This work characterizes transgenic lines with altered expression of calmodulin (CaM) N-methyltransferase to demonstrate that the methylation status of CaM plays a role in CaM-mediated signaling pathways. The findings add a new level of complexity to our understanding of CaM signaling mechanisms in plants.
Abstract
Calmodulin N-methyltransferase (CaM KMT) is an evolutionarily conserved enzyme in eukaryotes that transfers three methyl groups to a highly conserved lysyl residue at position 115 in calmodulin (CaM). We sought to elucidate whether the methylation status of CaM plays a role in CaM-mediated signaling pathways by gene expression analyses of CaM KMT and phenotypic characterization of Arabidopsis thaliana lines wherein CaM KMT was overexpressed (OX), partially silenced, or knocked out. CaM KMT was expressed in discreet spatial and tissue-specific patterns, most notably in root tips, floral buds, stamens, apical meristems, and germinating seeds. Analysis of transgenic plants with genetic dysfunction in CaM KMT revealed a link between the methylation status of CaM and root length. Plants with suppressed CaM methylation had longer roots and CaM KMT OX lines had shorter roots than wild type (Columbia-0). CaM KMT was also found to influence the root radial developmental program. Protein microarray analyses revealed a number of proteins with specificity for methylated forms of CaM, providing candidate functional intermediates between the observed phenotypes and the target pathways. This work demonstrates that the functionality of the large CaM family in plants is fine-tuned by an overarching methylation mechanism.
INTRODUCTION
Calmodulin (CaM) is a small (148-residue), highly conserved, ubiquitous, calcium (Ca2+) binding protein (Klee and Vanaman, 1982; Chin and Means, 2000; Yamniuk and Vogel, 2004). As the central transducer of Ca2+ signaling, CaM binds to proteins involved in the regulation of an array of cellular processes, including gene transcription, muscle contraction, cell survival, and neurotransmitter disease (Klee and Vanaman, 1982; Chin and Means, 2000; Yamniuk and Vogel, 2004). In most organisms, CaM is posttranslationally modified by trimethylation of Lys-115, but the functional significance of this modification remains largely unknown. Of ∼300 known protein interactors with CaM, only four from a limited number of species have been examined for the effects of Lys-115 methylation on binding or activity. Methylation of CaM decreases activation of plant NAD kinase (NADK; Roberts et al., 1986), and may decrease the affinity of CaM for cyclic nucleotide phosphodiesterase (Marshak et al., 1984), but it has no effect on plant Glu carboxylase (Oh and Yun, 1999) or myosin light-chain kinase activity (Roberts et al., 1984). A recent study demonstrated that CaM methylation affects the conformational dynamics of CaM upon binding of Ca2+, as well as the thermal stability of the apoprotein form of CaM (Magnani et al., 2012).
Earlier reports suggest CaM activity could be regulated via methylation because the methylation state of CaM was observed to vary in a tissue-specific and developmentally specific pattern in Pisum sativum (pea) roots (Oh and Roberts, 1990) and according to the growth phase (logarithmic versus stationary) of Daucus carota (carrot) cells in suspension culture (Oh et al., 1992). Several studies have attempted to elucidate the role of CaM methylation in vivo by expression of genetically altered forms of CaM where Lys-115 was replaced with an unmethylatable Arg residue. In tobacco (Nicotiana tabacum) plants, constitutive overexpression of a CaM Lys-115-Arg mutant resulted in plants with reduced growth (short internodes), seed production, and pollen viability (Roberts et al., 1992). Later studies confirmed that these plants also had increased NADPH levels from hyperactivated CaM-dependent NADK (see above) and associated increases in reactive oxygen species (Harding et al., 1997). However, a similar study in Arabidopsis thaliana found that the lack of trimethylation of CaM had no effect on its repression of cold-regulated gene (COR) expression (Townley and Knight, 2002). In addition, Gallus gallus domesticus (chicken) cell lines expressing a CaM Lys-115-Arg mutant protein do not show any alterations in growth (Panina et al., 2012). A relatively rare gene deletion syndrome in Homo sapiens (humans) includes partial deletion of the gene that codes for the enzyme responsible for CaM methylation (Parvari et al., 2001, 2005; Parvari and Hershkovitz, 2007; Chabrol et al., 2008; Magnani et al., 2010). Lymphoblastoid cells from patients with this deletion syndrome have hypomethylated forms of CaM, and comparative phenotypic analyses of these individuals revealed several disorders including mild-to-moderate mental retardation, cytochrome c oxidase deficiency, and muscle weakness (Magnani et al., 2010; Magen et al., 2012). Collectively, the existing studies on the possible significance of CaM methylation suggest that there may be specific developmental events or tissues, or both, wherein methylation plays an important role, but there are certainly instances where CaM methylation is not a factor in regulating CaM activity. However, in these previous studies, the expression profile or gene sequence of CaM was altered along with the genetic perturbation of its methylation state, and overexpression of genetically altered forms of CaM may not necessarily reveal the role of methylation. A main obstacle to studies focusing on the methylation of CaM has been the lack of identification of genes responsible for the methylation activity. With the discovery of CaM N-methyltransferase (CaM KMT,previously known as C2orf34 in humans) and its identification as a highly conserved single-copy gene in all eukaryotic species (Magnani et al., 2010), we selected Arabidopsis as a model organism in which to explore the role of CaM methylation at a whole-organism level.
In this study, we elucidate the role of CaM KMT in CaM-mediated signaling pathways, and we characterize the CaM KMT promoter, which displays spatial and temporal regulation. CaM KMT is expressed at early stages in development, in some specialized plant organs, and appears to be involved in plant development and hormone as well as stress signaling pathways. This work also provides a global analysis of Arabidopsis proteins that recognize the methylation state of CaM.
RESULTS
Tissue-Specific Expression of Arabidopsis CaM KMT
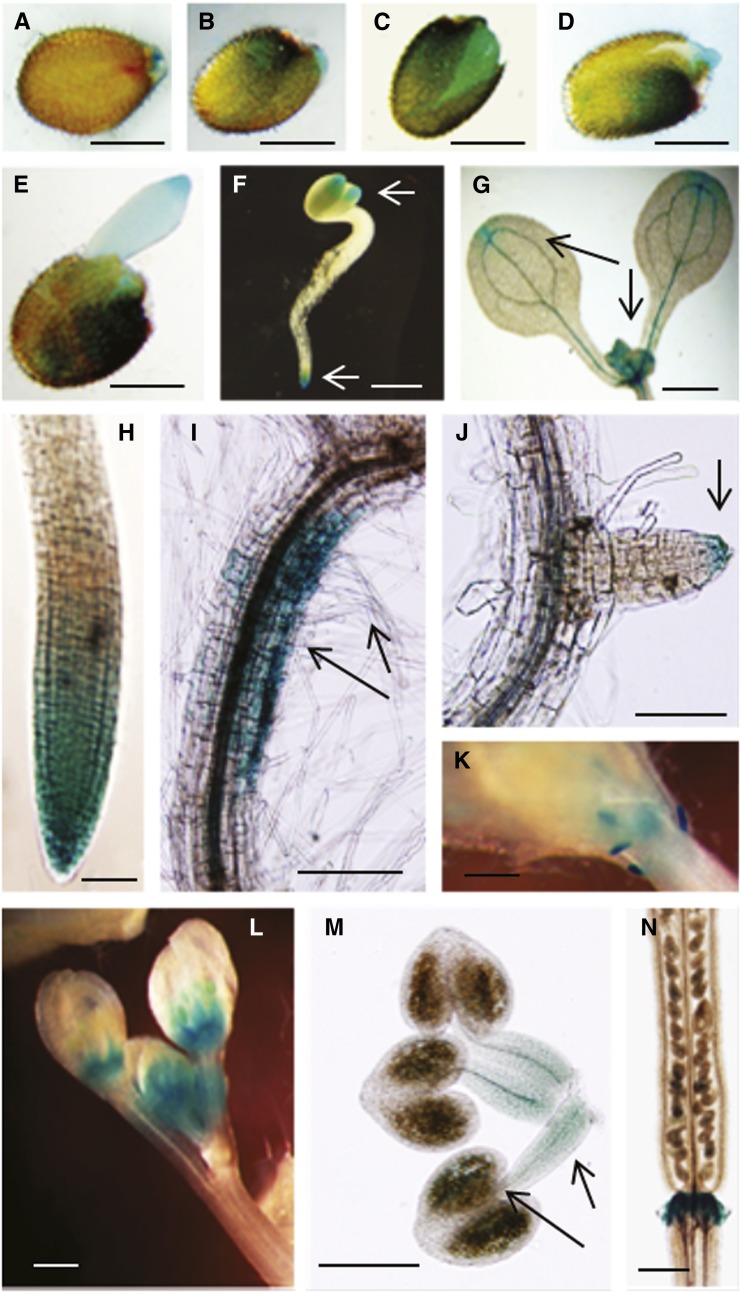
To aid in elucidating the functional role of Arabidopsis CaM KMT, we analyzed the expression of the endogenous CaM KMT gene, as well as the activity of its promoter fused with the reporter gene β-glucuronidase (GUS). Transcript abundance for endogenous CaM KMT in seedlings grown on Arabidopsis growth medium (AGM) was maximal at the cotyledonary leaf stage and then decreased up to the eight-leaf stage as determined by quantitative real-time PCR (qRT-PCR; see Supplemental Figure 1 online). For promoter expression analysis, several Arabidopsis transgenic lines were generated with the CaM KMTPro:GUS construct (see Supplemental Figure 2 online). The T2 generation CaM KMTPro:GUS plants showed differential GUS expression in vegetative and reproductive tissues (Figure 1). GUS expression varied with time after imbibition of the seed (Figures 1A to 1E). After stratification, GUS expression was observed in the micropylar end of the seed (Figure 1A), and 1 d after stratification, strong GUS expression appeared in the endosperm region and in the testa (Figures 1B to 1D). Two days after stratification, significant GUS expression was observed in the endosperm and emerging radicle (Figure 1E). In young seedlings, GUS expression was found in the root and cotyledon tips (Figure 1F). At a later stage of growth, faint expression was still observed in the tip of the cotyledons, but strong expression was seen in the shoot apical meristematic region (Figure 1G). Older seedlings of CaM KMTPro:GUS plants showed GUS expression at the primary root tip, at the basal side of root curvature, and at the lateral root tip (Figures 1H to 1J). In addition, GUS expression driven by the CaM KMT promoter was found in very young primordia, floral buds, and stamens (Figures 1K to 1M). In stamens, maximum expression was observed in the anther containing immature pollen, although it was also noted in the filament (Figure 1M). In siliques, there was faint GUS expression in developing seeds and strong expression in the abscission zone (Figure 1N). The CaM KMTPro:GUS data largely corroborated microarray expression data retrieved from the Genevestigator expression database (see Supplemental Figure 3 online). Interestingly, these results suggest that the CaM KMT promoter is active in tissues where auxin plays a major role in signaling during plant growth and development.
Figure 1.
Histochemical GUS Staining Showing Spatial and Tissue-Specific Expression Driven by the Arabidopsis CaM KMT Promoter.
(A) to (E) GUS expression in Arabidopsis (T2 generation seed harboring CaM KMTPro:GUS construct) seed at various times after imbibition after stratification. After surface sterilization, seed was stratified at 4°C for 2 d. GUS staining of seed immediately after stratification (A), 1 d after stratification (B) to (D), and 2 d after stratification (E).
(F) GUS expression in the cotyledon and young root tip as indicated by arrows.
(G) GUS expression in apical meristem and cotyledon tip as indicated by arrows.
(H) GUS expression in mature root tip.
(I) GUS expression in downward side of root curvature and in root hairs as indicated by arrows.
(J) GUS expression at the tip of young lateral root as indicated by arrow.
(K) and (L) GUS expression in young leaf primordia (K) and in floral buds (L).
(M) GUS expression in stamens with filaments and anther containing pollen grain sacks as indicated by arrows.
(N) GUS expression in the abscission zone at the base of the silique. Bar in (A) to (F) = 0.5 mm; bar in (G), (K), (L), and (N) = 1 mm; bar in (H) and (J) = 100 µm; bar in (I) and (M) = 200 µm.
Expression Profiling and Phenotypic Study of the CaM KMT Knock Out, Partially Silenced, and Overexpressed Lines
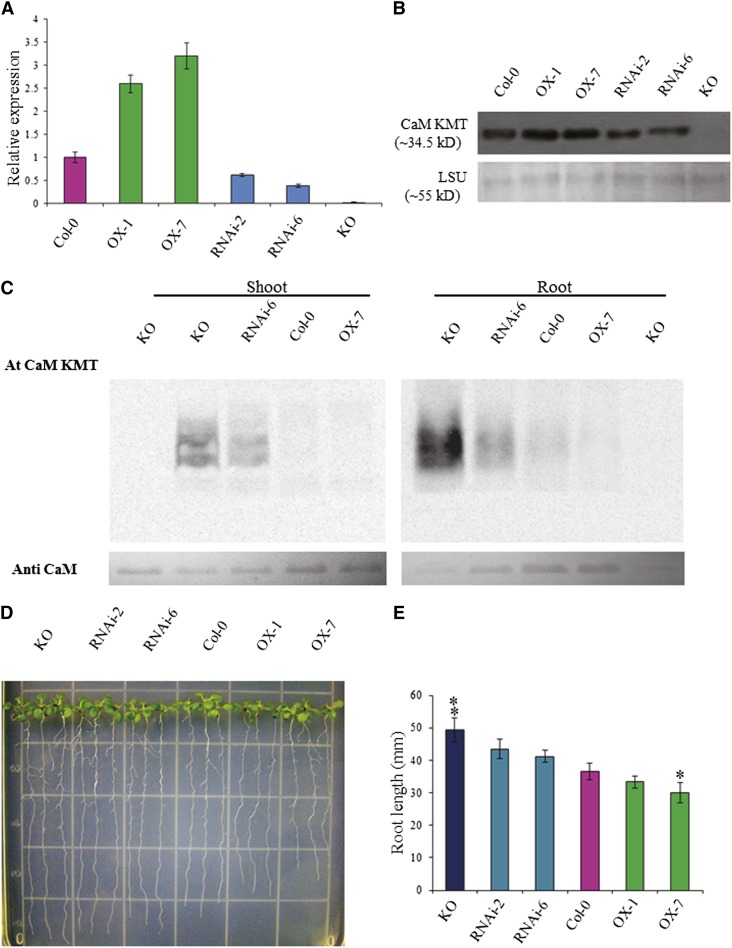
To evaluate the effect of CaM methylation in plants, we generated seven independent CaM KMT–overexpressed (OX) transgenic lines and seven lines with downregulated expression (RNAi) in the Columbia-0 (Col-0) background of Arabidopsis (see Supplemental Figures 4A and 4B online). An Arabidopsis T-DNA insertion line for CaM KMT (Alonso et al., 2003), designated as Knock Out (KO), was also characterized (see Supplemental Figure 5 online). After initial screening using qRT-PCR of the T2 generation lines, we selected two independent representative lines for further analysis, along with the Arabidopsis CaM KMT KO and wild type (Col-0) plants. Transcript analysis by qRT-PCR demonstrated that the relative expression of the CaM KMT transcript was between 2.5 and 3 times higher in the OX lines OX-1 and OX-7, respectively, than in Col-0 (Figure 2A). The two RNAi lines, RNAi-2 and RNAi-6, showed 60 and 40%, respectively, of the CaM KMT transcript of Col-0, whereas negligible expression was detected in the KO line compared with Col-0 (Figure 2A). Immunoblot analyses revealed that CaM KMT protein accumulation in the different lines was similar to their respective transcript expression profiles (Figure 2B). CaM was hypomethylated in the KO and RNAi-6 lines, with the maximal hypomethylation in KO (Figure 2C). Neither Col-0 nor the OX-7 line exhibited hypomethylated CaM (Figure 2C). Notably, root samples showed more hypomethylated CaM compared with shoot samples. Because of this difference in the methylation state of CaM between shoot and root tissues of Arabidopsis, we analyzed the root morphology and root growth of the various lines. Four-day-old seedlings were grown vertically for an additional 6 d in AGM. The roots of the KO line were significantly longer than those of Col-0, whereas OX-7 showed reduced root growth compared with Col-0 (Figures 2D and 2E).
Figure 2.
Molecular and Phenotypic Analysis of T2 Generation CaM KMT OX, RNAi, and KO Mutant Lines.
(A) CaM KMT transcript in seedlings of Col-0, OX-1, OX-7, RNAi-2, RNAi-6, and KO lines was detected by qRT-PCR. Data are the mean ± sd of three independent experiments with different biological samples, with four replicates for each sample in every experiment.
(B) CaM KMT protein accumulation in seedlings of Col-0, OX-1, OX-7, RNAi-2, RNAi-6, and KO lines as detected by immunoblotting with a CaM KMT-specific antibody. The bottom panel shows the accumulation of the Rubisco LSU as a loading control.
(C) Methylation state of calmodulin in Col-0, OX-7, RNAi-6, and KO lines as detected by incubation of 20 µg total plant protein from shoots and roots of 4-week-old seedlings with (+) or without (−) Escherichia coli–expressed Arabidopsis CaM KMT protein in the presence of [3H-methyl]-S-Adenosyl methionine. Samples were subjected to SDS-PAGE, and after transfer to a polyvinylidene difluoride membrane, radiolabeled methyl group incorporation was detected using a phosphor screen.
(D) Phenotypic analysis of root growth of seedlings of Col-0, OX-1, OX-7, RNAi-2, RNAi-6, and KO lines. Four-day-old seedlings were placed on AGM and allowed to grow vertically for an additional 6 d and then photographed. Three representative seedlings are shown for each line.
(E) The root length of seedlings of the Col-0, OX-1, OX-7, RNAi-2, RNAi-6, and KO lines were measured at 10 d of growth after stratification. After 4 d of growth, the seedlings were transferred and grown in AGM plates vertically for another 6 d. Data presented are the mean of at least 50 individual seedlings of each line ± sd. Asterisks and double asterisks indicate the significant deviation from Col-0 line at P < 0.005 and P < 0.001, respectively, using Student’s t test for pairwise comparison.
Phenotypic Analyses of Roots in Lines Expressing Different Levels of CaM KMT
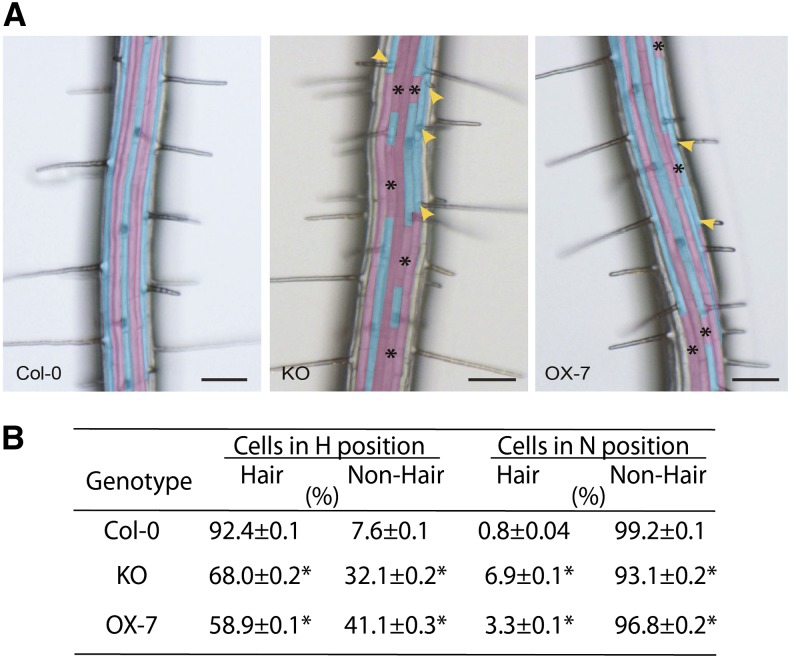
Because expression of CaM KMTPro:GUS was observed in the meristematic zone of growing roots, and the OX, RNAi, and KO CaM KMT lines showed differences in root length, we sought to elucidate whether the OX or KO lines of CaM KMT possess any further difference in root morphology. Examination of the root epidermal layer of KO versus Col-0 (wild-type), as well as OX (OX-7) versus Col-0, revealed ectopic root hair (H) cells in atrichoblast cell files (Figure 3A) and the presence of nonhair (N)cells in trichoblast cell files (Figure 3A). Quantification of the frequency of H and N cells in the epidermal layer suggested a minor scrambling of cell fate determination (Figure 3B). Furthermore, the KO (34 ± 0.2 se, n = 1352, P > 0.05, Mann–Whitney U test) and OX-7 (38 ± 0.2 se, n = 1506, P < 0.05, Mann–Whitney U test) lines had comparably more epidermal cells per unit cell area than Col-0 (28 ± 0.2 se, n = 1100). To explore this further, we used laser scanning confocal microscopy to examine propidium iodide–stained longitudinal optical sections of the root meristematic zone in 5-d-old vertically grown seedlings. Visual examination of these samples was consistent with a minor increase in the number of epidermal cells in KO compared with the OX and Col-0 (see Supplemental Figure 6A online). In agarose-embedded hand sections from the root elongation zone (∼1–2 mm from the root cap) stained with Calcofluor white (see Supplemental Figure 6B online), an increased number of epidermal cells in KO and OX-7 roots was observed. Despite the potential for ontogenic variance arising from hand-cut sections and the potential to section longitudinally on trichoblast versus atrichoblast cell files (which display unequal size), the results were consistent with some scrambling of N and H cell identity and a difference in the number of cells. Although there was a modest increase in the number of epidermal cells in KO and OX-7 lines as compared with Col-0, an overall decrease in the number of root hairs per unit area was observed for KO and OX-7 lines (see Supplemental Figure 7B online). This is consistent with the decrease in H cells and increase in N cells in trichoblast cell files of KO and OX-7 lines (Figure 3B).
Figure 3.
Genetic Alteration of CaM KMT Expression Alters H versus N Epidermal Cell Fate determination in Arabidopsis roots.
(A) Single-frame micrographs of roots of wild-type (Col-0), T-DNA insertion KO into CaM KMT mutant (KO), and CaM KMT overexpression line (OX-7) with false color cyan showing H cells and magenta showing N cells. Ectopic N cells (asterisks) form in an otherwise H cell file and ectopic H cells (yellow arrowheads) form in an otherwise N cell file in KO and OX-7 lines. Bar = 100 μm.
(B) Statistical assessment demonstrates a decrease in H cells in KO and OX-7 lines when compared with Col-0. An increase in ectopic N cells is observed in KO and OX-7 lines (n = 40 seedlings for each genotype). After vernalization and germination, individual Arabidopsis seedlings were grown on sterile one-half-strength MS-agar medium vertically for 5 days in light (24 h). Asterisks represent significant difference from the mean at P < 0.05 using the Mann–Whitney U test.
Visual inspection of the root hair length distributions revealed a significant decrease in root hair lengths for KO when compared with OX and Col-0 lines (see Supplemental Figures 7A and 7C online).
CaM KMT-Mediated Methylation of CaM Is Involved in Auxin Signaling
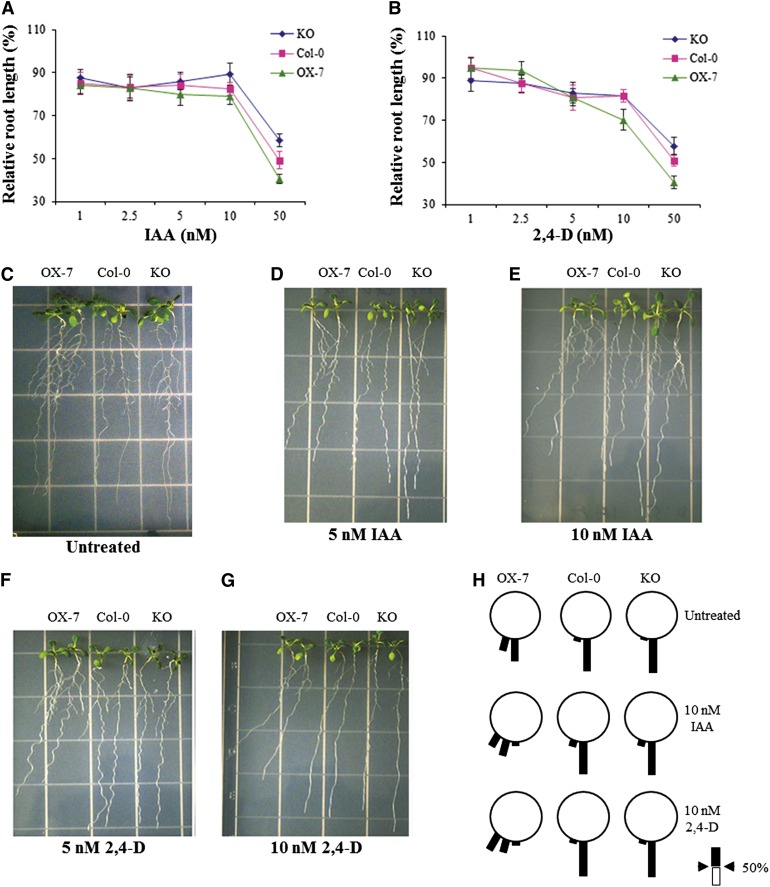
GUS expression driven by the CaM KMT promoter was observed mostly in those tissues where auxin plays a prominent role in growth and development. In addition, different CaM KMT OX or KO lines showed significant morphological differences in their root anatomy, root growth, and root hair distribution. The CaM KMT KO and OX-7 lines together with Col-0 plants were grown on medium containing different concentrations of indole-3-acetic acid (IAA) and 2,4-dichlorophenoxyacetic acid (2,4-D), to examine whether methylation of CaM is a key component of auxin signaling. In auxin-containing medium, the KO plants maintained root growth to a greater extent compared with the other lines. The CaM KMT KO seedlings were significantly less inhibited, as measured by relative root length, by 10 and 50 nM IAA (P < 0.005 and P < 0.001, respectively) as compared with the OX-7 seedlings (Figure 4A). Similarly, the KO seedlings responded less to those same concentrations of 2,4-D as measured by relative root length (P < 0.001) compared with the OX-7 seedlings (Figure 4B). In addition, in the absence of hormones, the root growth of all seedlings was gravitropic, but after 7 days of growth in 5 and 10 nM IAA, the OX-7 lines showed agravitropic root growth in a dose-dependent manner (Figures 4C to 4E). Similarly, at the same concentrations of 2,4-D, the roots of the OX-7 seedlings were more agravitropic compared with those of the Col-0 and KO seedlings (Figures 4F and 4G). Analysis of the percentage of roots among 12 classes designated by 15° increments from vertical showed that the CaM KMT OX-7 seedlings showed maximal agravitropic root curvature compared with the Col-0 and CaM KMT KO seedlings when grown in the presence of 10 nM 2,4-D, and that the root angle deviation from vertical in the OX-7 seedlings was more evident when treated with 2,4-D than with IAA (Figures 4H). In contrast, neither the Col-0 nor the CaM KMT KO seedlings deviated significantly from vertical root growth in different auxin media.
Figure 4.
Effect of Auxins on Root Growth of Col-0, CaM KMT KO mutant, and CaM KMT Overexpression Seedlings.
(A) and (B) Root growth assay in the presence of auxins (IAA and 2,4-D). Four-day-old germinated seedlings of Col-0 (pink squares), CaM KMT KO mutant (blue diamond), and OX-7 (green triangles) were transferred to medium containing the indicated concentrations of IAA (A) and 2,4-D (B), and grown vertically for additional 7 d in a chamber (16-h-light/8-h-dark cycle at 22°C). Values are presented as percentage of root length of hormone-treated seedlings relative to that of untreated seedlings for each line. Each data point represents the mean ± sd of 12 to 15 seedlings.
(C) to (G) Root growth phenotype under IAA and 2,4-D treatment. Four-day-old seedlings of the different lines were grown vertically in AGM without (untreated) or with indicated concentrations of IAA and 2,4-D for an additional 7 d. Two representative seedlings are shown for each line.
(H) The percentage of roots within each class of root angle for Col-0, OX-7, and KO seedlings in the absence (untreated) or presence of 10 nM IAA and 10 nM 2,4-D. The angles were grouped into 12 classes (each class is 15° from vertical in either direction) and expressed as percentage of roots with that class of root angle in a wheel diagram. Scale indicates 50% root growth in the given class.
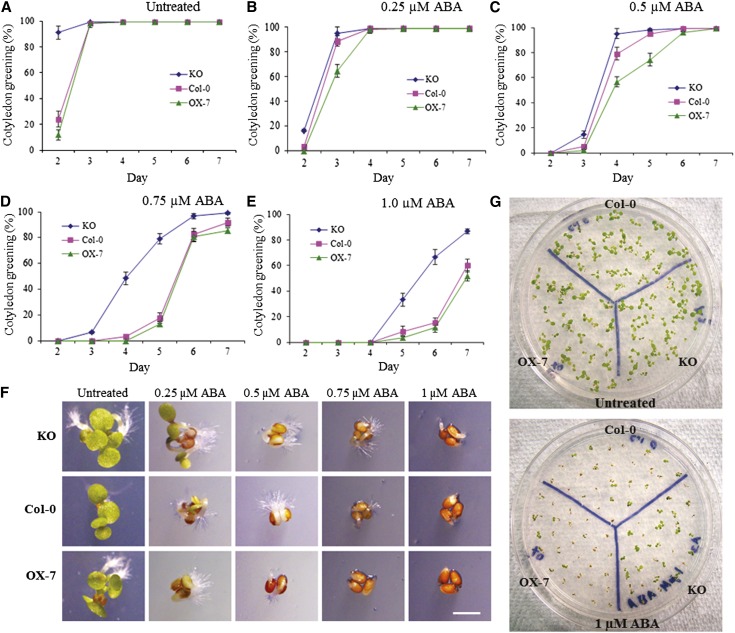
Effect of Abscisic Acid on Germination, Cotyledon Greening, and Growth in Different CaM KMT Lines
Abscisic acid (ABA) is a stress hormone, and during environmental stress conditions it can play a crucial role in different signaling pathways. To investigate the role of CaM methylation status in ABA signaling during stress, the CaM KMT KO and OX-7 seedlings were germinated on AGM supplemented with different concentrations of ABA (0.25, 0.5, 0.75, and 1 µM). No significant differences were observed among the seeds from the different lines during germination, but even without ABA treatment, the significantly more KO seedlings had green cotyledons at the first point of observation compared with either Col-0 or the OX-7 (Figure 5A). At 0.25 µM ABA, no large differences in the number of seedlings with green cotyledons were observed among the different lines, but as ABA concentrations were increased, the OX-7 and Col-0 seedlings turned green to a lesser extent and took longer to do so compared with the KO line (Figures 5B to 5E). Three days after stratification, KO seedlings clearly grew faster under different ABA stress conditions compared with both Col-0 and OX-7 seedlings (Figure 5F). After 7 days under the greatest ABA (1 µM) stress, more of the germinated seedlings of the KO lines proceeded with development in comparison with the other two lines (Figure 5G).
Figure 5.
The CaM KMT KO Mutant Shows Increased Resistance to ABA Stress during Early Seedling Growth.
(A) to (E) Cotyledon greening percentage of Col-0 (squares), CaM KMT KO mutant, and OX-7 seedlings under indicated concentrations of ABA. Seeds were placed on AGM without (A) and with 0.25 (B), 0.5 (C), 0.75 (D), and 1 µM (E) ABA. After stratification at 4°C for 2 d, the plates were transferred to a growth chamber (16-h-light/8-h-dark cycle at 22°C), and cotyledon greening was measured. For the phenotypic measurement, the seedlings having green cotyledons were scored each day from days 2 through 7 after stratification. All assays were repeated three times using ∼50 seeds for each experiment. Data represent mean ± sd (n = 3).
(F) Phenotypic differences observed among Col-0, KO, and OX-7 lines grown without (untreated) or with the indicated concentrations of ABA. The seedlings were digitally imaged under a microscope 3 d after stratification. Bar = 1 mm.
(G) Plant development of seedlings of Col-0, KO, and OX-7 lines under ABA stress. After stratification, seeds were grown in AGM without (untreated) or with 1 µM ABA, grown for additional 7 d, and then photographed.
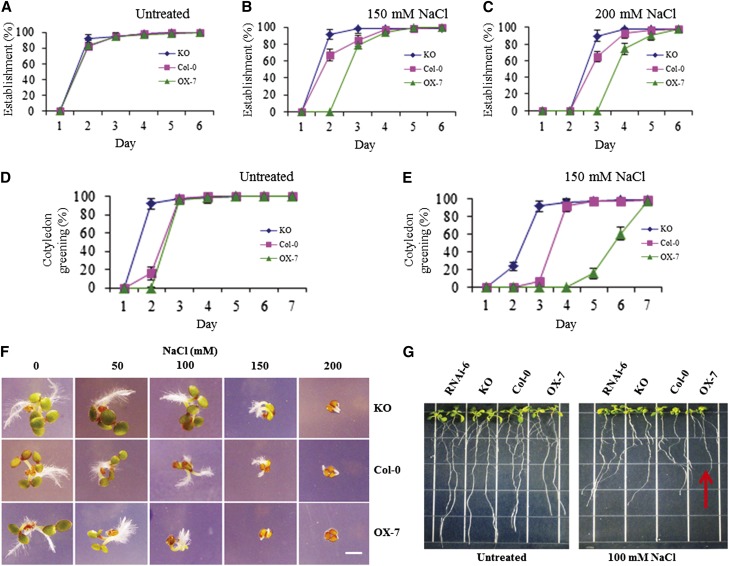
Effect of Salt Stress on Germination and Early Seedling Growth
The ABA stress signaling cascade can occur after or during salt stress (Zhu, 2002). To determine whether methylation of CaM is involved in a salt stress signaling network, we analyzed the CaM KMT KO and OX-7 lines for their response to NaCl stress during early seedling growth. As shown in Figure 6, the germination of OX-7 seeds in different concentrations of salt was delayed compared with that of Col-0 seeds, which was also delayed relative to the KO seeds, although all seeds reached 100% germination by day 6 regardless of salt concentration. At 150 mM NaCl, the cotyledon greening of the OX-7 seedlings started on day 5 compared with days 3 and 2 for the Col-0 and KO seedlings, respectively (Figure 6E). Progression of photomorphogenesis under these conditions demonstrated that the KO and OX-7 seedlings were less and more sensitive, respectively, to NaCl stress compared with the Col-0 seedlings (Figure 6F). Even at later growth stages, the 4-week-old CaM KMT OX-7 plants were more sensitive to salt stress (300 mM for 3 d), whereas the effects on Col-0 and KO were essentially indistinguishable (see Supplemental Figure 8 online). To elucidate the effect of NaCl stress on root growth, 4-d-old CaM KMT RNAi, KO, and OX-7 seedlings were placed on medium containing 100 mM NaCl for an additional 6 days. The OX-7 seedlings were stunted compared with Col-0 seedlings, and the growth inhibition was comparatively less in both the KO and RNAi seedlings (Figure 6G).
Figure 6.
The CaM KMT KO Mutant Shows Increased Resistance to Salt Stress during Germination and Early Seedling Growth.
(A) to (C) Germination of seeds of the mutant KO lines under NaCl stress conditions occurred earlier than for Col-0 and CaM KMT OX-7 lines. Seeds were placed on AGM without (A) or with 150 (B) and 200 mM (C) NaCl, and germination was measured after the indicated period. After stratification at 4°C for 2 d, the plates were transferred to a growth chamber (16-h-light/8-h-dark cycle at 22°C). For the phenotypic measurement of germination, seeds with a protruding radicle were scored daily from days 1 through 6 after stratification. All assays were repeated three times using ∼50 seeds for each experiment. Values are mean ± sd (n = 3).
(D) and (E) Cotyledon greening under NaCl stress of the mutant KO and OX-7 seedlings occurred sooner and later, respectively, than that of Col-0. Col-0, KO, and OX-7 seeds were placed on AGM without (D) or with 150 mM NaCl (E), and cotyledon greening was measured after the indicated period. After stratification at 4°C for 2 d, the plates were transferred to a growth chamber (16-h-light/8-h-dark cycle at 22°C) and subsequently analyzed. The seedlings with green cotyledons were scored daily from days 1 through 7 after stratification. All assays were repeated three times using ∼50 seeds for each experiment. Values are mean ± sd (n = 3).
(F) Phenotypes of seedlings of the Col-0, KO, and OX-7 lines grown without (0) or with the indicated concentrations of NaCl. Photos were taken 3 d after stratification. Bar = 1 mm.
(G) Effect of salt stress on root growth of seedlings of the different lines. After stratification, the Col-0, KO, RNAi-6, and OX-7 seeds were sown in AGM for 4 d, and subsequently two seedlings of each type were grown vertically in AGM without or with 100 mM NaCl for an additional 7 d before being photographed. OX-7 lines showed stunted growth and reduced lateral root formation (indicated by red arrow). The experiment was repeated at least three times with similar results.
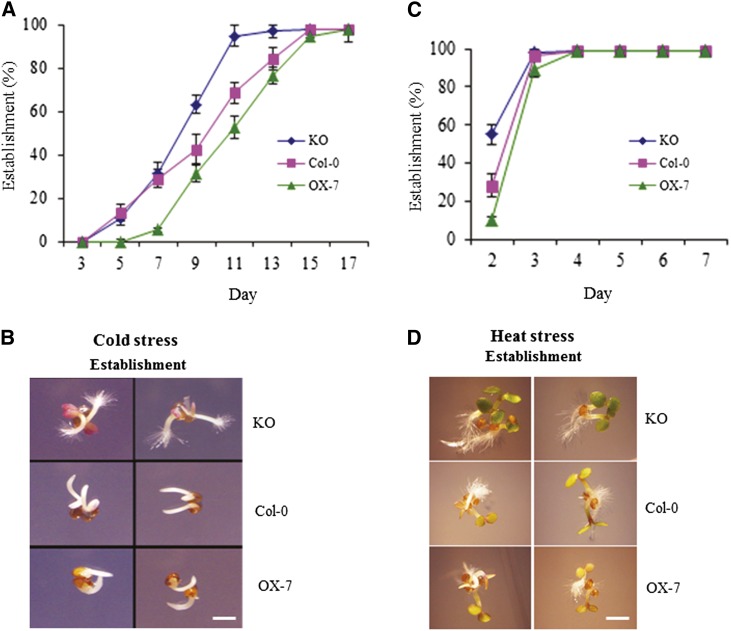
Effect of Cold and Heat on Germination and Early Seedling Growth of CaM KMT KO and OX Seedlings
Seeds from Col-0, CaM KMT KO, and OX-7 lines were stratified and then stressed with cold or heat to evaluate the possible role of CaM methylation in early growth of plants under other abiotic stresses. During continuous cold stress, the germination percentage of each line was measured daily. The seeds of CaM KMT KO lines reached 100% germination sooner compared with Col-0, and the OX-7 seeds were delayed beyond those of Col-0 (Figure 7A). Seed establishment under 20 d of cold stress clearly demonstrated that the development of the KO seedlings was faster compared with that of the other two lines (Figure 7B). A 2-h heat stress did not significantly affect the germination of CaM KMT KO and OX-7 seedlings (Figure 7C), but the subsequent photomorphogenesis of the KO seedlings was advanced compared with Col-0 and OX-7 seedlings (Figure 7D). Under field conditions, plants can face a number of stresses in combination. When the CaM KMT KO, Col-0, and CaM KMT OX plants were subjected to both cold and dehydration for 2 weeks, the KO seedlings were more tolerant compared with Col-0 and OX-7 lines (see Supplemental Figure 9 online), as indicated by the severity of leaf wilting.
Figure 7.
The CaM KMT KO Mutant Seeds Germinate Sooner under Cold Stress and Advance Further in Photomorphogenesis after Heat Stress.
(A) KO seeds germinated sooner under cold stress. Col-0, KO, and CaM KMT OX-7 seeds were sown on AGM and stratified at 4°C for 2 d, followed by growth at 4°C under continuous light. For the phenotypic measurement of germination, seeds with a protruding radical were scored each day after stratification. All assays were repeated three times using ∼50 seeds for each experiment. Values are mean ± sd (n = 3).
(B) Phenotypic observations show that the establishment of KO seedlings is faster compared with that of Col-0 and OX-7 seedlings under cold stress. The seedlings were digitally imaged after 20 d of stratification.
(C) Effect of heat stress on seed germination. After stratification, a heat stress consisting of 2 h at 42°C was followed by growth in a chamber (16-h-light/8-h-dark cycle at 22°C), and scoring followed the same procedure as in (A).
(D) Phenotypic observations show that photomorphogenesis after heat stress was advanced in KO seedlings compared with the Col-0 and OX-7 lines. Images were taken after 3 d of stratification and heat stress treatment as in (C). Bar in (B) and (D) = 1 mm.
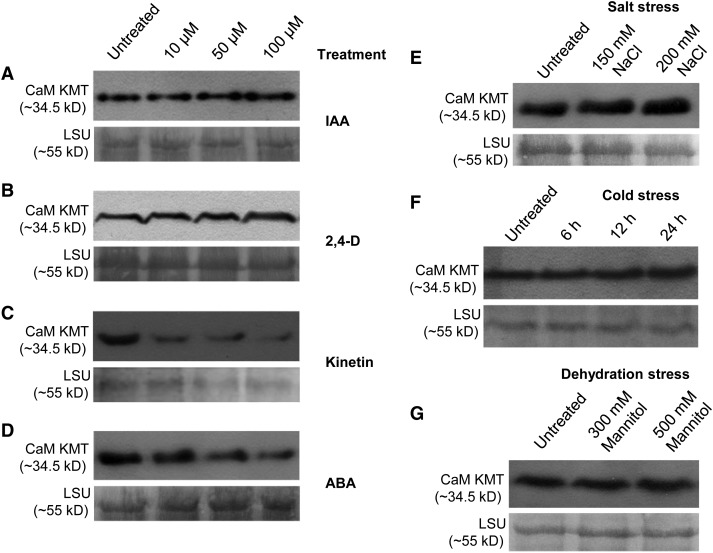
CaM KMT Protein Accumulation Is Influenced by Phytohormones and Environmental Stress
The accumulation of CaM KMT protein under different phytohormone treatment and stress conditions was examined using immunoblotting. After 6-h treatment with IAA or 2,4-D (10, 50, or 100 µM), CaM KMT accumulated in a concentration-dependent manner (Figures 8A and 8B). By contrast, CaM KMT amounts decreased in a dose-dependent manner in Col-0 seedlings during kinetin or ABA treatment for 6 h (Figures 8C and 8D). To examine further whether CaM KMT is upregulated at later stages of ABA treatment, we analyzed protein accumulation after 6, 12, and 24 h of ABA (10 µM) treatment (see Supplemental Figure 10 online). Although more CaM KMT protein accumulated after 12 h of ABA treatment than after 6 h, accumulation at neither 12 nor 24 h was greater than the untreated controls. After treatment of seedlings with 150 or 200 mM NaCl for 6 h, CaM KMT protein increased compared with untreated controls (Figure 8E). After 6 h of cold stress, CaM KMT accumulation was relatively unaffected, but amounts of protein did increase with longer times of imposed stress (12 and 24 h; Figure 8F). Similarly, the greater, but not the lesser, dehydration stress (500 versus 300 mM mannitol, respectively) increased CaM KMT protein accumulation relative to the untreated controls (Figure 8G).
Figure 8.
CaM KMT Protein Accumulation Under Different Phytohormone Treatments and Stress Conditions.
(A) to (D) CaM KMT protein amounts were detected in Col-0 seedlings treated with 0 (untreated), 10, 50, and 100 µM IAA (A), 2,4-D (B), Kinetin (C), and ABA (D).
(E) CaM KMT protein accumulation under 0 (untreated), 150, and 200 mM NaCl stress.
(F) Detection of CaM KMT protein without (untreated) and after 6, 12, and 24 h of cold stress.
(G) CaM KMT protein accumulation after dehydration stress using 0 (untreated), 300, and 500 mM mannitol.
(A) to (G) Three-week-old Arabidopsis Col-0 seedlings were subjected to different phytohormone, salt, and mannitol treatments along with untreated controls (water) for 6 h; for cold stress treatment, the seedlings were kept at 4°C for 6, 12, and 24 h. Total protein extracted from equal amounts of tissue from untreated and treated plants was separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane, which was probed with anti-CaM KMT antibody. In all cases, the bottom panel shows the loading control (Rubisco LSU) using Ponceau S stain.
Identification of Arabidopsis Proteins with Differential Binding Affinity for Methylated Versus Unmethylated Forms of CaM
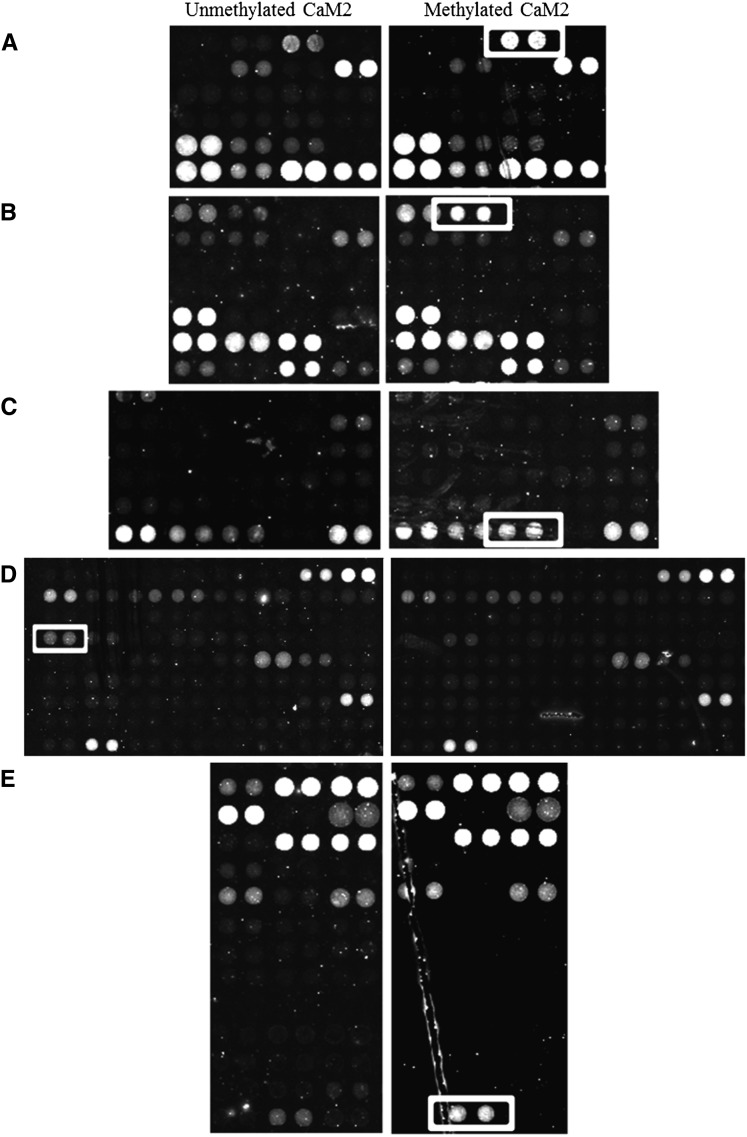
Arabidopsis-specific protein chips (AtProteinChip 2) were used to screen for CaM binding proteins capable of distinguishing between methylated and unmethylated forms of CaM2. CaM2 probes (methylated and unmethylated) bound with different levels of affinity to a large number of the 10,000 proteins spotted on the chip, both in the presence (see Supplemental Figure 11 online) or absence (data not shown) of calcium, although fewer protein binding partners were visible on the chip when using the apoprotein CaM2 probe (absence of calcium). Differential binding was observed between methylated and unmethylated CaM2 in the presence of calcium, with four proteins in particular appearing to be very specific for the methylated form (Figures 9A to 9C and 9E). Two of these proteins belong to the germin-like protein family, GERMIN-LIKE PROTEIN9 (GLP9) and GLP10 (Figures 9A and 9B). GLP10 also bound to methylated CaM2 in the presence of EGTA, whereas no signal was visible for GLP10 when unmethylated CaM2 was used as a probe (see Supplemental Figure 12 online). Two members of the nuclear-encoded chloroplast ribonucleoprotein family (CP28A and CP33B) were also identified as CaM2 binding partners in the presence of calcium, although their affinities for methylation were opposite. Specifically, the 28-kD protein (CP28A) was found to be highly specific for methylated CaM2 (Figure 9C), whereas the 33-kD protein (CP33B) seemed to preferentially bind unmethylated CaM2, despite a much weaker signal compared with CP28A (Figure 9D). Finally, a Xyl isomerase protein exhibited stronger binding to methylated CaM2 compared with unmethylated CaM2 (Figure 9E). Other Arabidopsis proteins previously described as hypothetical or known CaM binding proteins were also identified in our screen including At1g56410 and At1g27460 (NO POLLEN GERMINATION RELATED1) (Reddy et al., 2002; Golovkin and Reddy, 2003), At5G17330 (glutamate decarboxylase 1) (Zik et al., 1998), and At1G18840 (Abel et al., 2005).
Figure 9.
Binding of unmethylated CaM2 and methylated CaM2 to Arabidopsis proteins in the presence of calcium. Differential binding on Arabidopsis protein arrays (AtProteinChip 2) to unmethylated CaM2 (left panels) and methylated CaM2 (right panels) is indicated by white boxes. The surrounding spots serve as indicators of the uniformity of binding to CaM2 independent of the methylation status. All spots were printed in duplicate. The Arabidopsis protein binding partners identified were: (A) AT4G14630 (GLP9), (B) AT3G62020 (GLP10), (C) AT1G60000 (CP28A), (D) AT2G35410 (CP33B), and (E) AT5G57655 (Xyl isomerase).
DISCUSSION
CaM KMT Expression Patterns Correlate with Auxin Signaling Pathways
Our analysis of the CaM KMTPro:GUS fusion transgenic lines demonstrated that the CaM KMT promoter was active in a tissue and developmental pattern similar to that of auxin signaling. The promoter activity of CALMODULIN BINDING TRANSCRIPTION ACTIVATOR1, which is involved in auxin signaling and stress responses (Galon et al., 2010), shows some overlap with the expression pattern we found for CaM KMT. In addition, similar promoter activity at the base of the silique was described for the auxin-regulated gene involved in organ size (Hu et al., 2003). This type of tissue-specific expression pattern was also reported during monitoring of free auxin production (Aloni et al., 2003), with free auxin production highest in the young primordia, young leaf, and root tips. Gravity-stimulated auxin signal asymmetry in roots (Ottenschläger et al., 2003) is very similar to the gravitropic GUS expression driven by the CaM KMT promoter in the root curvature. In addition, the maximal CaM KMT transcript abundance in the cotyledonary stage (see Supplemental Figure 1 online) hints at the involvement of CaM KMT in apical meristem development. Together, these expression profile results suggest that CaM KMT is mostly expressed in young regenerating tissue and has some role in the auxin signaling network.
CaM Methylation Status Influences Root Growth and Morphology
Earlier studies in pea plants found that the CaM methylation level is lower in the root tip, apical root segment, and young lateral roots compared with the mature or differentiated root tissue, stem, and leaves (Oh and Roberts, 1990). In this study, we found that CaM in root samples from the CaM KMT KO and RNAi lines was hypomethylated (Figure 2C), whereas the absence of any significant [3H-methyl] group incorporation into CaM from Col-0 or CaM KMT OX lines was an indication that the CaM in those lines was completely methylated. Root length was modestly increased in CaM KMT KO seedlings and decreased in CaM KMT OX seedlings compared with Col-0 (Figures 2D and 2E). Ca2+/CaM is involved in root growth (Björkman and Leopold, 1987; Liu et al., 1992), but how methylation of CaM regulates root growth and development is unclear. Surprisingly, both CaM KMT KO and OX lines displayed increased numbers of epidermal cells (see Supplemental Figure 6 online). Furthermore, scrambling of the cell fate was evident in both genotypes (Figure 3).
SCRAMBLED, a leucine-rich repeat receptor-like kinase (LRR-RLK), affects the cell-type specification of epidermal cells in Arabidopsis roots (Kwak and Schiefelbein, 2008). Because SCRAMBLED lacks a functional kinase domain, its activity is predicted to be reliant on an unknown ligand that is delivered in a position-dependent manner (Kwak and Schiefelbein, 2008; De Smet et al., 2009). Could this ligand be a methylated or unmethylated version of CaM? Other small peptides play important roles in plant development, for instance, the CLAVATA3/EMBRYO SURROUNDING REGION–related proteins necessary for meristem maintenance (Mitchum et al., 2008). A report by Roberts et al. (1992) points to unmethylated forms of CaM as having deleterious effects on plant growth through a possible NADK as a target. Therefore, it is plausible that the hypomethylated form of CaM (in the case of CaM KMT KO) and the hypermethylated form of CaM (in the case of CaM KMT OX) both cause deleterious effects to plant developmental pathways in a similar manner.
There were clear differences in root hair length between the CaM KMT KO and OX seedlings compared with the Col-0 plants (see Supplemental Figure 7 online). Root hairs are similar to pollen tubes in that they exhibit tip growth, mediated by polarized secretion and endocytosis that maintains the tubular structure of the root hair or pollen tube. There is a higher concentration of both free cytosolic calcium and CaM at the pollen tube apex (Rato et al., 2004), and this apical calcium and CaM follow a similar oscillatory pattern with the growth of the pollen tube (Rato et al., 2004). In root hairs, tip-focused cytosolic calcium that oscillates with growth is known to play a pivotal role in vesicle exocytosis and tip growth (Cárdenas, 2009). However, whether CaM functions in root hair growth as in pollen tube growth is unknown, let alone what effects the methylation state of CaM might have. This study points to a mechanism wherein the methylation state of CaM, in both hypomethylated and hypermethylated forms, affects root hair growth.
CaM KMT Affects Auxin Signaling and Root Gravitropism2
The role of auxin flux and auxin basipetal transport in root growth has been extensively characterized (Ottenschläger et al., 2003). CaM plays a role in both auxin signaling (Yang and Poovaiah, 2000) and root gravity sensing (Björkman and Leopold, 1987), but how methylation of CaM might be involved in these processes is unclear. The root growth of the CaM KMT OX seedlings was hypersensitive to high concentrations of auxin (Figure 4), whereas seedlings of both the CaM KMT KO line and the auxin mutant line aux1-7 (Biswas et al., 2007) show less sensitivity to IAA and 2,4-D. In addition, the root hair distribution pattern of the CaM KMT KO seedlings was similar to that of the auxin mutant aux1-7 and aux1-22 lines (Pitts et al., 1998; Jones et al., 2009). The root growth of CaM KMT OX seedlings became agravitropic in the presence of exogenous auxin (Figure 4), an effect that was more evident with 2,4-D than with IAA. A similar differential effect of 2,4-D and IAA was also observed in a study related to the root growth of an antiauxin-resistant mutant (Biswas et al., 2007) and might be because of the partially different signaling pathways for 2,4-D and IAA (Rahman et al., 2006). In this study, CaM KMTPro:GUS roots showed gravitropic GUS expression in the root curvature (Figure 1I). Interestingly, methyl jasmonate treatment led to little difference in root growth among seedling from all lines (see Supplemental Figure 13 online). A number of auxin-related factors (putative small auxin-up RNA [SAUR_B] and IAA31) bind to several CaM isoforms and CaM-related proteins in Arabidopsis (Popescu et al., 2007). Our data, together with the previous findings, support the idea that the methylation status of CaM is involved in root gravitropism and its interaction with auxin may also play a crucial role in gravitropic root growth.
CaM KMT KO Lines Are Resistant to ABA Stress
The role of ABA in seed dormancy and germination is well established, and ABA biosynthesis, as well as the ABA signaling pathway, is triggered during salt stress (Magnan et al., 2008). Our results show that constitutive and tissue-indiscriminant methylation of CaM is negatively correlated with ABA stress (Figure 5). In maize leaves, CaM functions in the nitric oxide synthase–mediated nitric oxide production involved in the ABA-induced antioxidant defense pathway (Sang et al., 2008). In addition, CaM-dependent protein kinase functions in ABA-induced antioxidant defense response (Xu, 2010). In Arabidopsis, mutant plants of calmodulin-like protein9 exhibit increased sensitivity to ABA, NaCl, and KCl during germination (Magnan et al., 2008). Calcineurin B-Like 9, a calcium censor, also has been shown to act as a negative regulator in the ABA signaling network (Pandey et al., 2004). These observations suggest that there may be specific interactions between methylated CaM and different stress signaling pathways, and that the CaM KMT KO lines might be less sensitive to ABA stress because of the absence of the methylated form of CaM.
CaM Methylation Status Influences Early Growth and Development under Abiotic Stress Conditions
The CaM KMT KO lines displayed increased resistance to salt and cold stress, whereas the CaM KMT OX lines exhibited increased sensitivity (Figures 6 and 7). CaM has been reported to be involved in responses to different stresses (Pardo et al., 1998; Delumeau et al., 2000), as well as in plant recovery after stress (Gu et al., 2004). In terms of salt stress, calcineurin, a Ca2+/CaM-dependent protein phosphatase, functions in the associated signal transduction pathway (Pardo et al., 1998), and analysis of the salt overly sensitive Arabidopsis mutant revealed that a Ca2+/CaM-dependent protein kinase pathway is involved in salt stress signaling (Xiong et al., 2002). Ca2+-dependent protein kinase genes are also activated during different abiotic stresses (Xiong et al., 2002). CaM plays a significant role during cold (Townley and Knight, 2002) and heat stress (Larkindale and Knight, 2002), but exactly how CaM methylation is involved in that signaling is unclear, even from our results. Recently, differential binding of Arabidopsis CaM and CaM-related proteins has been documented, and possible targets include kinases, F-box proteins, WRKY factors, and TGA factors (Popescu et al., 2007). Another study reported that a large number of calcium-dependent protein kinases are involved in salt, cold, heat, and other abiotic stresses signaling either through ABA-dependent or ABA-independent pathways (Das and Pandey, 2010). Based on earlier observations and our data, it can be hypothesized that CaM methylation plays a regulatory role in the response of Arabidopsis plants to salt, heat, and cold stress.
Differential Regulation of Expression of CaM KMT in Different Phytohormone Signaling Networks
We found that auxin upregulates the expression of CaM KMT. By contrast, the expression of CaM KMT was downregulated after kinetin treatment (Figure 8C). The upregulation of CaM KMT expression in response to salt, cold, and dehydration stresses (Figure 8E to 8G) clearly suggests that it is involved in the stress signaling cascade. However, CaM KMT expression was initially downregulated by ABA and subsequently recovered (Figure 8D, see Supplemental Figure 10 online). It is possible that the involvement of CaM KMT in stress signaling is ABA independent, or that there is some feedback inhibition between the CaM KMT and ABA signaling. Regardless, the results implicate the methylation status of CaM as an important factor in hormone and stress signaling pathways. Given the phenotypes of the lines with altered CaM KMT expression under various conditions, it is tempting to speculate that CaM methylation may be a key integrator, not only of endogenous hormone signaling during normal root growth and development, but also of various environmental stressors. Our data support a hypothesis of methylation as a rheostat-like control mechanism for coordination of the multifaceted Ca2+/CaM signaling networks involved in growth and development, as well as environmental responses.
Arabidopsis Proteins That Read the Methylation Status of CaM May Transmit This Information to Downstream Signaling Pathways
The phenotypes observed for the CaM KMT OX, RNAi, and KO Arabidopsis lines suggest that the methylation status of CaM can have a rather broad influence on biotic/abiotic stress signaling pathways, as well as developmental processes in roots. Identification of proteins that can read the methylation status of CaM represents a first step in understanding methylation-dependent CaM signaling pathways, and several such proteins were identified in this article. Among these were germin-like proteins GLP9 and GLP10, which belong to a family of ubiquitous plant-specific proteins involved in a multitude of physiological and developmental processes, and play a significant role during abiotic and biotic stresses (Dunwell et al., 2008). Arabidopsis GLP9 is involved in NaCl stress, and its expression is increased during NaCl stress (Jiang et al., 2007). A GLP from the halophyte Artiplex lentiformis is differentially regulated by ABA (Tabuchi et al., 2003), and the expression of a GLP from Citrus limonia is induced in roots under cold stress (Boscariol-Camargo et al., 2007) as described earlier (Dunwell et al., 2008). GLPs are also responsible for growth and development, as well as cellular anatomy. GLP1 from rice affects plant height, disease resistance, and maintenance of cell dimensions (Banerjee and Maiti, 2010). Genes encoding GLPs are overexpressed in Prunus salicina fruit when treated with auxins (El-Sharkawy et al., 2010), and another member from Arabidopsis (GLP4) binds auxins in vitro (Yin et al., 2009). GLP10 has not been characterized to date, but it shows high similarity (more than 80% of the amino acid sequence) to plasmodesmata GLP (AT1G02335), which has been recently described to be involved in root development (Ham et al., 2012). Plants overexpressing plasmodesmata GLP are characterized by inhibition of primary root growth and compensatory increased development of lateral roots. This phenotype resembles that of CaM KMT OX-7, suggesting that the methylation status of CaM may influence its binding to GLP10 with important consequences on root development. In Arabidopsis, 29 GLP proteins have been identified, but only 9 are present on the protein chip; among these GLP family members, only GLP5 (besides GLP9 and GLP10) binds CaM, but its interaction was not influenced by the methylation status of CaM. Considering that GLPs show a broad range of biochemical properties, occurrence, and localization, it is possible that other members of this vast family could also interact differentially with methylated versus unmethylated forms of CaM. Thus, the phenotypes described in this article could be a consequence of methylation-dependent binding of CaM to GLPs affecting the activation or compartmentalization of these proteins with subsequent effects on stress resistance and root development.
We also identified differential binding of methylated versus unmethylated CaM to two chloroplast ribonucleoproteins, CP28A and CP33B (Ohta et al., 1995; Lorkovic and Barta, 2002; Ruwe et al., 2011), which might be responsible for the activation or regulation of different targets in multiple signaling pathways during transcription or RNA processing. Proteomics studies have detected both proteins in the chloroplast stroma (SubCellular Proteomic Database: Tanz et al., 2013; Plant Proteome Database: Sun et al., 2008), although a CaM isoform for this organelle has yet to be identified. Cp33 has not been characterized, but cp29 and cp33 together with cp31 have been identified in Arabidopsis (Ohta et al., 1995), because of their homology to tobacco chloroplast RNA binding proteins. Tobacco chloroplast ribonucleoproteins function as stabilizing factors for ribosome-free mRNAs, and some of them are involved in pre-tRNA splicing (Nakamura et al., 1999). Arabidopsis CP33B and both isoforms of CP28A from alternatively spliced transcripts have a high probability of containing a calmodulin binding site. Experimental evidence of the presence of multiple Ser and Thr phosphorylation sites on CP33B (Zulawski et al., 2013) suggests that there is dynamic regulation of the function of this protein. CAS is a chloroplast-localized protein, and when a cas1 mutant plant was treated with the defense-elicitor flg22, Cp29 was upregulated (Nomura et al., 2012). The significance of the interaction between these RNA binding proteins and the methylation status of CaM is not immediately obvious, nor is how that interaction may relate to the phenotypes observed in this study. Likewise, the increased affinity of Xyl isomerase for methylated CaM does not indicate what role it may play or how it relates to the studies presented in this article.
It was expected that the protein array experiment would identify NADK (three isoforms NADK1, NADK2, and NADK3), whose activity is known to be influenced by the methylation of CaM (Roberts et al., 1986), and CaM KMT (AT4G35987) itself, because it uses CaM as a substrate. However, only NADK3 was found during probing with methylated or unmethylated CaM2, and it did not display differential binding. Interestingly, none of the proteins identified in this study as showing differential binding to methylated versus unmethylated forms of CaM are known to bind CaM. This may be because of the fact that the majority of CaM binding studies conducted to date use bacterially expressed forms of CaM (unmethylated) to understand and identify CaM binding partners. Because other studies have already demonstrated differential binding specificities of CaM isoforms, as well as of CaM-like proteins (Popescu et al., 2007), it is not unreasonable to speculate that other posttranslational modifications of CaM including phosphorylation and acetylation may also influence the diversity of CaM binding partners.
Conclusions
The findings reported in this study add a new level of complexity to our understanding of CaM signaling mechanisms in plants, and also identify new methylation-dependent CaM binding proteins. Indeed, by analogy to Lys methylation and demethylation in histones (Black et al., 2012), the methylation status of CaM could be dynamic and a consequence of interplay between CaM KMT and Lys demethylases, giving rise to entirely new areas of research. Furthermore, the binding partners identified in this study that recognize methylated forms of CaM likely contain new methyl-Lys binding domains, because none of the currently known domains such as chromo domains, ankyrin repeats, Plant Homeo Domain, or Tudor domains could be identified in these sequences. Given the ability of CaM to localize to the nucleus, it is tempting to speculate that widespread alterations in developmental processes, such as the root architecture described in this article, could be a consequence of the inability of nonmethylated CaM to interact with nuclear methyl-Lys binding domain proteins, followed by influences on chromatin structure and gene expression.
METHODS
Plant Material
All transgenic lines were derived from Arabidopsis thaliana Col-0 ecotype. Seven independent lines each of CaM KMT OX and RNAi were generated and screened both by qRT-PCR and immunoblot analysis. From the T2 generation, two lines from each of these genotypes were used for further analyses. After a KO (SALK_138607) homozygote was identified, the plants, including wild type (Col-0) and the other transgenics, were grown in a growth chamber (16-h-light/8-h-dark cycle at 22°C) for seed germination, seed production (using an Arasystem [Lehle Seeds] to prevent cross-pollination), and other analyses.
Germination and Seedling Growth under Stress Conditions
Arabidopsis seeds were surface sterilized (70% ethanol for 3 min, 30% Clorox for 20 min, and five water washes) and plated on AGM composed of one-half-strength Murashige and Skoog salts, 1% [w/v] Suc, and 0.5 g/L MES, pH 5.8. For synchronous germination, all seeds were stratified by incubation in the dark at 4°C for 2 d (Biswas et al., 2007). Other than the cold and heat stress treatments, all stratified seeds were placed in a growth chamber (16-h-light/8-h-dark cycle at 22°C) for seed establishment experiments. For cold stress after stratification, the plated seeds were kept at 4°C but transferred to continuous light for 20 d; for heat stress, the stratified seeds were placed at 42°C in the dark for 2 h and then kept in the growth chamber for further analysis.
Root Growth Assays under Phytohormone Treatment and Stress Conditions
For root growth analysis upon treatment with different phytohormones, the CaM KMT KO, RNAi, OX, and Col-0 seeds were surface sterilized, stratified, and plated on square plates containing AGM in a growth chamber. After 4 d of growth, the plantlets were transferred to AGM supplemented with different concentrations of IAA, 2,4-D, and methyl jasmonate for an additional 7 d. The relative root length was calculated by comparing root length on media with respective phytohormones with that without any growth regulators following an earlier procedure (Biswas et al., 2007). For pictorial presentation of the auxin-mediated gravitropic response on root growth, 4-d-old plants from the CaM KMT KO, CaM KMT OX, and Col-0 lines were grown in AGM with and without supplementation of 2,4-D and IAA for additional 7 d; then data were recorded. For root growth assay under salt stress, the 4-d-old vertically grown seedlings were transferred to AGM with and without 100 mM NaCl for an additional 7 d, and data were recorded.
Histochemical GUS Staining and Microscopic Analysis
Histochemical GUS staining to localize the distribution of GUS activity in plants was performed according to an earlier protocol (Jefferson et al., 1987) following some modifications (Dey and Maiti, 1999). For microscopic analysis and documentation of the CaM KMTPro:GUS plants and germinated seedlings from different Arabidopsis lines, an Olympus SZX-ILLB2-100 microscope with an attached camera was used. For ABA, salt, and heat stress, early seedling growth photographs were taken after 3 d of stratification. For cold stress treatments, photographs were taken after 20 d of stratification. Measurements for root epidermal patterning and root hair lengths were performed at the “root hair zone” described by Schiefelbein and Somerville (1990) from 5-d-old seedlings of Col-0, CaM KMT KO, and CaM KMT OX. This zone was approximately 1 vertical millimeter from the root tip. Images were acquired on a stereomicroscope (Olympus MVX10) with 2X zoom using image acquisition software cellSens (Olympus). Percentage emergence of H and N cells in H and N cell files was analyzed statistically using Prism4 (GraphPad) to obtain significance using Mann–Whitney U test. Root hair length measurements were obtained using the line tool (ImageJ) and statistically analyzed in Prism4 for obtaining the frequency distributions and significance using Mann–Whitney U test. Root hair lengths from at least 500 individual root hairs from the root hair zone from all the genotypes were measured. To obtain the optical longitudinal sections of roots from Col-0, CaM KMT KO, and CaM KMT OX lines, we imaged propidium iodide–stained seedlings on a confocal microscope (Olympus Fluoview 1000) equipped with lasers ranging from 405 to 633 nm. Propidium iodide staining was performed as previously described (Lee and Schiefelbein, 2002). In brief, 10 to 12 five-day-old seedlings from each line were immersed in 10 μg/ml propidium iodide for a few seconds, washed, and mounted on a glass slide to examine under a confocal microscope. Propidium iodide was excited using DsRed filter setting on the Olympus Fluoview software. Radial patterning of cortical and epidermal cells was examined in agarose-embedded handmade cross sections. Cross sections were prepared as described previously (Galway et al., 1994) and were from the maturation zone of 5-d-old seedlings. The sections were stained with Calcofluor white (Sigma-Aldrich) and imaged using an Olympus MVX-10 stereomicroscope using the 4′,6-diamidino-2-phenylindole filter and Prior light source (Olympus).
Construction of Plant Expression Vectors and Plant Transformation
CaM KMTPro:GUS Construct
The CaM KMTPro fragment was isolated from Arabidopsis genomic DNA by PCR amplification (see Supplemental Tables 1 and 2 online). The promoter fragment (921-bp), after sequence verification, was cloned into pKYLX71-GUS vector (Schardl et al., 1987) at its unique corresponding sites.
CaM KMT OX Construct
A synthetic DNA fragment of general structure 5′-XhoI-native CaM KMT-SstI-3′ corresponding to the cDNA sequence of the Arabidopsis CaM KMT gene was generated (GeneArt; see Supplemental Table 1 online). For the downregulated expression vector an Arabidopsis CaM KMT RNAi construct of general structure 5′- HindIII-CaM KMT was designed and the corresponding synthetic DNA fragment generated (GeneArt; see Supplemental Table 1). Plant expression vectors were mobilized in Agrobacterium tumefaciens strain C58C1:pGV3850 by freeze-thaw methods. Arabidopsis plants Col-0 ecotype were transformed with the engineered Agrobacterium using the floral dip method (Clough and Bent, 1998). Independent Arabidopsis transgenic plants were screened in the presence of kanamycin (30 mg/L). Regenerated kanamycin-resistant plants were grown in a growth chamber (16-h-light/8-h-dark cycle at 22°C).
RNA Isolation and qRT-PCR
Total RNA was extracted from frozen plant samples using the Plant RNeasy extraction kit (Qiagen) following the manufacturer’s instructions. The quantitative measurement of the CaM KMT transcript was done by qRT-PCR using gene-specific primers (see Supplemental Tables 1 and 2 online) following an earlier protocol (Suttipanta et al., 2011). The qRT-PCR was performed with four replicates, and it was repeated with three biological samples. The transcript levels were measured following the comparative threshold cycle method (Applied Biosystems bulletin). ACTIN gene-specific primers (see Supplemental Table 2 online) were used to normalize the amount of total mRNA in all samples (Remans et al., 2008).
Determination of CaM Methylation Status
The recombinant CaM KMT assay that determines the relative methylation status of CaM as indicated by the incorporation of [3H-methyl] groups followed an earlier reported protocol (Magnani et al., 2010), with some minor modifications. For different plant extracts (shoot and root samples), 20 µg total protein was used in the assay. The reactions were performed at room temperature (22–24°C) for 18 h. As a negative control, shoot and root protein extracts were used from the KO line without adding CaM KMT enzyme.
Protein Expression and Immunoblotting
The Arabidopsis CaM KMT protein was expressed in Escherichia coli as described earlier (Magnani et al., 2010). The bacterial-expressed CaM KMT protein was used to raise anti-CaM KMT antibodies in rabbits for subsequent use in immunoblotting. Protein extraction was performed on equal amounts of tissue from 3-week-old Arabidopsis plants of different lines, and immunoblotting was performed using anti-Arabidopsis CaM KMT antisera as primary antibody (1:5,000) and horseradish peroxidase as secondary antibody following an earlier protocol (Smalle et al., 2003). To analyze the accumulation of Arabidopsis CaM KMT protein under different phytohormone and stress conditions (other than cold stress), 3-week-old seedlings were subjected to different treatments for 6 h, along with untreated controls, and immunoblotting was performed using a similar protocol as described earlier. For time-course analysis of CaM KMT under cold stress and 10 µM ABA treatment, 3-week-old seedlings were subjected to different treatments for 6, 12, and 24 h, and CaM KMT protein was detected in immunoblots following the earlier protocol. In all immunoblot experiments, the Rubisco large subunit (LSU) was detected by Ponceau S staining and used as loading control.
Arabidopsis Protein Array Probing
The DNA sequence coding for isoform 2 of CaM was cloned into pET28b (Merck Millipore) using NotI and XhoI restrictions sites, which introduces a single N-terminal Cys residue (see Supplemental Tables 1 and 2 online). This plasmid was then expressed as described previously (Magnani et al., 2012), using coexpression with CaM KMT to generate a fully methylated form of CaM2 and expression without CaM KMT to generate the unmethylated form of CaM2. The two forms of CaM2 were purified and biotinylated using EZ-link iodoacetyl-PEG2-biotin reagent (Pierce) following the manufacturer’s protocol. Protein arrays (AtProteinChip 2) purchased from the ABRC were blocked with 30 mL 1X Tris-buffered saline, 1% BSA, 0.1% Tween 20, and then incubated for 1 h 30 min in the presence of methylated or unmethylated forms of CaM at a concentration of 2 μg/μL in probing buffer (1X Tris-buffered saline, 5 mM MgCl2, 0.05% Triton X-100, 5% glycerol, 1% BSA, and 0.5 mM DTT). CaCl2 or EGTA were added to the probing solution to a final concentration of 1 mM. The arrays were washed twice with 20 mL probing buffer and incubated for 30 min in the dark with 5 mL probing buffer containing 1 μg/ml streptavidin conjugated with Alexa Fluor 647 (Life Technologies). Two additional washes with 20 mL probing buffer were performed before drying the arrays and imaging them on Axon Genepix 4100A scanner (Molecular Devices). Except for the imaging, all steps were performed at 4°C.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or Gen Bank under the following locus identifiers: CaM KMT (At4g35987); CaM KMT RNAi construct (DQ672337); pKM24 (HM036220); CaM2 (At2g41110); GLP9 (At4g14630); GLP10 (At3g62020); CP28A (At1g60000); CP33B (At2g35410); xylose isomerase protein (At5g57655); NADK1 (At3g21070); NADK2 (At1g21640); NADK3 (At1g78590).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Profiling of the Arabidopsis CaM KMT Transcript: Arabidopsis CaM KMT–Specific Transcript Levels in Arabidopsis Seedlings at Different Early Growth Stages.
Supplemental Figure 2. Schematic Diagram of the CaM KMTPro:GUS Construct.
Supplemental Figure 3. Analysis of Arabidopsis CaM KMT in the Public Expression Database.
Supplemental Figure 4. Constructs Prepared for Overexpression and Downregulation of Expression of CaM KMT in Arabidopsis.
Supplemental Figure 5. CaM KMT Gene Structure in wild-type and KO Arabidopsis Lines.
Supplemental Figure 6. Increase in Epidermal Cells in Longitudinal and Cross Sections of Roots from T-DNA Insertion KO and OX Lines.
Supplemental Figure 7. Root Hair Lengths Are Affected as a Result of Genetic Alterations of CaM KMT Expression.
Supplemental Figure 8. CaM KMT Overexpressing Plants Are More Sensitive to Salt Stress.
Supplemental Figure 9. The Leaves of CaM KMT KO Mutant Plants Were More Turgid after Cold and Dehydration Stress Than Those of Col-0, Which Were More Turgid Than Those of CaM KMT OX-7.
Supplemental Figure 10. CaM KMT Protein Accumulation after Treatment with 10 µM ABA for the Indicated Periods.
Supplemental Figure 11. Binding of Unmethylated CaM2 and Methylated CaM2 to Arabidopsis Proteins Arrayed on a PATH Slide (AtProteinChip 2) in the Presence of Calcium.
Supplemental Figure 12. Binding of Unmethylated apoCaM2 and Methylated apoCaM2 to Arabidopsis Proteins Arrayed on a PATH Slide (AtProteinChip 2).
Supplemental Figure 13. Root Growth Assay with Methyl Jasmonate.
Supplemental Table 1. Constructs Used in This Work.
Supplemental Table 2. Primer Sequences Used in This Work.
Acknowledgments
This work was supported by a grant from the Kentucky Tobacco Research and Development Center to I.B.M. and R.L.H., and by Kentucky Agriculture Experiment Station Hatch Project KY011030 to R.L.H.
AUTHOR CONTRIBUTIONS
All authors contributed to designing the research and writing the article. J.B., R.M., M.N., and L.M.D. conducted the experiments.
References
- Abel S., Savchenko T., Levy M. (2005). Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol. Biol. 5: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloni R., Schwalm K., Langhans M., Ullrich C.I. (2003). Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216: 841–853 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Banerjee J., Maiti M.K. (2010). Functional role of rice germin-like protein1 in regulation of plant height and disease resistance. Biochem. Biophys. Res. Commun. 394: 178–183 [DOI] [PubMed] [Google Scholar]
- Biswas K.K., Ooura C., Higuchi K., Miyazaki Y., Van Nguyen V., Rahman A., Uchimiya H., Kiyosue T., Koshiba T., Tanaka A., Narumi I., Oono Y. (2007). Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2,4-dichlorophenoxyacetic acid in Arabidopsis roots. Plant Physiol. 145: 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björkman T., Leopold A.C. (1987). Effect of inhibitors of auxin transport and of calmodulin on a gravisensing-dependent current in maize roots. Plant Physiol. 84: 847–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.C., Whetstine J.R. (2012). LOX out, histones: A new enzyme is nipping at your tails. Molecular cell 46: 243–244 [DOI] [PubMed] [Google Scholar]
- Boscariol-Camargo R.L., Berger I.J., Souza A.A., Amaral A.M.d., Carlos E.F., Freitas-Astúa J., Takita M.A., Targon M.L.P.N., Medina C.L., Reis M.S., Machado M.A. (2007). In silico analysis of ESTs from roots of Rangpur lime (Citrus limonia Osbeck) under water stress. Genet. Mol. Biol. 30 (suppl. 3): 906–916 [Google Scholar]
- Cardenas L. (2009). New findings in the mechanisms regulating polar growth in root hair cells. Plant signaling & behavior 4: 4–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabrol B., Martens K., Meulemans S., Cano A., Jaeken J., Matthijs G., Creemers J.W. (2008). Deletion of C2orf34, PREPL and SLC3A1 causes atypical hypotonia-cystinuria syndrome. J. Med. Genet. 45: 314–318 [DOI] [PubMed] [Google Scholar]
- Chin D., Means A.R. (2000). Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 10: 322–328 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Das R., Pandey G.K. (2010). Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr. Genomics 11: 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delumeau O., Renard M., Montrichard F. (2000). Characterization and possible redox regulation of the purified calmodulin-dependent NAD+ kinase from Lycopersicon pimpinellifolium. Plant Cell Environ. 23: 1267–1273 [Google Scholar]
- De Smet I., Voss U., Jurgens G., Beeckman T. (2009). Receptor-like kinases shape the plant. Nature Cell Biology 11: 1166–1173 [DOI] [PubMed] [Google Scholar]
- Dey N., Maiti I.B. (1999). Structure and promoter/leader deletion analysis of mirabilis mosaic virus (MMV) full-length transcript promoter in transgenic plants. Plant Mol. Biol. 40: 771–782 [DOI] [PubMed] [Google Scholar]
- Dunwell J.M., Gibbings J.G., Mahmood T., Saqlan Naqvi S.M. (2008). Germin and Germin-like Proteins: Evolution, structure, and function. Crit. Rev. Plant Sci. 27: 342–375 [Google Scholar]
- El-Sharkawy I., Mila I., Bouzayen M., Jayasankar S. (2010). Regulation of two germin-like protein genes during plum fruit development. J. Exp. Bot. 61: 1761–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y., Aloni R., Nachmias D., Snir O., Feldmesser E., Scrase-Field S., Boyce J.M., Bouché N., Knight M.R., Fromm H. (2010). Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232: 165–178 [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Golovkin M., Reddy A.S. (2003). A calmodulin-binding protein from Arabidopsis has an essential role in pollen germination. Proc. Natl. Acad. Sci. USA 100: 10558–10563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu R., Fonseca S., Puskás L.G., Hackler L., Jr, Zvara Á., Dudits D., Pais M.S. (2004). Transcript identification and profiling during salt stress and recovery of Populus euphratica. Tree Physiol. 24: 265–276 [DOI] [PubMed] [Google Scholar]
- Ham B.-K., Li G., Kang B.-H., Zeng F., Lucas W.J. (2012). Overexpression of Arabidopsis plasmodesmata germin-like proteins disrupts root growth and development. Plant Cell 24: 3630–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding S.A., Oh S.H., Roberts D.M. (1997). Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO J. 16: 1137–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Xie Q., Chua N.H. (2003). The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15: 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Yang B., Harris N.S., Deyholos M.K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58: 3591–3607 [DOI] [PubMed] [Google Scholar]
- Jones A.R., Kramer E.M., Knox K., Swarup R., Bennett M.J., Lazarus C.M., Leyser H.M.O., Grierson C.S. (2009). Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 11: 78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee C.B., Vanaman T.C. (1982). Calmodulin. Adv. Protein Chem. 35: 213–321 [DOI] [PubMed] [Google Scholar]
- Kwak S.H., Schiefelbein J. (2008). A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Current Biology: 18: 1949–1954 [DOI] [PubMed] [Google Scholar]
- Larkindale J., Knight M.R. (2002). Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 128: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.M., Schiefelbein J. (2002). Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Jiang W., Li M. (1992). Effects of trivalent and hexavalent chromium on root growth and cell division of Allium cepa. Hereditas 117: 23–29 [Google Scholar]
- Lorkovic Z.J., Barta A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Research 30: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magen S., Magnani R., Haziza S., Hershkovitz E., Houtz R., Cambi F., Parvari R. (2012). Human calmodulin methyltransferase: Expression, activity on calmodulin, and Hsp90 dependence. PLoS ONE 7: e52425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnan F., Ranty B., Charpenteau M., Sotta B., Galaud J.-P., Aldon D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56: 575–589 [DOI] [PubMed] [Google Scholar]
- Magnani R., Dirk L.M., Trievel R.C., Houtz R.L. (2010). Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat Commun 1: 43. [DOI] [PubMed] [Google Scholar]
- Magnani R., Chaffin B., Dick E., Bricken M.L., Houtz R.L., Bradley L.H. (2012). Utilization of a calmodulin lysine methyltransferase co-expression system for the generation of a combinatorial library of post-translationally modified proteins. Protein Expr. Purif. 86: 83–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshak D.R., Clarke M., Roberts D.M., Watterson D.M. (1984). Structural and functional properties of calmodulin from the eukaryotic microorganism Dictyostelium discoideum. Biochemistry 23: 2891–2899 [DOI] [PubMed] [Google Scholar]
- Mitchum M.G., Wang X., Davis E.L. (2008). Diverse and conserved roles of CLE peptides. Current Opinion in Plant Biology 11: 75–81 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Ohta M., Sugiura M., Sugita M. (1999). Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett. 460: 437–441 [DOI] [PubMed] [Google Scholar]
- Nomura H., et al. (2012). Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3: 926. [DOI] [PubMed] [Google Scholar]
- Oh S.-H., Roberts D.M. (1990). Analysis of the state of posttranslational calmodulin methylation in developing pea plants. Plant Physiol. 93: 880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.H., Yun S.J. (1999). Effects of various calmodulins on the activation of glutamate decarboxylase and nicotinamide adenine dinucleotide kinase isolated from tobacco plants. Agriculture Chemistry and Biotechnology 42: 19–24 [Google Scholar]
- Oh S.H., Steiner H.Y., Dougall D.K., Roberts D.M. (1992). Modulation of calmodulin levels, calmodulin methylation, and calmodulin binding proteins during carrot cell growth and embryogenesis. Arch. Biochem. Biophys. 297: 28–34 [DOI] [PubMed] [Google Scholar]
- Ohta M., Sugita M., Sugiura M. (1995). Three types of nuclear genes encoding chloroplast RNA-binding proteins (cp29, cp31 and cp33) are present in Arabidopsis thaliana: presence of cp31 in chloroplasts and its homologue in nuclei/cytoplasms. Plant Mol. Biol. 27: 529–539 [DOI] [PubMed] [Google Scholar]
- Ottenschläger I., Wolff P., Wolverton C., Bhalerao R.P., Sandberg G., Ishikawa H., Evans M., Palme K. (2003). Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 100: 2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey G.K., Cheong Y.H., Kim K.-N., Grant J.J., Li L., Hung W., D’Angelo C., Weinl S., Kudla J., Luan S. (2004). The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panina S., Stephan A., la Cour J.M., Jacobsen K., Kallerup L.K., Bumbuleviciute R., Knudsen K.V., Sánchez-González P., Villalobo A., Olesen U.H., Berchtold M.W. (2012). Significance of calcium binding, tyrosine phosphorylation, and lysine trimethylation for the essential function of calmodulin in vertebrate cells analyzed in a novel gene replacement system. J. Biol. Chem. 287: 18173–18181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo J.M., et al. (1998). Stress signaling through Ca2+/calmodulin-dependent protein phosphatase calcineurin mediates salt adaptation in plants. Proc. Natl. Acad. Sci. USA 95: 9681–9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvari R., Hershkovitz E. (2007). Chromosomal microdeletions and genes’ functions: A cluster of chromosomal microdeletions and the deleted genes’ functions. Eur. J. Hum. Genet. 15: 997–998 [DOI] [PubMed] [Google Scholar]
- Parvari R., Brodyansky I., Elpeleg O., Moses S., Landau D., Hershkovitz E. (2001). A recessive contiguous gene deletion of chromosome 2p16 associated with cystinuria and a mitochondrial disease. Am. J. Hum. Genet. 69: 869–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvari R., Gonen Y., Alshafee I., Buriakovsky S., Regev K., Hershkovitz E. (2005). The 2p21 deletion syndrome: Characterization of the transcription content. Genomics 86: 195–211 [DOI] [PubMed] [Google Scholar]
- Pitts R.J., Cernac A., Estelle M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16: 553–560 [DOI] [PubMed] [Google Scholar]
- Popescu S.C., Popescu G.V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S.P. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. USA 104: 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A., Nakasone A., Chhun T., Ooura C., Biswas K.K., Uchimiya H., Tsurumi S., Baskin T.I., Tanaka A., Oono Y. (2006). A small acidic protein 1 (SMAP1) mediates responses of the Arabidopsis root to the synthetic auxin 2,4-dichlorophenoxyacetic acid. Plant J. 47: 788–801 [DOI] [PubMed] [Google Scholar]
- Rato C., Monteiro D., Hepler P.K., Malho R. (2004). Calmodulin activity and cAMP signalling modulate growth and apical secretion in pollen tubes. The Plant Journal for Cell and Molecular Biology 38: 887–897 [DOI] [PubMed] [Google Scholar]
- Reddy V.S., Ali G.S., Reddy A.S. (2002). Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem. 277: 9840–9852 [DOI] [PubMed] [Google Scholar]
- Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. (2008). Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227: 1343–1349 [DOI] [PubMed] [Google Scholar]
- Roberts D.M., Burgess W.H., Watterson D.M. (1984). Comparison of the NAD kinase and myosin light chain kinase activator properties of vertebrate, higher plant, and algal calmodulins. Plant Physiol. 75: 796–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.M., Rowe P.M., Siegel F.L., Lukas T.J., Watterson D.M. (1986). Trimethyllysine and protein function. Effect of methylation and mutagenesis of lysine 115 of calmodulin on NAD kinase activation. J. Biol. Chem. 261: 1491–1494 [PubMed] [Google Scholar]
- Roberts D.M., Besl L., Oh S.H., Masterson R.V., Schell J., Stacey G. (1992). Expression of a calmodulin methylation mutant affects the growth and development of transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 89: 8394–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H., Kupsch C., Teubner M., Schmitz-Linneweber C. (2011). The RNA-recognition motif in chloroplasts. Journal of Plant Physiology 168: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Sang J., Zhang A., Lin F., Tan M., Jiang M. (2008). Cross-talk between calcium-calmodulin and nitric oxide in abscisic acid signaling in leaves of maize plants. Cell Res. 18: 577–588 [DOI] [PubMed] [Google Scholar]
- Schardl C.L., Byrd A.D., Benzion G., Altschuler M.A., Hildebrand D.F., Hunt A.G. (1987). Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61: 1–11 [DOI] [PubMed] [Google Scholar]
- Schiefelbein J.W., Somerville C. (1990). Genetic Control of Root Hair Development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J., Kurepa J., Yang P., Emborg T.J., Babiychuk E., Kushnir S., Vierstra R.D. (2003). The pleiotropic role of the 26S proteasome subunit RPN10 in Arabidopsis growth and development supports a substrate-specific function in abscisic acid signaling. Plant Cell 15: 965–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D., van Wijk K.J. (2009). PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Research 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttipanta N., Pattanaik S., Kulshrestha M., Patra B., Singh S.K., Yuan L. (2011). The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 157: 2081–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T., Kumon T., Azuma T., Nanmori T., Yasuda T. (2003). The expression of a germin-like protein with superoxide dismutase activity in the halophyte Atriplex lentiformis is differentially regulated by wounding and abscisic acid. Physiol. Plant. 118: 523–531 [Google Scholar]
- Tanz S.K., Castleden I., Hooper C.M., Vacher M., Small I., Millar H.A. (2013). SUBA3: A database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Research 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townley H.E., Knight M.R. (2002). Calmodulin as a potential negative regulator of Arabidopsis COR gene expression. Plant Physiol. 128: 1169–1172 [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.): S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S. (2010). Abscisic acid activates a Ca2+-calmodulin-stimulated protein kinase involved in antioxidant defense in maize leaves. Acta Biochim. Biophys. Sin. (Shanghai) 42: 646–655 [DOI] [PubMed] [Google Scholar]
- Yamniuk A.P., Vogel H.J. (2004). Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27: 33–57 [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B.W. (2000). Molecular and biochemical evidence for the involvement of calcium/calmodulin in auxin action. J. Biol. Chem. 275: 3137–3143 [DOI] [PubMed] [Google Scholar]
- Yin K., Han X., Xu Z., Xue H. (2009). Arabidopsis GLP4 is localized to the Golgi and binds auxin in vitro. Acta Biochim. Biophys. Sin. (Shanghai) 41: 478–487 [DOI] [PubMed] [Google Scholar]
- Zhu J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zik M., Arazi T., Snedden W.A., Fromm H. (1998). Two isoforms of glutamate decarboxylase in Arabidopsis are regulated by calcium/calmodulin and differ in organ distribution. Plant Mol. Biol. 37: 967–975 [DOI] [PubMed] [Google Scholar]
- Zulawski M., Braginets R., Schulze W.X. (2013). PhosPhAt goes kinases–Searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Research 41: D1176–D1184 [DOI] [PMC free article] [PubMed] [Google Scholar]