Similar to tuning your radio to play one station out of all of the others on the airwaves, calcium binding proteins transduce calcium signatures into specific downstream responses. The expression and subcellular localization of calcium (Ca2+) binding proteins are developmentally and spatially regulated, creating the tuner that allows each subcellular compartment to receive locally generated Ca2+ signals. In radios, tuning to a station allows you hear its broadcast; in plants, transducing Ca2+ signals influences biotic and abiotic stress responses, as well as normal growth and development. Calmodulin (CaM) serves as the main transducer of Ca2+ signals in eukaryotes, although plants have evolved additional Ca2+ binding proteins (reviewed in Perochon et al., 2011). Upon binding Ca2+, CaM interacts with a range of proteins that regulate specific Ca2+-responsive processes. It has long been known that posttranslational trimethylation of CaM can affect its function (Roberts et al., 1986), but this aspect of Ca2+-CaM signaling has generally been overlooked. New work from Banerjee et al. (pages 4493–4511) demonstrates that trimethylation plays a role in several CaM-dependent processes.
CaM N-methyltransferase (CaM KMT), which trimethylates CaM at Lys-155 in eukaryotes (Magnani et al., 2010), is encoded by a single gene in Arabidopsis thaliana. Banerjee et al. took advantage of this to explore the global role of CaM trimethylation. They found that CaM KMT expression is highest in early seedling development, particularly in tissues where auxin signaling is active. Using transgenic lines with altered expression of CaM KMT and, therefore, altered levels of CaM methylation, the authors found that the biggest changes in CaM methylation were observed in the roots, which also displayed differences in length, epidermal cell fate, and epidermal cell number.
Analysis of the same lines provided evidence that CaM trimethylation plays a role in auxin responsiveness as well as ABA signaling and responses to salt, cold, and heat stress. Consistent with this, CaM KMT protein accumulation was regulated by auxin, abscisic acid, and abiotic stresses. The authors went on to identify several proteins that differentially bind methylated versus unmethylated CaM (see figure), suggesting that these binding partners could be responsible for differential responses to the methylated or methylated forms of CaM.
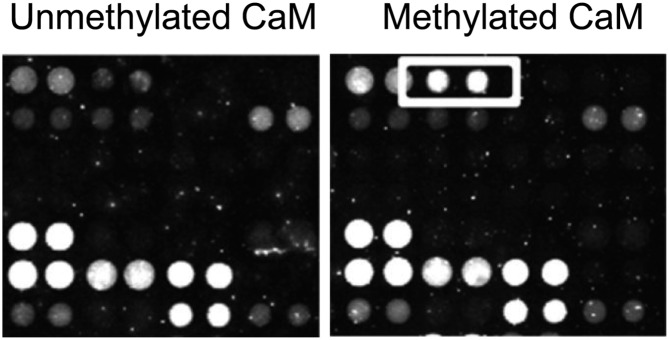
Differential binding of methylated versus unmethylated CaM probe on an Arabidopsis protein chip. The boxed spots preferentially bind methylated CaM in the presence of Ca2+. (Reprinted from Banerjee et al. [2013], Figure 9.)
This work serves to return CaM methylation to the limelight and emphasizes that it should be considered in examinations of Ca2+ signaling. The possibility that methylation-sensitive and methylation-insensitive binding partners of CaM coexist in the same subcellular compartment adds a level of complexity to the potential responses to Ca2+, wherein Ca2+ binding is enough to activate some CaM-mediated responses, but Ca2+ along with CaM methylation is required for others. It seems that CaM methylation could turn the cellular radio into a stereo.
References
- Banerjee J., Magnani R., Nair M., Lynnette D.M., DeBolt S., Maiti I.B., Houtz R.L. (2013). Calmodulin-mediated signal transduction pathways in Arabidopsis are fine-tuned by methylation. Plant Cell 25: 4493–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani R., Dirk L.M.A., Trievel R.C., Houtz R.L. (2010). Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 1: 43. [DOI] [PubMed] [Google Scholar]
- Perochon A., Aldon D., Galaud J.-P., Ranty B. (2011). Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 93: 2048–2053 [DOI] [PubMed] [Google Scholar]
- Roberts D.M., Rowe P.M., Siegel F.L., Lukas T.J., Watterson D.M. (1986). Trimethyllysine and protein function. Effect of methylation and mutagenesis of lysine 115 of calmodulin on NAD kinase activation. J. Biol. Chem. 261: 1491–1494 [PubMed] [Google Scholar]


