Abstract
The autoimmune process in rheumatoid arthritis depends on activation of immune cells, which utilize intracellular kinases to respond to external stimuli such as cytokines, immune complexes, and antigens. CD4+ T cells comprise a large proportion of the inflammatory cells that invade the synovial tissue and may therefore be a cell type of pathogenic importance. Both methotrexate and infliximab are effective in the treatment of inflammatory arthritis; however, the biological effects triggered by these treatments and the biochemical mechanisms underlining the cell response are still not fully understood. Thus, in this study the global metabolic changes associated with methotrexate or infliximab treatment of isolated human CD4+ T cells were examined using gas chromatography/mass spectrometry or liquid chromatography/mass spectrometry. In total 148 metabolites involved in selective pathways were found to be significantly altered. Overall, the changes observed are likely to reflect the effort of CD4+ cells to increase the production of cellular reducing power to offset the cellular stress exerted by treatment. Importantly, analysis of the global metabolic changes associated with MTX or infliximab treatment of isolated human CD4+ T cells suggested that the toxicity associated with these agents is minimal when used at clinically relevant concentrations.
Keyword: rheumatoid arthritis, autoimmunity disease, apoptosis, T cells
Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune disease, characterized by infiltration of the synovium, a structure that lines the joints, with inflammatory cells. The thickened synovium forms a lesion called pannus that slowly destroys the cartilage and the bone adjacent to the joint. Both the small joints of the hands and feet and the larger joints such as the knees and wrists become damaged in RA, ultimately leading to severe functional impairment. More aggressive forms of RA also involve extra-articular tissues, causing lung inflammation, splenomegaly with cytopenia, skin nodules, and vasculitis. Therefore, treatment of RA aims at stopping the inflammatory response before permanent damage occurs.
The first-line treatment, in inflammatory arthritis, is methotrexate (MTX)1 a potent antimetabolite drug that interferes with the metabolism of folic acid by inhibition of DHFR (dihydrofolate reductase). MTX, as compared with anti-inflammatory drugs that merely alleviate temporarily the symptoms of joint inflammation, changes the course of the disease, retarding or even preventing the development of bone erosions. Since tumor necrosis factor-α (TNF-α) plays a central role in the pathogenesis of RA,2 anti-TNF-α drugs (e.g., infliximab is an immunoglobulin G-1 [IgG1] antibody that binds to soluble and cell membrane-bound TNF-α with its inactivation3-9) are frequently used in forms of RA that are resistant to traditional therapeutic approaches and have acquired a prominent place in the management of rheumatologic conditions and psoriasis. Thus, both MTX and infliximab are efficacious in the treatment of inflammatory arthritis, though these 2 drugs have very different mechanisms of action and safety profiles.10 They have a good safety profile; however, with increasing use of these agents, adverse events have been reported. Some of these adverse drug reactions can be similar to the underlying disease for which they are given, making management of these conditions challenging.11 For example, anti-TNF therapy is used for the treatment of psoriasis, but psoriasis form lesions are sometimes observed in patients receiving therapy. Similarly, anti-TNF therapy is used for the treatment of rheumatologic diseases, but arthralgias and arthritis are sometimes observed in patients receiving anti-TNF agents.11
In the last few years, several lines of evidence suggested that several metabolic changes in cells that participate in inflammation are needed to obtain polarization and activation of cells as lymphocytes.12-15 Inflammation is triggered in response to tissue injury or infection, and mounting an inflammatory response is an energy-intensive process. The products of inflamed tissues that provoke inflammation can activate T cells.16-18 Indeed, T cells are prime examples of how cell metabolism can be dramatically altered to support the specific needs and functions of each cell state. A central feature that allows this flexibility in metabolism is direct regulation of metabolic pathways by cell-extrinsic signals that drive T-cell survival, growth, and proliferation.19-33 In each case, if metabolism fails to match the demands of the cell, cell function is impaired, or cells can undergo apoptosis.34-44 Conversely, excess metabolism may prevent apoptosis, exacerbate cell function, thus promote T-cell hyper-reactivity, and lead to autoimmunity and inflammatory diseases.34,45 Thus, it is critical to appreciate how T-cell metabolism is regulated, and how alterations in cell metabolism impacT-cell function and fate.
The aim of this study was to use a metabolomic strategy to investigate and characterize global biochemical profiles observed in human CD4+ T cells following administration of 2 doses of MTX or infliximab. This has revealed underlying biochemical phenomena associated with the treatment, and these data, in the longer term, will provide insights that help the development of a better understanding of the disease state. Clinically relevant concentrations of the drugs were used, allowing us to also investigate potential toxic off-target effects.
Results
Global biochemical profiles were determined in human CD4+ T cells and compared across treatment groups. Human CD4+ cells were isolated and treated with 2 different doses of MTX (10 nM or 100 nM), IgG1K control or infliximab (1 μg/ml) for 24 h before harvest for global metabolic profiling. Global biochemical profiles, by either gas chromatography/mass spectrometry (GC/MS) or liquid chromatography/mass spectrometry (LC/MS/MS), were performed as described,46,47 and a total of 148 metabolites were identified (Table 1; Table S1) and involved in the pathways described below.
Table 1. Metabolic changes.
| Statistical comparisons: Welch 2-sample t test | |||||
|---|---|---|---|---|---|
|
Significantly altered biochemicals |
Methotrexate |
|
|
|
|
|
10 nM Control |
100 nM Control |
Infliximab IgG1K |
IgG1K Control |
Infliximab Control |
|
| Total biochemical: P ≤ 0.05 |
0 |
5 |
0 |
0 |
3 |
| Biochemicals (↑↓) |
0|0 |
4|1 |
0|0 |
0|0 |
1|2 |
| Total biochemical: 0.05 < P < 0.10 |
6 |
1 |
3 |
2 |
2 |
| Biochemicals (↑↓) | 5|1 | 1|0 | 1|2 | 2|0 | 2|0 |
Welch 2-sample t-test was used to determine whether the means of the two populations are different. A total of 148 metabolites were identified. Red indicates an increase; green indicates a reduction. Original data are shown in Table S1.
Glucose metabolism
Glucose plays an important role in T-cell energy metabolism. The levels of maltose (Fig. 1A), maltotriose (Fig. 1B), and maltotetraose (Fig. 1C) intermediates involved in glycogen metabolism showed a non-statistically significant downward trend in the MTX-treated groups, consistent with a possible alteration in glycogenolysis. No significant change was observed in mannose-6-phosphate levels (Fig. 1D). In comparison to the IgG1K group, the above metabolites showed an upward trend in the infliximab-treated group, suggesting an opposite, but subtle, impact of MTX and infliximab on glycogen metabolism.
Figure 1. Glycogen metabolism. Biochemicals involved in glycogen metabolism showed consistent fluctuation in response to MTX treatment consistent with subtle disturbance in glycogen metabolism. In comparison to the IgG1K control group, the infliximab-treated group showed an opposite trend as those demonstrated by the MTX group. The effect of MTX and infliximab exposure for 24 h is shown on maltose (A), maltotriose (B), maltotetraose (C), mannose-6-phpsphate (D). See “Materials and Methods” for technical protocols. Relative values are indicated over control. Ctrl, control; Infxmb, infliximab.
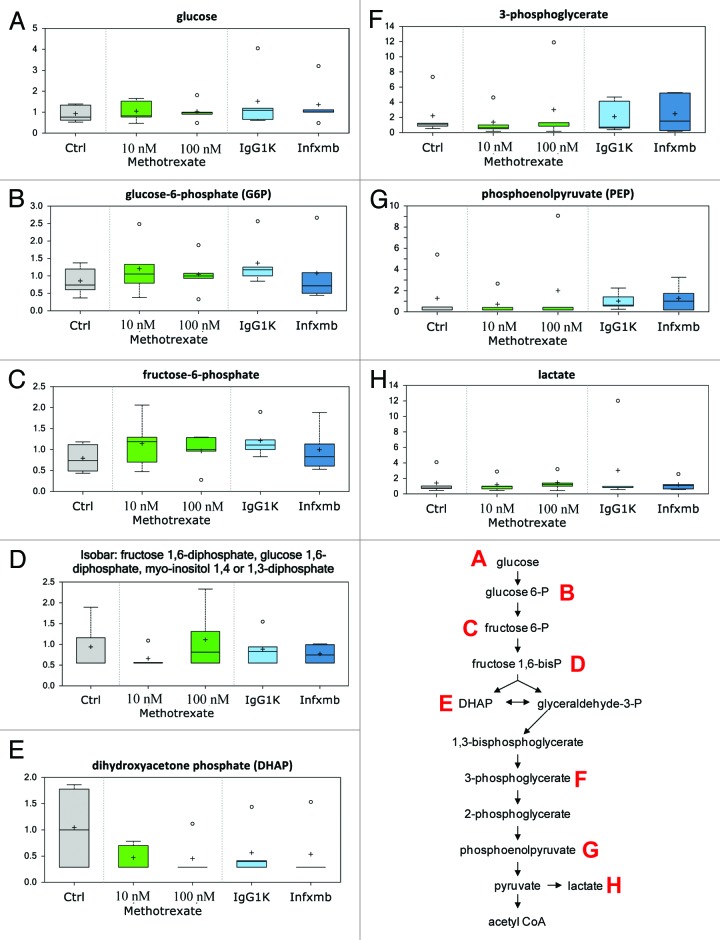
Glycolytic intermediates prior to phosphofructose kinase 1 (PFK1) activity in glycolysis (glucose, Fig. 2A, glucose 6-phosphate, Fig. 2B, and fructose 6-phosphate, Fig. 2C) showed an upward trend, while those downstream of PFK1 activity (fructose 1,6-diphosphate, Fig. 2D, dihydroxyacetone phosphate [DHAP, Fig. 2E], 3-phosphoglycerate, Fig. 2F, phosphoenolpyruvate [PEP, Fig. 2G] and lactate, Fig. 2H) demonstrated a downward trend in response to 10 nM MTX treatment in comparison to the control. This observation could suggest a subtle decrease in glycolysis and inhibition of PFK1 by MTX. However, the above fluctuation pattern in glycolytic intermediates was absent with 100 nM MTX or infliximab treatment.
Figure 2. Glycolysis. Low dose of MTX caused fluctuation in the biochemicals involved in glycolysis. Biochemicals prior to phosphofructokinase 1 (PFK1) activity showed an upward trend while those downstream showed a downward trend indicative of an inhibition of glycolysis. However, these trends were absent with higher dose of MTX. Infliximab did not elicit much change in the biochemicals involved in glycolysis. The effect of MTX and infliximab exposure for 24 h is shown on glycolytic metabolites: glucose (A), glucose-6-phosphate (B), fructose-6-phosphate (C), fructose 1,6 diphosphate (D), dihydroxyacetone phosphate (E), 3-phosphoglycerate (F), phosphoenolpyruvate (G), lactate (H). See Figure 1 for legend code.
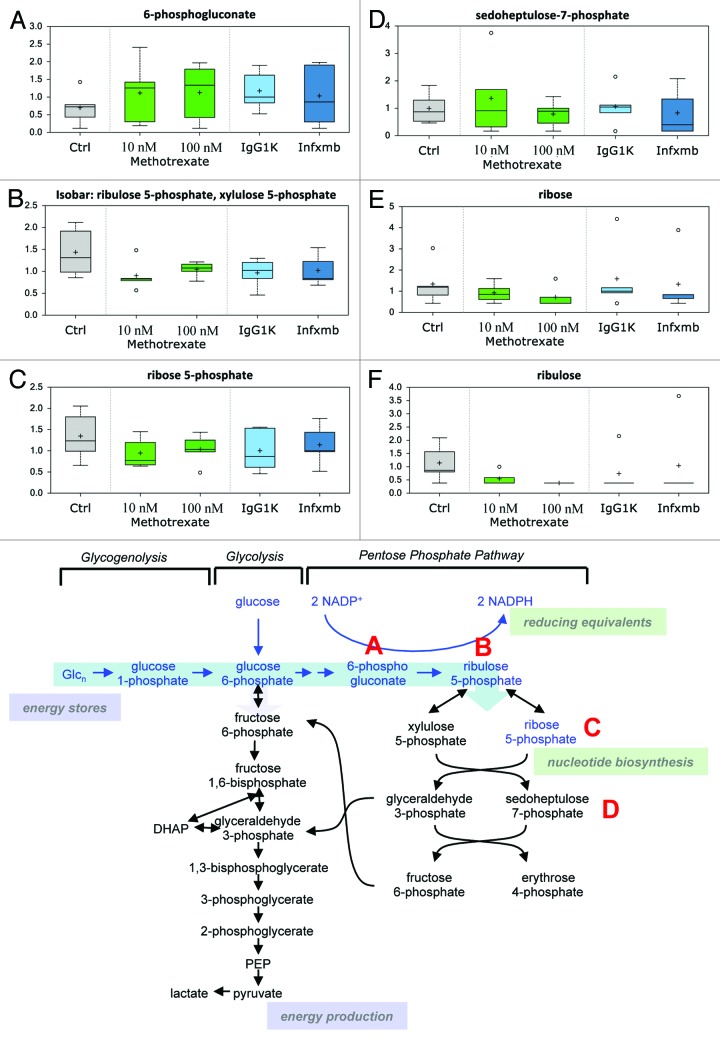
MTX treatment resulted in an increase in the levels of 6-phosphogluconate (Fig. 3A), the metabolite generated from the committed step of pentose phosphate pathway (PPP) catalyzed by glucose 6-phosphate dehydrogenase possibly indicative of a subtle increase in the PPP activity. This may reflect the effort of CD4+ cells to increase the production of cellular reducing power (NADPH) to offset the cellular stress exerted by MTX. Other biochemicals involved in the PPP, ribulose 5-phosphate (Fig. 3B), ribose 5-phosphate (Fig. 3C), sedoheptulose 7-phosphate (Fig. 3D), ribose (Fig. 3E), and ribulose (Fig. 3F), also demonstrated subtle fluctuations supportive of a possible disturbance in the PPP.
Figure 3. Pentose phosphate pathway. Many biochemicals involved in the PPP showed subtle fluctuations that may suggest perturbation in the PPP in response to MTX. The effect of MTX and infliximab exposure for 24 h is shown on 6-phosphogluconate (A), ribulose-5-phosphate (B), ribose-5-phosphate (C), sedoheptulose-7-phosphate (D), ribose (E), ribulose (F). See Figure 1 for legend code.
Inflammation
12-HETE (Fig. 4A), an eicosanoid from arachidonate produced by lipoxygenase is a proinflammatory mediator implicated in the pathogenesis of psoriasis. Compared with their corresponding controls, both 100 nM MTX and infliximab caused a non-statistically significant reduction in the levels of 12-HETE, indicative of a possible and subtle inhibition of inflammation. Within cells, glycerophospholipids represent key reservoirs for polyunsaturated fatty acids (PUFAs), especially arachidonate, an important precursor for various inflammatory mediators. In this study, the levels of phosphatidylethanolamine (Fig. 4B–D) and phosphatidylcholine (Fig. 4E–G), 2 major lysolipids generated from membrane phospholipids via the function of various families of phospholipase A (PLA) displayed an upward trend with both doses of MTX treatments in relation to the control indicative of a possible upregulation of PLA activity in response to MTX. In support of this notion, docosapentaenoate (n3DPA; 22:5n3, Fig. 4H), docosahexaenoate (DHA; 22:6n3. Fig. 4I), and arachidonate (20:4n6, Fig. 4J) also showed an upward trend with 100 nM MTX. Compared with the IgG1K group, infliximab treatment elicited an upward trend in the levels of ω-6 PUFAs (arachidonate, Fig. 4J, and dihomo-linoleate [20:2n6, Fig. 4K]), but failed to show any consistent trend in the levels of ω-3 PUFAs and lysolipids.
Figure 4. Inflammation. 12-HETE showed a non-statistically significant downward trend in the 100 nM MTX group in relation to the control and the 10 nM MTX group that could suggest an inhibition of inflammation by the higher dose of MTX treatment. A similar trend was observed between the infliximab-treated group in relation to the IgG1K group. PUFAs, mainly w-3 showed a subtle and upward trend with 100 nM MTX in comparison to the control and 10 nM MTX while w-6 showed subtle downward trending with infliximab treatment in comparison to the IgG1K control group. Lysolipids showed a consistent upward trend with MTX treatment in comparison to the control suggesting a subtle fluctuation in the activity of PLA. However, changes in lysolipids with infliximab treatment were not consistent. The effect of MTX and infliximab exposure for 24 h is shown on indicated lipids (A–K). See Figure 1 for legend code.
Nucleotide metabolism
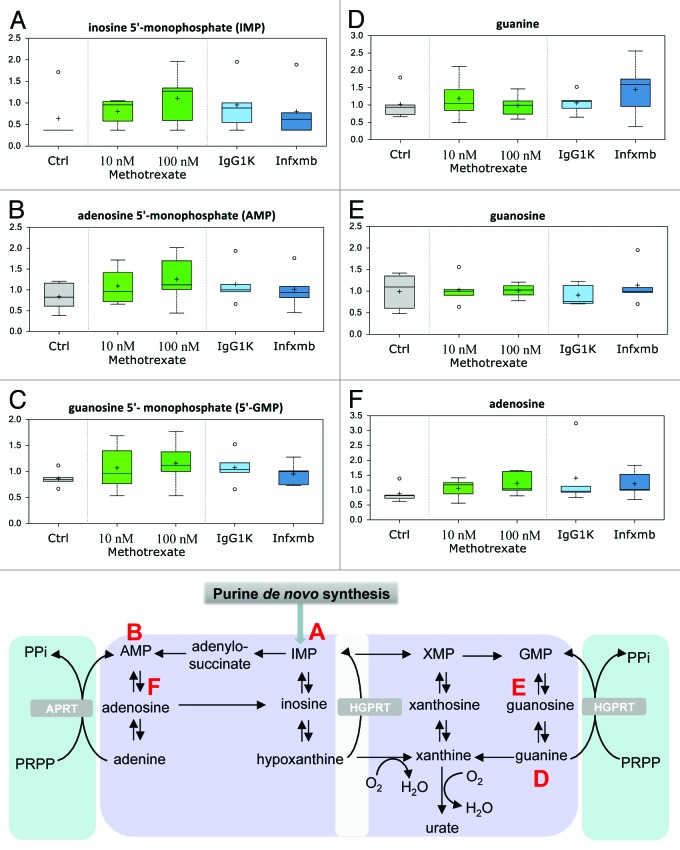
Purine nucleotides (inosine 5′-monophosphate [IMP, Fig. 5A], adenosine 5′-monophosphate [AMP, Fig. 5B], and guanosine 5′-monophosphate [GMP, Fig. 5C]) showed a consistent upward trend with both doses of MTX treatment in relation to the control, while infliximab elicited a decrease in relation to the IgG1K group. This could be a reflection of subtle changes in DNA/RNA metabolism and/or purine nucleotide de novo biosynthesis. It is intriguing that MTX treatment was associated with a general increase in the levels of purine nucleotides (Fig. 5D and E), since MTX has been shown to inhibit DHFR, the enzyme involved in the generation of N10-formal tetrahydrofolate from folate to support purine de novo biosynthesis. It has been reported in the literature that MTX interferes with purine metabolism, resulting in accumulation of adenosine, which was also observed in this study (Fig. 5F).
Figure 5. Purine metabolism. Purine nucleotides and adenosine showed a subtle upward fluctuation with MTX treatment. This could be a reflection of increased DNA/RNA breakdown, increased de novo biosynthesis or reduced breakdown of nucleotides. See Figure 1 for legend code.
Apoptosis
Finally, in order to determine whether there were cytotoxic effects associated with the treatments, the induction of apoptosis was evaluated by measuring phosphatidylserine externalization by flow cytometry using annexin VFITC as a probe. Cells were treated with 10 and 100 nM MTX, for 24 h. In control cells a large majority of cells (97%) showed a low affinity for annexin VFITC; the remaining 2%, corresponding to basal cell death, had mean fluorescence intensity in the range indicated below for treated cells. Indeed, after treatment with MTX, the labeled cells, corresponding to apoptotic cells, were 5%, and 6% for 10 and 100 nM MTX treatments, respectively (Fig S1A). There was no difference in apoptosis in cells treated with infliximab (Fig S1B). The data show that there was a very modest increase of apoptosis in cell death between cells MTX-treated and untreated cells within the time frame used. The data were also confirmed by propidium iodide (not shown).
Discussion
RA shows invasion by immune cells in the synovial tissues of the affected joints.48,49 Primarily the hands and feet are affected, but other joints may be progressively involved. Patients with RA experience a progressive destruction of the affected joints, leading to disability. CD4+ T cells play a central role in immune protection.50 They do so through their capacity to help B cells make antibodies, to induce macrophages to develop enhanced microbicidal activity, to recruit neutrophils, eosinophils, and basophils to sites of infection and inflammation, and, through their production of cytokines and chemokines, to orchestrate the full panoply of immune responses.51-53 However, abnormal activation of T cells was seen as the critical event in most organ-specific autoimmune diseases. For T-cell homeostasis and adaptive immunity, it is critical that T-cell metabolism adequately meets and is suited to the needs of specific T-cell functions. If metabolism fails to support T-cell demands, autophagy,54-63 cell cycle arrest,64,65 and apoptosis66-90 via Bcl-2 family proteins may ensue. Conversely, excess metabolism may promote T-cell hyper-reactivity and autoimmunity. A greater understanding of how these pathways are regulated and function in lymphocytes becomes increasingly important with the availability and interest in the therapeutic use of drugs that alter metabolism.91-101 Through metabolic manipulation, it may be possible to extend survival of lymphocytes and ultimately alter their differentiation. Conversely, it may be possible to target metabolic pathways and mediators to control the formation of T-cell lineages or to suppress T-cell responses by blocking specific metabolic pathways essential for T-cell growth and proliferation. This approach has proved promising in cancer therapy, and given the similarities of cancer cell metabolism with that of activated lymphocytes, it is likely that a similar strategy may provide a new approach to modulation of the immune system.
Both MTX and infliximab are effective in the treatment of inflammatory arthritis; however, while there is a very good response of some patients to a biological therapy, there is also a complete lack of response in a large number of other patients.102 Since the biological effects of these treatment and the mechanisms underlining the cell response are still not well understood, a more detailed understanding of the biochemical changes in T cells is required to elucidate toxicity pathways, the oxidative stress effects, and the response mechanisms triggered by this treatment. Therefore, the goal of this study was to examine the potential effects of these drugs on the metabolome by analyzing the global metabolic changes associated with MTX or infliximab treatment of isolated human CD4+ T cells. Cells were isolated and treated with either MTX or infliximab plus related controls and collected 24 h post-treatment.
The numbers of statistically significant changes in biochemicals associated with the treatment groups compared with the corresponding controls were limited and below the estimated numbers of false discovery, suggesting that the treatment effect failed to overcome the inherent biological noise associated with the study. Principal component analysis (PCA) supported the above conclusion by showing no clear separation among the treatment groups. However, the PCA demonstrated clustering among different CD4+ cell preparations, suggesting a confounding effect from different sources of CD4+ cells (Fig S2A). It is worth pointing out that MTX was detected in the 2 MTX treatment groups and was absent in the controls and infliximab-treated groups (Fig S2B). In addition, the relative concentrations of MTX in the 2 MTX groups reflected the doses used in the experiment confirming the platform performance.
In summary, the data presented herein on the global metabolic changes associated with MTX or infliximab treatment of isolated human CD4+ T cells, demonstrate that these drugs have a minimal toxic effects at clinically relevant concentrations. The biochemicals involved in a number of pathways showed some promising and subtle trends that did not reach statistical significance. Treatment of CD4+ cells with MTX was associated with subtle alterations in purine and glucose metabolism manifested in the fluctuations in biochemicals involved in glycogenolysis, glycolysis, and PPP. Both MTX and infliximab at the doses used in this study may exert subtle impact on inflammation demonstrated by the consistent trends observed in certain PUFA species and lysolipids. These observations provide some insights into the potential mechanisms involved in mediating the anti-inflammatory effects of MTX and infliximab. Since the metabolic profile for individual patients is highly dynamic compared with, for example, genes and protein levels, it would be worth studying how these metabolites correlate with disease activity. It is possible that the metabolite profile of individual patients could be used as a tool for predicting the disease course, and thereby facilitates optimal treatment.
Materials and Methods
Isolation of cells
Venous blood from healthy donors was collected by venepuncture into heparinized tubes (BD Biosciences). The donors, all males, were healthy volunteers recruited through advertisement by the University of Rome “Tor Vergata”, Rome, Italy. Their mean age was 24.0 y (range, 22–28 y of age). Peripheral blood mononuclear cells (PBMC) were isolated by density-gradient centrifugation with Ficoll Paque (Amersham Biosciences). After washing, the PBMC were then depleted of monocytes by MACS separation columns with CD4 microbeads (Miltenyi Biotec): CD4+ cells passed through the column and were collected, washed, and suspended in RPMI 1640 (Gibco) culture medium. Ethical approval was obtained from all participants that gave informed consent.
Cell culture
The cell cultures were maintained at 37 °C and 5% CO2 in RPMI 1640 medium supplemented with Glutamax I (Life Technologies) supplemented with 10% fetal calf serum (FCS), 1% of penicillin/streptomycin, 2% sodium pyruvate, 0.05 mM 2-mercaptoethanol, and 5 mM HEPES.
Reagents preparation and treatment
MTX and infliximab were prepared according to the manufacturers’ recommended protocol. Control reagent was a human IgG1K (Sigma). MTX at 10 nM and 100 nM concentrations or infliximab was added to the cell cultures. The total number of cell was divided in 5 different groups (5 groups of 5 samples each), and each group contained: (1) human CD4+ cells as control; (2) human CD4+ cells treated with MTX at the dosage of 10 nM; (3) human CD4+ cells treated with MTX at the dosage of 100 nM; (4) human CD4+ cells treated with infliximab at the dosage of 1 µg/ml; (5) human CD4+ cells treated with IgG1k at the dosage of 1 µg/ml as control of infliximab. After 24 h incubation, cells were harvested and prepared for metabolomic and other analysis.
Flow cytometry analysis
Flow cytometry was performed as described in Tucci et al.103 Phosphatidylserine exposure was evidenced using annexin VFITC added at a final concentration of 140 nM. Incubation at room temperature was allowed to proceed for 10 min in the dark before data acquisition. Samples were analyzed using a FACScan flow cytometer (Becton Dickinson).
Metabolomic analysis
Samples were immediately stored at −80 °C and, at the time of analysis, were extracted and prepared for analysis using a standard metabolic solvent extraction method, as described in references 29 and 30. Briefly, the extracted samples were split into equal parts for analysis by gas chromatography/mass spectrometry (GC/MS) or liquid chromatography/mass spectrometry (LC/MS/MS) platforms. Also included were several technical replicate samples created from a homogeneous pool containing a small amount of all study samples.
The LC/MS portion of the platform was based on a Waters ACQUITY UPLC and a Thermo-Finnigan LTQ mass spectrometer, which consisted of an electrospray ionization source and linear ion-trap mass analyzer. The sample extract was split into 2 aliquots, dried, and then reconstituted in acidic or basic LC-compatible solvents, each of which contained 11 or more injection standards at fixed concentrations. One aliquot was analyzed using acidic positive ion optimized conditions and the other using basic negative ion optimized conditions in 2 independent injections using separate dedicated columns. Extracts reconstituted in acidic conditions were gradient eluted using water and methanol, both containing 0.1% formic acid, while the basic extracts, which also used water/methanol, contained 6.5 mM ammonium bicarbonate. The MS analysis alternated between MS and data-dependent MS2 scans using dynamic exclusion.
The samples destined for GC/MS analysis were re-dried under vacuum desiccation for a minimum of 24 h prior to being derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide (BSTFA). The GC column was 5% phenyl and the temperature ramp is from 40 °C to 300 °C in a 16 min period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. The instrument was tuned and calibrated for mass resolution and mass accuracy on a daily basis. The information output from the raw data files was automatically extracted.
For ions with counts greater than 2 million, an accurate mass measurement could be performed. Accurate mass measurements could be made on the parent ion as well as fragments. The typical mass error was less than 5 ppm. Ions with less than 2 million counts require a greater amount of effort to characterize. Fragmentation spectra (MS/MS) were typically generated in a data-dependent manner, but if necessary, targeted MS/MS could be employed, such as in the case of lower level signals. Compounds were identified by comparison with library entries of purified standards or recurrent unknown entities. Identification of known chemical entities was based on comparison with metabolic library entries of purified standards. The combination of chromatographic properties and mass spectra gave an indication of a match to the specific compound or an isobaric entity.
Data quality: Instrument and process variability
Instrument variability was determined by calculating the median relative standard deviation (RSD) for the internal standards that were added to each sample prior to injection into the mass spectrometers. Overall process variability was determined by calculating the median RSD for all endogenous metabolites (i.e., non-instrument standards) present in 100% of the matrix samples, which are technical replicates of pooled client samples.
Metabolite summary and significantly altered biochemicals
The metabolic analysis, as from above, comprises a total of 148 named biochemicals. Following normalization to total protein determined by Bradford assay, log transformation, and imputation with minimum observed values was performed for each compound. Welch 2-sample t tests were then used to identify biochemicals that differed significantly between experimental groups. A summary of the numbers of biochemicals that achieved statistical significance (P ≤ 0.05), as well as those approaching significance (0.05 < P < 0.1) is shown as a table and a supplementary figure in the “Results” section.
An estimate of the false discovery rate (Q value) is calculated to take into account the multiple comparisons that normally occur in metabolic-based studies. For example, when analyzing 200 compounds, we would expect to see about 10 compounds meeting the P ≤ 0.05 cut-off by random chance. The Q value describes the false discovery rate; a low Q value (Q < 0.10) is an indication of high confidence in a result. While a higher Q value indicates diminished confidence, it does not necessarily rule out the significance of a result.
Supplementary Material
Acknowledgments
This work has been supported by the Medical Research Council, United Kingdom; MIUR, MinSan/IDI-IRCCS (RF73, RF57), ACC12, AIRC (2011-IG11955), AIRC 5xmille (MCO #9979) to GM.
Authors’ Contributions
MSC has performed the human sample collection, elaborated the scientific primary data, handled and contributed to the first draft of the manuscript; PT has elaborated the scientific results and written the manuscript. AEW, EC, and GM conceived the project, supervised the study and have written the manuscript.
Glossary
Abbreviations:
- RA
rheumatoid arthritis
- MTX
methotrexate
- DHFR
dihydrofolate reductase
- PFK1
phosphofructose kinase 1
- DHAP
dihydroxyacetone phosphate
- PEP
phosphoenolpyruvate
- PPP
pentose phosphate pathway
- PLA
phospholipase A
- TNF-α
tumour necrosis factor-α
- GC/MS
gas chromatography/mass spectrometry
- LC/MS/MS
liquid chromatography/mass spectrometry
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/cc/article/26067
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26067
s
- 1.Weinblatt ME, Trentham DE, Fraser PA, Holdsworth DE, Falchuk KR, Weissman BN, Coblyn JS. Long-term prospective trial of low-dose methotrexate in rheumatoid arthritis. Arthritis Rheum. 1988;31:167–75. doi: 10.1002/art.1780310203. [DOI] [PubMed] [Google Scholar]
- 2.Chimenti MS, Ballanti E, Perricone C, Cipriani P, Giacomelli R, Perricone R. Immunomodulation in psoriatic arthritis: focus on cellular and molecular pathways. Autoimmun Rev. 2013;12:599–606. doi: 10.1016/j.autrev.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Maini R, St Clair EW, Breedveld F, Furst D, Kalden J, Weisman M, Smolen J, Emery P, Harriman G, Feldmann M, et al. ATTRACT Study Group Infliximab (chimeric anti-tumour necrosis factor alpha monoclonal antibody) versus placebo in rheumatoid arthritis patients receiving concomitant methotrexate: a randomised phase III trial. Lancet. 1999;354:1932–9. doi: 10.1016/S0140-6736(99)05246-0. [DOI] [PubMed] [Google Scholar]
- 4.Weinblatt ME, van Riel PL. Targeted therapies: summary clinical trials working group. Ann Rheum Dis. 2006;65(Suppl 3):iii89. doi: 10.1136/ard.2006.060939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu YT, Tan HL, Huang Q, Sun XJ, Zhu X, Shen HM. zVAD-induced necroptosis in L929 cells depends on autocrine production of TNFα mediated by the PKC-MAPKs-AP-1 pathway. Cell Death Differ. 2011;18:26–37. doi: 10.1038/cdd.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takata M, Nakagomi T, Kashiwamura S, Nakano-Doi A, Saino O, Nakagomi N, Okamura H, Mimura O, Taguchi A, Matsuyama T. Glucocorticoid-induced TNF receptor-triggered T cells are key modulators for survival/death of neural stem/progenitor cells induced by ischemic stroke. Cell Death Differ. 2012;19:756–67. doi: 10.1038/cdd.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geering B, Simon HU. A novel signaling pathway in TNFα-induced neutrophil apoptosis. Cell Cycle. 2011;10:2821–2. doi: 10.4161/cc.10.17.16747. [DOI] [PubMed] [Google Scholar]
- 8.Christofferson DE, Li Y, Hitomi J, Zhou W, Upperman C, Zhu H, Gerber SA, Gygi S, Yuan J. A novel role for RIP1 kinase in mediating TNFα production. Cell Death Dis. 2012;3:e320. doi: 10.1038/cddis.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts NJ, Zhou S, Diaz LA, Jr., Holdhoff M. Systemic use of tumor necrosis factor alpha as an anticancer agent. Oncotarget. 2011;2:739–51. doi: 10.18632/oncotarget.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emery P, Breedveld FC, Hall S, Durez P, Chang DJ, Robertson D, Singh A, Pedersen RD, Koenig AS, Freundlich B. Comparison of methotrexate monotherapy with a combination of methotrexate and etanercept in active, early, moderate to severe rheumatoid arthritis (COMET): a randomised, double-blind, parallel treatment trial. Lancet. 2008;372:375–82. doi: 10.1016/S0140-6736(08)61000-4. [DOI] [PubMed] [Google Scholar]
- 11.Cleynen I, Vermeire S. Paradoxical inflammation induced by anti-TNF agents in patients with IBD. Nat Rev Gastroenterol Hepatol. 2012;9:496–503. doi: 10.1038/nrgastro.2012.125. [DOI] [PubMed] [Google Scholar]
- 12.O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–55. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 13.Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, McCormick LL, Fitzgerald P, Chi H, Munger J, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–82. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Ren G, Huang Y, Su J, Han Y, Li J, Chen X, Cao K, Chen Q, Shou P, et al. Mesenchymal stem cells: a double-edged sword in regulating immune responses. Cell Death Differ. 2012;19:1505–13. doi: 10.1038/cdd.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alonso R, Mazzeo C, Rodriguez MC, Marsh M, Fraile-Ramos A, Calvo V, Avila-Flores A, Merida I, Izquierdo M. Diacylglycerol kinase α regulates the formation and polarisation of mature multivesicular bodies involved in the secretion of Fas ligand-containing exosomes in T lymphocytes. Cell Death Differ. 2011;18:1161–73. doi: 10.1038/cdd.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tischner D, Wiegers GJ, Fiegl H, Drach M, Villunger A. Mutual antagonism of TGF-beta and Interleukin-2 in cell survival and lineage commitment of induced regulatory T cells. Cell Death Differ. 2012;19:1277–87. doi: 10.1038/cdd.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paulsen M, Valentin S, Mathew B, Adam-Klages S, Bertsch U, Lavrik I, Krammer PH, Kabelitz D, Janssen O. Modulation of CD4+ T-cell activation by CD95 co-stimulation. Cell Death Differ. 2011;18:619–31. doi: 10.1038/cdd.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madapura HS, Salamon D, Wiman KG, Lain S, Klein G, Klein E, Nagy N. p53 contributes to T-cell homeostasis through the induction of pro-apoptotic SAP. Cell Cycle. 2012;11:4563–9. doi: 10.4161/cc.22810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, Roodman DG, Windle JJ, Zhang X, Lu B. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012;19:144–52. doi: 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Szymczak-Workman AL, Gravano DM, Workman CJ, Green DR, Vignali DA. Preferential control of induced regulatory T-cell homeostasis via a Bim/Bcl-2 axis. Cell Death Dis. 2012;3:e270. doi: 10.1038/cddis.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunkle A, Dzhagalov I, He YW. Cytokine-dependent and cytokine-independent roles for Mcl-1: genetic evidence for multiple mechanisms by which Mcl-1 promotes survival in primary T lymphocytes. Cell Death Dis. 2011;2:e214. doi: 10.1038/cddis.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal A, Su H, Chen J, Osann K, Agrawal S, Gupta S. Increased IL-21 secretion by aged CD4+T cells is associated with prolonged STAT-4 activation and CMV seropositivity. Aging (Albany NY) 2012;4:648–59. doi: 10.18632/aging.100490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Denes A, Lopez-Castejon G, Brough D. Caspase-1: is IL-1 just the tip of the ICEberg? Cell Death Dis. 2012;3:e338. doi: 10.1038/cddis.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leverrier S, Salvesen GS, Walsh CM. Enzymatically active single chain caspase-8 maintains T-cell survival during clonal expansion. Cell Death Differ. 2011;18:90–8. doi: 10.1038/cdd.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramakrishnan P, Kahn DA, Baltimore D. Anti-apoptotic effect of hyperglycemia can allow survival of potentially autoreactive T cells. Cell Death Differ. 2011;18:690–9. doi: 10.1038/cdd.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramaswamy M, Cruz AC, Cleland SY, Deng M, Price S, Rao VK, Siegel RM. Specific elimination of effector memory CD4+ T cells due to enhanced Fas signaling complex formation and association with lipid raft microdomains. Cell Death Differ. 2011;18:712–20. doi: 10.1038/cdd.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cippà PE, Kraus AK, Lindenmeyer MT, Chen J, Guimezanes A, Bardwell PD, Wekerle T, Wüthrich RP, Fehr T. Resistance to ABT-737 in activated T lymphocytes: molecular mechanisms and reversibility by inhibition of the calcineurin-NFAT pathway. Cell Death Dis. 2012;3:e299. doi: 10.1038/cddis.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarkar A, Donkor MK, Li MO. T-cell- but not tumor cell-produced TGF-β1 promotes the development of spontaneous mammary cancer. Oncotarget. 2011;2:1339–51. doi: 10.18632/oncotarget.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verdino P, Wilson IA. JAML and CAR: Two more players in T-cell activation. Cell Cycle. 2011;10:1341–2. doi: 10.4161/cc.10.9.15294. [DOI] [PubMed] [Google Scholar]
- 30.Nakajima T, McGee H, Finn PW. CDK4: Regulatory functions related to lymphocytes. Cell Cycle. 2011;10:1527. doi: 10.4161/cc.10.10.15524. [DOI] [PubMed] [Google Scholar]
- 31.Beier UH, Wang L, Hancock WW. Combination of isoform-selective histone/protein deacetylase inhibitors improves Foxp3+ T-regulatory cell function. Cell Cycle. 2012;11:3351–2. doi: 10.4161/cc.21876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patsoukis N, Sari D, Boussiotis VA. PD-1 inhibits T-cell proliferation by upregulating p27 and p15 and suppressing Cdc25A. Cell Cycle. 2012;11:4305–9. doi: 10.4161/cc.22135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbosa CMV, Leon CMMP, Nogueira-Pedro A, Wasinsk F, Araújo RC, Miranda A, Ferreira AT, Paredes-Gamero EJ. Differentiation of hematopoietic stem cell and myeloid populations by ATP is modulated by cytokines. Cell Death Dis. 2011;2:e165. doi: 10.1038/cddis.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs SR, Herman CE, Maciver NJ, Wofford JA, Wieman HL, Hammen JJ, Rathmell JC. Glucose uptake is limiting in T-cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008;180:4476–86. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cham CM, Gajewski TF. Glucose availability regulates IFN-gamma production and p70S6 kinase activation in CD8+ effector T cells. J Immunol. 2005;174:4670–7. doi: 10.4049/jimmunol.174.8.4670. [DOI] [PubMed] [Google Scholar]
- 36.Chandra RK. Numerical and functional deficiency in T helper cells in protein energy malnutrition. Clin Exp Immunol. 1983;51:126–32. [PMC free article] [PubMed] [Google Scholar]
- 37.Agostini M, Tucci P, Melino G. Cell death pathology: perspective for human diseases. Biochem Biophys Res Commun. 2011;414:451–5. doi: 10.1016/j.bbrc.2011.09.081. [DOI] [PubMed] [Google Scholar]
- 38.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–20. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandenabeele P, Melino G. The flick of a switch: which death program to choose? Cell Death Differ. 2012;19:1093–5. doi: 10.1038/cdd.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013;••• doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 41.Tucci P. Caloric restriction: is mammalian life extension linked to p53? Aging (Albany NY) 2012;4:525–34. doi: 10.18632/aging.100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newbold A, Vervoort SJ, Martin BP, Bots M, Johnstone RW. Induction of autophagy does not alter the anti-tumor effects of HDAC inhibitors. Cell Death Dis. 2012;3:e387. doi: 10.1038/cddis.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melino G, Knight RA, Nicotera P. How many ways to die? How many different models of cell death? Cell Death Differ. 2005;12(Suppl 2):1457–62. doi: 10.1038/sj.cdd.4401781. [DOI] [PubMed] [Google Scholar]
- 44.Tóth K, Sarang Z, Scholtz B, Brázda P, Ghyselinck N, Chambon P, Fésüs L, Szondy Z. Retinoids enhance glucocorticoid-induced apoptosis of T cells by facilitating glucocorticoid receptor-mediated transcription. Cell Death Differ. 2011;18:783–92. doi: 10.1038/cdd.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rathmell JC, Elstrom RL, Cinalli RM, Thompson CB. Activated Akt promotes increased resting T-cell size, CD28-independent T-cell growth, and development of autoimmunity and lymphoma. Eur J Immunol. 2003;33:2223–32. doi: 10.1002/eji.200324048. [DOI] [PubMed] [Google Scholar]
- 46.Tucci P, Porta G, Agostini M, Dinsdale D, Iavicoli I, Cain K, Finazzi-Agró A, Melino G, Willis AE. Metabolic effects of TiO2 nanoparticles, a common component of sunscreens and cosmetics, on human keratinocytes. Cell Death Dis. 2013;4e:549. doi: 10.1038/cddis.2013.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tucci P, Porta G, Agostini M, Antonov A, Garabadgiu AV, Melino G, Willis AE. Rapamycin regulates biochemical metabolites. Cell Cycle. 2013;12 doi: 10.4161/cc.25450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. 2003;423:356–61. doi: 10.1038/nature01661. [DOI] [PubMed] [Google Scholar]
- 49.Chimenti MS, Graceffa D, Perricone R. Anti-TNFα discontinuation in rheumatoid and psoriatic arthritis: is it possible after disease remission? Autoimmun Rev. 2011;10:636–40. doi: 10.1016/j.autrev.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 50.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T-cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348–57. [PubMed] [Google Scholar]
- 51.Blanchard Z, Malik R, Mullins N, Maric C, Luk H, Horio D, Hernandez B, Killeen J, Elshamy WM. Geminin overexpression induces mammary tumors via suppressing cytokinesis. Oncotarget. 2011;2:1011–27. doi: 10.18632/oncotarget.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wogan GN, Dedon PC, Tannenbaum SR, Fox JG. Infection, inflammation and colon carcinogenesis. Oncotarget. 2012;3:737–8. doi: 10.18632/oncotarget.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez-Outschoorn UE, Whitaker-Menezes D, Lin Z, Flomenberg N, Howell A, Pestell RG, Lisanti MP, Sotgia F. Cytokine production and inflammation drive autophagy in the tumor microenvironment: role of stromal caveolin-1 as a key regulator. Cell Cycle. 2011;10:1784–93. doi: 10.4161/cc.10.11.15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–80. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hofius D, Munch D, Bressendorff S, Mundy J, Petersen M. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 2011;18:1257–62. doi: 10.1038/cdd.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Denton D, Nicolson S, Kumar S. Cell death by autophagy: facts and apparent artefacts. Cell Death Differ. 2012;19:87–95. doi: 10.1038/cdd.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.D’Eletto M, Farrace MG, Rossin F, Strappazzon F, Giacomo GD, Cecconi F, Melino G, Sepe S, Moreno S, Fimia GM, et al. Type 2 transglutaminase is involved in the autophagy-dependent clearance of ubiquitinated proteins. Cell Death Differ. 2012;19:1228–38. doi: 10.1038/cdd.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pagliarini V, Wirawan E, Romagnoli A, Ciccosanti F, Lisi G, Lippens S, Cecconi F, Fimia GM, Vandenabeele P, Corazzari M, et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 2012;19:1495–504. doi: 10.1038/cdd.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ling LU, Tan KB, Lin H, Chiu GNC. The role of reactive oxygen species and autophagy in safingol-induced cell death. Cell Death Dis. 2011;2:e129. doi: 10.1038/cddis.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drullion C, Trégoat C, Lagarde V, Tan S, Gioia R, Priault M, Djavaheri-Mergny M, Brisson A, Auberger P, Mahon FX, et al. Apoptosis and autophagy have opposite roles on imatinib-induced K562 leukemia cell senescence. Cell Death Dis. 2012;3:e373. doi: 10.1038/cddis.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rambold AS, Lippincott-Schwartz J. Mechanisms of mitochondria and autophagy crosstalk. Cell Cycle. 2011;10:4032–8. doi: 10.4161/cc.10.23.18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Criollo A, Chereau F, Malik SA, Niso-Santano M, Mariño G, Galluzzi L, Maiuri MC, Baud V, Kroemer G. Autophagy is required for the activation of NFκB. Cell Cycle. 2012;11:194–9. doi: 10.4161/cc.11.1.18669. [DOI] [PubMed] [Google Scholar]
- 63.Cianfanelli V, Cecconi F. Autophagy-dependent NFκB regulation. Cell Cycle. 2012;11:436–7. doi: 10.4161/cc.11.3.19224. [DOI] [PubMed] [Google Scholar]
- 64.Wang X, Zeng L, Wang J, Chau JFL, Lai KP, Jia D, Poonepalli A, Hande MP, Liu H, He G, et al. A positive role for c-Abl in Atm and Atr activation in DNA damage response. Cell Death Differ. 2011;18:5–15. doi: 10.1038/cdd.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walter BM, Nordhoff C, Varga G, Goncharenko G, Schneider SW, Ludwig S, Wixler V. Mss4 protein is a regulator of stress response and apoptosis. Cell Death Dis. 2012;3:e297. doi: 10.1038/cddis.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hui KKW, Kanungo AK, Elia AJ, Henderson JT. Caspase-3 deficiency reveals a physiologic role for Smac/DIABLO in regulating programmed cell death. Cell Death Differ. 2011;18:1780–90. doi: 10.1038/cdd.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Eto K, Goto S, Nakashima W, Ura Y, Abe SI. Loss of programmed cell death 4 induces apoptosis by promoting the translation of procaspase-3 mRNA. Cell Death Differ. 2012;19:573–81. doi: 10.1038/cdd.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang S, Xia P, Shi L, Fan Z. FADD cleavage by NK cell granzyme M enhances its self-association to facilitate procaspase-8 recruitment for auto-processing leading to caspase cascade. Cell Death Differ. 2012;19:605–15. doi: 10.1038/cdd.2011.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wickman G, Julian L, Olson MF. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012;19:735–42. doi: 10.1038/cdd.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.García-Sáez AJ. The secrets of the Bcl-2 family. Cell Death Differ. 2012;19:1733–40. doi: 10.1038/cdd.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jouan-Lanhouet S, Arshad MI, Piquet-Pellorce C, Martin-Chouly C, Le Moigne-Muller G, Van Herreweghe F, Takahashi N, Sergent O, Lagadic-Gossmann D, Vandenabeele P, et al. TRAIL induces necroptosis involving RIPK1/RIPK3-dependent PARP-1 activation. Cell Death Differ. 2012;19:2003–14. doi: 10.1038/cdd.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Graber TE, Holcik M. Distinct roles for the cellular inhibitors of apoptosis proteins 1 and 2. Cell Death Dis. 2011;2:e135. doi: 10.1038/cddis.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mühlethaler-Mottet A, Flahaut M, Bourloud KB, Nardou K, Coulon A, Liberman J, Thome M, Gross N. Individual caspase-10 isoforms play distinct and opposing roles in the initiation of death receptor-mediated tumour cell apoptosis. Cell Death Dis. 2011;2:e125. doi: 10.1038/cddis.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Badmann A, Keough A, Kaufmann T, Bouillet P, Brunner T, Corazza N. Role of TRAIL and the pro-apoptotic Bcl-2 homolog Bim in acetaminophen-induced liver damage. Cell Death Dis. 2011;2:e171. doi: 10.1038/cddis.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barbone D, Ryan JA, Kolhatkar N, Chacko AD, Jablons DM, Sugarbaker DJ, Bueno R, Letai AG, Coussens LM, Fennell DA, et al. The Bcl-2 repertoire of mesothelioma spheroids underlies acquired apoptotic multicellular resistance. Cell Death Dis. 2011;2:e174. doi: 10.1038/cddis.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yivgi-Ohana N, Eifer M, Addadi Y, Neeman M, Gross A. Utilizing mitochondrial events as biomarkers for imaging apoptosis. Cell Death Dis. 2011;2:e166. doi: 10.1038/cddis.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Osawa T, Davies D, Hartley JA. Mechanism of cell death resulting from DNA interstrand cross-linking in mammalian cells. Cell Death Dis. 2011;2:e187. doi: 10.1038/cddis.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Teng X, Cheng WC, Qi B, Yu TX, Ramachandran K, Boersma MD, Hattier T, Lehmann PV, Pineda FJ, Hardwick JM. Gene-dependenT-cell death in yeast. Cell Death Dis. 2011;2:e188. doi: 10.1038/cddis.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Knight RA, Melino G. Cell death in disease: from 2010 onwards. Cell Death Dis. 2011;2:e202. doi: 10.1038/cddis.2011.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seervi M, Joseph J, Sobhan PK, Bhavya BC, Santhoshkumar TR. Essential requirement of cytochrome c release for caspase activation by procaspase-activating compound defined by cellular models. Cell Death Dis. 2011;2:e207. doi: 10.1038/cddis.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gonzalez-Halphen D, Ghelli A, Iommarini L, Carelli V, Esposti MD. Mitochondrial complex I and cell death: a semi-automatic shotgun model. Cell Death Dis. 2011;2:e222. doi: 10.1038/cddis.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kober AMM, Legewie S, Pforr C, Fricker N, Eils R, Krammer PH, Lavrik IN. Caspase-8 activity has an essential role in CD95/Fas-mediated MAPK activation. Cell Death Dis. 2011;2:e212. doi: 10.1038/cddis.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song S, Choi K, Ryu SW, Kang SW, Choi C. TRAIL promotes caspase-dependent pro-inflammatory responses via PKCδ activation by vascular smooth muscle cells. Cell Death Dis. 2011;2:e223. doi: 10.1038/cddis.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salomoni P, Dvorkina M, Michod D. Role of the promyelocytic leukaemia protein in cell death regulation. Cell Death Dis. 2012;3:e247. doi: 10.1038/cddis.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong C, Anderson DJ, Lee EF, Fairlie WD, Ludlam MJC. Direct visualization of Bcl-2 family protein interactions using live cell fluorescent protein redistribution assays. Cell Death Dis. 2012;3:e288. doi: 10.1038/cddis.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Richard A, Tulasne D. Caspase cleavage of viral proteins, another way for viruses to make the best of apoptosis. Cell Death Dis. 2012;3:e277. doi: 10.1038/cddis.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Esposito F, Tornincasa M, Federico A, Chiappetta G, Pierantoni GM, Fusco A. High-mobility group A1 protein inhibits p53-mediated intrinsic apoptosis by interacting with Bcl-2 at mitochondria. Cell Death Dis. 2012;3:e383. doi: 10.1038/cddis.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 88.Korah J, Falah N, Lacerte A, Lebrun JJ. A transcriptionally active pRb-E2F1-P/CAF signaling pathway is central to TGFβ-mediated apoptosis. Cell Death Dis. 2012;3:e407. doi: 10.1038/cddis.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giam M, Okamoto T, Mintern JD, Strasser A, Bouillet P. Bcl-2 family member Bcl-G is not a proapoptotic protein. Cell Death Dis. 2012;3:e404. doi: 10.1038/cddis.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pobezinskaya YL, Liu Z. The role of TRADD in death receptor signaling. Cell Cycle. 2012;11:871–6. doi: 10.4161/cc.11.5.19300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.van Leeuwen IM, Laín S. Pharmacological manipulation of the cell cycle and metabolism to protect normal tissues against conventional anticancer drugs. Oncotarget. 2011;2:274–6. doi: 10.18632/oncotarget.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burger JA, Hoellenriegel J. Phosphoinositide 3′-kinase delta: turning off BCR signaling in Chronic Lymphocytic Leukemia. Oncotarget. 2011;2:737–8. doi: 10.18632/oncotarget.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ganapathy-Kanniappan S, Kunjithapatham R, Geschwind JF. Glyceraldehyde-3-phosphate dehydrogenase: a promising target for molecular therapy in hepatocellular carcinoma. Oncotarget. 2012;3:940–53. doi: 10.18632/oncotarget.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zha X, Sun Q, Zhang H. mTOR upregulation of glycolytic enzymes promotes tumor development. Cell Cycle. 2011;10:1015–6. doi: 10.4161/cc.10.7.15063. [DOI] [PubMed] [Google Scholar]
- 95.Lock R, Debnath J. Ras, autophagy and glycolysis. Cell Cycle. 2011;10:1516–7. doi: 10.4161/cc.10.10.15434. [DOI] [PubMed] [Google Scholar]
- 96.Blagosklonny MV. Rapamycin-induced glucose intolerance: hunger or starvation diabetes. Cell Cycle. 2011;10:4217–24. doi: 10.4161/cc.10.24.18595. [DOI] [PubMed] [Google Scholar]
- 97.Capparelli C, Guido C, Whitaker-Menezes D, Bonuccelli G, Balliet R, Pestell TG, Goldberg AF, Pestell RG, Howell A, Sneddon S, et al. Autophagy and senescence in cancer-associated fibroblasts metabolically supports tumor growth and metastasis via glycolysis and ketone production. Cell Cycle. 2012;11:2285–302. doi: 10.4161/cc.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.MacFarlane M, Robinson GL, Cain K. Glucose--a sweet way to die: metabolic switching modulates tumor cell death. Cell Cycle. 2012;11:3919–25. doi: 10.4161/cc.21804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bluemlein K, Grüning NM, Feichtinger RG, Lehrach H, Kofler B, Ralser M. No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget. 2011;2:393–400. doi: 10.18632/oncotarget.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luo W, Semenza GL. Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. 2011;2:551–6. doi: 10.18632/oncotarget.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bluemlein K, Glückmann M, Grüning NM, Feichtinger R, Krüger A, Wamelink M, Lehrach H, Tate S, Neureiter D, Kofler B, et al. Pyruvate kinase is a dosage-dependent regulator of cellular amino acid homeostasis. Oncotarget. 2012;3:1356–69. doi: 10.18632/oncotarget.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ballanti E, Perricone C, di Muzio G, Kroegler B, Chimenti MS, Graceffa D, Perricone R. Role of the complement system in rheumatoid arthritis and psoriatic arthritis: relationship with anti-TNF inhibitors. Autoimmun Rev. 2011;10:617–23. doi: 10.1016/j.autrev.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 103.Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, Muller PA, Dötsch V, Kehrloesser S, Sayan BS, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci U S A. 2012;109:15312–7. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.