SUMMARY
The formation of an activated cis-3-cyclohexylpropenoic acid by Plm1, the first extension module of the phoslactomycin PKS, is proposed to occur through an L-3-hydroxyacyl-intermediate as a result of ketoreduction by an A-type ketoreductase (KR). Here, we demonstrate that the KR domain of Plm1 (PlmKR1) catalyzes the formation of an L-3-hydroxyacyl product. The crystal structure of PlmKR1 revealed a well ordered active site with a nearby Trp residue characteristic of A-type KRs. Structural comparison of PlmKR1 with B-type KRs that produce D-3-hydroxyacyl intermediates revealed significant differences. The active site of cofactor-bound A-type KRs is in a catalysis-ready state, whereas cofactor-bound B-type KRs are in a pre-catalytic state. Furthermore, the closed lid loop in substrate-bound A-type KRs restricts active site access from all but one direction, which is proposed to control the stereochemistry of ketoreduction.
INTRODUCTION
Bacterial type I polyketide synthases (PKS) are large, modular, multifunctional proteins that catalyze the biosynthesis of various polyketides in a series of decarboxylative condensation reactions (Hopwood and Sherman, 1990; Khosla, 1997; Smith and Tsai, 2007). Minimally, each module contains three domains required for the extension of the growing polyketide; an acyl carrier protein (ACP), an acyltransferase (AT), and a ketosynthase (KS). Modules may also possess one or more β-carbon processing domains, which include a ketoreductase (KR), a dehydratase (DH), and an enoylreductase (ER) that progressively modify a β-keto group to a hydroxyl, a double bond and a single bond, respectively. The number of extension modules within a PKS, the degree of β-carbon processing, the starter unit and extender unit (typically malonyl-, methylmalonyl-, and ethylmalonyl-CoA) give rise to structurally diverse and complex polyketides, which are notable for their diverse pharmaceutical applications.
The KR domain catalyzes the NADPH-dependent reduction of a β-keto group to give a hydroxyl group. KRs are classified as A- or a B-type depending upon the stereochemistry of ketoreduction. In addition, some KRs catalyze epimerization of an α-substituent and thus can be further sub-classified. Distinct sequence motifs have been correlated with known stereochemistry displayed by type I polyketide KR domains, allowing one to predict the stereochemical outcome of ketoreduction by an uncharacterized KR with the natural substrate. A-type KRs contain a conserved tryptophan residue (“W” motif) and generate a L-3-hydroxyacyl intermediate whereas B-type KRs contain a conserved Leu-Asp-Asp (“LDD” motif) sequence motif located near the active site and generate D-3-hydroxyacyl intermediate (Caffrey, 2003; Keatinge-Clay, 2007; Keatinge-Clay and Stroud, 2006; Reid et al., 2003; Valenzano et al., 2009).
Detailed studies of EryKR1 (B-type), EryKR2 (A-type), EryKR5 (A-type) and EryKR6 (B-type) from the erythromycin PKS demonstrated that ketoreduction, which is analogous to fatty acid biosynthesis, always involves the stereospecific transfer of the 4-pro-S hydride from NADPH to the substrate (McPherson et al., 1998; Yin et al., 2001). These findings suggest that cofactor binding and hydride transfer are identical regardless of KR type, thus differences in the stereochemistry of ketoreduction between the A- and B-type KRs must stem from the different modes of substrate binding. One hypothesis is that the A-type and B-type KRs guide the substrate into the active site from opposite directions thereby presenting different faces of the carbonyl carbon of the β-keto to the NADPH, which would result in opposite β-hydroxyl stereochemistries (Keatinge-Clay, 2007). This is supported by the observation that the conserved A-type (W) and B-type (LDD) sequence motifs are located on opposite sides of the active site cleft (Keatinge-Clay, 2007). Additionally, previous studies suggest that substrate functional groups may help to orient the substrate to produce the desired stereochemistry (Zheng et al., 2010).
To date, crystal structures have been reported for five PKS KRs (two A-type and three B-type KRs) that catalyze ketoreduction (Keatinge-Clay, 2007; Valenzano et al., 2009; Zheng et al., 2012; Zheng et al., 2010) and for one KR that catalyzes epimerization of an α-methyl group but not ketoreduction (Zheng and Keatinge-Clay, 2011). A clear understanding of the mechanism by which A-type and B-type KRs produce opposite hydroxyl group stereochemistries has yet to emerge from these structures.
Double bonds in polyketides are generally established by the successive action of the ketoreductase-dehydratase (KR-DH) didomain. The DH removes the hydroxyl group generated by the KR domain, producing an olefin with either a cis or trans configuration. Most unsaturated polyketides generated by modular PKSs contain trans double bonds that are formed by DH dehydration of a D-3-hydroxyacyl intermediate produced by a B-type KR domain (Caffrey, 2003; Reid et al., 2003; Wu et al., 2005). Double bonds of the cis configuration are less common but nonetheless occur in a number of polyketides and arise through at least two mechanisms. In some cases, such as epothilone (Tang et al., 2003), borrelidin (Vergnolle et al., 2011) and hypothemycin (Reeves et al., 2008) biosynthesis, cis double bonds are formed by an enzyme-catalyzed isomerization of trans double bonds. In other pathways, such as the phoslactomycin (Alhamadsheh et al., 2007), rifamycin (Schupp et al., 1998), and fostriecin (Kong et al., 2013) synthases, it has been postulated that cis double bonds arise from the DH-catalyzed dehydration of an L-3-hydroxyacyl intermediate, which is formed by an A-type KR domain (Reid et al., 2003). The correlation of an A-type KR with DH-catalyzed cis double bond formation led to the postulate that the stereochemical outcome of the KR-catalyzed ketoreduction is key in establishing the configuration of the double bond (Reid et al., 2003) Although no biochemical evidence, to date, supports this hypothesis, it is consistent with the structures of DH domains where the narrow active site is better suited to binding an L-3-hydroxyacyl substrate in a pre-cis conformation and a D-3-hydroxyacyl substrate in a pre-trans conformation (Akey et al., 2010; Keatinge-Clay, 2008).
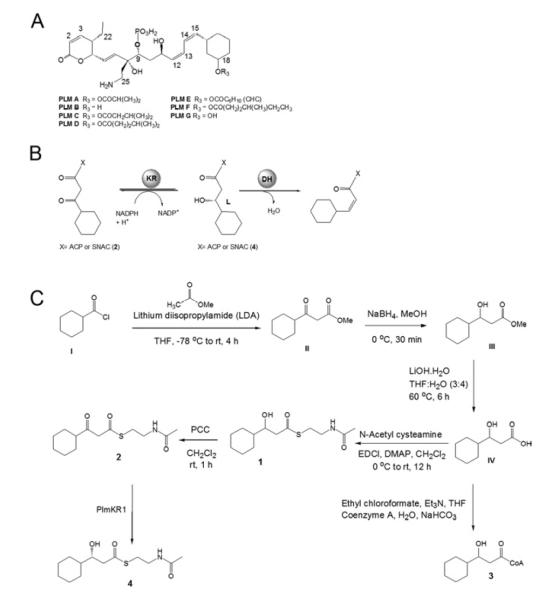
Phoslactomycins (PLMs, Figure 1A) are phosphorylated polyketide natural products produced by Streptomyces sp. HK-803 and a number of other Streptomyces that exhibit antifungal, antitumor and antiviral activity (Fushimi et al., 1989a; Fushimi et al., 1989b). The unusual architecture of PLMs is exemplified by the presence of three cis (Z) (Δ14,15, Δ12,13, Δ2,3) and one trans (E) (Δ6,7) double bonds (Alhamadsheh et al., 2007; Fushimi et al., 1989a; Fushimi et al., 1989b; Stampwala et al., 1983). Module 1 of the phoslactomycin PKS (Plm1) contains the loading domain and the first extension module and is responsible for the generation of the Δ14,15 cis double bond of PLM. In previous studies a Δplm1 deletion mutant was constructed and shown to be unable to produce PLM (Alhamadsheh et al., 2007). However, when Δplm1 was grown in the presence of cis-3-cyclohexylpropenoic N-acetylcysteamine (SNAC) thioester, production of PLM was restored. Conversely, the trans isomer failed to restore PLM production to the Δplm1 mutant (Alhamadsheh et al., 2007). These findings demonstrate that the activated cis-3-cyclohexylpropenoic acid is the diketide intermediate passed from Plm1 to Plm2. Therefore, the stereochemistry is directly established by the DH-KR didomain of Plm1 and not from a subsequent isomerization step (Alhamadsheh et al., 2007). The generation of the proposed cis-3-cyclohexylpropenoic acid diketide intermediate by Plm1 is thus predicted to occur via a L-3-hydroxyacyl intermediate as a result of ketoreduction by an A-type KR (Figure 1B).
Figure 1.
(A) General structure of Phoslactomycins (PLMs). (B) Proposed reaction catalyzed by the Plm1 KR-DH didomain. (C) Synthetic approaches to generate substrates for PlmKR1.
Here we examine the stereochemical outcome and control of ketoreduction by the KR domain from module 1 of the PLM PKS (PlmKR1) at a biochemical and structural level. We successfully cloned and purified PlmKR1 as an individual domain and carried out kinetic and stereochemical characterization studies using SNAC thioester derivatives of the natural 3-ketocyclohexylpropionic and 3-hydroxycyclohexylpropionic acid substrates. We demonstrate that PlmKR1-catalyzed reduction yields an L-3-hydroxyacyl product. Additionally, the crystal structure of PlmKR1 was solved to 1.68 Å resolution and revealed structural differences between A-type and B-type KRs that may be important in achieving stereochemical control.
RESULTS and DISCUSSION
Stereochemical Determination of the 3-Hydroxyl Group
The production of cis double bonds was hypothesized to occur through the DH-catalyzed dehydration of the L-3-hydroxyacyl intermediate generated by the KR (Alhamadsheh et al., 2007), but this has not been experimentally verified. Experimental validation came by demonstrating that the incubation of the stand-alone PlmKR1 with 3-ketocyclohexylpropionyl-SNAC (2) resulted in the exclusive formation of the predicted L-3-hydroxyacyl thioester product 4 as established by Mosher ester analysis. The L configuration at C-3 was deduced from the Δδ values obtained by subtracting the chemical shift (δR) of each proton of the (R)-MTPA ester (4a) from that (δS) of the (S)-MTPA ester (4b) (Figure S1A). In fact, all the protons lying on the left- and right-hand sides of the plane containing the ester group of MTPA had negative and positive Δδ values, respectively, as a consequence of the shielding effect of the Mosher’s acid phenyl ring. In the (R)–MTPA ester (4a), the phenyl (Ph) group and thioester moiety are in the same plane as shown in Figure S1B, thus Ph group shields the thioester moiety and the resultant chemical shifts of the thioester moiety side protons appear in the upfield region relative to (S)–MTPA ester (4b). Conversely in the (S)–MTPA ester (4b), the Ph group and cyclohexyl moiety are in the same plane as shown in Figure S1B, thus Ph group shields the cyclohexyl moiety and the resultant chemical shifts of cyclohexyl moiety side protons appear in upfield region relative to 4a. Since the natural diketide substrate of PlmKR1, 3-ketocyclohexylpropionic acid as its SNAC thioester (2), was used in this study, we do not expect the observed stereochemical outcome to differ if examined in a modular context or for a substrate tethered to the appropriate ACP. Thus, the PlmKR1 domain reduces the 3-ketocyclohexylpropionyl substrate to the L-3-hydroxycyclohexylprionyl product, as predicted. In order to generate the 3-L isomer, PlmKR1 must catalyze the transfer of the 4-pro-S hydride from NADPH to the si face at C3 of the substrate (McPherson et al., 1998; Yin et al., 2001).
Enzyme Activity
The results stemming from the analysis of PlmKR1 activity with 1, 2, and 4 (obtained from the PlmKR1 catalyzed conversion of 2 and stereoconfiguration confirmed by Mosher ester analysis) are summarized in Table 1. Substrate solubility issues prevented the use of substrate concentrations well above Km. However, approximate kcat/Km values were obtained from the linear fit of the limiting case of the Michaelis-Menten model (v = (kcat/Km)[S][E]o) for low substrate concentration (where [S]<<Km).
TABLE 1.
Kinetic parameters of PlmKR1
| Substrate |
kcat/Km (min−1mM−1)a,b |
|---|---|
| DL-3-Hydroxycyclohexylpropionyl-SNAC (1) | 0.23± 0.03 |
| 3-Ketocyclohexylpropionyl-SNAC (2) | 10.1± 0.3 |
| L-3-Hydroxycyclohexylpropionyl-SNAC (4) | 0.53± 0.01 |
Solubility issues prevented the use of substrate concentrations well above Km. However, approximate kcat/Km values were obtained from the linear fit of the limiting case of the Michaelis-Menten model (v = (kcat/Km)[S][E]o) for low substrate concentration (where [S]<<Km). The approximate kcat/Km values were calculated, from the average of minimally three sets of triplicate
Data reported as mean ± standard deviation.
See Figure 1 for structures of 1, 2 and 4
Incubating PlmKR1 with 3-ketoacyl-SNAC thioester 2 provided an approximate kcat/Km of 10.1 ± 0.3 min−1mM−1. The reaction was reversible but the reverse direction proceeded slower with a calculated kcat/Km of 0.23 ± 0.03 min−1mM−1 when using DL-3-hydroxycyclohexylpropionyl-SNAC (1) as a substrate. When L-3-hydroxycyclohexylpropionyl-SNAC (4) was incubated with PlmKR1, an approximate kcat/Km of 0.53 ± 0.01 min−1mM−1 was obtained, which is ~2.3 fold higher than that for 1. When the coenzyme-A (CoA) derivative of DL-3-hydroxycyclohexylpropionyl (3) was analyzed as a substrate, very little activity (< 1 % compared to the SNAC derivative) was observed.
The 3-hydroxyacyl thioester 1 was prepared as a racemic mixture and it is unclear if the D-isomer functions as a substrate for PlmKR1. The greater kinetic efficiency using the L-isomer (4) suggests the D-isomer is either not a substrate or a very poor one. Comparing the extent of NADP+ conversion by PlmKR1 under limiting substrate (either 4 or 1) led to the same conclusion. If PlmKR1 only accepts the L-isomer 4 as a substrate then approximately twice as much NADP+ conversion should be observed with the L-isomer as with the same concentration as the presumed racemic mixture. In the reductive direction using limiting 2 (0.1 mM) and a 2-fold excess of NADPH (0.2 mM), the reaction went to completion (Figure S2A). It is possible to obtain reversal of this reaction by using either 1 or 4 (0.25 mM) with a much larger excess (12-fold) of NADP+ (2 mM). Under these conditions approximately 21.0 ± 0.5 μM of NADPH was formed when PlmKR1 was incubated with L-isomer 4 whereas only 13.1 ± 0.5 μM NADPH was generated when the racemic mixture 1 was utilized as a substrate (Figure S2B).
Confirmation of the PlmKR1 reaction products in the reductive and oxidative direction was achieved by LC-MS analysis (Figure S3). Incubating PlmKR1 in the presence of 2 (broad m/z peak eluting at 20.5 min) (Figure S3B and S3C) and NADPH resulted in the formation of an ion that had the same m/z (274, [M+H]+ and 296, [M+Na]+) and retention time (19.5 min) as 1 (Figure S3F and S3G). The reaction appeared to go to completion under the conditions implemented (Figure S3E). Conversely, incubating PlmKR1 with 1 resulted in < 20% conversion of the substrate to a single product that had the same m/z (272, [M+H]+ and 294, [M+Na]+) and retention time as 2 (data not shown).
Structure of PlmKR1
The excised KR domain used for crystallization comprised residues 1814-2248 of Plm1. The elution profile from a gel filtration column indicated that the excised KR domain was monomeric. The crystal structure was determined to 1.68 Å using molecular replacement with the B-type KR from the first module of the erythromycin PKS (EryKR1) (Keatinge-Clay and Stroud, 2006) as a search model. The binary complex of PlmKR1 with NADP+ crystallized in space group P21 with two polypeptide chains in the asymmetric unit (Table 2). The final crystallographic model of PlmKR1 was complete excepting a few residues: Chain A residues 14-16 and 147-153 and Chain B residues 147-152.
TABLE 2.
Crystallographic Data and Refinement Statistics.
| Plm1 KR | |
|---|---|
| Data Collection | |
| Space group | P21 |
| Cell dimensions | |
| a,b,c (Å) | 45.2, 123.4, 79.9 |
| α, β, γ (°) | 90, 90.27, 90 |
| X-ray source | APS 23ID-B |
| Wavelength (Å) | 1.033 |
| dmin (Å) | 1.68 (1.74-1.68)a |
| Rsymm (%) | 0.077 (.457) |
| Ave I/σI | 23.5 (3.9) |
| Completeness (%) | 96.9 (94.3) |
| Average | |
| redundancy | 4.6 (4.8) |
| Refinement b | |
| Data range (Å) | 50-1.68 |
| No. reflections | 85,333 |
| Rwork/Rfree | 0.16/0.19 |
| Polypeptide Chains | 2 |
| Number of Atoms | |
| Protein | 6479 |
| Water | 1008 |
| Ligand | 96 |
| RMS Deviations | |
| Bond length (Å) | 0.012 |
| Bond angle (Å) | 1.4 |
| B-factors (Å2) | |
| Protein | 13.4 |
| Water | 26 |
| Ligand | 9.34 |
| Ramachandran plot | |
| Allowed (%) | 100 |
| Outliers (%) | 0 |
Values in parentheses pertain to the outermost shell of data.
The final structure is deposited in the PDB with accession code 4HXY.
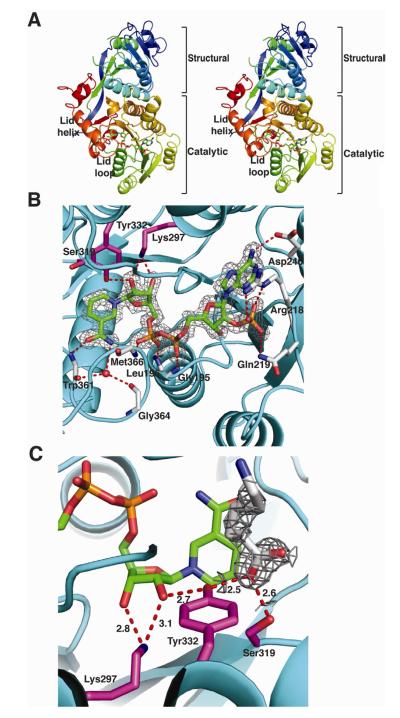
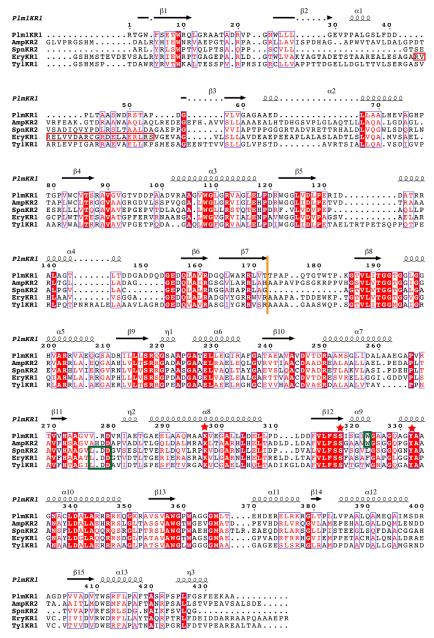
The PlmKR1 contains 2 domains: an N-terminal structural domain (KRstruc) and a C-terminal catalytic domain (KRcat) (Figure 2A and 2B). Both domains have the familiar Rossmann fold common to the short chain dehydrogenase/reductase enzymes (SDR). However, the structural domain contains a truncated Rossmann fold and other features that prevent NADPH binding, consistent with its presumed role to stabilize the catalytic domain. The lengths of PKS KRstruc domains vary. KRstruc of PlmKR1 and of the second-module KR from the spinosyn PKS (SpnKR2) are significantly shorter than the structural domains in previously reported KR crystal structures (Figure 3) (Zheng et al., 2012). Specifically, when compared with EryKR1, PlmKR1 KRstruc lacks a β-strand and an α-helix and contains two truncated β-strands and two truncated α-helices (Figure 3 and S4).
Figure 2. Structure of PlmKR1.
(A) Trace of the polypeptide chain from N- (blue) to C-terminus (red). The mobile lid helix and lid loop of the catalytic domain are labeled in the stereo image. The NADP+ cofactor is shown as sticks with atomic coloring (blue N, red O, yellow S and orange P) and green C.
(B) PlmKR1 active site and NADP+ density. Side chains of the catalytic residues (Tyr 332, Ser319, Lys297) are shown as sticks with magenta C. The NADP+ cofactor is shown as in A with the 2Fo-Fc electron density contoured at 1 σ. The residues that interact with NADP+ are shown as sticks with grey C. The conserved Trp332 of A-type KRs is shown with light red C. The ribbon diagram of the polypeptide has an orange structural domain and cyan catalytic domain.
(C) Density for 6-aminohexanoic acid. The 2Fo-Fc electron density for the 6-aminohexanoic acid is shown contoured at 1 σ. The 6-aminohexanoic acid is shown as sticks with grey C. Hydrogen bonds between active site residues, NADP+ nicotinamide ribose, and 6-aminohexanoic acid are shown in red. The catalytic residues are shown and colored as in B. The NADPH cofactor is shown as in A.
Figure 3. Structure based sequence alignment.
Structure-based multiple sequence alignment of PlmKR1 (accession AAQ84156), AmpKR2 (37% sequence identity to PlmKR1, accession AAK73513), SpnKR2 (32% sequence identity to PlmKR1, accession AAG23265), EryKR1 (35% identity to PlmKR1, accession AAB84070), TylKR1 (41% identity to PlmKR1, accession AAB66504). The red box indicates the location of β-strand and α-helix in EryKR1, which PlmKR1 lacks. Additionally, dotted lines in secondary structure elements indicate truncations of these elements in PlmKR1 when compared to the other KRs. Orange line indicates the end of the structural domain and start of the catalytic domain. Green boxes indicate the location of the W motif for A-type KRs (PlmKR1 and AmpKR2) and LDD motif for (B-type KRs). Red stars indicate the location of the active site residues. The sequence alignment was generated with MUSCLE (Edgar, 2004) and ESPript (Gouet et al., 1999) was used to generate secondary structure annotations.
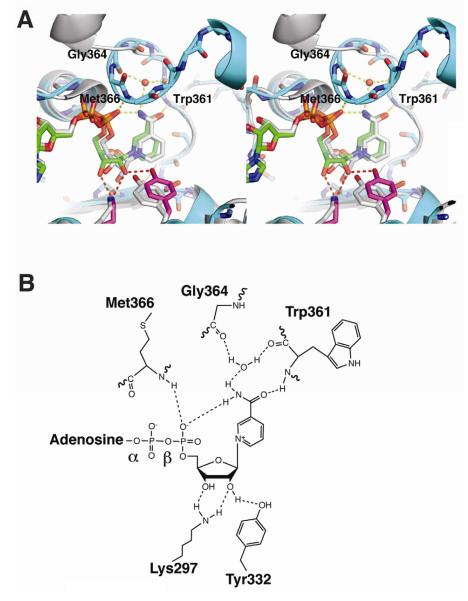
The KRcat domain contains a Rossmann fold with a bound NADP+ cofactor (Figure 2B). Clear density was observed for the entire NADP+ cofactor, which forms multiple contacts with the protein. Specifically, Asp246 hydrogen bonds to the adenine N6 amine. Arg218 and Gln219 hydrogen bond to the adenosine 2′-phosphate. The backbone amides of Gly195 (of the conserved GXGXXG dinucleotide binding motif), Leu196, and Met366 form hydrogen bonds to the diphosphate. Catalytic Lys297 and Tyr332 hydrogen bond to the nicotinamide ribose hydroxyl groups. The backbone amide of Trp361 is hydrogen bonded to the nicotinamide carbonyl. Finally, the nictinamide amide forms a water-mediated hydrogen bond to the backbone carbonyls of Trp361 and Gly364.
Active site structure
All features of the PlmKR1 active site are well defined by electron density and well ordered. The active site is comprised of the NADP+ cofactor and the conserved catalytic residues Tyr332, Ser319, and Lys297. The three amino acids are common to all PKS KRs and to members of the larger SDR family (Oppermann et al., 2003). According to the widely accepted model for SDR catalysis (Oppermann et al., 2003), NADPH transfers a hydride ion to the substrate carbonyl carbon atom, and the catalytic Tyr hydroxy donates a proton to the carbonyl oxygen atom. The Lys amino group is thought to assist catalysis through a hydrogen-bond donor-acceptor network that orients the Tyr hydroxy proton towards the substrate and also stabilizes the resulting Tyr hydroxylate. The PlmKR1 active site is structured as expected for this mechanism. The Lys amino group donates a proton to the nicotinamide ribose 2′-hydroxy. The ribose 2′-hydroxy donates a proton to the Tyr hydroxy, thereby directing the Tyr hydroxy proton towards the substrate site (Figure 2C, 4A, and 4B). The same Lys-ribose-Tyr hydrogen-bonding network occurs in cofactor-substrate ternary complexes of well-studied SDR enzymes, such as UDP-galactose 4-epimerase, and is thought to be essential to catalysis (Thoden et al., 1997). The active site Ser hydroxy may orient the substrate for catalysis (Oppermann et al., 2003).
Figure 4. Comparison of A-type and B-type KRs.
(A) Stereo view of PlmKR1 (cyan) and SpnKR2 superposition (white, PDB code: 3SLK) (RMSD = 1.187 Å). The active site tyrosine and the NADP(H) are in different positions in the A-type (PlmKR1, cyan) and B-type (SpnKR2, white) KRs. In PlmKR1, the hydrogen bond network of Lys297, ribose 2′-hydroxy and Tyr332 is shown as red dashes, and the hydrogen bond network of nicotinamide amide, lid loop, and NADP+ β-phosphate as yellow dashes. The PlmKR1 catalytic residues are drawn in stick form with magenta C and the NADPH cofactor with green C, and the EryKR1 NADPH was omitted because of disorder.
(B) Schematic of the hydrogen bond network of NADP+ bound to the A-type PlmKR1. In cofactor complexes of B-type KRs, only the hydrogen bonds of Lys to ribose 2′-hydroxy and of nicotinamide amide to NADP(H) β-phosphate have been observed.
In PKS KRs, a mobile helix and loop adjacent to the cofactor nicotinamide, the “lid helix” and “lid loop”, move upon cofactor and/or substrate binding (Keatinge-Clay, 2007; Keatinge-Clay and Stroud, 2006; Zheng et al., 2010). In PlmKR1, the lid helix (residues 369-378) and lid loop (residues 361-368) are well ordered and adjacent to the nicotinamide ring (Figure 2A, 5A, and 5B).
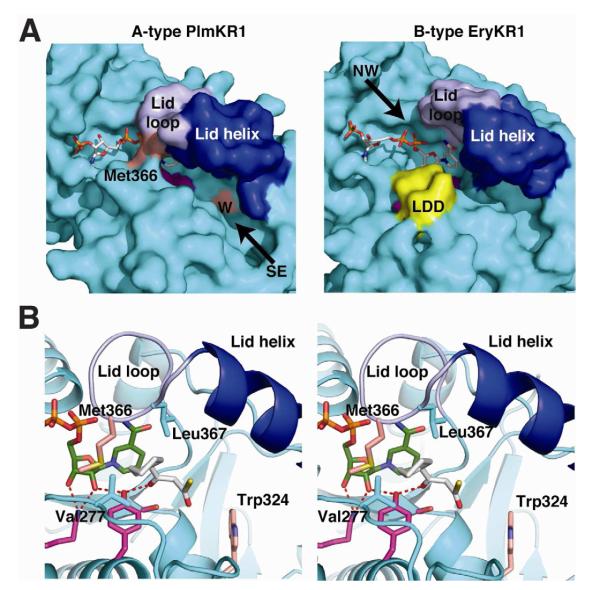
Figure 5. Substrate entry into A-type and B-type KRs.
(A) Active site channel of A-type PlmKR1 (left) and B-type EryKR1 (right). The surfaces of the catalytic domains are cyan, with blue lid helices, light blue lid loops, and magenta active site residues. The cofactors are shown in stick form with white C. With NADP+ and substrate bound, the A-type PlmKR1 active site is ordered and is accessible from the “southeast” direction where the W motif (light red surface) is located. The Met366 side chain (light red surface) blocks access to the active site from the “northwest” direction. With NADP+ bound, the B-type EryKR1 active site is open with access from either direction. The surface of the LDD motif is shown in yellow.
(B) Model of substrate binding to PlmKR1. A 3-ketocyclohexylpropionyl thioester (white C) was modeled by hand into the A-type PlmKR1 active site based on the position of 6-aminohexanoic acid in the crystal structure. The thioester sulfur indicates the phosphopantetheinyl-ACP attachment site. The Met 366 side chain (light red C) blocks access from the northwest direction. Hydrophobic amino acids (Val 277 and Leu 367) in the proposed cyclohexane site are shown in cyan. The hydrogen-bonding network between active site residues (Lys, Tyr, magenta C) are shown as red dashes. NADP+ is shown with green C and the W motif Trp324 with light red C. Other coloring as in A.
Substrate binding site
Electron density between the nicotinamide ring and catalytic Tyr332 in one of the PlmKR1 molecules in the asymmetric unit was interpreted as a 6-aminohexanoic acid additive to the crystallization solution (Figure 2C). The carboxylate carbon of 6-aminohexanoic acid is positioned above C4 of the nicotinamide ring and a carboxylate oxygen is hydrogen bonded to active site Tyr332 and Ser319. Presumably 6-aminohexanoate is not a PlmKR1 substrate because hydride transfer to the carboxylate anion is disfavored. The active site of AmpKR2, another A-type KR, also contains a carboxylic acid ligand, malic acid (Zheng et al., 2010). In both PlmKR1 and AmpKR2, the ligand carboxylate is bound between the NADPH nicotinamide and the catalytic Tyr, with hydrogen bonds to Tyr and Ser side chains as expected for KR substrates.
Stereochemical control in A-type and B-type KRs
Conserved sequence motifs in each type of PKS KR, the W motif for A-type and the LDD motif for B-type, are strongly correlated with stereochemical outcome of the KR reaction (Caffrey, 2003; Reid et al., 2003). These motifs are proposed to control, by an unknown mechanism, the direction of entry of the phosphopantetheinylated substrate into the active site and thereby control which face of the β-ketone is accessible to the NADPH cofactor for hydride transfer (Keatinge-Clay, 2007). The W motif (Trp324) in the PlmKR1 sequence (Figure 3) and the exclusive formation of the L-3-hydroxyacyl product demonstrate that PlmKR1 is an A-type KR (Figure 4C). However, the position of Trp324 in the PlmKR1 structure provides no insights into its role in substrate binding or the stereospecificity of ketoreduction. Furthermore, mutagenesis of the A- and B-type sequence motifs in AmpKR2 (Zheng et al., 2010) and in a full module from the erythromycin PKS (Kwan et al., 2011) demonstrated that the motifs are not the sole determinants of hydroxy group stereochemistry. Therefore, to identify other structural features that control stereochemical outcome, we compared PlmKR1 to the four previously reported Type 1 PKS KR structures. The comparison included three A-type KRs, PlmKR1, AmpKR2, (Zheng et al., 2010) and AmpKR11 (PDB code 4DI7), and three B-type KRs, EryKR1 (Valenzano et al., 2009), TylKR1 (Keatinge-Clay, 2007) and SpnKR2 (Zheng et al., 2012). Structures with bound NADP(H) cofactor are available for all but TylKR1, and views of the KR both with and without cofactor are reported for AmpKR2 and EryKR1. In addition, structures of A-type KRs were reported with a substrate mimic bound (AmpKR2 and PlmKR1) and without a substrate mimic bound (AmpKR2 and AmpKR11).
The most striking difference between A-type and B-type KR structures is the ordering of the active site. The nicotinamide ribose in A-type KRs is bound in a well ordered and catalysis-ready conformation with a Lys–2′-O-ribose–Tyr hydrogen-bonding network and a ribose C2′-endo pucker (Figure 2C, 4A, and 4B). The hydrogen bond network does not exist in cofactor-bound B-type KRs, where the ribose is poorly ordered (EryKR1) or is in a position with the 2′ hydroxy directed away from the catalytic Tyr (SpnKR2). We examined electron densities for ribose in the B-type KR crystal structures to determine whether the ribose could be rebuilt to form the hydrogen-bonding network, but this was not compatible with the electron density. Additionally, in B-type KRs with bound cofactor, the catalytic Tyr is shifted ~2 Å away from the catalysis-ready position in A-type KRs (Figure 4A). The pre-catalytic position of the Tyr in cofactor bound B-type KRs is identical to the Tyr position in cofactor-free structures of both A-type and B-type KRs (AmpKR2 3MJC, TylKR1). Thus, cofactor-bound A-type KRs have a fully formed active site, whereas cofactor-bound B-type KRs do not.
In A-type KRs with a substrate mimic in the active site, PlmKR1 and AmpKR2 (PDB codes 4HXY and 3MJS), the lid helix is well ordered and the lid loop closes over the bound cofactor and forms an intricate set of hydrogen bonds with the β-phosphate and nicotinamide base (Figure 4A and 4B). These specific contacts position the lid loop close to the phosphate-binding loop (amino acids 194-196 in PlmKR1), with conserved Gly365 only 4 Å from Gly195. The nicotinamide amide forms an intramolecular hydrogen bond with β-phosphate, which is also hydrogen bonded with the Met366 amide. The nicotinamide carbonyl is hydrogen bonded to the lid loop through the Trp361 amide. A well-ordered water molecule further organizes the lid loop through hydrogen bonds with the Trp361 carbonyl, the nicotinamide amide and the Gly364 carbonyl (Figure 4A and 4B). Finally, the Met366 side chain extends into the active site towards the nicotinamide ring and catalytic Tyr332 (Figure 5A and 5B). These protein-cofactor contacts were observed in PlmKR1, AmpKR2, and AmpKR2 mutants (3MJT and 3MJV), in which substrate mimics or unmodeled electron density were in the presumed substrate site.
For A-type KR structures with no evidence of binding in the substrate site (PlmKR1 molecule B, AmpKR2 PDB code 3MJE and AmpKR11 PDB 4DI7), the lid loop and helix have weaker density or are disordered. Nevertheless, even with an unoccupied substrate site, A-type KR active sites have a catalysis-ready Lys–2′-O-ribose–Tyr hydrogen-bonding network.
Based on our detailed structure comparison, the active site of A-type KRs bind cofactor with a Lys–2′-O-ribose–Tyr hydrogen-bonding network that is ready for catalysis. In most of these structures, the lid loop is closed, extending Met366 into the active site. The Met side chain blocks access to the nicotinamide and catalytic Tyr from one direction, which we designate “northwest”, while a “southeast” entry is accessible (Figure 5A). Previous studies of A-type KRs demonstrated that this methionine is essential for catalysis as a mutation to alanine abolishes activity (Zheng et al., 2010). We modeled the PlmKR1 substrate in the active site based on the positions of 6-aminohexanoate in the PlmKR1 crystal structure (Figure 2C) and malate in an AmpKR2 structure (Zheng et al., 2010) (Figure 5B). A carboxyl oxygen of these ligands is hydrogen bonded to the catalytic Tyr and the carboxyl carbon is near the NADPH hydride position, as expected for KR substrates. Substrate entry from the southeast places the appropriate face of the β-ketone towards the nicotinamide for A-type ketoreduction (Figure 5B). The Trp side-chain nitrogen of the W motif in A-type KRs points into the southeast entry channel where it may help orient the substrate phosphopantetheine by hydrogen bonding.
The available structural data indicate that cofactor binds loosely to B-type KRs. For example, the TylKR1 crystal structure has no electron density for the cofactor that was present in crystallization solutions (Keatinge-Clay, 2007). The nicotinamide and ribose are poorly ordered in the EryKR1 structure (Keatinge-Clay and Stroud, 2006) and the lid loop is open so that substrates could access the active site from both the northwest and southeast (Figure 5A). In SpnKR2, the lid loop and helix open completely to form a “domain swap” with another molecule in the crystal lattice (Zheng et al., 2012). As in the A-type KRs, substrate binding to B-type KRs is expected to assist in active-site formation by ordering the lid loop and helix. However in contrast to A-KRs, the catalytic Lys-2′-O-ribose-Tyr hydrogen bonding network in B-type KRs appears to form only after substrate binding. We conclude that productive binding and substrate-assisted active-site formation occur only when the substrate enters from the northwest direction, placing the appropriate face of the β-ketone towards the nicotinamide for B-type ketoreduction. The LDD motif of B-type KRs is near the catalytic Tyr (Figure 5A) and may contribute to substrate-assisted assembly of the active site.
Our analysis of the structures provides an explanation of how the A-type KRs restrict the direction of access to the active site through progressive ordering of the lid loop and helix upon cofactor and substrate binding, thereby controlling the stereochemical outcome of ketoreduction. The W motif appears to play no role in facilitating formation of an ordered lid loop, nor the LDD motif in preventing its formation. The W motif in A-type KRs is too far (10 Å) from the active site to have a direct effect (Figure 5). The LDD motif of B-type KRs is near enough, but its position does not directly block a closed lid loop. In fact, the side chains of the lid-loop Met and the LDD Leu could interact if the lid loop were to close in B-type KRs. The analogous hydrophobic contact exists in the A-type PlmKR1 between Met366 and Val277 (analogous to the LDD Leu) (Figure 5B). Thus, like other authors (Kwan et al., 2011; Zheng et al., 2010), we conclude that KR stereochemical control does not arise solely from the W and LDD motifs.
Our interpretation of the structures is in agreement with the direction-of-access hypothesis proposed by Keatinge-Clay (Keatinge-Clay, 2007). As different conformations of the enzyme control the direction of access, the mechanism of stereochemical control must also include aspects of protein dynamics. The lid loop likely populates several conformations with or without bound cofactor. As each crystal structure captures one or a few well-populated states, we conclude that the loop-closed conformations captured in the crystal structures of PlmKR1 and AmpKR2 predominate in cofactor-bound A-type KRs. However, experimental factors, such as introduction of non-natural substrates or site-directed mutagenesis, can alter the distribution of conformations within the population and thus the stereochemical outcome (Zheng et al., 2010).
SIGNIFICANCE
Collectively, the results presented here demonstrate that PlmKR1 is an A-type KR that catalyzes the stereospecific reduction of the β-keto intermediate to generate an L-configured alcohol. Such findings are consistent with the hypothesis that cis double bonds arise from the syn dehydration of L-3-hydroxyacyl intermediates by the DH domain. Furthermore, detailed structural analysis of A-type and B-type KRs demonstrates that formation of a catalysis-ready active site is cofactor-assisted in the A-type KRs, and apparently substrate-assisted in the B-type KRs. Closing of the lid loop upon cofactor and substrate binding in the A-type KRs restricts substrate access to the active site to a direction that is compatible with the observed stereochemical outcome of ketoreduction. These results add important new information to the understanding of stereochemical control in PKS KRs.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The generation of the expression construct for the stand-alone KR domain, pPlm1KR, is discussed in the Supplemental Information. Plasmid pPlm1KR was used to transform E. coli BL21(DE3) cells and the transformant was cultured at 37 °C to A600 = 0.8-1 in 0.5 L of TB media containing 0.1 mg/mL ampicillin. Cultures were incubated for 1 h at 20 °C, induced with 0.2 mM IPTG, and allowed to express for approximately 18 h. Cells were harvested by centrifugation at 5,670 × g for 25 min at 4 °C and stored at −80 °C.
The recombinant His6-tagged PlmKR1 fusion protein was purified by His trap column outlined in the manufacture’s manual (GE Healthcare). To remove the His6-tag, fractions containing the recombinant protein were pooled and incubated for 4 h at room temperature with TEV protease (30:1 molar ratio of protein to protease) and 2 mM dithiothreitol (DTT). After dialyzing overnight in buffer A, the cleavage reaction was loaded onto a His trap column to separate untagged PlmKR1 from His6-tagged TEV protease. The untagged PlmKR1 was collected in the flow-through and was purified further by size exclusion chromatography (HiPrep 16/60 Sephacryl S200 HR equilibrated with 50mM Tris pH 7.5, 300mM NaCl, 10%glycerol). The resulting protein was >90% pure, based on SDS-PAGE analysis (data not shown) and concentrated to 10 mg/mL, flash frozen in liquid nitrogen, and stored at −80 °C.
Synthesis of 3-hydroxy- and 3-ketocyclohexylpropionyl SNAC and CoA Thioesters (1,2, and 3) (Figure 1C)
General
The synthetic strategies used in the generation of SNAC thioesters of 3-ketocyclohexylpropionic acid (2) and DL-3-hydroxycyclohexylpropionic acid (1), and DL-3-hydroxycyclohexylpropionl-CoA (3) are based on methods described elsewhere (Bonnett et al., 2011; Eustáquio et al., 2009). Reagents were purchased from Aldrich and used without further purification. All reactions were carried out under argon/nitrogen atmosphere using dry solvents under anhydrous conditions, unless otherwise noted. Reactions were monitored by thin layer chromatography (TLC) carried out on 0.25 mm silica gel (60, F-254) plates using the following visualization techniques: UV light, iodine vapor and/or coated with 10% H2SO4 in MeOH and subsequent charring on hot plate. Chromatography was executed with silica gel (230-400 mesh) using mixtures of ethyl acetate and n-hexane as eluents. 1H NMR spectra were recorded on Bruker AMX-400 MHz spectrometer in CDCl3 , 13C NMR spectra were recorded at 100 MHz in CDCl3. Chemical shifts are reported as values in ppm relative to CHCl3 (7.26) in CDCl3 and TMS was used as internal standard.
Synthesis of methyl-3-ketocyclohexylpropionate (II)
Methyl acetate (10.1 g, 136.9 mmol) was added to a stirred solution of lithium diisopropylamide (LDA, 76 mL of 1.8 M solution in THF, 136.9 mmol) in THF (60 mL) at −78 °C. The solution was stirred at −78 °C for 15 min after which cyclohexanecarbonyl chloride (I, 10.0 g, 68.5 mmol) was added. The reaction was allowed to warm to room temperature, diluted with 1N HCl, and extracted with ethyl acetate (3 × 100 mL). The combined organic extracts were dried over anhydrous Na2SO4 and the solvents were removed under reduced pressure. The crude product was purified by silica gel column chromatography (10% EtOAc/n-hexane) to give the desired product II (10.5 g, 83%). 1H NMR (CDCl3, 400 MHz) δ 3.72 (s, 3H), 3.50 (s, 2H), 2.46 (m, 1H), 1.89–1.65 (m, 5H), 1.39–1.18 (m, 5H).
Synthesis of methyl-3-hydroxycyclohexylpropionate (III)
To a stirred solution of II (5.0 g, 27.2 mmol) in anhydrous methanol (15 mL) at 0 °C was added slowly NaBH4 (1.0 g, 27.2 mmol) and the reaction mixture was stirred at 0 °C for 30 min. The resulting solution was quenched with 1N HCl, extracted with ethyl acetate (3 × 100 mL) and the combined extracts were dried over anhydrous Na2SO4, and concentrated under reduced pressure. The crude product was seperated by silica gel column chromatography (30% EtOAc/n-hexane) to give the compound III (3.5 g, 69%). 1H NMR (CDCl3, 400 MHz) δ 3.77 (m, 1H), 3.71 (s, 3H), 2.88 (d, J = 4.2 Hz, 1H), 2.53 (dd, J = 3.0, 16.2 Hz, 1H), 2.43 (dd, J = 9.5, 16.2 Hz, 1H), 1.89–1.64 (m, 5H), 1.37 (m, 1H), 1.26–0.99 (m, 5H).
Synthesis of 3-hydroxycyclohexylpropionic acid (IV)
To a stirred suspension of III (1.0 g, 5.37 mmol) in a mixture of THF (15 mL) and water (20 mL) was added LiOH.H2O (0.86 mg, 21.5 mmol). The reaction mixture was stirred at 60 °C for 6 h after which it was cooled to 0 °C and carefully acidified to pH 2-3 with 1N HCl. The reaction mixture was extracted with ethyl acetate (3 × 50 mL) and the combined extracts were dried over anhydrous Na2SO4, and concentrated under reduced pressure. The crude product was purified by silica gel column chromatography (60% EtOAc/n-hexane) to give the desired product IV (0.87 g, 94%). 1H NMR (CDCl3, 400 MHz) δ 7.45 (br s, 1H), 3.81 (m, 1H), 2.55 (dd, J = 3.0, 16.2 Hz, 1H), 2.45 (dd, J = 9.5, 16.2 Hz, 1H), 1.87–1.38 (m, 5H), 1.24 (m, 1H), 1.21–0.98 (m, 5H); 13C NMR (CDCl3, 100 MHz) δ 177.9, 72.4, 43.0, 38.5, 28.7, 28.2, 26.3, 26.1, 25.9.
Synthesis of 3-hydroxycyclohexylpropionoyl N-acetylcysteamine (SNAC) thioester (1)
A solution of carboxylic acid IV (650 mg, 3.8 mmol) in anhydrous CH2Cl2 (10 mL) was cooled to 0 °C for 15 min. To this solution was added N-acetyl cysteamine (540 mg, 4.53 mmol) followed by 4-(N,N-dimethylamino)pyridine (DMAP, 115 mg, 0.25 mmol), and N-(3-dimethylamino-propyl)-N′-ethylcarbodiimide hydrochloride (EDCl, 866 mg, 4.53 mmol). The mixture was allowed to warm to room temperature and stirred overnight. Saturated aqueous NH4Cl solution (30 mL) was added and the organic phase was separated. The aqueous phase was extracted with diethyl ether (3 × 30 mL) and the combined organic phases were dried over anhydrous Na2SO4 and concentrated under reduced pressure. The residue was purified by silica gel column chromatography (60% EtOAc/n-hexane) to give the desired product 1 (940 mg, 91%). 1H NMR (CDCl3, 400 MHz) δ 5.85 (br s, 1H), 3.83 (m, 1H), 3.44 (m, 2H), 3.05 (m, 2H), 2.75 (dd, J = 3.0, 15.4 Hz, 1H), 2.68 (dd, J = 9.4, 15.4 Hz, 1H), 2.56 (d, J = 4.2 Hz, 1H), 1.97 (s, 3H), 1.78–1.67 (m, 5H), 1.42 (m, 1H), 1.25–0.99 (m, 5H); 13C NMR (CDCl3, 100 MHz) δ 200.1, 170.4, 72.9, 48.4, 43.3, 39.3, 28.9 (2C), 28.1, 26.3, 26.1, 26.0, 23.2.
Synthesis of 3-ketocyclohexylpropionoyl N-acetylcysteamine (SNAC) thioester (2)
To a stirred solution of 1 (200 mg, 0.73 mmol) in dry CH2Cl2 (15 mL) was added pyridinium chlorochromate (PCC, 236 mg, 1.1 mmol) and the mixture was stirred at room temperature under argon atmosphere for 1 h. Then the reaction was worked up by the addition of ether (20 mL) and the resulting mixture was filtered through a silica gel pad and washed with ether (2 × 30 mL). Combined organic phases were concentrated under reduced pressure and the residue was purified by silica gel column chromatography (50% EtOAc/n-hexanes) to give the desired product 2 (185 mg, 93%). Note. Compound 2 exists in keto-enol tautomerism. 1H NMR (CDCl3, 400 MHz) δ 12.65 (s, 0.35H-enol form), 6.24 (br s, 1H), 5.44 (s, 0.35H-enol form), 3.74 (s, 1.3H), 3.47 (m, 2H), 3.08 (m, 2H), 2.45 (m, 0.63H), 2.07 (m, 0.51H-enol form), 1.98 (s, 3H), 1.89–1.66 (m, 5H), 1.35 (m, 1H); 13C NMR (CDCl3, 100 MHz) δ 205.4, 194.5, 192.6, 181.2, 170.6, 170.4, 97.3, 55.1, 51.3, 43.4, 39.9, 39.2, 29.9, 29.2, 28.1 (2C), 27.8, 27.7, 25.8, 25.7, 25.6 (3C), 25.4, 25.3, 23.2.
Synthesis of 3-hydroxycyclohexylpropionoyl CoA thioester (3) from compound IV
To a stirred solution of compound IV (25 mg, 0.145 mmol) in anhydrous THF (5 mL), was added triethylamine (25 μL, 0.145 mmol) followed by ethyl chloroformate (15 μL, 0.145 mmol) under nitrogen at 0 °C. The reaction mixture was stirred at same temperature for additional 45 min. The stirring was then stopped to allow the resulting solid to precipitate. The clear supernatant was added slowly to a solution of hydrated coenzyme A (18 mg, 0.024 mmol) and NaHCO3 (12 mg, 0.145 mmol) in dd H2O (2 mL). The reaction mixture was stirred at room temperature for 4 h after which the THF was removed under reduced pressure. The aqueous solution was acidified to pH 3 using 1N HCl and then extracted with ethyl acetate (3 × 5 mL) to remove excess compound IV. The aqueous solution was then lyophilized and the resulting solid was washed with MeOH to recover the desired compound 3 (11 mg) as a white solid. 1H NMR (CD3OD, 400 MHz) δ 8.67 (s, 1H), 8.39 (s, 1H), 6.10 (br s, 1H), 4.75 (s, 1H), 4.49 (s, 1H), 4.29 (m, 2H), 4.03 (m, 3H), 3.82 (br s, 1H), 3.65 (m, 3H), 3.47 (m, 4H), 2.98 (m, 2H), 2.72 (m, 1H), 2.63 (m, 1H), 2.44 (m, 2H), 1.89–1.65 (m, 5H), 1.45–0.99 (m, 6H), 1.01 (s, 3H), 0.90 (s. 3H).
PlmKR1 Activity Assay
Activity assays for recombinant PlmKR1 were conducted spectrophotometrically by monitoring the change in absorbance at 340 nm, due to the formation or consumption of NADPH (ε340 = 6220 M−1cm−1). Assays were performed in the presence of 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10% glycerol, 1 mM TCEP, variable substrate (0-2 mM suspended in DMSO to give a final concentration of 1%) and either 0.2 mM NADPH or 1 mM NADP+ in a total volume of 0.5 mL. Reactions were initiated by the addition of protein (1.5 or 7 μM) and allowed to proceed at 25 °C for 10 min, taking readings every 15 sec. The approximation of kcat/Km values were calculated, from the average of minimally three sets of triplicate, using the limiting case of the Michaelis-Menten equation (v = (kcat/Km)[S][E]o) for low substrate concentration (where [S]<<Km).
The extent of NADP(H) conversion was also monitored over the course of 2 h. PlmKR1 (10 μM) was incubated in the presence of 0.25 mM 1 or 4 and 2 mM NADP+ in the oxidative direction and 0.1 mM 2 and 0.2 mM NADPH in the reductive direction.
LC-MS Analysis
Reactions were performed at 25 °C under the conditions described above in the presence of 0.5 mM NADP(H), 0.1 mM of either 2 or 1 and 5 μM protein in a total volume of 100 μL. Aliquots of 20 μL were taken at specific time points and analyzed by LC-MS in the positive mode on a Bruker MicroTOF-Q mass spectrometer equipped with an Agilent 1200 Series LC system using a Discovery HS C18 (250 × 2.1, 5 micron, Supelco) column at a flow rate of 0.3 mL/min. The following solvent gradient between Buffer A (0.05% formic acid in water) and Buffer B (0.05% formic acid in acetonitrile) was used in the analysis: (time, % Buffer B): 0 min, 20%; 30 min, 80%; 35 min, 80%; 38 min, 20%; and 45 min 20%.
Stereochemical Determination of the 3-Hydroxyl Group
PlmKR1 (0.5 μM) was incubated in the presence of 10 mM NADPH and 3 mM 2 (~400 mg) in a final reaction volume of 500 mL. The reaction was monitored at 235 nm by HPLC on Discovery HS C18 (250 × 4.6, 5 micron, Supelco) column at a flow rate of 1 mL/min using the solvent system described above. After incubating for 6 h the reaction appeared to have gone to completion as determined by HPLC and LC-MS analysis (data not shown). The resulting reaction product 4 was isolated by ethyl acetate extraction (2 × 500 mL). Organic extracts containing the reaction product were combined and dried under vacuo to obtain the desired product 4 (375 mg, 94%). The absolute configuration at C-3 of 4 was elucidated by applying the modified Mosher’s method (Dale and Mosher, 1973; Ohtani et al., 1989).
Synthesis of MTPA esters of 4 (Figure S5)
To a stirred solution of 4 (4.0 mg) in CH2Cl2 (1 mL) was added DMAP (a spatula tip), and (S)-MTPA chloride (15 μL), then maintained at room temperature under stirring overnight. After removal of the solvent, the reaction mixture was purified by preparative TLC (50% EtOAc/n-hexane) to obtain the (R)-MTPA ester (4a, 3.0 mg) in a pure state. Using (R)-MTPA chloride, the same procedure afforded the (S)-MTPA ester 4b in the same yield. (R)-MTPA ester (4a): 1H NMR (CDCl3, 400 MHz) δ 7.44 (m, 2H), 7.33 (m, 3H), 5.85 (br s, 1H), 5.29 (m, 1H), 3.42 (s, 3H), 3.3198 (m, 2H), 2.9242 (m, 2H), 2.8010 (dd, J = 8.5, 15.7 Hz, 1H), 2.7026 (dd, J = 3.6, 15.7 Hz, 1H), 1.87 (s, 3H), 1.6273 (m, 6H-overlaped), 1.0410 (m, 5H-overlaped); (S)-MTPA ester (4b): 1H NMR (CDCl3, 400 MHz) δ 7.50 (m, 2H), 7.33 (m, 3H), 5.86 (br s, 1H), 5.30 (m, 1H), 3.48 (s, 3H), 3.3401 (m, 2H), 2.9661 (m, 2H), 2.8363 (dd, J = 6.5, 15.5 Hz, 1H), 2.7502 (dd, J = 3.6, 15.5 Hz, 1H), 1.87 (s, 3H), 1.5534 (m, 6H-overlaped), 0.9585 (m, 5H-overlaped).
Crystallization
PlmKR1 was crystallized by vapor diffusion in 4 μL drops containing equal volumes of protein stock (10 mg/mL in 20 mM Tris (pH 7.5), 100 mM NaCl, 1mM NADP+) and well solution (200 mM calcium acetate, 39% polyethylene glycol 3350, 100 mM bis-tris propane (pH 6.5), 4% 6-aminohexanoic acid). Crystals grew within 48 h at 4 °C, and were harvested in loops, cryo-protected with the corresponding well solution containing 20% glycerol, and flash-cooled in liquid nitrogen.
Data Collection and Structure Determination
Data collected at GM/CA beamline 23ID-B at the Advanced Photon Source (APS), Argonne National Laboratory (Argonne, IL) were processed using HKL2000 (Otwinowski, 1997). Phases for PlmKR1 were determined by molecular replacement using Phenix (Adams et al., 2010) with the KR from module 1 of the erythromycin biosynthetic gene cluster (EryKR1, PDB code: 2FR0) (Keatinge-Clay and Stroud, 2006) as the search model. COOT (Emsley and Cowtan, 2004) was used for model building and REFMAC5 (Murshudov et al., 1997) of the CCP4 (Collaborative Computational Project, 1994) suite for refinement. Structures were validated by MOLPROBITY (Davis et al., 2004). Sequence alignments were done with the MUSCLE alignment tool (Edgar, 2004) and molecular figures were prepared with PyMOL (http://www.pymol.org/). The final structure is deposited in the PDB with accession code 4HXY.
Supplementary Material
HIGHLIGHTS.
PlmKR1 is an A-type KR that catalyzes the formation of an L-configured alcohol
Structure analysis reveals a cofactor–assisted formation of a catalysis-ready state
Conformation of the lid loop controls the substrate access to the active site
ACKNOWLEDGEMENTS
This work was supported, in whole or part by National Institutes of Health grant R01 AI051629 to KAR and DK42303 to JLS. Mass spectrometric analysis was carried out at the Portland State Bioanalytical Mass Spectrometer Facility which was supported in part by a Grant from the NSF (Grant #: 0741993).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental information includes four figures, one table, and Supplemental Experimental Procedures and can be found with this article at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey DL, Razelun JR, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Crystal structures of dehydratase domains from the curacin polyketide biosynthetic pathway. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhamadsheh MM, Palaniappan N, DasChouduri S, Reynolds KA. Modular polyketide pynthases and cis double bond formation: Establishment of activated cis-3-cyclohexylpropenoic acid as the diketide intermediate in phoslactomycin biosynthesis. J Am Chem Soc. 2007;129:1910–1911. doi: 10.1021/ja068818t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnett SA, Papireddy K, Higgins S, del Cardayre S, Reynolds KA. Functional characterization of an NADPH dependent 2-alkyl-3-ketoalkanoic acid reductase involved in olefin biosynthesis in Stenotrophomonas maltophilia. Biochemistry. 2011;50:9633–9640. doi: 10.1021/bi201096w. [DOI] [PubMed] [Google Scholar]
- Caffrey P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. Chembiochem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project. N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dale JA, Mosher HS. Nuclear magnetic resonance enantiomer regents. Configurational correlations via nuclear magnetic resonance chemical shifts of diastereomeric mandelate, O-methylmandelate, and alpha-methoxy-alpha-trifluoromethylphenylacetate (MTPA) esters. J Am Chem Soc. 1973;95:512–519. [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Eustáquio AS, McGlinchey RP, Liu Y, Hazzard C, Beer LL, Florova G, Alhamadsheh MM, Lechner A, Kale AJ, Kobayashi Y, et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-l-methionine. Proc Natl Acad Sci USA. 2009;106:12295–12300. doi: 10.1073/pnas.0901237106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushimi S, Furihata K, Seto H. Studies on new phosphate ester antifungal antibiotics phoslactomycins. II. Structure elucidation of phoslactomycins A to F. J Antibiot (Tokyo) 1989a;42:1026–1036. doi: 10.7164/antibiotics.42.1026. [DOI] [PubMed] [Google Scholar]
- Fushimi S, Nishikawa S, Shimazu A, Seto H. Studies on new phosphate ester antifungal antibiotics phoslactomycins. I. Taxonomy, fermentation, purification and biological activities. J Antibiot (Tokyo) 1989b;42:1019–1025. doi: 10.7164/antibiotics.42.1019. [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Hopwood DA, Sherman DH. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genetics. 1990;24:37–62. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay A. Crystal Structure of the Erythromycin Polyketide Synthase Dehydratase. Journal of Molecular Biology. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides. Chem Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- Keatinge-Clay AT, Stroud RM. The structure of a ketoreductase determines the organization of the beta-carbon processing enzymes of modular polyketide synthases. Structure (Oxford, UK) 2006;14:737–748. doi: 10.1016/j.str.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Khosla C. Harnessing the biosynthetic potential of modular polyketide synthases. Chem Rev. 1997;97:2577–2590. doi: 10.1021/cr960027u. [DOI] [PubMed] [Google Scholar]
- Kong R, Liu X, Su C, Ma C, Qiu R, Tang L. Elucidation of the Biosynthetic Gene Cluster and the Post-PKS Modification Mechanism for Fostriecin in Streptomyces pulveraceus. Chemistry & biology. 2013;20:45–54. doi: 10.1016/j.chembiol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Kwan DH, Tosin M, Schlager N, Schulz F, Leadlay PF. Insights into the stereospecificity of ketoreduction in a modular polyketide synthase. Org Biomol Chem. 2011;9:2053–2056. doi: 10.1039/c1ob00022e. [DOI] [PubMed] [Google Scholar]
- McPherson M, Khosla C, Cane DE. Erythromycin biosynthesis: The β-ketoreductase domains catalyze the stereospecific transfer of the 4-pro-S hydride of NADPH. J Am Chem Soc. 1998;120:3267–3268. [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ohtani I, Kusumi T, Ishitsuka MO, Kakisawa H. Absolute configurations of marine diterpenes possessing a xenicane skeleton. An application of an advanced mosher’s method. Tetrahedron Lett. 1989;30:3147–3150. [Google Scholar]
- Oppermann U, Filling C, Hult M, Shafqat N, Wu X, Lindh M, Shafqat J, Nordling E, Kallberg Y, Persson B, Jornvall H. Short-chain dehydrogenases/reductases (SDR): The 2002 update. ChemBiol Interact. 2003;143-144:247–253. doi: 10.1016/s0009-2797(02)00164-3. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor V. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Reeves CD, Hu Z, Reid R, Kealey JT. Genes for the biosynthesis of the fungal polyketides hypothemycin from Hypomyces subiculosus and radicicol from Pochonia chlamydosporia. Appl Environ Microbiol. 2008;74:5121–5129. doi: 10.1128/AEM.00478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- Schupp T, Toupet C, Engel N, Goff S. Cloning and sequence analysis of the putative rifamycin polyketide synthase gene cluster from Amycolatopsis mediterranei. FEMS Microbiology Letters. 1998;159:201–207. doi: 10.1111/j.1574-6968.1998.tb12861.x. [DOI] [PubMed] [Google Scholar]
- Smith S, Tsai S-C. The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampwala SS, Bunge RH, Hurley TR, Willmer NE, Brankiewicz AJ, Steinman CE, Smitka TA, French JC. Novel antitumor agents CI-920, PD 113,270 and PD 113,271. II. Isolation and characterization. J Antibiot (Tokyo) 1983;36:1601–1605. doi: 10.7164/antibiotics.36.1601. [DOI] [PubMed] [Google Scholar]
- Tang L, Ward S, Chung L, Carney JR, Li Y, Reid R, Katz L. Elucidating the mechanism of cis double bond formation in epothilone biosynthesis. J Am Chem Soc. 2003;126:46–47. doi: 10.1021/ja030503f. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Hegeman AD, Wesenberg G, Chapeau MC, Frey PA, Holden HM. Structural analysis of UDP-sugar binding to UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:6294–6304. doi: 10.1021/bi970025j. [DOI] [PubMed] [Google Scholar]
- Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. The biochemical basis for stereochemical control in polyketide biosynthesis. J Am Chem Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle O, Hahn F, Baerga-Ortiz A, Leadlay PF, Andexer JN. Stereoselectivity of isolated dehydratase domains of the borrelidin polyketide synthase: Implications for cis double bond formation. Chembiochem. 2011;12:1011–1014. doi: 10.1002/cbic.201100011. [DOI] [PubMed] [Google Scholar]
- Wu J, Zaleski TJ, Valenzano C, Khosla C, Cane DE. Polyketide double bond biosynthesis. Mechanistic analysis of the dehydratase-containing module 2 of the picromycin/methymycin polyketide synthase. J Am Chem Soc. 2005;127:17393–17404. doi: 10.1021/ja055672+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Gokhale R, Khosla C, Cane DE. Erythromycin biosynthesis. The 4-pro-S hydride of NADPH is utilized for ketoreduction by both module 5 and module 6 of the 6-deoxyerythronolide B synthase. Bioorg Med Chem Lett. 2001;11:1477–1479. doi: 10.1016/s0960-894x(00)00529-1. [DOI] [PubMed] [Google Scholar]
- Zheng J, Gay DC, Demeler B, White MA, Keatinge-Clay AT. Divergence of multimodular polyketide synthases revealed by a didomain structure. Nat Chem Biol. 2012;8:615–621. doi: 10.1038/nchembio.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Keatinge-Clay AT. Structural and Functional Analysis of C2-Type Ketoreductases from Modular Polyketide Synthases. Journal of Molecular Biology. 2011;410:105–117. doi: 10.1016/j.jmb.2011.04.065. [DOI] [PubMed] [Google Scholar]
- Zheng J, Taylor CA, Piasecki SK, Keatinge-Clay AT. Structural and functional analysis of A-type ketoreductases from the amphotericin modular polyketide synthase. Structure (Oxford, UK) 2010;18:913–922. doi: 10.1016/j.str.2010.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.