Short-term cold stress delays flowering by activating the floral repressor FLOWERING LOCUS C (FLC). This study shows that the cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1) acts as a chromatin remodeling factor for FLC regulation under short-term cold stress by antagonizing the actions of FVE and its interacting partner histone deacetylase 6.
Abstract
Exposure to short-term cold stress delays flowering by activating the floral repressor FLOWERING LOCUS C (FLC) in Arabidopsis thaliana. The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1) negatively regulates cold responses. Notably, HOS1-deficient mutants exhibit early flowering, and FLC expression is suppressed in the mutants. However, it remains unknown how HOS1 regulates FLC expression. Here, we show that HOS1 induces FLC expression by antagonizing the actions of FVE and its interacting partner histone deacetylase 6 (HDA6) under short-term cold stress. HOS1 binds to FLC chromatin in an FVE-dependent manner, and FVE is essential for the HOS1-mediated activation of FLC transcription. HOS1 also interacts with HDA6 and inhibits the binding of HDA6 to FLC chromatin. Intermittent cold treatments induce FLC expression by activating HOS1, which attenuates the activity of HDA6 in silencing FLC chromatin, and the effects of intermittent cold are diminished in hos1 and fve mutants. These observations indicate that HOS1 acts as a chromatin remodeling factor for FLC regulation under short-term cold stress.
INTRODUCTION
Seasonal fluctuations in ambient temperature affect various aspects of plant growth and development. In particular, low temperatures significantly limit plant propagation, reproduction, and geographical distribution. Therefore, plants have evolved a variety of adaptive mechanisms to increase freezing tolerance after exposure to low but nonfreezing temperatures, which is known as cold acclimation (Chinnusamy et al., 2007; Zhu et al., 2007).
During the cold acclimation process, a group of genes encoding transcriptional regulators, such as C-repeat/dehydration-responsive element binding factors (CBFs), is induced, and the encoded proteins activate downstream genes that trigger cold responses. Cold induction of the CBF genes is mediated by INDUCER OF CBF EXPRESSION1 (ICE1), a cold-activated MYC transcription factor (Chinnusamy et al., 2003; Lee et al., 2005). Cold responses occur within minutes, and plants are cold acclimated within a few days after exposure to cold temperatures (Sung and Amasino, 2005; Medina et al., 2011).
Floral transition is profoundly affected in the process of cold acclimation (Chinnusamy et al., 2007; Zhu et al., 2007). The effects of cold stress on flowering time have been investigated more precisely by intermittent cold treatments, in which plants are exposed to 4°C for several hours during the day until flowering (Kim et al., 2004). Intermittent cold treatments delay flowering and induce FLOWERING LOCUS C (FLC) gene expression by approximately threefold (Kim et al., 2004; Seo et al., 2009). By contrast, prolonged exposure to cold temperatures promotes flowering, which is known as vernalization, by suppressing the FLC gene (Sung and Amasino, 2005; Kim et al., 2009). The short-term cold stress response would provide an adaptive strategy that prevents precocious flowering under short-term temperature fluctuations, which often occur during early spring and late autumn. In this regard, the cold stress response apparently differs from vernalization, which mediates a prolonged cold effect to promote flowering.
Consistent with the role of FLC in flowering initiation under cold stress, the flowering of FLC-deficient mutants is not influenced by intermittent cold treatments (Seo et al., 2009). It has recently been shown that cold signaling components that mediate the cold acclimation response, such as ICE1 and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (HOS1), are not involved in vernalization (Bond et al., 2011; Lazaro et al., 2012). In addition, transgenic plants overexpressing CBF genes exhibit late flowering with moderate induction of FLC expression (Gilmour et al., 2004; Seo et al., 2009), supporting the signaling linkage between cold response and flowering time.
The effects of cold stress on flowering initiation are also mediated by FVE, which belongs to the autonomous flowering pathway. FVE negatively regulates the expression of the FLC gene and several cold-responsive genes in Arabidopsis thaliana (Kim et al., 2004). Consequently, loss-of-function fve mutants show enhanced freezing tolerance and late flowering, and flowering is not delayed by intermittent cold treatments in the mutants (Ausín et al., 2004; Kim et al., 2004).
The FVE protein is an Arabidopsis homolog of the human retinoblastoma-associated protein 46/48 that constitutes histone deacetylase (HDAC) corepressor complexes (Ausín et al., 2004; Gu et al., 2011). FVE induces histone deacetylation and transcriptional silencing of various genetic loci, including FLC, which are targeted by small interfering RNAs and antisense RNAs associated with HDA6 (Gu et al., 2011; Pazhouhandeh et al., 2011; To et al., 2011). FVE also interacts with a chromatin repressive complex, which is similar to Polycomb repressive complex2 (PRC2), to modulate the deposition of the H3K27 trimethylation (H3K27Me3) that acts as a repressive chromatin mark in FLC repression (Pazhouhandeh et al., 2011). Therefore, FVE appears to mediate cold stress signals in the regulation of FLC expression by forming multiprotein complexes with various chromatin remodeling factors.
The Arabidopsis REALLY INTERESTING NEW GENE finger E3 ligase HOS1 is a negative regulator of cold-responsive genes, such as CBF and COLD-REGULATED genes (Ishitani et al., 1998; Lee et al., 2001; Dong et al., 2006). Under cold stress, HOS1 triggers the degradation of the ICE1 transcription factor, a direct upstream activator of the CBF3 gene (Chinnusamy et al., 2003; Dong et al., 2006). Although the inactivation of the HOS1 gene blocks cold-induced degradation of ICE1, the overexpression of HOS1 accelerates ICE1 degradation even at normal temperatures (Dong et al., 2006). Interestingly, loss-of-function hos1 mutants exhibit early flowering, and FLC expression is reduced in the mutants (Ishitani et al., 1998; Lee et al., 2001). It is therefore envisioned that HOS1 plays a role in cold regulation of flowering in an FLC-dependent manner. Notably, recent studies have shown that HOS1 regulates flowering time in response to ambient temperature changes and intermittent cold treatments (Jung et al., 2012a; Lee et al., 2012). However, it remains largely unknown how HOS1 regulates FLC expression.
In this article, we demonstrate that HOS1 regulates FLC transcription at the chromatin level, under cold stress, through interactions with FVE and HDA6. HOS1 binds to FLC chromatin and inhibits the association of HDA6 with FLC chromatin, resulting in FLC induction and delayed flowering. However, HOS1 does not degrade either HDA6 or FVE. Our data support the notion that HOS1 plays a role in the modulation of FLC chromatin and provide an insight into how cold acclimation response is linked with Arabidopsis flowering time control.
RESULTS
HOS1 Regulates FLC Transcription in Arabidopsis Flowering
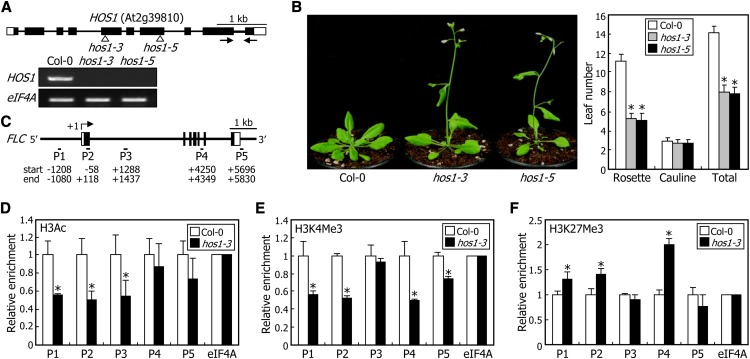
To elucidate the molecular mechanisms by which HOS1 regulates FLC expression, we isolated two mutant alleles of the HOS1 gene in Columbia-0 (Col-0) background (Figure 1A). Both the hos1-3 and hos1-5 loss-of-function mutants exhibited early flowering under long days (LDs) (Figure 1B), as observed with the hos1-1 mutant in C24 background (Ishitani et al., 1998; Lee et al., 2001). The loss-of-function mutants initiated flowering at a total leaf number of 8, whereas Col-0 plants initiated flowering at a total leaf number of 14 under our assay conditions. In addition, FLC expression was reduced in the mutants but elevated approximately twofold in HOS1-overexpressing transgenic plants (see Supplemental Figure 1 online), showing that HOS1 is correlated with FLC expression.
Figure 1.
HOS1 Regulates FLC Transcription at the Chromatin Level.
(A) Isolation of hos1 mutants. In the hos1-3 and hos1-5 mutants, one copy of the T-DNA element is inserted in the 5th and 7th exons of the HOS1 gene, respectively (top panel). Black boxes represent exons, and white boxes represent untranslated regions. Black arrows indicate the primer pair used for RT-PCR. HOS1 expression was examined by RT-PCR using total RNA samples extracted from 10-d-old whole plants grown on MS-agar plates (bottom panel). The eIF4A gene was used as the RNA quality control.
(B) Flowering times of hos1 mutants. Five-week-old plants grown in soil under LDs were photographed (left panel). The leaf numbers of 20 plants were averaged for each plant genotype and statistically treated using a Student’s t test (*P < 0.01) (right panel). Bars indicate se.
(C) Structure of the FLC gene. The sequence regions marked by P1 to P5 indicate regions examined in the ChIP assays. Numbers indicate residue positions relative to the transcription start site. Black boxes indicate exons, and white boxes denote untranslated regions.
(D) to (F) Relative levels of histone modifications in FLC chromatin. ChIP assays were performed using anti-H3Ac (D), anti-H3K4 trimethylation (E), and anti-H3K27Me3 (F) antibodies. Plants grown on MS-agar plates for 10 d under LDs were used for chromatin preparation. An eIF4A DNA fragment was used for normalization. Four measurements were averaged for each plant genotype and statistically treated using a Student’s t test (*P < 0.01). Bars indicate se.
[See online article for color version of this figure.]
FLC expression is epigenetically regulated in response to various endogenous and environmental cues (He, 2009; Jarillo et al., 2009). We therefore performed chromatin immunoprecipitation (ChIP) assays on FLC chromatin using five sets of primer pairs covering the FLC genomic DNA (Figure 1C). It was found that H3 acetylation (H3Ac) and H3K4 trimethylation, which are marks for active gene expression (He, 2009; Jarillo et al., 2009), were reduced in the hos1-3 mutant (Figures 1D and 1E). By contrast, H3K27Me3, which is a repressive chromatin mark (He, 2009; Jarillo et al., 2009), was elevated in the mutant (Figure 1F), which is consistent with the low-level expression of FLC in the mutant. These observations suggest that HOS1 regulates FLC transcription at the chromatin level.
HOS1 Binding to FLC Chromatin Is Induced by Cold Stress
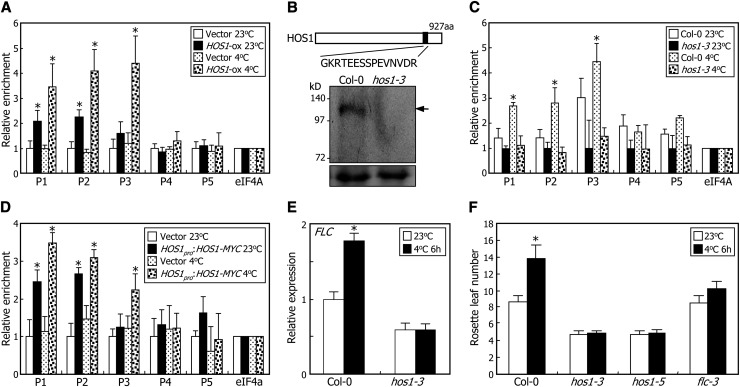
Our data suggested that HOS1 would bind directly to FLC chromatin. To examine this, we performed ChIP assays with or without cold treatments, using transgenic plants overexpressing the HOS1-MYC gene fusion (HOS1-ox), in which the MYC-coding sequence was fused in frame to the 3′ end of the HOS1 gene and driven by the cauliflower mosaic virus (CaMV) 35S promoter. The results showed that HOS1 binds to FLC chromatin primarily in the P1, P2, and P3 sequence regions and HOS1 binding is dramatically elevated at 4°C (Figure 2A).
Figure 2.
Chromatin Binding of HOS1 Induces FLC Expression under Cold Stress.
For the ChIP assays, biological triplicates were averaged for each measurement and statistically treated using a Student’s t test (*P < 0.01). Bars indicate se.
(A) ChIP assays of HOS1 binding to FLC chromatin using MYC-specific antibody. ChIP assays were conducted as described in Figure 1, using an anti-MYC antibody. The HOS1-ox transgenic plants grown on MS-agar plates at 23°C for 10 d were further grown at either 23°C or 4°C for 2 d before harvesting plant materials. The genomic DNA sequences examined in the ChIP assays were identical to those described in Figure 1C. Transgenic plants harboring the 6xMYC-pBA vector alone were used as control plants.
(B) Schematic illustration of anti-HOS1 antibody production. A peptide region covering residues 796 to 810 of HOS1 protein was selected for the epitope (black box). Nuclear proteins extracted from Col-0 plants and hos1-3 mutants were used for examination of anti-HOS1 antibody specificity. HOS1 proteins were immunologically detected using the anti-HOS1 antibody. Arrow indicates endogenous HOS1 protein. Parts of Coomassie blue–stained gels are shown at the bottom as loading control. aa, amino acids.
(C) ChIP assays of HOS1 binding to FLC chromatin using HOS1-specific antibody. ChIP assays were conducted as described in Figure 1, using an anti-HOS1 antibody. Chromatin extracts from Col-0 and hos1-3 plants were used for the ChIP assays. The conditions for the cold treatments were identical to those described in (A).
(D) ChIP assays using HOS1pro:HOS1-MYC transgenic plants. ChIP assays were performed using an anti-MYC antibody as described in Figure 1. The conditions for the cold treatments were identical to those described in (A). Transgenic plants harboring the 6xMYC-pBA vector were used as control plants.
(E) Effects of intermittent cold on FLC expression. Plants treated with intermittent cold for 6 h at dawn for 15 d after germination were used for extraction of total RNA. Levels of FLC mRNA were determined by qRT-PCR. Biological triplicates were averaged for each plant genotype and statistically treated using a Student’s t test (*P < 0.01). Bars indicate se.
(F) Effects of intermittent cold on flowering time. Plants were treated with intermittent cold for 6 h at dawn, each day until flowering. Rosette leaves of 15 plants were counted for each plant genotype and statistically treated using a Student’s t test (*P < 0.01). Bars indicate se.
Because the ectopic expression of the HOS1 gene may cause potential artifacts, we generated a HOS1-specific antibody for the ChIP assays. The epitope for the antibody is a peptide sequence that differs from any other sequence of annotated Arabidopsis proteins (Figure 2B). To verify the specificity of the anti-HOS1 antibody, we performed protein gel blot analysis of the HOS1-ox transgenic plants. Transgenic plants overexpressing the MYC-FCA gene fusion (FCA-ox) were used as control (Jung et al., 2012b). Using the anti-MYC antibody, HOS1 and FCA proteins were detected in the transgenic plants. However, protein gel blot analysis using the anti-HOS1 antibody showed that HOS1 proteins were detected in both the HOS1-ox and FCA-ox transgenic plants, whereas FCA proteins were not detected (see Supplemental Figure 2 and Supplemental References 1 online). In addition, HOS1 proteins were not detected in the hos1-3 plants (Figure 2B), showing that the anti-HOS1 antibody detects specifically HOS1 proteins.
Next, we conducted ChIP assays using the HOS1-specific antibody with cold-treated plants. The results showed that the HOS1 proteins were enriched in the P1, P2, and P3 sequence regions in the cold-treated plants (Figure 2C), as observed in Figure 2A. To further examine the HOS1 binding to FLC chromatin, the MYC-coding sequence was fused in frame to the 3′end of the HOS1 gene with the native promoter sequence consisting of ∼2.8 kb upstream of the transcription start site (Lee et al., 2001), and the fusion construct was transformed into Col-0 plants. ChIP–quantitative PCR (qPCR) assays of the transgenic plants revealed that HOS1 binding to FLC chromatin was detectably elevated in the cold-treated plants (Figure 2D), confirming that HOS1 binds directly to FLC chromatin in planta.
We also examined whether the cold induction of HOS1 binding to FLC chromatin is associated with FLC expression. The hos1-3 mutant was treated with intermittent cold, and the FLC expression was analyzed by quantitative real-time RT-PCR (qRT-PCR). Although FLC expression was induced in Col-0 plants after intermittent cold treatments, the inductive effects of intermittent cold disappeared in the hos1-3 mutant (Figure 2E). Consistent with this observation, flowering initiation was not delayed in the hos1-3 and hos1-5 mutants treated with intermittent cold, as observed in the null allele of the FLC gene (Figure 2F; Seo et al., 2009; Lee et al., 2012). These observations indicate that the HOS1 binding to FLC chromatin induces FLC expression under short-term cold stress.
HOS1 Interacts with FVE in the Nucleus
We next determined how HOS1 regulates FLC expression. HOS1 is an E3 ubiquitin ligase that triggers the degradation of the ICE1 protein through the 26S proteasome pathway (Dong et al., 2006). Therefore, one possibility is that HOS1 targets the negative regulator(s) of FLC expression.
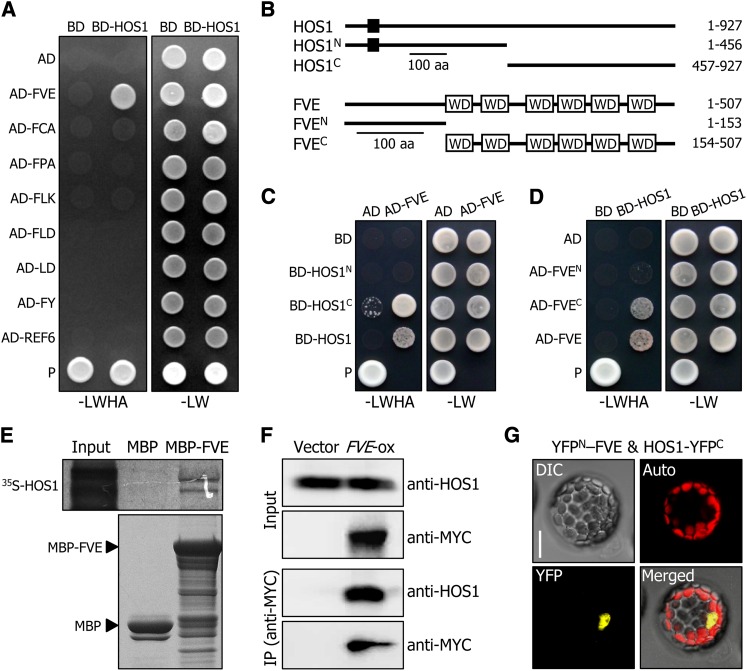
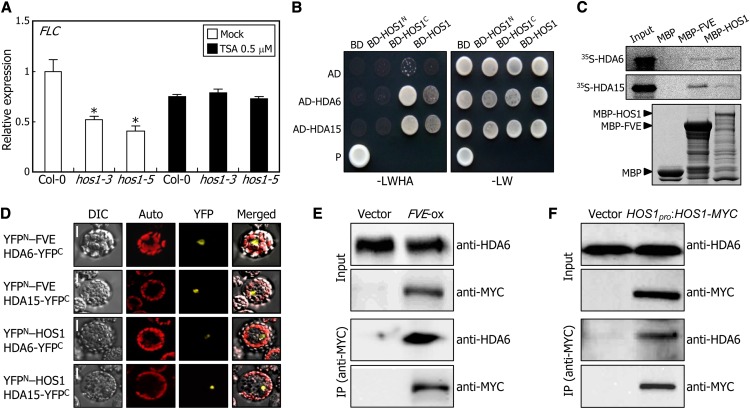
We first examined whether HOS1 interacts with the components of the autonomous flowering pathway that negatively regulate FLC expression (He, 2009; Jarillo et al., 2009). Yeast two-hybrid assays showed that HOS1 interacts with FVE (Figure 3A). By contrast, no obvious interactions were detected between HOS1 and other components. To determine the protein domains responsible for the HOS1–FVE interactions, we generated several deletions of the HOS1 and FVE proteins (Figure 3B), and the deletions were coexpressed in Saccharomyces cerevisiae AH109 cells. We found that the C-terminal region of the HOS1 protein, covering residues 457 to 927, is responsible for the interaction between HOS1 and ICE1 (Dong et al., 2006), and the C-terminal region of the FVE protein (residues 154 to 507), containing a series of WD40 domains, mediates the HOS1–FVE interaction (Figures 3C and 3D). The HOS1-FVE interaction was also verified by in vitro pull-down assays using a recombinant MBP-FVE fusion protein prepared in Escherichia coli cells and using in vitro–translated 35S-labeled HOS1 polypeptides (Figure 3E).
Figure 3.
HOS1 Interacts with FVE in the Nucleus.
(A) Interactions of HOS1 with FVE in yeast cells. Autonomous flowering pathway genes and the HOS1 gene were fused in frame to the GAL4 activation domain (AD)–coding and GAL4 DNA binding domain (BD)–coding sequences, respectively. The HOS1–FVE interactions were assayed by cell growth on selective medium. –LWHA indicates Leu, Trp, His, and Ade dropout plates. –LW indicates Leu and Trp dropout plates. P, positive control.
(B) Deletion constructs of HOS1 and FVE. Numbers indicate residue positions. Black box, REALLY INTERESTING NEW GENE finger domain. WD, WD40 domain. aa, amino acids.
(C) and (D) Interactions of HOS1 with FVE in yeast cells. The HOS1 (C) or FVE (D) constructs were fused with GAL4 BD or GAL4 AD, respectively. Yeast two-hybrid assays were performed as described in (A).
(E) In vitro pull-down assays. Recombinant MBP-FVE fusion and in vitro–translated 35S-labeled HOS1 polypeptides were used (top panel). Part of a Coomassie blue–stained gel is shown (bottom panel). The input represents 5% of the radiolabeled protein used in the assays.
(F) Coimmunoprecipitation assays. Total proteins were extracted from transgenic plants overexpressing a FVE-MYC gene fusion (FVE-ox) grown on MS-agar plates for 6 d. Protein complexes were immunoprecipitated using an anti-MYC antibody and detected immunologically using an anti-MYC or anti-HOS1 antibody. Transgenic plants harboring the 6xMYC-pBA vector alone were used as control plants.
(G) BiFC assays. The YFPN-FVE and HOS1-YFPC constructs were coexpressed transiently in Arabidopsis protoplasts and visualized by differential interference contrast microscopy and fluorescence microscopy. Bar = 20 μm.
To test whether HOS1 interacts with FVE in vivo, coimmunoprecipitation assays were performed using transgenic plants overexpressing a FVE-MYC gene fusion (FVE-ox), in which the MYC-coding sequence was fused in frame to the 3′ end of the FVE gene under the control of the CaMV 35S promoter. Total protein extracts were immunoprecipitated using an anti-MYC antibody, and the precipitated protein complexes were analyzed immunologically using an anti-MYC or anti-HOS1 antibody. Whereas a band having the expected size of the HOS1 protein was detected in the protein extracts of the FVE-ox transgenic plants, no such band was detected in the extracts of control plants (Figure 3F), indicating that HOS1 interacts with FVE in vivo.
Bimolecular fluorescence complementation (BiFC) assays were also employed to examine the HOS1-FVE interaction in planta. The HOS1 gene sequence fused in frame to the 5′ end of a gene sequence encoding the C-terminal half of yellow fluorescent protein (HOS1-YFPC) and the FVE gene sequence fused in frame to the 3′ end of a gene sequence encoding the N-terminal half of YFP (YFPN-FVE) were transiently coexpressed in Arabidopsis protoplasts. It was found that HOS1 interacts with FVE in the nucleus (Figure 3G), where chromatin modifications occur (He, 2009; Jarillo et al., 2009).
FVE interacts with DAMAGED DNA BINDING1 (DDB1) proteins, which are core subunits of the E3 ligase complex containing CULLIN4 (CUL4), via the Trp-Asp-X-Arg motif in the C-terminal region of FVE (Pazhouhandeh et al., 2011). We therefore examined whether the Trp-Asp-X-Arg motif is required for the HOS1–FVE interaction. The Arg (R) residue in the Trp-Asp-X-Arg motif was mutated to a His (H) residue (R323H) (see Supplemental Figure 3A online), a mutation known to strongly reduce the FVE–DDB1 interaction (Pazhouhandeh et al., 2011). Yeast two-hybrid assays showed that the R323H mutation does not disrupt the HOS1–FVE interaction (see Supplemental Figure 3B online), indicating that the HOS1–FVE interaction is structurally different from the FVE–DDB1 interaction.
HOS1 Regulation of FLC Chromatin Is Mediated by FVE
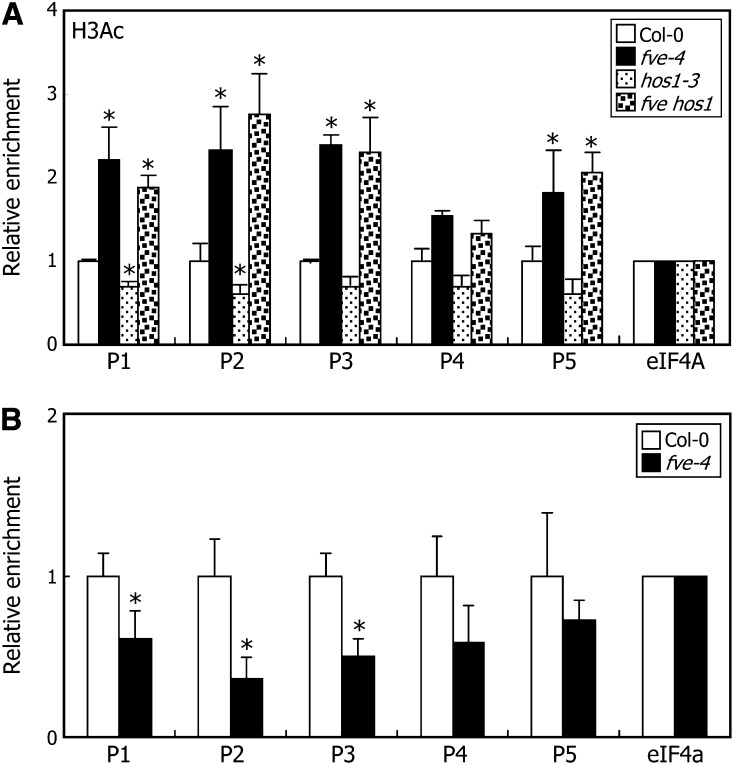
FVE regulates FLC expression at the chromatin level (Ausín et al., 2004; Pazhouhandeh et al., 2011). We found that HOS1 interacts with FVE and regulates FLC expression by binding to FLC chromatin. To study how HOS1 is functionally linked with FVE in the chromatin regulation of FLC expression, we crossed the early-flowering hos1-3 mutant with the late-flowering FVE-deficient fve-4 mutant to produce an fve hos1 double mutant and examined the flowering phenotypes of the double mutants. The fve hos1 double mutant flowered later than the hos1-3 mutant (see Supplemental Figures 4A and 4B online). Consistent with the flowering phenotypes, the levels of FLC mRNA were significantly higher in the double mutant than in the hos1-3 mutant (see Supplemental Figure 4C online). In addition, ChIP assays of FLC chromatin revealed that the level of H3Ac in the double mutant was similar to that in the fve-4 mutant but markedly higher than that in the hos1-3 mutant (Figure 4A). These observations show that the HOS1-mediated transcriptional regulation of FLC expression is at least in part mediated by FVE.
Figure 4.
HOS1 Regulation of FLC Transcription Requires FVE.
Whole plant materials grown on MS-agar plates at 23°C under LDs for 10 d were used for ChIP assays. Biological quadruplicates were averaged for each measurement and statistically treated using a Student\x{2019}s t test (*P < 0.01). Bars indicate se.
(A) Relative levels of H3Ac in FLC chromatin. ChIP assays were conducted using an anti-H3Ac antibody.
(B) Binding of HOS1 to FLC chromatin in the fve-4 mutant. ChIP assays were conducted using an anti-HOS1 antibody.
We next examined whether HOS1 binding to FLC chromatin is influenced by FVE. ChIP-qPCR assays using an anti-HOS1 antibody revealed that HOS1 binding to FLC chromatin is reduced by ∼50% in the fve-4 mutant (Figure 4B), indicating that FVE is required for the efficient binding of HOS1 to FLC chromatin.
The level of FLC mRNA in the fve-4 mutant was comparable to that in the fve hos1 double mutant (see Supplemental Figure 4C online). However, the double mutant flowered earlier than the fve-4 mutant (see Supplemental Figure 4B online), suggesting that HOS1 regulation of flowering time does not entirely depend on FLC gene expression (see Discussion).
FVE Is Not Ubiquitinated by HOS1
Under cold stress, nuclear-localized HOS1 proteins ubiquitinates the ICE1 transcription factor, which is then degraded through the proteasome-mediated pathway (Dong et al., 2006). We found that HOS1 interacts with FVE, suggesting that HOS1 may ubiquitinate FVE.
We first examined whether the stability of the FVE protein is altered by cold treatments. The FVE-ox transgenic plants were exposed to 4°C, and total protein extracts were prepared for protein gel blot analysis. It was found that the levels of FVE protein were uninfluenced by cold treatments (see Supplemental Figure 5A online). We next performed in vivo ubiquitination assays. The ICE1 protein was also included in the assays for comparison. Although a high molecular weight polypeptide band corresponding to the polyubiquitinated form of the ICE1-MYC fusion was detected in the cold-treated transgenic plants overexpressing the ICE1-MYC gene fusion, no polyubiquitinated forms of the FVE-MYC fusion was detected in the cold-treated FVE-ox transgenic plants (see Supplemental Figure 5B online). These observations indicate that FVE is not ubiquitinated by HOS1 under cold stress.
FVE binds to FLC chromatin (Jeon and Kim, 2011; Pazhouhandeh et al., 2011). We therefore performed ChIP assays to examine whether cold treatments influence the binding of FVE to FLC chromatin. Col-0 plants and FVE-ox transgenic plants were cold treated, and chromatin preparations were subjected to ChIP assays using the primer pairs employed in the ChIP assays to study the binding of HOS1 to FLC chromatin (Figure 1C). We found that the cold treatments did not affect the FVE binding to FLC chromatin (see Supplemental Figure 5C online), showing that the binding of FVE to FLC chromatin was not influenced by HOS1-mediated cold stress signals.
HOS1 Interacts with HDA6 in the Nucleus
FVE acts as an integral component of multiple chromatin-modifying complexes that mediate transcriptional repression (Ausín et al., 2004; Gu et al., 2011). FVE associates with HDA6 to form protein complexes that bind to FLC chromatin, leading to transcriptional silencing by histone deacetylation (Gu et al., 2011). Our data indicate that HOS1 binding to FLC chromatin is mediated by FVE. We therefore postulated that HOS1 interacts with HDA6 and other HDAC enzymes to modulate their association with FLC chromatin.
We first employed trichostatin A (TSA), a potent inhibitor of class I and II mammalian HDACs (Taunton et al., 1996; Richon et al., 2000), to examine whether HDAC activity is related to the HOS1 regulation of FLC expression. The levels of FLC mRNA were examined in plants grown for 10 d on one-half-strength Murashige and Skoog (MS)–agar plates containing 0.5 μM TSA. Notably, the low level of FLC expression in the hos1 mutants was recovered to a level comparable to that observed in Col-0 plants after TSA treatments (Figure 5A), indicating that the HDACs are involved in the HOS1-mediated activation of FLC expression.
Figure 5.
HOS1 Interacts with HDA6 in the Nucleus.
(A) Effects of TSA on FLC expression. Plants were grown for 10 d on MS-agar plates containing 0.5 μM TSA. The levels of FLC mRNA were determined by qRT-PCR. Biological triplicates were averaged for each plant genotype and statistically treated (Student’s t test, *P < 0.01). Bars indicate se.
(B) Interactions of HOS1 with HDA6 and HDA15 in yeast cells. The HDA6 and HDA15 genes were fused in frame to the GAL4 AD-coding sequence. The HOS1 constructs used were as described in Figure 3B.
(C) In vitro pull-down assays. Recombinant MBP-FVE and MBP-HOS1 fusions as well as in vitro–translated, 35S-labeled HDA6 and HDA15 polypeptides were used (top and middle panels, respectively). Part of a Coomassie blue–stained gel is shown (bottom panel).
(D) BiFC assays. Partial YFP fusion constructs were coexpressed transiently in Arabidopsis protoplasts and visualized by differential interference contrast microscopy (DIC) and fluorescence microscopy. Bars = 20 μm.
(E) and (F) in vivo interaction of HDA6 with FVE (E) and HOS1 (F). Total proteins extracted from the FVE-ox and HOS1pro:HOS1-MYC transgenic plants grown for 6 d were immunoprecipitated with anti-MYC agarose beads. The FVE, HOS1, and HDA6 proteins were detected immunologically using an anti-MYC or anti-HDA6 antibody.
We next performed yeast two-hybrid assays to examine whether HOS1 interacts directly with HDAC proteins. Of the HDACs assayed, HDA6 and HDA15 interacted strongly with both FVE and HOS1 (see Supplemental Figures 6A and 6B online). The assays using truncated HOS1 forms revealed that the HOS1–HDAC interactions occurred via the C-terminal region of HOS1 (Figure 5B), like the HOS1–FVE interaction (Figures 3B and 3C). HDA5 and HDA9 also interacted moderately with FVE and HOS1 in yeast cells (see Supplemental Figures 6A and 6B online).
The interactions of HDA6 and HDA15 with FVE and HOS1 were also examined by in vitro pull-down assays using in vitro–translated, 35S-labeled HDAC polypeptides and recombinant MBP-FVE or MBP-HOS1 fusion proteins. HDA6 and HDA15 interacted with both FVE and HOS1 (Figure 5C), consistent with the results obtained with yeast cells. We conducted BiFC assays using Arabidopsis protoplasts to examine whether the interactions occur in vivo. Strong reconstituted YFP signals were detected in the nuclei of cells coexpressing the YFPN-FVE and HDAC-YFPC fusions as well as the YFPN-HOS1 and HDAC-YFPC fusions (Figure 5D), demonstrating that the interactions of HDA6 and HDA15 with FVE and HOS1 occur in the nucleus.
Coimmunoprecipitation assays were also employed to confirm the in vivo interactions of FVE and HOS1 with HDA6 using total proteins extracted from FVE-ox or HOS1pro:HOS1-MYC transgenic plants. We found that HDA6 proteins were immunoprecipitated by both FVE- and HOS1-MYC proteins coupled with an anti-MYC antibody (Figures 6E and 6F). However, no HDA6 protein was immunoprecipitated from total proteins extracted from control plants expressing only MYC. These observations demonstrate that both FVE and HOS1 physically interact with HDA6 in vivo.
Figure 6.
Cold Stress Attenuates Binding of HDA6 to FLC Chromatin.
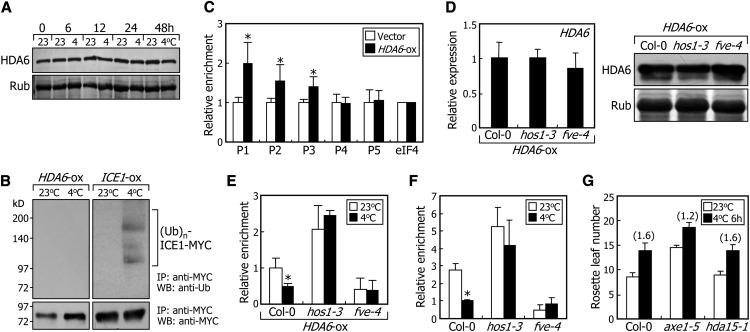
(A) Effects of cold stress on HDA6 protein stability. Ten-day-old transgenic plants overexpressing a HDA6-MYC gene fusion under the control of the CaMV 35S promoter (HDA6-ox) were exposed to 4°C for the indicated time. Whole plants were harvested for preparation of protein extracts. The levels of HDA6 were compared immunologically using an anti-MYC antibody. Parts of Coomassie blue–stained gels containing the ribulose-1,5-bis-phosphate carboxylase/oxygenase protein are shown at the bottom.
(B) Effects of cold stress on HDA6 ubiquitination. Ten-day-old HDA6-ox transgenic plants were used in the ubiquitination assays. Total protein was immunoprecipitated with an anti-MYC antibody and analyzed immunologically using anti-MYC and anti-ubiquitin antibodies. Transgenic plants overexpressing the ICE1-MYC gene fusion (ICE1-ox) were also analyzed for comparison. IP, immunoprecipitation assay. WB, protein gel blot analysis.
(C) Binding of HDA6 to FLC chromatin. The HDA6-ox transgenic plants were grown on MS-agar plates for 2 weeks before harvesting of plant materials. ChIP assays were performed using an anti-MYC antibody, as described in Figure 1. Biological triplicates were averaged and statistically treated using Student’s t test (*P < 0.01). Bars indicate se.
(D) Relative levels of HDA6 protein and HDA6 mRNA in HDA6-ox transgenic plants in different genetic backgrounds. The mRNA levels were examined by qRT-PCRs (left panel). Biological triplicates were averaged and statistically treated (t test, *P < 0.01). Bars indicate se. The levels of HDA6 were determined immunologically using an anti-MYC antibody (right panel).
(E) Effects of cold stress on HDA6 binding to FLC chromatin. ChIP assays were performed as described in Figure 1, using an anti-MYC antibody. Chromatin extracts from plant materials described in (D) were used. Ten-day-old plants grown on MS-agar plates at 23°C were further grown at either 23 or 4°C for 2 d before harvesting of plant materials. The P1 sequence region of the FLC chromatin, as shown in Figure 1C, was used in the ChIP assays. Biological triplicates were averaged and statistically treated (t test, *P < 0.01). Bars indicate se.
(F) Binding of HDA6 to FLC chromatin in the hos1-3 and fve-4 mutants. ChIP assays were performed as described in Figure 1, using an anti-HDA6 antibody. Chromatin extracts from Col-0 plants and hos1-3 and fve-4 mutants were used. Conditions for cold treatments were identical to those described in (E). Biological triplicates were averaged and statistically treated (t test, *P < 0.01). Bars indicate se.
(G) Effects of intermittent cold on flowering time. Intermittent cold treatments and measurements of flowering times were performed as described in Figure 2. The numbers in parentheses refer to the ratios of rosette leaf numbers with and without intermittent cold treatments.
HOS1-Mediated Cold Stress Signals Inhibit HDA6 Binding to FLC Chromatin
We found that HOS1 interacts with HDA6, an interacting partner of FVE in the repression of the FLC gene (Gu et al., 2011). Therefore, a question was whether HOS1 ubiquitinates HDA6. We examined whether HDA6 is degraded by the HOS1-mediated ubiquitination pathway using transgenic plants overexpressing HDA6-MYC gene fusions (HDA6-ox). The transgenic plants were exposed to 4°C, and the levels of HDA6-MYC fusion protein were analyzed immunologically using an anti-MYC antibody. The results showed that the levels of HDA6 protein were not influenced by cold treatments (Figure 6A). In addition, no visible ubiquitination of HDA6 was detected under identical conditions (Figure 6B), indicating that HOS1-mediated cold stress signals do not influence the stability of the HDA6 protein.
Both FVE and HDA6 bind to FLC chromatin (Jeon and Kim, 2011; Pazhouhandeh et al., 2011). We found that HOS1 also binds directly to FLC chromatin and interacts with FVE and HDA6. Therefore, one possibility was that HOS1 interfered with the binding of HDA6 to FLC chromatin. ChIP assays using the HDA6-ox transgenic plants revealed that HDA6 binds to FLC chromatin in the P1, P2, and P3 sequence regions of FLC genomic DNA (Figure 6C), to which HOS1 also binds (Figures 2A, 2C, and 2D), suggesting that HOS1 may compete with HDA6 for binding to FLC chromatin.
To examine this possibility, we transformed the HDA6-MYC gene fusion into Col-0 plants and the hos1-3 and fve-4 mutants. We identified several HDA6-ox transgenic plants suitable for ChIP assays, in which the levels of HDA6 mRNA and HDA6 protein in both the mutant backgrounds were similar to those in Col-0 background (Figure 6D, left and right panels, respectively).
We next conducted ChIP assays of the binding of HDA6 to FLC chromatin using the HDA6-ox transgenic plants in different genetic backgrounds. At normal temperatures, the HDA6 binding to FLC chromatin was lower in the fve-4 background but higher in the hos1-3 background compared with that in Col-0 background (Figure 6E). These results are consistent with the high expression of FLC and delayed flowering in the fve-4 mutant as well as the low expression of FLC and accelerated flowering in the hos1 mutant (see Supplemental Figure 4C online). Under cold stress, the association of HDA6 with FLC chromatin decreased by ∼50% in Col-0 background (Figure 6E). This result is consistent with the elevated FLC expression and delayed flowering under cold stress (Figures 2E and 2F). However, the effects of cold stress on the HDA6 binding to FLC chromatin were largely dampened in both the hos1-3 and fve-4 mutant backgrounds. ChIP assays using a HDA6-specific antibody also showed that the HDA6 binding to FLC chromatin was not affected by cold treatments in the hos1-3 and fve-4 mutant backgrounds (To et al., 2011; Figure 6F).
ChIP assays of the binding of HDA6 to FLC chromatin were also performed using chromatin extracts prepared from HOS1-ox transgenic plants. The HDA6 binding to FLC chromatin was reduced by ∼50% in the HOS1-ox transgenic plants (see Supplemental Figure 7 online). By contrast, the binding of HOS1 to FLC chromatin was unaffected in the axe1-5 mutant (see Supplemental Figure 8 online). Altogether, these observations unequivocally demonstrate that cold stress signals influence the HDA6 binding to FLC chromatin via FVE and HOS1. It is also evident that HOS1 suppresses the HDA6 binding to FLC chromatin
Our data suggested that HDA6 is involved in the regulation of flowering time via HOS1-mediated cold signaling. To examine this, the axe1-5 mutant was treated with intermittent cold until flowering. Intermittent cold treatments delayed flowering by ∼60% in Col-0 plants but only 20% in the axe1-5 mutant (Figure 6G), showing that the effects of cold stress on the HDA6 binding to FLC chromatin is physiologically important in flowering time control. By contrast, the effects of intermittent cold on flowering initiation were similar in the hda15-1 mutant and Col-0 plants, showing that the HDA15 function appears somewhat different from that of HDA6 in flowering time control under cold stress.
DISCUSSION
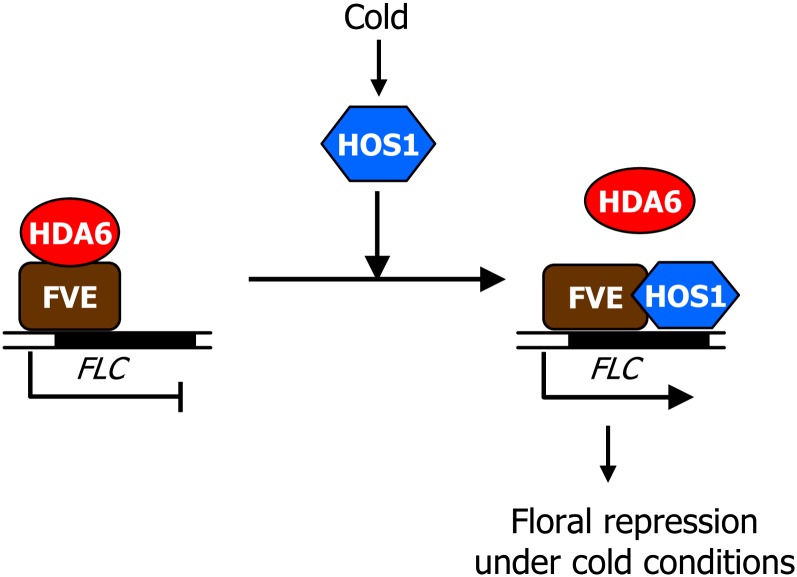
We report a HOS1-mediated molecular mechanism that links cold signaling with the onset of flowering in Arabidopsis. In this working scenario, HOS1, which is otherwise a well-known E3 ubiquitin ligase that attenuates cold responses (Dong et al., 2006), acts as a chromatin remodeling factor that regulates the dynamics of FLC chromatin under short-term cold stress (Figure 7). Under cold stress, HOS1 binds to FLC chromatin in an FVE-dependent manner to limit the chromatin accessibility of HDA6, resulting in activation of FLC chromatin and delayed flowering.
Figure 7.
Model of HOS1 Function in Arabidopsis Flowering.
Under normal conditions, the FVE-HDA6 complex silences FLC chromatin. Under cold stress, HOS1 strongly binds to FLC chromatin and interacts with FVE and HDA6 to dissociate HDA6 from FLC chromatin, resulting in the activation of FLC chromatin and delayed flowering.
[See online article for color version of this figure.]
HOS1-Mediated Activation of FLC Chromatin under Short-Term Cold Stress
Our data depict that cold-activated HOS1 prevents HDA6 from binding to FLC chromatin to activate FLC expression, perhaps by destabilizing the FVE-HDA6 complex (Figure 7). This scheme is in agreement with the late-flowering phenotype of the fve-4 and axe1-5 mutants as well as the early-flowering phenotype of the hos1 mutants.
It is remarkable that FVE interacts with HOS1, an E3 ubiquitin ligase that mediates the cold-induced degradation of ICE1 (Chinnusamy et al., 2003; Dong et al., 2006), providing a distinct role of FVE in cold regulation of FLC chromatin. HOS1 interacts with HDA6 and binds to specific sequence regions within FLC chromatin to which HDA6 also binds. We therefore concluded that HOS1 inhibits the binding of HDA6 to FLC chromatin. We propose that the multiple interactions among HOS1, FVE, and HDA6 enzymes serve as a molecular web that integrates cold stress signals into the flowering genetic network at the FLC locus.
FVE is a component of a large multiprotein complex consisting of at least 16 protein subunits (Jeon and Kim, 2011). Our data showed that FVE interacts with at least four HDACs: HDA5, HDA6, HDA9, and HDA15. FVE is also associated with several H3K4 demethylases, such as FLD, LDL1, LDL2, and LDL3, which repress FLC expression (He et al., 2003; Jiang et al., 2007). Previous data and the data presented here support that FVE serves as a platform for the dynamic assembly of chromatin-modifying complexes involved in the epigenetic regulation of various loci, including the FLC gene.
Our data support that FVE acts as a platform for binding of HDA6 and HOS1 to FLC chromatin. Given the induction of FLC expression and delayed flowering in the HDA6-deficient mutant (Gu et al., 2011; Yu et al., 2011), which is in contrast with the effects of hos1 mutation on FLC expression and flowering time, it is unlikely that HOS1 and HDA6 bind to FLC chromatin as parts of protein complexes. Instead, they would compete with each other for chromatin binding in response to temperature changes.
FVE interacts with the CUL4-DDB1 E3 ligase complex, which plays a role in photomorphogenesis and flowering time control (Pazhouhandeh et al., 2011). The CUL4-DDB1 complex, in association with a CURLY LEAF–containing PRC2-like complex, epigenetically represses the FLC and FT genes. However, the CUL4-DDB1 enzyme complex does not seem to be related to the HOS1–FVE interaction. First, mutations of the CUL4 gene induce FLC expression (Pazhouhandeh et al., 2011); by contrast, FLC expression is repressed in the hos1 mutants. Second, both CUL4 and HOS1 require FVE for regulating FLC expression (Pazhouhandeh et al., 2011; see Supplemental Figure 4 online). However, though the binding of FVE to FLC chromatin requires CUL4, binding is not influenced by HOS1-mediated cold stress signals (see Supplemental Figure 5C online). Finally, the Trp-Asp-X-Arg motif of FVE, which is required for the interaction of FVE with DDB1 proteins (Pazhouhandeh et al., 2011), is not essential for the HOS1–FVE interaction (see Supplemental Figure 3B online). It is therefore likely that HOS1 acts independently of the CUL4-DDB1 complex in the transcriptional silencing of the FLC gene. Therefore, it is proposed that FVE incorporates two different environmental cues into the flowering genetic networks at the chromatin level: light signals via the CUL4-DDB1 complex and cold stress signals via HOS1.
Our work provides a molecular mechanism by which HOS1, together with FVE and HDA6, regulates FLC transcription in response to cold stress. Besides FLC, FLC gene homologs and several transposable elements, which are silenced by RNA-directed DNA methylation, are targeted by the FVE-HDA6 complex (Gu et al., 2011; Pazhouhandeh et al., 2011; To et al., 2011). Therefore, it is possible that the HOS1-FVE-HDA6 regulatory circuit may also play a role in maintaining the silencing of FLC gene homologs and transposable elements. Genome-wide transcript profiling and epigenome analysis of the loss-of-function hos1 mutant will be greatly helpful for understanding how HOS1 modulates the transcriptional regulation of FLC gene in Arabidopsis.
HOS1 as a Chromatin Remodeling Factor in Flowering Time Control
HOS1 activation of FLC expression is important for suppression of flowering initiation under short-term cold stress. However, the early flowering phenotype of the hos1 mutants at normal temperatures is not likely to be related to the activities of FLC. Although the level of FLC mRNA in the fve-4 mutant was comparable to that in the fve hos1 double mutant (see Supplemental Figure 4C online), the double mutant flowered earlier than the fve-4 mutant (see Supplemental Figure 4B online). In addition, mutations of FLC and its gene homologs in Col-0 background have minimal effect on flowering at normal temperatures (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001; Figure 2F). It is therefore envisioned that the HOS1-mediated regulation of flowering time is exerted via two distinct mechanisms: modulation of FLC chromatin and regulation of yet undefined flowering time gene(s) distinct from FLC.
The floral integrator FT is a potential target of HOS1-mediated chromatin regulation. The CUL4-DDB1 E3 ligase complex, which interacts with FVE (Pazhouhandeh et al., 2011), is necessary for maintaining the repression of FT and FLC chromatin (Pazhouhandeh et al., 2011). The CUL4-DDB1 E3 ligase complex also negatively regulates the abundance of CONSTANS (CO) (Chen et al., 2010), an upstream activator of FT expression (Onouchi et al., 2000; Samach et al., 2000). It has recently been reported that HOS1 interacts with CO to regulate CO abundance in photoperiodic flowering (Jung et al., 2012a; Lazaro et al., 2012), supporting the notion that HOS1 influence the protein stability of CO. Alternatively, HOS1 may regulate the status of CO and FT chromatin through interactions with FVE and HDACs.
We did not find any evidence for ubiquitination and degradation of HOS1-interacting partners under both normal and cold temperature conditions. However, it is still possible that the E3 ligase activity of HOS1 is involved in cold regulation of FLC expression. Most studies of the epigenetic systems mediated by E3 ubiquitin ligases in Arabidopsis have been focused on the monoubiquitination of H2A or H2B histone derivatives. H2A monoubiquitination is mediated by PRC1-like E3 ubiquitin ligase complexes (Bratzel et al., 2010). H2B monoubiquitination is regulated by two E3 ligases, HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2 (Cao et al., 2008; Gu et al., 2009). In addition, the CUL4-DDB1 E3 ligase complex facilitates cellular responses to DNA damage by modulating the monoubiquitination of H2A, H3, and H4 (Kapetanaki et al., 2006; Wang et al., 2006). Future investigations include determining whether HOS1 plays a role in the monoubiquitination events of histone proteins and how these biochemical events affect the binding of HDA6 to FLC chromatin.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana lines used were in Col-0 background. The hos1-3 (SALK-069312), hos1-5 (SAIL-1211-D02), axe1-5 (Murfett et al., 2001), and hda15-1 (SALK-004027) mutants were isolated from a pool of T-DNA insertion lines deposited in the ABRC (Ohio State University). The fve-4 and flc-3 mutants have been described previously (Kim et al., 2004; Seo et al., 2009). To produce transgenic plants overexpressing the HOS1, FVE, HDA6, HDA15, and ICE1 genes, six copies of the MYC-coding sequence were fused in frame to the 3′ ends of the genes and driven by the CaMV 35S promoter using the myc-pBA vector (Seo et al., 2010), resulting in HOS1-ox, FVE-ox, HDA6-ox, HDA15-ox, and ICE1-ox plants, respectively. To generate HOS1pro:HOS1-MYC transgenic plants, a HOS1 genomic fragment, which contains a 2.8-kb promoter sequence upstream of the transcription start site (Lee et al., 2001), was subcloned into the modified myc-pBA vector, in which the CaMV 35S promoter was deleted. The PCR primers used for subcloning are listed in Supplemental Table 1 online. Agrobacterium tumefaciens–mediated Arabidopsis transformation was performed according to a modified floral dip method (Clough and Bent, 1998). Plants were grown in soil or on MS-agar plates in a controlled culture room at 23°C under LDs (16-h-light and 8-h-dark cycles).
Flowering Time Measurement and Intermittent Cold Treatment
Plants were grown in soil at either 23 or 4°C under LDs until flowering. The flowering times were determined by counting the number of rosette and cauline leaves at bolting. Fifteen to 20 plants were counted and averaged for each measurement. For intermittent cold treatments, plants were placed at 4°C for 6 h at dawn until flowering.
Analysis of Gene Transcript Levels
Gene transcript levels were examined by either RT-PCR or qRT-PCR. RT-PCR reactions were performed using the Applied Biosystems 2720 Thermal Cycler to verify absence of HOS1 gene expression in the hos1-3 and hos1-5 mutants. qRT-PCR reactions were performed in 96-well blocks with the Applied Biosystems 7500 real-time PCR system using the SYBR Green I master mix in a reaction volume of 20 μL. All qRT-PCR assays were performed in biological triplicates using total RNA samples extracted from three independent plant materials treated under identical conditions and gene-specific primer pairs listed in Supplemental Table 1 online. RNA preparation, data processing, and determination of reaction specificities were performed as described previously (Jung et al., 2012b).
Generation of the Anti-HOS1 Antibody
The HOS1-specific antibody was generated against a synthetic peptide covering residues 796 to 810 (GKRTEESSPEVNVDR) of the Arabidopsis HOS1 protein. The synthetic peptide was HPLC purified, conjugated to keyhole limpet hemocyanine, and injected into rabbits over an 8-week period (one primary injection and three boosting injections), as described previously (To et al., 2011). Two rabbits were used for immunization (AbClon). The antiserum obtained was affinity purified to remove nonspecific antibodies, increasing sensitivity and reducing background, and used for ChIP assays.
Protein Gel Blot Analysis
The FVE-, HDA6-, and HDA15-MYC gene fusions were overexpressed under the control of the CaMV 35S promoter in Col-0 plants. The resultant FVE-ox, HDA6-ox, and HDA15-ox transgenic plants were grown on MS-agar for 10 d and exposed to cold (4°C) for appropriate time periods. Whole plants were harvested for preparation of total protein extracts. Total proteins were separated by SDS-PAGE and transferred to a Millipore Immobilon-P membrane, and immunological analysis was performed to detect the fusion proteins using an anti-MYC antibody at 1:2000 dilution (Millipore).
Ubiquitination Assays
In vivo detection of ubiquitinated proteins was performed as described previously (Dong et al., 2006; Miura et al., 2011). Ten-day-old transgenic plants overexpressing the FVE-, HDA6-, HDA15-, and ICE1-MYC gene fusions were pretreated with 50 μM MG132 (Sigma-Aldrich), a potent 26S proteasome–specific inhibitor (Lee and Goldberg, 1998), for 24 h and then exposed to cold (4°C) for 15 h. The total protein extract was immunoprecipitated using an anti-MYC antibody coupled to agarose beads and analyzed immunologically using anti-MYC and antiubiquitin antibodies.
ChIP Assays
ChIP assays were performed using 10- or 14-d-old plants grown on MS-agar plates under LDs. Processing of plant materials and qPCR were performed as described previously (Yang et al., 2011). The PCR primers for qPCR were designed to amplify DNA fragments of 100 to 200 bp and listed in Supplemental Table 1 online. An eIF4A DNA fragment was used for normalization. To validate the ChIP-qPCR runs in the P1 to P5 sequence regions of FLC gene, we sequenced the ChIP-qPCR products (see Supplemental Data Set 1 online) using an Applied Biosystems 3730xl DNA analyzer at the National Instrumentation Center for Environmental Management (Seoul National University).
Protein–Protein Interaction Assays
Protein–protein interactions were examined by yeast two-hybrid assays using the BD Matchmaker system (Clontech) and in vitro pull-down assays, as described previously (Seo et al., 2012). To evaluate whether the protein–protein interactions occur in vivo, BiFC assays were performed using Arabidopsis mesophyll protoplasts, as described previously (Hong et al., 2011). Coimmunoprecipitation assays were also employed for the same purpose, as described previously (Song et al., 2012).
TSA Treatments
Plants were grown for 10 d on MS-agar plates containing 0.5 μM TSA, a potent inhibitor of class I and II mammalian HDACs (Taunton et al., 1996; Richon et al., 2000). Whole plants were used for extraction of total RNA. The levels of FLC mRNA were examined by qRT-PCR.
Statistical Analysis
The statistical significance between two means of measurements was determined using a Student’s t test with P values < 0.01.
Accession Numbers
Sequence data from this article can be obtained from the Arabidopsis Genome Initiative databases under the following accession numbers: eIF4A (At3g13920), FLC (At5g10140), FVE (At2g19520), HDA6 (At5g63110), HDA15 (At3g18520), HOS1 (At2g39810), and ICE1 (At3g26744).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. HOS1 Is Correlated with FLC Expression.
Supplemental Figure 2. Examination of Anti-HOS1 Antibody Specificity by Protein Gel Blot Assays.
Supplemental Figure 3. The Trp-Asp-X-Arg Motif Is Not Required for HOS1–FVE Interactions.
Supplemental Figure 4. FLC Expression in fve hos1 Double Mutant.
Supplemental Figure 5. Cold Stress Does Not Influence FVE Protein Stability.
Supplemental Figure 6. Interactions of FVE and HOS1 with Histone Deacetylases in Yeast Cells.
Supplemental Figure 7. Binding of HDA6 to FLC Chromatin in the HOS1-ox Transgenic Plant.
Supplemental Figure 8. HOS1 Binding to FLC Chromatin in the axe1-5 Mutant.
Supplemental Table 1. Primers Used in qRT-PCR, ChIP-qPCR, and Gene Cloning.
Supplemental Data Set 1. Sequencing Analysis of ChIP-qPCR Products.
Supplemental References 1. References for Supplemental Figures.
Acknowledgments
We thank Ju Yun for subcloning of expression constructs and Ilha Lee for providing fve-4 and flc-3 mutants. This work was supported by the Leaping Research (20120005600) and Global Research Lab (2012055546) Programs provided by the National Research Foundation of Korea, the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. 201203013055290010200) provided by the Rural Development Administration, and by a grant from the Agricultural R&D Promotion Center (309017-05-4-HD140), Korea Ministry for Food, Agriculture, Forestry, and Fisheries. It was also supported in part by the Human Frontier Science Program (RGP0002/2012). It was also supported in part by Japan Science and Technology Agency (JST), Core Research for Evolutionary Science and Technology (CREST) and grants from the RIKEN Research Institute.
AUTHOR CONTRIBUTIONS
C.-M.P. designed the experiments. J.-H.J., J.-H.P., and S.L. performed the experiments. C.-M.P. and J.-H.J. analyzed the data. T.K.T., J.-M.K., and M.S. generated and evaluated the anti-HDA6 antibody. C.-M.P. and J.-H.J. wrote the article.
Glossary
- HDAC
histone deacetylase
- H3K27Me3
H3K27 trimethylation
- Col-0
Columbia-0
- LDs
long days
- ChIP
chromatin immunoprecipitation
- H3Ac
H3 acetylation
- CaMV
cauliflower mosaic virus
- qPCR
quantitative PCR
- qRT-PCR
quantitative real-time RT-PCR
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- TSA
trichostatin A
- MS
Murashige and Skoog
References
- Ausín I., Alonso-Blanco C., Jarillo J.A., Ruiz-García L., Martínez-Zapater J.M. (2004). Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Bond D.M., Dennis E.S., Finnegan E.J. (2011). The low temperature response pathways for cold acclimation and vernalization are independent. Plant Cell Environ. 34: 1737–1748 [DOI] [PubMed] [Google Scholar]
- Bratzel F., López-Torrejón G., Koch M., Del Pozo J.C., Calonje M. (2010). Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr. Biol. 20: 1853–1859 [DOI] [PubMed] [Google Scholar]
- Cao Y., Dai Y., Cui S., Ma L. (2008). Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.H., Zhu D., Deng X.W. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.H., Hong X., Agarwal M., Zhu J.K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J.K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12: 444–451 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dong C.H., Agarwal M., Zhang Y., Xie Q., Zhu J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Fowler S.G., Thomashow M.F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54: 767–781 [DOI] [PubMed] [Google Scholar]
- Gu X., Jiang D., Wang Y., Bachmair A., He Y. (2009). Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57: 522–533 [DOI] [PubMed] [Google Scholar]
- Gu X., Jiang D., Yang W., Jacob Y., Michaels S.D., He Y. (2011). Arabidopsis homologs of retinoblastoma-associated protein 46/48 associate with a histone deacetylase to act redundantly in chromatin silencing. PLoS Genet. 7: e1002366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant 2: 554–564 [DOI] [PubMed] [Google Scholar]
- He Y., Michaels S.D., Amasino R.M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754 [DOI] [PubMed] [Google Scholar]
- Hong S.Y., Kim O.K., Kim S.G., Yang M.S., Park C.M. (2011). Nuclear import and DNA binding of the ZHD5 transcription factor is modulated by a competitive peptide inhibitor in Arabidopsis. J. Biol. Chem. 286: 1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Lee H., Stevenson B., Zhu J.K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarillo J.A., Piñeiro M., Cubas P., Martínez-Zapater J.M. (2009). Chromatin remodeling in plant development. Int. J. Dev. Biol. 53: 1581–1596 [DOI] [PubMed] [Google Scholar]
- Jeon J., Kim J. (2011). FVE, an Arabidopsis homologue of the retinoblastoma-associated protein that regulates flowering time and cold response, binds to chromatin as a large multiprotein complex. Mol. Cells 32: 227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Yang W., He Y., Amasino R.M. (2007). Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19: 2975–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Seo P.J., Ahn J.H., Park C.M. (2012b). Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J. Biol. Chem. 287: 16007–16016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Seo P.J., Park C.M. (2012a). The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287: 43277–43287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanaki M.G., Guerrero-Santoro J., Bisi D.C., Hsieh C.L., Rapić-Otrin V., Levine A.S. (2006). The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 103: 2588–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.H., Doyle M.R., Sung S., Amasino R.M. (2009). Vernalization: Winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25: 277–299 [DOI] [PubMed] [Google Scholar]
- Kim H.J., Hyun Y., Park J.Y., Park M.J., Park M.K., Kim M.D., Kim H.J., Lee M.H., Moon J., Lee I., Kim J. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Piñeiro M., Jarillo J.A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Henderson D.A., Zhu J.K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17: 3155–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Goldberg A.L. (1998). Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim J.J., Kim S.H., Cho H.J., Kim J., Ahn J.H. (2012). The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana. Plant Cell Physiol. 53: 1802–1814 [DOI] [PubMed] [Google Scholar]
- Medina J., Catalá R., Salinas J. (2011). The CBFs: Three Arabidopsis transcription factors to cold acclimate. Plant Sci. 180: 3–11 [DOI] [PubMed] [Google Scholar]
- Miura K., Ohta M., Nakazawa M., Ono M., Hasegawa P.M. (2011). ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. Plant J. 67: 269–279 [DOI] [PubMed] [Google Scholar]
- Murfett J., Wang X.J., Hagen G., Guilfoyle T.J. (2001). Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onouchi H., Igeño M.I., Périlleux C., Graves K., Coupland G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazhouhandeh M., Molinier J., Berr A., Genschik P. (2011). MSI4/FVE interacts with CUL4-DDB1 and a PRC2-like complex to control epigenetic regulation of flowering time in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 3430–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Kumimoto R.W., Wong B.J., Riechmann J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O.J., Nadzan G.C., Reuber T.L., Riechmann J.L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126: 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon V.M., Sandhoff T.W., Rifkind R.A., Marks P.A. (2000). Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA 97: 10014–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Scortecci K.C., Michaels S.D., Amasino R.M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26: 229–236 [DOI] [PubMed] [Google Scholar]
- Seo E., Lee H., Jeon J., Park H., Kim J., Noh Y.S., Lee I. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Park J.Y., Kim S.Y., Jeon J., Lee Y.H., Kim J., Park C.M. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61: 661–671 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Park M.J., Lim M.H., Kim S.G., Lee M., Baldwin I.T., Park C.M. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S., Amasino R.M. (2005). Remembering winter: Toward a molecular understanding of vernalization. Annu. Rev. Plant Biol. 56: 491–508 [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig C.A., Schreiber S.L. (1996). A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408–411 [DOI] [PubMed] [Google Scholar]
- To T.K., et al. (2011). Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genet. 7: e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhai L., Xu J., Joo H.Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. (2006). Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22: 383–394 [DOI] [PubMed] [Google Scholar]
- Yang S.D., Seo P.J., Yoon H.K., Park C.M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23: 2155–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.W., Liu X., Luo M., Chen C., Lin X., Tian G., Lu Q., Cui Y., Wu K. (2011). HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiol. 156: 173–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Dong C.H., Zhu J.K. (2007). Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 10: 290–295 [DOI] [PubMed] [Google Scholar]