The proper regulation and spatial organization of heterochromatin is crucial for a broad range of cellular processes, ranging from gene silencing to maintenance of genomic integrity and cell division. The most prominent heterochromatin domains are formed across repetitive sequences (major satellite repeats) around the centromere regions. Those pericentric heterochromatin regions can form clusters in interphase nuclei, termed chromocenters, which are well-discernible as DAPI-dense spots in the nucleus. The localization of chromocenters in the nucleus is not random. In most cell types chromocenters are attached to the nuclear lamina or to nucleoli.
Establishment of heterochromatin at pericentric regions is assured by a complex network of chromatin-organizing activities that establish a specific histone modification signature, leading to the recruitment of additional players that mediate chromatin compaction and organization. The major constitutive heterochromatin proteins are the histone methyltransferases Suv39h, Suv4-20h, and heterochromatin protein 1 (HP1) isoforms. These proteins act in a sequential pathway1 to establish the combinatorial H3K9me3 and H4K20me3 modification signature at pericentric heterochromatin. Recently, additional players acting upstream of Suv39h recruitment have been identified.2,3 Major satellite repeats contain binding sites for Pax3 and Pax9 transcription factors. Both proteins are necessary to repress noncoding transcripts from major satellite repeats and aid in the recruitment of Suv39h. An additional requirement for Suv39h enzymes to establish H3K9me3 at pericentric regions appears to be a pre-existing mono-methylation on lysine 9. Two cytoplasmic histone methyltransferases, Prdm3 and Prdm16, induce H3K9me1 in the cytoplasmic pool of H3. These premethylated histones are probably incorporated in repressive chromatin domains where the mono-methylation can be converted to H3K9me3 by Suv39h enzymes.
We have recently investigated the downstream components of the sequential pathway.4 This analysis was triggered by our finding that the core heterochromatin proteins show very different dynamic association with heterochromatin. HP1 proteins are dynamic components, whereas the histone methyltransferase Suv4-20h2 very stably associates with heterochromatin. We identified a small region in Suv4-20h2 (clamp domain) featuring several independent HP1 interaction sites to mediate the stable association with heterochromatin. Our data led us to hypothesize that multiple interactions with HP1 molecules through the clamp domain result in a synergistic stabilization of Suv4-20h2 with heterochromatin. The stable binding of Suv4-20h2 is crucial for proper establishment of a compact heterochromatin structure, as Suv4-20h-deficient cells (Suv4-20h dko) display defects in chromatin compaction and chromocenter organization (chromocenter scattering).4 These data suggest that Suv4-20h2 is a major regulator at different levels of chromatin organization to mediate condensed chromatin domains as well as long-range heterochromatin interactions. Interestingly, the cellular phenotypes of mutants in the upstream components of the heterochromatin pathway (Pax3/9, Prdm3/16, Suv39h) resemble the Suv4-20h mutant phenotype, indicating that processes involving Suv4-20h2 or downstream of Suv4-20h2 regulate heterochromatin organization. A general defect in cells lacking central heterochromatin components is improper chromosome segregation. We found that Suv4-20h2 is crucial for recruitment of the cohesin complex to pericentric heterochromatin. Cells lacking Suv4-20h2 display reduced levels of cohesin at heterochromatin, leading to reduced sister chromatid cohesion and chromosome segregation defects. We also observed these defects in cells lacking the upstream Suv39h enzymes, demonstrating that the heterochromatin pathway is important for proper establishment of pericentric cohesin levels. Notably, cohesin is not fully lost from heterochromatin in Suv39h or Suv4-20h dko cells, demonstrating the existence of additional complementary recruitment pathways that remain to be identified.
Alterations in heterochromatin organization have been observed in different human diseases. Enhanced heterochromatin formation occurs in cells with defective nuclear lamina in patients with Hutchinson-Gilford progeria syndrome.5 These findings suggested interplay between heterochromatin and lamina organization, which was recently supported by additional evidence from mouse models.6 Mice lacking lamin B receptor (LBR) or laminA/C show reduced peripheral heterochromatin around the nuclear lamina and chromocenter fusions in different cell types, suggesting that proper heterochromatin organization depends on an intact nuclear lamina. Interestingly, certain features of heterochromatin are necessary for interaction with the lamin network. We found that in Suv4-20h-deficient cells, the density of peripheral heterochromatin is reduced, indicating that Suv4-20h2 or H4K20me3 contribute to tethering heterochromatin to the nuclear lamina.4 It will thus be interesting to test if enhanced heterochromatin formation in progeria patients can be rescued by reducing Suv4-20h2 levels. An indication that reduced heterochromatin formation may at least partially rescue progeria-like phenotypes came from a recent publication showing that Suv39h1-deficiency resulted in increased survival in a mouse progeria model.7
Another disease with critical alterations in heterochromatin is cancer. We showed earlier that different human cancer types feature low levels of H4K20me3, indicating compromised heterochromatin organization.8 Interestingly, follow-up analyses in small cell lung cancer have revealed that low H4K20me3 levels correlate with a bad prognosis. Based on our data, we hypothesize that reduced H4K20me3 in cancer cells may result from lower expression levels or activity of Suv4-20h2, which would lead to reduced cohesin levels at pericentric heterochromatin. Whether this results in subsequently increased genomic instability and aggressiveness of the cancer cells remains to be demonstrated.
These examples show that a better understanding of the pathways that establish heterochromatin will increase our understanding of human diseases and may, in the future, allow the development of novel diagnostic or even interventional strategies.
Figure 1.
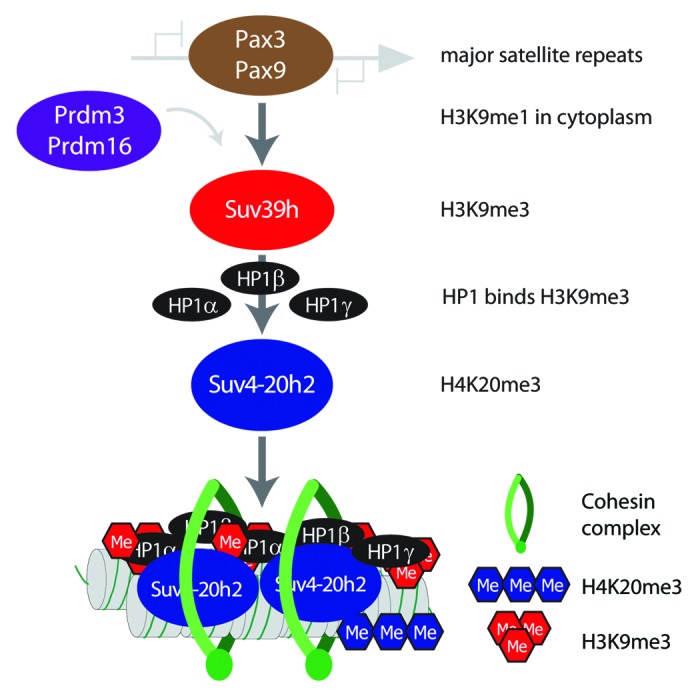
Pathway for heterochromatin establishment. Major satellite repeats can be recognized by 2 transcription factors (Pax3 and Pax9). Binding of these factors is crucial to mediate recruitment of Suv39h enzymes that establish H3K9me3 at pericentric heterochromatin. For full activity Suv39h enzymes require H3K9me1 as substrate, which is deposited at heterochromatin from a cytoplasmic pool generated by Prdm3 and Prdm16. Once H3K9me3 is established at heterochromatin, HP1 proteins can bind to this modification. Suv4-20h2 can then stably associate with regions of high HP1 occupancy and mediate H4K20me3. Suv4-20h2 has additional roles in mediating chromatin compaction and recruitment of cohesin.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26179
References
- 1.Schotta G, et al. Genes Dev. 2004;18:1251–62. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bulut-Karslioglu A, et al. Nat Struct Mol Biol. 2012;19:1023–30. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 3.Pinheiro I, et al. Cell. 2012;150:948–60. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 4.Hahn M, et al. Genes Dev. 2013;27:859–72. doi: 10.1101/gad.210377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shumaker DK, et al. Proc Natl Acad Sci U S A. 2006;103:8703–8. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solovei I, et al. Cell. 2013;152:584–98. doi: 10.1016/j.cell.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, et al. Nat Commun. 2013;4:1868. doi: 10.1038/ncomms2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraga MF, et al. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]


