This work shows that HOS1, previously characterized as a nuclear pore–associated E3 ubiquitin ligase, is required for nucleo-cytoplasmic mRNA export. This study demonstrates that this reduction in nucleo-cytoplasmic export by hos1, or mutations to other previously characterized nuclear pore–associated proteins, leads to altered RNA levels and rhythms, circadian clock function, and cold signaling.
Abstract
Cold acclimation has been shown to be attenuated by the degradation of the INDUCER OF CBF EXPRESSION1 protein by the E3 ubiquitin ligase HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1). However, recent work has suggested that HOS1 may have a wider range of roles in plants than previously appreciated. Here, we show that hos1 mutants are affected in circadian clock function, exhibiting a long-period phenotype in a wide range of temperature and light environments. We demonstrate that hos1 mutants accumulate polyadenylated mRNA in the nucleus and that the circadian defect in hos1 is shared by multiple mutants with aberrant mRNA export, but not in a mutant attenuated in nucleo-cytoplasmic transport of microRNAs. As revealed by RNA sequencing, hos1 exhibits gross changes to the transcriptome with genes in multiple functional categories being affected. In addition, we show that hos1 and other previously described mutants with altered mRNA export affect cold signaling in a similar manner. Our data support a model in which altered mRNA export is important for the manifestation of hos1 circadian clock defects and suggest that HOS1 may indirectly affect cold signaling through disruption of the circadian clock.
INTRODUCTION
Plants sense cold temperatures and mount responses to survive freezing, a process known as cold acclimation. The best characterized molecular genetic pathway regulating cold responses is the CBF (for C-repeat binding factor) pathway (Fowler and Thomashow, 2002; Zhu et al., 2004; Chinnusamy et al., 2010). In Arabidopsis thaliana, the expression of three of the CBF transcription factors CBF1, CBF2, and CBF3, is induced in response to low temperatures (Medina et al., 1999), and constitutive overexpression of these genes is sufficient to increase freezing tolerance (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Sharabi-Schwager et al., 2010). The CBF proteins bind to characterized cis-elements in cold-responsive genes to alter their expression and induce pathways enabling cold survival (Stockinger et al., 1997; Gilmour et al., 1998). Cold signaling can also affect CBF transcription through calcium signaling (Yang et al., 2010) and calmodulin-binding transcription factor activity (Doherty et al., 2009).
HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) has been proposed to be a key negative regulator of cold signaling (Ishitani et al., 1998; Lee et al., 2001; Dong et al., 2006a). HOS1 is a large protein that shares sequence similarity with Ring E3 ligases (Lee et al., 2001). In the nucleus, HOS1 attenuates the cold response by the degradation of INDUCER OF CBF EXPRESSION1 (ICE1), a Myc-like basic helix-loop-helix transcription factor that binds the promoter of CBF3 and is necessary and sufficient for the upregulation of CBF3 expression in response to cold treatment (Lee et al., 2001; Chinnusamy et al., 2003; Dong et al., 2006a; Miura et al., 2007; Lee et al., 2012). hos1 mutants show constitutive expression of cold-responsive genes, whereas HOS1 overexpression leads to a reduction in CBF expression and increased sensitivity to freezing (Ishitani et al., 1998; Lee et al., 2001; Dong et al., 2006a). In addition to a role in cold acclimation, HOS1 has more recently been shown to influence the abundance of the photoperiod sensor protein CONSTANS (CO; Jung et al., 2012; Lazaro et al., 2012) and to physically interact with components of the nuclear pore (Tamura et al., 2010).
The plant circadian clock is composed of an interlocking series of transcriptional negative feedback loops, and alterations to the expression of any one of the genes can have profound effects on the levels and timing of the others (Pokhilko et al., 2012). Loss-of-function mutations in CIRCADIAN CLOCK ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), the pseudo-response regulators PRR5 and PRR3, or TIMING OF CAB EXPRESSION1 (TOC1) shorten the period of the circadian clock, whereas loss of PRR9, PRR7, or ZEITLUPE (ZTL) lengthen period, and mutations in GIGANTEA (GI) and the evening complex components LUX ARRHYTHMO (LUX), EARLY FLOWERING3 (ELF3), or ELF4 result in a reduction in amplitude and rhythm robustness (Nagel and Kay, 2012). Loss of clock function not only alters the expression of the clock genes themselves, but also has physiological consequences, including altered flowering time (Imaizumi, 2010), growth regulation (Nozue et al., 2007), and plant fitness and vigor (Dodd et al., 2005). The circadian clock is also essential for mounting a cold response (Dong et al., 2011; Seo et al., 2012).
CBF genes themselves are rhythmically expressed (Harmer et al., 2000), and their cold induction is gated by the circadian clock (Fowler et al., 2005). Perturbation of the circadian clock results in altered cold responses in Arabidopsis and poplar (Populus spp; Nakamichi et al., 2009; Ibáñez et al., 2010; Dong et al., 2011), and cold temperatures profoundly affect clock function (Bieniawska et al., 2008). More interestingly, CCA1 and LHY can transduce cold signals to CBFs through a mechanism that promotes cold-regulated alternative splicing (Dong et al., 2011; James et al., 2012; Seo et al., 2012).
HOS1 interacts with Arabidopsis structural nuclear pore components and localizes to the nuclear envelope (Tamura et al., 2010; Lazaro et al., 2012), suggesting that HOS1 has a more general function in plant cells. Here, we show that HOS1 is necessary for normal circadian rhythms and for mRNA export out of the nucleus. We demonstrate that mRNA export is essential for correct periodicity of the circadian clock and that the effects of hos1 on cold-regulated gene expression and circadian periodicity are shared by nuclear pore mutants previously demonstrated to be required for normal mRNA export. Our results show that HOS1 affects cold signaling at least in part through altered mRNA export and that this may be mediated by changes in clock gene expression.
RESULTS
HOS1 Is Necessary to Maintain Wild-Type Circadian Clock Periodicity
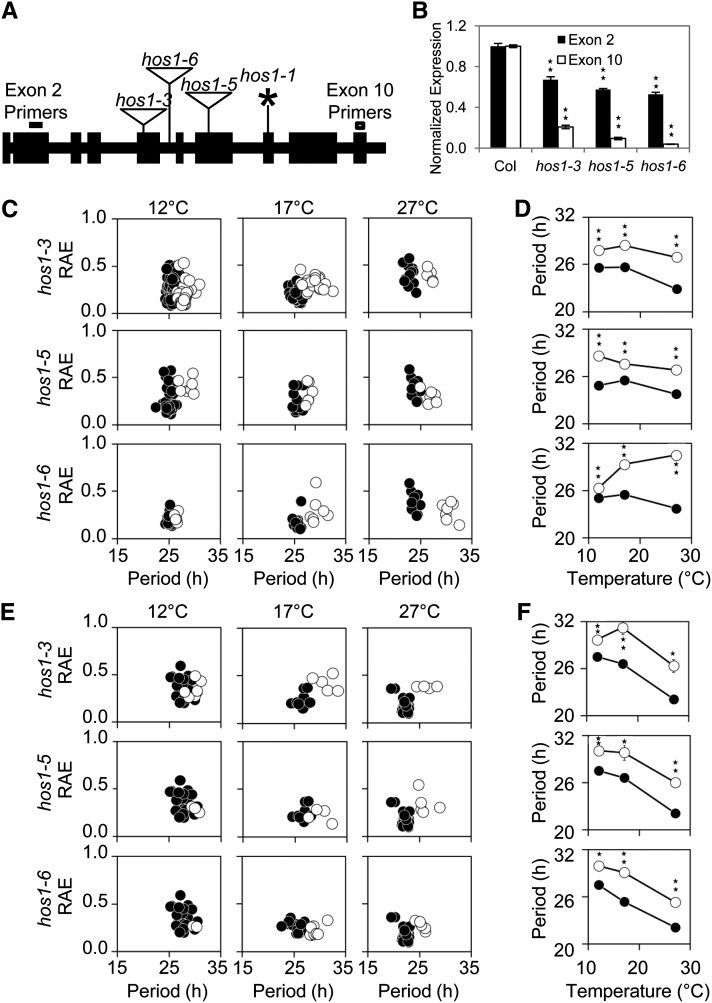
We screened hos1-3 and two novel alleles in Columbia (Col; designated hos1-5 and hos1-6; see Methods) for circadian-period defects using delayed fluorescence (Figure 1). Mutants harboring these alleles showed a decrease in the amount of HOS1 RNA (Figure 1B) and shared the previously described phenotypes of other hos1 mutants, including early flowering and increases in cold-responsive gene expression such as that of CBF3 (see Supplemental Figure 1 online; Lee et al., 2001; Lazaro et al., 2012). All three insertion lines exhibited significant lengthening of the clock period compared with the wild type (Figures 1C to 1F). The long-period hos1 mutant phenotype was observed when plants were grown at 12, 17, or 27°C in either constant red or blue light or in the dark (Figures 1C to 1F; see Supplemental Figure 2 online). Alleles of hos1 in C24 (hos1-1) and Landsberg erecta (hos1-2) also shared the long-period phenotype (see Supplemental Figure 3 online); therefore, the long-period phenotype of hos1 is common between all types of alleles and accessions investigated. To demonstrate that this phenotype was not specific to the delayed fluorescence assay, we transformed the hos1-3 mutant with luciferase (LUC) reporter lines under the control of the CHLOROPHYLL A/B BINDING PROTEIN2 (CAB2), COLD-REGULATED15a (COR15a), or COLD, CIRCADIAN RHYTHM, AND RNA BINDING2 (CCR2) promoter. Compared with the wild type, hos1-3 had a significantly longer period for all three reporter lines at all temperatures tested (see Supplemental Figure 4 online). hos1-3 mutants also showed a long-period phenotype for LHY:LUC, PRR9:LUC, PRR7:LUC, and TOC1:LUC (see Supplemental Figure 5 online). Therefore, HOS1 is required for the maintenance of circadian period in Arabidopsis, and this phenotype is independent of accession, temperature, light quality, or output measured.
Figure 1.
Mutations to HOS1 Result in a Long-Period Circadian Phenotype across a Range of Physiological Temperatures and Light Conditions.
(A) Schematic diagram showing positions of the hos1 mutations across the HOS1 locus. Exons are shown in black boxes and introns in black lines. The position of the hos1 mutations and allele designations are labeled. Triangles represent T-DNA insertions and the asterisk represents point mutations. The primer positions in exons 2 and 10 are marked to illustrate the positions of qPCR primers used to produce the data in Figure 1B.
(B) qPCR data showing reduced levels of HOS1 mRNA expression in each of the hos1 mutants studied. Primers for exon 2 (dark gray bars) are located upstream of and exon 10 (white bars) are located downstream of all hos1 mutations studied (see [A]). Seedlings were grown for 10 d before being harvested at dawn. Data are averages of biological triplicate ± se normalized for transcript abundance to UBQ10. Stars indicate significant difference to the respective wild type as measured by Student’s t test where one star indicates P ≤ 0.05 and two stars P ≤ 0.01.
(C) to (F) Delayed fluorescence assays comparing the wild type (Col) to the hos1 alleles. Seedlings were entrained for 7 d in 12L:12D white-light cycles at 22°C before transfer to experimental conditions of constant 12, 17, or 27°C and constant red light ([C] and [D]) or constant blue light ([E] and [F]). Delayed fluorescence was then measured in several hos1 mutants (white circles) compared with wild-type data (Col, black circles).
(C) and (E) Plots showing period estimations produced via mathematical analysis plotted versus RAE for the wild type (Col) versus the hos1 mutants. Data are from individual wells containing 10 to 20 seedlings.
(D) and (F) Plot illustrating the effect of temperature on the variable weighted mean of the period for the wild type (Col) versus hos1 mutants. Data are variable weighted mean ± se. Stars indicate significant difference to the respective wild type as measured by Student’s t test where one star indicates P ≤ 0.05 and two stars P ≤ 0.01.
hos1 Does Not Require Individual Clock Components for the Observed Long-Period Phenotype
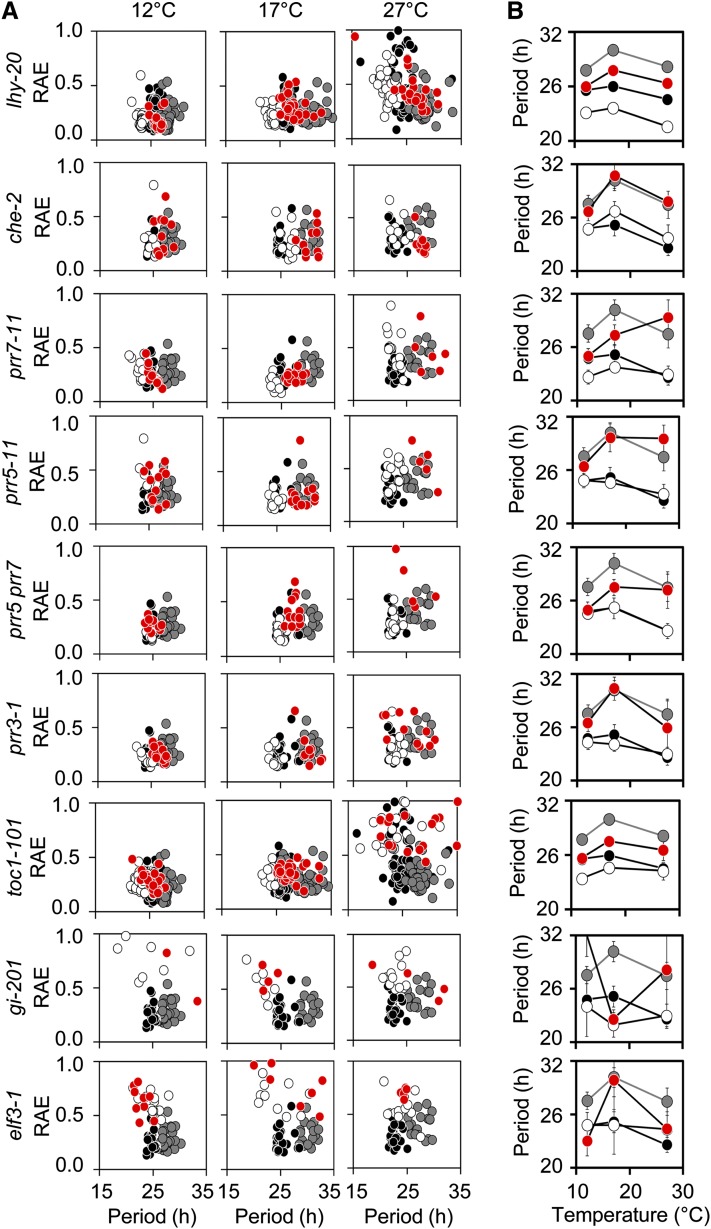
Previously, HOS1 has been shown to exhibit E3 ubiquitin ligase activity and to destabilize the ICE1 or CO proteins through proteasomal degradation targeted by ubiquitination (Dong et al., 2006a; Lazaro et al., 2012). In both of these cases, hos1 phenotypes are ascribed to the consequences of overaccumulation of the target protein. We took a genetic approach to determine whether HOS1 requires specific clock components for the long-period phenotype and screened double mutants between hos1 and eight core clock components for altered rhythms in delayed fluorescence at 12, 17, and 27°C in constant red light (Figure 2). We did not find any clock mutant with a period phenotype that was epistatic to hos1-3. Under these conditions, in cases where single clock mutants had little effect on period, double mutants had a period resembling the hos1-3 single mutant (cca1 hiking expedition2 [che-2], prr3-1, and prr5-11). Additive interactions were observed between hos1-3 and toc1-101, lhy-20, prr7-11, and prr7 prr5 double mutants, whereas elf3-1 and gi-201 led to arrhythmic plants in this assay with or without the additional hos1-3 mutation. Therefore, the hos1 long-period phenotype is not dependent on misregulation of these individual clock components.
Figure 2.
The Long-Period Phenotype in hos1 Does Not Require Selected Individual Clock Components.
(A) Delayed fluorescence assays comparing the wild type (Col, black circles) to hos1 (gray circles) and the single (white circles) and double (red circles) mutants. Seedlings were entrained for 16 d in 12L:12D white-light cycles at 22°C before transfer to experimental conditions of constant 12, 17, or 27°C and constant red light. Plots show period estimations produced via mathematical analysis plotted versus RAE. Data are from individual wells containing 10 to 20 seedlings.
(B) Plots illustrating the effect of temperature on the variable weighted mean of the period for the wild type (Col, black circles) to hos1 (gray circles) and the single (white circles) and double (red circles) mutants. Data are variable weighted mean ± se. Stars indicate significant difference to the respective wild type as measured by Student’s t test where one star indicates P ≤ 0.05 and two stars P ≤ 0.01.
We also tested whether hos1 mutants had altered hypocotyl elongation, a characteristic shared by multiple circadian clock mutants. In 12-h-light/12-h-dark (12L:12D) white-light cycles or various fluences of constant red or blue light, hos1 had consistently longer hypocotyls than the wild type, but when grown in the dark, the hos1 hypocotyls were shorter (see Supplemental Figures 6A to 6E online). Therefore, HOS1 is required for normal hypocotyl elongation in Arabidopsis. We then screened the double mutants for epistasis; all of the double mutant hypocotyls were longer than those of the corresponding single clock mutant in 12L:12D white-light cycles (see Supplemental Figure 6F online) or shorter when grown in the dark (see Supplemental Figure 6G online), suggesting that the hos1 hypocotyl phenotype cannot be ascribed to overaccumulation of one of these clock proteins. We were unable to isolate double mutants with cca1 (At2g46830), prr9 (At2g46790), or elf4 (At2g40080) mutants because of their genomic proximity to the HOS1 locus.
To demonstrate that the circadian and hypocotyl phenotypes are caused by the loss of HOS1, we created transgenic hos1-3 lines expressing translational fusions between HOS1 and cyan fluorescent protein (CFP) under the 35S promoter and demonstrated the HOS1-CFP fusion protein fully complemented both the hos1-3 hypocotyl and period phenotypes (see Supplemental Figure 7 online).
hos1 Mutants Show Protein and Promoter Rhythms Despite Poor mRNA Rhythm Robustness
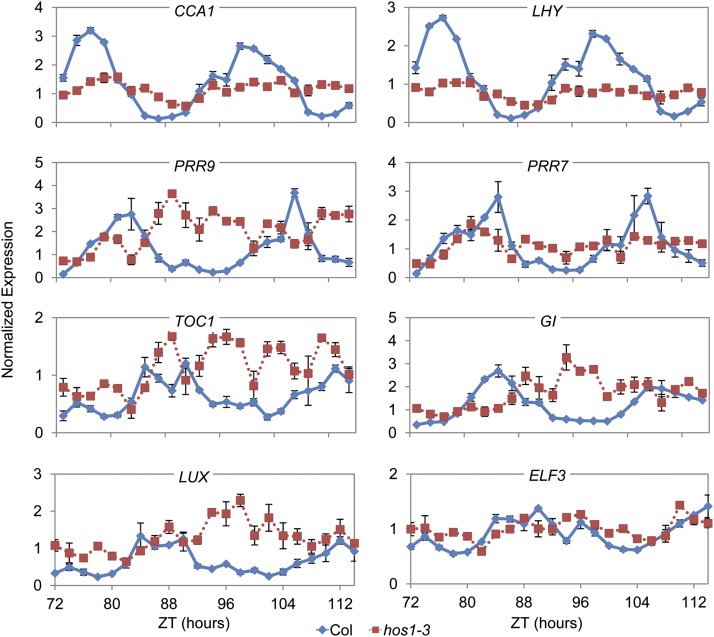
To determine if HOS1 is required for proper expression of the core components of the circadian clock, RNA from the wild type and hos1-3 was sampled every 2 h from seedlings growing in constant red light or 12L:12D red-light cycles (see Methods). Based on the analysis of previous long-period circadian clock mutants, we expected the period of clock gene expression to be lengthened and/or alterations in the amplitudes of expression for some genes. However, the effects of hos1-3 on clock gene expression were inconsistent with a typical long-period mutant (Figure 3; see Supplemental Figure 8 online). In hos1-3, the amplitude and rhythmicity of expression of the circadian clock genes was significantly affected after 2 d in constant conditions (Figure 3). Notable features under constant conditions were the reduction in amplitude of CCA1, LHY, and PRR7, whereas PRR9, TOC1, GI, LUX, and ELF3 expression showed little evidence of rhythmicity (Figure 3). In 12L:12D red-light cycles, the most prominent effect of hos1-3 was on the amplitude of expression, where, for instance, at 17°C the maximal expression of LHY, CCA1, and PRR5 was reduced and peak expression of PRR9, PRR7, TOC1, and ELF3 was increased (see Supplemental Figure 6 online). The phase of peak expression in diurnal conditions was largely unaffected in hos1-3 (see Supplemental Figure 8 online). Overall patterns of expression were similar when the plants were grown at 27, 22, 17, or 12°C (see Supplemental Figure 8 online). These analyses indicate that HOS1 is required not only for regulation of cold signaling (Ishitani et al., 1998; Lee et al., 2001; Dong et al., 2006a) and flowering time genes (Lazaro et al., 2012) but also for correct expression of the core circadian clock genes at all these temperatures and for robust mRNA rhythms in constant conditions.
Figure 3.
Under Constant Red Light, HOS1 Is Required for Correct Expression of Clock Genes.
Plants were grown for 7 d under 12L:12D white-light cycles at 22°C followed by 2 d in 12L:12D red-light cycles at 17°C before being transferred to constant red light at 17°C for sampling in biological triplicate at the Zeitgeber time (ZT) times indicated. The cDNA were analyzed by qPCR three times for each biological replicate. Data presented are the average of three biological repeats ± se. Transcript abundance levels were normalized to ACT7. Blue diamonds with solid lines are data from the wild type (Col), whereas red squares with dotted lines are data from hos1-3.
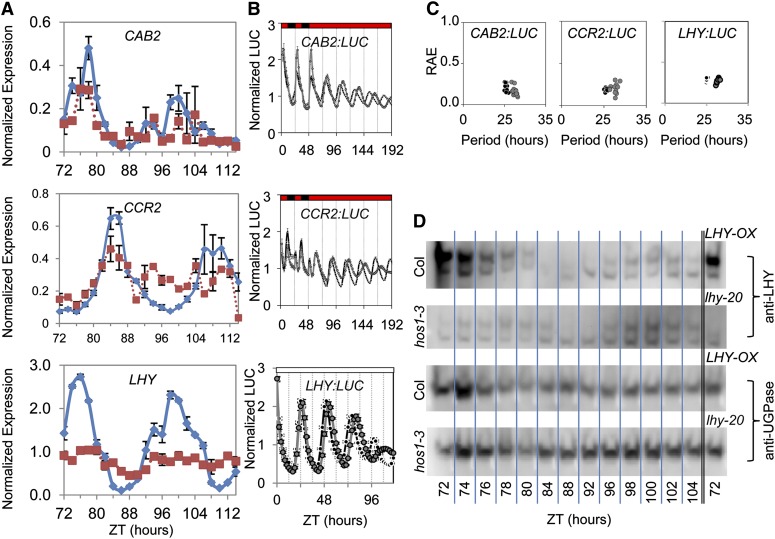
Transcript analysis showed that hos1 exhibited poor mRNA rhythms, whereas delayed fluorescence and promoter:LUC data were rhythmic, albeit with a long period and lowered amplitude. To determine if protein rhythms continue in the hos1 mutant, we measured LHY protein abundance as well as LHY target gene expression and promoter activity under conditions matching those for the clock gene mRNA study in Figure 3. Quantitative PCR (qPCR) revealed that endogenous CAB2 and CCR2 mRNA rhythms were markedly less robust with no detectable oscillations after 80 h under constant red light, in common with LHY (Figure 4A). By contrast, LHY:LUC, CAB2:LUC, and CCR2:LUC all oscillated with a long period in hos1-3 (Figures 4B and 4C). LHY protein levels oscillated with a long period (Figure 4D), and we observed reduced LHY protein levels in hos1-3 as would have been predicted by the observed reduction in LHY mRNA abundance (Figure 4A). Taken together, our evidence suggests that mRNA rhythms are most strongly affected by the hos1 mutation and that clock protein abundance and promoter activity exhibit more robust rhythms, although with a long period.
Figure 4.
hos1 Mutants Exhibit Long Period but Robust Protein and Promoter Rhythms Despite Poor mRNA Rhythm Robustness.
(A) The expression of endogenous CAB2, CCR2, and LHY in the wild type (Col) and hos1-3 in constant red light (see Methods). Data are the average of the three biological repeats ± se. Transcript abundance levels were normalized to ACT7. Samples were taken at the Zeitgeber time (ZT) indicated. Blue diamonds are data from Col, whereas red squares show data from hos1-3.
(B) Plots to show the normalized mean CAB2:LUC, CCR2:LUC, and LHY:LUC luminescence profiles for the wild type and hos1-3 mutant. Data are presented as means ± se. Transgenic seedlings containing the CAB2:LUC or CCR2:LUC reporter were entrained for 7 d in 12L:12D white-light cycles at 22°C before transferred to experimental conditions of constant 17°C and 2 d of 12L:12D red-light cycles followed by 6 d of constant red light. Transgenic seedlings containing the LHY:LUC reporter were entrained for 6 d in 12L:12D white-light cycles at 22°C before transferred to experimental conditions of constant white light at room temperature. hos1-3 (gray filled circles) is compared with wild-type data (black filled circles).
(C) Plots to show the period estimations for CAB2:LUC, CCR2:LUC, and LHY:LUC versus RAE for the wild type versus the hos1-3 mutant.
(D) Immunoblot analysis showing endogenous LHY levels in the wild type (Col) compared with hos1-3, with LHY-OX and lhy-20 lines as positive and negative controls, respectively. The blots were reprobed using an antibody against UGPase for the loading control. Plants were grown for 7 d under 12L:12D white-light cycles at 22°C followed by 2 d in 12L:12D red-light cycles at 17°C before being transferred to constant red light at 17°C for sampling at the Zeitgeber time (ZT) times indicated. These images are representative images from technical duplicates.
hos1 Accumulates Polyadenylated mRNA in the Nucleus
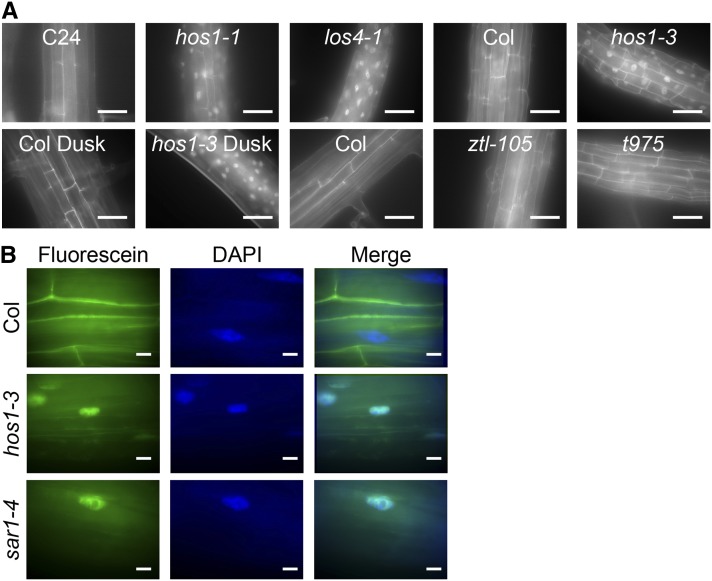
From our expression analysis of core clock genes, it is clear that hos1 alters clock gene mRNA amplitude. Therefore, we speculated that HOS1 may be playing a functional role in regulating the abundance of clock mRNA. In a proteomic screen, HOS1 interacts with the nuclear pore proteins RNA EXPORT FACTOR1–green fluorescent protein (GFP) and the nucleoporin Nup43-GFP (Tamura et al., 2010) and associates with the nuclear periphery (Lazaro et al., 2012). Nuclear pore complexes are required for the export of mRNA from the nucleus; mutating components of the nuclear pore complex results in abnormal mRNA export as shown by in situ hybridization against poly(A)-containing mRNA (Dong et al., 2006b; Parry et al., 2006; Wiermer et al., 2012). Therefore, we used in situ hybridization to detect poly(A)-containing mRNA to determine if HOS1 plays a role in nucleo-cytoplasmic mRNA export. Like the previously described los4-1 mutant (Gong et al., 2002, 2005), hos1 plants exhibited increased nuclear retention of poly(A)-containing RNA in the nuclei compared with the appropriate wild-type control (Figure 5). There was no difference in the in situ fluorescence when fixation was performed at dawn compared with dusk in either wild-type or hos1-3 samples, and no significant accumulation of signal was observed in another long-period mutant, ztl-105, or the arrhythmic toc1 prr5 prr7 prr9 quadruple mutant (Figure 5). Our data therefore suggest that while HOS1 is required for normal mRNA export, this occurs independently of the circadian clock.
Figure 5.
HOS1 Is Required for Nucleo-Cytoplasmic mRNA Export.
(A) In situ hybridization with a fluorescein-labeled oligo(dT) probe in wild-type, hos1, los4, ztl (long period), and toc1 prr9 prr7 prr5 (t975) quadruple mutant (arrhythmic) roots. Plants were fixed at dawn (unless otherwise indicated) after 2 weeks of growth under 12L:12D white-light cycles. Bars = 50 µm.
(B) In situ hybridization with a fluorescein-labeled oligo(dT) in wild-type, hos1, and sar1 roots counterstained with DAPI. Bars = 10 µm.
To determine if the increased signal observed in the in situ hybridization was due to retention of a small number of mRNA species to a high degree or a low-level retention of many genes, RNA sequencing was performed using samples harvested at dawn (see Methods). Of the total 15,070 genes present, 1308 were upregulated in hos1-3 compared with the wild type, and 126 were downregulated twofold or more (Table 1; see Supplemental Data Set 1 online). AmiGO analysis (Carbon et al., 2009) of these up- or downregulated genes showed that they fell into a wide range of Gene Ontology classifications (see Supplemental Data Set 2 online). This analysis shows that hos1 mutants exhibit a profound misregulation of a large number of genes, most of which are mildly upregulated. This effect is consistent with the expected consequences of transcripts being retained in the nucleus, as in eukaryotes mRNA degradation takes place in the cytoplasm (Tourrière et al., 2002). Of the 12 genes that make up the core circadian clock machinery (Pokhilko et al., 2012), seven genes were significantly upregulated and none were significantly downregulated in the RNA sequencing data set at dawn (Table 1; the numerical values for LHY, CCA1, and PRR9 were lower in hos1-3 compared with the wild type but failed significance testing). Using qPCR, we were able to verify that expression of PRR5, TOC1, LUX, ELF4, and ELF3 were upregulated in hos1-3 at dawn (Figure 6). The upregulation of these genes in hos1-3 was also observed in the diurnal data set in Supplemental Figure 8 online even though these genes were lowly expressed at dawn when this sampling occurred, as would be expected for evening-phased genes. These upregulated clock genes were predominantly evening-expressed transcriptional inhibitors, which may explain the fact that 71% of the genes downregulated in hos1-3 had circadian expression patterns (Table 1). Of the 1308 genes that were upregulated in hos1, 48.1% of those were classified as circadian as analyzed by Covington et al. (2008); this figure is not far from the calculated value of 41.4% circadian-expressed genes within the whole genome (Covington et al., 2008). Therefore, we suggest that loss of HOS1 directly causes upregulation of gene expression of a large percentage of the genome through mRNA retention but that the genes downregulated in hos1 are more likely to be affected indirectly through the perturbation of circadian function.
Table 1. Loss of HOS1 Results in Large Numbers of Upregulated Genes.
| Category | Upregulated in hos1-3 | Downregulated in hos1-3 |
|---|---|---|
| Total number of genes | 1308 | 126 |
| Expressed C+E union | 832 | 49 |
| Circadian C+E union | 400 | 35 |
| Genes circadian expressed (%) | 48.1 | 71.4 |
| Differentially expressed clock genes | PRR5, PRR3, TOC1, LUX, ELF3, ELF4, CHE | None |
Genes that are downregulated are also dominated by circadian-expressed genes. Analysis of RNA sequencing shows that reads map to 15,070 genes expressed in the hos1 mutant at dawn. C+E union refers to the list of circadian-expressed genes defined by Covington et al. (2008). Significant gene expression differences were calculated by Cufflinks (Trapnell et al., 2012).
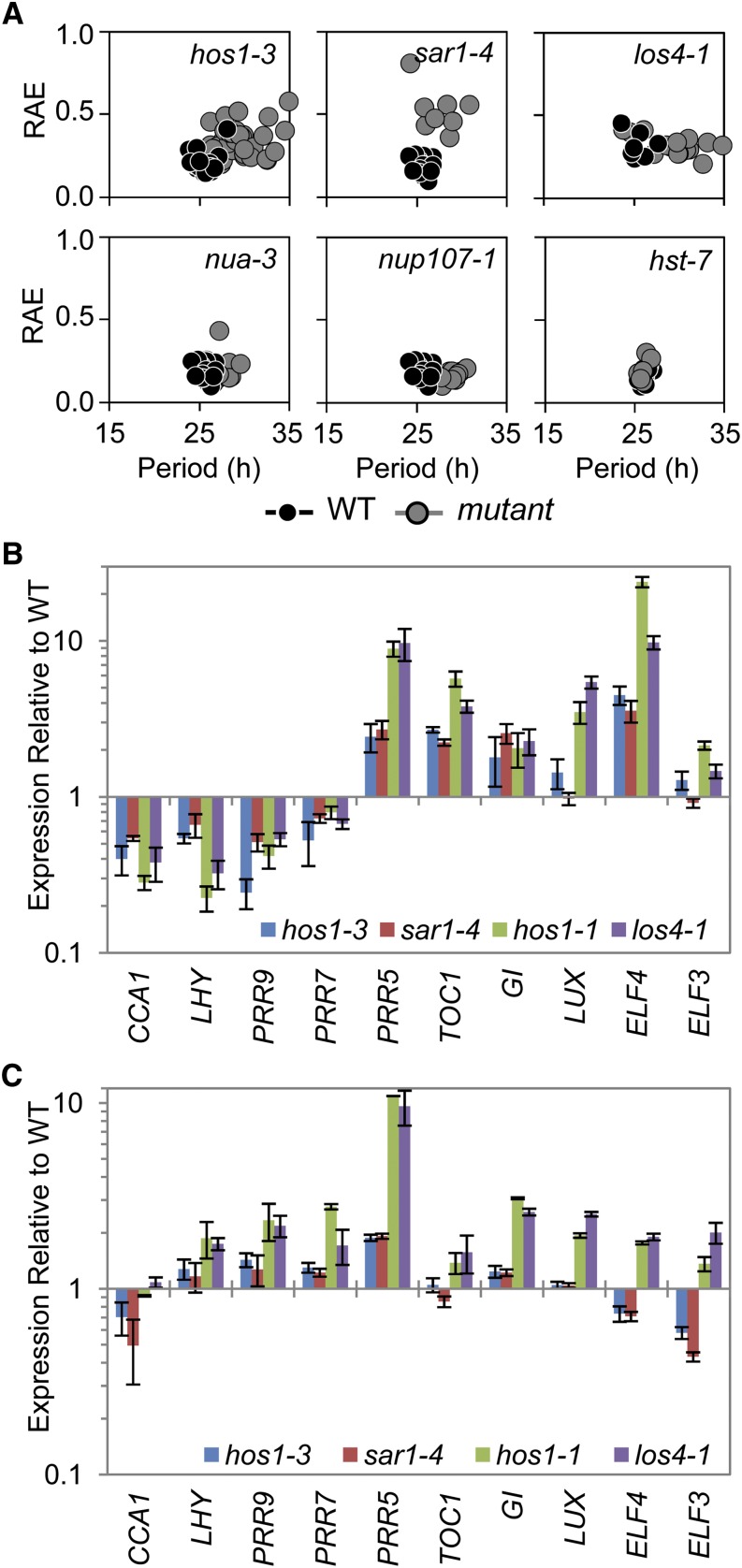
Figure 6.
Mutants Resulting in Altered mRNA Export Exhibit Long-Period Circadian Phenotypes and Alterations to Clock Gene Expression Similar to hos1.
(A) Delayed fluorescence was performed on a range of nuclear pore–associated mutants. Seedlings were grown in 12L:12D white-light cycles for 16 d prior to transferring to constant 17°C and constant red light for analysis. Each plot shows period estimations plotted versus RAE comparing the wild type (WT; black circles) and mutant (gray circles).
(B) and (C) RNA analysis comparing the expression of clock genes in hos1 alleles, sar1-4 and los4-1, to their appropriate wild types. Plants were grown for 2 weeks in 12L:12D white-light cycles and harvested in triplicate at dawn CT0 (B) or dusk CT12 (C). Data are shown relative to the wild-type values for comparison. Data are averages of biological triplicate ± se normalized for transcript abundance to TUB9 (B) or UBQ10 (C). Bar colors are as follows: hos1-3, blue; sar1-4, red; hos1-1, green; and los4-1, purple.
Normal Nucleo-Cytoplasmic mRNA Export Is Required for Circadian Periodicity and Expression of Clock Genes
To determine if the altered nuclear mRNA export is responsible for the long-period phenotype observed in hos1, we assessed the circadian period of four further lines in which nuclear pore function is compromised: suppressor of axr1 (sar1-4), low expression of osmotically responsive genes4 (los4-1), nuclear pore anchor (nua-3), and the recently identified nucleoporin mutant nup107-1. SAR1, LOS4, and NUA are all required for correct mRNA nuclear export (Cernac et al., 1997; Gong et al., 2002, 2005; Dong et al., 2006b; Parry et al., 2006; Xu et al., 2007), and NUP107 is a structural nucleoporin that is part of the evolutionarily conserved Nup107-Nup160 subcomplex with SAR1 (Tamura et al., 2010; Clever et al., 2012). In a manner resembling hos1 mutants, loss of any of these genes led to a long period, as measured by delayed fluorescence (Figure 6A). To demonstrate that the effect on period lengthening was specific to mRNA export, we tested the hasty mutant (hst-7), the Arabidopsis ortholog of the importin β-like proteins that are involved in nucleo-cytoplasmic transport of microRNAs (Bollman et al., 2003; Park et al., 2005). We found that the hst-7 mutant had a wild-type period (Figure 6A). In further support of the role of mRNA export in proper regulation of the circadian clock, the expression of the core circadian clock components was altered in a similar manner in los4-1 or sar1-4 at dawn (Figure 6B) or dusk (Figure 6C) compared with the appropriate wild type. Our results therefore show that period lengthening is a general consequence of disrupting nucleo-cytoplasmic mRNA export in plants and reveal that hos1 mutants have a period defect resulting from an RNA export defect.
Cold Signaling Is Similarly Perturbed by hos1, sar1, and los4
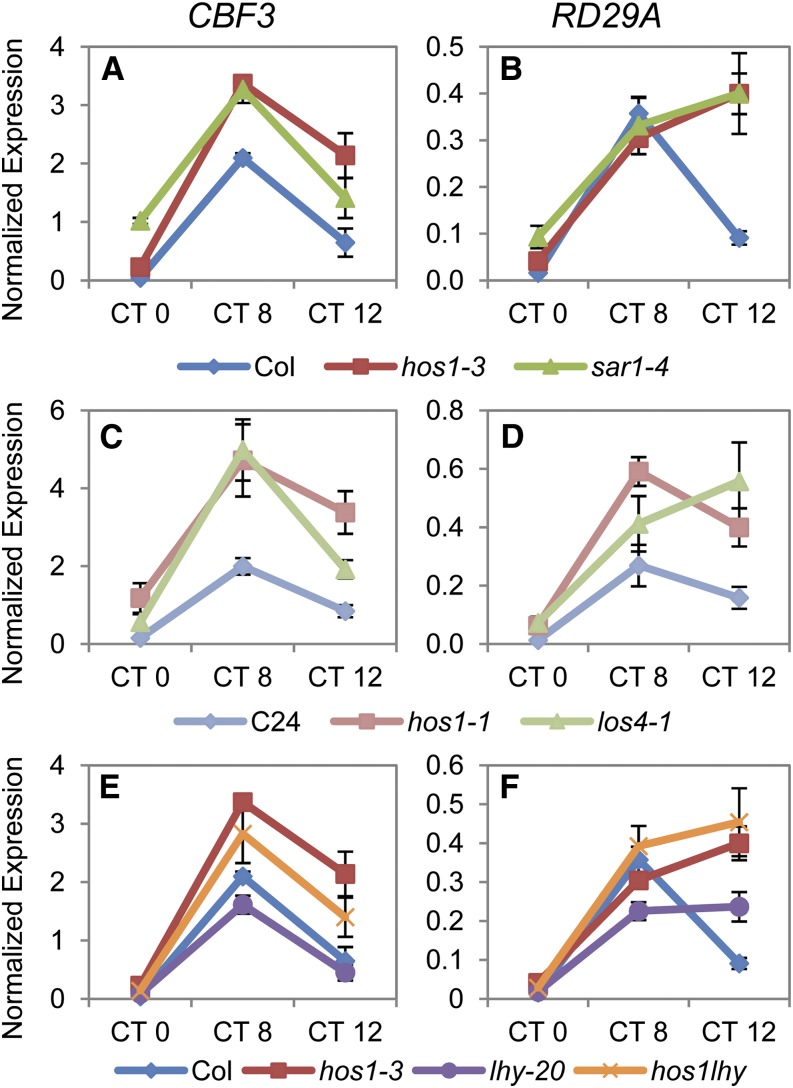
Although loss of HOS1, LOS4, or SAR1 have been shown to result in altered cold responses (Ishitani et al., 1998; Gong et al., 2002, 2005; Dong et al., 2006a, 2006b), these studies have not shown that mutations in these genes lead to similar effects on cold-responsive gene expression. One possibility is that timing of analyses may significantly affect the outcome of experiments, as the cold-signaling genes being monitored are under circadian regulation (Harmer et al., 2000; Kreps et al., 2002), and loss of these nuclear pore components alters circadian function (Figure 6). Therefore, to determine whether cold signaling was perturbed similarly in these mutants, we examined the expression of cold-signaling genes in these mutants in parallel. Expression of CBF3 and RESPONSIVE TO DESSICATION29A (RD29A) was assayed at 0, 8, and 12 h after dawn in seedlings grown in 12L:12D white-light cycles at 22°C (Figures 7A to 7D). The mutations in los4 or sar1 led to similar increases in the expression of CBF3 and its target gene RD29A as observed in hos1 when compared with their wild types. Therefore, when assayed in parallel, the hos1, sar1, and los4 mutants display comparable perturbations of cold-responsive gene expression.
Figure 7.
Expression of Cold Signaling Genes Analyzed in Parallel Indicates Similar Perturbations in hos1 as in sar1-4 and los4-1 and This Perturbation Does Not Require LHY.
Expression analysis of CBF3 ([A], [C], and [E]) and RD29A ([B], [D], and [F]) in the wild type (Col, dark-blue diamonds; C24, light-blue diamonds), hos1 alleles (hos1-3, red squares; hos1-1, pink squares), mRNA export mutants (sar1-4, dark-green triangles; los4-1, light-green triangles), lhy (lhy-20, purple circles), and hos1-3 lhy-20 double mutants (hos1lhy, orange Xs). Plants were grown for 2 weeks under 12L:12D white-light cycles and harvested at dawn (CT0), 8 h after dawn (CT8), or at dusk (CT12). Data are averages of biological triplicate ± se normalized for transcript abundance to UBQ10.
It has been recently demonstrated that differential splicing events occur within LHY in response to cold stress and that plants incubated at 4°C have an increased proportion of a nonfunctional isoform of LHY in which exon E5a is retained (James et al., 2012). We tested the hypothesis that altered cold signaling in hos1 mutants was due to misexpression of LHY by comparing CBF3 and RD29A expression in hos1 and hos1-3 lhy-20 double mutants that therefore lack both isoforms of LHY (Figures 7E and 7F). Expression of these genes in the double mutant was intermediate between that in lhy and hos1, showing that the mechanisms that regulate alternative splicing of LHY alone do not account for the increase in cold-induced gene expression in the hos1 mutant. This work shows that HOS1 affects cold signaling through a pathway that does not require LHY.
DISCUSSION
The circadian clock requires the rhythmic synthesis and degradation of mRNAs encoding transcriptional regulators in plants. Here, we show that loss of HOS1 results in period lengthening and the inability to maintain normal mRNA rhythms under constant conditions (Figures 1 and 3; see Supplemental Figures 1 to 5 online). The hos1 long-period phenotype is shared by four different mutants known to disrupt the nuclear pore complex and/or mRNA export (Figure 6), and we demonstrate that hos1 mutants constitutively accumulate polyadenylated mRNA in the nuclei (Figure 5). These data, together with the double mutant analysis (Figure 2; see Supplemental Figure 6 online), suggest that the hos1 circadian defect is due to the loss of nucleo-cytoplasmic mRNA transport, rather than to an interaction with a specific clock protein. Demonstrating the specificity of the nuclear defect, los4, sar1, and nua, which share hos1’s long-period phenotype (Figure 6A), are unable to export mRNA to the cytoplasm at normal rates (Figure 5; Gong et al., 2005; Parry et al., 2006; Xu et al., 2007), whereas the hst-7 mutant, which affects microRNA rather than mRNA export (Bollman et al., 2003), showed no circadian clock phenotype (Figure 6A). Therefore, we conclude that a nucleo-cytoplasmic mRNA export defect results in the circadian clock defect observed in hos1 mutants.
It is interesting to note that constitutive overexpression of HOS1 does not result in either circadian arrhythmia or a short period, as occurs for many clock components (reviewed in McClung, 2006); instead, HOS1 overexpression rescues the clock defect in hos1-3 to an approximately wild-type period (see Supplemental Figure 7 online). This observation is therefore not consistent with HOS1 playing a central role in the circadian clock. This work also shows that hos1 affects signaling and gene expression by mechanisms other than or in addition to the degradation of specific proteins such as ICE1 and CO (Dong et al., 2006a; Lazaro et al., 2012). Currently it is not clear whether the E3 ubiquitin ligase activity of HOS1 is necessary for its role in the promotion of mRNA export. Double mutant analysis suggests that HOS1 modifies the delayed fluorescence and hypocotyl elongation phenotypes observed in the clock genes analyzed (Figure 2; see Supplemental Figure 6 online), suggesting that HOS1 may not act on the circadian clock through one specific component. However, double mutants with all clock genes could not be isolated due to the close genomic proximity of HOS1 with CCA1, PRR9, and ELF4. Thus, it remains possible that hos1 could show a strong genetic interaction with these mutants.
Although the hos1 long-period phenotype is shared by mutants lacking four different nuclear pore-associated proteins, it is not clear precisely how disruption of mRNA export affects period. A simple explanation would be that altered export of clock gene RNAs results in a reduced ability to maintain period and rhythms of mRNA abundance. In hos1 mutants, gross changes in the transcriptome were revealed by RNA sequencing, affecting genes in multiple functional categories rather than specifically circadian- and cold-regulated processes. Because LHY protein rhythm and target-gene promoter activity appear more rhythmic than those of the corresponding mRNAs (Figure 4), our data strongly suggest that hos1 mutants are disrupted in circadian rhythms primarily because of alterations occurring at the RNA level. The best explanation for the unusual hos1 circadian phenotype is that because protein synthesis occurs in the cytoplasm, as long as a rhythm of mRNA export is maintained, is it possible to get long-period oscillations of promoter:LUC and protein levels without robust total mRNA rhythms. Furthermore, as we measured total mRNA, which includes RNA retained in the nucleus as well as that in the cytoplasm, it is possible that the population that is available for translation is sufficiently rhythmic in hos1 to drive oscillations. An effect of hos1 on circadian protein nuclear import is unlikely in our view, as Saccharomyces cerevisiae mutants defective in nuclear protein import are not viable: Indeed, early embryo-lethal pore mutants have been described in Arabidopsis (Braud et al., 2012).
Loss of HOS1, LOS4, or SAR1 have all been reported to result in altered cold responses; however, the effects of the mutations do not give consistent phenotypes, including two alleles of LOS4 with apparent opposite alterations to cold-signal transduction (Ishitani et al., 1998; Gong et al., 2002, 2005; Dong et al., 2006a, 2006b). Here, we show that when analyzed together, two hos1 alleles, los4 and sar1, all show similarly altered cold signaling; expression of CBF3 and RD29A are upregulated in each mutant when assayed 8 to 12 h after dawn (Figures 7A to 7D). Expression of cold-signaling genes, including CBF3 and RD29A, is under circadian control, with maximal expression occurring at 8 to 12 h after dawn (CT8 to CT12) (Harmer et al., 2000; Mockler et al., 2007). Because circadian function is disrupted in mutants with aberrant mRNA export (Figure 6A), and the circadian clock is required for normal cold responses (Dong et al., 2011; Seo et al., 2012), one possibility is that cold signaling is perturbed as a consequence of altered circadian function. Our analysis of hos1-3 lhy-20 double mutants reveals an intermediate effect on CBF3 and RD29A expression (Figures 7F and 7G), showing that if HOS1 does affect cold signaling in part through the clock, then this pathway is not solely dependent on LHY. Analysis of hos1 lhy cca1 triple mutants could confirm that cold-signaling defects in hos1 mutants do not result from the interaction between mRNA export and circadian clock function.
los4 and sar1 mutants share a suite of phenotypes with hos1 plants, including early flowering (see Supplemental Figure 1 online; Ishitani et al., 1998; Gong et al., 2005; Parry et al., 2006; Lazaro et al., 2012), defective cold signaling (see Supplemental Figure 1 online; Figure 7; Ishitani et al., 1998; Lee et al., 2001; Gong et al., 2005; Dong et al., 2006a, 2006b), and circadian rhythms (Figure 6). This suggests that these phenotypes share a common cause. Our data show that loss of HOS1 causes mRNA retention and this has large consequences for gene expression in hos1 mutants. Further studies of abiotic signaling pathways need to discriminate between the direct and indirect mechanisms through which alterations to these proteins result in the phenotypes and gene expression changes observed.
METHODS
Plant Materials
The genotypes and accessions described herein are as follows. The wild types are Col, C24 expressing RD29A:LUC (C24) (Ishitani et al., 1997), or Landsberg erecta. The hos1 mutants are hos1-1 (Ishitani et al., 1998), hos1-2 (Lazaro et al., 2012), hos1-3 (SALK_069312), hos1-5 (SALK_131629), and hos1-6 (SALK_052108). Circadian clock mutants used have been described and are lhy-20 (Michael et al., 2003), che-2 (Pruneda-Paz et al., 2009), prr9-10 (Ito et al., 2003), prr7-11 (Yamamoto et al., 2003), prr5-11 (Yamamoto et al., 2003), prr3-1 (Para et al., 2007), toc1-101 (Kaczorowski, 2004), gi-201 (Martin-Tryon et al., 2007), lux-5 (Hazen et al., 2005), elf3-1 (Hicks et al., 1996), and ztl-105 (SALK-069091) (Martin-Tryon et al., 2007). Previously published mRNA export mutants were obtained from the Nottingham Arabidopsis Stock Centre and have been previously described: sar1-4 (SALK_126801) (Parry et al., 2006), los4-1 (Gong et al., 2005), nua-3 (SAIL_505_H11) (Xu et al., 2007), nup107-1 (SALK_057072) (Wiermer et al., 2012, Clarkson et al., 1997), and hst-7 (Bollman et al., 2003).
Growth Conditions
For RNA extraction, hypocotyl measurements, confocal microscopy, and mRNA retention, experiments seeds were surface sterilized and stratified at 4°C for 2 to 4 d on Murashige and Skoog (MS) agar (Melford) and 0.9% agar (Sigma-Aldrich) with no Suc added to the media based on Macgregor et al. (2008). For the diurnal RNA measurements in Supplemental Figure 6 online, 0.5× MS and 1.2% agar were used. Unless noted, standard growth conditions were for 2 weeks in 12L:12D white-light cycles (80 µmol/m2/s) at 22°C.
For delayed fluorescence and luminescence techniques a chlorine gas–based methodology was used for sterilizing seeds (Clough and Bent, 1998). The seed was then sown in groups of 10 to 20 seedlings on microtiter plates containing MS media and 1.5% agar. After 2 d at 4°C, seedlings were grown for either 6 d for luciferase experiments or 15 d for delayed fluorescence experiments in 22°C under 12L:12D white-light cycles (80 µmol/m2/s white light). After this time in growth conditions, plates were transferred to experimental conditions at dawn of the 7th or 16th day.
DNA Constructs and Plant Transformation
The 35S:HOS1-CFP lines were created by cloning the full-length cDNA (long isoform according to Lee et al., 2012) using ligation-independent cloning from a pET-YSBLIC3C intermediate vector (Alzari et al., 2006) into a Cambia binary vector (pPCV-CFP-H) between a 2x35S promoter and a 3′ fusion to the CFP sequence. The HOS1 wild-type cDNA was amplified from plasmid available from the ABRC using HiTEL-HOS1 forward primer and HiTEL-HOS1 reverse primer as detailed in Supplemental Data Set 3 online. The COR15a:LUC transgene was made using InFusion cloning where 2.9 kb of the genomic region upstream of COR15a start was placed at the StuI site of a pPCV-based binary vector (LUC-NOS pPCV) (Tóth et al., 2001). The primers used were pCOR15A_F and pCOR15A_R (see Supplemental Data Set 3 online). The CAB2:LUC+ transgene (Hall et al., 2002) and the CCR2:LUC transgene (Doyle et al., 2002) were previously described. hos1-3 plants were transformed with these constructs using standard Agrobacterium tumefaciens–mediated transformation and selected on the appropriate antibiotics. Single insert, homozygous transgenic plants were used in all the assays.
Primers
All primers used for genotyping and qPCR are described in Supplemental Data Set 3 online. Unpublished primers were designed by hand or aided by SIGnAL iSct primers (http://signal.salk.edu/tdnaprimers.2.html) or Primer3 (Rozen and Skaletsky, 2000).
Luminescence and Delayed Fluorescence
Luciferase analysis was performed on 7-d-old seedlings, measuring luminescence every 2 h. Luciferase experimental conditions consisted of constant temperature of 12, 17, or 27°C and 2 d in 12L:12D red-light cycles followed by constant red light (30 µmol/m2/s). Luciferase experimental conditions for Supplemental Figure 4 online were room temperature in constant white light (40 µmol/m2/s). The delayed fluorescence technique was performed on 16-d-old seedlings, measuring delayed fluorescence every hour in 12, 17, or 27°C and constant red or blue light (30 µmol/m2/s) (Gould et al., 2009). Both luminescence and delayed fluorescence were measured and processed as previously described (Gould et al., 2006, 2009). Data analysis was performed on Biodare (http://www.biodare.ed.ac.uk) using BRASS (available from www.amillar.org). BRASS produces individual period estimates as well as relative amplitude errors (RAEs) via fast Fourier transform nonlinear least-squares analysis (Plautz et al., 1997).
Gene Expression Analysis
For Figure 1B, plants were grown using the standard plant growth conditions described above for 10 d and harvested in triplicate at dawn into liquid nitrogen. RNA was extracted using the standard protocols for Qiagen RNeasy plant mini kit, and 1 µg of RNA was converted into cDNA using with Invitrogen’s SuperScript II reverse transcriptase and oligo(dT) 12-18 primer. qPCR was performed on the resulting cDNA using Agilent Technologies Brilliant III Ultra-Fast SYBR Green qPCR master mix and ROX Passive Reference Dye on a Stratagene Mx3000P real-time PCR system with the recommended settings for SYBR Green (with dissociation curve). Data were analyzed with MxPro-Mx3005P v4.10 software. The transcript abundance levels were normalized to UBIQUITIN10 (UBQ10) (Gould et al., 2006).
For the constant light time-series data in Figure 3, seeds were grown for 7 d in 12L:12D white-light cycles (80 µmol/m2/s) at 22°C followed by 2 d in 12L:12D red-light cycles (30 to 40 µmol/m2/s) at 17°C before being transferred to constant red light at 17°C. Seedlings were harvested in triplicate at the times indicated and frozen in liquid nitrogen. RNA was extracted and a cDNA library prepared as in Figure 2B. The qPCR was set up using a liquid handling robot (Tecan Freedom Evo) and run in a LightCycler 480 system (Roche) using LightCycler 480 SYBR green master mix (Roche). Data were analyzed with the Roche LightCycler 480SW 1.5 using advance relative quantification based on the 2nd derivative max method. Each cDNA sample was assayed in triplicate. The transcript abundance levels were normalized to ACTIN7 (ACT7) (Hall et al., 2003).
For red diurnal time series data in Supplemental Figure 6 online, seedlings were grown as for Figure 3 but harvested during 12-h red light and 12-h dark cycles after a total of 13 d of growth into Ambion’s RNAlater and stored at 4°C overnight. Total RNA was extracted using GE Healthcare Illustra RNAspin 96 using a liquid handling robot (Tecan Freedom Evo). RNA was extracted from three biological repeats. Total RNA (1 µg) was reverse transcribed using the Invitrogen Superscript VILO cDNA synthesis kit according to the manufacturer’s instructions. qPCR analysis was done as in Figure 3.
For Figures 5 and 6, seedlings were grown for 14 d using the standard growth conditions described above and then harvested in triplicate into liquid nitrogen at the times indicated. RNA was extracted and cDNA synthesized as in Figure 2B. qPCR was performed as in Figure 2B. The transcript abundance levels were normalized to TUBULIN BETA-9 CHAIN (TUB9) (Koo et al., 2010) for the analysis in Figure 5B and UBQ10 (Gould et al., 2006) for the analysis in Figures 5C and 6.
RNA Sequencing and Analysis
Seeds were grown for 14 d using the standard plant growth conditions described above and harvested at dawn in triplicate. RNA was extracted using the standard protocols for Qiagen RNeasy plant mini kit with the additional steps for DNase treatment. Five micrograms of RNA was submitted to sequencing using standard protocols for Illumina TruSeq (version 3) technologies. The input material was spiked with ∼1% Ambion ERCC RNA spike-in mix. The resulting FASTQ files contained 262,407,174 reads for Col and 258,662,478 reads for hos1-3. These were then filtered using the fastq-mcf program from the ea-utils package (http://code.google.com/p/ea-utils/) with the parameters -x 0.01 -q 20 -p 10 -u. Spike-in reads were removed using Bowtie and the lower limit of detection quantified using custom scripts. The remaining reads were aligned using Tophat v1.4.1 and the following parameters: -I 10,000 -r 50–mate-std-dev 100 –library-type fr-unstranded. The TAIR10 reference sequence was used. The Cufflinks package was used to quantify gene and isoform abundance using the -G flag. The 17 cuffcompare and cuffdiff components of Cufflinks were used to quantify gene and isoform differential expression (Trapnell et al., 2012). A false discovery rate of 5% was used. All other settings were as per the default settings. Further analysis was performed using the cummeRbund package (Trapnell et al., 2012).
Hypocotyl Assays
Seeds were exposed to 2 d of 12L:12D white-light cycles to induce germination followed by 7 d at 22°C in either 12L:12D white-light cycles or constant darkness. The plates were scanned and hypocotyl lengths measured using ImageJ (http://rsbweb.nih.gov/ij/).
In Situ Hybridization
In situ hybridization was performed on plants grown in the standard growth conditions for 14 d and fixed at dawn or dusk of the 12L:12D white-light cycles. Whole-mount in situ hybridization was performed as described (Engler et al., 1994; Gong et al., 2005) with minor modifications. Microscopy was performed on the root tissues using an IX81 motorized inverted microscope (Olympus), equipped with a VS-LMS4 Laser-Merge-System with solid-state lasers (488 nm/70 mW; Visitron System). The 4′,6-diamidino-2-phenylindole (DAPI) was imaged with mercury lamp illumination of the IX81 microscope and standard DAPI filter sets (377 ± 50/406/447 ± 60). For Figure 5A, the ×40 lens and standard GFP illumination settings were used with equivalent exposure times. For Figure 5B, 5 μL of 10 µg/mL of DAPI in water was applied directly to the roots on a slide and plants were mounted in 25 μL of Vectashield. Images were taken using the ×100 lens with standard settings for the Epi488 laser and DAPI with equivalent exposure times across the images.
Protein Extraction and Immunoblotting
Protein was extracted using extraction buffer [50% ethylene glycol, 100 mM Tris-HCl, pH 8.5, 150 mM (NH4)2SO4, 10 mM EDTA, 60 mM NaHSO3, 1 mM DTT, Roche complete EDTA-free protease inhibitor cocktail, and 0.1 M phenylmethylsulfonyl fluoride]. The samples were ground in a tissue-lyzer for at least 2 min and centrifuged for 5 min at 4°C. The supernatant was removed to a new tube and kept at −80°C. Equal volumes of protein were loaded onto precast RunBlue SDS Gel 4-12% (17-wells Expedeon) and run according to the manufacturers’ guidelines. Protein transfer to Invitrogen’s iBlot polyvinylidene difluoride membrane was accomplished using Invitrogen’s iBlot protocol using Program P3 for a total of 8 min. Once transferred, the membrane was blocked overnight and incubated with anti-LHY antibody (Kim et al., 2003) for 2 h at room temperature and probed with the secondary antibody anti-rabbit IgG–horseradish peroxidase (Insight Biotechnology SC-2004) at a 1:3000 dilution. The blot was then reprobed for UGPase using Millipore’s SNAP ID Western Blotting protein detection system according to the manufacturer’s protocols. The anti-UGPase rabbit polyclonal serum (Agrisera AS05-086) was used at 1:6000. Enhanced chemiluminescence quantitation was performed with Millipore’s Immobilon horseradish peroxidase chemiluminescent substrate (Fisher Scientific). Detection was done using G-BOX iChemi chemical imager from Syngene set for 1-, 3-, 5-, and 10-min exposures using GeneSnap image acquisition software.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACT7 (At5g09810), CAB2 (At1g29920), CCA1 (At2g46830), CCR2 (At2g21660), CHE (At5g08330), ELF3 (At2g25930), GI (At1g22770), HOS1 (At2g39810), HST (At3g05040), LHY (At1g01060), LOS4 (At3g53110), LUX (At3g46640), NUA (At1g79280), NUP107 (At3g14120), PRR3 (At5g60100), PRR5 (At5g24470), PRR7 (At5g02810), PRR9 (At2g46790), RD29A (At5g52310), SAR1 (At1g33410), TOC1 (At5g61380), TUB9 (At4g20890), UBQ10 (At4g05320), and ZTL (At5g57360),
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Loss of HOS1 in Col Leads to Early Flowering and Altered Expression of CBF3.
Supplemental Figure 2. The hos1 Mutant Has a Long-Period Circadian Phenotype across a Range of Physiological Temperatures.
Supplemental Figure 3. Alleles of hos1 in Multiple Accessions Share the Long-Period Phenotype as Measured by Delayed Fluorescence.
Supplemental Figure 4. The hos1 Mutant Has a Long-Period Circadian Phenotype across a Range of Physiological Temperatures.
Supplemental Figure 5. The hos1 Mutant Has a Long-Period Circadian Phenotype for LHY:LUC, PRR9:LUC, PRR7:LUC, and TOC1:LUC.
Supplemental Figure 6. HOS1 Is Required for Wild-Type Hypocotyl Elongation and This Phenotype Does Not Require the Clock Mutants Analyzed.
Supplemental Figure 7. Constitutive Overexpression of HOS1-CFP in hos1-3 Compliments the Long-Hypocotyl and Long-Period Delayed Fluorescence Phenotypes of hos1-3.
Supplemental Figure 8. HOS1 Is Required for Correct Expression of Clock Genes at a Variety of Physiological Temperatures under 12L:12D Red-Light Cycles.
Supplemental Data Set 1. Genes Up- or Downregulated Two or More Fold in hos1 Compared with Col.
Supplemental Data Set 2. AmiGO Analysis.
Supplemental Data Set 3. Primer Sequences.
Acknowledgments
This work was funded by Biotechnology and Biological Science Research Council Grants BB/F005296/2 (to S.P.), BB/F005237/1 (to K.J.H.), and BB/F005318/1 (to A.J.H.). We thank Isabelle Carre for the gift of the LHY antibody.
AUTHOR CONTRIBUTIONS
D.R.M. created the stable 35S:HOS1-CFP Arabidopsis lines, isolated and verified the mutant lines from the stock centers, performed RNA extractions, real-time qPCR, hypocotyl assays, and in situ hybridizations, prepared samples for and did the analysis of data resulting from RNA sequencing, and crossed in and performed the analysis of clock gene:luciferase period in hos1 mutants, with assistance from S.B. and R.P. P.G. originally identified hos1-3 as having a clock phenotype and performed circadian delayed fluorescence and luciferase analysis for downstream reporter:luciferase transgenic lines in hos1 with assistance from J.Y. J.F. and J.G. performed RNA extractions and real-time PCR and created the reporter:luciferase transgenic lines in hos1, with assistance from G.S. and K.S. K.P. performed the bioinformatic analysis for the RNA sequencing. S.P. and A.J.H. designed experiments with contributions from A.J.M. and K.J.H. S.P., A.J.H., D.R.M., and P.G. cowrote the article with the assistance of J.F. and K.J.H. S.P., A.J.H., and K.J.H. were principal investigators on the listed grants.
Glossary
- Col
Columbia
- 12L:12D
12-h-light/12-h-dark
- CFP
cyan fluorescent protein
- qPCR
quantitative PCR
- GFP
green fluorescent protein
- MS
Murashige and Skoog
- RAE
relative amplitude error
- DAPI
4′,6-diamidino-2-phenylindole
References
- Alzari P.M., et al. (2006). Implementation of semi-automated cloning and prokaryotic expression screening: The impact of SPINE. Acta Crystallogr. D Biol. Crystallogr. 62: 1103–1113 [DOI] [PubMed] [Google Scholar]
- Bieniawska Z., Espinoza C., Schlereth A., Sulpice R., Hincha D.K., Hannah M.A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman K.M., Aukerman M.J., Park M.-Y., Hunter C., Berardini T.Z., Poethig R.S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493–1504 [DOI] [PubMed] [Google Scholar]
- Braud C., Zheng W., Xiao W. (2012). LONO1 encoding a nucleoporin is required for embryogenesis and seed viability in Arabidopsis. Plant Physiol. 160: 823–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S., AmiGO Hub. Web Presence Working Group (2009). AmiGO: Online access to ontology and annotation data. Bioinformatics 25: 288–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A., Lincoln C., Lammer D., Estelle M. (1997). The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development 124: 1583–1591 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V., Ohta M., Kanrar S., Lee B.-H., Hong X., Agarwal M., Zhu J.-K. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J.-K., Sunkar R. (2010). Gene regulation during cold stress acclimation in plants, In Plant Stress Tolerance, Vol. 639 of Methods in Molecular Biology, R. Sunkar, ed (Totowa, N.J.: Humana Press) pp. 39–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson W.D., Corbett A.H., Paschal B.M., Kent H.M., McCoy A.J., Gerace L., Silver P.A., Stewart M. (1997). Nuclear protein import is decreased by engineered mutants of nuclear transport factor 2 (NTF2) that do not bind GDP-Ran. J. Mol. Biol. 272: 716–730 [DOI] [PubMed] [Google Scholar]
- Clever M., Funakoshi T., Mimura Y., Takagi M., Imamoto N. (2012). The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus 3: 187–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Covington M.F., Maloof J.N., Straume M., Kay S.A., Harmer S.L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A.R. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Doherty C.J., Van Buskirk H.A., Myers S.J., Thomashow M.F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21: 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.-H., Agarwal M., Zhang Y., Xie Q., Zhu J.-K. (2006a). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103: 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.-H., Hu X., Tang W., Zheng X., Kim Y.S., Lee B.H., Zhu J.-K. (2006b). A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol. Cell. Biol. 26: 9533–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M.A., Farré E.M., Thomashow M.F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M.R., Davis S.J., Bastow R.M., McWatters H.G., Kozma-Bognár L., Nagy F., Millar A.J., Amasino R.M. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419: 74–77 [DOI] [PubMed] [Google Scholar]
- Engler J.A., Montagu M., Engler G. (1994). Hybridization in situ of whole-mount messenger RNA in plants. Plant Mol. Biol. Rep. 12: 321–331 [Google Scholar]
- Fowler S., Thomashow M.F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S.G., Cook D., Thomashow M.F. (2005). Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 137: 961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S.J., Zarka D.G., Stockinger E.J., Salazar M.P., Houghton J.M., Thomashow M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Gong Z., Dong C.-H., Lee H., Zhu J., Xiong L., Gong D., Stevenson B., Zhu J.-K. (2005). A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 17: 256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Lee H., Xiong L., Jagendorf A., Stevenson B., Zhu J.-K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99: 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould P.D., Diaz P., Hogben C., Kusakina J., Salem R., Hartwell J., Hall A. (2009). Delayed fluorescence as a universal tool for the measurement of circadian rhythms in higher plants. Plant J. 58: 893–901 [DOI] [PubMed] [Google Scholar]
- Gould P.D., Locke J.C., Larue C., Southern M.M., Davis S.J., Hanano S., Moyle R., Milich R., Putterill J., Millar A.J., Hall A. (2006). The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Bastow R.M., Davis S.J., Hanano S., McWatters H.G., Hibberd V., Doyle M.R., Sung S., Halliday K.J., Amasino R.M., Millar A.J. (2003). The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A., Kozma-Bognár L., Bastow R.M., Nagy F., Millar A.J. (2002). Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J. 32: 529–537 [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.-S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Hazen S.P., Schultz T.F., Pruneda-Paz J.L., Borevitz J.O., Ecker J.R., Kay S.A. (2005). LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. USA 102: 10387–10392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks K.A., Millar A.J., Carré I.A., Somers D.E., Straume M., Meeks-Wagner D.R., Kay S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274: 790–792 [DOI] [PubMed] [Google Scholar]
- Ibáñez C., Kozarewa I., Johansson M., Ogren E., Rohde A., Eriksson M. (2010). Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 153: 1823–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T. (2010). Arabidopsis circadian clock and photoperiodism: Time to think about location. Curr. Opin. Plant Biol. 13: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Lee H., Stevenson B., Zhu J.-K. (1998). HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M., Xiong L., Stevenson B., Zhu J.K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Matsushika A., Yamada H., Sato S., Kato T., Tabata S., Yamashino T., Mizuno T. (2003). Characterization of the APRR9 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 44: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- James A.B., Syed N.H., Bordage S., Marshall J., Nimmo G.A., Jenkins G.I., Herzyk P., Brown J.W., Nimmo H.G. (2012). Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24: 961–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.-H., Seo P.J., Park C.-M. (2012). The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J. Biol. Chem. 287: 43277–43287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczorowski, K. (2004). Mutants in Phytochrome-Dependent Seedling Photomorphogenesis and Control of the Arabidopsis Circadian Clock. (Berkeley, CA: University of California). [Google Scholar]
- Kim J.Y.Y., Song H.R.R., Taylor B.L., Carré I.A. (2003). Light-regulated translation mediates gated induction of the arabidopsis clock protein LHY. The EMBO J. 22: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo S.C., et al. (2010). Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box gene AGAMOUS-LIKE6. Plant J. 62: 807–816 [DOI] [PubMed] [Google Scholar]
- Kreps J.A., Wu Y., Chang H.-S., Zhu T., Wang X., Harper J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A., Valverde F., Piñeiro M., Jarillo J.A. (2012). The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24: 982–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Xiong L., Gong Z., Ishitani M., Stevenson B., Zhu J.K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo-cytoplasmic partitioning. Genes Dev. 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim S.H., Kim J.J., Ahn J.H. (2012). Alternative splicing and expression analysis of high expression of osmotically responsive genes1 (HOS1) in Arabidopsis. BMB Rep 45: 515–520 [DOI] [PubMed] [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor D.R., Deak K.I., Ingram P.A., Malamy J.E. (2008). Root system architecture in Arabidopsis grown in culture is regulated by sucrose uptake in the aerial tissues. Plant Cell 20: 2643–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Tryon E.L., Kreps J.A., Harmer S.L. (2007). GIGANTEA acts in blue light signaling and has biochemically separable roles in circadian clock and flowering time regulation. Plant Physiol. 143: 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R. (2006). Plant circadian rhythms. Plant Cell 18: 792–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J., Bargues M., Terol J., Pérez-Alonso M., Salinas J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael T.P., Salomé P.A., Yu H.J., Spencer T.R., Sharp E.L., McPeek M.A., Alonso J.M., Ecker J.R., McClung C.R. (2003). Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Miura K., Jin J.B., Lee J., Yoo C.Y., Stirm V., Miura T., Ashworth E.N., Bressan R.A., Yun D.-J., Hasegawa P.M. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19: 1403–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T.C., Michael T.P., Priest H.D., Shen R., Sullivan C.M., Givan S.A., McEntee C., Kay S.A., Chory J. (2007). The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Nagel D.H., Kay S.A. (2012). Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22: R648–R657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N., Kusano M., Fukushima A., Kita M., Ito S., Yamashino T., Saito K., Sakakibara H., Mizuno T. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nozue K., Covington M.F., Duek P.D., Lorrain S., Fankhauser C., Harmer S.L., Maloof J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Para A., Farré E.M., Imaizumi T., Pruneda-Paz J.L., Harmon F.G., Kay S.A. (2007). PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.Y., Wu G., Gonzalez-Sulser A., Vaucheret H., Poethig R.S. (2005). Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G., Ward S., Cernac A., Dharmasiri S., Estelle M. (2006). The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18: 1590–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz J.D., Straume M., Stanewsky R., Jamison C.F., Brandes C., Dowse H.B., Hall J.C., Kay S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H. (2000). Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Seo P.J., Park M.-J., Lim M.-H., Kim S.-G., Lee M., Baldwin I.T., Park C.-M. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharabi-Schwager M., Lers A., Samach A., Guy C.L., Porat R. (2010). Overexpression of the CBF2 transcriptional activator in Arabidopsis delays leaf senescence and extends plant longevity. J. Exp. Bot. 61: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straszer K., Hurt E. (2000). Yra1p, a conserved nuclear RNA-binding protein, interacts directly with mex67p and is required for mRNA export. The EMBO Journal 19: 410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth R., Kevei E., Hall A., Millar A.J., Nagy F., Kozma-Bognár L. (2001). Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 127: 1607–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Tazi J. (2002). mRNA degradation machines in eukaryotic cells. Biochimie 84: 821–837 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Cheng Y.T., Imkampe J., Li M., Wang D., Lipka V., Li X. (2012). Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 70: 796–808 [DOI] [PubMed] [Google Scholar]
- Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., Meier I. (2007a). NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell 19: 1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Sato E., Shimizu T., Nakamich N., Sato S., Kato T., Tabata S., Nagatani A., Yamashino T., Mizuno T. (2003). Comparative genetic studies on the APRR5 and APRR7 genes belonging to the APRR1/TOC1 quintet implicated in circadian rhythm, control of flowering time, and early photomorphogenesis. Plant Cell Physiol. 44: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Yang T., Chaudhuri S., Yang L., Du L., Poovaiah B.W. (2010). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285: 7119–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Shi H., Lee B.H., Damsz B., Cheng S., Stirm V., Zhu J.-K., Hasegawa P.M., Bressan R.A. (2004). An Arabidopsis homeodomain transcription factor gene, HOS9, mediates cold tolerance through a CBF-independent pathway. Proc. Natl. Acad. Sci. USA 101: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]