votra S, Kemphues K, Bretscher A, Pruyne D. Z-line formins promote contractile lattice growth and maintenance in striated muscles of C. elegans. J Cell Biol. 2012;198:87–102. doi: 10.1083/jcb.201202053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemphues KJ, Kusch M, Wolf N. Maternal-effect lethal mutations on linkage group II of Caenorhabditis elegans. Genetics. 1988;120:977–86. doi: 10.1093/genetics/120.4.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. doi: 10.1016/S0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 33.Gengyo-Ando K, Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;269:64–9. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, Landmann F, Zahreddine H, Rodriguez D, Koch M, Labouesse M. A tension-induced mechanotransduction pathway promotes epithelial morphogenesis. Nature. 2011;471:99–103. doi: 10.1038/nature09765. [DOI] [PubMed] [Google Scholar]
- 35.Westendorf JJ, Koka S. Identification of FHOD1-binding proteins and mechanisms of FHOD1-regulated actin dynamics. J Cell Biochem. 2004;92:29–41. doi: 10.1002/jcb.20031. [DOI] [PubMed] [Google Scholar]
- 36.Labouesse M. Epithelial junctions and attachments. WormBook. 2006;13:1–21. doi: 10.1895/wormbook.1.56.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koka S, Minick GT, Zhou Y, Westendorf JJ, Boehm MB. Src regulates the activity of the mammalian formin protein FHOD1. Biochem Biophys Res Commun. 2005;336:1285–91. doi: 10.1016/j.bbrc.2005.08.257. [DOI] [PubMed] [Google Scholar]
- 38.Boehm MB, Milius TJ, Zhou Y, Westendorf JJ, Koka S. The mammalian formin FHOD1 interacts with the ERK MAP kinase pathway. Biochem Biophys Res Commun. 2005;335:1090–4. doi: 10.1016/j.bbrc.2005.07.191. [DOI] [PubMed] [Google Scholar]
- 39.Chan PM, Lim L, Manser E. PAK is regulated by PI3K, PIX, CDC42, and PP2Calpha and mediates focal adhesion turnover in the hyperosmotic stress-induced p38 pathway. J Biol Chem. 2008;283:24949–61. doi: 10.1074/jbc.M801728200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koh CG, Tan EJ, Manser E, Lim L. The p21-activated kinase PAK is negatively regulated by POPX1 and POPX2, a pair of serine/threonine phosphatases of the PP2C family. Curr Biol. 2002;12:317–21. doi: 10.1016/S0960-9822(02)00652-8. [DOI] [PubMed] [Google Scholar]
- 41.Norman KR, Moerman DG. Alpha spectrin is essential for morphogenesis and body wall muscle formation in Caenorhabditis elegans. J Cell Biol. 2002;157:665–77. doi: 10.1083/jcb.200111051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bei Y, Hogan J, Berkowitz LA, Soto M, Rocheleau CE, Pang KM, et al. SRC-1 and Wnt signaling act together to specify endoderm and to control cleavage orientation in early C. elegans embryos. Dev Cell. 2002;3:113–25. doi: 10.1016/S1534-5807(02)00185-5. [DOI] [PubMed] [Google Scholar]
- 44.Mains PE, Sulston IA, Wood WB. Dominant maternal-effect mutations causing embryonic lethality in Caenorhabditis elegans. Genetics. 1990;125:351–69. doi: 10.1093/genetics/125.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 46.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 47.Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–94. doi: 10.1016/S0091-679X(08)61396-5. [DOI] [PubMed] [Google Scholar]
- 48.Costa M, Draper BW, Priess JR. The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev Biol. 1997;184:373–84. doi: 10.1006/dbio.1997.8530. [DOI] [PubMed] [Google Scholar]
- 49.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]