The lignin pathway is a favored target for improvement of lignocellulosic feedstocks because lignin affects enzymatic sugar release from cell walls. Using a combination of approaches, this article identifies candidate lignin pathway genes likely to be functionally involved in lignification in the dedicated energy crop switchgrass, as well as some expected candidates with questionable function.
Abstract
It is necessary to overcome recalcitrance of the biomass to saccharification (sugar release) to make switchgrass (Panicum virgatum) economically viable as a feedstock for liquid biofuels. Lignin content correlates negatively with sugar release efficiency in switchgrass, but selecting the right gene candidates for engineering lignin biosynthesis in this tetraploid outcrossing species is not straightforward. To assist this endeavor, we have used an inducible switchgrass cell suspension system for studying lignin biosynthesis in response to exogenous brassinolide. By applying a combination of protein sequence phylogeny with whole-genome microarray analyses of induced cell cultures and developing stem internode sections, we have generated a list of candidate monolignol biosynthetic genes for switchgrass. Several genes that were strongly supported through our bioinformatics analysis as involved in lignin biosynthesis were confirmed by gene silencing studies, in which lignin levels were reduced as a result of targeting a single gene. However, candidate genes encoding enzymes involved in the early steps of the currently accepted monolignol biosynthesis pathway in dicots may have functionally redundant paralogues in switchgrass and therefore require further evaluation. This work provides a blueprint and resources for the systematic genome-wide study of the monolignol pathway in switchgrass, as well as other C4 monocot species.
INTRODUCTION
Switchgrass (Panicum virgatum) is a perennial C4 species with high biomass yield. As a biofuel crop, it requires relatively minimal agronomic inputs and has broad cultivation adaptability (Bouton, 2007). Switchgrass biomass has been reported to produce more than five times more energy than the energy expended during production, harvesting, and processing, and the plant has significant environmental benefits (Schmer et al., 2008). The key step in the conversion of lignocellulosic biomass into renewable fuels is the release of fermentable sugars (saccharification) from the polysaccharides embedded in the cell walls. Because of the inherent recalcitrance of switchgrass to saccharification, switchgrass biomass requires a chemical pretreatment before polysaccharide hydrolysis for sugar release. The cost of processing to overcome recalcitrance has limited the commercialization of biomass-derived ethanol (Himmel et al., 2007; Pauly and Keegstra, 2008).
The presence of lignin in lignocellulosic biomass negatively affects the earlier described conversion steps (Chen and Dixon, 2007; Himmel et al., 2007; Pauly and Keegstra, 2008; Ding et al., 2012). Lignin is an aromatic heteropolymer that is mainly deposited in plant cell walls undergoing secondary thickening. It is synthesized from the monolignols 4-coumaryl, coniferyl, and sinapyl alcohols (see Supplemental Figure 1 online); these correspond, respectively, to the 4-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units in the lignin polymer that can be released and measured by thioacidolysis followed by gas chromatography–mass spectrometry (GC-MS) (Lapierre et al., 1985). Phenylpropanoid aldehydes, acetates, and esters (e.g., of 4-coumarate and ferulate) are also found in lignins (Withers et al., 2012). Examination of natural variation in alfalfa (Medicago sativa) (Dien et al., 2006), switchgrass (Dien et al., 2006; Shen et al., 2009b), canarygrass (Phalaris canariensis) (Dien et al., 2006), poplar (Populus trichocarpa) (Davison et al., 2006; Dien et al., 2006), and sorghum (Sorghum bicolor) (Dien et al., 2009) has shown that low lignin levels are associated with improved saccharification efficiency in vitro, and there is also a negative relationship between cell wall ester-linked 4-coumaric acid/ferulic acid ratio and sugar release efficiency in switchgrass (Shen et al., 2009b).
Downregulation of the lignin biosynthetic enzyme caffeic acid 3-O-methyltransferase (COMT) in switchgrass reduces recalcitrance and increases ethanol production by up to 38% using conventional biomass fermentation processes (Fu et al., 2011b). COMT downregulation also improves cell wall processing ability in sorghum, maize (Zea mays), and alfalfa with only small negative agronomic impacts (slightly increased lodging in some lines) (Fritz et al., 1990; Vignols et al., 1995; Guo et al., 2001b; Dien et al., 2009; Fu et al., 2011b). Overexpression of a lignin biosynthetic repressor (MYB4) in switchgrass can increase sugar release efficiency as much as threefold without acid pretreatment, and this translates to a more than twofold enhancement of ethanol yield (Shen et al., 2012, 2013).
Engineering of lignin content in switchgrass is limited by the lack of genetic information on the lignin biosynthetic pathway in this species, and in monocots in general; strategies to date have mainly relied on translation from results obtained in dicotyledonous plants, particularly model species and trees such as poplar, with a sequenced genome (Hu et al., 1999; Baucher et al., 2000, 2003; Halpin, 2004; Chen et al., 2006; Hamberger et al. 2007; Vanholme et al., 2008; Li et al., 2008; Simmons et al., 2010; Shi et al. 2010). Even though genomics-based descriptions of lignin pathway genes in monocots have been presented (Barrière et al., 2007; Lawrence and Walbot, 2007; Andersen et al., 2008), the corresponding enzymatic functions encoded by many of the gene candidates have yet to be verified biochemically or genetically.
To date, four monolignol pathway enzymes from switchgrass have been functionally characterized by biochemical or genetic approaches, or both. These are COMT (Fu et al., 2011b), cinnamyl alcohol dehydrogenase (CAD) (Fu et al., 2011a; Saathoff et al., 2011), 4-coumarate CoA ligase (4CL) (Xu et al., 2011), and cinnamoyl CoA reductase (CCR) (Escamilla-Treviño et al., 2010). Based on these studies, the monolignol pathway in switchgrass appears to be consistent with that defined in other species (see Supplemental Figure 1 online), although the early steps have yet to be experimentally confirmed because the candidate genes are not clear.
A number of resources are now available to facilitate the identification of gene candidates in tetraploid switchgrass. These include linkage maps constructed using a full-sib population of 238 plants and simple sequence repeat (SSR) and sequence tagged site (STS) markers (Okada et al., 2010); 61,585 high-quality ESTs from switchgrass Kanlow (Tobias et al., 2008); a collection of more than 10 million ESTs from switchgrass Alamo and switchgrass Kanlow (putative switchgrass unique transcript sequences [PviUTs]) (Zhang et al., 2013); a small EST collection from laser capture-microdissected vascular tissues (Srivastava et al., 2010); and an Affymetrix microarray chip with 122,400 probe sets representing ∼90% of the transcriptome and 10- to 15-fold coverage of the genome (Zhang et al., 2013).
Although large collections of EST sequences coupled to bioinformatic interrogation provide powerful tools for classifying switchgrass gene families (Shen et al., 2009a), simple biological systems for functionally studying lignification in switchgrass are currently lacking. Lignification of the cell wall is tightly regulated in different tissues during different developmental stages, even within different horizontal sections of a single internode of a tiller (Shen et al., 2009b). We have made use of this phenomenon by selecting the top, middle, and bottom sections of internode four (I4) at elongation stage four (E4) (Moore et al., 1991; Hardin et al., 2013) for transcriptomic analysis of candidate monolignol pathway genes. To provide additional predictive power, we have developed a switchgrass cell suspension system in which lignification can be induced. Cell-suspension cultures have been widely used as models for studying xylogenesis, lignin biosynthesis, and cell wall development. Plant hormones such as gibberellic acid, brassinolide (BL), and auxin can induce lignification during wood and fiber formation, as well as ectopically (Aloni et al., 1990; Sánchez-Rodríguez et al., 2010; Trupiano et al., 2012), and BL, methyl jasmonate, and auxin have been used to induce the formation of extracellular and cell wall–associated lignin in suspension cells (Kärkönen and Koutaniemi, 2010).
We report a combined phylogenetic and transcriptomic analysis, integrating data from both individual internode regions and lignifying suspension cells, to reveal candidate genes for lignin biosynthesis; predictions were then tested by gene silencing in transgenic plants. This approach confirmed the involvement in lignification of candidates for late pathway steps, while suggesting the possibility of functional redundancy in the early steps of the monolignol pathway in switchgrass.
RESULTS
The One Internode System for the Study of Lignification in Switchgrass
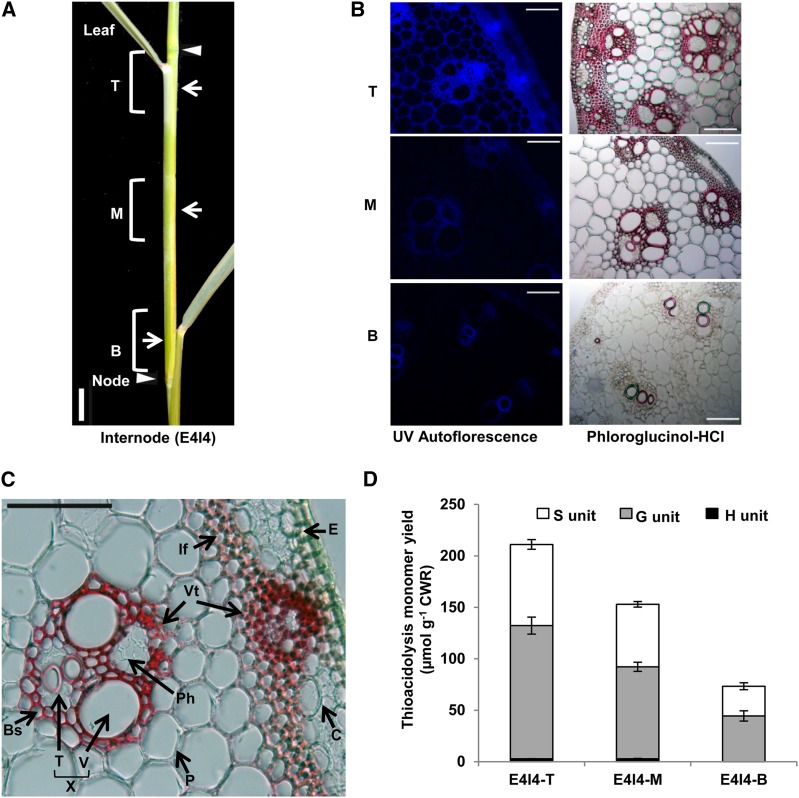
The mature whole tillers of switchgrass have more lignin than younger tillers, and the stem tissues in the whole tillers have relatively higher lignin content than other aboveground tissues such as leaves, leaf sheath, or inflorescence (Shen et al., 2009b). Even within one internode of a stem, different portions (top, middle, and bottom) have different lignin levels (Figures 1A and 1D) (Shen et al., 2009b). Also, different types of cells within the stem undergo lignification to different extents. For example, interfascicular fiber cells show more lignin staining than do parenchyma or cortex cells (Figures 1B and 1C). We chose fourth internodes (I4) of E4 stage tillers as biological material for transcriptomic analysis for several reasons. First, the E4 stage is the last tiller elongation stage before the reproductive stage under our greenhouse conditions, and the top, middle, and bottom sections showed the largest gradient of lignification, with the lignin content of the top section of E4I4 being about threefold higher than that of the bottom section (Figure 1D). Furthermore, the different types of lignified cells, such as interfascicular fibers and vascular elements, appear strikingly different within different sections of this one internode (Figure 1B).
Figure 1.
One Internode System for the Study of Gene Expression and Lignification in Switchgrass Stem Tissues.
(A) The I4 internodes with leaf and leaf sheath at E4 stage are shown. T, M, and B are the top, middle, and bottom sections of the individual internode, respectively. Bar = 1.0 cm.
(B) UV microscopy and phloroglucinol-HCl staining of the cross sections of top, middle, and bottom regions of I4 internodes at E4 developmental stage. Red color indicates the staining of lignin. Bar = 100 μm.
(C) Plant anatomy of switchgrass stem. Bs, bundle sheath; C, cortex; E, epidermis; If, interfascular fiber; P, parenchyma; Ph, phloem; T, tracheids; X, xylem; V, vessels; Vt, vascular tissue. Bar = 100 μm.
(D) Lignin content and composition in different portions of one internode as measured by thioacidolysis. Black bars represent H units; gray bars represent S units; white bars represent G units. Error bars represent se (n = 3).
Establishment of an Inducible Cell Suspension System for the Study of Lignification in Switchgrass
To increase our ability to predict lignin pathway gene candidates by their expression patterns, we developed a second system using switchgrass suspension cells. We first generated callus from immature inflorescences of switchgrass (Alamo), which is amenable to stable transformation. Details of the initiation and establishment of different types of cell culture (sandy, fine milky, and ultrafine) have been reported previously (Mazarei et al., 2011). The sandy cell culture type (see Supplemental Figures 2A to 2E online) was used throughout this study in view of its faster and more uniform growth. The basal level of lignin in these cultures was assessed by phloroglucinol-HCl staining (see Supplemental Figure 2F online, right panel), which detects cinnamaldehyde end groups in lignin (Pomar et al., 2002). No red coloration was observed compared with the nonstained samples (see Supplemental Figure 2F online, left panel), indicating that there is very little lignin in the cells. The cells were generally present in compact, small groups, mostly consisting of elongated cells and only occasionally round and slightly elongated cells (see Supplemental Figures 2G to 2I online).
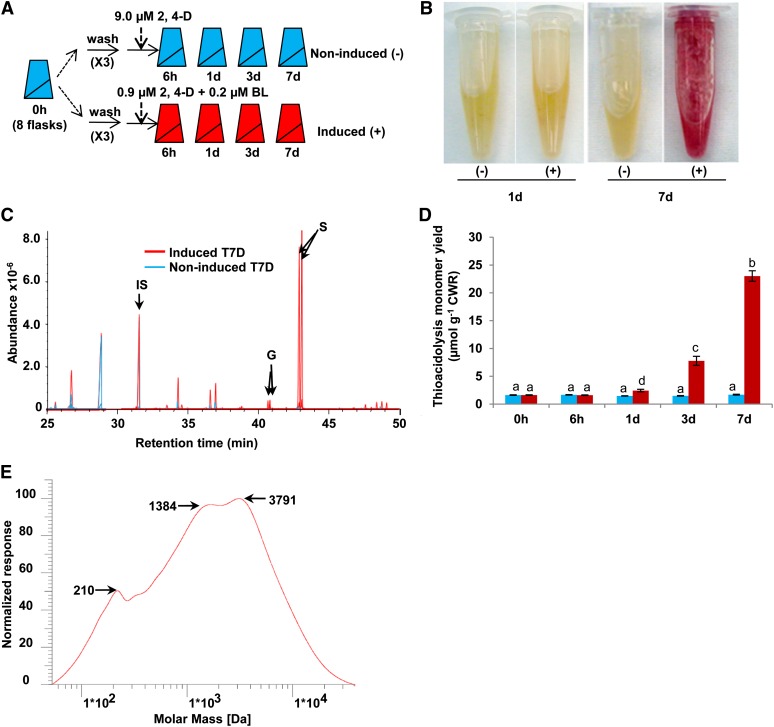
To induce lignification, we first tested the response of the cells to different combinations of BL and 2,4-D. Removal of 2,4-D from the medium induced lignin formation (see Supplemental Figure 3A online), but the cells grew poorly. A combination of 0.2 µM BL and 0.9 µM 2,4-D was finally selected because this supports cell growth (albeit slow) and the highest and most consistent induction of lignin (see Supplemental Figure 3B online). Suspension culture from one original flask was propagated into two flasks every 7 to 10 days, and after three subculture cycles, eight flasks of cells were used for induction (Figure 2A). A pooled sample was harvested from these eight flasks as the noninduced time zero sample (T0, initial control). The cells were then washed three times with liquid culture medium (without 2,4-D), and the medium adjusted to 9 µM 2,4-D (noninduced controls) or 0.9 µM 2,4-D + 0.2 µM BL (induced). Induced and noninduced samples were then harvested at 6 h, 1 d, 3 d, and 7 d (Figure 2A). Phloroglucinol-HCl staining of induced and noninduced cells at days 1 and 7 showed very strong staining only in the induced sample at day 7 (Figure 2B).
Figure 2.
An Inducible Cell Suspension System for the Study of Lignification in Switchgrass.
(A) Experimental design. Both induced and noninduced medium contains 4.4 µM 6-BA.
(B) Induced (+) and noninduced (−) suspension cells harvested at days 1 and 7 and stained with phloroglucinol. Red color indicates the presence of lignin.
(C) GC-MS (extracted ion-specific) chromatograms of thioacidolysis products from induced and noninduced day 7 suspension cells. IS, internal standard, docosane.
(D) Lignin accumulation in induced (red bars) and noninduced (blue bars) suspension cells as measured by thioacidolysis. Letters indicate significant differences at the P < 0.05 level. Error bars represent se (n = 3).
(E) GPC analysis of ball-milled lignin isolated from induced suspension cells. The average molecular mass is ∼2,958 D with a small amount of monomer present ∼210 D. Polystyrenes with molecular mass range of 580 to 196,000 D were used for establishing the calibration curve.
To determine the content and composition of the induced lignin in the suspension cells, we analyzed dried extracted cell wall residues by thioacidolysis followed by GC-MS. Predominantly S lignin monomers were detected, with trace amounts of G units and no H units (Figure 2C). Lignin began to accumulate after 2 to 3 days of treatment, and its levels increased ∼15-fold at day 7 compared with the induced sample at day 1 (Figure 2D). Only trace amounts of lignin were detected in the noninduced samples. To further confirm that the lignin synthesized in the suspension cells is polymeric, we performed gel permeation chromatography (GPC) analysis of ball-milled lignin isolated from induced suspension cells. The average molecular weight of the isolated lignin was ∼2,958 D, with a small amount of monomer present (Figure 2E). Based on the mass of syringyl alcohol (184 D), the lignin polymer has an average degree of polymerization of ∼16 units.
Annotation of Monolignol Biosynthetic Genes in Switchgrass by Sequence Comparison and Phylogenetic Analysis
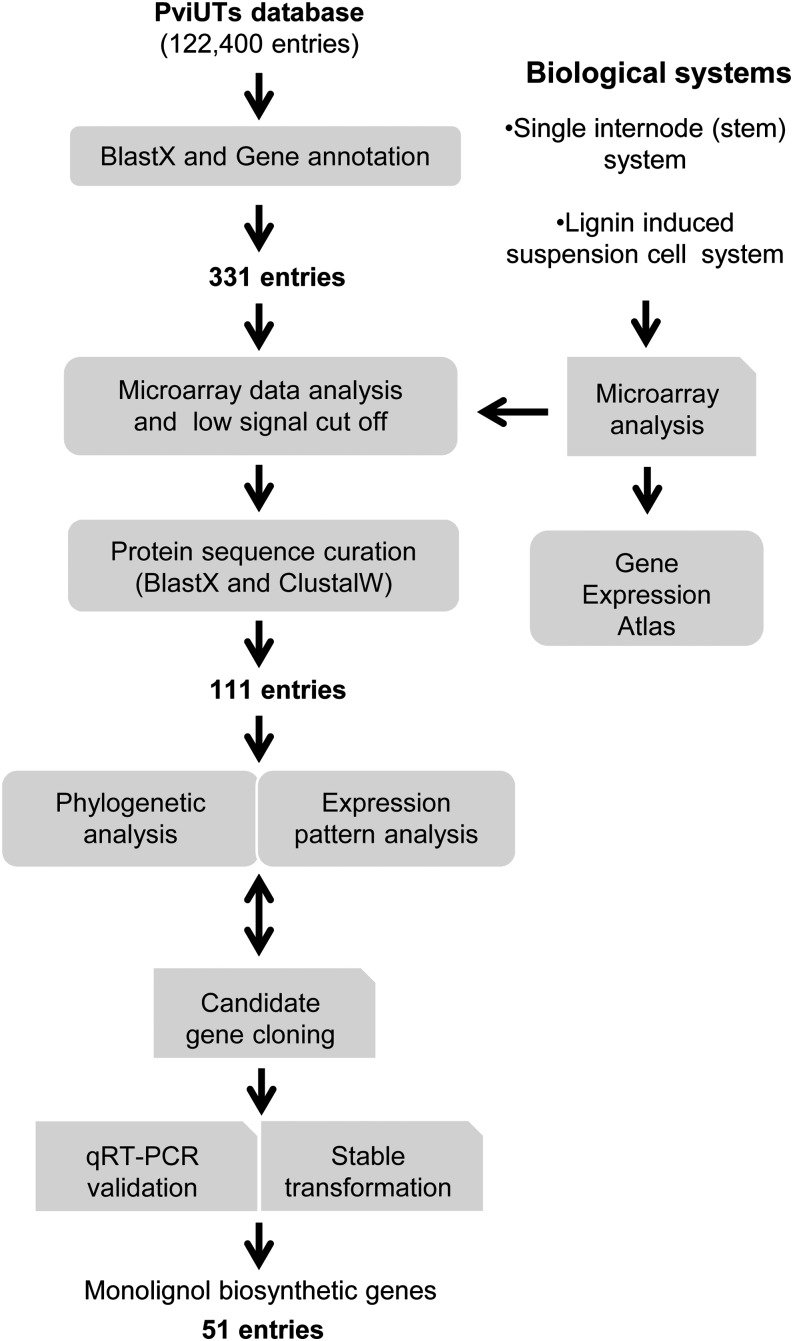
A combined genomic, bioinformatic, and genetic approach, illustrated in Figure 3, was used to derive monolignol biosynthetic pathway gene candidates in switchgrass. We first annotated all the available PviUTs that may be involved in phenylpropanoid/monolignol biosynthesis (Zhang et al., 2013). The PviUTs were analyzed by BlastX against the Tair10 Arabidopsis thaliana protein sequences (www.arabidopsis.org), and 331 candidates showed high sequence identity to the functionally characterized and annotated (http://cellwall.genomics.purdue.edu/index.html) monolignol pathway enzymes l-phenylalanine ammonia-lyase (PAL), 4-coumaroyl shikimate 3′-hydroxylase (C3′H), cinnamate 4-hydroxylase (C4H), ferulate 5-hydroxylase (F5H), 4CL, CCR, hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase (HCT), caffeoyl-CoA 3-O-methyltransferase (CCoAOMT), COMT, or CAD (see Supplemental Data Set 1 online).
Figure 3.
Scheme Showing the Approach for Selection and Validation of Monolignol Pathway Genes in Switchgrass.
The 331 and 111 entries are the expressed sequence tag candidates shown in Supplemental Table 1 and Supplemental Data Set 2 online. PviUTs, switchgrass unique transcript sequences (http://switchgrassgenomics.noble.org/index.php).
To narrow down the gene candidates, we checked their expression levels by conducting microarray analysis of different segments of the E4I4 internode and of noninduced and induced cell suspension cultures. The one internode system had been integrated into the recently published switchgrass Gene Expression Atlas (Zhang et al., 2013) for studying transcript levels in different tissues, and the suspension cell data will be integrated into the atlas in future upgrades. On the basis of this preliminary microarray analysis (signal cutoff of 100; see Supplemental Data Set 1 online, column M), 155 candidates were selected for further analysis. Protein sequences of these 155 PviUTs were translated based on the National Center for Biotechnology (NCBI) BlastX program and manually curated to correct reading frames. Based on ClustalW alignment (http://www.ebi.ac.uk/Tools/msa/clustalw2/), short and redundant sequences were removed (Harrison and Langdale, 2006) (see Supplemental Data Set 1 online, column N) and 111 PviUTs were retained as the most reliable candidates for further investigation (Figure 3).
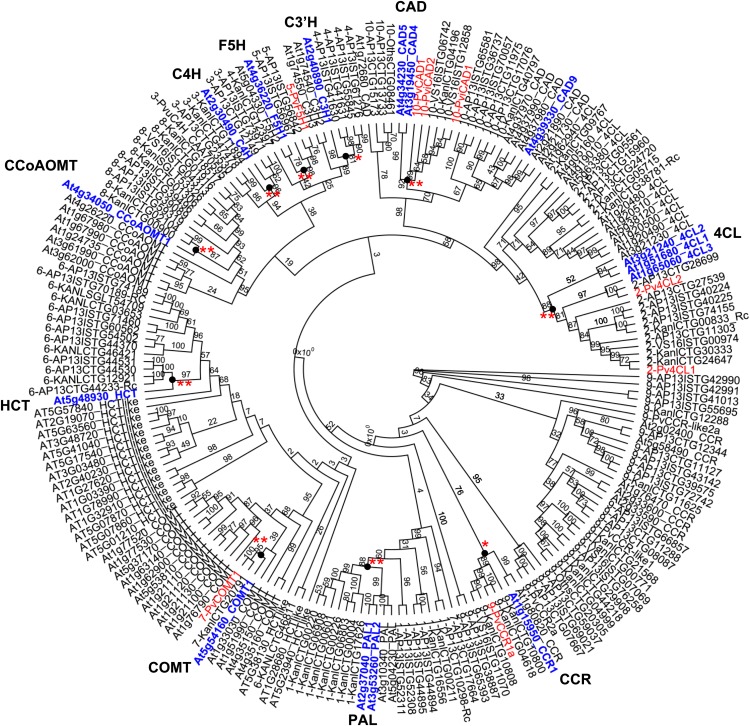
We next performed a phylogenetic analysis of these 111 candidate protein sequences with 81 reference protein sequences from Arabidopsis (http://cellwall.genomics.purdue.edu/index.html) and 12 switchgrass monolignol biosynthetic protein sequences from the published literature. The 204 protein sequences were aligned by ClustalW, and the phylogenetic tree was built by PHYML (phylogenetic estimation using maximum likelihood version 2.0.10) with default settings integrated in Geneious Pro 5.5.6 software (Biomatters) (Figure 4). The phylogenetic tree showed the presence of switchgrass genes corresponding to all 10 known monolignol biosynthetic enzymes, as well as “like-genes” (closely related genes that may not have the ascribed function). Each family forms a distinct cluster, with the C4H, C3′H, and F5H proteins grouped into a cytochrome P450 superfamily cluster (Figure 4).
Figure 4.
Phylogenetic Analysis of Switchgrass and Arabidopsis Monolignol Pathway Gene Candidates.
The phylogenetic tree was built by PHYML (maximum likelihood version 2.0.10) with default settings integrated in Geneious Pro 5.5.6 software (Biomatters). Numbers by the branches are bootstrap values. Names in blue are the genetically characterized Arabidopsis lignin biosynthetic genes. Names in red are the published switchgrass genes listed in Supplemental Data Set 2 online. Genes that closely cluster with known Arabidopsis monolignol biosynthetic genes and form a well-defined phylogeny cluster (marked by black dots) are considered as having intermediate evidence (bootstrap value >75, one asterisk) or strong evidence (bootstrap value >85, two asterisks) in support of candidate selection. The CLUSTAL sequence alignment for the sequences analyzed in this figure is presented in Supplemental Data Set 3 online, and higher resolution phylogenetic analyses of each gene family (with 1000 bootstrap replicates) are presented in Supplemental Figure 4.
The protein sequences of the 111 putative PviUTs were also used to BlastP search against the The Arabidopsis Information Resource 10 protein database and published switchgrass protein sequences that correspond to genes known to be involved in the monolignol biosynthetic pathway (see Supplemental Data Set 2 online). We also mapped these putative PviUTs onto the current switchgrass draft genome through the BLASTN program provided by Phytozome V9.0 (http://www.phytozome.net; see Supplemental Data Set 2 online). The Arabidopsis genes that are involved in monolignol biosynthesis with genetic support are At PAL1 (At2g37040) and PAL2 (At3g53260) (Rohde et al., 2004); C4H (At2g30490) (Schilmiller et al., 2009); 4CL1, 4CL2, and 4CL3 (Lee et al., 1997; Yang et al., 2011); C3′H1(Franke et al., 2002; Abdulrazzak et al., 2006); HCT (Hoffmann et al., 2004); F5H1 (Meyer et al., 1998); COMT1 (Goujon et al., 2003); CCoAOMT1 (Do et al., 2007); CCR1(Jones et al., 2001); CAD4, CAD5, and CAD9 (Kim et al., 2004; Eudes et al., 2006; Sibout et al., 2005). The PviUTs that correspond to switchgrass genes that have been functionally characterized genetically are Pv 4CL1, 4CL2, COMT1, CAD1, PviCAD1, and PviCAD2 (Fu et al., 2011a,b; Saathoff et al., 2011; Xu et al., 2011), and CCR1 has been functionally characterized biochemically but not genetically (Escamilla-Treviño et al., 2010).
PviUTs that were classified with the known lignin biosynthetic genes in Arabidopsis were scored as having strong (bootstrap value ≥85), intermediate (85 > bootstrap value ≥75), and weak (bootstrap value <75) support for candidate selection (Figure 4, see Supplemental Data Set 2 online). All of the functionally characterized switchgrass monolignol pathway genes showed highest sequence identity and clustered most closely to the corresponding Arabidopsis genes (bootstrap value ≥75); these phylogeny clades are marked with asterisks in Figure 4. Note that Figure 4 compares a large number of genes from different families, and the number of bootstrap replicates was therefore limited to 100 to allow the analysis to be completed within a reasonable timeframe. Higher resolution trees with 1000 bootstrap replicates for the same members of each individual gene family are presented in Supplemental Figure 4 online.
Transcript Profiling through Microarray Clustering Analysis
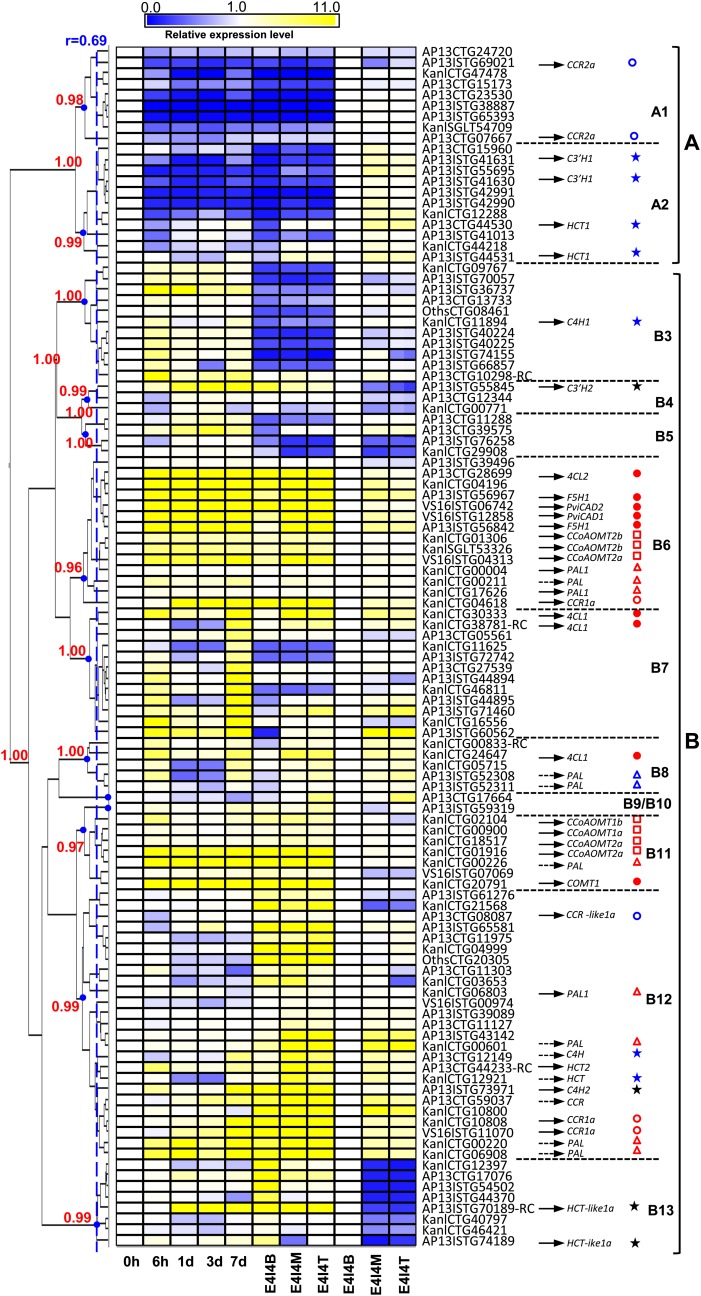
To combine a second criterion for narrowing down the candidate PviUTs, we checked their transcript abundance patterns by clustering using Spotfire software (TIBCO Software) with approximately unbiased (AU) P value support calculated by pvclust (Suzuki and Shimodaira, 2006) (Figure 5). First, the data for induced cell cultures and the E4 single internode samples (a comparison of the top, middle and bottom portions of the internode, where lignification is highest at the top and lowest at the bottom; Figure 1D; Shen et al., 2009b) were normalized by setting the nonlignified cell culture T0 values as 1. Then, we included a second expression pattern by normalizing the relative expression of middle and top portions to the bottom section of the E4I4 internode (Figure 5) The expression data clustered into 13 groups, with a correlation coefficient (r) cutoff of 0.69 with AU P values higher than 0.95 (Figure 5). Clearly, with a few exceptions, the transcript abundance patterns fall into two major groups. Genes in group A (A1 and A2) are generally downregulated, whereas genes in group B (B3–B13) are generally upregulated in the induced suspension cells (Figure 5). Some genes, such as those in the middle cluster of group B12, are repressed in the suspension cells but are quite highly expressed in the internode samples, whereas the genes in groups B3, B4, and B5 show moderate induction in the suspension cells but have very low expression in the internode samples. The B6, B11, and B12 clusters of genes generally show moderate to strong induction or expression in both the suspension cells and the internode samples (Figure 5).
Figure 5.
Clustering of Expression Patterns of Putative Phenylpropanoid/Monolignol Biosynthetic Genes in Switchgrass, as Determined by Microarray Analysis.
Normalization for clustering was done in two ways: first, the expression levels in the top and middle of the fourth stem internode and in induced suspension cells at different times postinduction were normalized to the time zero value for the cell cultures; second, the values for the middle and top of the internode were normalized to the value for the bottom of the internode. B, M, and T are bottom, middle, and top sections of the E4I4 internode, respectively. Black arrows indicate the cloned ESTs that have the highest sequence identity to the corresponding PviUTs in the database. Yellow and blue colors indicate increased or reduced expression levels, respectively, relative to those in the samples (T0 culture or bottom internode) used for normalization (expression value = 1). The heat map of transcript-level patterns was generated using MultiExperiment Viewer software (Saeed et al., 2003) with AU P value (red numbers on the left) support calculated by pvclust (Suzuki and Shimodaira, 2006). At a correlation coefficient (r) cutoff of 0.69 with AU P values higher than 0.95, the expression patterns are clustered into 13 groups (A1 to B13). The expression patterns of different genes are indicated by different-colored shapes: F5H1, COMT1, CAD1, PviCAD1, PviCAD2, 4CL1, and 4CL2 (red dots); CCR and CCR-like (red and blue circles); PAL (red and blue triangles); CCoAOMT (red squares); C4H, C3′H, and HCT that are not well induced in the suspension cells (blue stars); C3′H2 and HCT-like1a (black stars). Gene names indicated by dotted lined arrows are the candidates that have not been cloned. Cloned genes (solid lined arrows) are also shown in Supplemental Data Set 2 online. Because of allelic variation, genomic complexity, and EST assembly inaccuracy, one cloned gene may have several top hit PviUTs. When a PviUT has two probe sets on the switchgrass Affymetrix chip designed according to different regions of one EST (see Supplemental Table 1 online), only the probe set with the highest signal is presented in the heat map.
To check the effectiveness of the transcript profiling for identifying lignin biosynthetic genes, we checked the transcript-level patterns of PviUTs that had been mapped to the switchgrass genes that have been functionally characterized (see Supplemental Data Set 2 online). As expected, the expression patterns of the lignin-associated switchgrass F5H1, COMT1, CAD1, CAD2, 4CL1, and 4CL2 genes (Fu et al., 2011a,b; Saathoff et al., 2011; Xu et al., 2011) are mainly defined by groups B6, B7, B8, and B11, with strong induction in the suspension cells and high expression in internode samples (Figure 5, red dots). Therefore, we used them as reference genes for candidate selection. Genes in groups B6, B7, B8, and B11 were considered as strong candidates, and genes in groups A1, A2, and B3 to B5 were generally considered as weak candidates unless they were obvious outliers, in which case they were listed as candidates supported by intermediate evidence (Figure 5, see Supplemental Data Set 2 online).
Based on previous analysis of enzymatic activities, CCR1, but not CCR2 or the CCR-like proteins, is the major CCR involved in lignin biosynthesis in switchgrass (Escamilla-Treviño et al., 2010). This study confirmed this conclusion: the phylogenetic analysis shows that Pv CCR1, but not Pv CCR2 or Pv CCR-like1, clusters closely with the lignin-related At CCR1 (At1g15950) and At CCR2 (At1g80820) genes (Figure 4), and CCR1, but not CCR2 or CCR-like1, is highly induced in the cell cultures (Figure 5).
Seven PAL gene candidates cluster closely with At PAL1 (At2g37040) and At PAL2 (At3g53260) in the protein phylogeny analysis (Figure 4) and are strongly induced in the cell cultures and highly expressed in lignifying internodes (Figures 5, red and blue triangles). In contrast, expression of the switchgrass genes most similar to functionally characterized Arabidopsis C4H (AP13CTG12149), C3′H (AP13ISTG41630 and AP13ISTG41631), and HCT (AP13CTG44530, AP13ISTG44531, and KanlCTG12921; see Supplemental Data Set 2 online) was either repressed in the lignifying cell cultures or only slightly induced at day 7 (Figure 5, blue stars), although lignification begins between days 1 and 3 (Figure 2C). Transcript levels of HCT1 and C3′H1 were higher in the middle and top sections (higher lignin level) than in the bottom section within the E4I4 internode, similar to the pattern of CAD, F5H1, CCR1, and 4CL (Figure 5C). However, C4H (KanlCTG11894), C3′H (AP13ISTG41630 and AP13ISTG4163), and HCT (AP13CTG44530 and AP13CTG44531) had lower expression levels in the developing stem sections (only expressed in the bottom section for AP13CTG44531) compared with the nonlignified T0 cell culture samples (Figure 5); in contrast, CCR1, F5H1, CAD1, 4CL1, and COMT1 had higher expression levels in all the developing stem sections than in T0 cell culture samples (Figure 5).
Cloning and Sequence Analysis of Additional Lignin Pathway Gene Candidates
The top PviUT candidates selected by phylogenetic analysis for Pv C4H, C3′H, and HCT genes showed a different expression pattern from that of the known lignification-associated genes. To rule out the possibility that these microarray signals do not correctly reflect the genes’ transcript levels because of problems with the probe design, we attempted to clone most of the candidates for further validation by detailed sequence analysis and real-time quantitative RT-PCR (qRT-PCR).
The candidate PviUTs for PAL, C4H, C3′H, HCT, and CCoAOMT were KanlCTG00220, KanlCTG06908, KanlCTG00601, KanlCTG00226, KanlCTG06803, KanlCTG00004, KanlCTG17626 (PAL); AP13ISTG73971, AP13CTG12149, KanlCTG11894 (C4H); AP13ISTG41630, AP13ISTG41631, AP13ISTG55845, AP13ISTG61276 (C3′H); AP13ISTG74189, KanlCTG12921, AP13CTG44530, AP13ISTG44531, AP13CTG44233-Rc, AP13ISTG70189-Rc (HCT); and KanlCTG00900, KanlCTG02104 (CCoAOMT). We attempted to clone these candidate genes either by 3′RACE (rapid amplification of cDNA ends) or by full-length EST cloning based on direct PCR amplification from switchgrass Alamo cDNAs with primers (see Supplemental Table 1 online) designed according to the PviUT sequences. The cloning of the top candidates for PAL, C3′H, HCT, and CCoAOMT was successful but failed for AP13CTG12149 (C4H). All cloned genes were confirmed by sequencing; their accession numbers are listed in Supplemental Data Set 2 online.
Because there are few reports on early monolignol pathway biosynthetic genes in monocot species, we performed further sequence analysis through protein sequence alignment and phylogenetic analysis for C4H, C3′H, HCT, and CCoAOMT genes (or -like genes) with an expanded list of gene orthologues from other monocot species, as described later.
C4H
The cloned EST for AP13ISTG73971 (named Pv C4H2) is 80% identical to the functionally characterized At C4H (At2g30490) and Mt C4H from Medicago truncatula (Sewalt et al., 1997) and 84% identical to Ptr C4H1 and Ptr C4H2 from P. trichocarpa (Chen et al., 2011). Pv C4H1 is 76% identical to At C4H, Mt C4H, and Ptr C4H1 and Ptr C4H2. The phylogenetic tree shows that Pv C4H1, together with several putative C4Hs from maize, rice, wheat (Triticum aestivum), sorghum, and Brachypodium distachyon, is more closely clustered with At C4H than with Pv C4H2 (see Supplemental Figure 5 online). Protein sequence alignment showed that although both Pv C4H1 and Pv C4H2 have the conserved substrate recognition sites (SRSs) of C4H/CYP73A5 enzymes, including SRS1 (Ser100ArgThrArgAsnValValPheAspIlePheThrGlyLysGlyGlnAspMetValPheThrValTyr122), SRS2 (Leu216SerGlnSerPheGluTyrAsnTyr224), SRS4 (Ile299ValGluAsnIleAsnValAlaAlaIleGluThrThrLeuTrpSer314), and SRS5 (Arg368MetAlaIleProLeuLeuValProHis377), Pv C4H2 is more similar in these conserved regions to At C4H, Mt C4H, and Ptr C4Hs (see Supplemental Figure 6 online). Nevertheless, both Pv C4H1 and Pv C4H2 belong to the class I C4Hs that are believed to catalyze the conversion of trans-cinnamic acid to p-coumaric acid (Sewalt et al., 1997) and that have diverse functions in stress or fungal elicitor responses (Ralston et al., 2001), vascular differentiation (Nedelkina et al., 1999), and lignin biosynthesis (Blee et al., 2001). Because the expression of C4H1 is very low in stem tissue (Figure 5), we predict that C4H2 is the main C4H gene involved in lignin biosynthesis in switchgrass stems.
C3′H
The cloned EST for AP13ISTG55845 is 67% identical to At C3′H (At2g40890), 78% identical to Pv C3′H1, and 99% identical to the predicted AP13ISTG55845 database sequence, and we named the gene Pv C3′H2. The phylogenetic analysis indicated that Pv C3′H2 and Pv C3′H1, together with maize C3′H (Barrière et al., 2007) and wheat C3′H1 and C3′H2 (http://urgi.versailles.inra.fr/Projects/Genoplante), form a distinct C3′H clade, although both have between 60 and 70% protein identity to At C3′H1 (At2g40890; see Supplemental Figure 7 online). It is possible that both C3′H1 and C3′H2 are involved in the shikimate shunt pathway (see Supplemental Figure 1 online). Because the expression of C3′H1 is very low in stems (Figure 5), C3′H2 is predicted to be the main C3′H gene involved in lignification of switchgrass stems.
HCT
The phylogenetic analysis of HCT gene candidates was more complex (see Supplemental Figure 8 online). Pv HCT1 and Pv HCT2 cluster together with At HCT (At5g48930), Pr HCT (ABO52899), maize HCT candidates HCT1 and HCT2 (Barrière et al., 2007), and wheat HCT candidates HCT1 and HCT2. Hydroxycinnamoyl CoA:quinate hydroxycinnamoyl transferases (HQTs) from artichoke, tobacco, and tomato formed a separate HQT cluster. Interestingly, there are no HCT or HCT-like proteins from switchgrass that cluster with dicot HQT proteins (see Supplemental Figure 8 online). We aligned all the HCT and HCT-like protein sequences with known HQTs to determine whether there were any conserved HQT motifs (Sonnante et al., 2010) in these sequences. The alignment showed that all the proteins have the conserved motifs for the acyl transferase family: HXXXDG and DFGWG (see Supplemental Figure 9 online); however, no conserved motifs that have been identified specifically for dicot HQTs (Sonnante et al., 2010) (Supplemental Figure 9 online) are found in these HCT-like proteins. The protein sequence translated from the cloned full-length EST of AP13ISTG70189-Rc was 93% identical to AP13ISTG74189, 87% identical to AP13ISTG70189-Rc, 36% identical to At HCT and Pv HCT2, and 37% identical to Pv HCT1. Because the protein sequence identity to functionally characterized HCTs is so low, we named this gene HCT-Like1a (HCT-L1a). HCT-L1a is grouped more closely with Os HCT4 (Os06g0185500) than with typical HCT/HQT enzymes such as Os HCT1 and Os HCT2 (Kim et al., 2012) (see Supplemental Figure 9 online). Os HCT4 was recently characterized as a hydroxycinnamoyltransferase that mediates the trans-esterification of glycerol and shikimic acid in the presence of hydroxycinnamoyl-CoA (Kim et al., 2012). The biochemical properties of the proteins encoded by the switchgrass HCT and HCT-like genes, as well as the C3′H1 and C3′H2 genes, are presented elsewhere (Escamilla-Treviño et al., 2013). Purified recombinant Pv HCT proteins prefer shikimic acid as acyl acceptor, whereas HCL-L1a proteins prefer quinic acid. Although HCT-L1a can catalyze formation of both 4-coumaroyl quinate and caffeoyl quinate, it cannot catalyze the formation of the corresponding shikimate esters (Escamilla-Treviño et al., 2013).
CCoAOMT
EST sequencing and gene cloning from rice and maize suggest that three Os CCoAOMTs (COA1, COA20, and COA26) (Zhao et al., 2004) and five Zm CCoAOMTs (CCoAOMT1– CCoAOMT5) (Barrière et al., 2007) may be involved in lignin biosynthesis. Our phylogenetic analysis showed that the Pv CCoAOMT1a and Pv CCoAOMT1b candidates share high sequence similarity to Os COA1, Zm CCoAOMT1, and Zm CCoAOMT2, whereas Pv CCoAOMT2a and Pv CCoAOMT2b have higher similarity to Os COA20 and Zm CCoAOMT5 (see Supplemental Figure 10 online). Pv CCoAOMT1a, Pv CCoAOMT1b, Pv CCoAOMT2a, and Pv CCoAOMT2b are all induced in the lignifying suspension cells (Figures 5) and were considered as potential candidates for involvement in lignin biosynthesis, although they fall into two distinct clusters on the phylogenetic tree (see Supplemental Figure 10 online). Further sequence alignment showed that all the conserved motifs including the three putative S-adenosyl-l-methionine binding motifs (A, B, and C) and CCoAOMT signature motifs (D, E, F, G, and H) (Joshi and Chiang, 1998; Zhao et al., 2004) are present in these PviUTs (see Supplemental Figure 11 online). The Zm CCoAOMT4 sequence is not aligned well with these sequences and most of these motifs are not found, so we removed it from the alignment.
qRT-PCR–Based Transcript Analysis of Lignin Pathway Gene Candidates
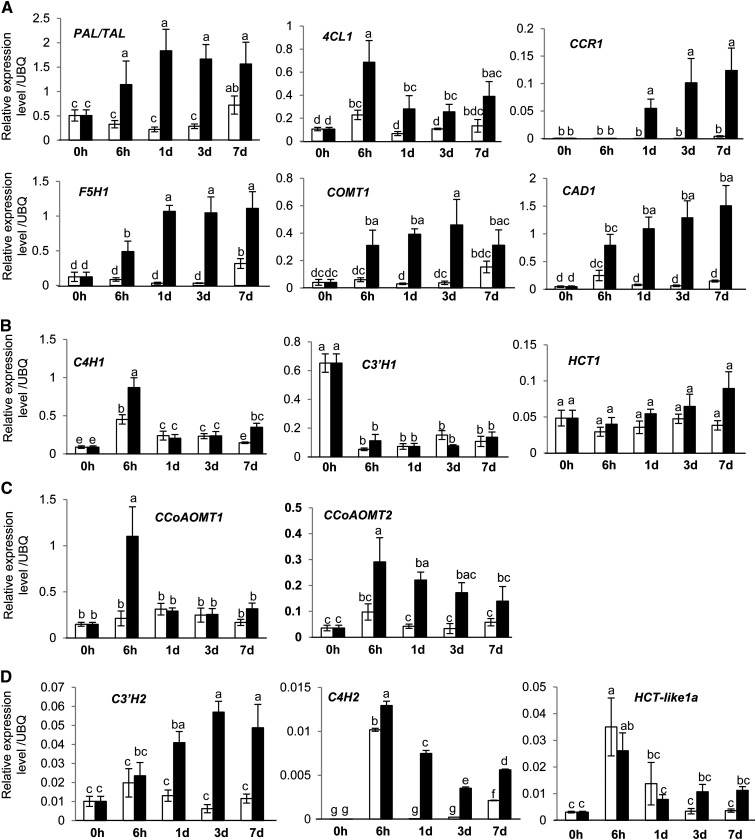
We next confirmed the transcript levels of the candidate monolignol pathway genes in the induced cell cultures by qRT-PCR analysis. Transcripts encoding PAL1, 4CL1, COMT1, CCR1, F5H1, and CAD1 were strongly and coordinately induced during the period of lignin accumulation. Transcript levels of COMT1, F5H1, and CCR1 increased by ∼13-, 32-, and 275-fold, respectively, within 24 h of induction (Figure 6A). However, the genes encoding C4H1 and the enzymes of the shikimate shunt (HCT1 and C3′H1), all catalyzing early steps in the currently accepted monolignol pathway (see Supplemental Figure 1 online), were not significantly induced over the time course (Figure 6B).
Figure 6.
Candidate Gene Evaluation through qRT-PCR analysis.
qRT-PCR analysis of transcript levels in induced and control cell suspension cultures. Open bars represent noninduced samples; closed bars represent induced samples. Data are means ± se (n = 4). Letters indicate significant differences at the P ≤ 0.05 level.
(A) Genes that are strongly induced in coordination with lignin appearance.
(B) Genes with weak induction, inconsistent induction pattern, or repression.
(C) CCoAOMT1 and CCoAOMT2.
(D) C3′H2, C4H2, and HCT-like1a. UBQ, ubiquitin.
When compared with the nonlignified T0 cell culture samples, both CCoAOMT1 and CCoAOMT2 have higher expression levels in the induced cell culture samples, but when compared with the relevant noninduced controls at each time point, only CCoAOMT2 is induced over the entire time course, although the difference from controls is not significant at later time points (Figure 6C). Furthermore, CCoAOMT2 expression declines after day 1, opposite to the trend of lignin accumulation (Figure 2D).
The transcript levels of C4H2, C3′H2, and HCT-L1a in the suspension cell system were also confirmed by qRT-PCR analysis; expression of C4H2 and HCT-L1a was higher in induced than in control cells at days 3 and 7, but it did not parallel the expression of CCR, F5H, COMT, and CAD, whereas C3′H2 was consistently induced through days 1 to 7 (Figure 6D).
Recently, a caffeoyl shikimate esterase (CSE, encoded by At1g52760) was reported to convert caffeoyl shikimate to caffeic acid during lignin biosynthesis in Arabidopsis. CSE, together with 4CL active toward caffeic acid, bypasses the conversion of caffeoyl shikimate to caffeoyl CoA, hence rewriting part of the pathway (Vanholme et al. 2013). Putative CSE homologues were identified in the switchgrass PviUTs database through BLAST search. We have also shown that caffeoyl shikimate (but not caffeoyl quinate) can be hydrolyzed to caffeic acid by crude protein extracts of switchgrass stem tissues (Escamilla-Treviño et al., 2013), suggesting that a similar route to caffeoyl CoA could also exist in switchgrass. Phylogenetic analysis revealed putative CSE/CSE-like PviUTs (AP13ITG63270, KanlowCTG44303, and AP13ITG70675) that grouped into a CSE clade with At CSE (see Supplemental Figure 12A online). Among them, AP13ITG63270 has the highest expression level in different tissues of switchgrass based on analysis of the gene atlas (see Supplemental Figure 12B online). However, qRT-PCR analysis showed that expression of AP13ITG63270 in suspension cells is repressed during lignification (see Supplemental Figure 12C online). Whether a CSE or CSE-like gene might be involved in lignification in switchgrass therefore remains unclear.
Functional Analysis in Support of the Above Predictions
Based on the earlier analyses, the switchgrass 4CL1, 4CL2, F5H, COMT1, CAD1, and CCR1 genes are expressed in a manner consistent with an involvement in lignification, and this has been verified genetically, for all except F5H and CCR1, through independent studies in which the genes have been either downregulated or overexpressed in transgenic switchgrass (Fu et al., 2011a,b; Saathoff et al., 2011; Xu et al., 2011). In contrast, although strong candidates based on sequence analysis, AP13ISTG52308 and AP13ISTG52311 (for PAL), AP13CTG12149 (for C4H1), AP13ISTG41630 (for C3′H1), and KanlCTG12921, AP13CTG44530, and AP13ISTG44531 (for HCT) do not have the expected transcript appearance in the suspension cell system, the internode system, or both (Figure 5). We therefore predict that these genes may not be, by themselves, critical for lignin biosynthesis (at least in stem). Pv HCT-L1a, CCoAOMT1 and CCoAOMT2 are also questionable candidates based on qRT-PCR validation. The more likely candidates are PAL1, C4H2 (AP13ISTG73971), C3′H2 (AP13ISTG55845), and HCT2 (AP13CTG44233-Rc).
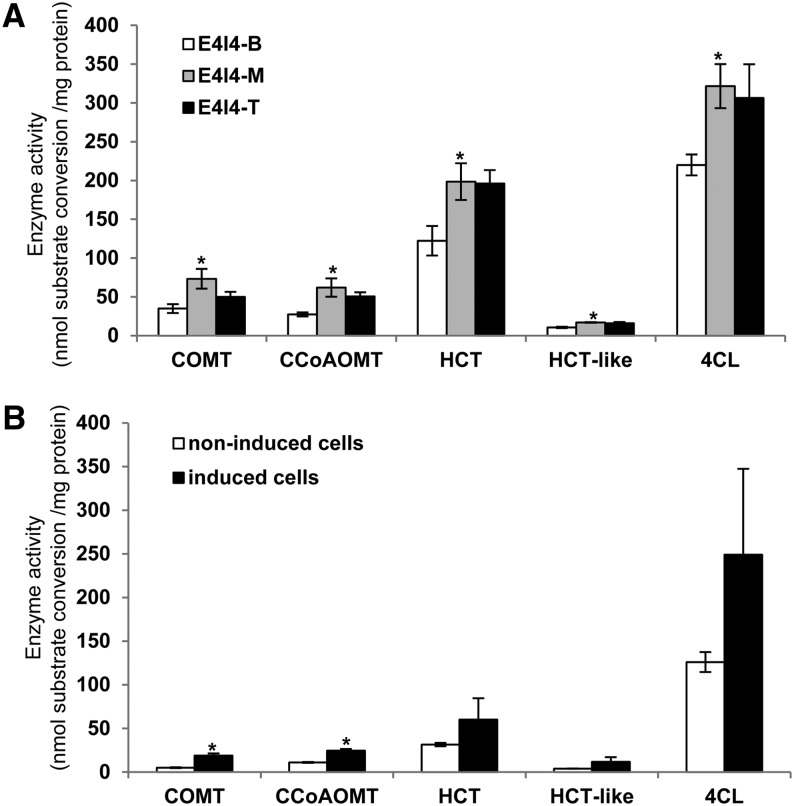
Switchgrass is a complex tetraploid species. A particular enzyme may be encoded by homologous genes, and different alleles of these genes (Escamilla-Treviño et al., 2010). To obtain an overview of the overall expression of potential lignin pathway enzymes in switchgrass internodes and induced cell cultures, we measured total extractable enzyme activities of a group of selected enzymes, including two predicted to be involved in lignin biosynthesis by our transcript analyses (4CL, COMT) and two that were questionable (HCT and CCoAOMT). Crude protein extracts were prepared from the bottom, middle, and top sections of E4I4 internodes, as well as 7-d noninduced and induced suspension cells, and incubated with optimized concentrations of the relevant substrates and cofactors. Extractable enzyme activities of COMT and CCoAOMT were significantly higher in the middle internode sections than in the top or bottom sections, and in induced suspension cells compared with noninduced cells (Figure 7). A similar trend was found for HCT activity converting 4-coumaroyl CoA to its shikimate (HCT) or quinate (HCT-like) esters in the internode samples, but not in the 7-d suspension cell samples where the variation in induced samples meant that the values were not statistically different from controls (Figure 7).
Figure 7.
Extractable Enzyme Activities in Crude Protein Extracts from Switchgrass Internode Sections and Suspension Cells.
Data show total extractable activities of 4CL, CCoAOMT, HCT, and HCT-like (HQT) enzymes in crude protein extracts for different internode sections (A) and suspension cells (B). Substrates used for the assays were 4-coumaric acid, CoA, and ATP for 4CL; 5-hydroxyconiferaldehyde and S-adenosyl-l-methionine for COMT; caffeoyl CoA and S-adenosyl-L-methionine for CCoAOMT; 4-coumaroyl CoA and shikimate for HCT; 4-coumaroyl CoA and quinate for HCT-like. T, M, and B are the top, middle, and bottom sections of the individual internode as shown in Figure 1A. The noninduced and induced culture samples were collected at day 7 as shown in Figure 2A. Data are means ± se (n = 3). Asterisks indicate values that were determined by the Student’s t test to be significantly different (P < 0.05) from their equivalent control (bottom sections and noninduced cells).
Taken together, the transcript level and enzyme activity data suggest that the shikimate shunt and subsequent conversion of caffeoyl CoA to feruloyl CoA is a feasible route for lignification in switchgrass, but that the genes most orthologous to those identified as playing a role in lignification in dicot species may not by themselves be necessary or sufficient. To confirm which gene(s) is/are responsible for supporting these pathways requires genetic evidence by analysis of mutants (difficult in an outcrossing tetraploid) or knockdown lines generated through stable transformation. Furthermore, because of potential genetic redundancy, full functional validation of selected candidate genes means that it may be necessary to downregulate multiple gene forms. To begin this analysis, we targeted four genes for downregulation: CCR1, which was predicted to be a strong candidate by our analyses, and C4H1, CCoAOMT1, and CCoAOMT2, which, although showing the highest sequence identity to functionally characterized C4H and CCoAOMT genes involved in lignification in dicots, did not pass our criteria for critical involvement in lignification. Downregulation of the target genes was achieved by RNA interference (RNAi) through Agrobacterium tumefaciens–mediated stable transformation. The construction of plasmids and the transformation process are illustrated in Supplemental Figure 13 online.
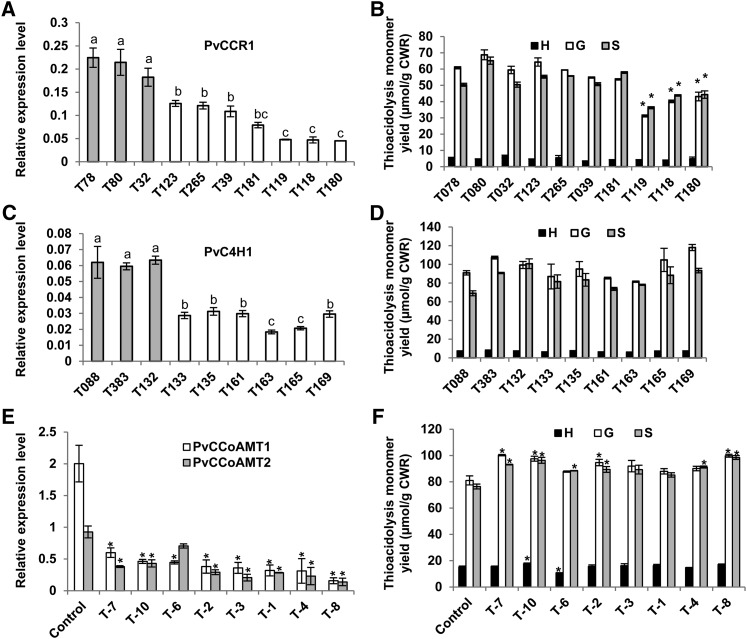
Downregulation of CCR1 transcripts by ∼75% reduced the lignin thioacidolysis yields by ∼25% for both G and S units (Figures 8A and 8B), consistent with the involvement of CCR1 as an enzyme of monolignol biosynthesis. In contrast, downregulation of C4H1 transcripts in lines T163 and T165 by ∼70% (Figure 8C) led to no significant change in lignin content or composition, as compared with the relevant controls (Figure 8D). Furthermore, downregulation of CCoAOMT1 and CCoAOMT2 transcripts by up to 90% in lines 4 and 8 (Figure 8E) led to no significant reduction (line 4) or even a slight increase (line 8) of lignin content (Figure 8F). These data contrast strongly with the clearly reduced lignin phenotypes in switchgrass with reduced expression of CCR1 (Figures 8A and 8B) or 4CL1, COMT1, or CAD (Fu et al., 2011a,b; Saathoff et al., 2011; Xu et al., 2011).
Figure 8.
RNAi-Mediated Silencing of Potential Monolignol Pathway Genes in Switchgrass.
(A, C, E) Transcript levels of the target genes in the C4H1-, CCR1-, and CCoAOMT-RNAi lines. (B, D, F) Corresponding lignin contents measured by thioacidolysis. Black bars represent H units; gray bars represent S units; white bars represent G units. Data are means ± se (n = 3). T78, T80, and T32 are the controls for CCR1-RNAi lines and T88, T88, and T132 are the controls for C4H1-RNAi lines. The controls for CCoAMT-RNAi lines are labeled as control, and the bars show average data from three clonally derived biological replicates. Letters indicate significant differences at the P ≤ 0.05 level. Asterisks indicate values that were determined by Student’s t test to be significantly different from their equivalent control (P < 0.05).
Discussion
Sequence-Based Classification of Potential Monolignol Biosynthetic Genes in Switchgrass.
Because of the complexity of the tetraploid switchgrass genome, classifying potential monolignol biosynthetic genes is challenging. Genome-wide EST/genome sequence-based classification and annotation have provided solid foundations for genetic studies of the gene families involved in lignin biosynthesis in Arabidopsis, maize, Populus, and Eucalyptus grandis (Raes et al., 2003; Barrière et al., 2007; Hamberger et al. 2007; Shi et al. 2010; Paiva et al., 2011). However, combined approaches including enzyme biochemistry, determination of tissue-specific expression patterns, forward genetic screening of mutants, and targeted reverse genetics by overexpression, antisense suppression, or RNAi are required to confirm functions in lignification. Although there are more than 10 million PviUTs from switchgrass (Alamo) and switchgrass (Kanlow) (Zhang et al., 2013), and more than 300 putative ESTs have been annotated as monolignol biosynthetic genes, only a few genes (4CL, COMT, CAD, and F5H) have been functionally characterized through reverse genetics in switchgrass. We have developed a gene function prediction system based on phylogenetic analysis, transcript level pattern analysis, and qRT-PCR validation. By this approach, we were able to identify, from 122,400 ESTs, 51 candidates from 10 gene families that are predicted to be involved in the monolignol biosynthesis pathway. The lignin-inducible suspension cell and the one internode-stem systems proved useful tools for such predictions. Among these predictions, half of the gene candidates have been proved correct by functional analysis. To confirm the remaining candidates, more genetic evidence through stable transformation of RNAi constructs with gene-specific or gene family–specific targets is needed.
Biological Systems for Studying Monolignol Biosynthesis in Switchgrass
In our switchgrass cell culture system, essentially nonlignified cells could be induced to lignify by supplying BL and reducing the 2,4-D concentration. No tracheary element differentiation or production of extracellular (i.e., released to the culture medium) lignins were observed in this system; in this respect, it behaved similarly to a previously described cell culture system in pine (Eberhardt et al., 1993) and is unlike other systems reported in zinnia (Fukuda and Komamine, 1982), pine (Nose et al., 1995), and Arabidopsis (Pauwels et al., 2008) in which deposition of cell wall–associated lignin was either accompanied by tracheary element differentiation or release of lignin to the extracellular medium. Compared with the stems, the induced suspension cells have lignin enriched in S units, but this lignin is, however, of a similar degree of polymerization to lignin isolated from whole-plant tissues. Lignin polymers rich in S units are also found in fiber cells. The genetic regulation of monolignol composition in different tissues is poorly understood; our system provides a tool for understanding the genetic and metabolic regulation of S-lignin biosynthesis that may possibly apply to fiber development in switchgrass tillers. However, it is important to bear in mind that the cell suspension lignin appears to be mostly deficient in G units, and switchgrass fibers do contain significant amounts of G-lignin. Therefore, we have used both the cell suspension system and the stem internode section system to make predictions. Maize cell culture systems have been used previously to study cell wall cross-linking through ferulic acid or p-coumaric acids (Grabber et al., 1995; Fry et al., 2000; Kerr and Fry, 2004; Grabber et al., 2009), and it will be interesting to pursue such features of cell wall biosynthesis in the switchgrass cell culture system.
The Routes for Monolignol Biosynthesis in Switchgrass
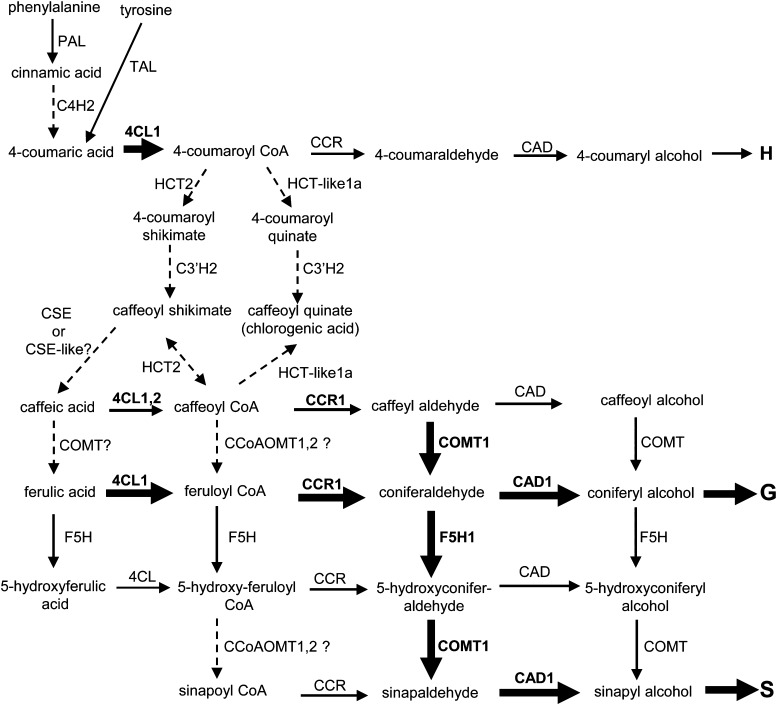
Based on previous publications and these studies, we can draw a pathway to monolignols in switchgrass that highlights both known and questionable features (Figure 9). The later steps of monolignol biosynthesis through CCR, COMT, F5H, and CAD appear to operate similarly in switchgrass to the dicot model. However, many of the early steps will need further evaluation (Figure 9).
Figure 9.
Proposed Monolignol Biosynthesis Pathway in Switchgrass Stems.
Black arrows show reactions that are likely to occur in vivo; thicker lines indicate the routes that are most strongly supported by both biochemical and genetic evidence. Gene names with specific numbers are from our current prediction (see Supplemental Data Set 2 online). C4H1, C3′H1, and HCT1 are not strongly supported for involvement in monolignol biosynthesis in stem according to their tissue expression patterns. Dotted lines are routes that need further confirmation with genetic evidence. This model is based on the results of this study and others (Escamilla-Treviño et al., 2010, 2013; Fu et al., 2011a, b; Saathoff et al., 2011; Vanholme et al., 2013; Xu et al., 2011).
4CL seems to play a critical role in the formation of hydroxycinnamyl CoAs for lignification in switchgrass, as in dicot models. Switchgrass 4CL enzymes have highest affinity toward 4-coumaric and ferulic acids, and RNAi-mediated downregulation of Pv4CL1 in switchgrass leads to a significant reduction in lignin content (Xu et al., 2011), consistent with the CoA activation step depicted in Figure 9.
According to the traditional route, 4-coumaroyl CoA is then converted to caffeoyl CoA by the shikimate/quinate shunt enzymes HCT/HQT and C3′H. These enzyme activities are observed in crude protein extracts of switchgrass internode samples (Escamilla-Treviño et al., 2013). Because our study identified no ESTs homologous to HQT, and the most obvious ortholog of dicot lignin pathway HCT has transcript-level patterns inconsistent with involvement in lignification in either the cell culture or internode systems, it is possible that the “shikimate/quinate shunt” pathway may operate differently (by alternative HCT or HCT-like genes) in switchgrass compared with dicots. There is significantly more caffeoyl quinate (chlorogenic acid) present in the bottom sections of one internode than in the middle or top sections in switchgrass (Shen et al., 2009b; Escamilla-Treviño et al., 2013), and this correlates with the expression pattern of HCT-like 1a in internode samples. In vitro enzyme kinetic studies of recombinant C3′H, HCT, and HCT-like1a enzymes have confirmed the potential for the operation of a shikimate shunt pathway in switchgrass. However, the formation of quinate esters is catalyzed by HCT-like proteins instead of traditional HQTs as found in dicots, and formation of caffeoyl CoA or caffeic acid from chlorogenic acid could not be demonstrated (Escamilla-Treviño et al., 2013) (Figure 9).
Several alternative routes have been proposed for conversion of 4-coumarate moieties to caffeate moieties in plants, although in most cases the biochemical data have yet to be supported by genetic studies. Zn2+-dependent direct conversion of 4-coumaroyl CoA to caffeoyl CoA (Kneusel et al., 1989), and 3-hydroxylation of 4-coumaraldehyde and 4-coumaryl alcohol (Weng et al., 2010) have yet to be explored in switchgrass. Other possible routes, for example, through hydroxylation of hydroxycinnamate glycosides (Tanaka and Kojima, 1991; Niggeweg et al., 2004), also require evaluation. A recent study in Arabidopsis suggests an alternative route from caffeoyl shikimate to caffeic acid catalyzed by a CSE (Vanholme et al. 2013). Switchgrass appears to possess at least three CSE genes, one of which is highly expressed in stem tissue along with high extractable enzyme activity (Escamilla-Treviño et al., 2013). However, it is not induced in lignifying cell cultures, and further genetic evidence will be necessary to confirm involvement of this enzyme in lignification in monocots. Switchgrass 4CL is less active with caffeic acid, but the kinetics (Xu et al., 2011) do not rule out a role in CoA activation of caffeate generated by a CSE (Vanholme et al., 2013).
Our transcript-level data argue against a direct involvement of CCoAOMT1 and CCoAOMT2 in lignin biosynthesis in switchgrass, although our enzyme assays from switchgrass stem and suspension cells indicate the conversion of caffeoyl CoA to feruloyl CoA in crude extracts. It is possible that more than one CCoAOMT or CCoAOMT-like gene is involved redundantly, although knockdown of both CCoAOMT1 and CCoAOMT2 by ∼90% results in no reduction in lignin content in switchgrass. Although an alternative route, from caffeoyl CoA to coniferaldehyde through CCR and COMT or CCoAOMT and CCR has been proposed in Medicago (Zhou et al., 2010), the downregulation of CCoAOMT in dicot species (alfalfa) still yields low lignin phenotypes (Guo et al., 2001a; Marita et al., 2003). Similar alternative routes may also exist in switchgrass (Figure 9) because COMT1 shows very high affinity for caffeyl aldehyde (Fu et al., 2011b), and CCR1 and CCR2 show relative high affinities for caffeoyl CoA (Escamilla-Trevino et al., 2010); if functional, these routes might dilute the contributions of CCoAOMT to lignin biosynthesis. Alternatively, it is possible that downregulation of CCoAOMT by greater than 90% is necessary to reveal a phenotype in switchgrass.
Although more and more EST, genomic, and transcriptomic data are becoming available for the plant science community, handling and interpreting these large-scale data sets can be challenging. It is also challenging to study complex pathways such as monolignol biosynthesis in a tetraploid, outcrossing species such as switchgrass. Our data show clear similarities, and also point to potential differences, between the pathways for monolignol synthesis in dicots and monocots. We cannot rule out the possibility that several homologous candidate genes may function redundantly at a single step, and work is in progress to address this possibility. In addition to the direct transgenic loss-of-function approach initiated in this work, computer modeling (Lee et al., 2012) and forward and reverse genetic approaches in simpler model systems such as rice and Brachypodium may also prove useful for elucidating the sequence of reactions in the pathway. It will be interesting to determine whether differences in the lignin biosynthetic pathway between monocots and dicots are associated with the different patterns of vascular development between these two classes, with the cylindrical arrangement of vascular bundles arising from the activity of a secondary vascular cambium in the dicots as compared with the scattering of the bundles throughout the stem in the monocots.
METHODS
Establishment and Induction of Switchgrass Cell Suspension Cultures
Embryogenic callus, formed from in vitro-developed inflorescences of nodal segments of lowland-type switchgrass (Panicum virgatum) cv Alamo (2n = 4× = 36), was used for generating cell suspension cultures as described previously (Mazarei et al., 2011). Alamo cell cultures were propagated and maintained in liquid culture medium (Murashige and Skoog medium + 9 µM 2,4-D + 4.4 µM 6-benzylaminopurine [6-BA]) every 7 to 10 d. Cultures were washed and resuspended in medium containing 0.2 µM BL, 0.9 µM 2,4-D, and 4.4 µM 6-BA to induce lignification. Aliquots of the cultures were harvested at 0 h, 6 h, 1 d, 3 d, and 7 d after transfer. Lignin formation was confirmed by staining a small aliquot of the culture with phloroglucinol-HCl and by performing thioacidolysis analysis of cell wall residues at 7 d postinduction.
Cell Imaging and Phloroglucinol-HCl staining
Switchgrass internode samples were collected in the greenhouse and immediately frozen in liquid nitrogen. Different internode sections were cut with a Leica CM 1850 cryostat (Leica Microsystems) at −20°C. Samples were then immersed in phloroglucinol solution (0.5% w/v in ethanol) for 1 min, then in 50% HCl for 2 min. Excess solution was wiped away with paper towels, and samples were transferred to 50% glycerol solution. Photographs were taken using a Nikon DXM 1200 color camera attached to a Nikon Microphot-FX microscope system with ACT-1 software (Nikon Instruments).
RNA Isolation and qRT-PCR
RNA isolation and real-time qRT-PCR were performed as described previously (Shen et al., 2012). qRT-PCR primers used for transcript-level validation are listed in Supplemental Table 1 online. Four and three biological replicates were used for qRT-PCR analysis of suspension cell samples and transgenic switchgrass lines, respectively. cDNA samples were diluted 20-fold, and 2 μL diluted samples were used as the qRT-PCR templates. PCRs were performed with Power SYBR Green PCR Master Mix (Life Technologies) in an optical 384-well plate with an ABI PRISM 7900 HT Sequence Detection System (Applied Biosystems).
Measurement of Lignin Content and Composition
Cell wall residues were prepared by sequentially extracting switchgrass suspension cells and whole tillers from transgenic plants with chloroform/methanol (1:1), 100% methanol, 50% methanol and water for three times each at room temperature. Fifteen (tiller samples) and 30 (suspension cells) mg of lyophilized samples were used for lignin analysis. The methods used for determining total lignin content by thioacidolysis followed by GC-MS were as described previously (Shen et al., 2009b).
GPC Analysis of Lignin
Switchgrass suspension cell samples were ball milled and then extracted with dioxane/water (9:1 v/v) for 24 h. The extracts were combined and the solvent removed with rotary evaporation under reduced pressure. The isolated suspension cell lignin sample was dissolved in a mixture of acetic anhydride/pyridine (1:1 v/v) and stirred at room temperature for 24 h. After acetylation, ethanol was added to the reaction mixture and the solvent removed with a rotary evaporator. The addition and removal of ethanol was repeated until all the solvents were removed from the sample. The acetylated lignin was dissolved in tetrahydrofuran, and GPC analysis was performed on a GPC SECurity 1200 system (Polymer Standards Service) featuring Agilent HPLC 1200 components equipped with four Waters Styragel columns (HR1, HR2, HR4, and HR6) and a UV detector (270 nm). Tetrahydrofuran was used as the mobile phase, and the flow rate was 1.0 mL/min. Standard narrow polystyrene samples were used for establishing the calibration curve. Data collection and processing were performed using Polymer Standards Service WinGPC Unity software (Build 6807).
Phylogenetic Analysis
All the phylogenies were reconstructed using Genenious (Biomatters) software with multiple sequence alignment and phylogeny tree building plugins. For the data in Figure 4, multiple protein sequence alignments (see Supplemental Data Set 3 online) were performed using the ClustalW alignment default settings. The phylogenetic tree was built by Geneious Pro 5.5.6 with PHYML (maximum likelihood version 2.0.10) plugin (Nuin et al., 2006) with the JTT model, 100 replicates of bootstrap analyses, estimated proportion of invariable sites, four rate categories, estimated gamma distribution parameter, and optimized tree topology, length, and substitution rate with best network network interface (NNI) and subtree pruning and regrafting (SPR) topology search. For all the phylogenetic trees in the supplementary figures, multiple protein sequence alignments (see Supplemental Data Set 4 online) were performed using the MAFFT version 7.017 program (Katoh et al., 2002), using the L-INS-i algorithm with BLOSUM62 scoring matrix at 1.53 gap open penalty and 0.123 offset value. The phylogenetic trees were built by Geneious 7.0.2 with PHYML (maximum likelihood version 2.2.0) plugin (Nuin et al., 2006) with the JTT model, 1,000 replicates of bootstrap analyses, estimated proportion of invariable sites, four rate categories, estimated gamma distribution parameter, and optimized tree topology, length, and substitution rate with best NNI and SPR topology search.
DNA Microarray Analysis
A custom-designed switchgrass cDNA chip Pvi_cDNAa520831 (Affymetrix) was designed based on PviUT version 1.2 sequences as described previously (Zhang et al., 2013). Total RNA for microarray analysis was isolated from induced and noninduced suspension cells using an RNeasy Mini kit according to the manufacturer’s protocol (Qiagen). RNA was cleaned and concentrated using the RNeasy MinElute Cleanup Kit (Qiagen), and 0.5 µg purified RNA was used for microarray analysis. Probe labeling, hybridization, and scanning were conducted according to the manufacturer’s instructions with the IVT Express Kit (Affymetrix). The raw microarray data from the Gene Atlas and suspension cells were normalized and analyzed together. Data normalization between chips was conducted using Robust Multichip Average (Irizarry et al., 2003). Because there were no mismatch probes to gauge the reliability of the hybridization signal, only genes with maximum expression level greater than 100 were kept for further analysis. At this expression level, the majority of genes were considered to be above the background cutoff of 32. If all the signal readings from the different plant tissues and suspension cell samples were less than 100, the hybridization signal was considered too low and the candidate was discarded (these were usually short sequences). The raw and processed microarray data for suspension cells and single internode samples have been integrated into the Switchgrass Gene Expression Atlas (Zhang et al., 2013) at the Switchgrass Functional Genomics Server (http://switchgrassgenomics.noble.org).
Gene Cloning
Primers were designed according to the switchgrass EST database (hosted at http://switchgrassgenomics.noble.org/blast/blast.php) to clone the predicted genes. The open reading frames were amplified by pfu PCR with KOD Hot Start DNA Polymerase (EMD Chemicals). Stem cDNAs synthesized from total RNA using Superscript III reverse transcriptase (Invitrogen) were used as template. The PCR products were gel-purified using a Qiagen Gel Extraction Kit (Qiagen), cloned using the pENTR/D-TOPO Cloning Kit (Invitrogen), and transformed into E. coli strain DH5α competent cells for sequencing. All primers used for cloning are listed in Supplemental Table 1 online.
Switchgrass Tissue Culture and Transformation
Agrobacterium tumefaciens–mediated switchgrass transformation was performed based on a previously published protocol (Xi et al., 2009). pANIC vector (Mann et al., 2012) was used for switchgrass transformation. Young switchgrass inflorescences from the ST2 line (cv Alamo) were used as explants for stable transformation. The ST2 line was specifically selected for its high tissue culture response. Because the ST2 line is vegetatively propagated by tillers, all the lines are in the same genetic background. This is an important point for a highly heterozygous outcrossing species.
Determination of Monolignol Pathway Enzyme Activities
The substrates or products of the 4CL, HCT, and CCoAOMT enzymes, namely, 4-coumaroyl CoA, caffeoyl CoA, and feruloyl CoA, were synthesized as described previously (Stöckigt and Zenk, 1975). 5-Hydroxyconiferaldehyde (COMT substrate) was synthesized as described previously (Chen et al., 2001), whereas sinapaldehyde, 4-coumaric acid, CoA, S-(5′-adenosyl)-l-methionine chloride dihydrochloride, shikimic acid, and quinic acid were purchased from Sigma-Aldrich. Dithiothreitol was purchased from Roche, and 4-coumaroyl shikimate was kindly provided by Dr. John Ralph (University of Wisconsin-Madison).
From 0.13 to 0.25 g switchgrass internode tissue or 0.5 to 0.8 g cell culture was ground very finely using a freezing mill, and the protein extract prepared as described previously (Gallego-Giraldo et al., 2011), to prepare crude protein extracts for enzyme activity assay.
The enzyme activities assayed were HCT/HQT, using 4-coumaroyl CoA as substrate with either shikimic acid or quinic acid as acyl acceptor to generate 4-coumaroyl shikimate or 4-coumaroyl quinate, respectively; 4CL, to produce 4-coumaroyl CoA from 4-coumaric acid and CoA; CCoAOMT, using caffeoyl CoA to form feruloyl CoA; and COMT, using 5-hydroxyconiferaldehyde as substrate generating sinapaldehyde as product.
For determination of HCT activity using shikimic acid as acyl acceptor, 4.2 to 10.3 µg crude protein extracts were incubated at 30°C for 15 min with 100 mM sodium phosphate buffer pH 7.5, 500 µM shikimic acid, 500 µM dithiothreitol, and 60 µM of the acyl donor substrate (4-coumaroyl CoA) in a final volume of 100 µL. Similarly, for determination of HQT activity using quinic acid, 8.4 to 21.5 µg protein was incubated at 30°C for 30 min using the same buffer and dithiothreitol concentration with 500 µM quinic acid and 100 µM 4-coumaroyl CoA in a final volume of 100 µL. The reactions were stopped by adding 10 μL glacial acetic acid, and products were injected onto an HPLC with a reverse-phase C18 column (Spherisorb 5µ ODS2; Waters) and separated in a step gradient using 1% phosphoric acid in water as solvent A and acetonitrile as solvent B. To quantify products, we measured peak areas and converted them to units of quantity using calibration curves that were constructed with authentic 4-coumaroyl shikimate standard. Because 4-coumaroyl quinate is not commercially available, the same 4-coumaroyl shikimate standard was used for quantification of HQT activities.
For determination of 4CL activity, crude protein extracts (4.2–10.3 µg) were incubated at 30°C for 30 min with 100 mM Tris-HCl buffer pH 7.5, 3 mM MgCl2, 5 mM ATP, 500 µM CoA, and 60 µM 4-coumaric acid in a final volume of 100 µL. The reactions were terminated by adding 10 μL glacial acetic acid. Reaction products were analyzed by HPLC as described earlier. Calibration curves were constructed with an authentic standard of 4-coumaroyl CoA.
For determination of CCoAOMT activity, 4.2 to 10.3 µg of crude protein extracts was incubated at 30°C for 30 min with 100 mM sodium phosphate buffer pH 7.5, 0.6 mM MgCl2, 2 mM dithiothreitol, 0.2 mM S-(5′-adenosyl)-l-methionine chloride dihydrochloride, and 60 µM caffeoyl CoA in a final volume of 50 µL. For COMT activity assays, 8.4 to 21.5 µg crude protein extracts was incubated at 30°C for 30 min with 100 mM sodium phosphate buffer pH 7.2, 0.6 mM MgCl2, 2 mM dithiothreitol, 1 mM S-(5′-adenosyl)-l-methionine chloride dihydrochloride, and 0.5 mM 5-hydroxyconiferaldehyde in a final volume of 50 µL. The reactions were stopped by adding 5 μL glacial acetic acid, and products were analyzed by HPLC as described earlier. Feruloyl CoA was used to construct the calibration curve for CCoAOMT activity assays, and sinapaldehyde was used to construct the calibration curve for COMT activity assays.
Statistical Analysis
Statistical analysis was done by Student's t test (Microsoft Office Excel 2007). The multiple comparisons were done by Duncan grouping with the SAS 9.3 software (SAS Institute).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers for switchgrass genes: JX845715, JX845715, and JX845715 (PAL1), JX845712 (C4H1), JX845713 (C4H2), AB723823 (C3′H1a), AB723824 (C3′H2a), AB723825 (CCoAOMT1a), JX845716 (CCoAOMT1b), AB723826 (CCoAOMT2a), JX845717 (CCoAOMT2b), AB723827 and AB723827 (HCT1a), KC696573 (HCT2a), JX845714 and JX845714 (HCT-Like1a), AB608019 and AB608019 (F5H1). Arabidopsis gene accession numbers are: At2g37040 (PAL1), At3g53260 (PAL2), At2g30490 (C4H), At1g51680 (4CL1), At3g21240 (4CL2), At1g65060 (4CL3), At2g40890 (C3′H1), At5g48930 (HCT), At4g36220 (F5H1), At5g54160 (COMT1), At4g34050 (CCoAOMT1), At1g15950 (CCR1), At3g19450 (CAD4), At4g34230 (CAD5), and At4g39330 (CAD9).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Possible Monolignol Biosynthesis Pathways in Plants.
Supplemental Figure 2. Establishment of the Switchgrass Suspension Cell System.
Supplemental Figure 3. Optimizing Hormone Concentrations for Lignin Induction in Switchgrass Cell Suspension Cultures.
Supplemental Figure 4. Phylogenetic Analysis of Different Monolignol Biosynthetic Gene Groups from Figure 4.
Supplemental Figure 5. Phylogenetic Analysis of Switchgrass Putative C4H Proteins.
Supplemental Figure 6. CLUSTAL Sequence Alignment for C4H Proteins.
Supplemental Figure 7. Phylogenetic Analysis of Switchgrass C3′H and C3′H-Like Proteins.
Supplemental Figure 8. Phylogenetic Analysis of HCT and HCT-Like Proteins.
Supplemental Figure 9. CLUSTAL Sequence Alignment for HCT and HCT-Like Proteins.
Supplemental Figure 10. Phylogenetic Analysis of Switchgrass Putative CCoAOMT Proteins.
Supplemental Figure 11. CLUSTAL Sequence Alignment for CCoAOMT Proteins.
Supplemental Figure 12. Analysis of Putative CSE/CSE-Like Genes in Switchgrass.
Supplemental Figure 13. Transgene Constructs and Stable Transformation of Switchgrass for Functional Characterization of Putative Monolignol Biosynthetic Genes.
Supplemental Table 1. Sequences of the Primers Used in This Work.
Supplemental Data Set 1. Three Hundred and Thirty-One Putative Monolignol Biosynthesis PviUTs in Switchgrass.
Supplemental Data Set 2. Predicted Switchgrass Monolignol Biosynthetic Genes with Their Arabidopsis Orthologues (111 accessions).
Supplemental Data Set 3. CLUSTAL Sequence Alignment for Proteins Listed in Figure 4.
Supplemental Data Set 4. MAFFT Sequence Alignment for Proteins Listed in the Supplemental Figures.
Acknowledgments
This work was supported by the BioEnergy Sciences Center, a U.S. Department of Energy Bioenergy Research Center, through the Office of Biological and Environmental Research in the Department of Energy Office of Science. We thank Yanbin Yin, Jiyi Zhang, Yinbin Ge, and Nick Krom for excellent assistance with EST sequence annotation and bioinformatics; Jin Nakashima for assistance with cell imaging; Mohamed Bedair for assistance with GC-MS analysis; Tui Ray for technical support with qRT-PCR analysis; Debra Mohnen and Ivana Gelineo-Albersheim for helpful discussions concerning inducible cell cultures; and Mingyi Wang and Xiaolan Rao for critical reading of the article.
AUTHOR CONTRIBUTIONS
H.S. and R.A.D. designed the research; H.S., M.M., H.H., L.E.-T., C.F., Y.P., M.R.R., Y.T., X.X., L.J., G.L., T.H., and F.C. performed research; H.S., Y.T., A.J.R., C.N.S., Z.-Y. W. and R.A.D. analyzed data; H.S. and R.A.D. wrote the article.
References
- Abdulrazzak N, et al. (2006). A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 140: 30–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R., Tollier M.T., Monties B. (1990). The role of auxin and gibberellin in controlling lignin formation in primary phloem fibers and in xylem of Coleus blumei stems. Plant Physiol. 94: 1743–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.R., Zein I., Wenzel G., Darnhofer B., Eder J., Ouzunova M., Lübberstedt T. (2008). Characterization of phenylpropanoid pathway genes within European maize (Zea mays L.) inbreds. BMC Plant Biol. 8: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrière Y., Riboulet C., Méchin V., Maltese S., Pichon M., Cardinal A., Lapierre C., Lübberstedt T., Martinant J.P. (2007). Genetics and genomics of lignification in grass cell walls based on maize as model species. Genes Genomes Genomics 1: 133–156 [Google Scholar]
- Baucher M., Van Montagu M., Boerjan W. (2000). Improvement of wood quality for the pulp and paper industry by genetic modification of lignin biosynthesis in poplar. Dev. Plant. Genet. Breed. 5: 215–221 [Google Scholar]
- Baucher M., Halpin C., Petit-Conil M., Boerjan W. (2003). Lignin: Genetic engineering and impact on pulping. Crit. Rev. Biochem. Mol. Biol. 38: 305–350 [DOI] [PubMed] [Google Scholar]
- Blee K., Choi J.W., O’Connell A.P., Jupe S.C., Schuch W., Lewis N.G., Bolwell G.P. (2001). Antisense and sense expression of cDNA coding for CYP73A15, a class II cinnamate 4-hydroxylase, leads to a delayed and reduced production of lignin in tobacco. Phytochemistry 57: 1159–1166 [DOI] [PubMed] [Google Scholar]
- Bouton J.H. (2007). Molecular breeding of switchgrass for use as a biofuel crop. Curr. Opin. Genet. Dev. 17: 553–558 [DOI] [PubMed] [Google Scholar]
- Chen F., Kota P., Blount J.W., Dixon R.A. (2001). Chemical syntheses of caffeoyl and 5-OH coniferyl aldehydes and alcohols and determination of lignin O-methyltransferase activities in dicot and monocot species. Phytochemistry 58: 1035–1042 [DOI] [PubMed] [Google Scholar]
- Chen F., Dixon R.A. (2007). Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen F., Srinivasa Reddy M.S., Temple S., Jackson L., Shadle G.L., Dixon R.A. (2006). Multi-site genetic modulation of monolignol biosynthesis suggests new routes for formation of syringyl lignin and wall-bound ferulic acid in alfalfa (Medicago sativa L.). Plant J. 48: 113–124 [DOI] [PubMed] [Google Scholar]
- Chen H.-C., Li Q., Shuford C.M., Liu J., Muddiman D.C., Sederoff R.R., Chiang V.L. (2011). Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc. Natl. Acad. Sci. USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison B.H., Drescher S.R., Tuskan G.A., Davis M.F., Nghiem N.P. (2006). Variation of S/G ratio and lignin content in a Populus family influences the release of xylose by dilute acid hydrolysis. Appl. Biochem. Biotechnol. 129–132: 427–435 [DOI] [PubMed] [Google Scholar]
- Dien B.S., Jung H.-J.G., Vogel K.P., Casler M.D., Lamb J.F.S., Iten L., Mitchell R.B., Sarath G. (2006). Chemical composition and response to dilute-acid pretreatment and enzymatic saccharification of alfalfa, reed canarygrass, and switchgrass. Biomass Bioenergy 30: 880–891 [Google Scholar]
- Dien B.S., Sarath G., Pedersen J.F., Sattler S.E., Chen H., Funnell-Harris D.L., Nichols N.N., Cotta M.A. (2009). Improved sugar conversion and ethanol yield for forage sorghum (Sorghum bicolor L. Moench) lines with reduced lignin contents. Bioenergy Res. 2: 153–164 [Google Scholar]
- Ding S.-Y., Liu Y.-S., Zeng Y., Himmel M.E., Baker J.O., Bayer E.A. (2012). How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 338: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Do C.-T., Pollet B., Thévenin J., Sibout R., Denoue D., Barrière Y., Lapierre C., Jouanin L. (2007). Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Eberhardt T.L., Bernards M.A., He L., Davin L.B., Wooten J.B., Lewis N.G. (1993). Lignification in cell suspension cultures of Pinus taeda. In situ characterization of a gymnosperm lignin. J. Biol. Chem. 268: 21088–21096 [PubMed] [Google Scholar]
- Escamilla-Treviño L.L., Shen H., Uppalapati S.R., Ray T., Tang Y., Hernandez T., Yin Y., Xu Y., Dixon R.A. (2010). Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties. New Phytol. 185: 143–155 [DOI] [PubMed] [Google Scholar]
- Escamilla-Treviño L.L., Shen H., Hernandez T., Yin Y.-B., Xu Y., Dixon R.A. (November 5, 2013). Early lignin pathway enzymes and routes to chlorogenic acid in switchgrass (Panicum virgatum L.). Plant Mol. Biol. doi/10.1007/s11103-013-1252-y [DOI] [PubMed] [Google Scholar]
- Eudes A., Pollet B., Sibout R., Do C.T., Séguin A., Lapierre C., Jouanin L. (2006). Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225: 23–39 [DOI] [PubMed] [Google Scholar]
- Franke R., Humphreys J.M., Hemm M.R., Denault J.W., Ruegger M.O., Cusumano J.C., Chapple C. (2002). The Arabidopsis REF8 gene encodes the 3-hydroxylase of phenylpropanoid metabolism. Plant J. 30: 33–45 [DOI] [PubMed] [Google Scholar]
- Fritz J.O., Moore K.J., Jaster E.H. (1990). Digestion kinetics and cell wall composition of brown midrib sorghum X sudangrass morphological components. Crop Sci. 30: 213–219 [Google Scholar]
- Fry S.C., Willis S.C., Paterson A.E. (2000). Intraprotoplasmic and wall-localised formation of arabinoxylan-bound diferulates and larger ferulate coupling-products in maize cell-suspension cultures. Planta 211: 679–692 [DOI] [PubMed] [Google Scholar]
- Fu C., Xiao X., Xi Y., Ge Y., Chen F., Bouton J., Dixon R.A., Wang Z.Y. (2011a). Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. Bioenergy Res. 4: 153–164 [Google Scholar]
- Fu C., Mielenz J.R., Xiao X., Ge Y., Hamilton C.Y., Chen F., Bouton J., Foston M., Dixon R.A., Wang Z.-Y. (2011b). Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc. Natl. Acad. Sci. USA 108: 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda H., Komamine A. (1982). Lignin synthesis and its related enzymes as markers of tracheary-element differentiation in single cells isolated from the mesophyll of Zinnia elegans. Planta 155: 423–430 [DOI] [PubMed] [Google Scholar]
- Gallego-Giraldo L., Escamilla-Trevino L., Jackson L.A., Dixon R.A. (2011). Salicylic acid mediates the reduced growth of lignin down-regulated plants. Proc. Natl. Acad. Sci. USA 108: 20814–20819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon T., Sibout R., Pollet B., Maba B., Nussaume L., Bechtold N., Lu F., Ralph J., Mila I., Barrière Y., Lapierre C., Jouanin L. (2003). A new Arabidopsis thaliana mutant deficient in the expression of O-methyltransferase impacts lignins and sinapoyl esters. Plant Mol. Biol. 51: 973–989 [DOI] [PubMed] [Google Scholar]
- Grabber J.H., Hatfield R.D., Ralph J., Zoń J., Amrhein N. (1995). Ferulate cross-linking in cell walls isolated from maize cell suspensions. Phytochemistry 40: 1077–1082 [Google Scholar]
- Grabber J.H., Mertens D.R., Kim H., Funk C., Lu F., Ralph J. (2009). Cell wall fermentation kinetics are impacted more by lignin content and ferulate cross-linking than by lignin composition. J. Sci. Food Agric. 89: 122–129 [Google Scholar]
- Guo D., Chen F., Inoue K., Blount J.W., Dixon R.A. (2001a). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa. impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13: 73–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D., Chen F., Wheeler J., Winder J., Selman S., Peterson M., Dixon R.A. (2001b). Improvement of in-rumen digestibility of alfalfa forage by genetic manipulation of lignin O-methyltransferases. Transgenic Res. 10: 457–464 [DOI] [PubMed] [Google Scholar]
- Halpin C. (2004). Investigating and manipulating lignin biosynthesis in the postgenomic era. Adv. Bot. Res. 41: 63–106 [Google Scholar]
- Hamberger B., Ellis M., Friedmann M., de Azevedo Souza C., Barbazuk B., Douglas C.J. (2007). Genome-wide analyses of phenylpropanoid-related genes in Populus trichocarpa, Arabidopsis thaliana, and Oryza sativa: The Populus lignin toolbox and conservation and diversification of angiosperm gene families. Can. J. Bot. 85: 1182–1201 [Google Scholar]
- Hardin C.F., Fu C., Hisano H., Xiao X., Shen H., Stewart C.N., Jr, Parrott W., Dixon R.A., Wang Z.-Y. (2013). Standardization of switchgrass sample collection for cell wall and biomass trait analysis. Bioenergy Res. 6: 755–762 [Google Scholar]
- Harrison C.J., Langdale J.A. (2006). A step by step guide to phylogeny reconstruction. Plant J. 45: 561–572 [DOI] [PubMed] [Google Scholar]
- Himmel M.E., Ding S.Y., Johnson D.K., Adney W.S., Nimlos M.R., Brady J.W., Foust T.D. (2007). Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315: 804–807 [DOI] [PubMed] [Google Scholar]
- Hoffmann L., Besseau S., Geoffroy P., Ritzenthaler C., Meyer D., Lapierre C., Pollet B., Legrand M. (2004). Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 16: 1446–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W.-J., Harding S.A., Lung J., Popko J.L., Ralph J., Stokke D.D., Tsai C.-J., Chiang V.L. (1999). Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat. Biotechnol. 17: 808–812 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jones L., Ennos A.R., Turner S.R. (2001). Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis. Plant J. 26: 205–216 [DOI] [PubMed] [Google Scholar]
- Joshi C.P., Chiang V.L. (1998). Conserved sequence motifs in plant S-adenosyl-L-methionine-dependent methyltransferases. Plant Mol. Biol. 37: 663–674 [DOI] [PubMed] [Google Scholar]
- Kärkönen A., Koutaniemi S. (2010). Lignin biosynthesis studies in plant tissue cultures. J. Integr. Plant Biol. 52: 176–185 [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr E.M., Fry S.C. (2004). Extracellular cross-linking of xylan and xyloglucan in maize cell-suspension cultures: The role of oxidative phenolic coupling. Planta 219: 73–83 [DOI] [PubMed] [Google Scholar]
- Kim I.A., Kim B.G., Kim M., Ahn J.H. (2012). Characterization of hydroxycinnamoyltransferase from rice and its application for biological synthesis of hydroxycinnamoyl glycerols. Phytochemistry 76: 25–31 [DOI] [PubMed] [Google Scholar]
- Kim S.J., Kim M.R., Bedgar D.L., Moinuddin S.G.A., Cardenas C.L., Davin L.B., Kang C.H., Lewis N.G. (2004). Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneusel R.E., Matern U., Nicolay K. (1989). Formation of trans-caffeoyl-CoA from trans-4-coumaroyl-CoA by Zn2+-dependent enzymes in cultured plant cells and its activation by an elicitor-induced pH shift. Arch. Biochem. Biophys. 269: 455–462 [DOI] [PubMed] [Google Scholar]
- Lapierre C., Monties B., Rolando C. (1985). Thioacidolysis of lignin: Comparison with acidolysis. J. Wood Chem. Technol. 5: 277–292 [Google Scholar]
- Lawrence C.J., Walbot V. (2007). Translational genomics for bioenergy production from fuelstock grasses: Maize as the model species. Plant Cell 19: 2091–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Meyer K., Chapple C., Douglas C.J. (1997). Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Escamilla-Treviño L., Dixon R.A., Voit E.O. (2012). Functional analysis of metabolic channeling and regulation in lignin biosynthesis: A computational approach. PLoS Comput. Biol. 8: e1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Weng J.-K., Chapple C. (2008). Improvement of biomass through lignin modification. Plant J. 54: 569–581 [DOI] [PubMed] [Google Scholar]
- Mann D.G.J., Lafayette P.R., Abercrombie L.L., King Z.R., Mazarei M., Halter M.C., Poovaiah C.R., Baxter H., Shen H., Dixon R.A., Parrott W.A., Neal Stewart C., Jr (2012). Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10: 226–236 [DOI] [PubMed] [Google Scholar]
- Marita J.M., Ralph J., Hatfield R.D., Guo D., Chen F., Dixon R.A. (2003). Structural and compositional modifications in lignin of transgenic alfalfa down-regulated in caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase. Phytochemistry 62: 53–65 [DOI] [PubMed] [Google Scholar]
- Mazarei M., Al-Ahmad H., Rudis M.R., Joyce B.L., Stewart C.N., Jr (2011). Switchgrass (Panicum virgatum L.) cell suspension cultures: Establishment, characterization, and application. Plant Sci. 181: 712–715 [DOI] [PubMed] [Google Scholar]
- Meyer K., Shirley A.M., Cusumano J.C., Bell-Lelong D.A., Chapple C. (1998). Lignin monomer composition is determined by the expression of a cytochrome P450-dependent monooxygenase in Arabidopsis. Proc. Natl. Acad. Sci. USA 95: 6619–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K.J., Moser L.E., Vogel K.P., Waller S.S., Johnson B.E., Pedersen J.F. (1991). Describing and quantifying growth stages of perennial forage grasses. Agron. J. 83: 1073–1077 [Google Scholar]
- Nedelkina S., Jupe S.C., Blee K.A., Schalk M., Werck-Reichhart D., Bolwell G.P. (1999). Novel characteristics and regulation of a divergent cinnamate 4-hydroxylase (CYP73A15) from French bean: Engineering expression in yeast. Plant Mol. Biol. 39: 1079–1090 [DOI] [PubMed] [Google Scholar]
- Niggeweg R., Michael A.J., Martin C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22: 746–754 [DOI] [PubMed] [Google Scholar]
- Nose M., Bernards M.A., Furlan M., Zajicek J., Eberhardt T.L., Lewis N.G. (1995). Towards the specification of consecutive steps in macromolecular lignin assembly. Phytochemistry 39: 71–79 [DOI] [PubMed] [Google Scholar]
- Nuin P.A., Wang Z., Tillier E.R. (2006). The accuracy of several multiple sequence alignment programs for proteins. BMC Bioinformatics 7: 471 http://www.biomedcentral.com/1471–2105/7/471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Lanzatella C., Saha M.C., Bouton J., Wu R., Tobias C.M. (2010). Complete switchgrass genetic maps reveal subgenome collinearity, preferential pairing and multilocus interactions. Genetics 185: 745–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva J.A, et al. (2011). Advancing Eucalyptus genomics: Identification and sequencing of lignin biosynthesis genes from deep-coverage BAC libraries. BMC Genomics 12: 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly M., Keegstra K. (2008). Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 54: 559–568 [DOI] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomar F., Merino F., Barceló A.R. (2002). O-4-Linked coniferyl and sinapyl aldehydes in lignifying cell walls are the main targets of the Wiesner (phloroglucinol-HCl) reaction. Protoplasma 220: 17–28 [DOI] [PubMed] [Google Scholar]
- Raes J., Rohde A., Christensen J.H., Van de Peer Y., Boerjan W. (2003). Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 133: 1051–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston L., Kwon S.T., Schoenbeck M., Ralston J., Schenk D.J., Coates R.M., Chappell J. (2001). Cloning, heterologous expression, and functional characterization of 5-epi-aristolochene-1,3-dihydroxylase from tobacco (Nicotiana tabacum). Arch. Biochem. Biophys. 393: 222–235 [DOI] [PubMed] [Google Scholar]
- Rohde A., et al. (2004). Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16: 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saathoff A.J., Sarath G., Chow E.K., Dien B.S., Tobias C.M. (2011). Downregulation of cinnamyl-alcohol dehydrogenase in switchgrass by RNA silencing results in enhanced glucose release after cellulase treatment. PLoS ONE 6: e16416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed A.I., et al. (2003). TM4: A free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Sánchez-Rodríguez C., Rubio-Somoza I., Sibout R., Persson S. (2010). Phytohormones and the cell wall in Arabidopsis during seedling growth. Trends Plant Sci. 15: 291–301 [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.-K., Humphreys J., Ruegger M.O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Schmer M.R., Vogel K.P., Mitchell R.B., Perrin R.K. (2008). Net energy of cellulosic ethanol from switchgrass. Proc. Natl. Acad. Sci. USA 105: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt V.J.H., Ni W., Blount J.W., Jung H.G., Masoud S.A., Howles P.A., Lamb C., Dixon R.A. (1997). Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol. 115: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Yin Y., Chen F., Xu Y., Dixon R. (2009a). A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenergy Res. 2: 217–232 [Google Scholar]
- Shen H., Fu C., Xiao X., Ray T., Tang Y., Wang Z., Chen F. (2009b). Developmental control of lignification in stems of lowland switchgrass variety Alamo and the effects on saccharification efficiency. Bioenergy Res. 2: 233–245 [Google Scholar]
- Shen H., He X., Poovaiah C.R., Wuddineh W.A., Ma J., Mann D.G.J., Wang H., Jackson L., Tang Y., Stewart C.N., Jr, Chen F., Dixon R.A. (2012). Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 193: 121–136 [DOI] [PubMed] [Google Scholar]
- Shen H., et al. (2013). Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol. Biofuels 6: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Sun Y.-H., Li Q., Heber S., Sederoff R., Chiang V.L. (2010). Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 51: 144–163 [DOI] [PubMed] [Google Scholar]
- Sibout R., Eudes A., Mouille G., Pollet B., Lapierre C., Jouanin L., Séguin A. (2005). CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons B.A., Loqué D., Ralph J. (2010). Advances in modifying lignin for enhanced biofuel production. Curr. Opin. Plant Biol. 13: 313–320 [DOI] [PubMed] [Google Scholar]
- Sonnante G., D’Amore R., Blanco E., Pierri C.L., De Palma M., Luo J., Tucci M., Martin C. (2010). Novel hydroxycinnamoyl-coenzyme A quinate transferase genes from artichoke are involved in the synthesis of chlorogenic acid. Plant Physiol. 153: 1224–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A.C., Palanichelvam K., Ma Y., Steele J., Blancaflor E.B., Tang Y. (2010). Collection and analysis of expressed sequence tags derived from laser capture microdissected switchgrass (Panicum virgatum L. Alamo) vascular tissues. Bioenergy Res. 3: 278–294 [Google Scholar]
- Stöckigt J., Zenk M.H. (1975). Chemical syntheses and properties of hydroxycinnamoyl-coenzyme A derivatives. Z. Naturforsch. C. 30: 352–358 [DOI] [PubMed] [Google Scholar]
- Suzuki R., Shimodaira H. (2006). Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Tanaka M., Kojima M. (1991). Purification and characterization of p-coumaroyl-D-glucose hydroxylase of sweet potato (Ipomoea batatas) roots. Arch. Biochem. Biophys. 284: 151–157 [DOI] [PubMed] [Google Scholar]
- Tobias C.M., Sarath G., Twigg P., Lindquist E., Pangilinan J., Penning B.W., Barry K., McCann M.C., Carpita N.C., Lazo G.R. (2008). Comparative genomics in switchgrass using 61,585 high-quality expressed sequence tags. Plant Genome 1: 111–124 [Google Scholar]
- Trupiano D., Di Iorio A., Montagnoli A., Lasserre B., Rocco M., Grosso A., Scaloni A., Marra M., Chiatante D., Scippa G.S. (2012). Involvement of lignin and hormones in the response of woody poplar taproots to mechanical stress. Physiol. Plant. 146: 39–52 [DOI] [PubMed] [Google Scholar]
- Vanholme R., Morreel K., Ralph J., Boerjan W. (2008). Lignin engineering. Curr. Opin. Plant Biol. 11: 278–285 [DOI] [PubMed] [Google Scholar]
- Vanholme R., et al. (2013). Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vignols F., Rigau J., Torres M.A., Capellades M., Puigdomènech P. (1995). The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J.K., Akiyama T., Bonawitz N.D., Li X., Ralph J., Chapple C. (2010). Convergent evolution of syringyl lignin biosynthesis via distinct pathways in the lycophyte Selaginella and flowering plants. Plant Cell 22: 1033–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers S., Lu F., Kim H., Zhu Y., Ralph J., Wilkerson C.G. (2012). Identification of grass-specific enzyme that acylates monolignols with p-coumarate. J. Biol. Chem. 287: 8347–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Ge Y., Wang Z.Y. (2009). Genetic transformation of switchgrass. Methods Mol. Biol. 581: 53–59 [DOI] [PubMed] [Google Scholar]
- Xu B., Escamilla-Treviño L.L., Sathitsuksanoh N., Shen Z., Shen H., Zhang Y.H., Dixon R.A., Zhao B. (2011). Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol. 192: 611–625 [DOI] [PubMed] [Google Scholar]
- Yang J., Chen F., Yu O., Beachy R.N. (2011). Controlled silencing of 4-coumarate:CoA ligase alters lignocellulose composition without affecting stem growth. Plant Physiol. Biochem. 49: 103–109 [DOI] [PubMed] [Google Scholar]
- Zhang J.-Y., et al. (2013). Development of an integrated transcript sequence database and a gene expression atlas for gene discovery and analysis in switchgrass (Panicum virgatum L.). Plant J. 74: 160–173 [DOI] [PubMed] [Google Scholar]
- Zhao H., Sheng Q., Lü S., Wang T., Song Y. (2004). Characterization of three rice CCoAOMT genes. Chin. Sci. Bull. 49: 1602–1606 [Google Scholar]
- Zhou R., Jackson L., Shadle G., Nakashima J., Temple S., Chen F., Dixon R.A. (2010). Distinct cinnamoyl CoA reductases involved in parallel routes to lignin in Medicago truncatula. Proc. Natl. Acad. Sci. USA 107: 17803–17808 [DOI] [PMC free article] [PubMed] [Google Scholar]