Abstract
The Bacillus genus is a large heterogeneous group in need of an efficient method for species differentiation. To determine the current validity of a sequence-based method for identification and provide contemporary data, PCR and sequencing of a 500-bp product encompassing the V1 to V3 regions of the 16S rRNA gene were undertaken using 65 of the 83 type strains of this genus. This region proved discriminatory between most species (70.0 to 100% similarity), the exceptions being clinically relevant B. cereus and B. anthracis as well as nonpathogenic B. psychrotolerans and B. psychrodurans. Consequently, 27 type and clinical strains from the B. cereus group were used to test alternate targets (rpoB, vrrA, and the 16S-23S spacer region) for identification. The rpoB gene proved the best alternate target, with a conserved 4-nucleotide difference between B. cereus and B. anthracis. The high 16S rRNA gene sequence similarities between some strains demonstrated the need for a polyphasic approach to the systematics of this genus. This approach is one focus of the Ribosomal Differentiation of Medical Microorganisms mandate. Accordingly, the 16S rRNA gene sequences generated in this study have been submitted for inclusion into its publicly accessible, quality-controlled database at http://www.ridom_rdna.de/.
The Bacillus genus is an extensive heterogeneous group encompassing 83 validly described species to date (http://www.bacterio.cict.fr/b/bacillus.html). Many species in this taxon are of major clinical importance, such as the B. cereus group (comprised of B. cereus, B. anthracis, B. thuringiensis, B. mycoides, and B. weihenstephanensis), but unfortunately, members of this group share a great deal of morphological and biochemical similarities (3, 8, 16). In contrast, the environmental and nonpathogenic species of this genus exhibit a wide range of physiology, DNA base content, and nutritional requirements (2, 4, 15). Since the biochemical approach for species identification can be tedious, expensive, and inaccurate, a rapid, definitive method is greatly needed. Molecular procedures are increasingly being used for rapid species identification. However, some methods used for this genus such as restriction digests of a target gene (i.e., 16S rRNA gene) (11) or randomly amplified polymorphic DNA analysis (22) are limiting in discriminating between a large group of species (6). Sequencing of the 16S rRNA gene and select housekeeping genes has shown to be particularly useful, generating large public sequence databases due to the tangible, exact nature of sequence data. With the increasing use of these methods and decreased expense of running sequencing reactions after the initial equipment investment, more laboratories are relying on sequence data for species identification (21).
A previous study using the 16S rRNA gene for rapid identification of the Bacillus genus was undertaken by Goto et al. (6). At this time, the validity of using a hypervariable region (nucleotides [nt] 70 to 344) of the gene was proven adequate to discriminate between all the species except between B. cereus and B. anthracis and between B. mojavensis and B. atrophaeus. However, new sequence data were only acquired for 19 of the species, with the rest obtained from preexisting sequences available from the National Center for Biotechnology Information GenBank. The GenBank nucleotide database is well known for the non-quality-controlled nature of its data, including base errors, ambiguous base designation, and incomplete, short sequences. Several recent studies have examined the problems surrounding the use of non-quality-controlled databases such as GenBank and the Ribosomal Database Project for identification purposes and have shown the benefits of standardized, maintained databanks that include subsidiary information, such as Ribosomal Differentiation of Medical Microorganisms (RIDOM) (7, 21).
With the available data on this genus incomplete and the many problems associated with public database use for similarity searches, a fragment of the 16S rRNA gene (Escherichia coli nt 54 to 510) for species of the Bacillus genus was sequenced for submission to RIDOM. Current sequence technologies allow the acquisition of unambiguous, error-free data for definitive identification. This is only one of many collaborative ongoing efforts to collect quality-controlled sequence data for RIDOM for free access to others. Second, alternate sequence targets for identification of the closely related B. cereus group were reviewed and tested for inclusion into RIDOM.
MATERIALS AND METHODS
A total of 65 of 83 Bacillus type strains were currently available for this study (Table 1). The partial 16S rRNA gene sequence (corresponding to primers for E. coli 16S rRNA positions 8 to 27 and 536 to 518) (21) was determined using standard 16S rRNA gene primers for PCR and sequencing. For the members of the B. cereus clade, rpoB gene amplification and sequencing were undertaken with previously published primers (positions 1482 to 1500 and positions 2281 to 2300 of the B. subtilis rpoB gene) (17). Both forward and reverse strands were sequenced using standard procedures of cycle sequencing with an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems).
TABLE 1.
Bacillus species type strains used in this study
| Species | Identifiera (accession no.) |
|---|---|
| B. agaradhaerens | DSM 8721T |
| B. alcalophilus | DSM 485T (X76436) |
| B. amyloliquefaciens | ATCC 23350T |
| B. anthracis | ATCC 14578T |
| B. arseniciselenatis | ATCC 700614T |
| B. atrophaeus | DSM 7264T |
| B. azotoformans | DSM 1046T |
| B. badius | ATCC 14574T |
| B. benzoevorans | DSM 5391T |
| B. carboniphilus | LMG 19001T |
| B. cereus | ATCC 14579T |
| B. chitinolyticus | DSM 11030T |
| B. circulans | ATCC 4513T |
| B. clarkii | DSM 8720T |
| B. clausii | DSM 8716T |
| B. coagulans | ATCC 7050T |
| B. cohnii | DSM 6307T |
| B. decolorationis | LMG 19507T (AJ315075) |
| B. edaphicus | T7 / AF006076 |
| B. ehimensis | DSM 11029T |
| B. endophyticus | 2DTT (AF295302) |
| B. fastidiosus | DSM 91T |
| B. firmus | ATCC 14575T |
| B. flexus | DSM 1320T |
| B. fumarioli | LMG 19448T |
| B. funiculus | NAF001/AB049195 |
| B. fusiformis | DSM 2898T |
| B. gibsonii | DSM 8722T |
| B. halmapalus | DSM 8723T |
| B. haloalkaliphilus | DSM 5271T |
| B. halodenitrificans | DSM 10037T |
| B. halodurans | DSM 497T |
| B. halophilus | DSM 4771T |
| B. horikoshii | DSM 8719T |
| B. horti | DSM 12751T |
| B. infernus | DSM 10277T |
| B. insolitus | ATCC 23299T |
| B. vallismortis | DSM 11031T |
| B. vedderi | DSM 9768T |
| B. vulcani | DSM 13174T |
| B. weihenstephanensis | DSM 11921T |
| B. tusciae | DSM 2912T |
| B. jeotgali | YKJ-10T (AF221061) |
| B. laevolacticus | DSM 442T |
| B. lentus | ATCC 10840T |
| B. lichenformis | ATCC 14580T |
| B. luciferensis | LMG 19422T (AJ419629) |
| B. megaterium | ATCC 14581T |
| B. methanolicus | C1 (X64465) |
| B. mojavensis | DSM 9205T |
| B. mucilaginosus | AF006077 |
| B. mycoides | ATCC 6462T |
| B. naganoensis | DSM 10191T |
| B. nealsonii | FO-092 (AF234863) |
| B. neidei | BD-87 (AF169520) |
| B. niacini | DSM 2923T |
| B. okuhidensis | GTC854/AB047684 |
| B. oleronius | DSM 9356T |
| B. pallidus | DSM 3670T |
| B. pseudalcaliphilus | DSM 8725T |
| B. pseudofirmus | DSM 8715T |
| B. pseudomycoides | DSM 12442T |
| B. psychrodurans | DSM 11713T (AJ277984) |
| B. psychrosaccharolyticus | DSM 6T |
| B. psychrotolerans | DSM 11706T (AJ277983) |
| B. pumilus | ATCC 7061T |
| B. pycnus | NRS-1691 (AF169531) |
| B. schlegelii | ATCC 43741T (AB042060) |
| B. selenitireducens | ATCC 700615T |
| B. silvestris | DSM 12223T |
| B. simplex | DSM 1321T |
| B. siralis | DSM 13140T |
| B. smithii | DSM 4216T |
| B. sonorensis | DSM 13779T |
| B. sphaericus | ATCC 14577T |
| B. sporothermodurans | DSM 10599T |
| B. subterraneus | —b |
| B. subtilis subsp. subtilis | ATCC 6051T |
| B. subtilis subsp. spizizenii | NRRL B-23049T |
| B. thermantarcticus | M1 (AJ493665) |
| B. thermoamylovorans | LMG 19084T |
| B. thermocloaceae | DSM 5250T |
| B. thuringiensis | ATCC 10792T |
Abbreviations: DSM, Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (German Collection of Microorganisms and Cell Cultures), Braunschweig, Germany; ATCC, American Type Culture Collection, Manassas, Va.; LMG, Belgian Coordinated Collections of Microorganisms, Laboratorium voor Microbiologie, Universiteit Gent (RUG), Ghent, Belgium; NRRL, Northern Regional Research Laboratory, U.S. Department of Agriculture, Peoria, Ill.
—, identifier not applicable; GenBank sequence not available.
Alignments and phylogenetic analysis of the 16S rRNA gene sequences completed in-house were performed using nucleotide sequences from position 54 to 510. For complete analysis of the genus, sequences of 17 Bacillus species that we were unable to obtain in this study were chosen from GenBank. Except for three species noted in Fig. 1, these sequences were deemed free of any questionable deletions, insertions, or ambiguous bases (accession numbers are noted in Table 1). In addition, one newly described species, B. subterraneus ATCC BAA 136T, did not have a 16S sequence available in GenBank. rpoB gene sequences were analyzed using a fragment from position 1821 to 1995 of the B. subtilis rpoB gene. Multiple alignments and the construction of a neighbor-joining phylogenetic tree subjected to a bootstrapping analysis of 1,000 simulations to assess topology were performed with Bionumerics (version 2.50; Applied Maths) default parameters. The sequences obtained from GenBank were highlighted in the tree to distinguish them from the strains sequenced in-house. Alicyclobacillus acidocaldarius (X60742) was used as the outgroup to compare our results with those of Goto et al. (6). The sequences determined in the study have been submitted to RIDOM to be available in the near future for similarity searches.
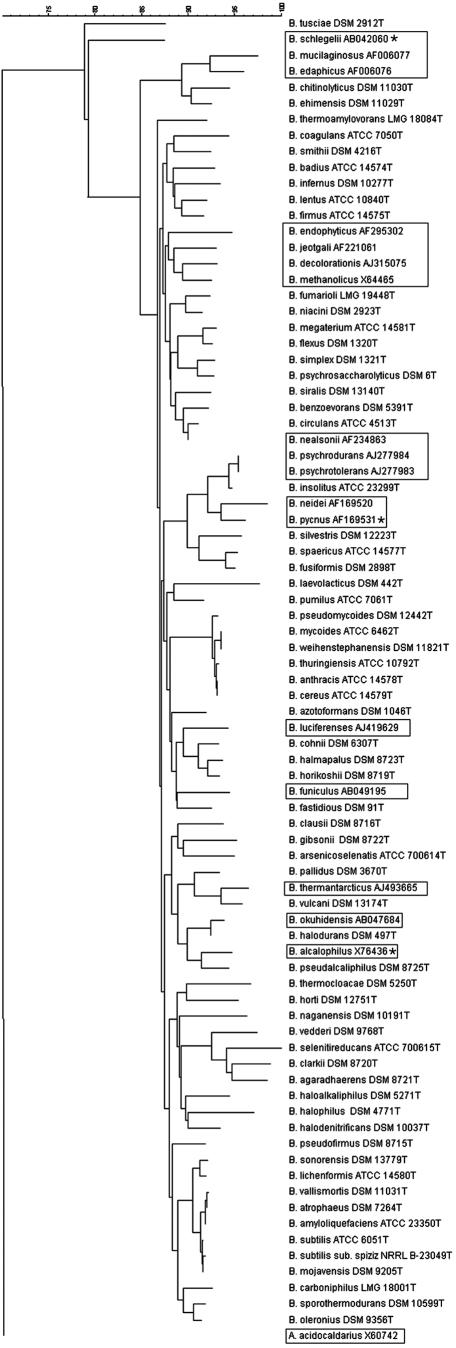
FIG.1.
Neighbor-joining phylogenetic tree based on the V1-V3 region of the 16S rRNA gene (E. coli nt 54 to 510) of Bacillus species used in this study. Sequences we were unable to obtain in this study were taken from GenBank (boxed). Three strains (*) had one ambiguous base pair (n). The branching pattern is rooted using A. acidocaldarius as the outlier. Created with Bionumerics (version 2.50).
RESULTS
Interspecies sequence identity results of the 16S rRNA gene sequences from bp 54 to 510, which includes hypervariable regions V1 to V3, demonstrated a range of 70.0 to 100% similarity (data not shown), with the closest related species (excluding the B. cereus clade) being two recently published environmental species, B. psychrotolerans and B. psychrodurans (1), which showed 100% identity. Within the B. subtilis group, between B. atrophaeus and B. vallismortis, as well as B. subtilis subsp. spizizenii and B. mojavensis, a 1-bp difference was observed (99.8% identity). B. atrophaeus and B. mojavensis have 100% sequence identity in the region used in previously published studies (nt 70 to 344) but can be differentiated due to a 3-bp difference in the V3 region. The most distantly related Bacillus species were B. tusciae and B. neidei, presumably due to several regions of deletions detected in B. tusciae. Use of this fragment of the gene for phylogenetic analysis shows similar clade assignments compared to phylogenetic trees constructed using the near complete 16S rRNA gene sequences as illustrated in previous publication (6) (Fig. 1).
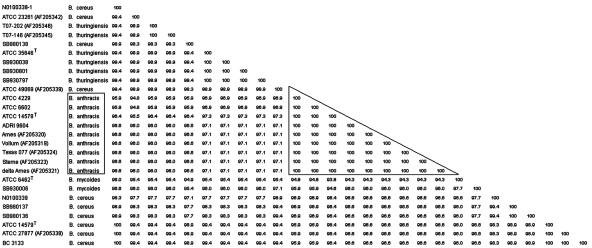
A review of current chromosomal targets for identification of the medically relevant B. cereus group prompted us to examine the use of the vrrA region (10), 16S-23S spacer region (4, 8), and the rpoB (17) gene for sequence-based identification. The vrrA region does not include a known, conserved housekeeping gene, and the variability observed is much more suitable for subtyping instead of identification (12). The 16S-23S spacer region shows a single base insertion difference between B. cereus and B. anthracis. The rpoB was the best alternate target, allowing discrimination between B. cereus and B. anthracis by a conserved 4-bp difference over a region of 175 bp in all isolates tested in this study as well as previous research (17). As illustrated in Fig. 2, the similarity index indicates 100% identity in therpoB sequences of B. anthracis, making it an ideal target for identification purposes.
FIG. 2.
B. cereus group members (clinical as well as type strains) used for rpoB gene analysis. The similarity matrix (pairwise comparison) and corresponding phylogenetic tree (neighbor joining) were created with Bionumerics (version 2.5).
DISCUSSION
A generally accepted concept in bacterial taxonomy is that the DNA base (GC) composition of species within a genus should not differ by more than 10 to 12 %mol G+C (15). Nonetheless, values within the Bacillus genus ranged from 33 to 65 %mol G+C in 1993, although many of the species did cluster at 40 to 50 %mol G+C (15). Subsequently, recent phylogenetic analyses have reclassified some of the Bacillus species into new genera, including Paenibacillus, Geobacillus, and Brevibacillus (4). Due to these recent advances, it has become increasingly difficult to classify species within the Bacillus genus, as many share similar physiology, metabolism, and morphology as well as highly conserved 16S rRNA genes. Fox et al. (5) indicate that a new species should be created when the organism has a sequence difference of 1.5% (over 1,000 bp) in conjunction with phenotypic differences. However, these studies on Bacillus globisporus and Bacillus psychrophilus demonstrated a 16S rRNA gene sequence similarity of 99.5%. These data revealed that although 16S rRNA gene sequences can be routinely used to identify and establish relationships between genera and well-resolved species, very recently diverged species may not be identified (5, 14).
It is important to note that ideally a polyphasic approach to the systematics of this genus (and all genera) should be practiced to fully understand and classify organisms, as a reliance on a singular molecular method such as 16S rRNA gene sequencing cannot account for slight evolutionary events and may “overspeciate” the genus of study (i.e., may subdivide the genus into too many species). In contrast, two species may exist with identical 16S rRNA sequences yet have phenotypic differences or may differ in clinical relevance. Therefore, in practice, a number of phenotypic and phylogenetic properties should be examined to establish taxonomic positions of groups of related strains as a strain or a species (20).
Several examples of applying a polyphasic approach to delineate a new species from a group of similar strains were observed within this genus, specifically among the recently or newly described species. B. psychrotolerans and B. psychrodurans are newly described psychrotolerant species that have 100% sequence identity with the region of the 16S rRNA gene chosen in this study, but they can be differentiated further downstream of the 16S rRNA gene, as well as by biochemical characteristics (1). This is also evident for members recently established within the B. subtilis group, i.e., B. atrophaeus and B. mojavensis, which can be differentiated by both a 3-nt difference in the region tested and phenotypic differences such as oxidase activity. Thus, in the case of a nontype strain of these two species with a possible 16S rRNA sequence polymorphism(s), testing for oxidase activity could support identification to the species level (18).
In contrast, other closely related organisms within this genus can share phenotypic properties as well but have been classified as different species based on DNA reassociation values. This is observed between B. subtilis subsp. subtilis and B. subtilis subsp. spizizenii, which share phenotypic profiles but are segregated based on DNA reassociation values of 58 to 69%, in addition to minor polymorphisms in the 16S rRNA gene between the type strains (13). Furthermore, B. mojavensis and B. subtilis subsp. spizizenii have only a 1-bp difference in the 16S rRNA gene and can only be distinguished from each other by sexual isolation, divergence in DNA sequences of the rpoB and gyrA genes, and fatty acid composition (13). These are a examples where reliance on only biochemical-based identification could lead to inaccurate identification of an organism.
The above discussion focuses on harmless saprophytes which are currently not of clinical importance, for which a rapid turnaround time to identification is less critical. However, B. cereus and B. anthracis, which can be extremely pathogenic, have 100% sequence identity across the entire 16S rRNA gene. The B. cereus group is highly homologous, as shown by genomic DNA-DNA hybridization, and the validity of classifying each as a species on the basis of pathogenicity has been questioned (9, 17). Although the species belonging to the B. cereus group can generally be differentiated from each other with conventional biochemical tests, such as capsular staining, motility, hemolysis, and observing the presence of intracellular para-crystalline formation (8, 9, 17), these tests are time-consuming and, in the case of genetically modified strains, may not even be useful for identification to the species level.
Although a recent publication by Sacchi et al. cites differences in the complete 16S rRNA gene (19), the single difference present over the entire 1,554-bp gene between B. anthracis and three B. cereus strains is a W (representing A or T) versus an A. This difference at bp 1146 of the gene (beyond the region examined in this study) may only be a reflection of base pair variation between multiple ribosomal operons in Bacillus species and not a true interspecies difference. The disadvantage of using this target for identification is twofold. First, the sequencing technology has to be PCR and not clone based in order to detect the “mixed” nucleotide caused by multiple ribosomal operons, and second, multiple primers would be necessary to obtain the complete sequence, which is not as rapid and unmistakable as using an alternate, smaller target with greater sequence variability. Several alternate chromosomal targets have been studied, although most suffer from inadequacy in some aspect, such as the Ba813 marker which has been detected in both B. cereus and B. thuringiensis (17). The vrrA region tested in this study has been noted as a possible credible method of distinguishing B. anthracis from B. cereus due to specific allele patterns defined for B. anthracis; however, only a limited amount of B. cereus and B. thuringiensis isolates were tested (12). Furthermore, as mentioned earlier, this target is useful primarily for subtyping and not for routine identification in a clinical laboratory. The use of a conserved, housekeeping gene necessary for the survival of the organism such as rpoB is a desirable alternative.
In conclusion, the Bacillus genus requires a polyphasic approach to definitive species identification, including alternate gene targets as well as chemotaxonomic and clinical information (20). RIDOM is attempting to fill this niche by means of a quality-controlled, error-free 16S rRNA gene sequence-based identification database that also includes both secondary targets (such as the 16S-23S spacer region, and possibly the rpoB gene in the near future) and ancillary information regarding phenotypical characteristics. Consequently, when newly described pathogenic Bacillus species that have 16S ribosomal DNA sequences almost identical or identical to those of preexisting species are validated, the accumulation of a variety of strain characteristics in such a database is critical in the establishment of taxonomic positions. From a clinical standpoint, rapid, presumptive identification to the level of a certain group is useful to confirm medical diagnosis and aid in further differentiation.
Acknowledgments
We thank the following for the kind donation of Bacillus strains: K. Bernard of Special Bacteriology, NML, for the ATCC strains; L. K. Nakamura for B. subtilis subsp. spizizenii; and J. S. Blum for B. arseniciselenatis and B. selenitireducens.
REFERENCES
- 1.Abd El-Rahman, H. A., D. Fritze, C. Sproer, and D. Claus. 2002. Two novel psychrotolerant species, Bacillus psychrotolerans sp. nov. and Bacillus psychrodurans sp. nov., which contain ornithine in their cell walls. Int. J. Syst. Evol. Microbiol. 52:2127-2133. [DOI] [PubMed] [Google Scholar]
- 2.Ash, C., J. A. E. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus Bacillus revealed by comparative analysis of small-subunit-ribosomal RNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 3.Ash, C., J. A. E. Farrow, M. Dorsch, E. Stackebrandt, and M. D. Collins. 1991. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16s rRNA. Int. J. Syst. Bacteriol. 41:343-346. [DOI] [PubMed] [Google Scholar]
- 4.Dong, X., and J. C. Cote. 2003. Phylogenetic relationships between Bacillus species and related genera inferred from comparison of 3′ end 16S rDNA and 5′ end 16S-23S ITS nucleotide sequences. Int. J. Syst. Evol. Microbiol. 53:695-704. [DOI] [PubMed]
- 5.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Evol. Microbiol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 6.Goto, K., T. Omura, Y. Hara, and Y. Sadaie. 2000. Application of the partial 16S rDNA sequence as an index for rapid identification of species in the genus Bacillus. J. Gen. Appl. Microbiol. 46:1-8. [DOI] [PubMed] [Google Scholar]
- 7.Harmsen, D., J. Rothganger, M. Frosch, and J. Albert. 2002. RIDOM: ribosomal differentiation of medical micro-organisms database. Nucleic Acids Res. 30:416-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrel, L. J., G. L. Anderson, and K. H. Wilson. 1995. Genetic variability of Bacillus anthracis and related species. J. Clin. Microbiol. 33:1947-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Anderson, K. H. Wilson, M. E. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joung, D. J., and J. C. Cote. 2002. Evaluation of ribosomal RNA gene restriction patterns for the classification of Bacillus species and related genera. J. Appl. Microbiol. 92:97-108. [DOI] [PubMed] [Google Scholar]
- 12.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakamura, L. K., M. S. Roberts, and F. M. Cohan. 1999. Relationship of Bacillus subtilis clades associated with strains 168 and W23: a proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int. J. Syst. Bacteriol. 49:1211-1215. [DOI] [PubMed] [Google Scholar]
- 14.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 15.Priest, F. G. 1993. Systematics and ecology of Bacillus, p. 369-373. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 16.Priest, F. G., M. Goodfellow, and C. Todd. 1988. A numerical classification of the genus Bacillus. J. Gen. Microbiol. 134:1947-1982. [DOI] [PubMed] [Google Scholar]
- 17.Qi, Y., G. Patra, X. Liang, L. E. Williams, S. Rose, R. J. Redkar, and V. G. DelVecchio. 2001. Utilization of the rpoB gene as a specific chromosomal marker for real-time PCR detection of Bacillus anthracis. Appl. Environ. Microbiol. 67:3720-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts, M. S., L. K. Nakamura, and F. M. Cohan. 1994. Bacillus mojavensis sp. nov., distinguishable from Bacillus subtilis by sexual isolation, divergence in DNA sequence, and differences in fatty acid composition. Int. J. Syst. Bacteriol. 44:256-264. [DOI] [PubMed] [Google Scholar]
- 19.Sacchi, C. T., A. M. Whitney, L. W. Mayer, R. Morey, A. Stiegerwalt, A. Boras, R. S. Weyant, and T. Popovic. 2002. Sequencing of 16S rRNA gene: a rapid tool for identification of Bacillus anthracis. Emerg. Infect. Dis. 8:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimot, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossello-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047 [DOI] [PubMed] [Google Scholar]
- 21.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. (Erratum, 40:2316, 2002.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki, K., T. Okubo, N. Inoue, and H. Shinano. 1997. Randomly amplified polymorphic DNA (RAPD) for rapid identification of spoilage bacterium Alicyclobacillus acidoterrestris. Biosci. Biotechnol. Biochem. 61:1016-1019. [Google Scholar]