Plastid-localized glutathione reductase2 regulates root growth and root meristem maintenance through modulation of its GSH redox status, resulting in regulation of downstream effectors of the auxin/PLETHORA pathway, as well as of some auxin/ PLETHORA-independent pathways. This function differs from that of glutathione reductase1, which is not essential for plant development.
Abstract
Glutathione is involved in thiol redox signaling and acts as a major redox buffer against reactive oxygen species, helping to maintain a reducing environment in vivo. Glutathione reductase (GR) catalyzes the reduction of glutathione disulfide (GSSG) into reduced glutathione (GSH). The Arabidopsis thaliana genome encodes two GRs: GR1 and GR2. Whereas the cytosolic/peroxisomal GR1 is not crucial for plant development, we show here that the plastid-localized GR2 is essential for root growth and root apical meristem (RAM) maintenance. We identify a GR2 mutant, miao, that displays strong inhibition of root growth and severe defects in the RAM, with GR activity being reduced to ∼50%. miao accumulates high levels of GSSG and exhibits increased glutathione oxidation. The exogenous application of GSH or the thiol-reducing agent DTT can rescue the root phenotype of miao, demonstrating that the RAM defects in miao are triggered by glutathione oxidation. Our in silico analysis of public microarray data shows that auxin and glutathione redox signaling generally act independently at the transcriptional level. We propose that glutathione redox status is essential for RAM maintenance through both auxin/PLETHORA (PLT)-dependent and auxin/PLT-independent redox signaling pathways.
INTRODUCTION
Glutathione (γ-l-glutamyl-l-cysteinylglycine) helps plants maintain a reducing cellular environment because of its high abundance (millimolar level in plant cells; Meyer et al., 2001; Queval et al., 2011) and low midpoint redox potential (–230 mV at pH = 7) (Foyer and Noctor, 2005). Glutathione is also considered an indicator of the cellular redox environment (Schafer and Buettner, 2001) and the hub of the complex antioxidant networks responsible for regulating active oxygen levels (Noctor and Foyer, 1998). For example, to prevent oxidative damage to the chloroplast, the finely-tuned photosynthetic machinery is maintained in a reducing environment in which reactive oxygen species are minimized by the glutathione-ascorbate cycle (Foyer and Halliwell, 1976). Through such a pathway, hydrogen peroxide (H2O2) is ultimately detoxified through the oxidation of reduced glutathione (GSH) into the oxidized form of glutathione, glutathione disulfide (GSSG); GSSG is then recycled by NADPH-dependent glutathione reductase (GR), thereby catalyzing the reduction of GSSG to GSH (Foyer and Noctor, 2005, 2011). Recently, glutathione was shown to have a role in NADPH-dependent glutathione/glutaredoxin (Grx) system (NGS)–dependent thiol redox signaling. The NGS, together with the NADPH-dependent thioredoxin (Trx) system (NTS), transfers a redox signal to target proteins with the reversible dithiol/disulfide exchange (S-S → 2HS), in a process known as thiol redox regulation (Buchanan and Balmer, 2005; Dietz, 2008; Rouhier et al., 2008; Dalle-Donne et al., 2009; Dietz and Pfannschmidt, 2011; Foyer and Noctor, 2011; Noctor et al., 2011). NGS-dependent thiol redox signaling in plants has been dissected (Dietz, 2008; Dietz and Pfannschmidt, 2011): glutathione, as a redox input element, accepts electrons from NADPH and transfers them to Grx (redox transmitter); subsequently, electrons enter target proteins, as in the case of deglutathionylation (Dalle-Donne et al., 2009). Through such signaling, the biological activity of a given protein (e.g., enzymatic activity) is tightly regulated, usually in response to environmental, physiological, and developmental cues. Therefore, it is not surprising that glutathione has roles in multiple processes, such as heavy metal detoxification, glyoxylase and formaldehyde metabolism, sulfur assimilation, cell cycle regulation, the pathogen response, and the Calvin-Benson and Krebs cycles (Rouhier et al., 2008; Noctor et al., 2011, 2012).
According to the Nernst equation, glutathione redox potential is associated with [GSH]2:GSSG under a given condition (Schafer and Buettner, 2001). Hence, glutathione redox status is determined by total glutathione levels and the GSH:GSSG ratio, which are strictly regulated by glutathione biosynthesis and oxidation/reduction, respectively. The enzyme that catalyzes the rate-limiting step of glutathione synthesis is γ-glutamylcysteine synthetase (γ-ECS), encoded by the single-copy gene GSH1 in Arabidopsis thaliana (May and Leaver, 1994). Knockout of GSH1 leads to embryonic lethality (Cairns et al., 2006). Notably, a GSH1 mutation root meristemless1 (rml1), which depletes glutathione levels to 2.7% of those observed in the wild type, produces rootless plants due to the G1/S transitional arrest of cells in the root apical meristem (RAM) (Cheng et al., 1995; Vernoux et al., 2000). Treatment with l-buthionine (S,R)-sulfoximine (BSO), a specific inhibitor of γ-ECS, significantly inhibits cell division in the RAM and subsequent root growth (Sánchez-Fernández et al., 1997; Koprivova et al., 2010). On the other hand, GR is the unique enzyme that catalyzes the reduction of GSSG to GSH. The Arabidopsis genome encodes two GR proteins: GR1 and GR2. GR1 is localized to the cytosol and peroxisomes (Marty et al., 2009; Kataya and Reumann, 2010), and GR2 is predicted to be chloroplast localized, according to The Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org/). It has been indicated that GR1 is not crucial for plant development under normal conditions, although the knockout of GR1 gives rise to a lower GSH:GSSG ratio (Marty et al., 2009). Nevertheless, a lack of GR2 confers embryonic lethality (Tzafrir et al., 2004), implying a crucial role for GR2 in plant development.
The potential link between thiol redox signaling and auxin signaling, which is the critical regulator of plant development (Benjamins and Scheres, 2008), has aroused great concern. It has been reported that BSO-triggered root inhibition is accompanied by a decrease in the expression of the auxin efflux facilitator PIN-formed (PIN) genes (Koprivova et al., 2010). Bashandy et al. (2010) observed reduced polar auxin transport (PAT) and auxin levels, as well as many auxin-related phenotypes, such as decreased apical dominance, pin-like inflorescence, reduced PAT, vasculature defects, and perturbed root development in the ntra ntrb cadmium sensitive2-1 (cad2-1) triple mutant. ntra ntrb is a double mutant of NADPH-Trx reductase in the NTS (Reichheld et al., 2005, 2007); cad2-1 is a weak GSH1 mutant allele that exhibits hypersensitivity to cadmium but is nearly aphenotypic under normal conditions (Howden et al., 1995; Cobbett et al., 1998). These findings indicate that the interplay between NTS and NGS is involved in auxin transport, biosynthesis, and/or signaling. It seems that NGS alone is sufficient to affect auxin signaling because the knockout of GRXS17, a Grx gene, leads to decreased PAT, a lower auxin response, temperature-dependent root growth inhibition, and agravitropism (Cheng et al., 2011).
Here, we report the essential role of the GR2-regulated glutathione redox status in RAM maintenance and root growth. We identify a GR2 mutant, miao, that displays strong RAM defects and root growth inhibition and describe GR2 as a widely expressed plastid-localized protein. Through the assessment of glutathione redox status in miao and pharmacological tests, we demonstrate that glutathione oxidation is responsible for the root phenotype observed in miao. Although the in silico analysis shows that auxin and glutathione redox signaling in whole seedlings are largely independent at the transcriptional level, a reduced glutathione environment is required for the downstream effectors of PLETHORA (PLT), the auxin-inducible master regulator of root development (Aida et al., 2004; Galinha et al., 2007), while an auxin/PLT-independent redox- signaling pathway is also important for RAM maintenance.
RESULTS
The miao Mutant Shows RAM Defects and Root Growth Inhibition
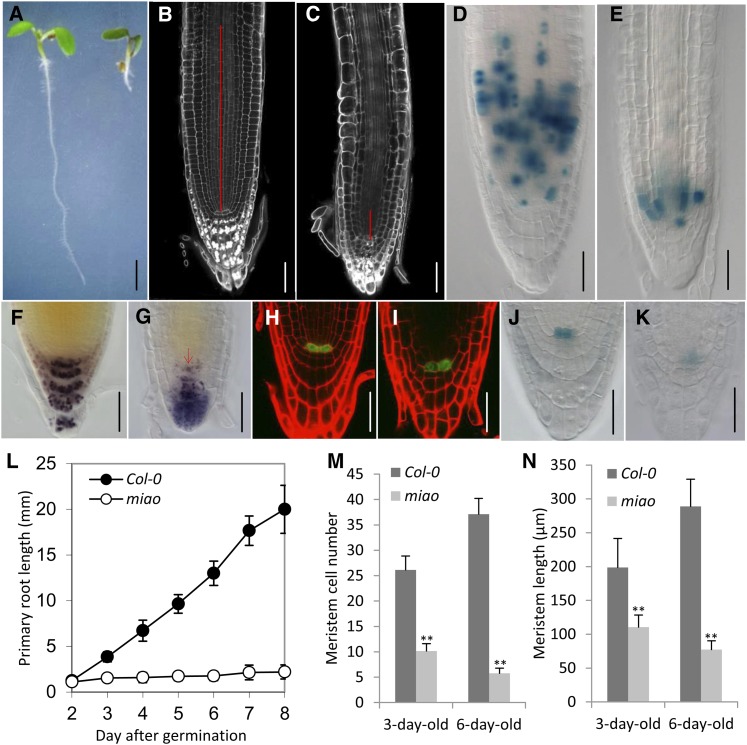
In an ethyl methanesulfonate mutagenesis screen of Arabidopsis Columbia-0 (Col-0), we isolated miao, a mutant displaying a short primary root (Figures 1A and 1L; see Supplemental Figures 1A and 1B online). After germination, the primary root growth of miao was strongly inhibited (Figure 1L), but adventitious roots emerged at 7 to 8 d after germination (DAG). In vitro–grown miao seedlings had dark-green cotyledons (Figure 1A), whereas soil-grown miao plants were smaller and dwarfed (see Supplemental Figures 1D, 1E, 1G, and 1H online). All F1 plants from the cross of miao with Col-0 exhibited the Col-0 phenotype, and in the F2 population, the miao phenotype segregated at a ratio of ∼3:1 (Col-0: miao, 201:72), indicating that miao corresponded to a single, recessive locus. Homozygous miao plants were further backcrossed with Col-0 to eliminate background mutations.
Figure 1.
Root Phenotype of miao Seedlings.
(A) Six-day-old Col-0 (left) and miao (right) growing on vertical one-half-strength MS medium. Bar = 2 mm.
(B) and (C) mPS-PI staining of roots of 6-d-old Col-0 (B) and miao (C). Red lines indicate the meristem. The boundary of the meristem is based on the first elongation of cortex cell files. Bars = 50 µm.
(D) and (E) Nomarski images showing CycB1;1pro:GUS expression in the RAM of 6-d-old roots of Col-0 (D) and miao (E). Bars = 50 µm.
(F) and (G) Nomarski images showing Lugol’s staining of roots in 6-d-old Col-0 (F) and miao (G) seedlings. Note the staining of the quiescent center and DSC in miao (arrow). Bars = 50 µm.
(H) and (I) CLSM images of WOX5pro:GFP expression in 6-d-old Col-0 (H) and miao (I). Red, PI staining; green, GFP. Bars = 50 µm.
(J) and (K) QC25:GUS expression in 6-d-old Col-0 (J) and miao (K). Bars = 50 µm.
(L) Primary root length of Col-0 and miao seedlings after germination on vertical one-half-strength MS medium. Data are the means ± sd; n = 15.
(M) and (N) RAM cell number and lengths of 3- and 6-d-old Col-0 and miao seedlings growing on vertical plates of one-half-strength MS medium. Data are the means + sd; n = 7. Differences between miao and Col-0 were analyzed using Student’s t tests. Double asterisks represent highly significant differences, P < 0.01.
The remarkable root growth inhibition phenotype of miao prompted us to examine its RAM architecture. Based on the RAM boundary, where the first cells in the cortex file are typically elongated (Casamitjana-Martínez et al., 2003; Dello Ioio et al., 2007), the modified pseudo-Schiff propidium iodide (mPS-PI) staining and Nomarski images showed that the RAM length (in micrometers) and cell number of the cortex cell file of miao were decreased to ∼27 and ∼15%, respectively, compared with Col-0 (Figures 1B, 1C, 1M, 1N, and 2B). The RAM of miao completely collapsed 7 to 8 DAG (see Supplemental Figure 2 online). In line with the reduced RAM length of miao, the expression of CYCLIN B1;1 (CycB1;1)pro:GUS (for β-glucuronidase), a reporter for the G2/mitotic phase of the cell cycle (Colón-Carmona et al., 1999), was restricted to a small number of proximal meristem cells (Figures 1D and 1E). However, the expression level of CycB1;1pro:GUS in the RAM of miao was not remarkably altered when considering the decreased length of the RAM (Figures 1D and 1E), indicating repressed mitosis and/or enhanced endocycle at the transition from cell proliferation to elongation in the root of miao.
Figure 2.
MIAO Encodes GR2.
(A) Map-based cloning of MIAO/GR2. MIAO encodes AT3G54660 (GR2), localized at BAC-T5N23, chromosome 3. Boxes indicate exons, including the open reading frame (dark gray) and 5′ and 3′ untranslated regions (light gray). Lines between boxes represent introns. Arrow shows the site of the mutation that converted C to T at the nucleotide 449 bp downstream of ATG. This mutation leads to a Ser/Phe shift at the 150th amino acid on GR2.
(B) Nomarski images of root tips in Col-0 (left), miao (middle), and plants complemented with GR2pro:gGR2 (right). Solid lines indicate the meristem. Bars = 50 µm.
(C) GR activity in total protein extracts of whole seedlings of 3-, 5-, and 7-d-old Col-0 and miao. Data are the mean values + sd (n = 3). Significant differences determined by Student's t tests are denoted by double asterisks, P < 0.01.
[See online article for color version of this figure.]
We next examined whether the RAM defects in miao were associated with changes in quiescent center (QC) properties and stem cell formation. After staining starch granules with Lugol’s solution, we observed that starch granules accumulated in the QC and neighboring distal stem cells (DSCs), indicating the differentiation status of these cells (Figures 1F and 1G). This observation was further resolved using a modification of mPS-PI staining, which showed starch granules in both the QC and DSC in miao (see Supplemental Figure 3 online). Using the QC identity markers QC25:GUS (Sabatini et al., 1999) and WUSCHEL-related homeobox5 (WOX5)pro:GFP (for green fluorescent protein) (Blilou et al., 2005), we observed that QC25:GUS expression was strongly reduced and that WOX5pro:GFP expression was only slightly reduced in miao (Figures 1H to 1K), indicating a partial loss of QC identity.
MIAO Encodes GR2
We used map-based cloning to identify the mutated MIAO gene. Initial mapping localized MIAO to the right arm of chromosome III, between the molecular markers ciw4 and nga6. Subsequent fine-scale mapping using PCR-based molecular markers narrowed MIAO to an ∼19-kb region, between positions 951 and 28,593 bp in BAC T5N23 (Figure 2A). All genes in this region of the miao genome were sequenced and compared with the gene sequences of Col-0. One cytosine-to-thymine mutation at the 449th nucleotide of the At3g54660 coding region was detected. At the protein level, this base change translates to a replacement of Ser-150 by Phe (Figure 2A). Annotated by TAIR as GR2, At3g54660 is one of two genes encoding GR in the Arabidopsis genome. GR belongs to the pyridine nucleotide-disulphide oxidoreductase family. GR2 contains 11 exons and 10 introns. The full-length open reading frame is 1698 bp long and encodes a 565–amino acid protein. GR2 shows 47.8% identity to GR1. The Conserved Domain Database (Marchler-Bauer et al., 2009) reveals that GR2 contains one pyridine nucleotide-disulphide oxidoreductase domain from amino acid 431 to 540 and one Rossmann-fold NAD(P)(+) binding domain between residues 262 and 334. In addition, the sequence contains a pyridine nucleotide-disulphide oxidoreductase domain from residue 132 to 142, where Cys-135 and Cys-140 can form the active disulfide bonds (http://prosite.expasy.org/).
Homozygous miao plants were transformed with a binary vector containing a 5617-bp genomic GR2 DNA fragment from Col-0, which included the 2745-bp genomic coding region, as well as the 2215-bp upstream and 657-bp downstream flanking sequences. This DNA fragment rescued the short-root and RAM defects, as well as the dwarf phenotype, of miao (Figure 2B; see Supplemental Figure 1 online), confirming that the GR2 mutation is responsible for the miao phenotype. GR2pro:GR2-Venus and GR2pro:GR2-GUS also completely rescued the miao phenotype (see Supplemental Figure 4 online); hence, these constructs were used for further GR2 localization (see below).
To determine the effect of the GR2 mutation on GR activity, we assayed total GR activity in vivo. Total protein was extracted from the whole seedlings of Col-0 and miao lines, and GR activities were measured using 1.0 mM GSSG as the substrate and 0.2 mM NADPH as the reducing power. As shown in Figure 2C, the total GR activity at 3, 5, and 7 DAG decreased to 43, 52, and 59% of that of Col-0, respectively, in miao extracts. It was previously reported that GR1 accounts for 40 to 65% of the total GR activity in Arabidopsis leaf tissue (Marty et al., 2009; Mhamdi et al., 2010), suggesting that the activity of GR2 in miao is seriously impaired.
GR2 Is Widely Expressed in Plants and Localizes to Plastids
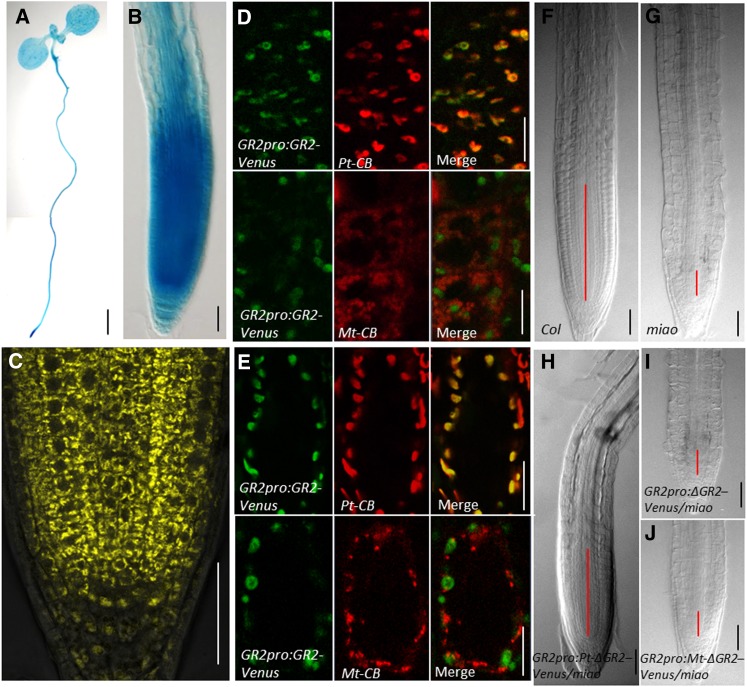
Histochemical staining of GR2pro:GR2-GUS transgenic plants showed that GR2 was expressed throughout the whole seedling (Figure 3A). Strong GR2pro:GR2-GUS and GR2pro:GR2-Venus expression was observed in the RAM (Figures 3B and 3C), in line with the RAM defects and root growth inhibition of the GR2 mutant miao (Figure 1; see Supplemental Figures 2 and 3 online). In addition, GR2pro:GR2-GUS was widely expressed in various organs and tissues (e.g., the stele of the mature root zone and the lateral roots, leaves, mature flowers, and siliques) (see Supplemental Figure 5 online), suggesting that GR2 is involved in pleiotropic developmental processes.
Figure 3.
The Expression and Plastid Localization of GR2 Partially Rescue RAM Defects of miao.
(A) and (B) Light micrographs showing histochemical staining of GR2pro:GR2-GUS in 6-d-old whole seedlings (A) and root tips (B). For GUS staining, 50 mg/L of 5-bromo-4-chloro-3-indolyl-β-glucuronic acid was used at 37°C for 90 min for whole seedlings (A) and 20 min for root tips (B). Bars = 10 mm in (A) and 50 µm in (B).
(C) CLSM image of GR2pro:GR2-Venus expression in RAM. Bar = 50 µm.
(D) and (E) GR2pro:GR2-Venus (Venus, green) is colocalized with the plastid marker, Pt-CB (CFP, red), but not the with the mitochondria marker, Mt-CB (CFP, red), in the roots (D) and hypocotyl cells (E). Bars = 10 µm in (D) and 20 µm in (E).
(F) to (J) Nomarski images of root tips from 6-d-old Col-0 (F), miao (G), GR2pro:Pt-ΔGR2-Venus/miao (H), GR2pro:ΔGR2-Venus /miao (I), and GR2pro:Mt-ΔGR2-Venus/miao (J) transgenic plants. Truncated GR2 without the original N-terminal targeting sequence GR2pro:ΔGR2-Venus was fused with either a plastid targeting signal, generating GR2pro:Pt-ΔGR2-Venus, or a mitochondria targeting signal, generating GR2pro:Mt-ΔGR2-Venus. These constructs were transformed into the miao mutant mediated by Agrobacterium. For each transformation, at least 15 independent lines exhibited similar phenotypes. Red lines indicate the meristem. The meristem boundary is based on the first elongation of cortex cell files. Bars = 50 µm.
In contrast with the cytosol and peroxisome localization of GR1 (Marty et al., 2009; Kataya and Reumann, 2010), GR2 is predicted to be a plastidic protein by TAIR and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html) (Small et al., 2004). Moreover, ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) reveals that the first 73 N-terminal amino acids of GR2 constitute the plastid/chloroplast localization signal. However, previous fluorescence microscopy studies on transiently transformed soybean (Glycine max) suspension cells claimed that GR2 could localize to both plastids and mitochondria (Chew et al., 2003). To further define the subcellular localization of GR2, GR2pro:GR2-Venus transgenic plants were crossed with either a plastid-cyan fluorescent protein (CFP) marker line, Pt-CB, or a mitochondria-CFP marker line, Mt-CB. Pt-CB and Mt-CB marker lines were generated by plant transformation with binary vectors, Pt-CB or Mt-CB, which were constructed by the fusion of the CFP with the chloroplast targeting signal of the tobacco (Nicotiana tabacum) ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) small subunit and the mitochondria targeting signal of Saccharomyces cerevisiae COX4, respectively (Nelson et al., 2007). The confocal microscopy images revealed that GR2pro:GR2-Venus colocalized with Pt-CB but not with Mt-CB in the root and hypocotyl cells (Figures 3D and 3E), demonstrating the plastid localization of GR2. In addition, our coexpression of 35Spro:GR2-Venus and Pt-CB or Mt-CB in two transient transformation systems (i.e., protoplast transformation mediated by polyethylene glycol [PEG] and seedling transformation mediated by Agrobacterium tumefaciens; see Supplemental Figure 6 online) confirmed the plastid localization of GR2. Abundant plastids in RAM were shown by the plastid marker Pt-CB (see Supplemental Figure 7 online). The high density of plastids in the RAM probably reflects the high activity of these cells, considering that many crucial housekeeping metabolic reactions, for example, glycolysis (Muñoz-Bertomeu et al., 2009) and the oxidative pentose phosphate pathway (Kruger and von Schaewen, 2003), occur in plastids. However, other subcellular localizations resulting from alternative splicing or translation initiation in different temporal and spatial contexts could not be excluded.
To test the importance of the subcellular localization of GR2 to root growth and RAM maintenance, we generated GR2pro:ΔGR2-Venus by truncating the first 73 amino acids of the N-terminal region of GR2, which were a predicted plastid localization signal. The plastid targeting signal from the tobacco Rubisco small subunit and the mitochondria targeting signal from S. cerevisiae COX4 were inserted into the N terminus of GR2pro:ΔGR2-Venus, giving rise to GR2pro:Pt-ΔGR2-Venus and GR2pro:Mt-ΔGR2-Venus, respectively. These vectors were then transformed into miao plants. For each transformation, at least 15 independent lines were identified, all showing similar phenotypes. The transformations of both GR2pro:ΔGR2-Venus and GR2pro:Mt-ΔGR2-Venus failed to rescue the root defects, but GR2pro:Pt-ΔGR2-Venus conferred partial recovery of the root phenotype of miao (Figures 3F to 3J; see Supplemental Figure 8 online). The only partial recovery was likely due to the lower expression of GR2pro:Pt-ΔGR2-Venus, compared with that of GR2pro:GR2-Venus, in the RAM (see Supplemental Figure 9 online), the result of an unknown mechanism caused by the truncation of 219 bp (encoding 73 amino acids) and/or insertion of 237 bp (encoding 79 amino acids) at the 5′ end of GR2. Nevertheless, our findings demonstrate that GR2 is localized to plastids and that its plastid localization is essential to root growth and RAM maintenance.
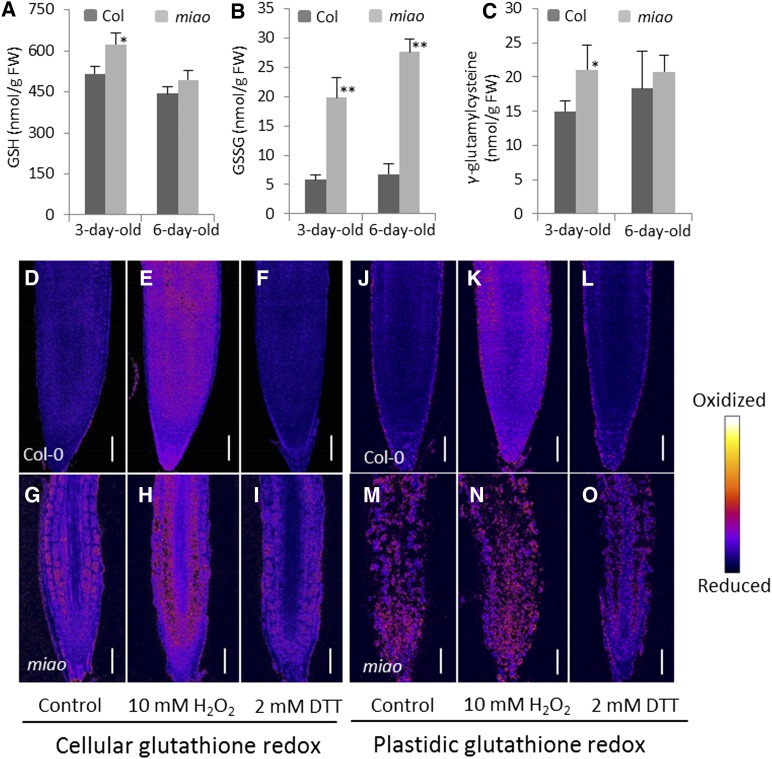
GR2 Mutation in miao Affects Glutathione Redox Status
Given that GR regulates the GSH:GSSG ratio by catalyzing the reduction of GSSG, we measured the GSH:GSSG ratio in the miao mutant. Glutathione and its precursor, γ-glutamylcysteine (γ-EC), were extracted from whole seedlings of Col-0 and miao and quantified by reverse-phase HPLC (Schupp and Rennenberg, 1988; Strohm et al., 1995). GSH and γ-EC levels were slightly higher in miao (Figures 4A and 4C). Notably, miao had a steep increase in GSSG (3.4-fold in 3-d-old seedlings and 4.1-fold in 6-d-old seedlings) (Figure 4B). Hence, the GSH:GSSG ratios of 3- and 6-d-old miao dropped to ∼35 and ∼31%, respectively, of those of Col-0. GRX1-reduction oxidation sensitive GFP (GRX1-roGFP2) was developed as an indicator of glutathione redox status (Meyer et al., 2007; Gutscher et al., 2008). GRX1-roGFP2 can be excited at 405 and 488 nm in its oxidized and reduced states, respectively; thus, the 405:488 ratio reflects the glutathione redox status in vivo. To restrict this measurement to plastids, we fused a chloroplast-targeting signal of the tobacco Rubisco small subunit (Nelson et al., 2007) to the N terminus of GRX1-roGFP2, giving rise to Pt-GRX1-roGFP2. Ratiometric imaging showed that in both Col-0 and miao, treatment with H2O2 increased the 405:488 ratio, whereas treatment with the thiol-reducing agent DTT decreased the 405:488 ratio (Figures 4D to 4O). Consistent with the lower ratio of GSH:GSSG in miao, a higher 405:488 ratio was observed with both GRX1-roGFP2 and Pt-GRX1-roGFP2 markers in the root tip of miao (Figures 4D, 4G, 4J, and 4M), indicating an oxidized glutathione status. Notably, in the defective RAM of miao, glutathione appeared to be more oxidized as indicated by Pt-GRX1-roGFP2 than by GRX1-roGFP2 (Figures 4G and 4M), corroborating the plastid localization of GR2.
Figure 4.
Glutathione Content and Redox Status in the miao Mutant.
(A) to (C) The content of GSH (A), GSSG (B), and the GSH precursor γ-glutamylcysteine (C) in whole seedlings, measured by reverse-phase HPLC. Data are the means + sd (n = 3). A single asterisk indicates significant differences from the Col-0, P < 0.05 (Student’s t test). Double asterisks indicate highly significant differences from the Col-0, P < 0.01 (Student’s t test).
(D) to (I) Cellular glutathione redox in the root of Col-0 ([D] to [F]) and miao ([G] to [I]) expressing GRX1-roGFP2.
(J) and (O) Plastidic glutathione redox in roots of Col-0 ([J] to [L]) and miao ([M] to [O]) expressing Pt-GRX1-roGFP2.
From (D) to (O), control and samples receiving a 2-min treatment of 10 mM H2O2 or 2 mM DTT were detected at 505 to 530 nm by CLSM with excitation at 405/488 nm. The ratio of 405/488 nm was used for in situ ratiometric imaging. Bars = 50 µm.
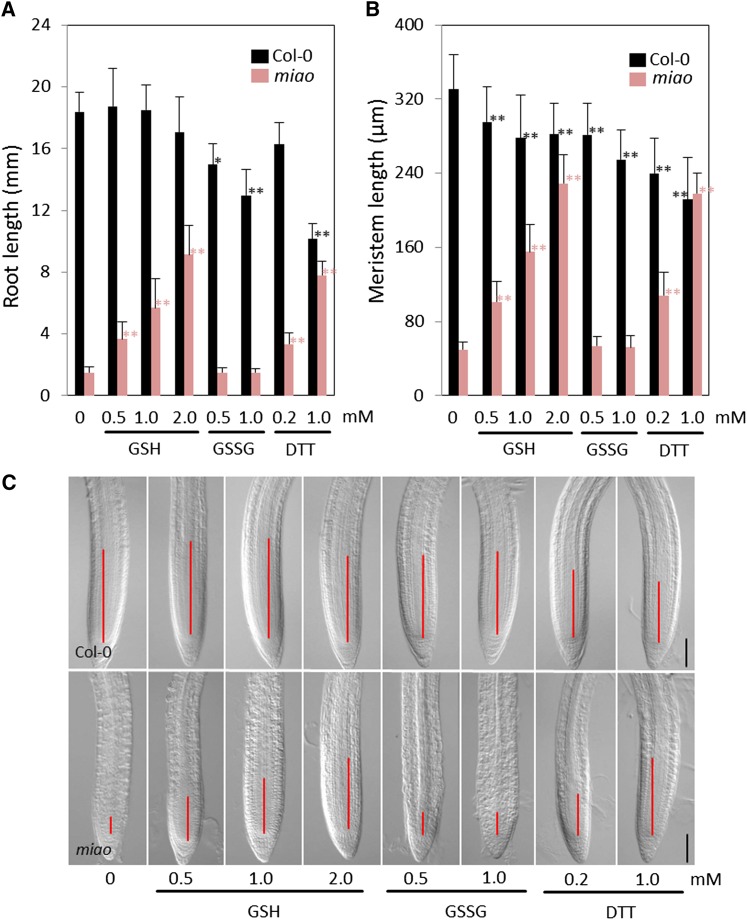
Rescue of miao Mutant Root Phenotypes by Exogenous GSH and DTT
The increased oxidation of glutathione in miao prompted us to test whether exogenous thiol-reducing agents were able to rescue the root phenotype. The seedlings of Col-0 and miao were germinated and grown on one-half-strength Murashige and Skoog (MS) medium supplemented with GSH, GSSG, or DTT. At 6 DAG, GSH significantly enhanced the root and RAM lengths of miao in a dose-dependent manner (Figure 5), whereas the oxidized form of GSH, GSSG, had no effect on miao roots (Figure 5). Moreover, treatments of miao with 0.2 and 1.0 mM DTT also caused root length to increase by 2.2 and 5.2 times and the RAM length to increase by 1.5 and 4.3 times, respectively (Figure 5). These results confirm that the RAM defects and root growth inhibition in miao are triggered by glutathione oxidation.
Figure 5.
Root Defects of the miao Mutant Are Recovered by GSH and the Reducing Agent DTT.
(A) and (B) Length of primary roots (A) and RAM (B) of 6-d-old Col-0 and miao grown on vertical plates of one-half-strength MS medium supplemented with GSH, GSSG, or DTT. Data are the means + sd; n = 20. Differences between treatments and controls were analyzed using Student's t test. A single asterisk represents significant differences (P < 0.05); double asterisks represent highly significant differences (P < 0.01).
(C) Nomarski images of root tips of 5-d-old Col-0 and miao seedlings grown on vertical one-half-strength MS medium supplemented with various concentrations of GSH, GSSG, or DTT. Solid lines indicate meristem. The boundary of the meristem is based on elongation of cortex cell files. Bars = 100 µm.
[See online article for color version of this figure.]
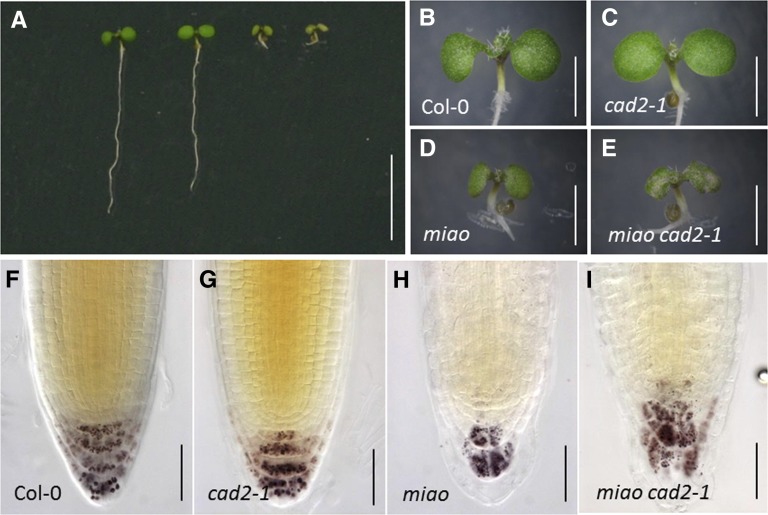
Phenotypes of the miao cad2-1 Mutant
Glutathione redox status is affected by both the GSH:GSSG ratio and the total glutathione level. To elucidate the possible genetic interactions between glutathione biosynthesis and reduction, we crossed miao with a weak GSH1 mutant allele, cad2-1, which has only 15 to 35% of the glutathione found in the wild type and shows cadmium sensitivity but displays nearly normal seedling development under control conditions (Howden et al., 1995; Cobbett et al., 1998). From the F2 generation, mutants homozygous for miao, but heterozygous for cad2-1, were selected and self-pollinated. In the F3 population, the phenotypes segregated, with ∼75% of the seedlings exhibiting the miao phenotype and the others showing cotyledon bleaching (Figures 6A to 6E), a characteristic feature of oxidative-stress sensitivity. The seedlings with cotyledon bleaching exhibited more severe QC/DSC differentiation and more disorganized columella-cell tiering than miao (Figures 6H and 6I). The development of these seedlings was arrested at ∼10 DAG, and the double homozygous plants for both miao and cad2-1 were not found in the next generation. Taken together, miao cad2-1 double mutants displayed RAM defects and postembryonic lethality, probably caused by oxidative stress, suggesting that the combination of these two plastidic proteins, GR2 and γ-ECS, plays a critical role in plant development by regulating glutathione redox status.
Figure 6.
The Phenotype of the miao cad2-1 Double Mutant.
(A) Six-day-old Col-0, cad2-1, miao, and miao cad2-1 (from left to right) seedlings growing on vertical one-half-strength MS medium. Bar = 10 mm.
(B) to (E) Cotyledons of Col-0 (B), cad2-1 (C), miao (D), and miao cad2-1 (E). Bars = 2 mm.
(F) to (I) Nomarski images showing Lugol’s staining of RAM (F), cad2-1 (G), miao (H), and miao cad2-1 (I). Bars = 50 µm.
In addition, the miao gr1-2 double mutant showed the same phenotype as miao at different development stages, and no additive effects were observed (see Supplemental Figure 10A online). However, our glutathione measurement showed that miao gr1-2 had remarkably higher GSSG levels and lower GSH:GSSG ratios than the miao or gr1-2 single mutants (see Supplemental Figure 10B online). These results, together with the known differences in subcellular localization, suggest that the functions of GR2 and GR1, in terms of GSSG reduction, are not redundant. We also observed that GSH content was increased by ∼20% in the miao gr1-2 seedlings compared with the miao or the gr1-2 seedlings (see Supplemental Figure 10B online); this increase was likely a result of the redox regulation of γ-ECS activity (Hicks et al., 2007; Gromes et al., 2008).
In Silico Analysis of Gene Expression under BSO and Auxin Treatment
Auxin is well known as a crucial regulator of plant development (Benjamins and Scheres, 2008). In an attempt to disclose the relationship between glutathione-dependent redox and auxin signaling, we performed a comparative in silico analysis of gene expression in whole seedlings under BSO-mediated glutathione oxidation and auxin treatments, using publicly available data from Arabidopsis seedlings (Paponov et al., 2008; Koprivova et al., 2010). To select genes that were differentially regulated by BSO, we used a false discovery rate of q < 0.05 and a 1.5-fold change cutoff and identified 425 upregulated and 170 downregulated genes. Of the 425 upregulated genes, 47 were upregulated under at least one of the conditions of indole-3-acetic acid (IAA) application (see Supplemental Figure 11 and Supplemental References 1 online). By contrast, only three of these genes were upregulated by auxin under three or more conditions of auxin application, indicating their robust auxin response. None of the 47 upregulated genes belongs to a classical auxin-responsive family (e.g., Auxin/IAA, Gretchen Hagen3, or SMALL AUXIN UPREGULATED RNA [SAUR]). Of these 47 genes, 21 were upregulated only under extremely high levels of IAA application (10 µM), indicating that this gene group is more closely linked with stress responses than with specific features of auxin signaling. This finding was confirmed by an analysis of the promoter sequences of this group of IAA-induced genes, whose promoters lacked the auxin-responsive element (P = 0.946). The W-box promoter (P < 0.00001) and DEHYDRATION RESPONSE ELEMENT B1A/C-REPEAT BINDING FACTOR3 (P < 0.001) cis-elements were overrepresented in this group of genes, which again strongly indicates a link to stress responses. Of the 170 genes downregulated by BSO, 18 were upregulated under at least one selected auxin application condition. Of these 18 genes, only five could be considered truly robust auxin-responsive genes because they were upregulated under at least five of seven selected conditions of auxin application (see Supplemental Figure 12 online). Among the five genes, we selected IAA29 and SAUR23, the classic auxin-responsive genes for investigating gene expression in miao by quantitative RT-PCR (qRT-PCR). However, miao and Col-0 seedlings had similar expression levels of IAA29 and SAUR23 (see Supplemental Figure 13 online). This discrepancy may result from the differences in the extent of GSH oxidation and/or the means by which glutathione oxidation occurs (i.e., total glutathione decrease and GSSG accumulation). Nevertheless, because the node of genes with overlapping expression under IAA treatment and glutathione oxidation mediated by BSO is small, we conclude that glutathione-dependent redox and auxin signaling act nearly independently in whole seedlings at the transcriptional level.
Notably, one of the PLT genes, PLT1, was weakly downregulated (fold change 0.83, q = 0.029) after 6 d of treatment with 1.25 mM BSO. PLT is a crucial regulator in root development (Aida et al., 2004; Galinha et al., 2007). Auxin can induce the expression of PLT (Aida et al., 2004); conversely, PLT regulates PIN, which stabilizes the auxin gradient by PAT (Blilou et al., 2005; Galinha et al., 2007). This prompted us to investigate the potential interaction between glutathione-dependent redox signaling and the auxin/PLT pathway (see below).
GR2 Mutation Affects Auxin/PLT Pathway
We first found that levels of free IAA were slightly but significantly (Student’s t test, P < 0.05, n = 4) lower (by ∼10%) in 6-d-old miao seedlings (9.7 ng/g fresh weight [FW]) than in wild-type seedlings (10.7 ng/g FW). YUCCA-dependent auxin biosynthesis may account for the lower auxin levels in miao, as the expression of YUCCA3 and YUCCA4 in miao seedlings was decreased to 22 and 68% of wild-type levels, respectively, according to our qRT-PCR results (see Supplemental Figure 14 online). By contrast, the expression of several other auxin biosynthesis–related genes, such as TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS1 (TAA1), NITRILASE1 (NIT1) and NIT2, was unchanged in miao (see Supplemental Figure 14 online). The slightly lower IAA content in miao is different from the strong decrease (threefold reduction) observed in ntra ntrb cad2-1 (Bashandy et al., 2010), while the auxin level was not significantly affected in the ntra ntrb and cad2-1 mutants (Bashandy et al., 2010). These results suggest that severe glutathione oxidation by GR2 mutation could trigger a slight decrease in auxin levels and that the combination of NGS- and NTS-dependent redox signaling is necessary for auxin homeostasis.
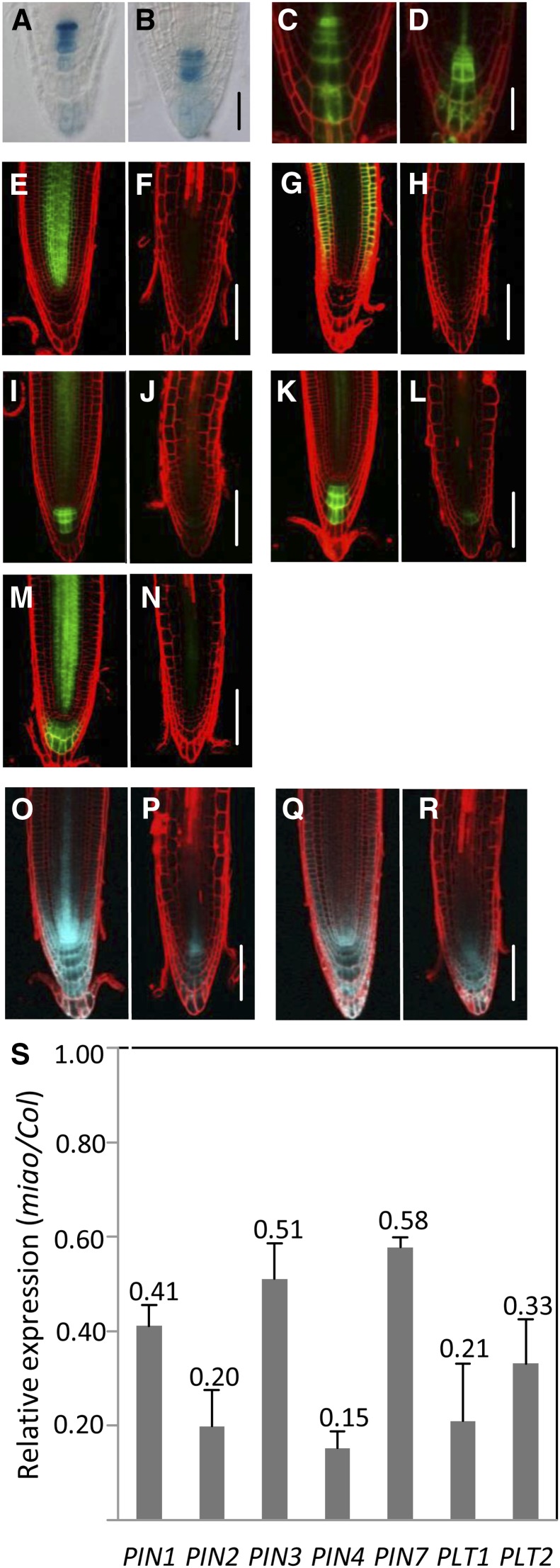
The auxin maximum indicated by DR5:GUS and DR5rev:GFP in QC was considerably lower in miao than in Col-0 (Figures 7A to 7D). Moreover, the auxin maximum in miao was localized to the first tier of columella cells, rather than to the QC, as observed in Col-0 (Figures 7B and 7D), suggesting that the auxin response and/or gradient in the stem cell niche was strongly perturbed. This auxin perturbation may have been responsible for the alteration of QC differentiation and the partial loss of QC identity (Figures 1F to 1K) (Sabatini et al., 1999; Blilou et al., 2005). PIN-mediated PAT establishes and substantiates auxin gradients, thereby regulating root patterning and growth (Blilou et al., 2005). The expression levels of PIN1pro:PIN1-GFP, PIN2pro:PIN2-GFP, PIN3pro:PIN3-GFP, PIN4pro:PIN4-GFP, and PIN7pro:PIN7-GFP were severely reduced in the RAM of miao (Figures 7E to 7N). Lower expression of PIN2pro:GUS, PIN4pro:GUS, and PIN7pro:GUS was also observed in miao (see Supplemental Figure 15 online). The qRT-PCR results confirmed that the mRNA levels of these PINs in the root tips of miao were decreased (Figure 7S). These findings indicate that PIN expression is under transcriptional regulation by glutathione oxidation. Our results are in agreement with the previously observed reductions in PIN expression following application of the glutathione biosynthesis inhibitor BSO (Bashandy et al., 2010; Koprivova et al., 2010).
Figure 7.
GR2 Mutation Affects DR5, PIN, and PLT Expression.
(A) to (D) The expression patterns of DR5:GUS ([A] and [B]) and DR5rev:GFP ([C] and [D]) in the root tip of 6-d-old Col-0 ([A] and [C]) and miao ([B] and [D]). Bars = 50 µm.
(E) to (R) The expression patterns of PIN1pro:PIN1-GFP ([E] and [F]), PIN2pro:PIN2-eGFP ([G] and [H]), PIN3pro:PIN3-GFP ([I] and [J]), PIN4pro:PIN4-GFP ([K] and [L]), and PIN7pro:PIN7-GFP ([M] and [N]) in 6-d-old root tips of Col-0 ([E], [G], [I], [K], and [M]) and miao ([F], [H], [J], [L], and [N]). Bars = 100 µm.
(O) to (R) The expression of PLT1pro:CFP ([O] and [P]) and PLT2pro:CFP ([Q] and [R]) in 6-d-old root tips of Col-0 ([O] and [Q]) and miao ([P] and [R]). Bars = 100 µm.
(S) Relative expression of PIN1, PIN2, PIN3, PIN4, PIN7, PLT1, and PLT2 genes in 6-d-old miao, compared with that in Col-0, measured by real-time qRT-PCR. Total RNA was collected from the 2-mm root apex of seedlings. For each gene, the mRNA level was normalized to UBIQUITIN10 (AT4G05320). Data are the means + sd of three biological replicates.
From (C) to (R): red, PI staining; green, GFP; and cyan, CFP.
The expression of PLT1 and PLT2 was estimated by investigating the PLT1pro:CFP and PLT2pro:CFP marker lines. In the Col-0 background, the expression of PLT1pro:CFP and PLT2pro:CFP was high in the stem cell niche, reached an intermediate level in rapidly dividing cells, and was low in the elongated cells (Figures 7O and 7Q), consistent with previous observations (Galinha et al., 2007). The overall expression levels of PLT1pro:CFP and PLT2pro:CFP were decreased in the miao roots, and the PLT1 and PLT2 expression domains were restricted to the stem cell niche, the columella, and the proximate meristem cells (Figures 7P and 7R), corresponding to the reduced RAM of miao. The qRT-PCR results indicated that the transcriptional expression of PLT1 and PLT2 in the root tips of miao was decreased to 21 and 33% of wild-type values, respectively (Figure 7S). The perturbed DR5, PIN, and PLT expression indicates the impairment of the auxin/PLT pathway in the oxidized glutathione environment of the miao RAM.
Glutathione Oxidation Blocks Auxin/PLT Signaling
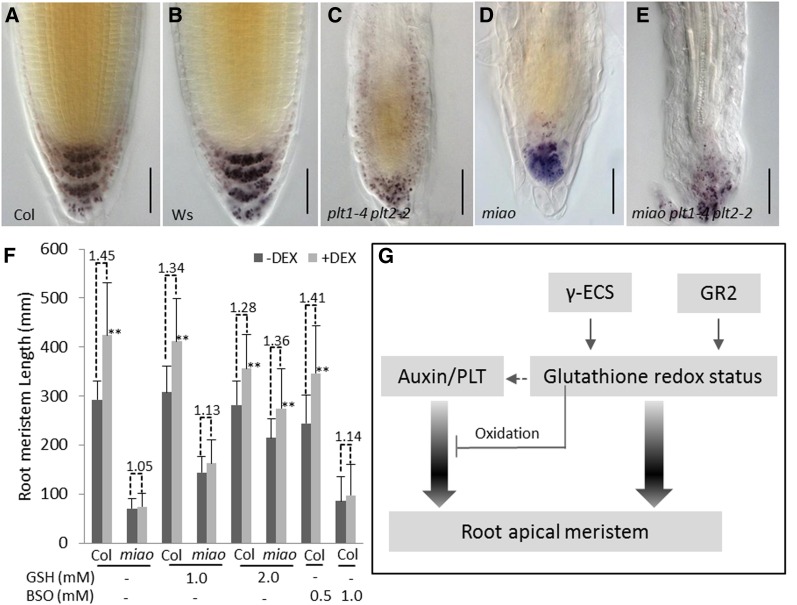
To elucidate the potential interaction between glutathione-dependent redox signaling and auxin/PLT signaling, the PLT double mutant plt1-4 plt2-2, which shows strong defects in the RAM and stem cell niche (Aida et al., 2004; Galinha et al., 2007), was crossed with miao. The plt1-4 plt2-2 miao triple mutant exhibited smaller seedling size, serious RAM defects, and more disorganized columella cells than plt1-4 plt2-2 or miao (Figures 8A to 8E; see Supplemental Figure 16 online). The additive phenotype of miao plt1-4 plt2-2 suggests that auxin/PLT and glutathione-dependent redox signaling represent different pathways in RAM maintenance (Figure 8G). Next, we introduced the dexamethasone (DEX)–inducible PLT2 overproduction transgenic line, 35Spro:PLT2-Glucocorticoid Receptor (GLR), into the miao background. Similar to a previous report (Galinha et al., 2007), PLT2 overproduction induced by DEX significantly (Student’s t test, P < 0.01) increased RAM length by 45% in the 35Spro:PLT2-GLR wild-type background, while PLT2 overproduction conferred no increase in RAM length in the miao background (Figure 8F; see Supplemental Figure 17 online). However, 35Spro:PLT2-GLR/miao did exhibit RAM expansion of 13 and 36% in response to DEX in the medium supplemented with 1.0 and 2.0 mM GSH, respectively. Moreover, the RAM expansion induced by DEX in the 35Spro:PLT2-GLR wild-type background was slightly inhibited by treatment with 0.5 mM BSO and more strongly inhibited by treatment with 1.0 mM BSO (Figure 8F; see Supplemental Figure 17 online). These results indicate that the oxidized glutathione environment disrupts auxin/PLT downstream effectors. In other words, a reduced glutathione environment is required for the auxin/PLT pathway to function in RAM maintenance.
Figure 8.
GSH Redox Is Required for the Auxin/PLT Pathway.
(A) to (E) Nomarski images showing Lugol’s staining of root tips of Col-0 (A), Ws (B), plt1-4 plt2-2 (C), miao (D), and miao plt1-4 plt2-2 (E). Bars = 50 µm.
(F) Five-day-old seedlings receiving treatments of 1.0 and 2.0 mM GSH or 0.5 and 1.0 mM BSO were transferred to new plates (same medium) either containing 1.5 µM DEX or no DEX for 24 h. Then, the RAM expansion of 35Spro:PLT2-GLR and 35Spro:PLT2-GLR/miao, upon DEX induction under different treatments, was recorded with Nomarski images. RAM length was counted based on the first elongation of cortex cell files. The ratios of DEX-induced RAM to non-DEX-induced RAM are given above the bars. Data are the means of 20 repetitions + sd. Double asterisks indicate highly significant differences (Student's t test, P < 0.01).
(G) Complex roles of glutathione redox status in RAM maintenance. Glutathione redox status is critically regulated by two plastidic proteins, γ-ECS and GR2. γ-ECS participates in glutathione synthesis, and GR2 regulates the GSH:GSSG ratio. The glutathione redox status maintains the RAM independently of the auxin/PLT pathway; however, glutathione oxidation markedly affects the auxin/PLT pathway and blocks the RAM expansion induced by PLT2 overexpression. A reduced glutathione environment is required for the downstream effectors of the auxin/PLT pathway.
[See online article for color version of this figure.]
Glutathione measurement with reverse-phase HPLC showed no significant differences in GSH and GSSG between the plt1-4 plt2-2 double mutant and the wild-type line (Wassilewskija [Ws] ecotype) (GSH and GSSG: 1048.1 + 72.2 nmol/g FW and 20.8 + 1.4 nmol/g FW in Ws; 902.5 + 24.3 nmol/g FW and 17.6 + 0.3 nmol/g FW in plt1-4 plt2-2, means + sd, n = 3, Student’s t test, P < 0.05), indicating that glutathione oxidation is not increased in the defective RAM of the PLT loss-of-function mutants. Furthermore, the application of 1.0 mM GSH, which was sufficient to rescue RAM defects in miao (Figure 5), did not rescue the defective RAM in plt1-4 plt2-2 (see Supplemental Figure 18 online), substantiating that the glutathione oxidization is not a mechanism of the RAM defects in the loss of function of PLT.
DISCUSSION
Plant root growth is highly plastic and responds to the changing environment. The in vivo redox status, influenced by the surrounding environment, is a direct and reliable signal for plant growth and development. As a major redox buffer to oxidants and, perhaps more importantly, a critical component in redox signaling, the small-molecule glutathione has received much attention. In this work, we reported that plastid-localized GR2 modulates the glutathione redox status by regulating the GSH:GSSG ratio, and we subsequently disclosed the essential and complex roles of glutathione redox status in RAM maintenance.
RAM Maintenance Is Dependent on the Glutathione Redox Environment
The multiple roles of glutathione in plants have been well documented in recent reviews (Rouhier et al., 2008; Noctor et al., 2011, 2012). Evidence from pharmacological and genetic manipulations of the glutathione biosynthesis enzyme γ-ECS indicates a critical role of glutathione in root growth (Cheng et al., 1995; Sánchez-Fernández et al., 1997; Vernoux et al., 2000; Koprivova et al., 2010). In this work, we characterized plastid-localized GR2, whose mutation led to glutathione oxidation attributable to GSSG accumulation in the miao mutant (Figures 4B, 4D to 4O). Thus, it is plausible to reason that the severe RAM defects and root growth inhibition in miao were caused by glutathione oxidation and not by the GSH content. Indeed, the root phenotype of miao could be rescued by both GSH and the thiol-reducing agent DTT (Figure 5), substantiating our conclusion. Our results provide evidence to support the previous hypothesis that redox regulation is implicated in RAM organization (Jiang and Feldman, 2005; De Tullio et al., 2010).
Many regulators, from transcription factors, phytohormones, and peptides to microRNAs, contribute to RAM development and organization (Aichinger et al., 2012; Perilli et al., 2012; Petricka et al., 2012). However, which targets are regulated by glutathione redox status in RAM development and maintenance? The identification of glutathione redox target proteins is certainly a challenging task, as thiol redox regulation is rapid and reversible in response to different environmental and developmental cues. The thiol redox regulation of a given protein is difficult to predict. Pioneering work has identified 94 putative targets of a Populus trichocarpa ☓ deltoides Grx via detection by liquid chromatography coupled to tandem mass spectrometry (Rouhier et al., 2005). Recently, 225 glutathionylated proteins were identified in a photosynthetic model organism, Chlamydomonas reinhardtii, by biotinylated glutathione and streptavidin affinity chromatography (Zaffagnini et al., 2012). These target proteins are involved in many biological processes and pathways, ranging from carbon, energy, and nitrogen metabolism to protein biogenesis and degradation (Zaffagnini et al., 2012). Interestingly, some targets were linked to cell cycle control and hormone signaling, and their corresponding putatively orthologous genes in Arabidopsis may participate in RAM maintenance. These data are valuable in establishing an overall scheme of the redox regulation network; however, additional genetic, physiological, and proteomic data are required to realize this aim.
Glutathione Redox Status in Plastids Is Critical to Plant Development
GR2 was shown to be localized to plastids in this work (Figures 3D and 3E; see Supplemental Figure 6 online). Previously, GR2 was identified as a gene required for embryo development (Tzafrir et al., 2004), and in this work, miao, the weak mutant allele of GR2, displayed RAM defects and root growth inhibition (Figure 1; see Supplemental Figure 1 online). By contrast, the other member of the GR family, the cytosolic/peroxisomal GR1, was not crucial for plant development, although the glutathione oxidation was also remarkable in the gr1 knockout mutant (Marty et al., 2009). Interestingly, the rate-limiting enzyme in glutathione synthesis, γ-ECS encoded by GSH1, was also localized to plastids (Wachter et al., 2005). The GSH1 null mutant was embryo lethal (Cairns et al., 2006), and another mutant allele, rml1, exhibited RAM defects and seedling lethality. Furthermore, the double mutant of these two plastid-localized genes, miao cad2-1, showed stronger RAM defects, seedling lethality, and cotyledon bleaching (Figure 6), whereas miao gr1-2 exhibited a similar phenotype to miao (see Supplemental Figure 10A online). These findings strongly suggest that the glutathione redox status in plastids has a critical role in plant development.
The plastid is an endosymbiotic organelle, originating from a cyanobacterium (Dyall et al., 2004). Many important metabolic pathways, such as carbon assimilation, nitrogen assimilation, starch metabolism, amino acid biosynthesis, and fatty acid biosynthesis, occur in plastids. Many enzymes in these metabolic pathways are regulated in a redox-dependent manner. One example is NADP-glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a key enzyme in glycolysis in both plastids and the cytosol, as well as in the Calvin-Benson cycle in chloroplasts. The activity of one chloroplastic GAPDH was activated by an f-type Trx, whose activity was inhibited by glutathionylation (Michelet et al., 2005); the activity of a Trx-independent chloroplastic GAPDH was also decreased by glutathionylation (Zaffagnini et al., 2007). In the recent identification of glutathionylated proteins in C. reinhardtii, proteins related to the Calvin-Benson cycle in chloroplasts were found to be major targets of glutathionylation (Zaffagnini et al., 2012). Perturbed metabolic pathways in plastids, affected by an unfavorable glutathione redox status, may give rise to the RAM and other developmental defects observed in the miao and rml1 mutants.
We observed significant decreases in the expression of the nuclear genes PIN and PLT in the oxidized glutathione environment of miao (Figures 7E to 7S; see Supplemental Figure 15 online). Moreover, we identified 425 genes that were upregulated and 170 that were downregulated, both by BSO treatment from the publicly available Arabidopsis data (Koprivova et al., 2010). The large number of genes regulated at the transcriptional level by l-buthionine (S,R)-sulfoximine (BSO) treatment could be a consequence of the plastid-to-nucleus retrograde signaling (Rodermel, 2001; Nott et al., 2006), although the secondary effects of glutathione oxidation cannot be excluded. In retrograde signaling, plastid-generated factors enter the nucleus and affect the expression of nuclear genes. It has been proposed that the redox status of photosynthetic electron transport components (e.g., the plastoquinone pool) and reactive oxygen species act as retrograde signals from the plastids to the nucleus (Rodermel, 2001; Nott et al., 2006). Recently, it was demonstrated that glutathione was transported across plastid envelopes into the cytosol by the CRT-LIKE TRANSPORTER (Maughan et al., 2010) and recruited into the nucleus to regulate the cell proliferation cycle through glutathione redox (Vivancos et al., 2010). Therefore, it is reasonable to hypothesize that glutathione redox status is a signal in plastid-to-nucleus retrograde signaling.
Complex Roles of glutathione Redox Status in the Auxin/PLT Pathway
Auxin has long been known as a crucial regulator of plant development (Benjamins and Scheres, 2008); however, the interaction between auxin and thiol redox signaling had remained elusive until recently. Through the characterization of the triple mutant ntra ntrb cad2-1, defective in NGS and NTS signaling, a link between cellular redox status and auxin signaling was demonstrated (Bashandy et al., 2010). In our work, the severe root defects observed in the miao RAM are reminiscent of those induced by the knockout of the auxin-induced regulator of root development, PLT (Aida et al., 2004; Galinha et al., 2007).
Indeed, the expression levels of components of the auxin/PLT pathway (DR5, PIN, and PLT) were notably decreased in miao root tips (Figure 7; see Supplemental Figure 15 online). The disruption of auxin/PLT may be a direct result of the strong GSH oxidation by the GR2 mutation, or more likely, a secondary effect of disturbed meristematic activity. In line with the independence between auxin and glutathione redox signaling in whole seedlings at the transcriptional level, as indicated by our in silico analysis, an unknown auxin/PLT-independent redox signaling pathway seems to be involved in RAM maintenance (Figure 8G), which is supported by the additive phenotypes of miao plt1-4 plt2-2 (Figures 8A to 8E; see Supplemental Figure 16 online). Nevertheless, PLT2 overexpression failed to induce RAM proliferation in miao or the BSO-treated plants. However, the failed RAM expansion response to PLT2 overexpression in miao was reversed by the application of GSH (Figure 8F; see Supplemental Figure 17 online), suggesting that the oxidized glutathione environment disrupts the downstream effectors of the auxin/PLT pathway (Figure 8G). We propose that a reduced glutathione environment is required for auxin/PLT function in RAM maintenance; however, to date, we have little information on the downstream effectors in the auxin/PLT pathway. The identification of more auxin/PLT downstream effectors will help us understand how the oxidized glutathione environment blocks the function of the auxin/PLT signaling pathway.
Taken together, glutathione redox status has a complex role in RAM maintenance, as shown in Figure 8G. The identification and characterization of NGS-dependent target proteins and intermediates via genetic, physiological, and proteomic approaches will provide insights into the role of the glutathione redox status in RAM development and maintenance. The GR2 mutant miao exhibits severe RAM defects but is viable and fertile and will therefore be a useful genetic tool in achieving this goal.
METHODS
Plant Materials and Growth Conditions
The marker lines and mutants used in this work have been described previously. CYCLIN B1;1pro:GUS (Colón-Carmona et al., 1999), QC25:GUS (Sabatini et al., 1999), WOX5pro:GFP (Blilou et al., 2005), GRX1-roGFP2 (Meyer et al., 2007; Gutscher et al., 2008), cad2-1 (Howden et al., 1995; Cobbett et al., 1998), gr1-2 (Marty et al., 2009), DR5:GUS (Ulmasov et al., 1999), DR5rev:GFP (Benková et al., 2003), PIN1pro:PIN1-GFP (Benková et al., 2003), PIN2pro:PIN2-eGFP (Men et al., 2008), PIN3pro:PIN3-GFP (Blilou et al., 2005), PIN4pro:PIN4-GFP (Blilou et al., 2005), PIN7pro:PIN7-GFP (Blilou et al., 2005), PIN2pro:GUS (Benková et al., 2003), PIN4pro:GUS (Ulmasov et al., 1999; Benková et al., 2003), PIN7pro:GUS (Benková et al., 2003), PLT1pro:CFP (Galinha et al., 2007), PLT2pro:CFP (Galinha et al., 2007), and 35Spro:PLT2-GLR (Galinha et al., 2007) mutants are ecotype Columbia-0 (Col-0), while the plt1-4 plt2-2 mutant (Aida et al., 2004) is ecotype Wassilewskija.
Surface-sterilized seeds were imbibed on one-half-strength Murashige and Skoog (MS) and 1.2% agar medium supplemented with 1% Suc at 4°C in the dark for 2 d and grown at 22°C with a 16-h-light (∼100 µmol m−2 s−1)/8-h-dark cycle. Seedlings were transferred to soil between 8 and 12 DAG and grown under the described conditions in a phytochamber. For glutathione treatment experiments, filter-sterilized GSH or GSSG stock solution (100 mM) was freshly prepared, and the pH was adjusted to 5.6 with 1 M potassium hydroxide (Potters et al., 2010). For BSO treatment experiments, filter-sterilized BSO stock solution (100 mM) was freshly prepared.
Map-Based Cloning of MIAO
For positional cloning of the mutated gene, miao (Arabidopsis thaliana Col-0 background) was crossed with Landsberg erecta to generate a large F2 mapping population. Cloning was performed according to published procedures (Lukowitz et al., 2000). Based on bulked segregant analysis, the mutated site was localized to a region between molecular markers ciw4 and nga6. For fine-scale mapping, simple allele-discriminating PCR (Bui and Liu, 2009) and derived cleaved amplified polymorphic sequence markers (Neff et al., 1998) were selected based on sequence polymorphisms between Col-0 and Landsberg erecta that were predicted by the Monsanto Arabidopsis Polymorphism and Ler Sequence Collection (http://www.Arabidopsis.org/browse/Cereon/index.jsp). The mutated site of miao was finally narrowed to a region between 951 and 28,593 bp of BAC-T5N23 by two markers, T5N23.951 (simple allele-discriminating PCR) and T5N23.28593 (derived cleaved amplified polymorphic sequence). The sequence information for the primers used for these two markers is shown in Supplemental Table 1 online.
Plasmid Construction and Plant Transformation
The sequence information for primers used to construct vectors is shown in Supplemental Table 2 online. GR2 genomic DNA containing the 2745-bp genomic coding region, as well as the 2215-bp upstream– and 657-bp downstream– flanking sequences, was amplified by PCR from the Col-0 genome. AscI and PacI sites were introduced using primer pair p353/p354, and the amplicon was cloned into the binary vector pMDC99, generating pMDC99-GR2pro:gGR2. GR2 cDNA with 5′-AscI was amplified, with primer pair p355/p405, by RT-PCR from the total RNA of Col-0; Venus with 3′-PacI was amplified with primer pair p406/p404 from the Venus/pCS2 vector. The GR2-Venus (with 10☓Ala as a linker) fragment was fused by overlapping PCR and then introduced into the AscI and PacI sites on the pMDC32 vector, giving rise to pMDC32-35Spro:GR2-Venus. To generate pMDC99-GR2pro:GR2-Venus, the GR2 promoter with 5′-AscI from −2215 to +26 bp of the coding region was amplified from pMDC99-GR2pro:gGR2, using primer pair p353/p432; the GR2-Venus-NOS-terminator fragment with 3′-EcoRI was amplified from 35Spro:GR2-Venus with primer pair p433/p422; their overlapping PCR-derived product was then cloned into the AscI and EcoRI sites of pMDC99, resulting in pMDC99-GR2pro:GR2-Venus. The GR2-GUS (with 10☓Ala as a linker) fragment was derived from the fusion of GR2 cDNA and GUS, which was amplified from pMDC162 with primer pair p426/p388. Next, the GR2 promoter with 5′-AscI, GR2-GUS, and the NOS terminator with 3′-EcoRI (amplified from 35Spro:GR2-Venus with primer pair p386/p422) were fused by overlapping PCR, and the product was cloned into the AscI and EcoRI sites of pMDC99, generating pMDC99-GR2pro:GR2-GUS. For pMDC99-GR2pro:ΔGR2-Venus, the ΔGR2-Venus fragment with 3′-EcoRI was amplified from GR2pro:GR2-Venus with primer pair p648/p422 and then fused with the GR2 promoter with 5′-AscI by primer pair p353/p647. To construct pMDC99-GR2pro:Mt-ΔGR2-Venus and pMDC99-GR2pro:Pt-ΔGR2-Venus, the pMDC99-GR2pro vector was first constructed via the insertion of the GR2 promoter fragment, which was amplified with primer pair p353/657, into the AscI and EcoRI sites of pMDC99. The Mt-ΔGR2-Venus-NOS-terminator fragment was amplified from 35Spro:GR2-Venus with primer pair p658/p422 and was then inserted in the EcoRI site of pMDC99-GR2pro, generating pMDC99-GR2pro:Mt-ΔGR2-Venus. Here, the mitochondrial targeting signal from yeast (Saccharomyces cerevisiae) COX4 (Nelson et al., 2007) was introduced in the 5′-p658 site (see Supplemental Table 2 online). The plastid targeting signal from the Rubisco small subunit of tobacco (Nicotiana tabacum) was amplified from the Pt-CB vector (Nelson et al., 2007) with primer pair p665/666, and the ΔGR2-Venus-NOS-terminator fragment was amplified with primer pair p667/422. The two fragments were fused by overlapping PCR, and the product was cloned into the EcoRI site of pMDC99-GR2pro, generating GR2pro:Pt-ΔGR2-Venus vectors. To construct Pt-GRX-RoGFP2, a fragment of GRX-RoGFP2 with 5′-BamHI and 3′-XmaI sites was amplified from the genome of GRX-RoGFP2 seedlings (Meyer et al., 2007), using p447/p448, and cloned into the downstream region of the plastid-targeting signal of Pt-CB by replacing the original CFP cassette (Nelson et al., 2007).
Plant transformation via floral dip, mediated by Agrobacterium tumefaciens GV3101, was performed according to Clough and Bent (1998).
Phenotypic Analysis
Seedlings were scanned and root lengths were measured using Image J (National Institutes of Health; http://rsb.info.nih.gov/ij). For Nomarski imaging of roots, seedlings were fixed and mounted in chloral hydrate solution (chloral hydrate:water:glycerol = 4:3:1). Images were recorded with an AxioVision microscope (Carl Zeiss Microimaging). RAM size was quantified with Image J, based on the region between the QC and the first elongating cell in the cortex cell file (Casamitjana-Martínez et al., 2003; Dello Ioio et al., 2007). mPS-PI staining of roots was performed essentially according to Truernit et al. (2008), but with major modifications. In brief, seedlings were fixed in 2% formaldehyde for 30 min and incubated in methanol for 15 min at room temperature. Seedlings were then rinsed with water and incubated in 1% periodic acid (H5IO6) at room temperature for 30 min; subsequently, the seedlings were rinsed with water and incubated in a reagent containing 100 mM sodium metabisulphite and 0.15 M hydrochloric acid with 5 µg/mL propidium iodide (PI) for 15 to 30 min, until the roots were visibly stained. The samples were rinsed in water, incubated in chloral hydrate solution for 5 min, and mounted in chloral hydrate solution for confocal laser scanning microscopy (CLSM) analysis. For Lugol’s staining, seedlings were incubated in Lugol’s solution (5 g iodide + 10 g potassium iodide in 85 mL of water) in the dark for 15 to 25 min and mounted in chloral hydrate solution for imaging (Carl Zeiss Microimaging).
GR Activity Assay
GR activity was measured essentially following the established procedure (Foyer and Halliwell, 1976; Donahue et al., 1997). Crude enzyme preparations were extracted from whole seedlings of Col-0 and miao (∼80 mg). After homogenization and centrifugation, the crude enzymes were extracted in 50 mM phosphate buffer, pH 7.0, 1 mM EDTA, 0.05% Triton, 2% polyvinylpolypyrrolidone, and 1 mM ascorbic acid. The assay mixture contained 0.1 M Tris buffer, pH 7.8, 2 mM ethylenediaminetetraacetic acid, 0.2 mM β-nicotinamide adenine dinucleotide phosphate, and 1.0 mM GSSG. The rate of NADPH oxidation was measured by the decreased in OD340 at 25°C. Total protein was quantified using a Bradford assay.
Quantification of GSH Content
The in vivo glutathione and γ-glutamylcysteine level was quantified by reverse-phase HPLC after derivatization with monobromobimane, according to established procedures (Schupp and Rennenberg, 1988; Strohm et al., 1995). Approximately 100 mg of fresh material from 3- and/or 6-d-old whole seedlings was used for the measurement.
Transient Transformation
Arabidopsis mesophyll protoplast isolation and transformation, mediated by PEG, were performed as previously described (Yoo et al., 2007). The transient transformation of Arabidopsis seedlings using Agrobacterium GV3101 was performed according to Marion et al. (2008); however, in our experiment, we used 20% PEG-4000 and 200 µM acetosyringone, instead of MgCl2 or Suc, as described in the original report, as the infiltration medium. With this minor modification, we achieved high transformation efficiencies in the hypocotyls and cotyledons. The fluorescent protein signals were recorded by CLSM.
Histochemical Analysis
Histochemical GUS assays were performed as previously described (Jefferson et al., 1987). Plant samples were fixed in cold 90% acetone for 15 min. The samples were subsequently incubated with the staining solution (100 mM phosphate buffer, pH 7.0, 10 mM EDTA, 50 mg/L 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 0.1% Triton X-100, 5 mM K3Fe[CN]6, and 0.5 mM K4Fe[CN]6) at 37°C for a period ranging from 20 min to 4 h. After staining, the samples were washed several times with 95% ethanol and were then incubated in water and mounted in chloral hydrate solution (chloral hydrate:water:glycerol = 4:3:1). The stained tissues were photographed with either an AxioVision microscope (Carl Zeiss Microimaging) for high magnification or an AxioVision stereomicroscope (Carl Zeiss Microimaging) coupled to an AxioCam MRc digital camera (Carl Zeiss Microimaging) for low magnification.
Confocal Microscopy
GFP, yellow fluorescent protein, CFP and PI fluorescence were photographed under a Zeiss LSM 510 microscope (Carl Zeiss Microimaging). For GFP/eGFP, Venus, CFP, and PI, the 488-, 514-, 458-, and 561-nm lines of the laser were used for excitation, and emission was detected at 505 to 530, 530 to 600, 465 to 520, and 590 to 635 nm, respectively. Zen2009 (Carl Zeiss Microimaging) was used to process and extract the images. For analyzing reduction oxidation-sensitive GFP, wavelengths of 405 and 488 nm were used for excitation, respectively, and emission was detected at 505 to 530 nm. The ratiometric image of 405/488 nm, which represents GSH redox status, was processed using Image J software (National Institutes of Health; http://rsb.info.nih.gov/ij) following previous procedures (Meyer et al., 2007; Gutscher et al., 2008).
In Silico Transcriptomic Analysis
The effect of BSO on the whole transcriptomic response in seedlings was analyzed using data from Koprivova et al. (2010). We used a log2 scale robust multiarray average procedure for background correction and normalization (Irizarry et al., 2003). An Empirical Bayes approach was applied using the Limma R package. A false discovery rate of q < 0.05 and a 1.5-fold change cutoff were used to select differentially regulated genes. The selected genes regulated by BSO were then compared with the differentially regulated genes from five data sets obtained by the application of IAA to the seedlings, as documented by Paponov et al. (2008). Athena (http://www.bioinformatics2.wsu.edu/cgi-bin/Athena/cgi/home.pl) (O’Connor et al., 2005) was used to analyze cis-regulatory elements in the 1000-bp upstream promoter sequences of differentially expressed genes.
Measurement of IAA Levels
Six-day-old wild-type and miao seedlings (∼60 mg FW) were used for IAA measurements. IAA extraction and measurements, using an Acquity Ultra Performance Liquid Chromatograph (Waters) and a triple quadruple tandem mass spectrometer (Quattro Premier XE; Waters), were performed according to published methods (Kojima et al., 2009; Zhou et al., 2010).
Quantitative Real-Time PCR
Gene expression was assessed by quantitative RT-PCR (qRT-PCR). Total RNA from 2-mm root tips of 6-d-old seedlings was extracted using the innuPREP Plant RNA kit (Analyticjena). The remaining DNA was digested by DNase I (Thermo Scientific) at 37°C for 30 min followed by the inactivation of DNase I with 5 mM EDTA at 65°C for 10 min. Reverse transcription was performed using 1 µg of total RNA and RevertAid H Minus Reverse Transcriptase (Thermo Scientific) according to the manufacturer’s instructions. qRT-PCR was performed with Maxima SYBR Green qPCR master mix (Thermo Scientific) on an ABI 7300 real-time PCR system (Applied Biosystems). The thermocycle program was as follows: 10 min of preincubation at 95°C followed by 35 cycles of 15 s at 95°C, 15 s at 56°C, and 30 s at 72°C. The gene-specific primers for qRT-PCR are listed in Supplemental Table 3 online. UBIQUITIN10 was used as a reference gene. The relative gene expression was analyzed using the Δ-Δ cycle threshold method. Three biological replicates and two technical replicates were used to evaluate gene expression.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: At3g24170 (GR1), At3g54660 (GR2), At4g23100 (GSH1), At4g37490 (CYCB1;1), At3g11260 (WOX5), At4g05320 (UBQ10), At1g73590 (PIN1), At5g57090 (PIN2), At1g70940 (PIN3), At2g01420 (PIN4), At1g23080 (PIN7), At3g20840 (PLT1), Atg51190 (PLT2), At4g32280 (IAA29), At5g18060 (SAUR23), At4g32540 (YUCCA1), At4g13260 (YUCCA2), At1g04610 (YUCCA3), At5g11320 (YUCCA4), At3g44310 (NIT1), At3g44300 (NIT2), and At1g70560 (TAA1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Phenotypes of miao and Complemented Plants.
Supplemental Figure 2. Root Tips of 8-d-Old Col-0 and miao.
Supplemental Figure 3. mPS-PI Staining of Root Tips of Col-0 and miao.
Supplemental Figure 4. Six-Day-Old Seedlings of GR2pro:GR2-Venus/miao and GR2pro:GR2-GUS/miao Transgenic Plants.
Supplemental Figure 5. GR2pro:GR2-GUS Is Widely Expressed in Various Tissues.
Supplemental Figure 6. Subcellular Localization of 35Spro:GR2-Venus in Transient Transformation Systems.
Supplemental Figure 7. The Expression of Pt-CB in Root Tips.
Supplemental Figure 8. Six-Day-Old GR2pro:ΔGR2-Venus/miao, GR2pro:Mt-ΔGR2-Venus/miao, and GR2pro:Pt-ΔGR2-Venus/miao Transgenic Seedlings.
Supplemental Figure 9. The Expression of GR2pro:GR2-Venus and GR2pro:Pt-ΔGR2-Venus in the Root Tips of 6-Day-Old miao.
Supplemental Figure 10. The Phenotype and Glutathione Levels in miao gr1-2 Double Mutants.
Supplemental Figure 11. Genes Upregulated by Auxin under Different Conditions and Upregulated by BSO.
Supplemental Figure 12. Genes Upregulated by Auxin under Different Conditions and Downregulated by BSO.
Supplemental Figure 13. The Relative Expression of IAA29 and SAUR23 in miao Seedlings.
Supplemental Figure 14. The Relative Expression of Auxin Biosynthesis–Related Genes in miao Seedlings.
Supplemental Figure 15. The Expression of PIN2pro:GUS, PIN4pro:GUS, and PIN7pro:GUS in Col-0 and miao.
Supplemental Figure 16. The Phenotype of the miao plt1-4 plt2-2 Mutants.
Supplemental Figure 17. 35Spro:PLT2-GLR and 35Spro:PLT2-GLR/miao RAM under GSH and BSO Treatments.
Supplemental Figure 18. GSH Treatment Failed to Rescue RAM Defects in the plt1-4 plt2-2.
Supplemental Table 1. Primers of SAP and dCAPS Markers Used for the Fine-Scale Mapping of the Mutated Gene in miao.
Supplemental Table 2. Primers Used for Vector Construction in This Study.
Supplemental Table 3. Primers Used for qRT-PCR in This Study.
Supplemental References 1. In Silico Analysis of Gene Expression and Mitochondria Targeting Signal.
Acknowledgments
We thank Andreas J. Meyer and Ben Scheres for providing seeds. We thank Peter Schopfer and other members of our team for helpful discussion about the work and critical reading of the article. We also thank Katja Rapp and Bernd Gross for excellent technical support. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 746), the Excellence Initiative of the German Federal and State Governments (EXC 294), Bundesministerium für Forschung und Technik (BMBF, SYSBRA; SYSTEC, PROBIOPA), Deutsches Zentrum für Luft und Raumfahrt (DLR 50WB1022), the Freiburg Initiative for Systems Biology, the European Union Framework 6 Program (AUTOSCREEN, LSHG-CT-2007-037897), and the National Natural Science Foundation of China (31320103910).
AUTHOR CONTRIBUTIONS
X.Y., X.L., and K.P. designed the research. X.Y. and X.L. performed the research. X.Y., T.P., X.L., I.P., K.P., and W.T. analyzed data. M.E., H. R., H.W., T.P., F.D., P.K., I.P., J.S., W.Z., and C.L. contributed reagents, materials, and analytical tools. X.Y. and K.P. wrote the article.
Glossary
- H2O2
hydrogen peroxide
- GR
GSH reductase
- NGS
NADPH-dependent GSH/glutaredoxin system
- NTS
NADPH-dependent thioredoxin system
- TAIR
The Arabidopsis Information Resource
- γ-ECS
γ-glutamylcysteine synthetase
- RAM
root apical meristem
- BSO
l-buthionine (S,R)-sulfoximine
- PAT
polar auxin transport
- Col-0
Columbia-0
- DAG
days after germination
- mPS-PI
modified pseudo-Schiff-propidium iodide
- QC
quiescent center
- DSC
distal stem cell
- CFP
cyan fluorescent protein
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
- PEG
polyethylene glycol
- MS
Murashige and Skoog
- IAA
indole-3-acetic acid
- qRT-PCR
quantitative RT-PCR
- FW
fresh weight
- DEX
dexamethasone
- Ws
Wassilewskija
- GAPDH
NADP-glyceraldehyde-3-phosphate dehydrogenase
- PI
propidium iodide
- CLSM
confocal laser scanning microscopy
References
- Aichinger E., Kornet N., Friedrich T., Laux T. (2012). Plant stem cell niches. Annu. Rev. Plant Biol. 63: 615–636 [DOI] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120 [DOI] [PubMed] [Google Scholar]
- Bashandy T., Guilleminot J., Vernoux T., Caparros-Ruiz D., Ljung K., Meyer Y., Reichheld J.P. (2010). Interplay between the NADP-linked thioredoxin and glutathione systems in Arabidopsis auxin signaling. Plant Cell 22: 376–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Scheres B. (2008). Auxin: The looping star in plant development. Annu. Rev. Plant Biol. 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Buchanan B.B., Balmer Y. (2005). Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Bui M., Liu Z. (2009). Simple allele-discriminating PCR for cost-effective and rapid genotyping and mapping. Plant Methods 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns N.G., Pasternak M., Wachter A., Cobbett C.S., Meyer A.J. (2006). Maturation of Arabidopsis seeds is dependent on glutathione biosynthesis within the embryo. Plant Physiol. 141: 446–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamitjana-Martínez E., Hofhuis H.F., Xu J., Liu C.M., Heidstra R., Scheres B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Cheng J.-C., Seeley K.A., Sung Z.R. (1995). RML1 and RML2, Arabidopsis genes required for cell proliferation at the root tip. Plant Physiol. 107: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.-H., Liu J.-Z., Liu X., Wu Q., Thompson S.M., Lin J., Chang J., Whitham S.A., Park S., Cohen J.D., Hirschi K.D. (2011). Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. J. Biol. Chem. 286: 20398–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew O., Whelan J., Millar A.H. (2003). Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J. Biol. Chem. 278: 46869–46877 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cobbett C.S., May M.J., Howden R., Rolls B. (1998). The glutathione-deficient, cadmium-sensitive mutant, cad2-1, of Arabidopsis thaliana is deficient in γ-glutamylcysteine synthetase. Plant J. 16: 73–78 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Dalle-Donne I., Rossi R., Colombo G., Giustarini D., Milzani A. (2009). Protein S-glutathionylation: A regulatory device from bacteria to humans. Trends Biochem. Sci. 34: 85–96 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682 [DOI] [PubMed] [Google Scholar]
- De Tullio M.C., Jiang K., Feldman L.J. (2010). Redox regulation of root apical meristem organization: Connecting root development to its environment. Plant Physiol. Biochem. 48: 328–336 [DOI] [PubMed] [Google Scholar]
- Dietz K.J. (2008). Redox signal integration: From stimulus to networks and genes. Physiol. Plant. 133: 459–468 [DOI] [PubMed] [Google Scholar]
- Dietz K.J., Pfannschmidt T. (2011). Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol. 155: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue J.L., Okpodu C.M., Cramer C.L., Grabau E.A., Alscher R.G. (1997). Responses of antioxidants to paraquat in pea leaves. Plant Physiol. 113: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall S.D., Brown M.T., Johnson P.J. (2004). Ancient invasions: From endosymbionts to organelles. Science 304: 253–257 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Halliwell B. (1976). Presence of glutathione and glutathione reductase in chloroplasts-proposed role in ascorbic-acid metabolism. Planta 133: 21–25 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2005). Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 17: 1866–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (2011). Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Gromes R., Hothorn M., Lenherr E.D., Rybin V., Scheffzek K., Rausch T. (2008). The redox switch of gamma-glutamylcysteine ligase via a reversible monomer-dimer transition is a mechanism unique to plants. Plant J. 54: 1063–1075 [DOI] [PubMed] [Google Scholar]
- Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P. (2008). Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 5: 553–559 [DOI] [PubMed] [Google Scholar]
- Hicks L.M., Cahoon R.E., Bonner E.R., Rivard R.S., Sheffield J., Jez J.M. (2007). Thiol-based regulation of redox-active glutamate-cysteine ligase from Arabidopsis thaliana. Plant Cell 19: 2653–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden R., Andersen C.R., Goldsbrough P.B., Cobbett C.S. (1995). A cadmium-sensitive, glutathione-deficient mutant of Arabidopsis thaliana. Plant Physiol. 107: 1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Feldman L.J. (2005). Regulation of root apical meristem development. Annu. Rev. Cell Dev. Biol. 21: 485–509 [DOI] [PubMed] [Google Scholar]
- Kataya A.R.A., Reumann S. (2010). Arabidopsis glutathione reductase 1 is dually targeted to peroxisomes and the cytosol. Plant Signal. Behav. 5: 171–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M., Kamada-Nobusada T., Komatsu H., Takei K., Kuroha T., Mizutani M., Ashikari M., Ueguchi-Tanaka M., Matsuoka M., Suzuki K., Sakakibara H. (2009). Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: An application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Mugford S.T., Kopriva S. (2010). Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Rep. 29: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Kruger N.J., von Schaewen A. (2003). The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 6: 236–246 [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Gillmor C.S., Scheible W.R. (2000). Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., et al. (2009). CDD: Specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37 (Database issue): D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion J., Bach L., Bellec Y., Meyer C., Gissot L., Faure J.-D. (2008). Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J. 56: 169–179 [DOI] [PubMed] [Google Scholar]
- Marty L., Siala W., Schwarzländer M., Fricker M.D., Wirtz M., Sweetlove L.J., Meyer Y., Meyer A.J., Reichheld J.P., Hell R. (2009). The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan S.C., et al. (2010). Plant homologs of the Plasmodium falciparum chloroquine-resistance transporter, PfCRT, are required for glutathione homeostasis and stress responses. Proc. Natl. Acad. Sci. USA 107: 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May M.J., Leaver C.J. (1994). Arabidopsis thaliana gamma-glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proc. Natl. Acad. Sci. USA 91: 10059–10063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men S., Boutté Y., Ikeda Y., Li X., Palme K., Stierhof Y.-D., Hartmann M.-A., Moritz T., Grebe M. (2008). Sterol-dependent endocytosis mediates post-cytokinetic acquisition of PIN2 auxin efflux carrier polarity. Nat. Cell Biol. 10: 237–244 [DOI] [PubMed] [Google Scholar]
- Meyer A.J., Brach T., Marty L., Kreye S., Rouhier N., Jacquot J.P., Hell R. (2007). Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J. 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Meyer A.J., May M.J., Fricker M. (2001). Quantitative in vivo measurement of glutathione in Arabidopsis cells. Plant J. 27: 67–78 [DOI] [PubMed] [Google Scholar]
- Mhamdi A., Hager J., Chaouch S., Queval G., Han Y., Taconnat L., Saindrenan P., Gouia H., Issakidis-Bourguet E., Renou J.P., Noctor G. (2010). Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 153: 1144–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L., Zaffagnini M., Marchand C., Collin V., Decottignies P., Tsan P., Lancelin J.M., Trost P., Miginiac-Maslow M., Noctor G., Lemaire S.D. (2005). Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proc. Natl. Acad. Sci. USA 102: 16478–16483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Cascales-Miñana B., Mulet J.M., Baroja-Fernández E., Pozueta-Romero J., Kuhn J.M., Segura J., Ros R. (2009). Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 151: 541–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff M.M., Neff J.D., Chory J., Pepper A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Noctor G., Foyer C.H. (1998). Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 249–279 [DOI] [PubMed] [Google Scholar]
- Noctor G., Mhamdi A., Chaouch S., Han Y., Neukermans J., Marquez-Garcia B., Queval G., Foyer C.H. (2012). Glutathione in plants: An integrated overview. Plant Cell Environ. 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Noctor G., Queval G., Mhamdi A., Chaouch S., Foyer C.H. (2011). Glutathione. The Arabidopsis Book 9: e0142, /10.1199/tab.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A., Jung H.S., Koussevitzky S., Chory J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57: 739–759 [DOI] [PubMed] [Google Scholar]
- O’Connor T.R., Dyreson C., Wyrick J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Paponov I.A., Paponov M., Teale W., Menges M., Chakrabortee S., Murray J.A.H., Palme K. (2008). Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol. Plant 1: 321–337 [DOI] [PubMed] [Google Scholar]
- Perilli S., Di Mambro R., Sabatini S. (2012). Growth and development of the root apical meristem. Curr. Opin. Plant Biol. 15: 17–23 [DOI] [PubMed] [Google Scholar]
- Petricka J.J., Winter C.M., Benfey P.N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potters G., Jansen M.A.K., Horemans N., Guisez Y., Pasternak T. (2010). Dehydroascorbate and glutathione regulate the cellular development of Nicotiana tabacum L. SR-1 protoplasts. In Vitro Cell. Dev. Biol. Plant 46: 289–297 [Google Scholar]
- Queval G., Jaillard D., Zechmann B., Noctor G. (2011). Increased intracellular H₂O₂ availability preferentially drives glutathione accumulation in vacuoles and chloroplasts. Plant Cell Environ. 34: 21–32 [DOI] [PubMed] [Google Scholar]
- Reichheld J.-P., Khafif M., Riondet C., Droux M., Bonnard G., Meyer Y. (2007). Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19: 1851–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichheld J.P., Meyer E., Khafif M., Bonnard G., Meyer Y. (2005). AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 579: 337–342 [DOI] [PubMed] [Google Scholar]
- Rodermel S. (2001). Pathways of plastid-to-nucleus signaling. Trends Plant Sci. 6: 471–478 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Lemaire S.D., Jacquot J.-P. (2008). The role of glutathione in photosynthetic organisms: Emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59: 143–166 [DOI] [PubMed] [Google Scholar]
- Rouhier N., Villarejo A., Srivastava M., Gelhaye E., Keech O., Droux M., Finkemeier I., Samuelsson G., Dietz K.J., Jacquot J.P., Wingsle G. (2005). Identification of plant glutaredoxin targets. Antioxid. Redox Signal. 7: 919–929 [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis D., Wolkenfelt H., Murfett J., Guilfoyle T., Malamy J., Benfey P., Leyser O., Bechtold N., Weisbeek P., Scheres B. (1999). An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463–472 [DOI] [PubMed] [Google Scholar]
- Sánchez-Fernández R., Fricker M., Corben L.B., White N.S., Sheard N., Leaver C.J., Van Montagu M, Inzé D., May M.J. (1997). Cell proliferation and hair tip growth in the Arabidopsis root are under mechanistically different forms of redox control. Proc. Natl. Acad. Sci. USA 94: 2745–2750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer F.Q., Buettner G.R. (2001). Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 30: 1191–1212 [DOI] [PubMed] [Google Scholar]
- Schupp R., Rennenberg H. (1988). Diurnal changes in the glutathione content of spruce needles (Picea abies L.). Plant Sci. 57: 113–117 [Google Scholar]
- Small I., Peeters N., Legeai F., Lurin C. (2004). Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4: 1581–1590 [DOI] [PubMed] [Google Scholar]
- Strohm M., Jouanin L., Kunert K.J., Pruvost C., Polle A., Foyer C.H., Rennenberg H. (1995). Regulation of glutathione synthesis in leaves of transgenic poplar (Populus tremula X P. alba) overexpressing glutathione synthetase. Plant J. 7: 141–145 [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I., Pena-Muralla R., Dickerman A., Berg M., Rogers R., Hutchens S., Sweeney T.C., McElver J., Aux G., Patton D., Meinke D. (2004). Identification of genes required for embryo development in Arabidopsis. Plant Physiol. 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T., Hagen G., Guilfoyle T.J. (1999). Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Wilson R.C., Seeley K.A., Reichheld J.P., Muroy S., Brown S., Maughan S.C., Cobbett C.S., Van Montagu M., Inzé D., May M.J., Sung Z.R. (2000). The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos P.D., Dong Y., Ziegler K., Markovic J., Pallardó F.V., Pellny T.K., Verrier P.J., Foyer C.H. (2010). Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 64: 825–838 [DOI] [PubMed] [Google Scholar]
- Wachter A., Wolf S., Steininger H., Bogs J., Rausch T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: Implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41: 15–30 [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zaffagnini M., Bedhomme M., Groni H., Marchand C.H., Puppo C., Gontero B., Cassier-Chauvat C., Decottignies P., Lemaire S.D. (2012). Glutathionylation in the photosynthetic model organism Chlamydomonas reinhardtii: A proteomic survey. Mol. Cell. Proteomics 11: 014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M., Michelet L., Marchand C., Sparla F., Decottignies P., Le Maréchal P., Miginiac-Maslow M., Noctor G., Trost P., Lemaire S.D. (2007). The thioredoxin-independent isoform of chloroplastic glyceraldehyde-3-phosphate dehydrogenase is selectively regulated by glutathionylation. FEBS J. 274: 212–226 [DOI] [PubMed] [Google Scholar]
- Zhou W., Wei L., Xu J., Zhai Q., Jiang H., Chen R., Chen Q., Sun J., Chu J., Zhu L., Liu C.-M., Li C. (2010). Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]