The expression of calmodulin isoforms with unique Ca2+ binding affinities could confer on plant cells an additional ability to translate the distinct Ca2+ signals that accompany various stimuli. Using isoforms of conserved and divergent calmodulin sequences, this study finds that differences in affinity and the effect of competing Mg2+ tunes the responsiveness of the isoforms to the appropriate Ca2+ signature.
Abstract
The discovery that plants contain multiple calmodulin (CaM) isoforms of variable sequence identity to animal CaM suggested an additional level of sophistication in the intracellular role of calcium regulation in plants. Past research has focused on the ability of conserved or divergent plant CaM isoforms to activate both mammalian and plant protein targets. At present, however, not much is known about how these isoforms respond to the signal of an increased cytosolic calcium concentration. Here, using isothermal titration calorimetry and NMR spectroscopy, we investigated the calcium binding properties of a conserved (CaM1) and a divergent (CaM4) CaM isoform from soybean (Glycine max). Both isoforms bind calcium with a semisequential pathway that favors the calcium binding EF-hands of the C-terminal lobe over those of the N-terminal lobe. From the measured dissociation constants, CaM4 binds calcium with a threefold greater affinity than CaM1 (Kd,Ca,mean of 5.0 versus 14.9 μM) but has a significantly reduced selectivity against the chemically similar magnesium cation that binds preferentially to EF-hand I of both isoforms. The implications of a potential magnesium/calcium competition on the activation of CaM1 and CaM4 are discussed in context with their ability to respond to stimulus-specific calcium signatures and their known physiological roles.
INTRODUCTION
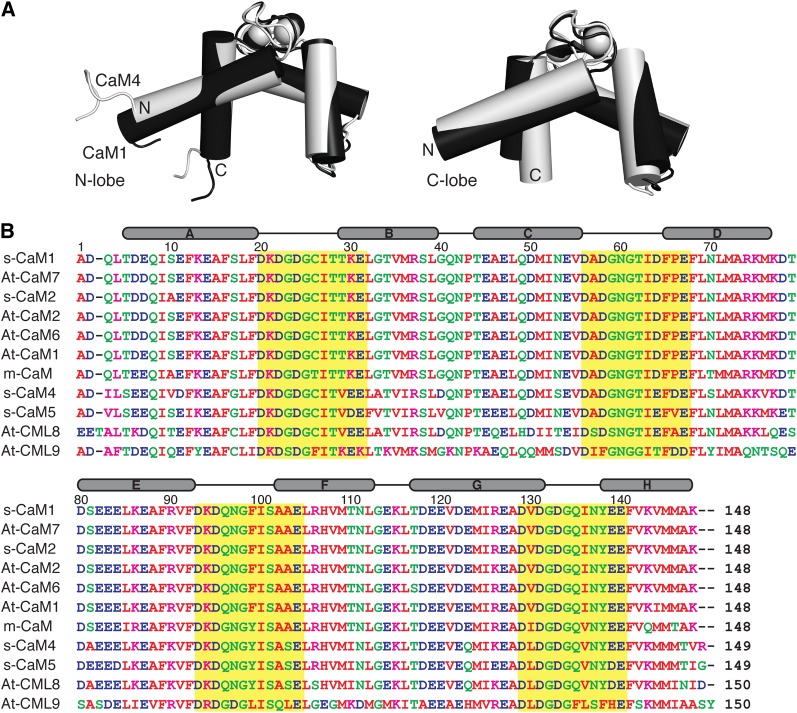
Ca2+ signaling systems are core regulators of the physiological and cellular processes of all eukaryotic organisms. In plants, a rapid and transient increase in the cytosolic and nuclear concentration of Ca2+ is observed in response to both abiotic (light, cold, heat, touch, wounding, drought, and oxidative stress) and biotic (phytohormones, pathogens, and symbiants) factors, triggering a myriad of cellular processes through which these organisms adapt to their environment (Reddy et al., 2011; Batistiĉ and Kudla, 2012). Analogous to other eukaryotes, this Ca2+ signal is intercepted via high-affinity Ca2+ binding proteins that regulate, in a Ca2+-dependent manner, the activity of their downstream targets. A major sensor of this Ca2+ signal in plant cells is the ubiquitous eukaryotic Ca2+ binding protein calmodulin (CaM) and the related CaM-like (CML) proteins (McCormack et al., 2005). Small, bilobal, and composed predominantly of α-helices, CaM contains two helix-loop-helix EF-hand Ca2+ binding motifs in each of its two globular domains (Figure 1A) (Gifford et al., 2007; Ishida et al., 2008). Upon binding Ca2+, each lobe of CaM undergoes a closed to open conformational change through which the side chains of buried hydrophobic amino acid residues, particularly Met residues, are exposed (Yamniuk and Vogel, 2005; Yamniuk et al., 2009; Gifford et al., 2011). This creates two hydrophobic pockets that drive target binding.
Figure 1.
Sequence Comparison and Ca2+-Bound Conformation of Soybean CaM1 and CaM4.
(A) Main-chain overlay of the N- and C-terminal lobes of Ca2+-CaM1 (black) and Ca2+-CaM4 (light gray) structures (Protein Data Bank codes: s-CaM1-N [2RO8], s-CaM1-C [2RO9], s-CaM4-N [2ROA], and s-CaM4-C [2ROB]). The structures overlay with a main-chain root mean squared deviation of 0.88 and 1.50 Å, respectively.
(B) Sequence alignment of conserved (s-CaM1/2, At-CaM1/2/6/7, and m-CaM) and divergent (s-CaM4/5) CaMs and select CMLs (At-CML8/9). The Ca2+ binding EF-loop is highlighted by a yellow box. The secondary structural elements are indicated above. The sequence conservation to s-CaM1 for each is as follows: At-CaM7 (0.99), s-CaM2 (0.98), At-CaM2 (0.98), At-CaM6 (0.98), At-CaM1 (0.97), m-CaM (0.90), s-CaM4 (0.79), s-CaM5 (0.79), At-CML8 (0.71), and At-CML9 (0.51).
Unlike animal cells, which have a single isoform of CaM encoded by three separate genes (Fischer et al., 1988), plants contain multiple CaM genes encoding several CaM isoforms with minor amino acid differences (Lee et al., 1995; Cho et al., 1998; Choi et al., 2002). These CaMs have varying degrees of sequence identity/similarity to the well-studied mammalian CaM (m-CaM) and represent conserved as well as more divergent CaMs. In addition to differing expression patterns that are modulated by developmental or environmental signals (Lee et al., 1995; Heo et al., 1999), in a number of studied plants, the various CaM isoforms selectively activate or inhibit CaM-dependent enzymes by virtue of a reciprocal target activation ability, and competition exists between the CaM isoforms for the CaM binding target proteins (Cho et al., 1998; Lee et al., 1999, 2000). In soybean (Glycine max), five known CaM genes encode four isoforms. Soybean CaM genes 1/3 and 2 (s-CaM1/3 and s-CaM2) encode CaMs very similar in sequence to the conserved plant CaMs identified in alfalfa (Medicago sativa), barley (Hordeum vulgare), and Arabidopsis thaliana, as well as to that found in mammals (90 identical to m-CaM) (Figure 1B). In contrast, s-CaM4 and s-CaM5 encode more novel CaMs (77 and 75% identical to m-CaM, respectively, both 79% identical to s-CaM1); s-CaM4 differs from s-CaM1 by 32 out of 149 amino acids with 15 of the substitutions being nonconserved exchanges. Structural studies on the conserved s-CaM1 and divergent s-CaM4 indicate that the two proteins adopt a highly similar protein fold in the Ca2+-bound state (Figure 1A) (Ishida et al., 2008), supporting mutational and biophysical experiments that point to the different activation abilities of the two isoforms as being due to very minor structural and chemical differences introduced by amino acid substitutions in the sequences of each protein (Kondo et al., 1999; Van Lierop et al., 2002; Yamniuk and Vogel, 2005). The sensitivity of CaM target proteins to the primary sequence of CaM underscores the potential for a sophisticated regulatory network given the diversity of CaMs and CaM binding proteins found in plants (Bouché et al., 2005).
An additional level of flexibility could be introduced into this signaling system by the different binding affinities of the various CaM isoforms for Ca2+, as this would modulate their ability to activate targets and hence create subsets of CaMs conditioned to decode particular Ca2+ signals. Here, we used biophysical techniques to investigate the Ca2+ binding properties of soybean CaM1 and CaM4 as well as the influence of the chemically similar magnesium cation (Mg2+) on the abilities of these proteins to bind Ca2+. We interpreted the observed Ca2+ binding abilities in the context of the specific physiological roles of these two CaM isoforms.
RESULTS AND DISCUSSION
Ca2+ Binds CaM1 and CaM4 through a Semisequential Binding Pathway Favoring the C-Terminal EF-Hands
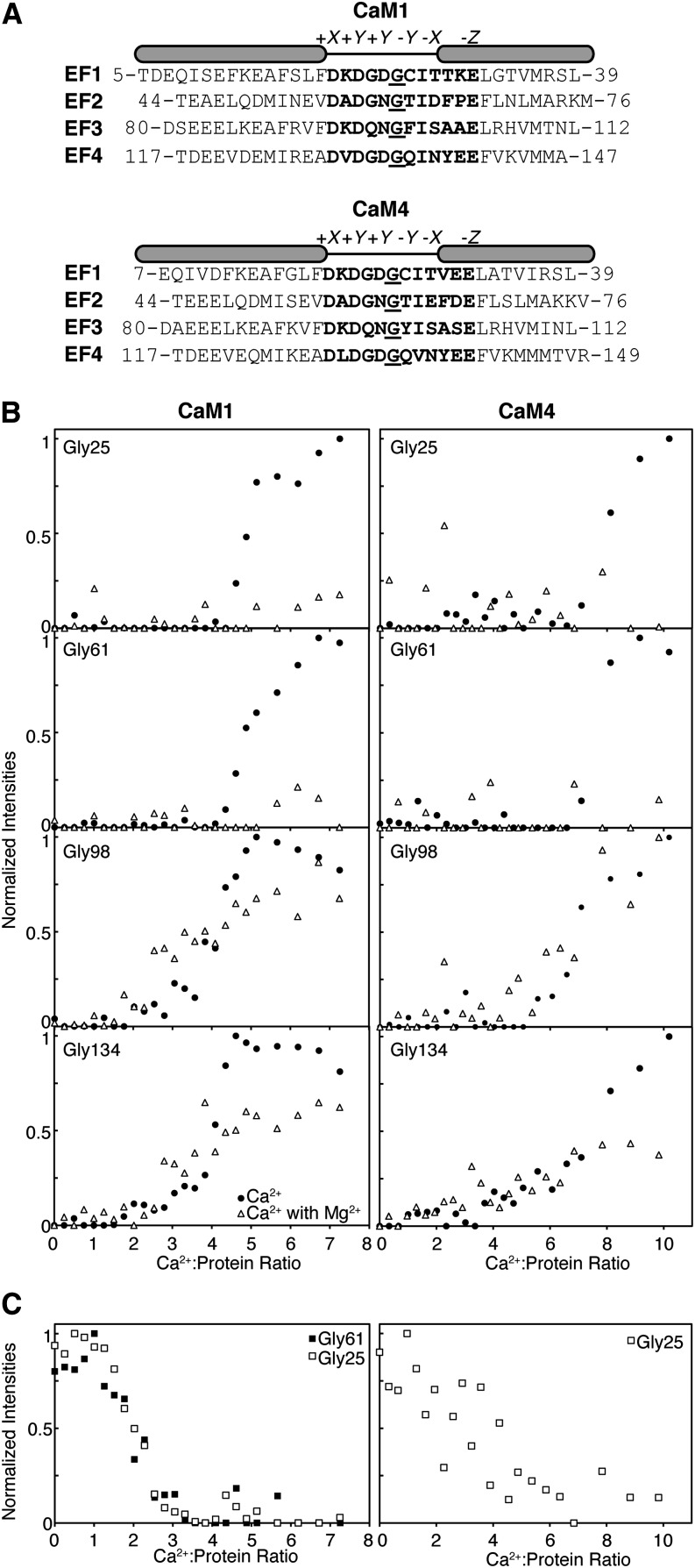
Analogous to other helix-loop-helix EF-hand Ca2+ binding proteins, the apo- and Ca2+-bound states of soybean CaM1 and CaM4 exist in distinct structural conformations (see Supplemental Figure 1 online). Readily observed in NMR spectra, the differences in conformation can be exploited to understand the binding pathway of the Ca2+ ions. Ca2+ titration of apo-CaM1 or apo-CaM4 was performed and monitored through rapidly acquired 1H-15N shift correlation experiments. In NMR spectroscopy, the spectra produced by the 1H-15N shift correlation class of experiments (including the [15N-1H]-HSQC [for heteronuclear single quantum coherence] or [15N-1H]-band-selective optimized-flip-angle short-transient heteronuclear multiple quantum coherence experiments used here) are very useful for studying protein–ligand interactions. In the [15N-1H]-HSQC spectra shown in Supplemental Figure 1 online, each dot represents the NMR signal (also known as a peak) from the amide (NH) group of each amino acid, and it is composed of the corresponding 1H and 15N chemical shift values. By comparing 1H-15N shift correlation spectra in the presence or absence of a ligand, protein–ligand interactions can be monitored by the broadening and/or position shift of each peak in the 1H-15N shift correlation spectrum. For many EF-hand proteins, including CaM1 and CaM4, strengthening of intraloop hydrogen bonds causes the main chain NH groups of homologous Gly residues found at the sixth position of the four Ca2+ binding EF-loops (Gly6: Gly-25, Gly-61, Gly-98, and Gly-134) to exhibit characteristic high (also described as downfield) 1H chemical shift values in their Ca2+-bound form (Figure 2A; see Supplemental Figure 1 online). The extent of this downfield shift places their corresponding NH peaks in the least crowded region of 1H-15N shift correlation spectra and enables them to act as indicators for the Ca2+-bound state of a given EF-hand (Mukherjee et al., 2007; Aravind et al., 2008). Figure 2B presents the plots of normalized Ca2+-bound Gly6 NH peak intensity as a function of the Ca2+:protein ratio for both CaM1 and CaM4. In the Ca2+ titration of both CaM isoforms, the Gly6 signals for the C-terminal lobe begin to reach their Ca2+-saturated chemical shift and intensity at Ca2+:protein ratios much lower than those of the N-terminal sites. The NH peaks of the Ca2+-bound C-terminal Gly6 residues of CaM1 (Gly-98 and Gly-134) develop simultaneously around a Ca2+:protein ratio of 2. By contrast, the peaks corresponding to the Ca2+-bound N-terminal Gly6 residues of this isoform (Gly-25 and Gly-61) develop together at a ratio of 4. The C-terminal Gly6 NH peaks of CaM4 show up at their Ca2+-saturated chemical shift almost together at a Ca2+:protein ratio near 4, while those of the N-terminal EF-hands appear together at a ratio of 7:1. The preference of Ca2+ for the C-terminal EF-hands is an equivalent binding pattern to that seen with m-CaM and is termed a semisequential binding pathway (Linse et al., 1991).
Figure 2.
Ca2+ Binds to the C-Terminal Lobe of CaM1 or CaM4 before the N-Terminal Lobe in Both the Absence and Presence of Saturating Mg2+.
(A) Alignment of the EF-hands of CaM1 and CaM4. The Ca2+-coordinating EF-loops are highlighted (bold), including the Gly6 used as a probe for cation binding (underlined). The positions of the amino acid ligands in Ca2+’s pentagonal bipyramidal coordinating geometry are indicated above.
(B) Plots of normalized main-chain NH peak intensities observed in 1H-15N shift correlation spectra for CaM1 or CaM4 EF-loop Gly6 residues as a function of the Ca2+:protein ratio in the absence and presence of MgCl2.
(C) Plots of normalized Gly6 NH peak intensities present in Mg2+-saturated 1H-15N shift correlation spectra as a function of Ca2+:protein ratio.
This experiment reports on the occupancy (through signal intensity) of the Ca2+-bound state of an EF-hand, not just the presence of a given Gly6 NH peak at its Ca2+-bound chemical shift. As a result, higher than stoichiometric Ca2+:protein ratios were required to reach saturation. Furthermore, the outcomes of the titrations, a gradual increase in intensity of the C-terminal Gly6 signals until saturation is observed versus the abrupt saturation of the N-terminal Gly6 peaks at higher stoichiometric ratios, reflect the different exchange rates through which Ca2+ binds to the two lobes of CaM1 and CaM4. Ca2+ binds in fast to intermediate exchange to the N-terminal lobe of both isoforms and in slow exchange to the C-terminal lobe. The type of exchange accompanying ligand binding is directly proportional to the affinity of the interaction and the discrepancy in exchange rates suggests that, for both isoforms, Ca2+ binds more weakly to the EF-hands of the N-terminal domain than to those of the C-terminal domain.
Ca2+ Binds apo-CaM4 with Greater Affinity than apo-CaM1
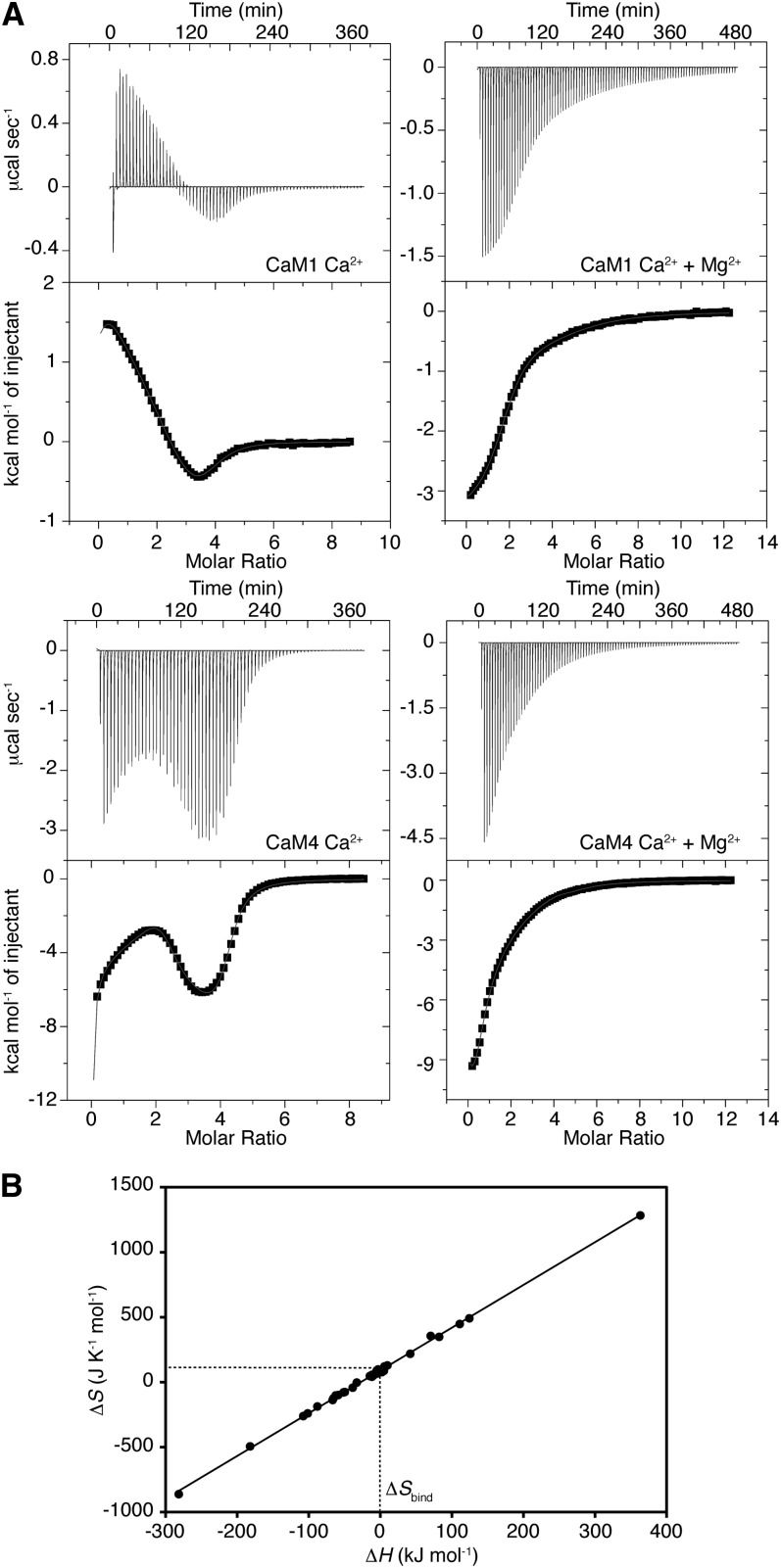
The energetics of Ca2+ binding to CaM1 and CaM4 were studied through isothermal titration calorimetry (ITC), a technique used to determine the thermodynamic parameters of an interaction by measuring the heat taken in (endothermic) or evolved (exothermic) when a ligand is titrated into a macromolecule (protein, DNA, etc.). Experimental data for the binding of Ca2+ to both CaM1 and CaM4 are presented in Figure 3A. The binding of Ca2+ to the apo-state of either isoform is a multiphasic process as both CaM1 and CaM4 display exothermic and endothermic Ca2+ binding sites. Data were fitted with the stoichiometry of Ca2+ binding fixed to four, following a procedure used previously for m-CaM and similar EF-hand-containing Ca2+ binding proteins (Gilli et al., 1998; Mukherjee et al., 2007; Dagher et al., 2011). Although the presence of auxiliary Ca2+ binding sites has been proposed for CaM (Lafitte et al., 1995), they are too weak to be measured by ITC due to the high concentration of Ca2+ required for saturation (Gilli et al., 1998). The values for the obtained Ca2+ dissociation constants (Kd1, Kd2, Kd3, and Kd4), corresponding to the strength of the first, second, third, and fourth Ca2+ binding site on CaM1 or CaM4, are presented in Supplemental Table 1 online. As the binding of Ca2+ is an electrostatic interaction, these values reflect the pH, buffer, and ion strength of the solutions used but are comparable to those previously reported for both m-CaM and a synthetic hybrid of animal and plant CaM (Linse et al., 1991; Gilli et al., 1998). The NMR titration data presented above allow us to assign the dissociation constants to specific lobes of CaM1 and CaM4: Kd1 and Kd2 belong to the C-terminal lobe of both isoforms, whereas Kd3 and Kd4 match the N-terminal lobes. CaM4 has an overall Ca2+ affinity thrice that of CaM1 (Kd,Ca,mean of 5.0 ± 0.8 μM versus 14.9 ± 1.8 μM, respectively) and binds Ca2+ with an 11 kJ mol−1 more favorable total Gibb’s free energy change (ΔG1,2,3,4), an affinity increase spread equally over both domains of CaM4 (Table 1). Furthermore, Ca2+ binds to the C-terminal EF-hands of CaM1 and CaM4 with an ∼10 kJ mol−1 greater affinity than those found in the N-terminal lobe of each isoform. This value is analogous to the interlobe disparity seen with m-CaM (Linse et al., 1991) and explains the Ca2+ binding preference and difference in exchange behavior observed in our NMR titration experiments (Figure 2B).
Figure 3.
Calorimetric Titration of CaM1 or CaM4 with Ca2+ in the Presence and Absence of MgCl2.
(A) Plots of kcal mol−1 of heat absorbed/released per injection of CaCl2 as a function of Ca2+:protein ratio in the absence and presence of 10 mM MgCl2. The least-squares fit of the data to a four site sequential binding model is given by the solid line.
(B) Enthalpy-entropy relationship for Ca2+ binding to the four EF-hands of CaM1 and CaM4. The data points represent the ΔH and ΔS for each Ca2+ binding site measured in 0, 0.5, 2, 5, 10, or 30 mM MgCl2. ΔSbind for Ca2+ can be determined from the y-intercept of the line described by ΔS = kΔH + ΔSintercept.
Table 1. Ca2+ Affinities and Free Energies of Binding for CaM1 or CaM4 in the Absence and Presence of Mg2+.
| CaM | Kd,N,ave (μM)a | Kd,C,ave (μM)b | Kd,ave (μM)c | ΔGN (kJ mol−1) | ΔGC (kJ mol−1) | ΔG1,2,3,4 (kJ mol−1) |
|---|---|---|---|---|---|---|
| s-CaM1 | 36.2 ± 7.4 | 6.2 ± 0.2 | 14.9 ± 1.8 | −51.8 ± 1.1 | −60.4 ± 0.1 | −112.2 ± 1.2 |
| + 10 mM Mg2+ | 107.8 ± 5.5 | 14.8 ± 0.6 | 39.8 ± 0.5 | −46.1 ± 0.2 | −56.1 ± 0.2 | −102.1 ± 0.0 |
| s-CaM4 | 13.0 ± 0.8 | 2.0 ± 0.5 | 5.0 ± 0.8 | −56.9 ± 0.7 | −66.3 ± 1.2 | −123.3 ± 1.8 |
| + 10 mM Mg2+ | 29.2 ± 6.6 | 34.1 ± 6.8 | 31.5 ± 6.7 | −52.8 ± 0.9 | −52.0 ± 1.0 | −104.9 ± 1.9 |
From our NMR data, sites 1 and 2 can be assigned to the C-terminal lobe and sites 3 and 4 to the N-terminal lobe.
Kd,N,ave = (K3K4)−1/2.
Kd,C,ave = (K1K2)−1/2.
Kd,ave = (K1K2K3K4)−1/4.
Ca2+ Binds to CaM1 and CaM4 with Different Energetic Driving Factors
The thermodynamic parameters that accompany the binding of each Ca2+ ion to the sites in CaM1 or CaM4 are presented in Table 2. As evidenced by the relationships presented in Methods (Equations 1 and 2), the strength of Ca2+’s interaction with each site is reflected in the magnitude of the decrease in free energy (−ΔG) and dictated by the accompanying changes in the system’s internal energy (also known as its enthalpy [ΔH]) and disorder (entropy [ΔS]). At a constant temperature (T = 30°C), a larger magnitude increase in ΔS or decrease in ΔH leads to higher affinity binding. The binding of Ca2+ to apo-CaM1 is an entropically driven process. The large, favorable value of the −TΔS term dominates Ca2+ binding to all four EF-hands of CaM1 and compensates for the endothermic and, thus, unfavorable ΔH that accompanies Ca2+ binding to all but one EF-hand (Ca2+ binding site 3; Table 2). Ca2+ binding to CaM4 is impelled by a different set of energetic factors: Two of the sites are enthalpically favorable but entropically unfavorable (sites 1 and 4), while two are enthalpically unfavorable and entropically favorable (sites 2 and 3).
Table 2. Thermodynamic Parameters for the Binding of Ca2+ to CaM1 or CaM4.
| CaM | No Mg2+ |
+ 10 mM Mg2+ |
||||
|---|---|---|---|---|---|---|
| ΔH (kJ mol−1)a | −TΔS (kJ mol−1) | ΔS (J K−1 mol−1)b | ΔH (kJ mol−1) | −TΔS (kJ mol−1) | ΔS (J K−1 mol−1) | |
| s-CaM1 | ||||||
| Site 1 | 5.6 ± 0.1 | −36.7 ± 0.1 | 121.1 ± 0.3 | −14.7 ± 0.2 | −14.3 ± 0.4 | 47.1 ± 1.4 |
| Site 2 | 9.8 ± 1.0 | −39.1 ± 1.1 | 129.2 ± 3.5 | −12.5 ± 0.1 | −14.6 ± 0.0 | 48.3 ± 0.1 |
| Site 3 | −11.0 ± 0.3 | −17.5 ± 0.2 | 57.9 ± 0.7 | 4.9 ± 1.2 | −26.5 ± 1.2 | 87.3 ± 4.0 |
| Site 4 | 0.5 ± 0.5 | −23.7 ± 0.7 | 78.2 ± 2.2 | −11.8 ± 0.8 | −12.6 ± 0.6 | 41.7 ± 2.1 |
| s-CaM4 | ||||||
| Site 1 | −101.6 ± 36.4 | 72.8 ± 36.8 | −240.2 ± 121.6 | −49.8 ± 3.3 | 22.9 ± 3.9 | −75.6 ± 12.8 |
| Site 2 | 70.3 ± 35.5 | −107.8 ± 37.1 | 355.9 ± 122.5 | −66.7 ± 7.1 | 41.5 ± 7.0 | −137.1 ± 23.0 |
| Site 3 | 124.3 ± 55.0 | −148.9 ± 53.5 | 491.3 ± 176.5 | 110.8 ± 2.7 | −135.7 ± 2.6 | 447.7 ± 8.7 |
| Site 4 | −182.0 ± 55.4 | 149.7 ± 54.6 | −494.0 ± 180.2 | −65.6 ± 4.7 | 37.6 ± 4.9 | −124.0 ± 16.0 |
From our NMR data, sites 1 and 2 can be assigned to the C-terminal lobe and sites 3 and 4 to the N-terminal lobe.
ΔH = ΔHbind + ΔHconf, where ΔHbind = 0.
ΔS = ΔSbind + ΔSconf, where ΔSbind = 90.6 J K−1 mol−1 (see text for details).
The protein–Ca2+ interaction is driven by two main thermodynamic mechanisms: (1) the desolvation of the cation as the water molecules that surround the cation are replaced by ligands from the protein (represented by ΔHbinding [ΔHbind] and ΔSbinding [ΔSbind]), and (2) the formation of favorable inter–amino acid residue contacts induced by a Ca2+-dependent protein conformational change (ΔHconformation [ΔHconf] and ΔSconformation [ΔSconf]) (Gilli et al., 1998). The ΔH and ΔS determined using ITC are composed as follows: ΔH = ΔHbind + ΔHconf and ΔS = ΔSbind + ΔSconf. As the same ligand coordination geometry surrounds Ca2+ in water and in EF-hand Ca2+ binding loops, ΔHbind can be approximated to zero and the measured ΔH corresponds primarily to ΔHconf (Table 2) (Gilli et al., 1998; Yamniuk et al., 2004; Mukherjee et al., 2007). It is possible to discriminate between ΔSbind and ΔSconf if we measure the thermodynamic parameters of the protein–Ca2+ interaction under different experimental conditions (Figure 3B). For both isoforms, plots of ΔH versus ΔS are linear under all conditions tested. This indicates that there is a direct relationship between the enthalpy and entropy changes observed upon Ca2+ binding, suggesting a clear example of enthalpy-entropy compensation (Grunwald and Steel, 1995). Taking into account this relationship and that ΔHbind = 0, ΔS can then be defined as ΔS = kΔH + ΔSintercept, where ΔSintercept is equal to the entropy of Ca2+ desolvation (ΔSbind). From our data, ΔSbind = 90.6 J K−1 mol−1, a value that agrees well with that obtained previously for a synthetic CaM (Gilli et al., 1998).
Using this information, we can identify the enthalpic and entropic contributions to the overall energetics of the Ca2+–CaM1/4 interaction and glean information on the driving factors for the differences in Ca2+ affinity of the two isoforms. The binding of Ca2+ to three of the four EF-hands of CaM1 is accompanied by unfavorable changes in ΔHconf (Table 2), a predictable outcome as the Ca2+-dependent helical rearrangements lead to a loss of interhelical interactions. By subtracting ΔSbind (90.6 J K−1 mol−1) from the entropy values listed in Table 2, we can calculate ΔSconf for Ca2+ binding to CaM1. Values of 31 and 39 J K−1 mol−1 for sites 1 and 2 suggest that Ca2+ binding to the C-terminal lobe of CaM1 induces a higher degree of conformational flexibility in the molecule. By contrast, the binding of this cation to the N-terminal EF-hands of CaM1 is accompanied by ΔSconf values of −33 and −12 J K−1 mol−1 for sites 3 and 4, respectively. This loss in entropy can be attributed to a combination of increased molecular rigidity and the Ca2+-induced exposure of hydrophobic target binding patches (see fluorescence spectroscopy data using 8-anilino-1-naphthalene sulfonic acid [ANS] below). Taken as a whole, our ITC data suggest the binding of Ca2+ to CaM1 is accompanied by energetically costly conformational changes and is therefore driven by the favorable desolvation of the Ca2+ cation (ΔSbind).
The energetics of the interaction of Ca2+ with CaM4 are in contrast with those of CaM1. The magnitudes of the measured enthalpy and calculated entropy changes are much higher, and Ca2+ binding to the first EF-hand of the C-terminal lobe and last EF-hand of the N-terminal lobe is accompanied by favorable changes in enthalpy and overall unfavorable changes in entropy (Table 2; ΔSconf = −331 and −585 J K−1 mol−1 for sites 1 and 4, respectively). These thermodynamic values likely reflect the induced structure formation observed for this isoform upon binding Ca2+ (see Supplemental Figure 1 online; Huang et al., 2010), a process accompanied by favorable changes in enthalpy due to electrostatic and hydrogen bond formation and a corresponding loss of flexibility and decrease in system entropy. The binding of Ca2+ to the partner EF-hands of these sites is enthalpically unfavorable and paired with an increase in conformational entropy (ΔSconf = 266 and 401 J K−1 mol−1 for sites 2 and 3, respectively), likely reflecting the loss of interhelical contacts as the buried hydrophobic side chains are exposed. Altogether, these ITC data suggest that CaM4’s increased Ca2+ affinity can be attributed to the structural instability of the apo-state of CaM4, and both energetically favorable conformational changes and favorable cation desolvation accompany Ca2+ binding to this isoform.
Mg2+ Binds to the EF-Loops of CaM1 and CaM4
Although the total Mg2+ content in a plant cell ranges from 15 to 25 mM, most Mg2+ cations are complexed with cellular components (phospholipids, phosphonucleotides, proteins, chlorophyll, etc.), and the free Mg2+ concentration is much lower, in the range of 0.4 to 0.5 mM (Waters, 2011). This concentration can rise to 1 mM or higher as a result of rapid expenditures of the Mg2+-chelator ATP and drop in pH that occur when the plant is stressed (Igamberdiev and Kleczkowski, 2001, 2011). The comparable chemical properties of Mg2+ and Ca2+ (similar atomic size; preference for carboxyl, carbonyl, or hydroxyl protein ligands) lead to a complicated case of cation selectivity as Ca2+ binding proteins must discriminate against a 102- to 104-fold excess of the Mg2+ cation (Gifford et al., 2007; Gifford and Vogel, 2013). Like many other EF-hand proteins, both CaM1 and CaM4 bind Mg2+, and 1H-15N shift correlation spectra indicate the Mg2+-bound conformations of the two isoforms are both folded and distinct from the apo- or Ca2+-bound structures (see Supplemental Figure 1 online; Huang et al., 2010).
Controversy has surrounded the location and number of Mg2+ binding sites in m-CaM, and Mg2+ has been proposed to act either as a direct competitor or as an allosteric regulator of Ca2+ binding to this CaM (Ohki et al., 1997; Gilli et al., 1998; Malmendal et al., 1999). The accepted model now considers Mg2+ to be a direct competitor of Ca2+ for the EF-hands of m-CaM, and NMR data, particularly the downfield shifted Gly6 NH signals, point to the Mg2+ binding sites being located within the EF-loop structure. The 1H-15N shift correlation spectrum of Mg2+-bound CaM1 or CaM4 show two downfield shifted Gly6 residues for CaM1 and one downfield shifted Gly6 residue for CaM4 (see Supplemental Figure 1 online). The assignment of these peaks in the Mg2+-CaM1 NMR spectrum as belonging to residues Gly-25 and Gly-61 of EF-hands I and II, respectively, was determined by conventional triple resonance experiments. A previous NMR signal assignment of Mg2+-CaM4 identifies the single downfield residue as Gly-25 of EF-hand I (Huang et al., 2010).
The relative strength of Mg2+’s affinity for the EF-hands of both isoforms can be inferred from peak quality in 1H-15N shift correlation spectra and signal saturation observed in NMR Mg2+ titrations. Much like that seen previously with soybean CaM4 (Huang et al., 2010), only the N-terminal domain of Mg2+-CaM1 gave rise to a near complete assignment of the NMR signals from the main chain atoms (85 and 92% for CaM1 and CaM4, respectively; compared with total assignments of 76 and 69%; see Supplemental Figure 2A online). By contrast, the C-terminal domains of both isoforms have a number of missing signals attributable to peak broadening. The majority of the missing signals are centered in or near EF-loops III and IV, suggesting that the weak interaction of Mg2+ with these C-terminal EF-hands is causing localized fluctuations in the protein conformation. Alternatively, the conformational stability observed for the N-terminal lobe of both proteins points to Mg2+ binding preferentially to EF-hands I and II. Of these two EF-hands, signal saturation at lower Mg2+ concentrations in an NMR titration experiment points to EF-hand I of apo-CaM1 as being the higher affinity Mg2+ interaction site (see Supplemental Figure 3 online). Although the Gly6 NH peak of Mg2+-CaM4 EF-hand II is not observed in 1H-15N shift correlation spectra, signals corresponding to other main chain NH groups of the EF-loop are present, and the loss of this peak likely reflects a weaker Mg2+ affinity at this site than EF-hand I (Huang et al., 2010). Interestingly, these results with plant CaMs differ from that seen with m-CaM in which again all four EF-hands bind Mg2+, but EF-hands I and IV do so with the strongest affinity (Ohki et al., 1997).
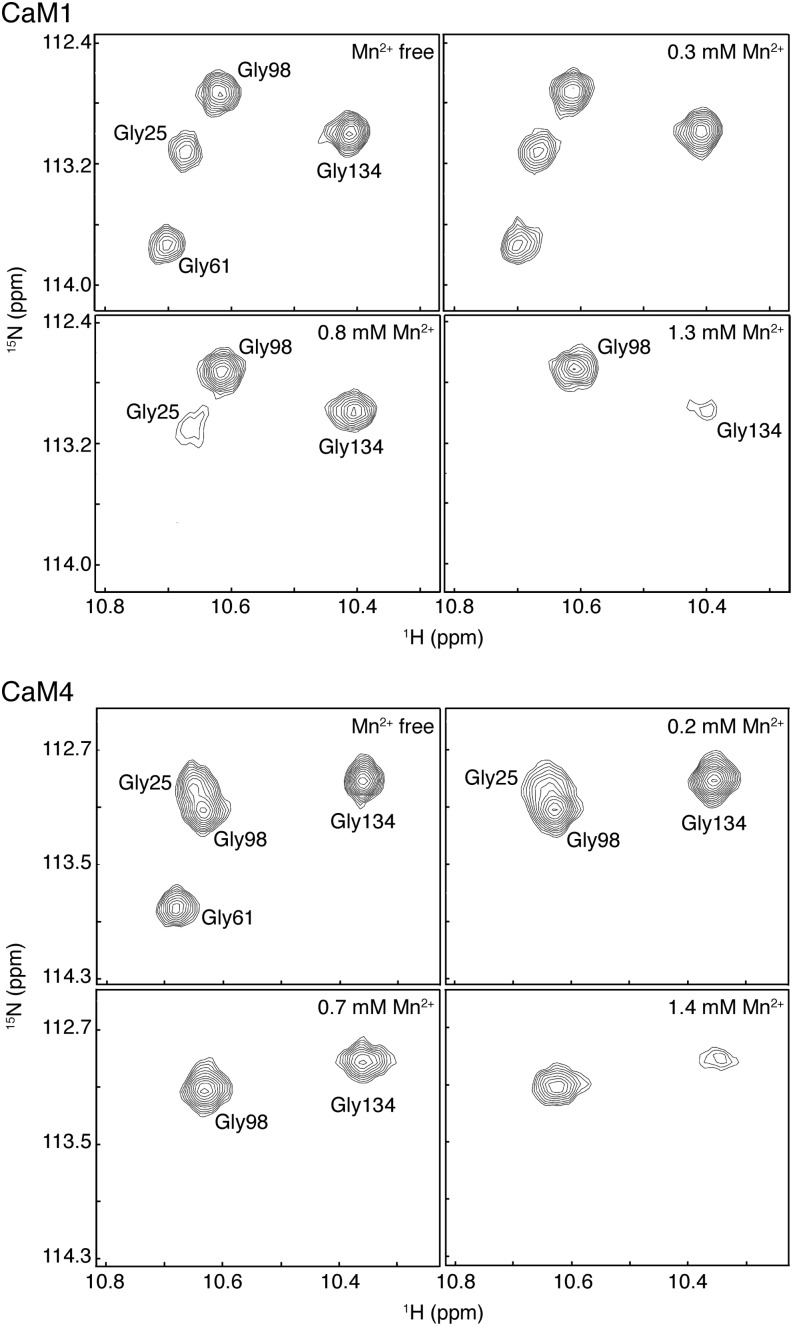
The preference of Mg2+ for the EF-hands of the N-terminal lobes of CaM1 and CaM4 as well as the localized binding of this cation to the EF-loops is supported by studies using the paramagnetic manganese cation (Mn2+). Mn2+ has an ionic radius close to that of Mg2+ (0.67 Å versus 0.72 Å, respectively, compared with 1.06 Å for Ca2+; Senguen and Grabarek, 2012), and both ions favor six ligands arranged in an octahedral geometry, but the unpaired nature of the nuclear spins in this cation cause the broadening/loss of nearby NMR signals (Aravind et al., 2008). At low Mn2+ concentrations and in the presence of Ca2+ and Mg2+, the N-terminal Gly6 NH peaks of Ca2+-bound CaM1 or CaM4 are observed to broaden and disappear first, indicating a ready interaction with these EF-hands (Figure 4). By contrast, no observable changes were seen for Gly-98 and Gly-134 of EF-hands III and IV at this low concentration. As the amount of Mn2+ is increased, the Gly-134 peak of both CaM1 and CaM4 is seen to broaden, while little change is observed for Gly-98, even at higher Mn2+ concentrations. This observation suggests that Mg2+ binds EF-III of both isoforms with the weakest affinity. In addition to showing the direct influence of the paramagnetic ion on the NMR signal of critical residues of the EF-loop, attributable to the presence of Ca2+, this competition experiment is also indicative of the relative Ca2+ binding strength of the various EF-loops. For both CaM1 and CaM4, Mn2+ appears to have the greatest affinity for EF-hand II, contrasting with Mg2+, which, as our results above suggest, binds the most strongly to EF-hand I. This discrepancy could be explained by the greater affinity of EF-hand I for Ca2+ compared with EF-hand II. Because of this increased Ca2+ affinity, Mn2+ likely cannot readily compete the Ca2+ off and binds EF-hand II first.
Figure 4.
Paramagnetic Effects of Mn2+ on the Ca2+-Bound Spectra of CaM1 or CaM4.
Selected regions of [15N-1H]-HSQC spectra recorded during the Mn2+ titration of 15N-labeled CaM1 or CaM4 showing Gly-25, Gly-61, Gly-98, and Gly-134 main-chain NH peaks at different Mn2+ concentrations. For these titrations, 509 and 706 μM CaM1 and CaM4, respectively, were used.
Mg2+ Stabilizes the Closed Conformation of CaM1 and CaM4
Ca2+ titration of Mg2+-bound CaM1 or CaM4 leads to the gradual disappearance of the Mg2+-associated peaks in 1H-15N shift correlation spectra and the concomitant appearance of those belonging to Ca2+-bound CaM1 or CaM4 (Figures 2B and C). This suggests both isoforms attain the same conformation after binding Ca2+ in the presence and absence of Mg2+, an observation predicated by the enthalpy-entropy compensation observed for Ca2+ binding to these isoforms under varying concentrations of Mg2+ (Figure 3B). Structural changes induced by the Mg2+ to Ca2+ transition can be probed by comparing the main chain 1H- and 15N-NMR chemical shifts of Mg2+- and Ca2+-bound CaM1 or CaM4 (see Supplemental Figure 2A online). In the presence of Mg2+, Ca2+ binding to the domains of both isoforms leads to drastic changes in helix orientation and EF-loop conformation, protein structural rearrangements manifested in main chain NH chemical shift perturbations (CSPs) of a magnitude much larger than that seen on average for the Ca2+-CaM:protein target interaction (Gifford et al., 2012). For both isoforms, the largest CSPs are observed to occur in the EF-loops, particularly in the loops’ N-terminal (Asp-20 [+X], Lys-21/Ala-57) and hinge regions (Ile-27, Thr-28/Ser-101 [−X]). Upon Ca2+ chelation, both of these regions play key roles in the induced closed-to-open conformational change (see Supplemental Figure 2B online; Grabarek, 2011). Differences here indicate an altered accommodation by the EF-loop ligands for the smaller Mg2+ and suggest that in the presence of this cation the EF-hands remain closed. The inability of Mg2+ to open the EF-hands of soybean CaM1 or CaM4 is analogous to that seen with Mg2+-bound m-CaM and S100G (a member of the S100 family of EF-hand proteins) (Andersson et al., 1997; Senguen and Grabarek, 2012), and is supported by previous structural work on the Mg2+-bound N-terminal domain of s-CaM4 [Supplemental Figure 2B online; (Huang et al., 2010)].
The stabilization of the “closed” state of all CaM1 and CaM4 EF-hands when bound to Mg2+ was further confirmed by fluorescence spectroscopy studies using the hydrophobic fluorophore ANS. ANS fluorescence is strongly dependent on the local environment and is significantly weaker in water where it occurs with a maximum wavelength of 515 nm (see Supplemental Figure 2C online). Because of the similarities with its fluorescence spectrum in water, we can conclude that neither apo- nor Mg2+-bound CaM1 or CaM4 interact significantly with ANS. By contrast, in the presence of Ca2+ and either isoform, the emission spectrum of ANS is marked by a considerable enhancement in the associated fluorescence and a blue shift of 37 nm. Both effects point to the exposure of large target binding hydrophobic pockets in this state, corroborated by the solution structures of both isoforms in the presence Ca2+ (Ishida et al., 2008). Taken together, our NMR and fluorescence spectroscopy data imply that although Mg2+ binds the Ca2+ binding EF-loops of CaM1 and CaM4, it does not mimic Ca2+, and these EF-hands remain functionally specific for the latter cation.
Mg2+ Acts as a Competitive Antagonist of Ca2+ Binding to the EF-Hands of CaM1 and as a Competitive Antagonist and Allosteric Activator of CaM4
As would be expected for a cation that competes with Ca2+ for the EF-loops, in many EF-hand proteins, the influence of Mg2+ often manifests itself as a decrease in the measured Ca2+ affinity through inhibitory effects on the kinetic on-rate of Ca2+ binding as Ca2+ association becomes dependent on Mg2+ dissociation (see Equation 3 in Methods; Trigo-Gonzalez et al., 1992; Ohki et al., 1997; Malmendal et al., 1999). Correspondingly, Ca2+ titrations of Mg2+-CaM1 or Mg2+-CaM4 monitored via 1H-15N shift correlation spectra point to higher concentrations of Ca2+ as being required for EF-hand saturation in the presence of 30 mM Mg2+ (Figure 2B). This result is particularly apparent for the N-terminal EF-hands of both isoforms, where at Ca2+:protein ratios at which saturation is observed in the absence of Mg2+, the Gly6 NMR signals have yet to approximate their Ca2+-bound chemical shift. Significantly higher Ca2+ concentrations are required to produce the Ca2+-bound chemical shift and intensity of the N-terminal Gly6 NH peaks, an unsurprising outcome as our titrations with Mn2+ and Mg2+ as well as CSP data point to the higher affinity Mg2+ binding sites as being located in this domain of both isoforms (Figure 4; see Supplemental Figure 2A online). Of note, despite Mg2+’s apparent effects on Ca2+ affinity, the Ca2+ binding preference of the EF-hands of both isoforms remains unchanged: the C-terminal sites are occupied before the those in the N terminus.
The influence of Mg2+ on the Ca2+ affinities of CaM1 and CaM4 was investigated using ITC (Figure 3A), and the Ca2+ dissociation constants in the presence of 10 mM Mg2+ are presented in Supplemental Table 1 online. The presence of Mg2+ decreases the overall Ca2+ affinity of both isoforms by 2.7- and 6.3-fold for CaM1 and CaM4, respectively (Table 1), and unfavorable changes in respective total free energy changes [ΔΔGCa(Mg)1,2,3,4] of 10.1 and 18.4 kJ mol−1 were observed under these conditions. On a domain-by-domain basis, the EF-hands of CaM1’s N-terminal lobe are more affected than those of the C-terminal lobe [ΔΔGCa(Mg)N = 5.7 kJ mol−1 versus ΔΔGCa(Mg)C = 4.3 kJ mol−1]. By contrast, the C-terminal EF-hands of CaM4 are more affected than those of the N-terminal domain [ΔΔGCa(Mg)N = 4.1 kJ mol−1 versus ΔΔGCa(Mg)C = 14.3 kJ mol−1], an unexpected finding as our NMR data point to the EF-hands of the N-terminal domain as containing the higher affinity Mg2+ binding sites for both isoforms (Figures 2B and 4; see Supplemental Figures 1 and 2A online). This discrepancy suggests Mg2+ acts not only as a competitive antagonist to Ca2+ binding to the N-terminal EF-hands of CaM4 but that it also serves as an allosteric activator, increasing the observed Ca2+ affinity of these EF-hands by folding the domain and thus paying the entropic cost in conformational energy of cation binding. As a result, the increased intrinsic Ca2+ affinity of the folded EF-hand pair in the presence of Mg2+ masks the competitive effect of the smaller cation and the ΔΔGCa(Mg) for Ca2+ binding to this domain in the absence and presence of Mg2+ is less than would be expected.
Also apparent from the presented ITC data is the effect of MgCl2 on the thermodynamic driving factors of Ca2+ binding to CaM1 and CaM4 (Figure 3A). Upon analysis, the much simpler and predominantly exothermic isotherms for Ca2+ binding to either isoform reflect the favorable ΔHconf that accompanies the binding of this cation to all but one EF-hand of CaM1 and CaM4 (Table 2). For CaM1, this process is accompanied by unfavorable changes in conformational entropy for all four EF-hands (ΔSconf values of −43.5, −42.3, −3.3, and −48.0 J K−1 mol−1 for the Ca2+ binding sites in order of affinity, respectively). Both trends can be explained by the conformational instability of Mg2+-bound CaM1 (see Supplemental Figure 2A online). The binding of Ca2+ first to the C-terminal and then the N-terminal EF-loops would stabilize the isoform’s conformation, leading to the formation of energetically favorable contacts (and, thus, a negative ΔHconf) but also the rigidification of CaM1 and the accompanying unfavorable changes in ΔSconf. Similarly, the interaction of Ca2+ with Mg2+-CaM4 is characterized by favorable changes in enthalpy and overall unfavorable changes in entropy for all but one EF-hand. As evidenced by the missing peaks in NMR spectra, the conformational instability of the C-terminal lobe of Mg2+-CaM4 is greater than that of Mg2+-CaM1 (see Supplemental Figure 2A online), and as a result, the adverse loss of molecular flexibility is higher, and unfavorable changes in conformational entropy (ΔSconf = −166.2, −227.7, 357.1, and −214.6 J K−1 mol−1, for the bindings sites, respectively) eclipse the favorable entropy of Ca2+ desolvation (90.6 J K−1 mol−1; see above). For both isoforms, one Ca2+ binding site remains enthalpically unfavorable and entropically favorable: site 3. From these characteristics, the identify of this site is likely EF-hand I as, in the presence of Mg2+, this site is well formed and structurally stable.
Competition from Free Mg2+ Affects the Activating Ca2+ Concentration of CaM1 and CaM4
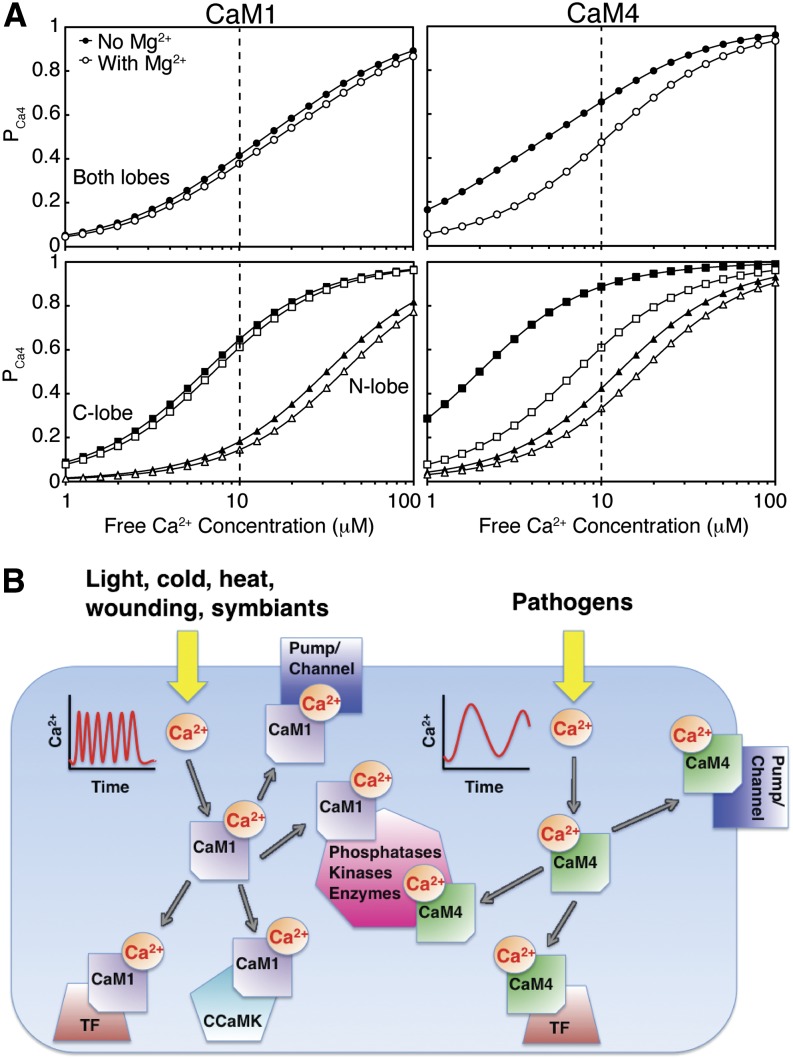
Unlike Ca2+, the binding of Mg2+ to these proteins is accompanied by a monophasic endothermic reaction (CaM1) or limited heat release (CaM4), hindering direct measurement of the Mg2+ binding affinities. However, as Mg2+ acts as a competitive antagonist of the Ca2+ binding EF-loops, the mean value of the Mg2+ dissociation constants, Kd,Mg,mean, can be determined from the effect this cation has on the measured Ca2+ dissociation constants (see Equation 5 in Methods). Calculated Kd,Mg,mean values of 6.3 and 2.0 mM for s-CaM1 and s-CaM4, respectively, agree with that previously determined for the N-terminal lobe of m-CaM (Malmendal et al., 1999) but are significantly lower than that reported for other EF-hand-containing proteins for which Mg2+ affinities as high as 10−6 M have been measured (Aravind et al., 2008). Under stress Mg2+ concentrations (1 mM Mg2+), competition from this cation decreases the Kd,Ca,mean,app by a factor of 1.2 and 1.7 for soybean CaM1 and CaM4, respectively. As a consequence, Mg2+ effectively lowers the population of the Ca2+ loaded state of CaM4 by a factor of 1.4 to 3.0 over the entire range of global cytosolic Ca2+ conditions (Figure 5A). By contrast, as a result of its increased selectivity for Ca2+, the drop in saturation for CaM1 is only a factor of 1.1 to 1.2. The effects of Mg2+ competition are most prominent for the C-terminal domain of CaM4, where the presence of this cation reduces the population of the Ca2+-loaded state by a factor of 1.5 to 3.7. In the absence of a CaM-binding domain that can modulate the observed Ca2+ affinity (Peersen et al., 1997; Gifford et al., 2007), under these conditions, no more than 40% of CaM1 or 50% of CaM4 molecules are ever saturated with Ca2+. The colocalization of CaM1 or CaM4 near a plasma membrane or organelle Ca2+ release channel could also increase the degree of saturation in either isoform as, due to the slow diffusion rate of this metal cation, the Ca2+ concentration in these microdomains can be orders of magnitude higher than that measured globally in the cytoplasm (Grant et al., 2000; Monshausen, 2012; Steinhorst and Kudla, 2013).
Figure 5.
Ca2+ Affinities and Selectivities of CaM1 and CaM4 Are Tuned to Their Physiological Roles.
(A) Populations of Ca2+-loaded CaM1 and CaM4 as a function of free [Ca2+] in the absence and presence of 1 mM Mg2+. The curves were calculated using the Adair equation: 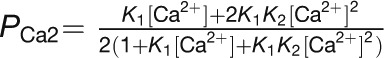 and
and 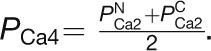 . Dashed line represents the upper limit of the global concentration of cytosolic free Ca2+.
. Dashed line represents the upper limit of the global concentration of cytosolic free Ca2+.
(B) Schematic representing the extracellular stimulants, approximate Ca2+ signature, and different intermolecular interactions of CaM1 and CaM4. TF, transcription factor; CCaMK, Ca2+- and Ca2+-CaM–dependent protein kinase.
The Ca2+ Affinities and Selectivities of CaM1 and CaM4 Are Tuned to Their Physiological Roles
Although all CaM isoforms are ubiquitously expressed in various plant tissues and show similar subcellular localization (Lee et al., 1995, 1999), cellular levels of CaM4 are much lower than that of CaM1 and, instead, rapidly and dramatically rise in specific response to pathogen elicitors or salt stress (Heo et al., 1999; Park et al., 2004b). By contrast, the same treatments have no effect on CaM1 expression levels. The role of CaM4 as a central component of plant host-defense pathways is beginning to emerge, and several target–protein interactions have been identified (Figure 5B) (Kondo et al., 1999; Park et al., 2004a; Alkharouf et al., 2006; Boursiac et al., 2010; Spalding and Harper, 2011). Ca2+ influx into the cytosol and nucleoplasm is an early plant cell response to pathogen attack, serving as a messenger to regulate specific downstream defense pathways (Ma et al., 2008; Reddy et al., 2011). In plant cells, although an increase in cytoplasmic Ca2+ concentration is an indicator of a wide variety of stimuli, each stimulus is accompanied by a Ca2+ signal of characteristic amplitude, duration, and oscillation frequency that is subsequently interpreted by Ca2+ binding proteins (McAinsh and Pittman, 2009; Batistič and Kudla, 2012). The sustained level of cytosolic/nuclear Ca2+ evoked by pathogen effectors differs considerably from the repetitive oscillation patterns seen during abiotic stresses or host–symbiant interactions (Lecourieux et al., 2002; Moscatiello et al., 2006; Dodd et al., 2010). During avirulent interactions, this continuous wave of Ca2+ is key to the induction of host-defense genes, one of which is CaM4, as well as the subsequent hypersensitive resistance response, the process of localized programmed cell death used to stop pathogen spread at a site of infection (Grant et al., 2000; Thuleau et al., 2013).
Our analysis suggests the differing sensitivities of CaM1 and CaM4 to both Ca2+ and Mg2+ would help to fine-tune each isoform to activate its required protein targets. Due to the constitutive high expression levels of CaM1/3, it is likely this isoform responds to the individualized, fast, spiking signals characteristic of abiotic stresses or root nodule host-symbiant relationships (McAinsh and Pittman, 2009; Wu and Jinn, 2012). Competition from Mg2+ does not significantly affect Ca2+ binding to CaM1, enabling this isoform to rapidly respond to the Ca2+ signal. By contrast, the reduced selectivity of CaM4 for the Mg2+ cation and the resulting Mg2+/Ca2+ competition would slow the response of this isoform to an increased cytoplasmic Ca2+ concentration as the association of Ca2+ is now dependent on Mg2+ dissociation (see Equation 3 in Methods). The effect of this Mg2+/Ca2+ competition on the responsiveness of CaM4 was observed previously in CaM-target enzyme activation assays. In MgCl2-containing experiments comparing the Ca2+-dependent activation ability of CaM1 and CaM4 on representative CaM-dependent enzymes (phosphodiesterase, CaM-dependent kinase II, or smooth muscle myosin light chain kinase), CaM4 displayed a reduced sensitivity to Ca2+, and, thus, activation ability, despite binding these same protein targets with a similar intermolecular affinity as CaM1 (Lee et al., 2000; Van Lierop et al., 2002; Yamniuk and Vogel, 2005), and Ca2+ with a higher affinity, as presented here. As a result of the influence of Mg2+, the energetics of Ca2+’s interaction with CaM4 are more favorable to the sustained increase in cytosolic Ca2+ concentration that accompanies host–pathogen interaction, and the higher Ca2+ affinity of CaM4 would mean it would successfully compete with CaM1 for the Ca2+ signal under these conditions, ensuring the CaM–target protein interactions required for pathogen resistance get precedence. The plant nitric oxide synthase (NOS) could be regulated in such a manner. Along with an elevated cytoplasmic Ca2+ concentration, the free radical nitric oxide is known to play a role in plant defense and the hypersensitive resistance response (Ma et al., 2008). The activation of animal NOS is Ca2+-CaM dependent, and biochemical and genetic evidence points to a similar regulatory mechanism for the plant enzyme (Ma et al., 2008). Plant NOS, recently identified and characterized in the green alga Ostreococcus tauri, is 45% similar to animal NOS and includes a CaM binding domain (Foresi et al., 2010). Both isoforms of soybean CaM bind animal NOS with equal strength, but this CaM-dependent enzyme is inhibited by s-CaM1 and activated by s-CaM4 (Cho et al., 1998; Yamniuk and Vogel, 2005). The hindering effects of a Mg2+/Ca2+ competition on the responsiveness of s-CaM4 would further tune the activation of NOS to be timed with the appropriate Ca2+ signal; in the absence of the sustained elevated cytoplasmic Ca2+ concentration that accompanies pathogen attack, the effects of this cation competition on CaM4 would facilitate CaM1 binding NOS, thus preventing enzyme activation.
In conclusion, we characterized the Ca2+ and Mg2+ binding abilities of two plant CaM isoforms: one representing a conserved CaM (s-CaM1) and one a divergent isoform (s-CaM4). Although determined in vitro, these binding constants represent the intrinsic Ca2+ affinities of these isoforms and thus are physiologically important. Our results suggest that in addition to having differential target regulation abilities, the different CaMs also have unique affinities for the activating Ca2+ cation. That these proteins would have differing affinities would not be readily predicted from their primary amino acid sequences. A high degree of identity/similarity exists between the CaMs of different plant species, and our findings can likely be extended to close homologs of soybean CaM1: s-CaM2 (alfalfa CaM, barley CaM, and At-CaM1/2/6/7) and s-CaM4 (s-CaM5, At-CML8, and antigens present in Chinese cabbage [Brassica rapa chinensis], rice [Oryza sativa], and tobacco [Nicotiana tabacum]) (Lee et al., 1995; Zielinski, 2002; Park et al., 2010) (Figure 1B). Because of their sessile nature it is perhaps not surprising that Ca2+ signaling pathways are more sophisticated in plants, and it appears that differing responsiveness toward the Ca2+ signal adds to the complexity of the disparity in target activation abilities already observed for these isoforms. Our knowledge of the plant stress response, of significant application in agriculture, relies on understanding the divergent nature of the CaM isoforms.
METHODS
Sequence Alignment
Amino acid sequences for m-CaM, CaMs, and select CMLs from soybean (Glycine max) and Arabidopsis thaliana were aligned with ClustalW2 using the default alignment parameters: Gonnet protein weight matrix, gap open and extension scores of 10 and 0.2, respectively, and a gap distance score of 5 (Larkin et al., 2007).
Protein Expression and Purification
CaM1 and CaM4 from soybean were overexpressed and purified from Escherichia coli strain BL21 (DE3), containing the pET-3d plasmid as described previously (Ishida et al., 2008). 15N-labeled or 13C,15N-labeled CaM1 and CaM4 were prepared in M9 media containing 0.5 g/L 15NH4Cl and 3 g/L of either unlabeled or 13C-labeled Glc. All proteins were purified to homogeneity by Ca2+-dependent phenyl-Sepharose chromatography (Ishida et al., 2008). The concentrations of CaM1 and CaM4 were determined using ε276 CaM1 = 1450 M−1 cm−1 and ε276 CaM4 = 2900 M−1 cm−1. For the biophysical studies described here, plastic ware and glass NMR tubes were rinsed with 1 M HCl, and Chelex-100–treated buffers and ultrapure grade chemicals from Sigma-Aldrich were employed in order to minimize Ca2+ contamination. Exogenous Ca2+ was removed from CaM1 and CaM4 samples using a Calcium Sponge S column (Molecular Probes).
Fluorescence Spectroscopy
Steady state ANS binding experiments were recorded on a Varian Cary Eclipse fluorescence spectrophotometer at 25°C with an excitation wavelength of 370 nm, and the fluorescence emission was recorded from 400 to 600 nm. The excitation and emission slit widths were 5 and 5 nm or 5 and 10 nm for CaM1 and CaM4, respectively. Samples contained 120 μM ANS and 20 μM CaM1 or CaM4 in a buffer containing 20 mM HEPES, pH 7.5, 100 mM KCl, and 1 mM DTT. For all experiments, the sample volume was 1 mL.
ITC
All ITC experiments were performed on a MicroCal VP-ITC microcalorimeter: 100 μM CaM1 or CaM4 was titrated with 5.1 mM CaCl2 in 20 mM HEPES, pH 7.5, 1 mM tris(2-carboxyethyl)phosphine, and either 100 mM KCl with 0, 0.5, or 2 mM MgCl2, 70 mM KCl with 10 mM MgCl2, or 10 mM KCl with 30 mM MgCl2. Prior to each experiment, the ITC sample cell was soaked with 10% EDTA to remove contaminating Ca2+ and subsequently washed extensively with Ca2+-free buffer. Data were fitted using the four sequential sites model in the MicroCal Origin software to determine the binding constant (K) and the ΔH associated with binding. The ΔH and K values were then used to calculate the ΔS through the following relationships:
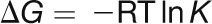 |
(1) |
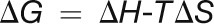 |
(2) |
The fitted K values are influenced by the kinetic on- (kon) and off-rate (koff) of Ca2+ binding through the relationship:
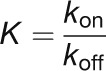 |
(3) |
and were converted to Kd values using:
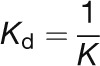 |
(4) |
The reported values represent the average and se of three independent titrations performed at 30°C. The strength of Mg2+ binding (Kd,Mg,mean) can be estimated from the effect of Mg2+ on the observed Ca2+ binding constants (KCa,mean,app at 10 mM MgCl2 versus KCa,mean at 0 mM MgCl2) using an equation describing the competition of two ions for a single site (Bryant, 1985):
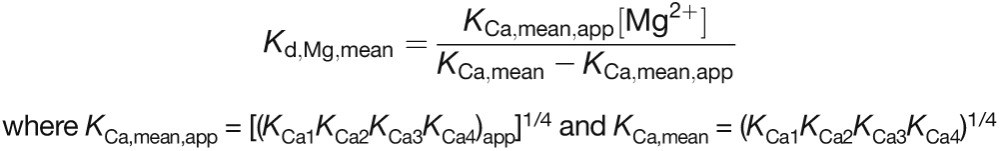 |
(5) |
where KCa,mean,app = [(KCa1KCa2KCa3KCa4)app]1/4 and KCa,mean = (KCa1KCa2KCa3KCa4)1/4
NMR Spectroscopy
All NMR experiments were performed on a Bruker Avance 500 MHz spectrometer equipped with a triple resonance inverse cryogenic probe. Main-chain NMR signal assignment of Mg2+-bound CaM1 was completed with two-dimensional [15N-1H]-HSQC and three-dimensional HNCACB, CBCACONH, HNCACO, and HNCO experiments using ∼0.5 mM 13C,15N-labeled CaM1, 100 mM KCl, 30 mM MgCl2, 10 mM DTT, and 0.5 mM 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) in 90% water/10% 2H2O), pH 6.8. The main-chain NMR signal assignments of Mg2+-CaM4, Ca2+-CaM1, and Ca2+-CaM4 were obtained from previous publications in which they were acquired under similar conditions (Ishida et al., 2008; Huang et al., 2010).
Ca2+ and Mg2+ titrations were performed with 40.4 and 40.6 mM stocks of CaCl2 and MgCl2, respectively. The protein concentration was ∼0.3 mM 15N-labeled CaM1 or CaM4 in 20 mM HEPES, pH 7.5, 100 mM KCl, 1 mM DTT, 90% water/10% 2H2O, and either 0 or 30 mM MgCl2. For each titration, an aliquot of the stock solution was added to the NMR tube containing the protein solution and mixed, and a two-dimensional [15N-1H]-selective optimized-flip-angle short-transient heteronuclear multiple quantum coherence experiment was recorded at 30°C (Schanda et al., 2005). The total experimental time for each spectrum was 3 min. [15N-1H]-HSQC spectra of apo-, Mg2+-, and Ca2+-bound CaM1 or CaM4 (both in the presence and absence of 30 mM MgCl2) were also acquired.
Mn2+ titrations were performed with ∼0.5 mM Ca2+-bound CaM1 or ∼0.7 mM CaM4 in 20 mM HEPES, pH 7.5, 100 mM KCl, 30 mM MgCl2, 5 mM DTT, 90% water/10% 2H2O, and 0.5 mM DSS, with a 16-fold molar excess of CaCl2. Mn2+ was titrated into the sample from a 100 mM MnCl2 stock solution. The titrations were performed at 30°C and monitored using [15N-1H]-HSQC spectra. No titration was performed beyond a Mn2+:protein ratio of 2 when the water peak started showing broadening due to the paramagnetic Mn2+ (Aravind et al., 2008).
Chemical shifts in the assignment spectra were referenced using DSS to obtain the 1H, 15N, and 13C chemical shifts. All spectra were processed using the program NMRPipe (Delaglio et al., 1995) and analyzed using the NMRView software (Johnson and Blevins, 1994). CSPs in the [15N-1H]-HSQC spectra of Ca2+- and Mg2+-bound CaM1 or CaM4 were calculated as the weighted averaged chemical shift difference of the 1H and 15N resonances according to the equation (Grzesiek et al., 1996):
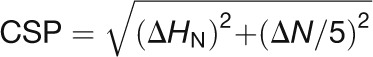 |
(6) |
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: s-CaM1 (NP_001238237.1), s-CaM4 (AAA341015.1), and m-CaM (AAD45181.1). The accession numbers for the amino acid sequences used in the sequence alignment can be found in Supplemental Table 2 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. [15N-1H]-HSQC Spectra of Ca2+-Bound, Mg2+-Bound, or Apo-15N-Labeled CaM1 or CaM4 with the Assigned EF-Hand Gly6 Peaks Indicated.
Supplemental Figure 2. Conformational Changes Induced upon Binding of Ca2+- to Mg2+-CaM1 or Mg2+-CaM4.
Supplemental Figure 3. Overlay of [15N-1H]-HSQC Spectra Monitoring the Mg2+ Titration of 0.2 mM 15N-Labeled CaM1.
Supplemental Table 1. Individual Dissociation Constants for Ca2+ Binding to CaM1 or CaM4 in the Absence and Presence of Mg2+.
Supplemental Table 2. Protein Names and IDs Used for the Sequence Alignment.
Supplemental References 1. References for Supplemental Figures 1-3.
Supplemental Data Set 1. Alignments Used to Generate the Sequence Alignment in Figure 1.
Acknowledgments
This research was supported by an operated grant from the Natural Sciences and Engineering Research Council of Canada. H.J.V. holds a scientist award from the Alberta Heritage Foundation for Medical Research.
AUTHOR CONTRIBUTIONS
J.L.G. designed the research, performed research, analyzed data, and wrote the article. M.J. analyzed data. J.M. performed research. H.I. contributed new analytical tools. H.J.V. designed the research and wrote the article.
Glossary
- CaM
calmodulin
- m-CaM
mammalian CaM
- HSQC
heteronuclear single quantum coherence
- ITC
isothermal titration calorimetry
- ANS
8-anilino-1-naphthalene sulfonic acid
- CSP
chemical shift perturbation
- NOS
nitric oxide synthase
- DSS
2,2-dimethyl-2-silapentane-5-sulfonate
References
- Alkharouf N.W., Klink V.P., Chouikha I.B., Beard H.S., MacDonald M.H., Meyer S., Knap H.T., Khan R., Matthews B.F. (2006). Timecourse microarray analyses reveal global changes in gene expression of susceptible Glycine max (soybean) roots during infection by Heterodera glycines (soybean cyst nematode). Planta 224: 838–852 [DOI] [PubMed] [Google Scholar]
- Andersson M., Malmendal A., Linse S., Ivarsson I., Forsén S., Svensson L.A. (1997). Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k. Protein Sci. 6: 1139–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind P., Chandra K., Reddy P.P., Jeromin A., Chary K.V., Sharma Y. (2008). Regulatory and structural EF-hand motifs of neuronal calcium sensor-1: Mg2+ modulates Ca2+ binding, Ca2+-induced conformational changes, and equilibrium unfolding transitions. J. Mol. Biol. 376: 1100–1115 [DOI] [PubMed] [Google Scholar]
- Batistič O., Kudla J. (2012). Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 1820: 1283–1293 [DOI] [PubMed] [Google Scholar]
- Bouché N., Yellin A., Snedden W.A., Fromm H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56: 435–466 [DOI] [PubMed] [Google Scholar]
- Boursiac Y., Lee S.M., Romanowsky S., Blank R., Sladek C., Chung W.S., Harper J.F. (2010). Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol. 154: 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant D.T. (1985). Quin 2: The dissociation constants of its Ca2+ and Mg2+ complexes and its use in a fluorimetric method for determining the dissociation of Ca2+-protein complexes. Biochem. J. 226: 613–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M.J., Vaghy P.L., Kondo R., Lee S.H., Davis J.P., Rehl R., Heo W.D., Johnson J.D. (1998). Reciprocal regulation of mammalian nitric oxide synthase and calcineurin by plant calmodulin isoforms. Biochemistry 37: 15593–15597 [DOI] [PubMed] [Google Scholar]
- Choi J.Y., et al. (2002). Identification of calmodulin isoform-specific binding peptides from a phage-displayed random 22-mer peptide library. J. Biol. Chem. 277: 21630–21638 [DOI] [PubMed] [Google Scholar]
- Dagher R., Peng S., Gioria S., Fève M., Zeniou M., Zimmermann M., Pigault C., Haiech J., Kilhoffer M.C. (2011). A general strategy to characterize calmodulin-calcium complexes involved in CaM-target recognition: DAPK and EGFR calmodulin binding domains interact with different calmodulin-calcium complexes. Biochim. Biophys. Acta 1813: 1059–1067 [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Dodd A.N., Kudla J., Sanders D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Fischer R., Koller M., Flura M., Mathews S., Strehler-Page M.A., Krebs J., Penniston J.T., Carafoli E., Strehler E.E. (1988). Multiple divergent mRNAs code for a single human calmodulin. J. Biol. Chem. 263: 17055–17062 [PubMed] [Google Scholar]
- Foresi N., Correa-Aragunde N., Parisi G., Caló G., Salerno G., Lamattina L. (2010). Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 22: 3816–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford J.L., Ishida H., Vogel H.J. (2011). Fast methionine-based solution structure determination of calcium-calmodulin complexes. J. Biomol. NMR 50: 71–81 [DOI] [PubMed] [Google Scholar]
- Gifford J.L., Ishida H., Vogel H.J. (2012). Structural insights into calmodulin-regulated L-selectin ectodomain shedding. J. Biol. Chem. 287: 26513–26527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford, J.L., and Vogel, H.J. (2013). EF-hand proteins and magnesium. In Encyclopedia of Metalloproteins, R.H. Kretsinger, V.N. Uversky, and E.A. Permyakov, eds (New York: Springer), pp. 775–783. [Google Scholar]
- Gifford J.L., Walsh M.P., Vogel H.J. (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 405: 199–221 [DOI] [PubMed] [Google Scholar]
- Gilli R., Lafitte D., Lopez C., Kilhoffer M., Makarov A., Briand C., Haiech J. (1998). Thermodynamic analysis of calcium and magnesium binding to calmodulin. Biochemistry 37: 5450–5456 [DOI] [PubMed] [Google Scholar]
- Grabarek Z. (2011). Insights into modulation of calcium signaling by magnesium in calmodulin, troponin C and related EF-hand proteins. Biochim. Biophys. Acta 1813: 913–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M., Brown I., Adams S., Knight M., Ainslie A., Mansfield J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23: 441–450 [DOI] [PubMed] [Google Scholar]
- Grunwald E., Steel C. (1995). Solvent reorganization and thermodynamic enthalpy-entropy compensation. J. Am. Chem. Soc. 117: 5687–5692 [Google Scholar]
- Grzesiek S., Bax A., Clore G.M., Gronenborn A.M., Hu J.S., Kaufman J., Palmer I., Stahl S.J., Wingfield P.T. (1996). The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 3: 340–345 [DOI] [PubMed] [Google Scholar]
- Heo W.D., Lee S.H., Kim M.C., Kim J.C., Chung W.S., Chun H.J., Lee K.J., Park C.Y., Park H.C., Choi J.Y., Cho M.J. (1999). Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. USA 96: 766–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Ishida H., Vogel H.J. (2010). The solution structure of the Mg2+ form of soybean calmodulin isoform 4 reveals unique features of plant calmodulins in resting cells. Protein Sci. 19: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev A.U., Kleczkowski L.A. (2001). Implications of adenylate kinase-governed equilibrium of adenylates on contents of free magnesium in plant cells and compartments. Biochem. J. 360: 225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev A.U., Kleczkowski L.A. (2011). Magnesium and cell energetics in plants under anoxia. Biochem. J. 437: 373–379 [DOI] [PubMed] [Google Scholar]
- Ishida H., Huang H., Yamniuk A.P., Takaya Y., Vogel H.J. (2008). The solution structures of two soybean calmodulin isoforms provide a structural basis for their selective target activation properties. J. Biol. Chem. 283: 14619–14628 [DOI] [PubMed] [Google Scholar]
- Johnson B.A., Blevins R.A. (1994). NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4: 603–614 [DOI] [PubMed] [Google Scholar]
- Kondo R., Tikunova S.B., Cho M.J., Johnson J.D. (1999). A point mutation in a plant calmodulin is responsible for its inhibition of nitric-oxide synthase. J. Biol. Chem. 274: 36213–36218 [DOI] [PubMed] [Google Scholar]
- Lafitte D., Capony J.P., Grassy G., Haiech J., Calas B. (1995). Analysis of the ion binding sites of calmodulin by electrospray ionization mass spectrometry. Biochemistry 34: 13825–13832 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lecourieux D., Mazars C., Pauly N., Ranjeva R., Pugin A. (2002). Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell 14: 2627–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Johnson J.D., Walsh M.P., Van Lierop J.E., Sutherland C., Xu A., Snedden W.A., Kosk-Kosicka D., Fromm H., Narayanan N., Cho M.J. (2000). Differential regulation of Ca2+/calmodulin-dependent enzymes by plant calmodulin isoforms and free Ca2+ concentration. Biochem. J. 350: 299–306 [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., et al. (1995). Identification of a novel divergent calmodulin isoform from soybean which has differential ability to activate calmodulin-dependent enzymes. J. Biol. Chem. 270: 21806–21812 [DOI] [PubMed] [Google Scholar]
- Lee S.H., et al. (1999). Competitive binding of calmodulin isoforms to calmodulin-binding proteins: Implication for the function of calmodulin isoforms in plants. Biochim. Biophys. Acta 1433: 56–67 [DOI] [PubMed] [Google Scholar]
- Linse S., Helmersson A., Forsén S. (1991). Calcium binding to calmodulin and its globular domains. J. Biol. Chem. 266: 8050–8054 [PubMed] [Google Scholar]
- Ma W., Smigel A., Tsai Y.C., Braam J., Berkowitz G.A. (2008). Innate immunity signaling: Cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 148: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmendal A., Linse S., Evenäs J., Forsén S., Drakenberg T. (1999). Battle for the EF-hands: Magnesium-calcium interference in calmodulin. Biochemistry 38: 11844–11850 [DOI] [PubMed] [Google Scholar]
- McAinsh M.R., Pittman J.K. (2009). Shaping the calcium signature. New Phytol. 181: 275–294 [DOI] [PubMed] [Google Scholar]
- McCormack E., Tsai Y.C., Braam J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10: 383–389 [DOI] [PubMed] [Google Scholar]
- Monshausen G.B. (2012). Visualizing Ca(2+) signatures in plants. Curr. Opin. Plant Biol. 15: 677–682 [DOI] [PubMed] [Google Scholar]
- Moscatiello R., Mariani P., Sanders D., Maathuis F.J. (2006). Transcriptional analysis of calcium-dependent and calcium-independent signalling pathways induced by oligogalacturonides. J. Exp. Bot. 57: 2847–2865 [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Mohan P.M., Chary K.V. (2007). Magnesium promotes structural integrity and conformational switching action of a calcium sensor protein. Biochemistry 46: 3835–3845 [DOI] [PubMed] [Google Scholar]
- Ohki S., Ikura M., Zhang M. (1997). Identification of Mg2+-binding sites and the role of Mg2+ on target recognition by calmodulin. Biochemistry 36: 4309–4316 [DOI] [PubMed] [Google Scholar]
- Park C.Y., et al. (2004a). Pathogenesis-related gene expression by specific calmodulin isoforms is dependent on NIM1, a key regulator of systemic acquired resistance. Mol. Cells 18: 207–213 [PubMed] [Google Scholar]
- Park H.C., et al. (2004b). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135: 2150–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.C., Park C.Y., Koo S.C., Cheong M.S., Kim K.E., Kim M.C., Lim C.O., Lee S.Y., Yun D.J., Chung W.S. (2010). AtCML8, a calmodulin-like protein, differentially activating CaM-dependent enzymes in Arabidopsis thaliana. Plant Cell Rep. 29: 1297–1304 [DOI] [PubMed] [Google Scholar]
- Peersen O.B., Madsen T.S., Falke J.J. (1997). Intermolecular tuning of calmodulin by target peptides and proteins: Differential effects on Ca2+ binding and implications for kinase activation. Protein Sci. 6: 794–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.S., Ali G.S., Celesnik H., Day I.S. (2011). Coping with stresses: Roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell 23: 2010–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schanda P., Kupce E., Brutscher B. (2005). SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 33: 199–211 [DOI] [PubMed] [Google Scholar]
- Senguen F.T., Grabarek Z. (2012). X-ray structures of magnesium and manganese complexes with the N-terminal domain of calmodulin: Insights into the mechanism and specificity of metal ion binding to an EF-hand. Biochemistry 51: 6182–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding E.P., Harper J.F. (2011). The ins and outs of cellular Ca(2+) transport. Curr. Opin. Plant Biol. 14: 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorst L., Kudla J. (2013). Calcium and ROS rule the waves of signaling. Plant Physiol. 163: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuleau P., Aldon D., Cotelle V., Brière C., Ranty B., Galaud J.P., Mazars C. (2013). Relationships between calcium and sphingolipid-dependent signalling pathways during the early steps of plant-pathogen interactions. Biochim. Biophys. Acta 1833: 1590–1594 [DOI] [PubMed] [Google Scholar]
- Trigo-Gonzalez G., Racher K., Burtnick L., Borgford T. (1992). A comparative spectroscopic study of tryptophan probes engineered into high- and low-affinity domains of recombinant chicken troponin C. Biochemistry 31: 7009–7015 [DOI] [PubMed] [Google Scholar]
- Van Lierop J.E., Wilson D.P., Davis J.P., Tikunova S., Sutherland C., Walsh M.P., Johnson J.D. (2002). Activation of smooth muscle myosin light chain kinase by calmodulin. Role of LYS(30) and GLY(40). J. Biol. Chem. 277: 6550–6558 [DOI] [PubMed] [Google Scholar]
- Waters B.M. (2011). Moving magnesium in plant cells. New Phytol. 190: 510–513 [DOI] [PubMed] [Google Scholar]
- Wu H.C., Jinn T.L. (2012). Oscillation regulation of Ca2+/calmodulin and heat-stress related genes in response to heat stress in rice (Oryza sativa L.). Plant Signal. Behav. 7: 1056–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamniuk A.P., Nguyen L.T., Hoang T.T., Vogel H.J. (2004). Metal ion binding properties and conformational states of calcium- and integrin-binding protein. Biochemistry 43: 2558–2568 [DOI] [PubMed] [Google Scholar]
- Yamniuk A.P., Vogel H.J. (2005). Structural investigation into the differential target enzyme regulation displayed by plant calmodulin isoforms. Biochemistry 44: 3101–3111 [DOI] [PubMed] [Google Scholar]
- Yamniuk A.P., Ishida H., Lippert D., Vogel H.J. (2009). Thermodynamic effects of noncoded and coded methionine substitutions in calmodulin. Biophys. J. 96: 1495–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski R.E. (2002). Characterization of three new members of the Arabidopsis thaliana calmodulin gene family: Conserved and highly diverged members of the gene family functionally complement a yeast calmodulin null. Planta 214: 446–455 [DOI] [PubMed] [Google Scholar]