This study shows that cryptochrome2 (CRY2) mediates light suppression of leaf senescence in soybean by interacting with the bHLH transcription factor CIB1. In the absence of blue light or CRY2 signal, CIB1 binds to the E-box DNA motif of the promoter of senescence-associated genes to promote leaf senescence. In blue light, CRY2 interacts with CIB1 to inhibit its DNA binding and leaf senescence.
Abstract
Cryptochromes are blue light receptors that regulate light responses in plants, including various crops. The molecular mechanism of plant cryptochromes has been extensively investigated in Arabidopsis thaliana, but it has not been reported in any crop species. Here, we report a study of the mechanism of soybean (Glycine max) cryptochrome2 (CRY2a). We found that CRY2a regulates leaf senescence, which is a life history trait regulated by light and photoperiods via previously unknown mechanisms. We show that CRY2a undergoes blue light–dependent interaction with the soybean basic helix-loop-helix transcription activator CIB1 (for cryptochrome-interacting bHLH1) that specifically interacts with the E-box (CANNTG) DNA sequences. Analyses of transgenic soybean plants expressing an elevated or reduced level of the CRY2a or CIB1 demonstrate that CIB1 promotes leaf senescence, whereas CRY2a suppresses leaf senescence. Results of the gene expression and molecular interaction analyses support the hypothesis that CIB1 activates transcription of senescence-associated genes, such as WRKY DNA BINDING PROTEIN53b (WRKY53b), and leaf senescence. CIB1 interacts with the E-box–containing promoter sequences of the WRKY53b chromatin, whereas photoexcited CRY2a interacts with CIB1 to inhibit its DNA binding activity. These findings argue that CIB-dependent transcriptional regulation is an evolutionarily conserved CRY-signaling mechanism in plants, and this mechanism is opted in evolution to mediate light regulation of different aspects of plant development in different plant species.
INTRODUCTION
Cryptochromes are the photolyase-related blue light receptors that regulate light responses and the circadian clock in all major evolutionary lineages, from microbials to plants and animals (Cashmore, 2003; Sancar, 2003; Chaves et al., 2011; H. Liu et al., 2011). The molecular mechanism of cryptochrome signal transduction in plants has been studied almost exclusively in the model plant Arabidopsis thaliana. The Arabidopsis genome encodes two cryptochromes, CRY1 and CRY2, which mediate primarily blue light suppression of hypocotyl elongation (Ahmad and Cashmore, 1993) and photoperiodic response of flowering time (Guo et al., 1998). Arabidopsis CRY1 and CRY2 are nuclear proteins that regulate photomorphogenic responses by at least two different mechanisms: proteolysis and transcription (Wu and Spalding, 2007; Yu et al., 2007; H. Liu et al., 2011). For example, photoexcited CRY2 physically interacts with SUPPRESSOR OF PHYTOCHROME A1 (SPA1) to suppress the activity of RING E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1, resulting in the accumulation of the CONSTANS and transcription of FLOWERING LOCUS T (FT) (Valverde et al., 2004; Endo et al., 2007; Jang et al., 2008; L.J. Liu et al., 2008; Zuo et al., 2011). In addition to SPA1, CRY2 also interacts with the basic helix-loop-helix (bHLH) transcription factor CIB1 (for cryptochrome-interacting bHLH1; H. Liu et al., 2008; Kennedy et al., 2010; Idevall-Hagren et al., 2012). CIB1 is a bHLH transcriptional factor that binds to the G-box (CACGTG) DNA motif in vitro, but heterodimerizes with other CIB1-related proteins that bind to the E-box (CANNTG) sequences to regulate transcription in vivo (H. Liu et al., 2008; Liu et al., 2013). The blue light–dependent CRY2–CIB1 interaction stimulates the transcriptional activation activity of CIB1 and transcription of the flowering integrator gene FT to promote floral initiation. Interestingly, the CRY-bHLH-E-box complex also exists in animals, presumably resulting from convergent evolution. For example, the bHLH proteins BMAL1 and CLOCK interact with cryptochromes to suppress E-box–driven transcription in mammals and zebra fish (Griffin et al., 1999; Kume et al., 1999; Shearman et al., 2000; Ishikawa et al., 2002; Zhang and Kay, 2010). However, it is unknown whether the CIB-dependent CRY signaling mechanism is evolutionarily conserved in plants or how important this mechanism is in other light responses or plant species.
Similar to the control of flowering time, leaf senescence is a life history trait regulated by not only developmental programs but also environmental conditions, such as light (Quirino et al., 2000; Lim et al., 2007; Zentgraf et al., 2010; Wu et al., 2012). For example, photoperiod-dependent regulation of the onset of leaf senescence has been reported in Xanthium pensylvanicum Wallr., aspen (Populus spp), and soybean (Glycine max) (Krizek et al., 1966; Keskitalo et al., 2005; Han et al., 2006). It has been reported that short-day (SD) photoperiods promote leaf senescence in soybean by a mechanism independent from the SD promotion of floral initiation (Han et al., 2006). Leaf senescence has been extensively investigated in Arabidopsis. It has been established that leaf senescence is governed by the developmental program via the actions of transcription regulators, such as WRKY DNA BINDING PROTEIN53 (WRKY53), for which the activities and expressions are modulated by phytohormones and environmental factors (Quirino et al., 2000; Lim et al., 2007; Zentgraf et al., 2010; Wu et al., 2012). However, how light regulates leaf senescence remains unclear. For example, depending on the experimental conditions, light, shade, and darkness have all been reported to promote leaf senescence in Arabidopsis, suggesting a complex mechanism of the light regulation of leaf senescence (Nooden et al., 1996; Weaver and Amasino, 2001; Lin and Wu, 2004; Parlitz et al., 2011). Multiple photoreceptors may act redundantly or antagonistically to regulate leaf senescence because none of the Arabidopsis photoreceptor mutants tested, including cry1 cry2 and hy2 hy3 double mutants, showed apparent defects in the light-dependent control of leaf senescence (Weaver and Amasino, 2001). Therefore, despite the fact that most of our current understanding of the developmental and hormonal controls of leaf senescence have resulted from molecular genetic studies of Arabidopsis, it remains elusive exactly which photoreceptors mediate light regulation of leaf senescence or how photoreceptors regulate leaf senescence in this model organism (Quirino et al., 2000; Lim et al., 2007; Zentgraf et al., 2010; Wu et al., 2012).
We previously reported a systematic analysis of cryptochromes in soybean (Zhang et al., 2008). The soybean genome encodes at least six cryptochromes, including four CRY1 (CRY1a, CRY1b, CRY1c, and CRY1d) and two CRY2 (CRY2a and CRY2b) proteins. We found that the rhythmic expression of CRY1a exhibits a strong correlation with the latitudinal cline in the photoperiod-dependent control of flowering time in soybean accessions and that CRY1a activates flowering in transgenic Arabidopsis, suggesting the involvement of CRY1a in the regulation of this life history trait in soybean (Zhang et al., 2008). The protein stability of CRY2a exhibits blue light–specific and ubiquitin-26S proteasome–dependent regulation, but its physiological functions were hitherto unclear. In this study, we investigated the function and action mechanism of CRY2a. We found that photoexcited CRY2a physically interacts with the bHLH transcription factor CIB1 to suppress its DNA binding activity and that CRY2a acts antagonistically with CIB1 to mediate light regulation of leaf senescence in soybean.
RESULTS
CRY2a Interacts with CIB1 in Response to Blue Light in Yeast and in Vitro
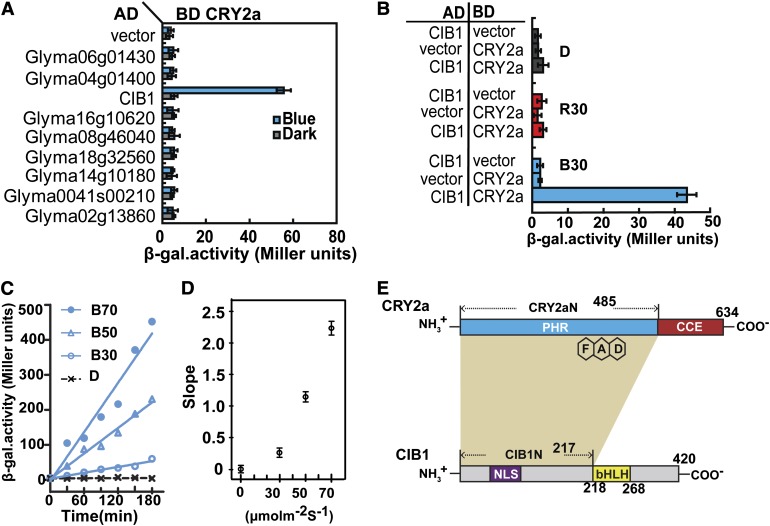
To test whether the CIB-dependent CRY-signaling mechanism is evolutionarily conserved in plants (H. Liu et al., 2008), we analyzed the soybean genome to identify bHLH proteins that are homologous to Arabidopsis CIB1. We cloned the cDNAs corresponding to nine of the 12 bHLH proteins that are closely related to Arabidopsis CIB1 (H. Liu et al., 2008; Schmutz et al., 2010) (see Supplemental Figure 1 online) and tested these bHLH proteins for possible interaction with CRY2a using yeast two-hybrid assay. Among the nine proteins tested by yeast two-hybrid assay with the auxotrophic reporter, only one (Glyma11g12450) that exhibits the highest amino acid sequence similarity to Arabidopsis CIB1 (see Supplemental Figure 1A online) showed obvious blue light–dependent interaction with CRY2a (see Supplemental Figure 2 online). We named this gene Gm-CIB1 and its translation product Gm-CIB1. For simplicity, here, we refer to GmCRY2a and GmCIB1 as CRY2a and CIB1, respectively. Identification of a soybean CIB implies that the origin of the CRY-CIB complex precedes at least the divergence of Cruciferae and Fabaceae. We investigated in more detail whether soybean CIB1 is a bona fide CRY2a-interacting protein by additional analyses (Figure 1; see Supplemental Figures 3 to 6 online). First, we confirmed the wavelength specificity of the CRY2a–CIB1 interaction in yeast cells (Figures 1A and 1B). Yeast cells irradiated with blue light at a fluence rate of 30 µmol m−2 s−1 showed appreciable reporter (β-galactosidase [β-gal]) activity after 120 min of irradiation (Figure 1B, B30), but no β-gal activity was detected in cells incubated in darkness (Figures 1B and 1D) or irradiated with red light of 30 µmol m−2 s−1 for the same time (Figure 1B, R30). Second, we showed that CRY2a exhibited a stronger interaction with CIB1 in response to a higher fluence rate of blue light (Figures 1C and 1D). A Jonckheere-Terpstra trend analysis of the results shown in Figure 1C confirms that the CRY2a–CIB1 interaction is fluence rate dependent (P = 0.003). To better understand the evolutionary history of the CRY-CIB complex, we tested whether Arabidopsis CRY2 interacts with soybean CIB-related proteins (see Supplemental Figure 3 online). Interestingly, in contrast with soybean CRY2a, which interacts with CIB1 but not with the other eight CIB-related proteins under the conditions tested, Arabidopsis CRY2 interacted with not only soybean CIB1, but also two more CIB-related soybean proteins (see Supplemental Figure 3A online). Consistent with its relatively lower specificity, Arabidopsis CRY2 also binds to soybean CIB1 with a relatively higher affinity than its soybean counterpart. It took ∼3 to 4 times longer for the CRY2a–CIB1 interaction to result in a similar level of reporter gene expression in yeast cells than the CRY2–CIB1 interaction (see Supplemental Figure 3B online). It is conceivable that other soybean cryptochromes might interact with other CIB proteins, but detailed relationships between soybean CRYs and CIBs remain to be determined. In the second experiment, we tested the blue light–dependent CRY2a–CIB1 interaction in vitro, using a light-responsive pull-down assay that we had previously established (Li et al., 2011). In this experiment, lysates of insect (Sf9) cells expressing CRY2a were mixed with lysates of insect cells expressing CIB1, and the CRY2a–CIB1 interaction was examined under different light conditions by a coimmunoprecipitation assay (Figure 2A). The results of this experiment show that CIB1 pulled down CRY2a from the reaction mixture irradiated with blue light, but not from the similar reaction mixture incubated in darkness, demonstrating again the blue light–dependent formation of the CRY2a-CIB1 complex.
Figure 1.
CRY2a Interacts with CIB1 in Response to Blue Light in Yeast Cells.
(A) β-Gal assay showing the interaction of CRY2a with CIB1 and other bHLH homologs in yeast cells treated with blue light (30 μmol m−2 s−1) or darkness. AD, activation domain; BD, binding domain.
(B) β-Gal assays showing the interaction of CRY2a and CIB1 in yeast cells treated with red light (R30, 30 μmol m−2 s−1), blue light (B30, 30 μmol m−2 s−1) or darkness (D) for 2 h. Yeast cells expressing various baits and preys are indicated. Means of three independent replicates and sd are shown ([A] and [B]).
(C) β-Gal assay showing the interaction of CRY2a and CIB1 in response to blue light of different fluence rates (D, darkness; B30, 30 μmol m−2 s−1; B50, 50 μmol m−2 s−1; B70, 70 μmol m−2 s−1) for the durations indicated. Increased β-gal activities of the indicated samples fitted by linear regression are shown.
(D) Slopes of linear regression curves of different fluence rates as shown in (C). The means (±sd) of three replicates of individual samples are plotted to show the metric of association kinetics in response to fluence rates of blue light (Jonckheere-Terpstra trend analysis by SPSS program, P = 0.003, n = 3).
(E) Schematic representation depicting the domains of CRY2a and CIB1 that are required for the CRY2a–CIB1 interaction (ocher shade).
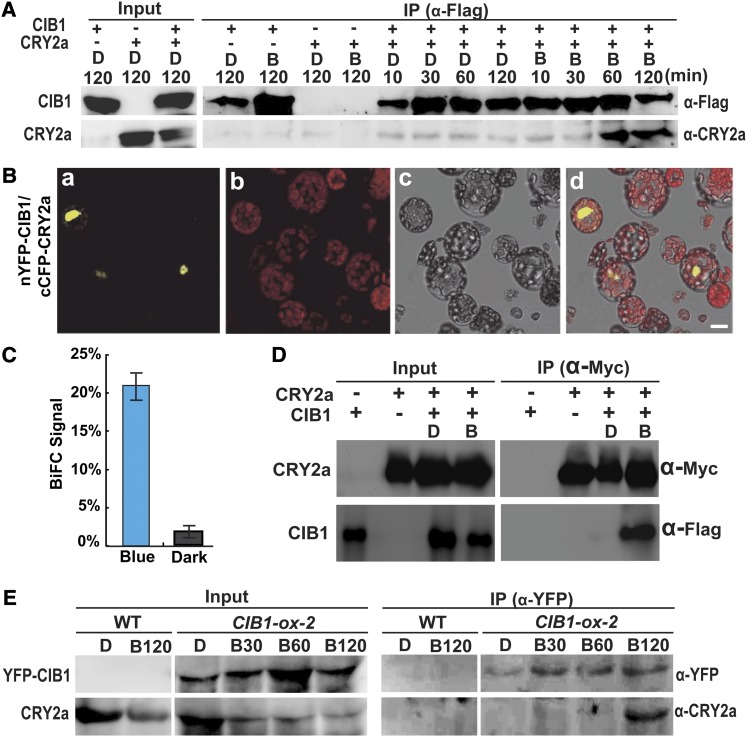
Figure 2.
CRY2a Interacts with CIB1 in Response to Blue Light in Vitro and in Plant Cells.
(A) A pull-down assay showing the blue light–dependent CRY2a–CIB1 interaction in vitro. Agarose beads conjugated with anti-Flag antibody (α-Flag) were mixed with the lysate of insect cells expressing 6His-CIB1-Flag (CIB1) and 6His-CRY2a (CRY2a). The mixture was treated with blue light (B, 22 μmol m−2 s−1) or darkness for the indicated durations. The bound proteins were eluted after washing and analyzed by immunoblots probed with anti-Flag antibody (α-Flag), stripped, and reprobed with anti-CRY2a antibody (α-CRY2a). IP, immunoprecipitation.
(B) BiFC assay showing the blue light–dependent CRY2a–CIB1 interaction in Arabidopsis protoplasts cotransfected with the plasmids encoding nYFP-CIB1 and cCFP-CRY2a. The mesophyll protoplasts of 4-week-old plants grown in LD (16 h light/8 h dark) conditions were cotransformed with plasmids encoding the indicated proteins, incubated for 12 h in the dark, and then transferred to blue light (22 μmol m−2 s−1) for 30 min prior to the confocal microscopy analysis. Image a, YFP fluorescence; image b, autofluorescence; image c, bright field; image d, merge of images a to c. Bar = 10 µm.
(C) The percentage of protoplasts that showed BiFC fluorescence signals was counted. Each sample contains at least 50 protoplasts. Means and sd (n = 3) are shown. P = 0.00026 (Student’s t test).
(D) Ex vivo coimmunoprecipitation assay showing blue light–dependent formation of the CRY2a-CIB1 complex in N. benthamiana. Young leaves were infiltrated with Agrobacteria harboring the plasmids encoding CIB1-Flag (CIB1) or CRY2a-Myc (CRY2a) as indicated, kept in continuous white light for 2 d, moved to darkness for 1 d, and then exposed to blue light (B; 22 μmol m−2 s−1) for 1 h or kept in darkness (D). The protein extracts were incubated with the agarose conjugated with anti-Myc antibody at 4°C for 60 min. Beads were collected and washed three times prior to the elution of immunoprecipitation products. Immunoblots of the total protein extracts (Input) and the IP product were performed using the anti-Myc antibody (α-Myc) and anti-Flag antibody (α-Flag), sequentially.
(E) Coimmunoprecipitation assays showing the blue light–dependent formation of the CRY2a-CIB1 complex in soybean. The wild-type (WT) soybean KN18 and a soybean transgenic line (line 2) overexpressing the Pro35S:YFP-CIB1 transgene (CIB1-ox-2) were grown in SD (8 h light/16 h dark) conditions for 2 weeks. Plants were transferred to darkness for 18 h and exposed to blue light (22 μmol m−2 s−1) for the time indicated periods of time (D, 0 min; B30, 30 min; B60, 60 min; B120, 120 min). Immunoblots of the protein extracts (Input) and the immunoprecipitation products using the agarose conjugated with anti-GFP antibody (α-GFP) were probed by anti-YFP antibody (α-YFP), stripped, and reprobed by anti-CRY2a antibody (α-CRY2a).
CRY2a Interacts with CIB1 in Response to Blue Light in Planta
We next examined the blue light–dependent CRY2a–CIB1 interaction in plant cells. In the first experiment, we used the bimolecular fluorescence complementation (BiFC) assay to test the CRY2a–CIB1 interaction in Arabidopsis protoplasts (Figures 2B and 2C; see Supplemental Figure 4 online). In this experiment, Arabidopsis protoplasts were cotransfected with two plasmids, which express the cCFP-CRY2a (C-terminal portion of CFP fusion with CRY2) or nYFP-CIB1 (N-terminal portion of YFP fusion with CIB1) fusion protein, respectively. The cotransfected protoplasts were incubated in darkness or illuminated with blue light, and the CRY2a–CIB1 interaction was examined and quantified by the percentage of cells exhibiting the BiFC signal. As expected, few (<3%) of the cotransfected protoplasts kept in the dark showed BiFC signals (Figure 2C; see Supplemental Figure 4 online). By contrast, when the cotransfected protoplasts were exposed to blue light for 30 min, more than 20% of the protoplasts showed BiFC signals (Figures 2B and 2C). This result demonstrates a blue light–dependent interaction between the cCFP-CRY2a and nYFP-CIB1 fusion proteins in Arabidopsis protoplasts (P = 0.00026, Student’s t test). The BiFC signals were detected primarily in the nucleus (Figures 2B and 2C; see Supplemental Figure 4 online), suggesting a function of the CRY2a-CIB1 complex in the nucleus. We then used both the BiFC assay and the yeast two-hybrid assay to map the interacting domains of CRY2a and CIB1. The results of both experiments indicate that, similar to the Arabidopsis CRY2–CIB1 interaction (H. Liu et al., 2008), the flavin adenine dinucleotide-containing photolyase domain of soybean CRY2a and the N-terminal region of soybean CIB1 are necessary and sufficient for the blue light–dependent physical interaction of these two proteins (Figure 1E; see Supplemental Figures 5 and 6 online).
We next examined whether CRY2a and CIB1 could form protein complexes in plants in response to blue light. We first tested for the presence of the blue light–dependent CRY2a-CIB1 protein complex in tobacco (Nicotiana benthamiana) leaves transiently expressing CRY2a and CIB1 (Figure 2C). Results of this experiment show that CRY2a and CIB1 form a protein complex in transfected tobacco leaves exposed to blue light but not in transfected tobacco leaves incubated in darkness. To test the CRY2a-CIB1 complex in soybean, we prepared transgenic soybean plants overexpressing yellow fluorescent protein (YFP)–tagged CIB1 and tested for the CRY2a-CIB1 complex by coimmunoprecipitation, using the anti-CRY2a and anti-YFP antibodies (Figure 2E). Results of this experiment demonstrate that the CRY2a-CIB1 protein complex is detected only in soybean plants exposed to blue light, but not in those kept in darkness. Taken together, we conclude that CIB1 is a blue light–specific CRY2a-interacting protein. Based on this finding, we hypothesize that CIB1 is a signaling partner of CRY2a and that the blue light– and CIB-dependent transcriptional regulation is an evolutionarily conserved mechanism of CRY signal transduction regulating light responses in plants.
CIB1 Is an E-Box DNA Binding Transcription Activator
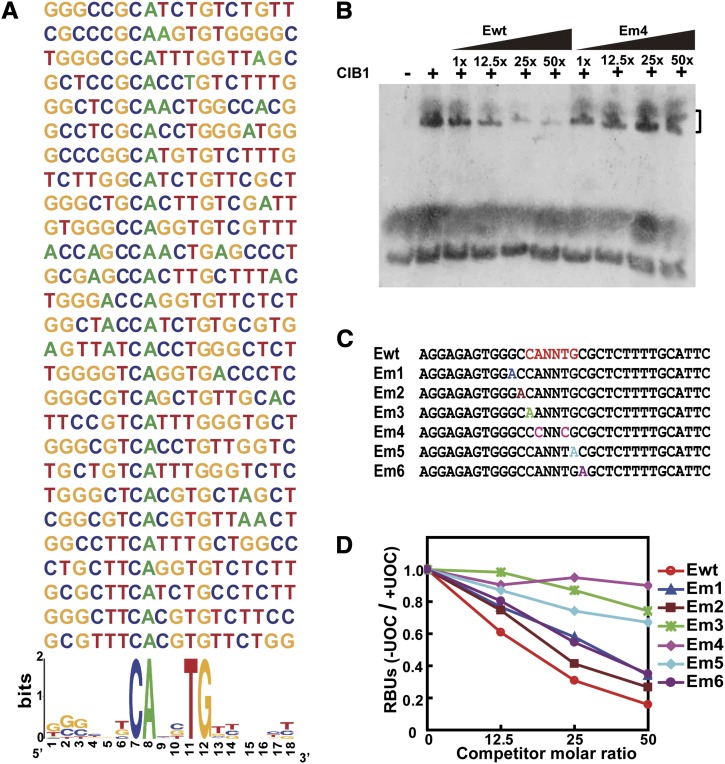
To test the above hypothesis, we investigated whether CIB1 is a sequence-specific DNA binding protein. We first used the random binding site selection assay to examine a possible CIB1–DNA interaction (H. Liu et al., 2008). This experiment shows that CIB1 preferentially binds to the E-box (CANNTG) DNA sequence in vitro (Figure 3A). We further analyzed the DNA binding specificity of CIB1 by the conventional oligonucleotide competition electrophoretic mobility shift assay (EMSA). Results of this experiment show that CIB1 binds to the E box sequences that contain arbitrary nucleotides at the two central variable positions of the sequence CANNTG (Figure 3B). Because the two central variable positions were not defined in this experiment, possible specificity of CIB1 to specific E-box sequences remain to be examined. Mutations of the invariable residues of the E-box (Em3, Em4, and Em5) effectively abolished the CIB1–DNA interaction, whereas mutations outside the invariable residues of the E box (Em1, Em2, and Em6) had little effect on the CIB1–DNA interaction (Figures 3C and 3D). These results demonstrate that CIB1 is a sequence-specific DNA binding protein.
Figure 3.
CIB1 Is a DNA Binding Protein Interacting with the E-Box (CANNTG) DNA Sequence.
(A) The alignment of DNA sequences selected by CIB1 via the random binding site selection assay (see Methods). Over 80% sequences selected by CIB1 contain the E-box element (CANNTG) (http://weblogo.berkeley.edu/).
(B) A competitive EMSA showing the interaction of CIB1 with the DIG-labeled E-box DNA. The CIB1–DNA interaction was completed by the unlabeled wild-type E-box (Ewt) or the mutant E-box (Em4) as shown in (C). Black wedges represent increasing amounts of competitors (12.5×, 25×, and 50× in molar excess).
(C) The DNA sequences of the wild-type E-box DNA (Ewt) and mutant E-box sequence (Em) competitors.
(D) A quantitative analysis of the competitive EMSA using the Ewt or Em competitors. Signals of the CIB1-bound probe in the presence of unlabeled oligonucleotide competitor (+UOC) are normalized by that in the absence of the unlabeled oligonucleotide competitor (-UOC) and presented as RBUs.
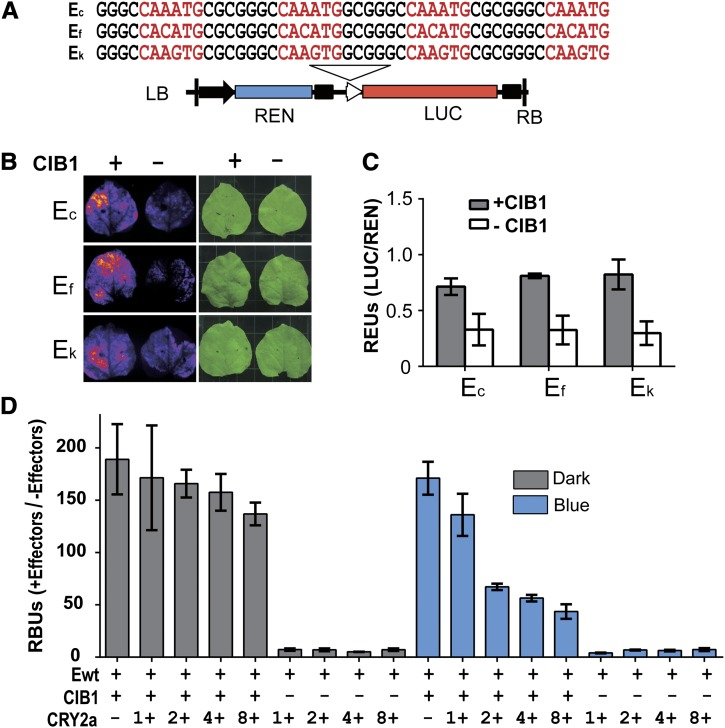
We next examined whether CIB1 might act as a transcription regulator in plant cells, using a dual luciferase in planta assay similar to that we previously reported (H. Liu et al., 2008). In this experiment, tobacco leaves were cotransformed with Agrobacterium tumefaciens strains harboring a plasmid expressing CIB1 or/and a plasmid expressing a dual luciferase reporter. Possible transcriptional regulatory activity of CIB1 was tested by its effect on the firefly luciferase (LUC) reporter gene driven by a hybrid promoter that contains the minimum 35S promoter and four copies of an E-box sequence (Figure 4A). Expression of the Renilla reniformis luciferase (REN) driven by the standard 35S promoter was used as the internal control (H. Liu et al., 2008). Results of this experiment show a CIB1-dependent stimulation of the expression of the reporter genes driven by three different E-box sequences (Figures 4B and 4C). These results support the hypothesis that CIB1 is an E-box–specific DNA binding protein that can act as a transcription activator in plant cells.
Figure 4.
CIB1 Is a Transcription Activator Regulated by CRY2a in Response to Blue Light.
(A) A diagram showing the structure of the E-box–driven dual-luciferase reporter gene and DNA sequence of the recombinant E-box elements. The DNA sequences (Ec, Ef, and Ek) containing four tandem-repeat E-box derived from the c, f, and k regions of WRKY53b chromatin (see Figure 7B). The 35S promoter (black arrow), 35S minimum promoter (white arrowhead), Renilla luciferase (REN), firefly luciferase (LUC), and T-DNA (left border [LB] and right border [RB] are indicated.
(B) Images showing the LUC activities of N. benthamiana leaves infiltrated with the Agrobacteria strain harboring the indicated reporter (Ec, Ef, or Ek), in the presence (+) or absence (−) of the cotransfecting Agrobacteria strain harboring the plasmid expressing CIB1. After Agrobacteria infiltration, the plants were kept in white light for 3 d before photographs were taken.
(C) Dual-luciferase assay of relative reporter activity of samples shown in (B). The relative LUC activities normalized to REN activity are presented as relative expression units (REUs). The sd is shown (n = 3). The P values of CIB1-dependent activation of the reporter expression of the Ec, Ef, or Ek recombinant promoters are 0.014, 0.003, or 0.006, respectively (Student’s t test).
(D) Results of EMSA assay showing the inhibitory effect of CRY2a on the DNA binding activity of CIB1 to E-box DNA in response to blue light. The E-box DNA (Ewt) was mixed with effectors, which are the insect cell lysates expressing 6His-CIB1-Flag (CIB1) fusion protein and increased amount (1 to 8×) of lysates of insect cells expressing 6His-CRY2a (CRY2a). The mixtures containing the indicated components were incubated under blue light (25 μmol m−2 s−1) or in darkness at 4°C for 2 h. The mixture was mixed with agarose beads conjugated with anti-Flag antibody and washed five times with binding buffer, and the bound DNA was eluted by elution buffer and subjected to quantitative PCR. RBUs are defined in Methods. The significance of the CRY2a-dependent effects of the affinity of CIB1 for DNA in the presence or absence of blue light are examined by the Jonckheere-Terpstra Trend test; P = 0.912 or 0.003 of the dark-treated or the light-treated samples, respectively.
Photoexcited CRY2a Inhibits the DNA Binding Activity of CIB1
We reasoned that because CIB1 interacts with both CRY2a and the E-box DNA, CRY2a may mediate a blue light modulation of the DNA binding activity of CIB1 to affect transcription and plant development. To test this hypothesis, we examined whether CRY2a affects the E-box DNA binding activity of CIB1 in vitro. Because CRY2a in the insect cell lysates was photochemically active and interacted with CIB1 in response to blue light (Figure 2A), we used this system to examine a possible CRY2a-dependent blue light effect on the CIB1–DNA interaction. In this experiment, the lysate of insect cells expressing the epitope (Flag)-tagged CIB1 fusion protein were mixed with the E-box DNA, in vitro, in the presence or absence of the lysate of insect cells expressing CRY2a. The mixture was incubated under blue light or in darkness, and epitope-tagged CIB1 was purified by affinity chromatography. The DNA copurified with CIB1 was measured by quantitative PCR (qPCR) (Figure 4D). Figure 4D shows that, in the absence of blue light, the addition of increased amounts of CRY2a-expressing insect cell lysate to the CIB1-DNA binding reaction resulted in a slight decrease of the CIB1–DNA interaction (Figure 4D, Dark). This minor change is statistically insignificant (P = 0.91, Jonckheere-Terpstra Trend test), which may result from a background level of nonspecific CRY2a–CIB1 interaction in the absence of light. Importantly, when aliquots of the same reaction mixture were illuminated with blue light, the E-box DNA copurified with the CIB1 protein decreased significantly (three- to fourfold) in response to the increased amount of lysate of insect cells expressing CRY2a (Figure 4D, Blue; P = 0.003, Jonckheere-Terpstra Trend test). This result argues strongly that the CIB1–CRY2a interaction lowers the affinity of CIB1 to the E-box DNA, supporting the hypothesis that the CRY2a mediates blue light inhibition of the DNA binding activity of CIB1, which may serve as a CRY signaling mechanism in soybean.
CRY2a and CIB1 Antagonistically Regulate Leaf Senescence
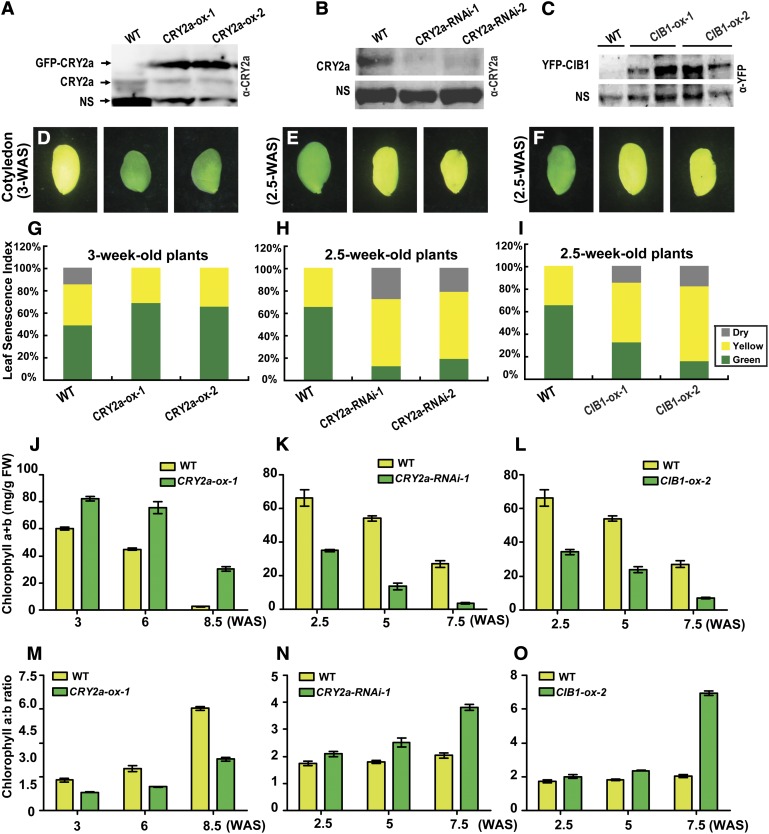
To investigate the physiological function of CRY2a and CIB1 in soybean, we prepared transgenic soybean plants that express the 35S:GFP-CRY2a, 35S:CRY2a-RNAi, or the 35S:YFP-CIB1 transgenes, respectively. These transgenic soybean lines are referred to as CRY2a-ox, CRY2a-RNAi, or CIB1-ox, respectively, in this report. Immunoblot analysis confirmed the expression of the recombinant CRY2a fusion protein in the independent CRY2a-ox lines (Figure 5A), reduced level of the endogenous CRY2a protein in the independent CRY2a-RNAi lines (Figure 5B), and expression of the recombinant CIB1 fusion protein in the independent CIB1-ox lines (Figure 5C). We also transformed soybean with the CIB1-RNAi constructs but failed to obtain transgenic lines that showed clearly reduced mRNA expression of CIB1. Independent transgenic soybean lines expressing the CRY2a-ox, CRY2a-RNAi, and CIB1-ox transgenes showed similar morphological phenotypes as the wild-type parents, except that the transgenic lines all exhibited altered leaf senescence phenotype when grown under long-day (LD) photoperiods or continuous illumination (Figures 5D to 5F and 6; see Supplemental Figures 7 to 16 online).
Figure 5.
CRY2a and CIB1 Regulate Leaf Senescence.
(A) to (C) Immunoblots showing the expression of GFP-CRY2a fusion protein or the endogenous CRY2a protein in CRY2a-ox plants, CRY2a-RNAi plants, and the wild-type (WT) controls or the expression of YFP-CIB1 fusion protein in CIB1-ox plants. Two independent lines of each genotype were examined. The total protein extracts were analyzed in a 10% SDS-PAGE gel for the immunoblot probed with α-CRY2a ([A] and [B]) or α-YFP antibodies. (C). The nonspecific bands (NS) recognized by the antibodies were used as the loading control.
(D) to (F) Images of representative cotyledons of the indicated lines showing different extents of senescence at the indicated growth stages.
(G) to (I) Cotyledons and unifoliolates were categorized into three groups according to their severities of senescence (green, nonsenescent; yellow, mildly senescent; gray, completely senescent) at the developmental stages indicated. The leaf senescence index is calculated as the percentage of each group with respect to the total leaf number of the individual plant (n ≥ 10).
(J) to (O) A comparison of the chlorophyll content (chlorophyll a+b) ([J] to [L]) or chlorophyll a/b ratio ([M] to [O]) of leaves of CRY2a-ox-1 ([J] and [M]), CRY2a-RNAi-1 ([K] and [N]), CIB1-ox-2 ([L] and [O]), and the control (wild type, WT). Mixed samples of two unifoliolates and the first two trifoliolates of a plant grown in continuous white light at the indicated development stages were collected for both measurements. The means and sd (n = 3) are shown.
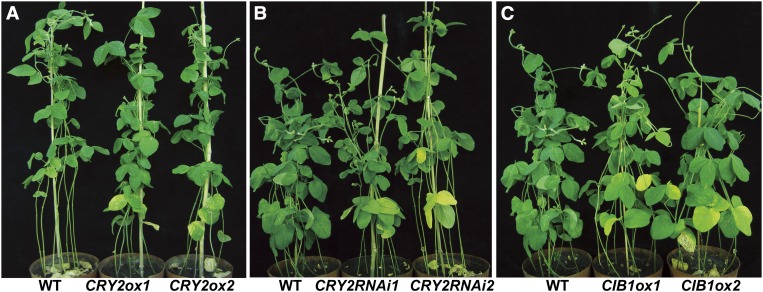
Figure 6.
The Leaf Senescence Phenotype of the Wild Type and the Respective Transgenic Soybean Plants.
(A) Transgenic soybean overexpressing CRY2a (CRY2ox) showed delayed leaf-senescence. The plants were grown in continuous light for 8.5 weeks. WT, the wild type.
(B) Transgenic soybean expressing CRY2a-RNAi (CRY2RNAi) showed accelerated leaf senescence. The plants were grown in continuous light for 7.5 weeks.
(C) Transgenic soybean overexpressing CIB1 (CIB1ox) showed accelerated leaf-senescence. The plants were grown in continuous light for 7.5 weeks.
Soybean leaf development is characterized by the emergence, growth, and senescence of three morphologically distinct types of leaves: cotyledons, unifoliolates, and progressively emerging trifoliolates. Although the CRY2a-ox, CRY2a-RNAi, and CIB1-ox transgenic soybean plants appeared normal in the emergence, growth, and morphology of all three types of leaves, the transgenic plants exhibited abnormal onset of leaf senescence of all three types of leaves. Specifically, the CRY2a-ox lines were delayed in leaf senescence, whereas the CRY2a-RNAi and CIB1-ox transgenic lines exhibited accelerated leaf senescence phenotype (Figures 5D to 5F and 6; see Supplemental Figures 7 to 16 online). The leaf senescence phenotypes of the respective transgenic lines are confirmed by the quantitative comparisons of the leaf senescence index (Figures 5G to 5I; see Supplemental Figures 7D to 7F online), chlorophyll content (Figures 5J to 5L), chlorophyll composition (Figures 5M to 5O), and photosynthesis rates (see Supplemental Figures 7G to 7I online) at different developmental stages. For example, more than 50% of cotyledons and unifoliolates were senescent (yellow or dead) in the wild-type plants at the age of 3 weeks after sewing (WAS), in comparison to ∼30% senescent cotyledons and unifoliolates in the CRY2a-ox lines of the same age (Figure 5G). By contrast, <40% of cotyledons and unifoliolates were senescent in the wild-type plants at 2.5 WAS, but 70 to 90% of cotyledons and unifoliolates were senescent in the CRY2a-RNAi and CIB1-ox lines of the same age, respectively (Figures 5H and 5I). Analyses of leaves at the later developmental stages of 6 to 8 WAS also demonstrated markedly delayed senescence of unifoliolates in the CRY2a-ox lines or accelerated senescence of unifoliolates in the CRY2a-RNAi and CIB1-ox lines (see Supplemental Figures 7D to 7F online). In comparison to the wild type, leaves (unifoliolates and trifoliolates) of the CRY2a-ox lines, which showed delayed senescence, accumulated more total chlorophyll (Figure 5J), had a relatively lower chlorophyll a/b ratio (Figure 5M), and had slightly higher photosynthesis rates (see Supplemental Figure 7G online). By contrast, leaves of the CRY2a-RNAi and CIB1-ox lines, which showed accelerated senescence, exhibited decreased total chlorophyll (Figures 5K and 5L), an increased chlorophyll a/b ratio (Figures 5N and 5O), and decreased photosynthesis rates (see Supplemental Figures 7H and 7I online), in comparison to the leaves of wild-type plants at the same age. Because leaf senescence causes chlorophyll breakdown, and chlorophyll a is the first chlorophyll breakdown product (Hörtensteiner, 2009), these results corroborate the visual senescence phenotypes of the respective transgenic lines. Taken together, results of those analyses argue strongly in favor of the hypothesis that CRY2a and CIB1 are negative and positive regulators of leaf senescence, respectively.
Soybean is a SD plant, for which the SD photoperiods not only stimulate floral initiation but also promote leaf senescence (Han et al., 2006). Interestingly, the altered leaf senescence phenotypes of the CRY2a-ox, CRY2a-RNAi, and CIB1-ox lines were only observed when plants were grown in LD photoperiods or continuous illumination, but not in plants grown in SD photoperiods (Figures 5 and 6; see Supplemental Figures 7 to 16 online). To further examine possible roles of CRY2a and CIB1 in the light regulation of leaf senescence in soybean, we adapted the dark-induced leaf senescence assay similar to that reported in Arabidopsis (Weaver and Amasino, 2001). In this experiment, detached leaves (unifoliolates and the first two trifoliolates) were placed in covered Petri dishes in darkness for up to 6 d, and the senescence phenotype of the wild type and transgenic lines were analyzed by imaging (see Supplemental Figure 17 online) and chlorophyll analyses (see Supplemental Figure 18 online). The detached leaves (unifoliolates and the first two trifoliolates) of the wild type exhibited signs of senescence within 4 d in the absence of light (see Supplemental Figure 17 online). Similar to the leaf senescence phenotype of the whole-plant assays described above, detached leaves of the CRY2a-ox lines showed delayed senescence in comparison to the wild type, whereas detached leaves of the CRY2a-RNAi and CIB1-ox transgenic lines exhibited accelerated senescence in this dark-induced senescence assay (see Supplemental Figure 17 online). Although senescence of undetached leaves of soybean plants grown in continuous light (Figures 5 and 6; see Supplemental Figures 7 to 16 online) and senescence of detached soybean leaves in darkness (see Supplemental Figures 17 and 18 online) may involve different mechanisms, the fact that the delayed senescence of the CRY2a-ox lines and accelerated senescence of the CRY2a-RNAi and CIB1-ox lines were observed in both light conditions support the hypothesis that CRY2a and CIB1 play important roles in the light regulation of leaf senescence.
CRY2a Mediates Blue Light Suppression of CIB1-Dependent Transcription and Leaf Senescence
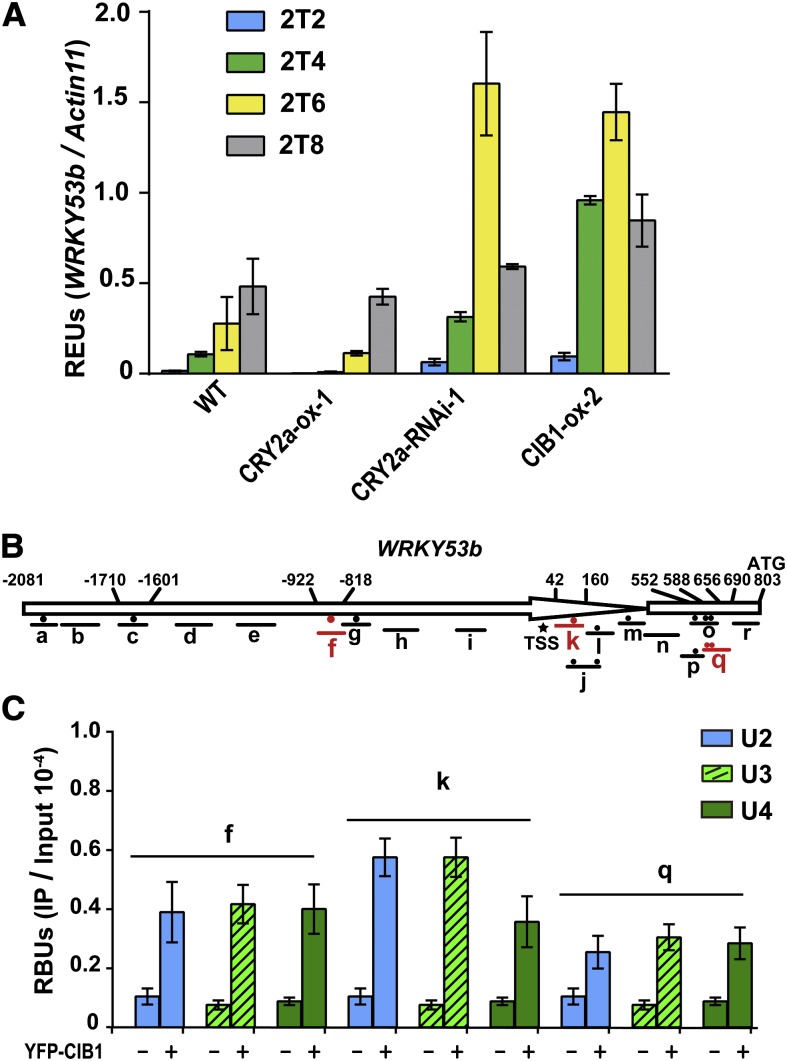
To investigate the mechanism of CRY2a- and CIB1-mediated light regulation of leaf senescence in soybean, we examined whether altered expression of the CRY2a and CIB1 proteins affected mRNA expression of the soybean genes structurally related to the senescence-associated genes (SAGs) found in other plants, especially Arabidopsis (Lim et al., 2007). SAGs are defined as genes for which the mRNA expression increases during senescence, and they often play roles in the process of leaf senescence (Quirino et al., 2000; Lim et al., 2007; Zentgraf et al., 2010; Wu et al., 2012). We surveyed more than a dozen soybean genes that are homologous to the known Arabidopsis SAGs. At least four soybean genes examined, WRKY53a, WRKY53b, SAG12, and PHEOPHORBIDE A OXYGENASE (PaO), showed clearly increased mRNA expression in aged or senescent leaves of wild-type soybean plants, so they are considered soybean SAGs (see Supplemental Figure 19 online). Arabidopsis WRKY53, SAG12, and PaO are known to encode key positive regulators of leaf senescence (Quirino et al., 2000; Lim et al., 2007; Zentgraf et al., 2010; Wu et al., 2012). In comparison to wild-type soybean, these soybean SAGs showed decreased expression in the CRY2a-ox lines, but increased expression in the CRY2a-RNAi and CIB1-ox lines (see Supplemental Figure 19 online). For example, mRNA expression of WRKY53b increased in mature leaves and peaked in senescent leaves in wild-type parents (Figure 7A; see Supplemental Figure 20 online). By contrast, the level of WRKY53b expression is suppressed in the CRY2a-ox lines (2T2, P = 0.007) but stimulated in the CRY2a-RNAi (2T6, P = 0.006) and CIB1-ox lines (2T6, P = 0.005) (Figure 7A), respectively. WRKY53b is particularly interesting to us because its counterpart in Arabidopsis not only promotes leaf senescence but also mediates environmental modulation of leaf senescence (Zentgraf et al., 2010). Although the hypothesis that WRKY53b acts as a positive regulator of leaf senescence in soybean remains to be tested genetically, the fact that WRKY53b exhibited an expression pattern that is not only characteristic of a SAG but also correlated with the different leaf senescence phenotype of the respective CRY2a or CIB1 transgenic lines supports such a hypothesis. Accordingly, we further hypothesize that CIB1 may activate WRKY53b expression to promote leaf senescence, whereas CRY2a interacts with CIB1 to suppress WRKY53b expression and leaf senescence.
Figure 7.
CIB1 Binds to the WRKY53b Chromatin to Promote WRKY53b mRNA Expression.
(A) Quantitative RT-PCR showing the RNA levels of WRKY53b in leaves of the indicated genotypes grown in continuous light. Relative expression units (REUs) were measured by normalization of the WRKY53b signal with that of the Actin11 control and are shown with the sd (n = 3). 2T2, 2T4, 2T6, or 2T8, the second trifoliolates collected from the plants at 2, 4, 6, or 8 WAS. The P values of differential expression of WRKY53b between the wild type (WT) and transgenic lines are 0.007, 0.046, 0.006, and 0.005 for CRY2a-ox (2 WAS), CRY2a-ox (6 WAS), CRY2a-RNAi (6 WAS), and CIB1-ox (6 WAS), respectively.
(B) A diagram depicting the predicted promoter (arrow) and the 5′ untranslated region (white box) regions of WRKY53b. Black or red circles indicate the positions of E-boxes (CANNTG). Different regions of the 2880-bp WRKY53b genomic DNA examined in the ChIP-qPCR reaction are indicated, with the short lines representing the region between the respective primer pairs used in the ChIP-qPCR reaction. Asterisk indicates the putative transcription start site, and numbers depict the position (bp) upstream (+) or downstream (−) of TSS.
(C) ChIP-qPCR analysis of samples collected from CIB1-ox-2 and wild-type plants at different developmental stages of the indicated chromatin region of WRKY53b. Plants were grown in continuous light. U2, U3, and U4 represent unifoliolates of the 2, 3, or 4 WAS stages. ChIP samples were prepared using anti-YFP antibody and subjected to quantitative PCR analysis. Results of ChIP-qPCR were quantified by normalization of the immunoprecipitation signal with the corresponding input signal. The sd is shown (n = 3). RBUs = PCR signal of immunoprecipitation reaction/PCR signal of input.
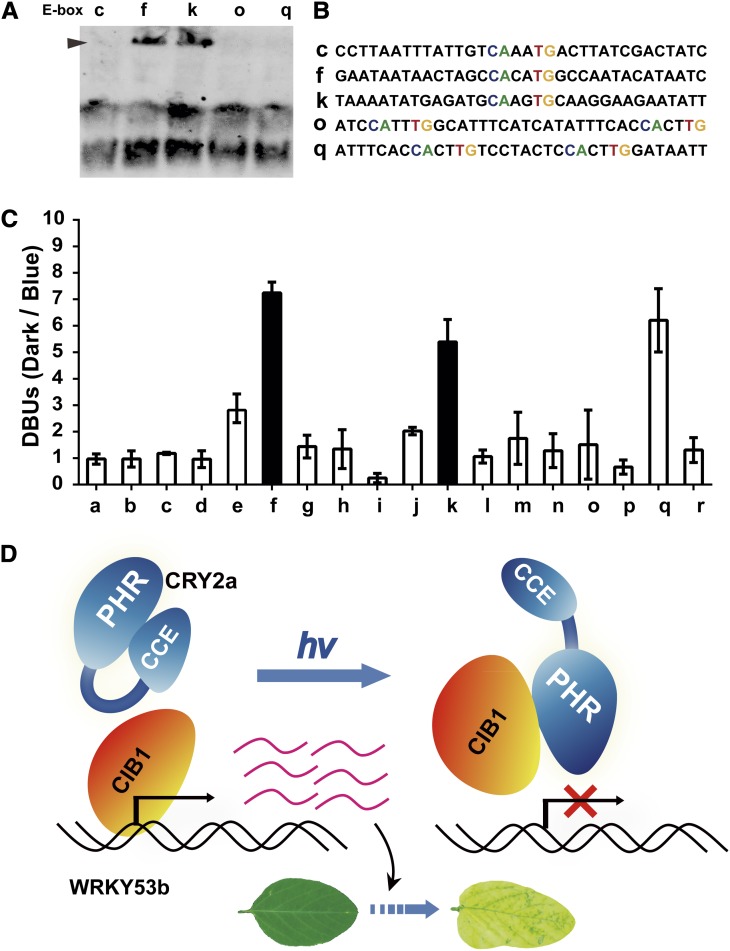
We reason that if CIB1 activates WRKY53b transcription to promote leaf senescence, it might do so by binding to the chromatin of soybean SAGs, such as WRKY53b, to affect their transcription. We evaluated this possibility by testing for an interaction between CIB1 and WRKY53b chromatin, using the chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) assay (Figure 7C; see Supplemental Figure 21 online). In this experiment, chromatins of wild-type soybean plants and transgenic soybean CIB1-ox plants expressing epitope-tagged CIB1 were subject to immunoprecipitation; the DNA signals amplified by PCR from the ChIP samples prepared from CIB1-ox plants were compared with those of the wild-type samples. Because only the CIB1-ox plants but not the wild-type plants express epitope-tagged CIB1, any PCR signal derived from the wild-type samples was considered background noise. Therefore, only the DNA regions amplified from the ChIP samples of the CIB1-ox plants to the extent that is significantly above the background levels detected in wild-type plants are considered putative CIB1 binding sites. We scanned the 2884-bp genomic sequence of WRKY53b upstream of the Adenine-Thymine-Guanine start codon, which contains at least 13 E-box sequences, by ChIP-qPCR, to search for possible CIB1 binding sites. We found that CIB1 binds to three different regions of WRKY53b chromatin, referred to as f, k, and q regions, which each contains at least one E-box sequence (Figures 7B and 7C). We then used EMSA to test whether CIB1 binds to the E-box DNA sequences corresponding to any of the three CIB1-interacting regions of WRKY53b chromatin. Figure 8A shows that CIB1 binds to the E-box DNA sequences corresponding to regions f and k of WRKY53b chromatin but does not bind to the DNA fragments that contain the E-box sequences derived from region q or the controls c and o. This result suggests that CIB1 may have a different affinity for different E-box sequences. More importantly, this experiment demonstrates that CIB1 binds to the f and k chromatin regions of WRKY53b by direct protein–DNA interactions. It is interesting that CIB1 binds to region q but not region o of the WRKY53b chromatin (Figure 7B), whereas it does not bind to the E-box DNA sequences derived from either region (Figures 8A and 8B). These results suggest that the E-box sequence is not the only determinant for the CIB1–chromatin interaction and that CIB1 may bind indirectly to region q of WRKY53b chromatin with the help of other transcription factor(s).
Figure 8.
Blue Light Suppresses the Interaction of CIB1 with Specific Regions of WRKY53b Chromatin.
(A) EMSA shows the direct interaction of CIB1 with the E-box sequences of the f and k regions of WRKY53b chromatin. See Figure 7 for the relative location of each region of the WRKY53b chromatin shown.
(B) The sequences of DNA probes used in (A).
(C) A comparison of the affinity of CIB1 for each region of the WRKY53b chromatin in response to blue light. Three-week-old plants grown in SD photoperiods (8 h light/16 h dark) were transferred to dark for 18 h, transferred to blue light (22 μmol m−2 s−1), or left in darkness until sample collection. The first trifoliolates were collected for ChIP analysis. DBUs were calculated by the formula: [IP of (CIB1/WT)/input of (CIB1/WT) of dark-treated sample]/[IP of (CIB1/WT)/input of (CIB1/WT) of blue light–treated sample], with sd (n = 3) shown. The light dependence of the interaction of CIB1 to the a, f, or k region of the WRKY53b chromatin has a P value of 0.8, 0.002, or 0.007, respectively (Student’s t tests). The f and k regions that show decreased interaction with CIB1 in response to blue light are highlighted by black. WT, the wild type.
(D) A working model depicting CRY2a-mediated blue light suppression of the CIB1-dependent activation of leaf senescence. PHR, photolyase homologous region; CCE, CRY C-terminal extension.
We next investigated whether the interaction of CIB1 with WRKY53b chromatin is affected by the age of leaves or light conditions. The ChIP-qPCR analyses of samples prepared from unifoliolates at different ages (2 to 4 WAS) showed little difference in the affinity of CIB1 to the WRKY53b chromatin in all three CIB1 binding sites (Figure 7C). Given that unifoliolates underwent senescence at around 3 WAS (see Supplemental Figures 7 to 16 online) and that CIB1 transgene expression showed no significant change in those leaves (see Supplemental Figure 19 online, CIB1), this result suggests that the affinity of CIB1 to WRKY53b chromatin is not significantly altered by the developmental or senescence programs. We next investigated whether blue light affects the CIB1–chromatin interaction. In this experiment, CIB1-ox plants were grown in a SD photoperiod, transferred to darkness for 18 h, and then exposed to blue light (light-treated) or left in darkness (dark-treated). The ChIP samples were prepared from unifoliolates of the light-treated and dark-treated plants by the anti-GFP (for green fluorescent protein) antibody and analyzed by quantitative PCR assays for the entire WRKY53b genomic sequence (see Supplemental Figure 21 online). To quantify possible light effects on the CIB1–chromatin interaction, we devised the parameter differential binding units (DBUs) to differentiate the relative binding of CIB1 to the individual WRKY53b chromatin regions in the light-treated and dark-treated plants. DBUs are calculated as the ratio of the ChIP signals of a specific WRKY53b region derived from the dark-treated CIB1-ox plants to that derived from light-treated CIB1-ox plants after normalization by the background signals of the wild-type samples (Figure 8B). Figure 8B shows that the CIB1 interaction with regions f or k of WRKY53b chromatin exhibited DBUs of ∼5 or 7, respectively, suggesting that the affinity of CIB1 to regions f and k of WRKY53b chromatin is approximately five- to sevenfold higher in dark-treated plants than in light-treated plants. By contrast, other regions of WRKY53b chromatin that do not interact with CIB1 exhibited DBUs of 1 to 3. Given that CIB1 directly interacts with the E-box DNA sequence of regions f and k of the WRKY53b chromatin (Figure 8A), that CRY2a mediates blue light inhibition of CIB1 interaction with the E-box DNA (Figure 4D), and that blue light does not suppress the expression of the CIB1 transgene expression (Figure 2E, Input), the blue light inhibition of the association of CIB1 to regions f (P = 0.002) and k (P = 0.007) of WRKY53b chromatin (Figure 8B) is most likely due to a CRY2a-mediated blue light suppression of the CIB1–DNA interaction. Interestingly, blue light also inhibits the indirect association of CIB1 with region q of WRKY53b chromatin that corresponds to the 5′-upstream untranslated region of the WRKY53b gene (Figure 8B; see Supplemental Figure 21 online). Taken together, these results support the hypothesis that CRY2a mediates blue light regulation of not only the CIB1–DNA interaction but also indirect CIB1-chromatin interaction in soybean.
DISCUSSION
We have shown in this study that a soybean CRY2 (CRY2a) undergoes blue light–specific interaction with soybean CIB1 (CIB1) (Figures 1 and 2). Phenotypic analyses of transgenic soybean expressing altered levels of the CRY2a and CIB1 proteins suggest that the CRY2-CIB1 complex plays an important role in the regulation of leaf senescence (Figures 5 and 6). Results of our analyses of blue light effects on the molecular interactions among CRY2a, CIB1, E-box DNA, and the WRKY53b chromatin (Figures 3, 4, 7, and 8) prompted us to propose a working hypothesis of the CIB1- and CRY2a-dependent regulation of leaf senescence (Figure 8). According to this hypothesis, CIB1 acts as a transcription activator that binds to the E-box DNA elements in the promoters of the SAGs, such as WRKY53b, to activate target gene transcription and leaf senescence in the absence of appropriate light signal. In response to blue light, photoexcited CRY2a interacts with CIB1 to suppress the DNA binding activity of CIB1, resulting in reduced transcription of SAGs, such as WRKY53b, and inhibition of leaf senescence. The CIB1-dependent signaling mechanism of CRY2a in soybean is reminiscent of the CIB1-dependent signaling mechanism of CRY2 in Arabidopsis (H. Liu et al., 2008). However, there are some notable differences between the two. First, regulation of flowering time is the primary physiological function of the CRY2-CIB1 complex in Arabidopsis, whereas regulation of leaf senescence is a major function of the CRY2a-CIB1 complex in soybean. Second, Arabidopsis CIB1 homodimers or heterodimers (with other CIB1-related proteins, such as CIB5) bind to the canonical (CACGTG) or noncanonical (CANNTG) E-box DNA sequences, respectively (H. Liu et al., 2008; Liu et al., 2013), whereas soybean CIB1 homodimers bind both canonical and non-canonical E box DNAs (Figures 3 and 8). Furthermore, although the photoexcited Arabidopsis CRY2 interacts with CIB1 to suppress reporter transcription in transient assays, it appears to stimulate the transcriptional activation activity of CIB1 (H. Liu et al., 2008), whereas photoexcited soybean CRY2a interacts with CIB1 to suppress the DNA binding and transcriptional activation activity of CIB1 in planta (Figures 4 and 8). These results argue that although the CIB-dependent CRY signaling mechanism is evolutionarily conserved, it mediates light regulation of different aspects of plant development by different mechanisms in different plant species.
The study described in this report demonstrates an important function and the underlying molecular mechanism of cryptochrome in light regulation of leaf senescence. Leaf senescence is a life history trait, which, like flowering time, is determined by developmental programs but regulated by the environmental factors, such as light and photoperiods. Arabidopsis has been the primary model organism for the study of the mechanism of photoreceptor signal transduction as well as the study of the mechanism controlling leaf senescence. However, the molecular mechanism underlying light regulation of leaf senescence has hitherto been unclear in all plant species, including Arabidopsis. The discoveries that CRY2a mediates blue light inhibition of CIB1 activity and leaf senescence are consistent with the hypothesis that CRY2a acts as a major photoreceptor responsible for the suppression of leaf senescence of soybean by LD photoperiods. However, it remains to be investigated whether WRKY53b acts as a positive regulator of leaf senescence in soybean like WRKY53 does in Arabidopsis, how many target genes other than WRKY53b are regulated by CIB1 and CRY2a, whether additional soybean cryptochromes are involved in the regulation of leaf senescence, and how the actions of cryptochromes and phytochromes are coordinated in soybean. Moreover, how CRY2a conveys the photoperiodic signal to the leaf senescence program also remains unclear. CRY2a protein is degraded in response to blue light, but the level of CRY2a protein shows surprisingly little change in young soybean seedlings under the photoperiodic conditions tested (Zhang et al., 2008). Therefore, additional studies are needed to further elucidate the mechanism underlying photoperiodic control of leaf senescence in soybean.
METHODS
Plant Materials and Soybean Transformation
An elite soybean (Glycine max) cultivar KN18 (Ken-nong 18) was used as the wild type in this study, which was obtained from the Soybean Germplasm Resources and Molecular Genetics (Chinese Academy of Agriculture Sciences, Beijing, China). Coding DNA sequence (CDS) of CIB1 (Glyma11g12450) and its related genes were amplified by PCR using the cDNA derived from young seedlings of KN18 as the template. The plasmid expressing 35S:YFP-CIB1 was prepared by cloning the CIB1 CDS into the pENSG-YFP vector using the Gateway system (Wenkel et al., 2006). The plasmid expressing 35S:GFP-CRY2a was prepared by cloning the CRY2a CDS into the pEGAD vector (Zhang et al., 2008). The plasmid expressing CRY2a-RNAi was prepared by cloning the sense and antisense fragments of CRY2a CDS (1210 to 1409 nucleotides) sequentially into the pFGC5941 vector to create a hairpin of the transcribed region (Kerschen et al., 2004). Primers used for the above constructs are described in Supplemental Table 1 online. Transgenic soybeans expressing 35S:GFP-CRY2a, 35S:CRY2a-RNAi, or 35S:YFP-CIB1 were prepared by Agrobacterium tumefaciens–mediated transformation using the cotyledon node method (Paz et al., 2006).
Recombinant Protein Expression and in Vitro Pull-Down Assays
CRY2a and CIB1-Flag were fused to the C terminus of the 6×His-tag, at the EcoRI and XhoI restriction sites of the vector pFastBacHTA (Invitrogen). The His-CRY2a and His-CIB1-Flag fusion proteins were expressed in Sf9 insect cells (Bac-to-Bac Baculovirus expression system; Invitrogen). The pull-down assays were performed as described before (Li et al., 2011), using lysates of insect cells expressing His-CRY2a and lysates of insect cells expressing His-CIB1-Flag. Protein concentration of lysate was measured using the Bradford method. Lysates were diluted to 1 μg/μL in lysis buffer (50 mM Tris, pH 7.8, 500 mM NaCl, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM DTT, 1 tablet/50 mL of protease inhibitor cocktail), and 500 μL of the diluted lysate samples of insect cells expressing His-CRY2a or His-CIB1-Flag was mixed and precleaned for 30 min with 30 μL of protein A/G beads (#A10001; Abmart). After a brief spin, the supernatants were mixed with 30 μL of suspensions of the agarose beads conjugated with anti-Flag antibody (#M20018; Abmart) and incubated with gentle rotation in darkness or under blue light (22 μmol m−2s−1) at 4°C for the indicated durations. After incubation, the agarose beads were collected by spinning at 1000 rpm for 3 min, washed with 1 mL of ice-cold wash buffer (50 mM Tris, pH 7.8, 500 mM NaCl, 0.1% Triton X-100, and 1 mM PMSF), and washed again three times with 200 μL of wash buffer. The agarose beads were suspended in 20 μL (4×) of SDS-PAGE loading buffer and boiled for 10 min. Ten microliters of the supernatants, or 0.2% of the input, were analyzed by SDS-PAGE for immunoblot analyses. The membranes were probed by anti-Flag antibody (Abmart) for the detection of the His-CIB1-flag protein, stripped, and probed again by anti-CRY2a antibody for the detection of His-CRY2a. The anti-CRY2a antibody was previously described (Zhang et al., 2008). The polyclonal rabbit anti-YFP antibody was generated using the His-YFP protein expressed and purified from Escherichia coli, which recognizes both YFP and GFP.
The ex Vivo Coimmunoprecipitation Assay
The ex vivo coimmunoprecipitation experiment was performed using samples prepared from the leaves of Nicotiana benthamiana infiltrated with Agrobacterium (strain GV3101) harboring pGWB17-CRY2a-Myc plasmid or pGWB11-CIB1-Flag plasmid as indicated. Prior to infiltration, Agrobacteria were grown overnight in 3 to 5 mL of Luria-Bertani medium (50 mg/L of kanamycin, 50 mg/L of gentamycin, and 50 mg/L of rifampicin) at 28°C until OD600 = 1.2, diluted 500- to 1000-fold in Luria-Bertani medium (10 mM MES, 20 μM acetosyringone, 50 mg/L of kanamycin, 50 mg/L gentamycin, and 50 mg/L of rifampicin), grown overnight (16 h) at 28°C, and then collected and resuspended in infiltration buffer (10 mM MES, 150 μM acetosyingone, and 10 mM MgCl2) to OD600 = 1.5 and incubated at room temperature for 4 h before infiltration. The strain harboring the pGWB17-CRY2a-Myc plasmid (CRY2a) or pGWB11-CIB1-Flag plasmid (CIB1) was either incubated alone or as a mixture with the other strain (at the CRY2a:CIB1 ratio of 1:1). Agrobacteria suspension in a 10-mL syringe (without the metal needle) was carefully press-infiltrated manually onto healthy leaves of 21-d-old N. benthamiana. The infiltrated plants were kept in continuous white light for 2 d, moved to darkness for 1 d, and then exposed to blue light (22 μmol m−2 s−1) for 1 h or kept in darkness. Equal amounts of sample (0.3 g) were collected under different treatments, ground in liquid nitrogen, and homogenized in 1 mL of extraction buffer (20 mM HEPES, pH 7.5, 40 mM KCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, and 1 tablet/50 mL of protease inhibitor cocktail). The protein extracts were incubated at 4°C for 15 min and centrifuged at 16,000g for 10 min. One milliliter of supernatant was precleared with 30 μL of protein A/G agarose at 4°C for 30 min. After a brief spin, the supernatants were mixed with 30 μL of suspensions of the agarose beads conjugated with anti-Myc antibody (#M20012; Abmart) and incubated at 4°C for 60 min. Beads were collected by spinning at 1500 rpm for 20 s and washed three times with the wash buffer (20 mM HEPES, pH 7.5, 40 mM KCl, and 0.1% Triton X-100). The proteins were eluted from the beads by mixing with 30 μL of 4× SDS-PAGE sample buffer, boiled for 5 min, and spun at 12,000 rpm for 5 min at room temperature. Ten microliters of supernatants were fractionated by 10% SDS-PAGE. Immunoblots were performed using the anti-Myc antibody (Abmart) for probing CRY2a-Myc and the anti-Flag antibody (Abmart) for probing CIB1-Flag, sequentially.
BiFC
The CDS of CRY2a, CRY2aN, CRY2aC, CIB1, CIB1N, and CIB1C were cloned into the pCCFP-GW or pNYFP-GW vector using a Gateway recombination system. Mesophyll protoplasts were isolated from the young leaves of Arabidopsis thaliana and transformed following the reported procedure (Yoo et al., 2007). Protoplasts were transfected with the plasmid DNA (Yoo et al., 2007). Samples were incubated for 12 to 14 h in darkness at 23°C, transferred to blue light (22 μmol m−2 s−1) for 30 min or kept in darkness, and then analyzed under confocal microscopy (Leica TCS SP2). The number of protoplasts showing clear nuclear BiFC fluorescence was counted for at least 50 cells per sample (n = 3) and presented as the percentage of cells exhibiting BiFC.
ChIP-qPCR Assays
The ChIP assay was performed as described with minor modifications (Saleh et al., 2008). Briefly, 4 g of plant tissues of soybean was immersed in 37 mL of cross-linking buffer (0.4 M Suc, 10 mM Tris-HCl, pH 8, 1 mM PMSF, 1 mM EDTA, and 1% formaldehyde) and vacuumed for 10 min, stopped by adding 2.5 mL of 2 M Gly and further vacuuming for 5 min. The plant tissues were washed three times in deionized water, frozen in liquid nitrogen, ground to powder, and suspended in 25 mL of precooled fresh nuclei isolation buffer (0.25 M Suc, 15 mM PIPES, pH 6.8, 5 mM MgCl2, 60 mM KCl, 15 mM NaCl, 1 mM CaCl2, 0.9% TritonX-100, 1 mM PMSF, 2 mg/mL of pepstatin A, and 1 tablet/50 mL of protease inhibitor cocktail). The homogenized slurry was filtered through cheesecloth prior to precipitation of nuclei by centrifugation. The isolated nuclei were suspended in nuclei lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, 1 mg/mL of pepstatin A, and 1 tablet/50 mL of protease inhibitor cocktail). The chromatin DNAs were sheared into 500-bp fragments by sonication. The CIB1-bound DNAs were immunoprecipitated using anti-GFP antibody, eluted, purified, and subjected to quantitative PCR analyses as previously described (H. Liu et al., 2008). The ChIP-qPCR results were provided as relative binding units (RBUs, IP/Input).
Plant Growth Conditions
Plants were cultured in environmentally controlled growth rooms with a defined photoperiod (SD, 8 h light/16 h dark; LD, 16 h light/ 8 h dark; or continuous light) at 25 to ∼28°C. Cool white fluorescent lights (TLD 18W/54; Philips) were used as a white light source (200 to ∼300 μmol m−2 s−1 above the plant canopy). The experiments using blue or red monochromatic light were performed in the blue-LED (436 ± 10 nm) or red-LED (658 ± 10 nm) growth chambers (Percival Scientific). Fluence rates were measured using a Li250 quantum photometer (Li-Cor).
Leaf Senescence Phenotype Analysis
Assays of leaf senescence phenotype were performed on plants grown in defined day lengths as indicated. The cotyledons, unifoliolates, and trifoliolates at different growth stages were photographed. For quantitative leaf senescence index analysis, cotyledons or unifoliolates were categorized into three groups according to their severities of senescence (green foliate, no senescence; yellow foliate, weak senescence; dry foliate, strong senescence) at different growth stages (n > 10). For the dark-induced leaf senescence assay, the unifoliolates and first trifoliolates of 3-week-old plants grown in continuous white light were detached, floated in 3 mM MES buffer, pH 5.7, and kept in darkness for the indicated durations.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed according to the manufacturer’s instructions (ProQues two-hybrid system with Gateway technology; Invitrogen). The CDS of soybean CRY2a, N-terminal domain of CRY2a (CRY2aN, 1 to 485 amino acids), C-terminal domain of CRY2a (CRY2aC, 486 to 634 amino acids), or Arabidopsis CRY2 were fused in frame with the CDS of the GAL4 DNA binding domain in the bait vector pDEST 32 (Invitrogen). The CDS of each soybean CIB, N-terminal domain of CIB1 (CIB1N, 1 to 217 amino acids), or C-terminal domain of CIB1 (CIB1C, 218 to 420 amino acids) were fused in frame with the CDS of the GAL4 transcription activation domain in the prey vector pDEST 22 (Invitrogen). The bait and prey plasmids were cotransformed into the yeast strain Saccharomyces cerevisiae MAV203 (Invitrogen). For the auxotrophic assay, yeast colonies were patched onto SD/-Leu/-Trp and SD/-Trp/-Leu/-His/-Ura plates, grown under blue light (25 μmol m−2 s−1), red light (30 μmol m−2 s−1), or in darkness at 28°C for 3 d. The β-gal assay was performed to quantify protein–protein interactions according to the manufacturer’s instructions, using chlorophenol red β-d-galactopyranoside as the substrate. Light treatment and calculation of the β-gal activity were performed as described previously (B. Liu et al., 2011).
Coimmunoprecipitation Assays
The coimmunoprecipitation experiments were performed as previously described with minor modifications (B. Liu et al., 2011; Zuo et al., 2011). Two-week-old plants grown in SD conditions were transferred to darkness for 18 h. The detached unifoliolates of the dark-treated plants were sliced into 2-mm strips and treated with MG132 (50 μM in 0.1% DMSO) for 3 h. Then, an equal amount of samples (0.5 g) was kept in darkness or exposed to blue light (22 μmol m−2 s−1) for various durations, then grinded in liquid nitrogen and homogenized in 1 mL of extraction buffer (20 mM HEPES, pH 7.5, 40 mM KCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF, and 1 tablet/50 mL of protease inhibitor cocktail). The protein extracts were incubated at 4°C for 15 min and centrifuged at 16,000g for 10 min. One milliliter of the supernatants was precleared with 30 μL of protein A/G agarose at 4°C for 30 min. After a brief spin, the supernatants were mixed with 30-μL suspensions of the agarose beads conjugated with anti-GFP antibody (MBL D153-8) and incubated at 4°C for 60 min. Beads were collected by spinning at 1500 rpm for 20 s and washed three times with wash buffer (20 mM HEPES, pH 7.5, 40 mM KCl, and 0.1% Triton X-100). The proteins were eluted from the beads by mixing with 30 μL of 4× SDS-PAGE sample buffer, boiled for 5 min, and spun at 12,000 rpm for 5 min at room temperature. Ten microliter supernatants were fractioned by 10% SDS-PAGE, and the membranes were probed by anti-YFP antibody for the detection of the YFP-CIB1, stripped, and probed again with anti-CRY2a antibody for the detection of CRY2a.
Measurement of Photosynthetic Rate and Chlorophyll Content
The plants were grown in continuous light, and the second trifoliolates of the indicated lines (n ≥ 10) were selected for the photosynthetic rate measurement from 31 to 43 d after sowing. Photosynthetic rate was measured following the manufacturer’s instructions (LICOR LI-6400 V4.0.1). For chlorophyll content analysis, 0.2 g of fresh sample (five individual leaves) of each indicated plant was frozen in liquid nitrogen, ground to power, mixed thoroughly with 20 mL of 80% acetone, and stored at −20°C for 1 h in darkness. Then the sample was centrifuged at 12,800g for 3 min and 1 mL of supernatants was measured for absorbance at 663 and 645 nm. Chlorophyll concentrations were calculated using the following formulas:
 |
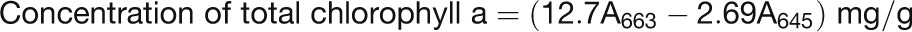 |
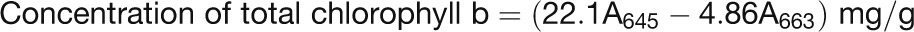 |
DNA Binding Assay
Random binding site selection was performed as previously described with some modifications. His-CIB1-Flag fusion protein expressed in Sf9 insect cells was bound to ProBond nickel-chelating resin (Novex) and incubated with a 64-bp double-stranded DNA probe containing 16 bp of random sequences at the center (see Supplemental Table 1 online) for 10 min at room temperature. After washing, the DNA bound to His-CIB1-Flag was eluted with 250 mM imidazole in native elution buffer as described in the user guide (Novex Document Part Number 25-0006). Prior to cloning the eluted DNA into the pEGM-T vector (Promega), eight rounds of binding and amplification were performed to enrich the target DNA probes. Competitive EMSA was performed as described previously (H. Liu et al., 2008).
To examine whether CRY2a affects the E-box DNA binding activity of CIB1, a quantitative PCR–based competitive DNA binding assay was performed. Briefly, the insect lysates expressing the His-CRY2a and His-CIB1-Flag fusion proteins were prepared in lysis buffer (50 mM Tris, pH 7.8, 500 mM NaCl, 0.5% Triton X-100, 1 mM PMSF, and 5 mM DTT). Five hundred microliters of each diluted cell lysate expressing His-CRY2a (1, 2, 4, or 8 μg/μL) was mixed with same volume of cell lysates expressing His-CIB1-Flag (1.5 μg/μL). Five hundred microliters of lysis buffer was mixed with 500 μL lysates expressing His-CRY2a (1 μg/μL) or His-CIB1-Flag (1.5 μg/μL) as a control. Then, each mixture or 1 mL of lysis buffer (negative control) was mixed with 25 μL of agarose beads conjugated with anti-Flag antibody (#M20018; Abmart) and kept in darkness or exposed to blue light (25 μmol m−2 s−1) at 4°C for 2 h . The beads were collected, mixed with 15 μL of Ewt DNA probe (60 pmol; see Supplemental Table 1 online), 10 μL of 5× DNA binding buffer (20% glycerol, 2.5 mM DTT, 250 mM KCl, 1 mg/mL of BSA, 50 mM Tris-HCl, pH 7.5, and 5 mM MgCl2), kept at room temperature for 15 min, and then washed five times with 1× DNA binding buffer. The immunoprecipitation products were eluted with 25 μL of elution buffer (0.2 M Gly, pH 2.5), neutralized immediately with 1 μL of Tris buffer (1.5 M, pH 9.0), and diluted 50-fold prior to quantitative PCR analysis. The RBU was calculated by the following formula: [RBU=2−ΔCt, ΔCt = Ct(+effector)−Ct(−effector); Ct (cycle threshold) represents the number of cycles required for the fluorescence signal to exceed background level].
Transient Transcription Dual-Luciferase Assays
A transient dual-luciferase assay to test the transcriptional activity of CIB1 was performed following a previously described method using N. benthamiana plants (Hellens et al., 2005) with some revisions. The effector plasmid, 35S:CIB1-Flag, was constructed by cloning CIB1 CDS into the pGWB11 vector using the attL × attR (LR) reaction (Invitrogen kit). The reporter plasmid, pGreen-E-LUC, encodes two luciferases: the firefly luciferase controlled by the recombinant E-box promoter and the Renilla luciferase controlled by the constitutive 35S promoter. The recombinant E-box promoter, which contains four copies of the wild-type E-box (Ec, Ef, or Ek) (Figure 4A), fused to the minimum 35S promoter was cloned into the vector pGreen-0800-LUC, which generated three different reporter plasmids: pGreen-Ec-LUC, pGreen-Ef-LUC, and pGreen-Ek-LUC. The sequences of the recombinant E-box promoters are included in Supplemental Table 1 online (dual-luciferase assay). Each pGreen-E-LUC reporter plasmid was transformed into Agrobacterium (strain AGL1) together with the helper plasmid pSoup-P19, which also encodes a repressor of cosuppression (Hellens et al., 2005). The Agrobacterium strain containing the reporter pGreen-E-LUC was used alone or mixed with the Agrobacterium strain containing the effector plasmid Pro35S:CIB1-Flag. Overnight cultures of Agrobacteria were collected by centrifugation, resuspended in the infiltration buffer (10 mM MES, 150 mM acetosyringone, and 10 mM MgCl2), and incubated at room temperature for 4 h before infiltration. The reporter strain was either incubated alone or as a mixture with the effector strain (at a reporter:effector ratio of 1:1). Agrobacteria suspension in a 10-mL syringe (without the metal needle) was carefully press-infiltrated manually onto healthy leaves of 21-d-old N. benthamiana. Plants were left under continuous white light for 3 d after infiltration, sprayed with luciferin (1 mM luciferin and 0.01% Triton X-100), and photographed using a charge-coupled device camera (Princeton Instruments). Leaf samples were collected for the dual-luciferase assay using a commercial kit (Promega; DLR reagents). Briefly, leaf discs infected with Agrobacteria were homogenized in 100 μL of passive lysis buffer. Eight microliters of crude extract was mixed with 40 μL of LUC assay buffer, and the LUC activity was measured using a multimode microplate reader (Berthold; TriStar LB941). Then, 40 μL of Stop and Glow buffer was added for the measurement of the REN activity. Multiple biological repeats (n ≥ 3) were performed for each sample.
mRNA Expression Analyses
Total RNAs were isolated using the Trizol kit (Invitrogen). cDNA was synthesized from 1 µg of total RNA using a SuperScript first-strand cDNA synthesis system (Invitrogen). LightCycler 480 SYBR Green I Master (Roche) was used for the quantitative PCR reaction. Briefly, the cDNA was diluted 50-fold, and 5 μL of diluted cDNA was used as template in a 20-μL quantitative PCR reaction, which was predenatured at 95°C for 5 min, followed by a 45-cycle program (95°C, 10 s; 60°C, 20 s; 72°C, 30 s per cycle). The mRNA level of Actin11 was used as the internal control. The quantitative PCR results shown are the average (±sd) of three biological repeats. All the primers used are described in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or Phytozome (http://www.phytozome.net/) databases under the following accession numbers: CRY2 (AT1G04400), CRY2a (Glyma20g35220), CIB1 (Glyma11g12450), Actin11 (Glyma18g52780), WKY53a (Glyma08g02580), WRKY53b (Glyma16g02960), SAG12 (Glyma11g20400), and PaO1 (Glyma12g08740).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree and Sequence Alignment of CIBs and Homologous Proteins.
Supplemental Figure 2. The Interaction of CRY2a with CIB1 or Homologous Proteins in Yeast Cells.
Supplemental Figure 3. The Interaction of Arabidopsis CRY2 with Soybean CIB1 or Homologs in Yeast Cells.
Supplemental Figure 4. Bimolecular Fluorescence Complementation Assays Showing the CRY2a–CIB1 Interaction in Arabidopsis Protoplasts.
Supplemental Figure 5. The N-Terminal Domain of CRY2a (CRY2aN) Undergoes a Blue Light–Dependent Interaction with CIB1.
Supplemental Figure 6. The N-Terminal Domain of CIB1 (CIB1N) Interacts with CRY2a in a Blue Light–Dependent Manner.
Supplemental Figure 7. Leaf Senescence Phenotype of the Wild Type and Each Transgenic Line Grown in Continuous Light.
Supplemental Figure 8. Leaf Senescence Phenotype of 4-Week-Old Transgenic Soybean Overexpressing CRY2a.
Supplemental Figure 9. Leaf Senescence Phenotype of 6-Week-Old Transgenic Soybean Overexpressing CRY2a.
Supplemental Figure 10. Leaf Senescence Phenotype of 8.5-Week-Old Transgenic Soybean Overexpressing CRY2a.
Supplemental Figure 11. Leaf Senescence Phenotype of 3-Week-Old Transgenic Soybean Expressing CRY2a-RNAi.
Supplemental Figure 12. Leaf Senescence Phenotype of 5-Week-Old Transgenic Soybean Expressing CRY2a-RNAi.
Supplemental Figure 13. Leaf Senescence Phenotype of 7.5-Week-Old Transgenic Soybean Expressing CRY2a-RNAi.
Supplemental Figure 14. Leaf Senescence Phenotype of 3-Week-Old Transgenic Soybean Overexpressing CIB1.
Supplemental Figure 15. Leaf Senescence Phenotype of 5-Week-Old Transgenic Soybean Overexpressing CIB1.
Supplemental Figure 16. Leaf Senescence Phenotype of 7.5-Week-Old Transgenic Soybean Overexpressing CIB1.
Supplemental Figure 17. Comparison of Dark-Induced Leaf Senescence Phenotype of the Wild Type and Each Transgenic Line.
Supplemental Figure 18. The Contents of Chlorophyll a+b (Blue Lines) and the Ratios of Chlorophyll a to b (Red Lines) in Each Transgenic Line.
Supplemental Figure 19. mRNA Levels of CRY2a, CIB1, and Potential Senescence-Associated Genes in the Wild Type and Each Transgenic Line.
Supplemental Figure 20. mRNA Levels of WRKY53b in Leaves of Different Transgenic Lines at Various Development Stages.
Supplemental Figure 21. ChIP-PCR Showing the Interaction of CIB1 and WRKY53b.
Supplemental Table 1. Oligonucleotide Primers Used in This Study.
Acknowledgments
This work is supported in part by the Ministry of Agriculture Transgenic Research Grant 2010ZX08010-002 to the Institute of Crop Science, Chinese Academy of Agricultural Sciences, by the National Natural Science Foundation of China (31171352 to B.L., 31371649 to H.L., and 31301346 to Y.M.), by the National Institutes of Health (GM56265 to C.L.), and by UCLA faculty research grants (to C.L.).
AUTHOR CONTRIBUTIONS
C.L. and B.L. designed the research. Y.M., H.L., and Q.W. performed the research. C.L. and B.L. wrote the article.
Glossary
- bHLH
basic helix-loop-helix
- SD
short-day
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- EMSA
electrophoretic mobility shift assay
- LD
long-day
- WAS
weeks after sewing
- SAG
senescence-associated gene
- ChIP-qPCR
chromatin immunoprecipitation quantitative PCR
- GFP
green fluorescent protein
- DBU
differential binding unit
- CDS
coding DNA sequence
- PMSF
phenylmethylsulfonyl fluoride
- β-gal
β-galactosidase
References
- Ahmad M., Cashmore A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Cashmore A.R. (2003). Cryptochromes: Enabling plants and animals to determine circadian time. Cell 114: 537–543 [PubMed] [Google Scholar]
- Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L.O., van der Horst G.T., Batschauer A., Ahmad M. (2011). The cryptochromes: Blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Endo M., Mochizuki N., Suzuki T., Nagatani A. (2007). CRYPTOCHROME2 in vascular bundles regulates flowering in Arabidopsis. Plant Cell 19: 84–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E.A., Jr, Staknis D., Weitz C.J. (1999). Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science 286: 768–771 [DOI] [PubMed] [Google Scholar]
- Guo H., Yang H., Mockler T.C., Lin C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Han T., Wu C., Tong Z., Mentreddy R.S., Tan K., Gai J. (2006). Postflowering photoperiod regulates vegetative growth and reproductive development of soybean. Environ. Exp. Bot. 55: 120–129 [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009). Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci. 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Idevall-Hagren O., Dickson E.J., Hille B., Toomre D.K., De Camilli P. (2012). Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA 109: E2316–E2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Hirayama J., Kobayashi Y., Todo T. (2002). Zebrafish CRY represses transcription mediated by CLOCK-BMAL heterodimer without inhibiting its binding to DNA. Genes Cells 7: 1073–1086 [DOI] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., Tucker C.L. (2010). Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods 7: 973–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschen A., Napoli C.A., Jorgensen R.A., Müller A.E. (2004). Effectiveness of RNA interference in transgenic plants. FEBS Lett. 566: 223–228 [DOI] [PubMed] [Google Scholar]
- Keskitalo J., Bergquist G., Gardeström P., Jansson S. (2005). A cellular timetable of autumn senescence. Plant Physiol. 139: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek D.T., McIlrath W.J., Vergara B.S. (1966). Photoperiodic, induction of senescence in xanthium plants. Science 151: 95–96 [DOI] [PubMed] [Google Scholar]
- Kume K., Zylka M.J., Sriram S., Shearman L.P., Weaver D.R., Jin X., Maywood E.S., Hastings M.H., Reppert S.M. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98: 193–205 [DOI] [PubMed] [Google Scholar]
- Li X., Wang Q., Yu X., Liu H., Yang H., Zhao C., Liu X., Tan C., Klejnot J., Zhong D., Lin C. (2011). Arabidopsis cryptochrome 2 (CRY2) functions by the photoactivation mechanism distinct from the tryptophan (trp) triad-dependent photoreduction. Proc. Natl. Acad. Sci. USA 108: 20844–20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P.O., Kim H.J., Nam H.G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Lin J.-F., Wu S.-H. (2004). Molecular events in senescing Arabidopsis leaves. Plant J. 39: 612–628 [DOI] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Liu B., Zhao C., Pepper M., Lin C. (2011). The action mechanisms of plant cryptochromes. Trends Plant Sci. 16: 684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. (2008). Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–1539 [DOI] [PubMed] [Google Scholar]
- Liu L.J., Zhang Y.C., Li Q.H., Sang Y., Mao J., Lian H.L., Wang L., Yang H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Li X., Li K., Liu H., Lin C. (2013). Multiple CIBs form heterodimers to mediate CRY2-dependent regulation of flowering-time in Arabidopsis. PLoS Genet. 9: e1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nooden L.D., Hillsberg J.W., Schneider M.J. (1996). Induction of leaf senescence in Arabidopsis thaliana by long days through a light-dosage effect. Physiol. Plant. 96: 491–495 [Google Scholar]
- Parlitz S., Kunze R., Mueller-Roeber B., Balazadeh S. (2011). Regulation of photosynthesis and transcription factor expression by leaf shading and re-illumination in Arabidopsis thaliana leaves. J. Plant Physiol. 168: 1311–1319 [DOI] [PubMed] [Google Scholar]
- Paz M.M., Martinez J.C., Kalvig A.B., Fonger T.M., Wang K. (2006). Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep. 25: 206–213 [DOI] [PubMed] [Google Scholar]
- Quirino B.F., Noh Y.-S., Himelblau E., Amasino R.M. (2000). Molecular aspects of leaf senescence. Trends Plant Sci. 5: 278–282 [DOI] [PubMed] [Google Scholar]
- Saleh A., Alvarez-Venegas R., Avramova Z. (2008). An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat. Protoc. 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Sancar A. (2003). Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103: 2203–2237 [DOI] [PubMed] [Google Scholar]
- Schmutz J., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Shearman L.P., Sriram S., Weaver D.R., Maywood E.S., Chaves I., Zheng B., Kume K., Lee C.C., van der Horst G.T., Hastings M.H., Reppert S.M. (2000). Interacting molecular loops in the mammalian circadian clock. Science 288: 1013–1019 [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Weaver L.M., Amasino R.M. (2001). Senescence is induced in individually darkened Arabidopsis leaves, but inhibited in whole darkened plants. Plant Physiol. 127: 876–886 [PMC free article] [PubMed] [Google Scholar]
- Wenkel S., Turck F., Singer K., Gissot L., Le Gourrierec J., Samach A., Coupland G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Spalding E.P. (2007). Separate functions for nuclear and cytoplasmic cryptochrome 1 during photomorphogenesis of Arabidopsis seedlings. Proc. Natl. Acad. Sci. USA 104: 18813–18818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.-Y., Kuai B.-K., Jia J.-Z., Jing H.-C. (2012). Regulation of leaf senescence and crop genetic improvement. J. Integr. Plant Biol. 54: 936–952 [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu X., Klejnot J., Zhao X., Shalitin D., Maymon M., Yang H., Lee J., Liu X., Lopez J., Lin C. (2007). Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19: 3146–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf U., Laun T., Miao Y. (2010). The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 89: 133–137 [DOI] [PubMed] [Google Scholar]
- Zhang E.E., Kay S.A. (2010). Clocks not winding down: Unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 11: 764–776 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Li H., Li R., Hu R., Fan C., Chen F., Wang Z., Liu X., Fu Y., Lin C. (2008). Association of the circadian rhythmic expression of GmCRY1a with a latitudinal cline in photoperiodic flowering of soybean. Proc. Natl. Acad. Sci. USA 105: 21028–21033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]