Using next-generation sequencing, this work reveals the complex genome-wide responses of rice roots and shoots to short-, medium-, and long-term phosphate deprivation as well as to recovery from phosphate starvation. In addition, analysis of miRNA expression profiles identified 20 miRNAs that were not previously associated with Pi starvation, thus paving the way for future research.
Abstract
Using rice (Oryza sativa) as a model crop species, we performed an in-depth temporal transcriptome analysis, covering the early and late stages of Pi deprivation as well as Pi recovery in roots and shoots, using next-generation sequencing. Analyses of 126 paired-end RNA sequencing libraries, spanning nine time points, provided a comprehensive overview of the dynamic responses of rice to Pi stress. Differentially expressed genes were grouped into eight sets based on their responses to Pi starvation and recovery, enabling the complex signaling pathways involved in Pi homeostasis to be untangled. A reference annotation-based transcript assembly was also generated, identifying 438 unannotated loci that were differentially expressed under Pi starvation. Several genes also showed induction of unannotated splice isoforms under Pi starvation. Among these, PHOSPHATE2 (PHO2), a key regulator of Pi homeostasis, displayed a Pi starvation–induced isoform, which was associated with increased translation activity. In addition, microRNA (miRNA) expression profiles after long-term Pi starvation in roots and shoots were assessed, identifying 20 miRNA families that were not previously associated with Pi starvation, such as miR6250. In this article, we present a comprehensive spatio-temporal transcriptome analysis of plant responses to Pi stress, revealing a large number of potential key regulators of Pi homeostasis in plants.
INTRODUCTION
P, in addition to being one of the most important macronutrients for all living organisms, is also one of the most limiting nutrients for plant growth and development. Although phosphorus (P) is often present in nonlimiting concentrations in the soil, the availability of Pi, the main source of P used by plants, is often hindered due to its low solubility in soil and high sorption capacity (Poirier and Bucher, 2002). As a consequence, application of Pi fertilizers has been the main alternative to overcome low soil Pi content to maintain and increase crop productivity. Yet, this process is not economically or environmentally sustainable as the price of phosphate rock, a nonrenewable resource, increases and run-off of excess Pi fertilizers damages the environment. Thus, studies to better understand the mechanisms involved in regulating Pi homeostasis in plants have been initiated to aid the development of crops with increased phosphorus use efficiency (Veneklaas et al., 2012; Wu et al., 2013).
Pi-limiting conditions trigger an array of adaptive responses at the morphological, physiological, biochemical, and molecular levels, such as reduced plant growth, altered root system architecture, and secretion of organic acids, phosphatases, and nucleases to increase Pi acquisition (Poirier and Bucher, 2002; Rouached et al., 2010; Chiou and Lin, 2011; Plaxton and Tran, 2011). With the emergence of microarray technology, several global transcriptome analyses have been performed in Arabidopsis thaliana (Misson et al., 2005; Morcuende et al., 2007; Müller et al., 2007; Bustos et al., 2010; Nilsson et al., 2010; Thibaud et al., 2010; Woo et al., 2012), and other plants, including rice (Oryza sativa; Wasaki et al., 2003, 2006; Zheng et al., 2009), greatly improving our knowledge of the complex mechanisms regulating Pi homeostasis (Chiou and Lin, 2011; Péret et al., 2011; Plaxton and Tran, 2011; Jain et al., 2012; Secco et al., 2012). Briefly, it appears that the responses of plants to Pi starvation are highly regulated at the transcript level and are largely conserved between Arabidopsis and rice, with the Myb transcription factor At-PHR1 (PHOSPHATE STARVATION RESPONSE1), and its paralogs At-PHL1 (PHR1-LIKE) and Os-PHR2 in Arabidopsis and rice, respectively, being key regulators in the Pi starvation signaling pathway (Rubio et al., 2001; Zhou et al., 2008; Bustos et al., 2010). Under Pi-limited conditions, At-PHR1 is sumoylated by At-SIZ1 (small ubiquitin-like modifier E3 ligase) (Miura et al., 2005), inducing the expression level of numerous genes, including the microRNA miR399, which leads to the degradation of its target PHO2 mRNA that encodes an E2 ubiquitin conjugase enzyme, which in turn negatively regulates genes involved in Pi responses. In Arabidopsis, PHO2 degrades PHO1 (PHOSPHATE1; Liu et al., 2012), a protein involved in Pi transfer from the roots to the shoots, thus modulating Pi homeostasis (Arpat et al., 2012). In addition, PHO2 indirectly affects the expression levels of some high-affinity Pi transporters, such as PHT1;8 and PHT1;9 (Aung et al., 2006; Bari et al., 2006; Hu et al., 2011). The PHO2-miRNA399 pathway is also regulated by target mimicry, where the non-protein-coding transcript INDUCED BY PHOSPHATE STARVATION1 (IPS1) is induced by Pi starvation and has a complementary region to miR399, thus ultimately inhibiting the action of miR399-charged silencing complexes on PHO2 mRNA (Franco-Zorrilla et al., 2007).
Although these studies contributed to our understanding of the global responses to Pi starvation, they were limited to the genes present on microarrays and mainly focused on a few time points and specific tissues, thus failing to capture the dynamic and genome-wide range of responses triggered by such a stress. In addition to mRNA regulation, microRNAs (miRNAs) function as key regulators of nutrient stress signaling (Chiou, 2007), with miR399 and miR827 regulating Pi homeostasis (Fujii et al., 2005; Chiou et al., 2006; Hsieh et al., 2009; Pant et al., 2009; Lin et al., 2010; Kant et al., 2011; Kuo and Chiou, 2011; Wang et al., 2012). Yet, to date, only a single study was performed to assess miRNA regulation in rice under Pi stress, using plants starved for a short period of time (5 d) and focusing only on miR399 and miR827 (Jeong et al., 2011), thus revealing the need for the identification of Pi-responsive miRNAs in rice.
The recent development of next-generation sequencing technology has enabled researchers to study whole transcriptomes at single-base resolution, in a multitude of samples/conditions, in a cost-effective manner. It is now possible to look at several levels of regulation, under a large number of conditions within a same study, thus providing an integrated understanding of the mechanisms involved in the regulation of Pi homeostasis. In addition, unlike microarrays, RNA sequencing (RNAseq) enables accurate quantification of mRNA and noncoding RNA expression, as well as revealing any previously unannotated transcripts and splicing isoforms (Trapnell et al., 2012). Using RNAseq to identify loci/isoforms associated with specific conditions, such as Pi deficiency, may lead to the discovery of key regulators of Pi homeostasis in rice (Trapnell et al., 2010).
In this study, we aimed to shed further light on the complex responses of rice plants to Pi stress using next-generation RNAseq coupled with a comprehensive time-course experiment, analyzing the early and late responses to Pi deprivation as well as Pi resupply. We generated a comprehensive and integrated data set that provides insight into the molecular responses of the rice transcriptome, and small RNAome, following Pi deprivation and resupply. This was accomplished with unprecedented resolution, thereby revealing key regulators of Pi homeostasis in plants.
RESULTS AND DISCUSSION
Experimental Design and Physiological Responses
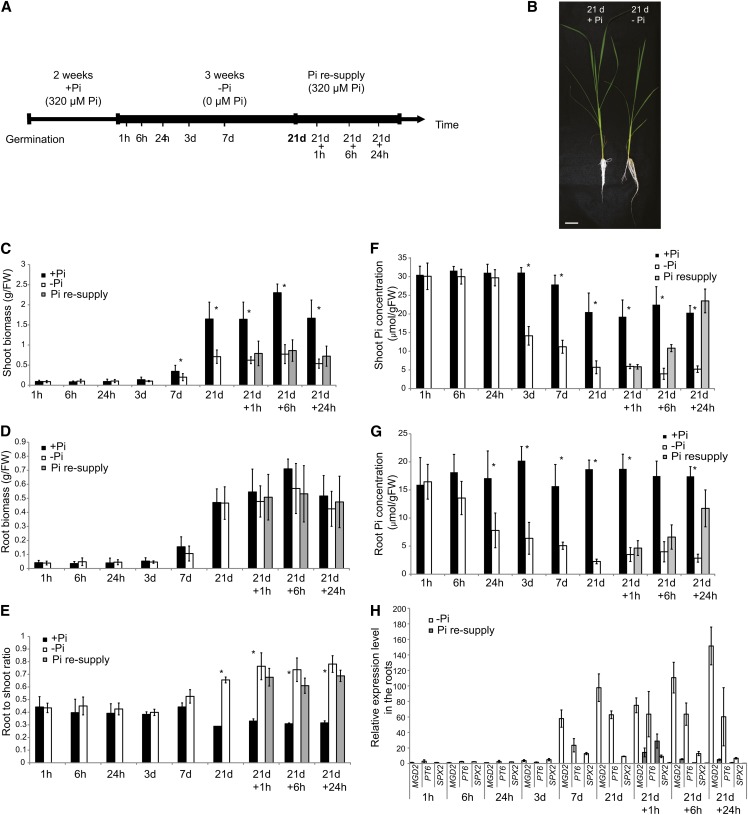
To better understand the complex mechanisms regulating Pi homeostasis in rice, we performed a time-course experiment, where pregerminated seedlings were grown hydroponically for 2 weeks on Pi-sufficient medium (0.32 mM Pi), before transferring half of the plants to Pi-deficient solution (0 mM Pi) for 21 d (Figure 1A). After 3 weeks of Pi deprivation treatment, half of these plants where then resupplied with Pi-sufficient media for up to 24 h. In total, nine time points were selected to cover short- and long-term responses to Pi deprivation as well as the effects of Pi resupply on Pi-starved plants. To confirm the effectiveness of the Pi deprivation, physiological and molecular analyses were performed. Growth analysis showed that the shoot biomass was significantly affected by 21 d of Pi deficiency, being 2.3-fold lower than that of shoots grown under Pi-sufficient conditions, but we observed no differences in root biomass (Figures 1B to 1D). Consequently, plants grown under Pi deprivation conditions showed a higher root-to-shoot biomass ratio compared with plants grown under Pi-sufficient conditions (Figures 1B and 1E), which is in accordance with previous studies (Reymond et al., 2006; Jiang et al., 2007). We observed significant differences in the root-to-shoot ratio between the two conditions as early as 7 d after initiating the Pi treatment, reaching more than 2.2-fold difference after 21 d of Pi starvation.
Figure 1.
Experimental Design and Physiological Responses of Rice Seedlings to Pi Deprivation and Resupply.
(A) Schematic representation of the experimental design. Seeds were germinated in water for 2 d and then transferred to a Pi-sufficient hydroponic (0.320 mM) solution for 2 weeks before being either transferred to Pi-deficient media (0 mM) or maintained in Pi-sufficient solution for 21 d. After 21 d of treatment, half of the Pi-starved plants were supplied with Pi-sufficient media for up to a day.
(B) Morphological appearance of rice seedling grown for 21 d on Pi-sufficient or -deficient media. Bar = 5 cm.
(C) to (G) Fresh weight (FW) ([C] and [D]) and Pi concentration ([F] and [G]) were assessed for shoots and roots. Root-to-shoot biomass ratio is represented (E).
(H) Relative expression of Pi starvation–responsive genes MGD2, PT6, and SPX2, in the roots. Expression levels are relative to the +Pi condition.
Errors bars are sd and n = 10, except for (H), where n = 3. Asterisk indicates statistically significant (P < 0.05) differences between samples grown under +Pi and –Pi conditions.
Pi concentration analysis revealed that both shoots and roots showed a gradual decrease in Pi content as the length of the treatment increased (Figures 1F and 1G). In the roots, Pi deprivation led to a 50% reduction in Pi concentration as early as 24 h after the initiation of the treatment, while it took 3 d to have similar effects in the leaves. Interestingly, even after 7 d of Pi treatment, the Pi concentration in both root and shoot was still twofold higher than that observed after 21 d of Pi starvation, suggesting that after 7 d of Pi deprivation, plants may not be completely starved (Figures 1F and 1G). The Pi concentration observed after 21 d of Pi starvation in the roots and shoots was similar to that reported in previous long-term Pi starvation studies (Wang et al., 2009a; Secco et al., 2010; Jia et al., 2011; Rouached et al., 2011). In addition, the shoot Pi concentration observed after 21 d of Pi starvation was similar to that of rice shoots when grown in P-poor soils (4.5 μmol/g fresh weight) (H. Shou, unpublished data). Pi resupply resulted in rapid increase of the internal Pi content in both roots and shoots. Indeed, within 6 h of Pi resupply, the shoot Pi concentration was already almost twice as high as that observed in plants grown under Pi starvation conditions and was similar to that of plants grown on Pi-sufficient media after 24 h of Pi resupply (Figure 1F).
Finally, we used quantitative RT-PCR (qRT-PCR) to assess the molecular responses of rice plants to Pi deprivation and resupply (Figure 1H). We measured expression of four known Pi starvation–induced marker genes, namely, the noncoding RNA IPS1 (Hou et al., 2005), the high-affinity Pi transporter PT6 (Ai et al., 2009), SPX2 (Wang et al., 2009b), and MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE2 (MGD2) involved in galactolipid synthesis (Kobayashi et al., 2009) in roots and shoots (Figure 1H; see Supplemental Figure 1 online). Expression of these Pi starvation–induced marker genes increased with the duration of Pi deprivation, reaching a maximum after 21 d of Pi starvation, before returning toward basal level after Pi resupply (Figure 1H). In the roots, IPS1 showed the strongest induction after long-term Pi starvation, being upregulated by more than 1000-fold, while MGD2 and PT6 were induced by more than 90 and 60 times, respectively. In the shoots, IPS1 and SPX2 displayed the highest induction upon long term Pi starvation, showing more than 60-fold the expression levels observed for plants grown under Pi-sufficient conditions (see Supplemental Figure 1 online).
Profiling the Short- and Medium-Term Responses to Pi Deprivation Using RNAseq
To analyze the effects of Pi status on the transcriptome of rice roots and shoots, we selected nine time points (Figure 1A) and used three biological replicates per condition for RNAseq, representing a total of 126 libraries (Figure 1A). Sequencing libraries were generated by first removing rRNAs and subsequently analyzing the remaining RNAs on an Illumina Hisequation 1000 using a 2X101 paired-end run. Overall, more than 3.6 billion paired-end reads passing filter were generated, and these were used for mapping onto the International Rice Genome Sequencing Project-1.0 (IRGSP-1.0) Nipponbare rice genome sequence. On average, each sample had 30 million paired-end reads, of which an average of 68 and 77% uniquely mapped to the rice genome (IRGSP-1.0) for root and shoot samples, respectively (see Supplemental Table 1 online). Thus, each sample had an average of 20 million uniquely mapped paired-end reads. The whole data set can be visualized at http://www.plantenergy.uwa.edu.au/annoj/Secco_2013.html using the AnnoJ genome browser as well as in the Rice Annotation Project Database (RAP-DB) genome browser.
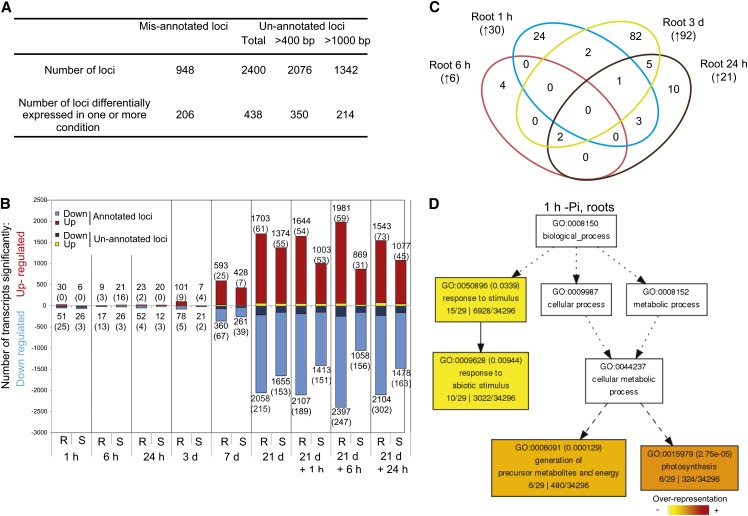
To search for unannotated transcripts and isoforms, we made a reference annotation–based transcript (RABT) assembly using the Cufflinks software (Trapnell et al., 2012). To do so, and to potentially identify loci and isoforms that are either tissue or condition specific, we used mapped reads corresponding to the 21-d samples (+Pi and –Pi, root and shoot). Comparison of the newly generated assembly to the original Michigan State University (MSU) Rice Genome Annotation Project assembly (v7) assembly led to the identification of putative misannotated loci and previously unannotated loci. Here, we consider misannotated loci as loci assembled by Cufflinks but spanning two or more loci annotated in MSU v7. This is often due to the incorrect annotation of the initial gene model or annotated genes being too close. By contrast, previously unannotated loci are an assembled group of reads not overlapping with any known gene model annotation from MSU v7, thus including potentially true novel loci, unannotated portions of annotated loci, such as untranslated regions (UTRs) or exons, or genes not annotated in MSU v7, such as rRNAs. In total, the RABT assembly identified 948 putative misannotated loci and 2400 previously unannotated loci (Figure 2A). Transcript abundance was then estimated by FPKM (for fragment per kilobase of exon per million fragments mapped) using Cuffdiff 2, a software known to robustly perform differential analysis using RNAseq data (Trapnell et al., 2012).
Figure 2.
Profiling the Transcriptome Response to Pi Stress Using RNAseq.
(A) Summary of the reference annotation–based transcript assembly and the corresponding mis- and unannotated loci identified.
(B) Number of significantly differentially expressed genes after varying length of Pi deprivation in roots (R) and shoots (S) using Cufflinks (P < 0.05, FDR < 0.05). Number in parenthesis represents the number of previously unannotated loci among the differentially expressed genes.
(C) Venn diagram representing the overlap of the early molecular responses in the roots upon Pi deprivation (1 h to 3 d). Number in parenthesis represents the number of genes upregulated by Pi deprivation.
(D) AgriGO representation of the overrepresented GO terms in the 30 genes upregulated by 1 h of Pi deprivation in the roots was generated using singular analysis enrichment (Fisher’s test, P < 0.05, FDR < 0.05). Number in parenthesis represents the FDR value.
Differential expression analysis showed that only a few genes were significantly differentially expressed in either root or shoot within 24 h to 3 d of Pi deprivation, respectively (P < 0.05, false discovery rate [FDR] < 0.05) (Figure 2B; see Supplemental Data Sets 1 to 3 online). In addition, these genes showed little overlap between the different early time points, suggesting either that the early steps of Pi deprivation sensing and signaling occur transiently or that these responses may be the consequences of nonspecific stresses due to the manipulation of the plants, and the Pi starvation sensing mechanism initiates later. Indeed, among the 30 genes upregulated by 1 h of Pi deprivation in the roots, only six of these were also induced after 6 h, 24 h, and 3 d of Pi deprivation (Figure 2C). Interestingly, several genes involved in photosynthesis and carbon fixation were specifically induced after 1 h of Pi deprivation in the roots and not in the control plants (see Supplemental Data Sets 1 and 3 online). This is intriguing, as with the hydroponic system used in this study, the roots were kept in the dark during the entire experiment, except for less than a minute during the transfer of plants between nutrient solution replacements, and special care was taken to avoid contamination from green photosynthesizing tissues. Gene Ontology (GO) analysis of this subset of genes revealed a significant overrepresentation of genes involved in photosynthesis and the generation of precursor metabolites and energy, associated with an overrepresentation of ribulose-bisphosphate carboxylase activity (Figure 2D). This set of genes was only upregulated at this very early time point (1 h) and was downregulated at the later time points. Among the 26 annotated genes downregulated by 1 h of Pi deprivation in the roots, several genes encoded functions involved in metal uptake, including two nicotianamine synthases, two metal transporters, and three thionins (see Supplemental Data Set 3 online). In the shoots, among the six significantly upregulated genes by 1 h of Pi deprivation, two genes encoded metallothioneins (LOC_Os12g38270 and LOC_Os12g38290). Metallothioneins affect metal tolerance and homeostasis and scavenge reactive oxygen species (Cobbett and Goldsbrough, 2002), which could be a mechanism to overcome the increase in certain ion concentration, such as iron, upon Pi starvation. Under Pi starvation, plants overaccumulated some ions, including iron (Misson et al., 2005; Hirsch et al., 2006; Lei et al., 2011), as a result of enhanced availability in the media.
After 6 h of Pi deprivation, several genes involved in iron homeostasis were upregulated in both roots and shoots. In roots, two putative vacuolar iron transporters (annotated as integral membrane proteins, LOC_Os04g45520 and LOC_Os09g23300) were among the six annotated upregulated genes, while in the shoots, two ferritins (LOC_Os11g01530 and LOC_Os12g01530) and one iron transporter (LOC_Os09g23300) were among the five induced annotated genes. The induction of iron storage genes and the repression of metal transporters (1 h in the roots) has been reported in Arabidopsis, and this is thought to be a response to iron overload due to low Pi (Misson et al., 2005; Thibaud et al., 2010). In Arabidopsis, a Pi starvation–responsive ferritin, Fer1, contains a P1BS element (Bournier et al., 2013), which is the binding site for PHR1, a major regulator of Pi starvation response. Further analysis confirmed that both PHR1 and its paralog, PHL1, could bind to Fer1, hence regulating the expression of Fer1 upon Pi starvation, demonstrating a direct link between iron and Pi homeostasis in Arabidopsis (Bournier et al., 2013). Promoter analysis of the genes involved in iron homeostasis that were induced by short-term Pi deprivation revealed that only LOC_OS12g01530, an ortholog of Arabidopsis Fer1, displayed a P1BS motif within the first two kilobases of its promoter, suggesting that the direct link between iron and Pi homeostasis observed in Arabidopsis is also conserved in rice. In addition, two genes (LOC_Os08g06010, a putative glycerol 3-phosphate permease, and LOC_Os03g40670, a putative glycerophosphoryl diester phosphodiesterase) are suggested to be involved in Pi remobilization (Cheng et al., 2011) and were also upregulated in the roots after 6 h of Pi starvation. The expression of these two genes gradually increased with the duration of Pi deprivation, reaching a maximum after 21 d and decreasing after Pi resupply, suggesting that this regulation is part of a continued response to Pi starvation. Within 24 h of Pi deprivation in the roots, 23 genes were upregulated, including two high-affinity Pi transporters, PT2 and PT4, which were significantly upregulated by more than 2.4- and 3.2-fold, respectively.
Medium-term Pi deprivation (3 d –Pi) led to a significant decrease in the internal Pi content by more than two- and threefold in both shoots and roots, respectively, but the number of differentially regulated genes remained moderately low, with only 28 and 179 genes being upregulated in the shoots and roots, respectively. Among the 179 significantly upregulated genes in the roots, several known Pi-responsive genes, including the high-affinity Pi transporters PT3 and PT6 as well as the noncoding RNA IPS1, were among the most induced genes, being upregulated by more than 11-fold. Surprisingly, in the shoots, medium-term Pi deprivation led to the upregulation of only three annotated genes, including IPS1 (Figure 2B; see Supplemental Data Set 3 online).
Profiling the Long-Term Responses to Pi Starvation Using RNAseq
Long-term Pi deprivation (7 d or more) resulted in severe changes in gene expression, with 3761 and 3029 genes (2654 and 1950, using a twofold cutoff) being significantly differentially expressed after 21 d of Pi starvation in roots and shoots, respectively (Figure 2B; see Supplemental Data Set 3 online). In the roots, genes such as the high-affinity Pi transporters PT3 and PT10, IPS1, MGD2, and a lipase (LOC_Os11g43760) were among the most induced genes, being induced more than 150-fold (Table 1; more than sevenfold [log2]). It is also worth noting that several genes, including a hydrolase (LOC_Os05g46460), a putative low-temperature and salt-responsive protein (LOC_Os06g44220), and PT9, were only expressed under Pi-deficient conditions (FPKM > 30 in –Pi, while in +Pi FPKM = 0) (Table 1). Among the genes most induced by long-term Pi starvation, several transcripts are not present on the rice Affymetrix arrays, including LOC_Os06g44220, encoding a putative low-temperature and salt-responsive protein, and thus constitutes potential key candidate genes involved in Pi homeostasis that have been missed by previous genome-wide studies. GO analysis of the subset of genes induced by 7 d of Pi deprivation in the roots revealed a significant overrepresentation (Fisher’s test, P < 0.05, FDR < 0.05) of genes involved in lipid metabolism, response to oxidative stress, and inorganic anion transport (see Supplemental Figure 2 online). To assess the response to long-term Pi starvation in rice, when the internal Pi levels are at a minimum, the PAGEMAN tool (Usadel et al., 2006) was used to identify overrepresented (Fisher’s test, P < 0.05) functions among the differentially expressed genes upon 21 d of Pi starvation in both roots and shoots (Figure 3A). Interestingly, most of the genes upregulated by Pi starvation in roots and shoots were overrepresented in the same functional categories. Only a limited number of functional categories were overrepresented. We also observed that the genes upregulated under Pi starvation were enriched in lipid metabolism, phenylpropanoid metabolism, cytochrome P450 and transport functions, and, more specifically, Pi transport, as previously reported (Misson et al., 2005; Morcuende et al., 2007). By contrast, genes downregulated by long-term Pi starvation were enriched in genes involved in photosynthesis and N metabolism in both roots and shoots. Also, the genes downregulated by Pi starvation in the shoots were enriched in the protein synthesis functional category, mainly through the overrepresentation of rRNAs. Despite using rRNA-depleted RNA to generate the sequencing libraries, we still detected reads originating from rRNA sequences. This phenomenon is not surprising as rRNAs are highly expressed genes; thus, removal of 95 to 99% of rRNA still results in a significant number of reads. Additionally, N metabolism, including ammonia and Asp metabolism as well as tetrapyrrole synthesis, were all also enriched in the downregulated genes.
Table 1. List of the 30 Most Induced Genes after 21 d of Pi Starvation in the Roots and Shoots.
| Gene ID | Description | +Pi (FPKM) | −Pi (FPKM) | Fold Change (Log2) |
|---|---|---|---|---|
| Shoot | ||||
| LOC_Os06g44220 | RCI2-9, a putative low-temperature and salt-responsive protein | 0 | 21.61 | N/A |
| LOC_Os04g32320 | Glycerophosphoryl diester phosphodiesterase family protein | 0 | 5.64 | N/A |
| LOC_Os03g29250 | SPX5* | 0.02 | 31.82 | 10.60 |
| LOC_Os06g03880 | Expressed protein | 0.02 | 3.24 | 7.61 |
| LOC_Os09g24520 | Pentatricopeptide | 0.27 | 37.25 | 7.13 |
| LOC_Os04g47330 | rho-GTPase–activating protein-related | 0.03 | 4.08 | 7.12 |
| LOC_Os11g43760 | Lipase class 3 family protein | 0.07 | 9.70 | 7.11 |
| LOC_Os08g20420 | MGD2* | 0.46 | 60.82 | 7.06 |
| LOC_Os11g13720 | Expressed protein | 0.08 | 6.92 | 6.50 |
| LOC_Os12g32640 | Haemolysin-III | 0.49 | 31.29 | 6.01 |
| LOC_Os09g24520 | Pentatricopeptide | 0.21 | 12.16 | 5.84 |
| LOC_Os03g19290 | Mitochondrial import inner membrane translocase subunit Tim17 | 1.18 | 64.66 | 5.77 |
| LOC_Os02g10780 | SPX2* | 3.37 | 147.99 | 5.46 |
| LOC_Os08g31670 | Transporter | 0.14 | 5.16 | 5.25 |
| LOC_Os07g42324 | Expressed protein | 3.45 | 125.43 | 5.19 |
| LOC_Os02g31030 | Glycerophosphoryl diester phosphodiesterase family protein* | 4.29 | 145.69 | 5.09 |
| LOC_Os05g46460 | Hydrolase, α/β fold family domain–containing protein | 1.65 | 53.93 | 5.03 |
| LOC_Os03g14180 | hsp20/α crystallin family protein | 0.20 | 6.48 | 4.99 |
| LOC_Os03g05334 | IPS1* | 12.00 | 352.94 | 4.88 |
| LOC_Os10g25310 | SPX3* | 0.43 | 11.51 | 4.74 |
| LOC_Os01g71180 | Pentatricopeptide repeat protein PPR1106-17 | 0.28 | 7.38 | 4.70 |
| LOC_Os03g63580 | Phosphoesterase family protein | 0.42 | 9.85 | 4.54 |
| LOC_Os01g52230 | Phosphoethanolamine/phosphocholine phosphatase* | 21.98 | 511.81 | 4.54 |
| LOC_Os04g01740 | Heat shock protein | 1.81 | 39.66 | 4.45 |
| LOC_Os04g10690 | PT5* | 0.30 | 6.25 | 4.38 |
| LOC_Os02g22020 | MYB family transcription factor | 1.27 | 25.39 | 4.32 |
| LOC_Os02g49970 | ZIM motif family protein, | 0.22 | 4.16 | 4.25 |
| LOC_Os08g17784 | Purple acid phosphatase* | 0.79 | 13.40 | 4.09 |
| LOC_Os03g44740 | Cytochrome P450 | 0.79 | 11.98 | 3.93 |
| LOC_Os07g47450 | Flowering promoting factor-like 1 | 0.51 | 7.69 | 3.90 |
| Root | ||||
| LOC_Os05g46460 | Hydrolase, α/β fold family domain–containing protein, | 0 | 90.69 | N/A |
| LOC_Os06g44220 | RCI2-9, a putative low-temperature and salt-responsive protein, | 0 | 41.45 | N/A |
| LOC_Os06g21920 | PT9* | 0 | 34.71 | N/A |
| LOC_Os09g24520 | Pentatricopeptide | 0 | 25.61 | N/A |
| LOC_Os09g29740 | β-Expansin precursor | 0 | 17.13 | N/A |
| LOC_Os06g21930 | Inorganic Pi transporter* | 0 | 9.04 | N/A |
| LOC_Os06g03880 | Expressed protein | 0 | 5.30 | N/A |
| LOC_Os06g03880 | Expressed protein | 0 | 3.65 | N/A |
| LOC_Os03g27680 | Conserved hypothetical protein | 0 | 3.18 | N/A |
| LOC_Os03g05334 | IPS1* | 1.30 | 4260.97 | 11.68 |
| LOC_Os10g30770 | PT3* | 0.09 | 106.10 | 10.14 |
| LOC_Os11g43760 | Lipase class 3 family protein | 0.02 | 19.77 | 9.94 |
| LOC_Os08g20420 | MGD2* | 0.18 | 115.99 | 9.36 |
| LOC_Os06g21950 | PT10* | 0.04 | 25.88 | 9.21 |
| LOC_Os01g50490 | Cytochrome P450 | 0.07 | 36.42 | 9.00 |
| LOC_Os09g28830 | SCP42, a putative Ser carboxypeptidase homolog | 0.02 | 6.80 | 8.20 |
| LOC_Os03g29250 | SPX5* | 0.47 | 125.40 | 8.06 |
| LOC_Os10g39380 | Asp proteinase nepenthesin-1 precursor | 0.27 | 57.56 | 7.72 |
| LOC_Os08g05690 | ATP binding cassette transporter, ATP binding protein | 0.05 | 8.98 | 7.38 |
| LOC_Os05g51240 | Hydrolase, α/β fold family domain–containing protein, | 0.03 | 4.00 | 7.24 |
| LOC_Os06g18820 | Ser/Thr kinase | 0.22 | 31.37 | 7.19 |
| LOC_Os03g55776 | Expressed protein | 0.02 | 3.28 | 7.17 |
| LOC_Os08g45000 | PT6* | 0.51 | 71.06 | 7.12 |
| LOC_Os07g42324 | Expressed protein | 2.29 | 317.36 | 7.11 |
| LOC_Os09g17970, LOC_Os09g17990 | – | 0.03 | 4.28 | 7.09 |
| LOC_Os10g23180 | Cytochrome P450 | 0.24 | 32.05 | 7.04 |
| LOC_Os12g11550 | Hypothetical protein | 0.05 | 7.10 | 7.03 |
| LOC_Os11g35930 | Chalcone synthase | 0.41 | 48.32 | 6.89 |
| LOC_Os04g27070 | Terpene synthase | 0.13 | 15.05 | 6.89 |
| LOC_Os04g49500 | U-box domain–containing protein | 0.05 | 5.81 | 6.86 |
Only annotated genes with FPKM > 3 (P < 0.05, FDR < 0.05) are shown. Asterisks indicate genes with known function in Pi homeostasis. N/A indicates that the corresponding genes were not expressed in +Pi conditions but were induced in −Pi. – indicates that no gene description is available for that locus, due to reads overlapping two loci.
Figure 3.
Profiling the Long-Term Responses to Pi Starvation and Pi Recovery Using RNAseq.
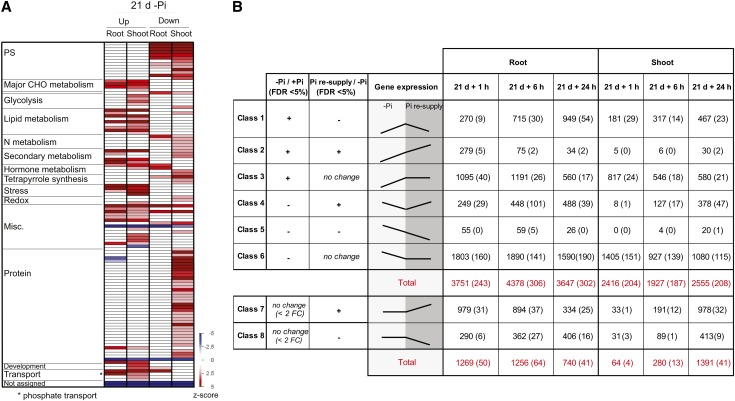
(A) PAGEMAN analysis showing significant over- and underrepresented functional categories in the roots and shoots after 21 d of Pi starvation. Fisher’s exact test was used to determine overrepresented functional categories, and scores are displayed as a heat map, where a z-score of 1.96 represents a P value of 0.05. PS, photosynthesis.
(B) Classification of the differentially regulated genes based on their expression pattern in response to Pi starvation and resupply and the corresponding number of genes present in each class (P < 0.05, FDR < 0.05). Number in parenthesis represents the number of previously unannotated loci among the differentially expressed genes. FC, fold change.
To compare the power of RNAseq to traditional microarray analyses, we assessed the proportion of differentially expressed genes identified by RNAseq in response to 21 d of Pi starvation in the roots having probe sets on the Affymetrix microarray chips (see Supplemental Figure 3 online). This analysis revealed that among the 3761 differentially expressed genes after 21 d of Pi starvation in the roots, 23% of these could not be detected by microarrays as a result of annotated genes lacking probe sets, genes being misannotated in MSU v7, or genes being previously unannotated; thus, RNAseq enabled the identification of a large number of key regulators of Pi homeostasis.
RNAseq Analysis of Pi Resupply to Pi-Starved Plants
To study the effects of Pi resupply, plants that were starved for 21 d were supplied with Pi sufficient media (0.32 mM Pi) for up to 24 h, allowing the classification of differentially expressed genes into eight groups according to their expression patterns in response to the Pi treatments (Figure 3B). This classification revealed genes sets that responded to Pi starvation and recovered after Pi resupply, genes that responded to Pi starvation but persisted in their response during Pi resupply (classes 2 and 5), genes differentially expressed under Pi starvation, but not responsive to Pi resupply (classes 3 and 6), and genes that were not affected by Pi starvation but responded to Pi resupply (classes 7 and 8) (see Supplemental Data Sets 4 and 5 online).
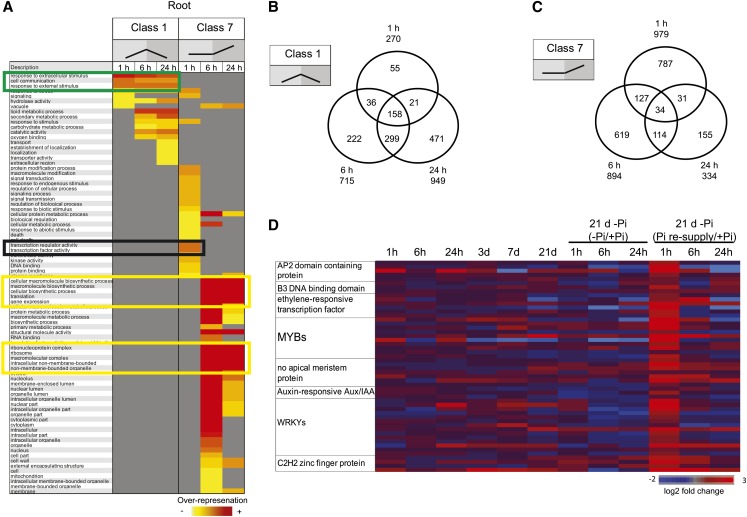
Classes 1 and 4 genes respond to both Pi starvation and Pi resupply and thus likely function in Pi homeostasis. The proportion of genes returning to basal expression levels after Pi refeeding increased with the duration of the Pi resupply in both roots and shoots. Indeed, while 24% (270 genes) of the upregulated genes in the roots had returned toward basal level after 1 h of Pi refeeding, 61% (949 genes) of them had returned toward basal level after 24 h of Pi resupply (Figure 3B). GO analyses (Fisher’s test, P < 0.05, FDR < 0.05) of the three time points of class 1 revealed an overrepresentation of similar functional categories (Figure 4A, green box), with the later time points having genes specifically enriched in lipid metabolism and catalytic activity (Figure 4A). Thus, as expected, the genes belonging to this class were highly conserved among the three time points (Figure 4B). Analysis of the 270 genes returning toward basal level after 1 h of Pi resupply revealed an overrepresentation of genes involved in the response to extracellular stimulus and cell communication (Figure 4A). Among this set of genes, several key genes involved in Pi homeostasis were present, including some PTs, SPX genes, and genes involved in lipid metabolism, such as MGD2 (Table 2). The presence of these genes, which are known to have key roles in Pi homeostasis, within this set (Table 2) suggests that the other genes in this group may have important roles in Pi homeostasis as well. A similar pattern was observed in the shoot, though to a lesser extent, with 1 h of Pi resupply resulting in 189 genes returning toward basal level. Among the 181 upregulated genes returning toward basal level, GO analyses (Fisher’s test, P < 0.05, FDR < 0.05) revealed an overrepresentation of genes involved in phosphatase activity and included key Pi homeostasis regulators, such as some PTs, SPX genes, and genes involved in lipid metabolism, such as MGD2.
Figure 4.
Detailed Temporal Analysis of Classes 1 and 7 in Response to Pi Resupply in the Roots.
(A) Schematic representation of the overrepresented gene ontologies in classes 1 and 7, after 1, 6, and 24 h of Pi resupply in the roots. Green box represents the set of functional categories that are similarly overrepresented in all three times points of class 1. Black box represents the main functional categories that are overrepresented specifically upon 1 h of Pi resupply (class 7), while the yellow boxes represent the main functional categories that are overrepresented upon 6 and 24 h of Pi resupply (class 7).
(B) and (C) Venn diagrams showing the overlap in response to Pi resupply in the roots for genes belonging to classes 1 (B) and 7 (C).
(D) Heat map representing the main transcription factors specifically induced by 1 h of Pi resupply in the roots.
Table 2. List of the 30 Most Induced Genes by 21 d of Pi Starvation (FPKM > 3) and Returning to Basal Level after 1 h of Pi Resupply in the Roots.
| RABT_id | Gene | Description | +Pi (FPKM) | −Pi (FPKM) | Pi Resupply (FPKM) |
|---|---|---|---|---|---|
| XLOC_017383 | LOC_Os12g12140 | Expressed protein | 0 | 6.49 | 0 |
| XLOC_042433 | LOC_Os06g44220 | OsRCI2-9, a putative low-temperature and salt-responsive protein | 0 | 29.98 | 8.59 |
| XLOC_054189 | – | – | 0 | 41.72 | 5.47 |
| XLOC_041420 | LOC_Os06g21930 | Inorganic Pi transporter | 0 | 4.21 | 0.41 |
| XLOC_045266 | – | – | 0 | 35.17 | 2.64 |
| XLOC_046358 | LOC_Os07g23850 | Glycosyl hydrolase | 0 | 6.59 | 0.02 |
| XLOC_041419 | LOC_Os06g21920 | PT9 | 0.01 | 18.41 | 1.61 |
| XLOC_007932 | LOC_Os10g30770 | PT3 | 0.07 | 71.77 | 35.79 |
| XLOC_024511 | – | – | 0.04 | 24.71 | 2.63 |
| XLOC_043797 | LOC_Os06g21950 | PT10 | 0.02 | 13.32 | 1.36 |
| XLOC_050832 | LOC_Os08g20420 | MGD2 | 0.19 | 83.25 | 16.09 |
| XLOC_012284 | LOC_Os11g43760 | Lipase class 3 family protein | 0.04 | 14.24 | 5.30 |
| XLOC_040283 | LOC_Os05g51240 | Hydrolase, α/β fold family domain–containing protein | 0.04 | 11.61 | 1.52 |
| XLOC_002206 | LOC_Os01g50490 | Cytochrome P450 | 0.11 | 24.13 | 7.03 |
| XLOC_025896 | LOC_Os03g29250 | SPX5 | 0.43 | 88.08 | 5.37 |
| XLOC_036619 | LOC_Os05g20030 | DUF617 domain–containing protein | 0.03 | 4.06 | 0.31 |
| XLOC_052016 | LOC_Os08g45000 | PT6 | 0.36 | 48.44 | 22.32 |
| XLOC_005551 | LOC_Os01g52230 | Phosphoethanolamine/phosphocholine phosphatase | 2.62 | 305.86 | 44.08 |
| XLOC_002788 | – | – | 2.02 | 191.95 | 114.17 |
| XLOC_005477 | LOC_Os01g50480 | SAM-dependent carboxyl methyltransferase | 0.08 | 6.31 | 0.71 |
| XLOC_047293 | LOC_Os07g42324 | Expressed protein | 2.39 | 186.97 | 12.17 |
| XLOC_050139 | LOC_Os08g05690 | ABC transporter, ATP binding protein | 0.10 | 7.47 | 2.61 |
| XLOC_056997 | LOC_Os09g24520 | Pentatricopeptide | 0.16 | 9.94 | 2.26 |
| XLOC_006405 | LOC_Os01g71420 | Ser/Thr protein phosphatase family protein | 0.44 | 25.60 | 5.54 |
| XLOC_050715 | LOC_Os08g17784 | Purple acid phosphatase | 0.55 | 25.67 | 8.49 |
| XLOC_008309 | LOC_Os10g39380 | Asp proteinase nepenthesin-1 precursor | 0.83 | 34.24 | 17.55 |
| XLOC_005627 | LOC_Os01g54270 | Transposon protein, putative, unclassified | 0.16 | 6.68 | 3.48 |
| XLOC_003476 | LOC_Os01g03630 | Multicopper oxidase domain–containing protein | 0.32 | 12.95 | 4.46 |
| XLOC_037170 | LOC_Os05g33140 | CHIT5, a chitinase family protein precursor | 0.23 | 9.25 | 0.87 |
| XLOC_053094 | LOC_Os08g23410 | Rubredoxin family protein | 0.11 | 3.81 | 0.72 |
| XLOC_012727 | LOC_Os11g05400 | Ser/Thr protein phosphatase family protein | 2.46 | 77.69 | 36.73 |
| XLOC_047706 | LOC_Os07g01540 | Ser/Thr protein phosphatase family protein | 0.75 | 23.56 | 8.97 |
Dash (–) indicates previously unannotated loci, where no gene name or description is available.
In parallel, the number of continuously positively regulated genes in the roots decreased from 24% (279 genes) of the total number of upregulated genes in the roots after 1 h of Pi refeeding to only 2% (34 genes) after 24 h of Pi resupply (Figure 3B). In the shoots, a limited number of genes showed a continuous response, with 30 genes induced by 24 h of Pi resupply, compared with Pi-starved plants. Classes 3 and 6 are more likely to represent genes that are not directly linked to Pi homeostasis as they are induced or repressed during Pi starvation but do not respond to Pi resupply. While the number of genes belonging to the persistent negative class remained similar independently of the duration of the Pi resupply, the numbers of genes belonging to the persistent positive response class decreased with the duration of Pi resupply (Figure 3B).
Finally, resupplying Pi to Pi-starved plants resulted in the differential expression of another class of genes, genes that were not affected by Pi starvation, but only responded to Pi resupply (classes 7 and 8) (see Supplemental Data Set 5 online). Indeed, 1 h of Pi resupply resulted in the differential expression of more than 1200 genes (789 when using a twofold cutoff) in the roots, with more than 900 (653 when using a twofold cutoff) being upregulated (Figure 3B). The differential expression of such a large number of genes after 1 h of treatment suggests that sensing external Pi occurs rapidly and triggers a large set of responses, unlike the one observed in response to Pi starvation, where only 30 genes were upregulated in the roots after 1 h of Pi starvation. After 24 h of Pi resupply, the number of upregulated genes in the roots was reduced by threefold, with only 334 genes being induced (Figure 3B). Interestingly, the overlap between the genes upregulated after 1 h of Pi resupply and the later time points was very low, with only 6% (65 genes) of the genes upregulated by 1 h of Pi resupply also being upregulated after 24 h of Pi resupply (Figure 4C). GO analyses of the genes upregulated in the three time points confirmed an evolution in the response to Pi refeeding (Figure 4A). Indeed, GO analysis of the 979 upregulated genes by 1 h Pi resupply revealed an overrepresentation (Fisher’s test, P < 0.05, FDR < 0.05) of genes involved in transcription factor and kinase activity, representing 20% of the upregulated genes (Figure 4A, black box). Among the various transcription factors induced by 1 h of Pi resupply, WRKYs and MYBs were the most represented with 14 and 10 members, respectively, and were significantly overrepresented (χ2 P < 0.05) (Figure 4D). However, longer Pi resupply resulted in the upregulation of genes involved in protein synthesis, with the translation functional category being the most significantly overrepresented (Figure 5A, yellow boxes).
Figure 5.
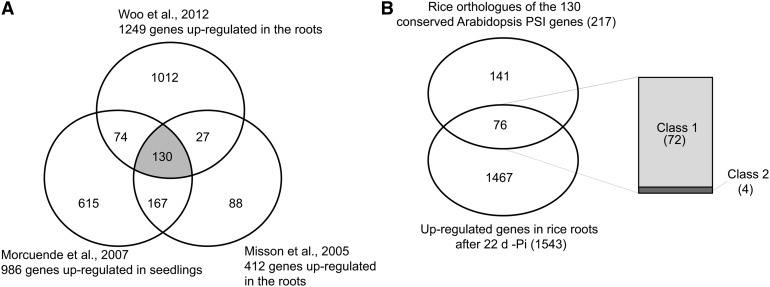
Comparative Analysis of the Pi Starvation–Responsive Genes in Rice and Arabidopsis.
(A) Identification of conserved Arabidopsis root Pi starvation–inducible (PSI) genes from three previous studies. Only genes significantly upregulated in all three studies were considered as being conserved PSI genes (color-coded in gray), resulting in the identification of 130 conserved PSI genes.
(B) Venn diagram showing the overlap between the rice orthologs of the 130 conserved Arabidopsis PSI genes identified using RiceDB, which integrates the InParanoid and Gramene orthology prediction methods, with the set of 1543 upregulated genes upon 22 d of Pi starvation in the roots. Among the 76 genes commonly regulated by Pi starvation in both species, right panel shows the distribution of those genes based on their response after 24 h of Pi resupply in rice (classes 1 and 2).
While the number of genes specifically responding to Pi resupply decreased in the roots with the length of the Pi resupply, an opposite trend was observed in the shoots, with 33 to 978 genes upregulated by 1 and 24 h of Pi resupply, respectively, suggesting that the shoot responds more slowly to Pi resupply than the roots (Figure 3B).
Identification of Conserved Pi Starvation–Induced Genes between Rice and Arabidopsis
To compare the responses of rice to long-term Pi starvation to those observed in other plants, we analyzed data from previous Arabidopsis genome-wide studies of long-term Pi starvation in roots or whole seedlings, including those of Misson et al. (2005), Morcuende et al. (2007), and Woo et al. (2012). From these data sets, we identified a set of conserved Arabidopsis genes induced in roots (Figure 5A). Overall, 130 genes were found to be upregulated by Pi starvation in the three studies, thus defining a set of core Arabidopsis Pi starvation–induced genes. Rice orthologs were then identified using riceDB (Narsai et al., 2013), which integrates the InParanoid (Ostlund et al., 2010) and Gramene (Liang et al., 2008) orthology prediction methods, resulting in the identification of 217 rice orthologs (Figure 5B). Interestingly, of the 217 rice orthologs, 76 genes were represented in our set of 1543 upregulated genes in the root after 21d + 24 h –Pi, thus defining a set of genes that are induced by Pi starvation in both rice and Arabidopsis. Among these 76 genes, 95% (72 genes) were genes that were responding to both Pi starvation and recovery in rice (class 1) and included genes such as the high-affinity transporters, genes from the PHO1 and SPX families, and some phosphatases (see Supplemental Data Set 6 online).
Expression Patterns of the PT and SPX Genes
Since several PT and SPX genes were among the genes that were highly upregulated by Pi starvation, and returning toward basal level of expression (class 1) as early as 1 h after Pi resupply, we decided to examine their regulation in response to Pi stress in more detail. Surprisingly, while these two gene families have been shown to have key roles in Pi homeostasis, very little is known regarding their regulation upon Pi stress and only a few members have been characterized in plants. Among the 13 high-affinity Pi transporters encoded by the rice genome, only four have been characterized, namely PT1, PT2, PT6, and PT8 (Ai et al., 2009; Jia et al., 2011; Sun et al., 2012). Similar observations can be made with the SPX gene family where only SPX1 and SPX3 have been characterized (Wang et al., 2009a, 2009b).
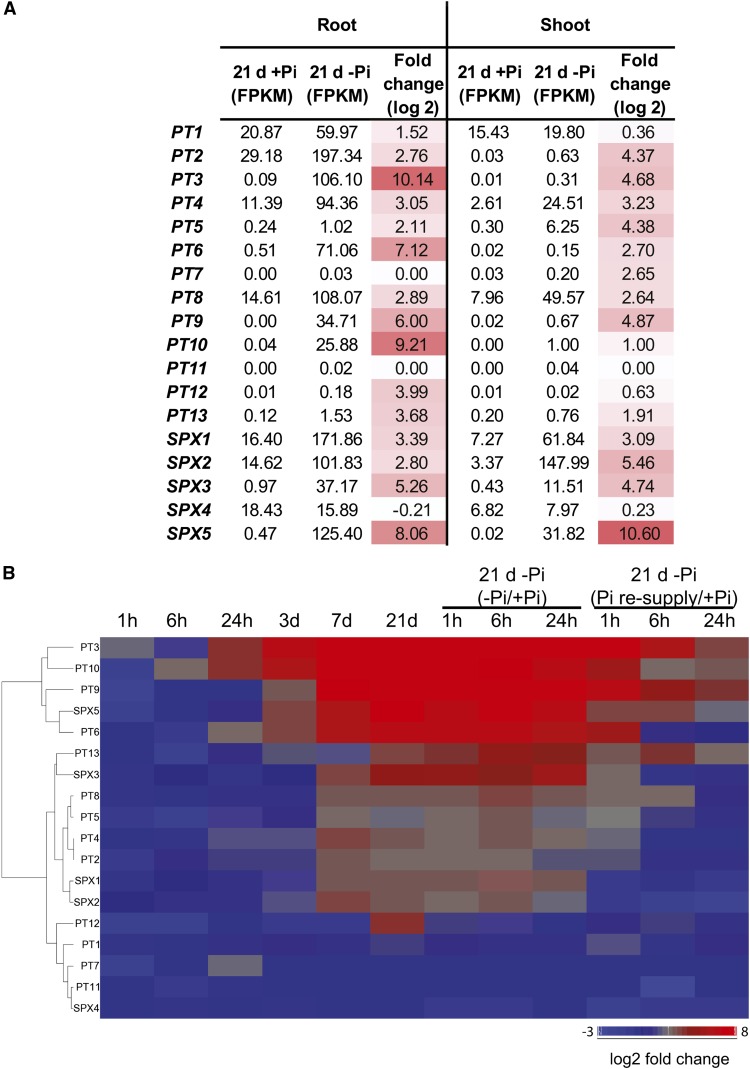
While most of the PTs are preferentially expressed in the roots, PT1, PT4, and PT8 were also expressed in the shoots (FPKM > 2), though to a lesser extent (Figure 6A). Changes in the expression of PTs in the shoots was largely unaffected by the first week of Pi deprivation, with only PT4 showing significant induction by more than 5.5-fold. Yet, after 21 d of Pi starvation, in addition to PT4, two others PTs were induced in the shoots, PT8 and PT5, with PT8 showing the highest expression level of all the PTs, with almost 50 FPKM observed. Pi resupply resulted in a return toward basal level as early as 1 h. In the roots, under Pi-sufficient conditions, PT1, PT2, PT4, and PT8 were the most expressed PTs (FPKM > 10), while the other PTs showed very little gene expression (FPKM < 0.5) (Figures 6A and 6B). After 21 d of Pi starvation, eight of the PTs displayed a high level of expression (FPKM > 25), with PT3 and PT10 showing the greatest induction in gene expression, being upregulated by more than 1000- and 500-fold and reaching more than 100 and 25 FPKM, respectively. Interestingly, PT3 and PT10 were already induced by more than 15-fold as early as 24 h of Pi deprivation. In opposition, 1 h of Pi resupply resulted in a return toward basal level of expression for all of the PTs, except for PT1, which still showed a slight 1.5-fold increase during that period. However, after 6 h of Pi resupply, PT1 was back to prestarvation levels.
Figure 6.
Expression Patterns of the High-Affinity Pi Transporters and SPX Genes.
(A) Schematic representation of the expression levels and fold changes of the high-affinity Pi transporters (PTs) and the SPX genes in response to 21 d of Pi starvation in roots and shoots.
(B) Hierarchical clustering of the expression levels of the SPX and PTs genes in response to Pi starvation and resupply in the roots.
Among the SPX genes, SPX1 and SPX4 showed the highest level of expression in the roots under Pi-sufficient conditions (FPKM > 15) (Figures 6A and 6B). Very little changes in gene expression could be observed during the first 3 d of Pi deprivation in the roots, with only SPX5 induced by 11-fold. After 21 d of Pi starvation, SPX1 displayed the highest expression level with more than 170 FPKM, corresponding to an induction of more than 10-fold (Figure 6B). However the greatest upregulation was observed for SPX5, which showed a 260-fold induction after 21 d of Pi starvation, and was associated with a FPKM of 125. In the shoots, all the SPX genes, with the exception of SPX4, were upregulated by Pi starvation, with SPX5 being the most upregulated gene showing more than a 1500-fold increase in expression. Interestingly, SPX2 was the most abundant SPX gene, with more than 140 FPKM after 21 d of Pi starvation.
Identification of Mis- and Unannotated Loci Involved in Pi Homeostasis
The generation of a reference annotation–based transcript assembly resulted in the identification of 948 misannotated loci and 2400 previously unannotated loci, when compared with the MSU v7 gene annotation, out of which 206 and 438 were significantly differentially expressed in at least one condition, respectively (Figure 2A; see Supplemental Data Set 7 online). While the aim of this study is not to generate an extensive list of loci or isoforms that require annotation, we provide a few examples of some previously unannotated loci/isoforms induced by Pi stress, highlighting the importance of improving the quality of the rice annotation to better understand the genome wide responses of rice to Pi stress.
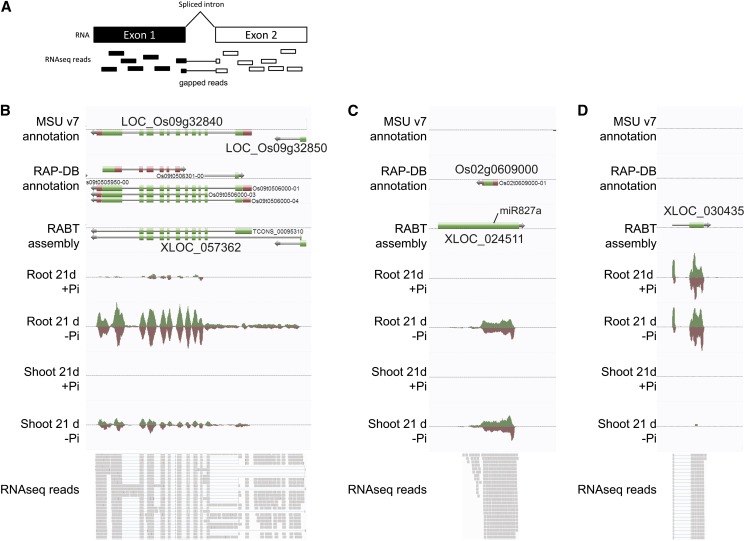
Among the 206 putative misannotated loci that were differentially expressed under Pi starvation were loci such as XLOC_057362 spanning two annotated loci, namely, LOC_Os09g32840, encoding a phosphatase and LOC_Os09g32850, encoding a putative nucleotide pyrophosphatase, which was induced by more than 80-fold after 21 d of Pi starvation in the roots (Figure 7B). The locus is supported by gapped reads, which arise from spliced transcripts, spanning the two original loci, and thus representing bona fide evidence of the presence of a single gene (Figure 7A). While some putative misannotated loci may be correctly annotated in other gene annotations, such as in the new release of RAP-DB (Kawahara et al., 2013; Sakai et al., 2013), some remain misannotated, such as XLOC_057362.
Figure 7.
Identification of Previously Unannotated Transcriptionally Active Loci.
(A) Graphical representation of gapped reads, which are reads mapping across an intron.
(B) Example of a putative misannotated locus, XLOC_057362, spanning two annotated genes, LOC_Os09g32840 and LOC_Os09g32850, and induced by Pi starvation.
(C) XLOC_024511, a locus not present in MSU v7, referred to as Os02g0609000 in RAP-DB, is highly expressed in both root and shoot under Pi starvation and is the precursor of miR827a.
(D) XLOC_030435, a loci neither annotated in MSU nor in RAP-DB, only expressed in the roots, and induced by Pi starvation. Top panels are screen captures from the AnnoJ genome browser (http://www.plantenergy.uwa.edu.au/annoj/Secco_2013.html), representing gene annotations from MSU v7, the RAP-DB IRGSP-1.00, and the RABT assembly as well as expression tracks. Bottom panels are screen captures from Integrative Genome Viewer representing individual RNAseq reads, where thick lines represents reads and thin lines represent gapped reads.
Among the 276 previously unannotated loci differentially regulated after 21 d of Pi deprivation, only 35 were significantly upregulated by more than twofold (with a FPKM > 3, and size >400 bp). Interestingly, 14 of these can now be found in the recently updated RAP-DB, including genes such as IPS2, genes encoding for various hypothetical proteins, and non-protein-coding proteins, five corresponded to rRNAs, three were fragments of known genes, and 10 appeared to be previously unannotated transcribed loci. This result also validates the quality of the RABT assembly generated. Among the genes recently added in the latest version of RAP-DB, Os02g0609000 (XLOC_024511) is of particular interest (Figure 7C). This transcript was one of the most highly induced genes in the roots with more than 1400-fold induction, as well as showing a 30-fold increase in the shoot. This locus also overlapped with the genomic location of O. sativa (osa)-miR827a, one of the key miRNAs involved in Pi homeostasis, which acts through the regulation of two target genes, SPX-MFS1 and SPX-MFS2 (Lin et al., 2010; Wang et al., 2012). This locus was considered to be the precursor of osa-miR827, but no cDNA or ESTs were reported for this locus so far (Lacombe et al., 2008), probably due to the fact that this gene is only expressed under Pi-deficient conditions. Furthermore, the transcript contains an open reading frame capable of coding for a protein of 10 kD (92 amino acids).
Among the previously unannotated loci identified, only three were differentially upregulated in the roots, namely, XLOC_054531, XLOC_030435, and XLOC_031078. XLOC_054531 showed the highest induction upon Pi starvation, with more than an 11-fold increase in gene expression observed after 21 d of Pi starvation. XLOC_030435, composed of two exons, appeared to be only expressed in the roots and was upregulated threefold after long term Pi starvation (Figure 7D). Analysis of the coding potential of these three mRNAs showed that they could encode small proteins, yet no homology was observed with known proteins in the National Center for Biotechnology Information database. To determine if mRNAs corresponding to some of the loci were likely to be translated into proteins, total RNA from roots grown under Pi-deficient conditions for 30 d was fractionated into polysomal and nonpolysomal fractions. Among the four genes tested, Os02g0609000, XLOC_054531, XLOC_030435, and XLOC_031078, only XLOC_030435 showed enrichment in the polysomal fraction, thus suggesting active translation (see Supplemental Figure 4 online). In opposition, transcripts from XLOC_031078 and Os02g0609000 were enriched in the nonpolysomal fraction (see Supplemental Figure 4 online), suggesting that these transcripts are likely to be noncoding. This finding is consistent with the role of Os02g0609000 being the precursor of miR827. In addition, sequence analysis of XLOC_031078 showed high sequence similarity to Os01g0554400, a non-protein-coding transcript.
In the shoots, XLOC_045298 and XLOC_018325 showed the highest increase in gene expression after 21 d of Pi starvation with more than 16-fold and eightfold increases seen, respectively. The expression level of most of these genes returned to basal level after Pi resupply for 24 h, suggesting that these genes are most likely involved in Pi homeostasis and are not responding as a consequence of secondary effects.
We also identified previously unannotated loci that specifically responded to Pi resupply, with 61 induced upon Pi resupply (with a FPKM > 3, more than twofold change cutoff, and size >400 bp). One hour of Pi resupply led to the upregulation of 29 of these loci, with XLOC_006597 showing the highest upregulation (50-fold increase) in roots (see Supplemental Figure 5A online). This locus was still the most upregulated previously unannotated locus after 6 h of Pi resupply, being induced by >20-fold. This locus is likely to be a misannotated gene, as it appears to span multiple genes, namely, LOC_Os01g18360, LOC_Os01g18370, and LOC_Os01g18375, with LOC_Os01g18360, encoding an auxin-responsive element, thus forming a much longer gene than initially predicted by MSU or RAP-DB. Additionally, XLOC_018878, which was the second most highly upregulated locus among the previously unannotated loci after 1 h of Pi resupply, was not in RAP-DB, and represents a small 500-nucleotide-long transcript with a single exon (see Supplemental Figure 5B online).
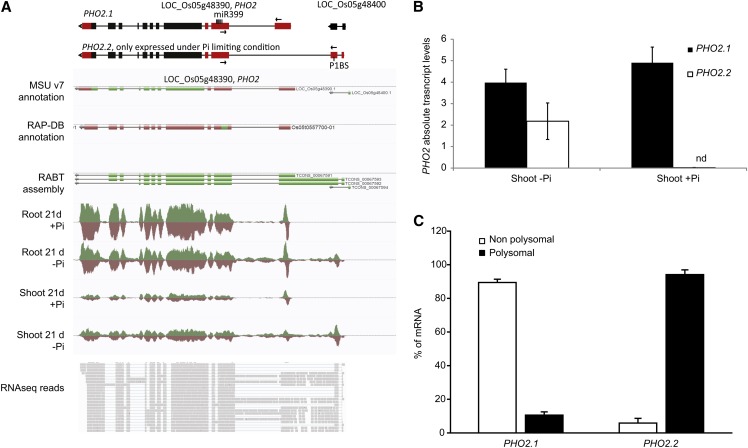
We also identified genes with alternative transcription start sites use upon Pi stress, using the differential promoter use parameter in Cufflinks. In this way, PHO2 (LOC_Os05g48390), one of the key regulators of Pi homeostasis (Figure 8A), was identified. PHO2 has an unusual gene structure, with a large 5′ UTR containing several binding sites for the Pi starvation–responsive miRNA, miR399 (Figure 8A). While PHO2 is not significantly regulated at the transcript level in the roots or shoots (see Supplemental Data Sets 1 and 2 online), an alternative isoform could be detected, specifically under Pi starvation in both root and shoot. We term these isoforms PHO2.1 and PHO2.2, with PHO2.2 being the alternative isoform we detected. PHO2.2 overlapped with LOC_Os05g48400, a gene annotated as an expressed protein. Yet, this locus is likely an artifact, as it is not present in the new release from RAP-DB, and gapped reads spanned the two annotated loci, suggesting that LOC_Os05g48400 is part of PHO2.2 (Figure 8A). To confirm the existence of this isoform, the sequences encoding the N termini of both isoforms were cloned and sequenced (see Supplemental Figure 6 online), revealing that the only difference between the isoforms resided within their 5′ UTRs, not altering the coding sequence of PHO2. Indeed, the first exon of the 5′ UTR of PHO2.1 was replaced by two upstream fragments in PHO2.2 (Figure 8A). Yet, both isoforms contained miR399 binding sites, suggesting that both isoforms are subject to miR399-mediated transcript cleavage (Figure 8A). In addition, sequence analysis of the N-terminal region of the two isoforms revealed the presence of the P1BS element in the second exon of the 5′ UTR of the PHO2.2 isoform. The P1BS element is the DNA motif to which the key transcription factor, PHR2, binds and is often found in Pi-responsive genes (Bustos et al., 2010), potentially explaining the fact that PHO2.2 is only expressed under Pi starvation. To further confirm this expression pattern, we performed qRT-PCR analyses on shoot samples, showing that PHO2.2 was specifically expressed upon Pi starvation (Figure 8B). In addition, to test whether the differences observed in the 5′ UTRs of both PHO2 transcript isoforms could result in altered rate of translation, their association with polysomes was determined using RNA fractionated onto a Suc gradient. PHO2.1 was enriched in the nonpolysomal fraction, but PHO2.2 was preferentially associated with the polysomes, suggesting that PHO2.2 is more actively translated into protein than isoform 1 (Figure 8C). The Pi starvation–induced mRNA isoform PHO2.2 is likely to constitute a new level of regulation of Pi homeostasis in rice, but further work will be required to identify the role of this isoform.
Figure 8.
Identification of a PHO2 Isoform Specifically Induced by Pi Deprivation.
(A) Top panel: A schematic representation of the established PHO2 isoform (PHO2.1) and the novel isoform (PHO2.2) specifically induced under Pi starvation. Bottom panel: A screen capture of the PHO2 locus after 21 d of Pi treatment (+Pi or −Pi) in AnnoJ. In the top panel, black boxes represent exons, and red boxes represent the 5′ and 3′ UTRs. Arrows represents primers used for qRT-PCR.
(B) Expression levels of the two PHO2 isoforms in the shoots after 21 d of Pi treatment. nd, not detected.
(C) Changes in the distribution of both PHO2 mRNA isoforms between polysomal or nonpolysomal fractions. RNA was extracted from roots grown for 30 d on Pi-deficient condition. The association with each fraction is shown as a linear plot of the percentage of RNA present in each fraction. Errors bars are sd, and n = 3.
Among the other unannotated isoforms showing specific induction under Pi starvation were genes such as an ATP binding cassette transporter (LOC_Os06g37364), a nucleotide pyrophosphatase (LOC_Os09g32840), and a cytochrome P450 (LOC_Os01g52560). Interestingly, unlike with PHO2, some isoforms were only expressed in the root, including the cytochrome P450, LOC_OS01g52560 and LOC_Os09g32840, which encodes a nucleotide pyrophosphatase.
Genome-Wide Analysis of the MiRNAs
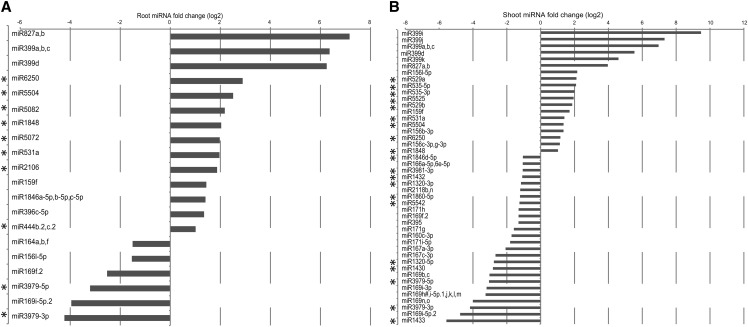
To increase our knowledge of the types of miRNAs involved in the long-term response to Pi starvation in rice, we generated and sequenced small RNAs libraries from roots and shoots of rice plants grown for 21 d under Pi-sufficient and -deficient conditions. On average, root and shoot samples had 13 and 60 million reads passing filter, respectively. Reads containing the 3′ adapter sequence were then trimmed and size selected (between 15 to 30 nucleotides long) before being analyzed with miRanalyzer (Hackenberg et al., 2009). We found that 58 to 90% of the reads aligned to the rice genome (IRGSP-1.0), but only 0.7 to 11.5% of the reads could be mapped to known miRNAs (see Supplemental Table 2 online). Distribution of the mapped small RNA reads revealed that the majority of the shoot small RNAs had a size of 21 and 24 nucleotides, while most of the root small RNAs reads had a length of 22 and 24 nucleotides (see Supplemental Figure 7 online). In addition, out of the 547 rice mature miRNAs tested by miRanalyzer, an average of 170 to 370 could be detected in the roots and shoots, respectively. MiRNA analysis revealed that osa-miR166 was the most abundant miRNA in roots and shoots, accounting for 80 and 50% of all miRNA reads, respectively. Differential expression analysis revealed that osa-miR827 and osa-miR399 were the most induced families in both root and shoot, with miR827 being the most induced miRNA after 21 d of Pi starvation, with more than a 140-fold increase seen in roots (Figures 9A and 9B; see Supplemental Data Set 8 online). These two miRNA families have been shown to be induced by Pi starvation in both Arabidopsis and rice (Aung et al., 2006; Bari et al., 2006; Lin et al., 2010; Jeong et al., 2011; Wang et al., 2012), with Arabidopsis miR399 negatively regulating genes such as PHO2 and the two Pi transporters Pht1;8 and Pht1;9 (Bari et al., 2006) and osa-miR827 regulating the expression of two putative Pi transporters, SPX-MFS1 and SPX-MFS2 (Lin et al., 2010; Wang et al., 2012), thus affecting Pi homeostasis. In addition to osa-miR399 and osa-miR827 families, 17 and 28 other miRNA families were differentially regulated upon 21 d of Pi starvation in root and shoot, respectively, corresponding to 33 unique Pi starvation–responsive miRNAs families (Figure 9). Comparison of these 33 rice miRNAs families to known Pi starvation–responsive miRNAs families identified in Arabidopsis and rapeseed (Brassica napus) (Hsieh et al., 2009; Pant et al., 2009; Kuo and Chiou, 2011) resulted in the identification of 20 miRNA families that were not previously associated with Pi stress (Figure 9). In addition, 80% of these Pi starvation–responsive miRNAs families were only found in rice (see Supplemental Table 3 online). Among these, miR6250 showed a seven- and two-fold induction upon Pi stress in the root and shoot, respectively (Figures 9A and 9B). Expression of osa-miR6250 can be induced by arsenite treatment (Liu, 2012), and arsenate, the oxidation product of arsenite, is a chemical analog of Pi and can be taken up by Pi transporters such as PT8 (Wu et al., 2011). It is thus temping to hypothesize that osa-miR6250 could be involved in the regulation of both Pi and arsenate homeostasis in rice. Yet, the only predicted target gene for osa-miR6250 is LOC_Os04g45570, encoding an expressed protein, which is not altered at the transcript level by Pi starvation.
Figure 9.
Identification of MiRNAs in Roots and Shoots after 21 d of Pi Starvation.
Differentially expressed miRNAs in response to 21 d of Pi starvation in the roots (A) and shoots (B) were identified using the miRanalyzer software.
Only significantly differentially expressed miRNAs with a fold change greater than 2 are shown (q value < 0.05). Asterisk indicates miRNAs that were not previously associated with Pi starvation.
Several miRNAs also showed dramatic decrease upon Pi starvation, such as osa-miR169, osa-miR3979, and osa-miR1433. Of note, the Arabidopsis ortholog of osa-miR169, miR169, as well as osa-miR3979 were recently shown to be involved in the nitrogen starvation response, being strongly downregulated by nitrogen starvation (Jeong et al., 2011; Zhao et al., 2011). Crosstalk between nitrate and Pi homeostasis is well documented in Arabidopsis (Kant et al., 2011), with miR827 and its target gene NLA being required to maintain nitrate-dependent Pi homeostasis. Thus, it is possible that other miRNAs could have a similar function in rice, being involved in both nitrate and Pi homeostasis.
In conclusion, this study presents a high-resolution genome-wide transcriptome analysis of the rice responses to Pi starvation and resupply in roots and shoots. Using RNAseq technology, we quantified expression of mRNAs and noncoding RNAs and determined the transcript structure, splicing patterns, and other posttranscriptional modifications of genes. We captured the full transcriptome dynamics associated with Pi stress in roots and shoots, which could not have been performed using traditional microarray technology because of a large number of genes not having probe sets or not being properly annotated. As a consequence, this study revealed the existence of potential key novel regulators of Pi homeostasis in rice. The RABT assembly also identified loci with different transcription start sites upon Pi stress, including PHO2, highlighting an additional level of complexity in the regulation of one of the key regulators of Pi homeostasis, through altered translation activity.
Classification of gene responses allowed us to disentangle the complex signaling pathways involved in Pi stress and identify genes that are directly involved in Pi deficiency and Pi resupply responses. So far, most of the previous studies focused on the genes directly involved Pi starvation, while neglecting the genes involved in response to resupplying Pi to Pi-starved plants. Yet, the latter condition is more comparable to plants grown in their natural environment, where they often confront Pi-limiting conditions and rarely encounter Pi-rich soil patches. Pi resupply induced many genes within a very short period of time, including transcription factors and kinases, suggesting the involvement of a numerous transcriptional and posttranslational modifications, ultimately leading to an increase in protein and amino acid synthesis.
Small RNAs regulate plant nutrition, including Pi homeostasis. Yet, only the study by Jeong et al. (2011) has addressed this question in rice, using plants Pi starved for a short period of time (5 d), thus potentially not detecting essential miRNAs that may be important later. Profiling miRNAs in plants starved for Pi for 21 d in root and shoot enabled us to confirm the well-characterized response of miR399 and miR827 to Pi starvation, but also to identify 20 miRNA families that were not previously associated with Pi starvation, including miR6250, that could be either directly involved in Pi homeostasis or involved in signaling crosstalk with other ions, such as arsenate.
This global analysis of the rice transcriptomic responses to Pi stress identifies candidate genes that can be further characterized to increase our knowledge of Pi homeostasis, bringing us one step closer to generating plants with increased Pi use and/or acquisition efficiency.
METHODS
Plant Materials and Growth Conditions
Rice (Oryza sativa cv Nipponbare) was used for all physiological experiments. Hydroponic experiments were performed under controlled conditions (day/night temperature of 30/22°C and a 12-h photoperiod, 200 µmol photons m−2 s−1), allowing 0.5 liters of hydroponic solution per plant. The hydroponic solution consisted of a modified solution as described by Yoshida et al. (1976), containing 1.425 mM NH4NO3, 0.513 mM K2SO4, 0.998 mM CaCl2, 1.643 mM MgSO4, 0.075 µM (NH4)6Mo7O24, 0.25 mM NaSiO3, 0.009 mM MnCl2, 0.019 µM H3BO3, 0.155 µM CuSO4, 0.152 µM ZnSO4, and 0.125 mM EDTA-Fe, with or without 0.323 mM NaH2PO4, resulting in the +Pi and −Pi conditions. The pH of the solution was adjusted to 5.5, and the solution was renewed every 3 d. Rice seeds were first pregerminated in tap water for 2 d before being transferred into the hydroponic solution, containing 0.323 mM Pi (+Pi) for 2 weeks. Half of the seedlings were then transferred to a solution lacking Pi (0 mM Pi) for 21 d before being resupplemented with 0.323 mM Pi for 24 h, while the other half of the seedlings continuously remained in +Pi conditions (control). During the resupply experiment, some rice seedlings were left in Pi-deficient media to serve as control. Roots and shoots were harvested separately at the following time points: 1 h, 6 h, 24 h, 3 d, 7 d, and 21 d after transfer to –Pi conditions, as well as 1, 6, and 24 h after Pi resupply. Furthermore, all experiment procedures such as media replacement and sample collection were performed at a similar time of the day (2 h after light) to minimize possible circadian effects.
Quantification of Pi Tissue Concentration
Determination of Pi in tissues was measured by releasing the cellular contents into water by repeated freeze-thaw cycles, or by incubation for 1 h at 85°C, and quantifying Pi by the molybdate assay according to the procedure of Ames (1966).
Total RNA Isolation and Library Preparation
The total RNA from the roots and shoots tissues was extracted using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. The integrity and quality of the total RNA was checked using NanoDrop 1000 spectrophotometer and formaldehyde-agarose gel electrophoresis. RNA was only used when the A260/A280 nm ratio was >1.8.
For RNAseq library synthesis (three biological replicates per condition and one plant per replicate), total RNA was first depleted of rRNA using the Ribo-Zero rRNA removal kit (Plant Leaf and Plant Seed/Root kits; Epicentre). To do so, 1 µg of total RNA from root samples was used as input for rRNA removal, while 2 µg of total RNA was used for shoot samples. Sequencing libraries were generated using the TruSeq RNA sample prep kit (Illumina). An average of eight libraries were multiplexed and loaded on each lane of the Illumina Hiseq flow cell v3. Sequencing was then performed on a Hisequation 1000, as a 2 × 101 paired-end run, according to the manufacturer’s instruction (Illumina).
For small RNAseq library generation (thee biological replicates per condition and one plant per replicate), 1 µg of total RNA was used as input for the Small RNA sample preparation kit (Illumina). In brief, specific 5′ and 3′ Illumina RNA adapters were sequentially ligated to small RNA molecules. Adapter-ligated molecules were then reverse transcribed and PCR amplified before being gel purified (corresponding to 15 to 30 nucleotides long). Libraries were then multiplexed (six libraries per lane) and sequenced for 51 cycles using the Illumina HiSequation 1000, as per the manufacturer’s instructions.
Polysome Gradient Preparation
Polysome isolation was performed as previously described (Jabnoune et al., 2013). Briefly, roots from plants starved for Pi for 30 d were harvested, frozen, and ground to powder in liquid nitrogen. To prepare cytoplasmic extracts, 150 mg of powder was combined with 1.2 mL of chilled polysome buffer (100 mM Tris-HCl, pH 8.4, 50 mM KCl, 25 mM MgCl2, 5 mM EGTA, 18 μM cycloheximide, 15.5 μM chloramphenicol, and 0.5% [v/v] Nonidet P-40). Debris was removed by centrifugation at 16,000g for 15 min at 4°C. Aliquots of the resulting supernatant were loaded on 20 to 50% (w/w) continuous Suc gradients and centrifuged at 32,000 rpm for 165 min in a Beckman SW41 rotor at 4°C. After centrifugation, fractions were collected from the bottom to the top of the gradient with continuous monitoring of the absorbance at 254 nm, resulting in a polysomal and nonpolysomal fraction. RNA was extracted using Trizol (Invitrogen) following the manufacturer’s instructions. A fixed volume of RNA samples corresponding to the polysomal and nonpolysomal fractions was then reversed transcribed and used for subsequent qRT-PCR.
Mapping of RNAseq Reads and Transcript Assembly and Abundance Estimation Using the Tuxedo Suite
All analyses were performed using the Cufflinks package (Trapnell et al., 2012), version 2.0.2 (Bowtie2 v2.0.0-beta7 and TopHat v2.0.3), and using the rice reference genome and gene model annotation file (GFF file) from MSU Rice Genome Annotation Project database v7 (http://rice.plantbiology.msu.edu/).
A Bowtie2 index was generated using the rice IRGSP-1.0 genome. To align the RNAseq reads to the genome, TopHat was run with the following options: –b2-sensitive –r 0 –mate-std-dev 100 –g 1 –G, allowing only one multihit and providing a gene model annotation file (MSU v7).
The resulting aligned reads were then used to create a RABT assembly using Cufflinks v 2.0.2. Cufflinks was run in the discovery mode, allowing the identification of previously unannotated genes, as described by Trapnell et al. (2012). For this purpose, only reads corresponding to samples originating from the 21-d time point (+Pi and –Pi) were used. Assemblies were then merged using Cuffmerge, using the MSU v7 gene model annotation file as the reference annotation, resulting in a new transcript assembly, used to quantify transcript abundance.
Finally, transcript abundance (FPKM) and identification of differentially expressed genes was performed using Cufflinks with the default parameters (P < 0.05 and 5% FDR cutoff) with the addition of –N -b, corresponding to upper quartile normalization, and running a bias detection and correction algorithm. Differential transcript abundance at all genes was calculated as the logarithm base-2 of the expression ratio (FPKMminus Pi/FPKMplus Pi and FPKMPi resupply/FPKMminus Pi).
Mapped reads were then visualized using AnnoJ genome browser (http://www.annoj.org/), accessible at http://www.plantenergy.uwa.edu.au/annoj/Secco_2013.html. Because of the large amount of tracks loaded by default in AnnoJ, users can remove tracks of low interest to increase the loading speed of the Web page. Three different gene annotations are provided: the MSU v7, the RAP-DB, and the RABT assembly.
qRT-PCR
cDNA was synthesized from 1 µg of total RNA using the Bio-Rad iScript reverse transcription kit, according to the manufacturer’s instructions. Transcript levels were assayed using the LightCycler 480 (Roche). Absolute expression levels were normalized to the housekeeping gene LOC_Os06g11170 (Narsai et al., 2010), encoding a nucleic binding protein (see Supplemental Table 4 online).
Small RNA Abundance and Estimation
To identify and remove the 3′ adapter sequence from raw reads, Cutadapt was used. For this, the first eight bases of the adapter sequence were used. Reads containing the adapter sequence were truncated up to the junction with the adaptor sequence. Reads were then filtered using the University of East Anglia Small RNA workbench to remove reads matching to known rice tRNAs and rRNAs, as well as remove reads that did not match to the rice genome (MSU v7) and discard reads with low complexity. Only reads between 19- and 24-nucleotides long were selected for further analyses. MiRNA abundance and differential expression analysis were performed using miRanalyzer (http://bioinfo2.ugr.es/miRanalyzer/miRanalyzer.php; Hackenberg et al., 2009), allowing zero mismatches to known miRNAs.
Accession Numbers
Illumina reads of all samples have been submitted to the Sequence Read Archive at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA097415. Supplemental Data Sets 1 to 4 are deposited in the DRYAD repository: http://doi.org/10.5061/dryad.4480g.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Patterns of Four Pi Deprivation–Induced Genes in Roots and Shoots.
Supplemental Figure 2. Overrepresented Functional Categories in the Roots after 7 d of Pi Starvation.
Supplemental Figure 3. Distribution of the Differentially Expressed Genes Detected by RNAseq Based on Their Presence on Affymetrix Microarrays.
Supplemental Figure 4. Distribution of mRNA of Some Mis- and Unannotated Loci and of Os02g0609000 to Polysomal and Nonpolysomal Fractions.
Supplemental Figure 5. Identification of Loci Induced by Pi Resupply.
Supplemental Figure 6. Alignment of the N Termini of PHO2.1 and PHO2.2 to gDNA.
Supplemental Figure 7. Distribution of the Mapped Small RNA Reads.
Supplemental Table 1. Mapping Summary Statistics of the RNAseq Reads.
Supplemental Table 2. Mapping Summary Statistics of the Small RNA Reads.
Supplemental Table 3. List of Identified Pi Starvation–Responsive miRNAs and Their Occurrence in Plant Species.
Supplemental Table 4. List of qRT-PCR Primers Used in This Study.
Supplemental Data Set 1. FPKM and Fold Changes of All Genes in Roots.
Supplemental Data Set 2. FPKM and Fold Changes of All Genes in Shoots.
Supplemental Data Set 3. FPKM and Fold Changes of Significantly Regulated Genes in Roots and Shoots.
Supplemental Data Set 4. List of Genes Forming Classes 1 to 6, in Roots and Shoots, with Expression Levels (FPKM).
Supplemental Data Set 5. List of Genes Forming Classes 7 and 8, in Roots and Shoots, with Expression Levels and Fold Changes.
Supplemental Data Set 6. List of the 76 Upregulated Genes in Both Rice and Arabidopsis Root upon Pi Starvation.
Supplemental Data Set 7. List of Putative Misannotated Loci, Loci with Alternative Transcription Start Sites, and Some of the Differentially Regulated Previously Unannotated Loci.
Supplemental Data Set 8. List of Differentially Regulated miRNAs in Roots and Shoots.
Acknowledgments
We thank Laurent Nussaume, Reena Narsai, and Oliver Berkowitz for critical reading of the article. This work was supported by the Australian Research Council Super Science Fellowship (FS100100022 to D.S.) and the Swiss National Foundation (31003A-138339 to Y.P.).
AUTHOR CONTRIBUTIONS
D.S. carried out all the experiments, analyses, and wrote the article. H.W. wrote scripts to facilitate the RNAseq analysis. J.W. participated in the design and writing of the article. H.S. and P.W. participated in the writing of the article. M.J. and Y.P. participated in polysomes isolation and transcript profiling. All authors read and approved the final article.
Glossary
- miRNA
microRNA
- RNAseq
RNA sequencing
- qRT-PCR
quantitative RT-PCR
- RAP-DB
Rice Annotation Project Database
- RABT
reference annotation–based transcript
- UTR
untranslated region
- FPKM
fragment per kilobase of exon per million fragments mapped
- FDR
false discovery rate
- GO
Gene Ontology
- MSU
Michigan State University
References
- Ai P., Sun S., Zhao J., Fan X., Xin W., Guo Q., Yu L., Shen Q., Wu P., Miller A.J., Xu G. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Ames B. (1966). Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8: 115–118 [Google Scholar]
- Arpat A.B., Magliano P., Wege S., Rouached H., Stefanovic A., Poirier Y. (2012). Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 71: 479–491 [DOI] [PubMed] [Google Scholar]
- Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournier M., Tissot N., Mari S., Boucherez J., Lacombe E., Briat J.F., Gaymard F. (2013). Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 288: 22670–22680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez-Pérez J., Solano R., Leyva A., Paz-Ares J. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., Zhou W., El Sheery N.I., Peters C., Li M., Wang X., Huang J. (2011). Characterization of the Arabidopsis glycerophosphodiester phosphodiesterase (GDPD) family reveals a role of the plastid-localized AtGDPD1 in maintaining cellular phosphate homeostasis under phosphate starvation. Plant J. 66: 781–795 [DOI] [PubMed] [Google Scholar]
- Chiou T.J. (2007). The role of microRNAs in sensing nutrient stress. Plant Cell Environ. 30: 323–332 [DOI] [PubMed] [Google Scholar]
- Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Lin S.I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Cobbett C., Goldsbrough P. (2002). Phytochelatins and metallothioneins: Roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53: 159–182 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., García J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Fujii H., Chiou T.J., Lin S.I., Aung K., Zhu J.K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Hackenberg M., Sturm M., Langenberger D., Falcón-Pérez J.M., Aransay A.M. (2009). miRanalyzer: A microRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 37 (Web Server issue): W68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J., Marin E., Floriani M., Chiarenza S., Richaud P., Nussaume L., Thibaud M.C. (2006). Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88: 1767–1771 [DOI] [PubMed] [Google Scholar]
- Hou X.L., Wu P., Jiao F.C., Jia Q.J., Chen H.M., Yu J., Song X.W., Yi K.K. (2005). Regulation of the expression of OsIPS1 and OsIPS2 in rice via systemic and local Pi signalling and hormones. Plant Cell Environ. 28: 353–364 [Google Scholar]
- Hsieh L.C., Lin S.I., Shih A.C., Chen J.W., Lin W.Y., Tseng C.Y., Li W.H., Chiou T.J. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Zhu C., Li F., Tang J., Wang Y., Lin A., Liu L., Che R., Chu C. (2011). LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 156: 1101–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M., Secco D., Lecampion C., Robaglia C., Shu Q., Poirier Y. (2013). A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. Plant Cell 25: 4166–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A., Nagarajan V.K., Raghothama K.G. (2012). Transcriptional regulation of phosphate acquisition by higher plants. Cell. Mol. Life Sci. 69: 3207–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong D.H., Park S., Zhai J., Gurazada S.G., De Paoli E., Meyers B.C., Green P.J. (2011). Massive analysis of rice small RNAs: Mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23: 4185–4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Ren H., Gu M., Zhao J., Sun S., Zhang X., Chen J., Wu P., Xu G. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C.F., Gao X.H., Liao L., Harberd N.P., Fu X.D. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Peng M., Rothstein S.J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Awai K., Nakamura M., Nagatani A., Masuda T., Ohta H. (2009). Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kuo H.F., Chiou T.J. (2011). The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 156: 1016–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe S., et al. (2008). Identification of precursor transcripts for 6 novel miRNAs expands the diversity on the genomic organisation and expression of miRNA genes in rice. BMC Plant Biol. 8: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M.G., Liu Y.D., Zhang B.C., Zhao Y.T., Wang X.J., Zhou Y.H., Raghothama K.G., Liu D. (2011). Genetic and genomic evidence that sucrose is a global regulator of plant responses to phosphate starvation in Arabidopsis. Plant Physiol. 156: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C.Z., et al. (2008). Gramene: A growing plant comparative genomics resource. Nucleic Acids Res. 36 (Database issue): D947–D953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.I., et al. (2010). Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 51: 2119–2131 [DOI] [PubMed] [Google Scholar]
- Liu Q.P. (2012). Novel miRNAs in the control of arsenite levels in rice. Funct. Integr. Genomics 12: 649–658 [DOI] [PubMed] [Google Scholar]
- Liu T.Y., Huang T.K., Tseng C.Y., Lai Y.S., Lin S.I., Lin W.Y., Chen J.W., Chiou T.J. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A.S., Raghothama K.G., Baek D., Koo Y.D., Jin J.B., Bressan R.A., Yun D.J., Hasegawa P.M. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 102: 7760–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R., Bari R., Gibon Y., Zheng W., Pant B.D., Bläsing O., Usadel B., Czechowski T., Udvardi M.K., Stitt M., Scheible W.R. (2007). Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Müller R., Morant M., Jarmer H., Nilsson L., Nielsen T.H. (2007). Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143: 156–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Devenish J., Castleden I., Narsai K., Xu L., Shou H., Whelan J. (October 21, 2013). Rice DB: An Oryza Information Portal linking annotation, sub-cellular location, function, expression, regulation and evolutionary information for rice and Arabidopsis. Plant J.http//dx..org/10.1111/tpj.12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R., Ivanova A., Ng S., Whelan J. (2010). Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 10: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L., Müller R., Nielsen T.H. (2010). Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol. Plant. 139: 129–143 [DOI] [PubMed] [Google Scholar]
- Ostlund G., Schmitt T., Forslund K., Köstler T., Messina D.N., Roopra S., Frings O., Sonnhammer E.L.L. (2010). InParanoid 7: New algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 38 (Database issue): D196–D203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B.D., Musialak-Lange M., Nuc P., May P., Buhtz A., Kehr J., Walther D., Scheible W.R. (2009). Identification of nutrient-responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B., Clément M., Nussaume L., Desnos T. (2011). Root developmental adaptation to phosphate starvation: Better safe than sorry. Trends Plant Sci. 16: 442–450 [DOI] [PubMed] [Google Scholar]
- Plaxton W.C., Tran H.T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Bucher M. (2002). Phosphate transport and homeostasis in Arabidopsis. The Arabidopsis Book 1: e0024, doi/10.1199/tab.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M., Svistoonoff S., Loudet O., Nussaume L., Desnos T. (2006). Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ. 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Rouached H., Arpat A.B., Poirier Y. (2010). Regulation of phosphate starvation responses in plants: Signaling players and cross-talks. Mol. Plant 3: 288–299 [DOI] [PubMed] [Google Scholar]
- Rouached H., Stefanovic A., Secco D., Bulak Arpat A., Gout E., Bligny R., Poirier Y. (2011). Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J. 65: 557–570 [DOI] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martín A.C., Iglesias J., Leyva A., Paz-Ares J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H., et al. (2013). Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 54: e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Baumann A., Poirier Y. (2010). Characterization of the rice PHO1 gene family reveals a key role for OsPHO1;2 in phosphate homeostasis and the evolution of a distinct clade in dicotyledons. Plant Physiol. 152: 1693–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Wang C., Arpat B.A., Wang Z., Poirier Y., Tyerman S.D., Wu P., Shou H., Whelan J. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Sun S., Gu M., Cao Y., Huang X., Zhang X., Ai P., Zhao J., Fan X., Xu G. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159: 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud M.C., Arrighi J.F., Bayle V., Chiarenza S., Creff A., Bustos R., Paz-Ares J., Poirier Y., Nussaume L. (2010). Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 64: 775–789 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B., Nagel A., Steinhauser D., Gibon Y., Bläsing O.E., Redestig H., Sreenivasulu N., Krall L., Hannah M.A., Poree F., Fernie A.R., Stitt M. (2006). PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneklaas E.J., Lambers H., Bragg J., Finnegan P.M., Lovelock C.E., Plaxton W.C., Price C.A., Scheible W.R., Shane M.W., White P.J., Raven J.A. (2012). Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 195: 306–320 [DOI] [PubMed] [Google Scholar]
- Wang C., Huang W., Ying Y., Li S., Secco D., Tyerman S., Whelan J., Shou H. (2012). Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol. 196: 139–148 [DOI] [PubMed] [Google Scholar]
- Wang C., Ying S., Huang H., Li K., Wu P., Shou H. (2009a). Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 57: 895–904 [DOI] [PubMed] [Google Scholar]
- Wang Z., Hu H., Huang H., Duan K., Wu Z., Wu P. (2009b). Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J. Integr. Plant Biol. 51: 663–674 [DOI] [PubMed] [Google Scholar]
- Wasaki J., et al. (2006). Transcriptomic analysis indicates putative metabolic changes caused by manipulation of phosphorus availability in rice leaves. J. Exp. Bot. 57: 2049–2059 [DOI] [PubMed] [Google Scholar]
- Wasaki J., et al. (2003). Transcriptomic analysis of metabolic changes by phosphorus stress in rice plant roots. Plant Cell Environ. 26: 1515–1523 [Google Scholar]
- Woo J., MacPherson C.R., Liu J., Wang H., Kiba T., Hannah M.A., Wang X.J., Bajic V.B., Chua N.H. (2012). The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 12: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Shou H., Xu G., Lian X. (2013). Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 16: 205–212 [DOI] [PubMed] [Google Scholar]
- Wu Z.C., Ren H.Y., McGrath S.P., Wu P., Zhao F.J. (2011). Investigating the contribution of the phosphate transport pathway to arsenic accumulation in rice. Plant Physiol. 157: 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S., Forno, D., Cock, J., and Gomez, K. (1976). Laboratory Manual for Physiological Studies of Rice, 3rd ed. (Manila, The Philippines: The International Rice Research Institute). [Google Scholar]
- Zhao M., Ding H., Zhu J.K., Zhang F.S., Li W.X. (2011). Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 190: 906–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Huang F., Narsai R., Wu J., Giraud E., He F., Cheng L., Wang F., Wu P., Whelan J., Shou H. (2009). Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 151: 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Jiao F., Wu Z., Li Y., Wang X., He X., Zhong W., Wu P. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]