Pathway engineering has been the predominant strategy used to enhance valuable natural products in plants. This study identifies sequestration mechanisms associated with pathway engineering outputs that enable the plastid organelle to adapt and accommodate newly synthesized precursors and products.
Abstract
Metabolic engineering of the carotenoid pathway in recent years has successfully enhanced the carotenoid contents of crop plants. It is now clear that only increasing biosynthesis is restrictive, as mechanisms to sequestrate these increased levels in the cell or organelle should be exploited. In this study, biosynthetic pathway genes were overexpressed in tomato (Solanum lycopersicum) lines and the effects on carotenoid formation and sequestration revealed. The bacterial Crt carotenogenic genes, independently or in combination, and their zygosity affect the production of carotenoids. Transcription of the pathway genes was perturbed, whereby the tissue specificity of transcripts was altered. Changes in the steady state levels of metabolites in unrelated sectors of metabolism were found. Of particular interest was a concurrent increase of the plastid-localized lipid monogalactodiacylglycerol with carotenoids along with membranous subcellular structures. The carotenoids, proteins, and lipids in the subchromoplast fractions of the transgenic tomato fruit with increased carotenoid content suggest that cellular structures can adapt to facilitate the sequestration of the newly formed products. Moreover, phytoene, the precursor of the pathway, was identified in the plastoglobule, whereas the biosynthetic enzymes were in the membranes. The implications of these findings with respect to novel pathway regulation mechanisms are discussed.
INTRODUCTION
Carotenoids are a large class of natural yellow, orange, and red pigments, which color fruits, flowers, birds, and crustacea (Hirschberg, 2001; Fraser and Bramley, 2004). Moreover, in higher plants, carotenoids act as free-radical scavengers (Demmig-Adams and Adams, 2002), ancillary photosynthetic pigments (Dall’Osto et al., 2007), and precursors of phytohormones (Auldridge et al., 2006). Dietary carotenoids with biological antioxidant properties have been intensively studied with respect to their potential in alleviating age-related diseases in humans (Fraser and Bramley, 2004; Krinsky and Johnson, 2005). These health-promoting properties, along with their ability to act as natural colorants, have created intense biotechnological interest to increase or alter their contents in foodstuffs and to develop new, renewable sources that are capable of competing with chemical synthesis, which for many carotenoids is the present production method of choice.
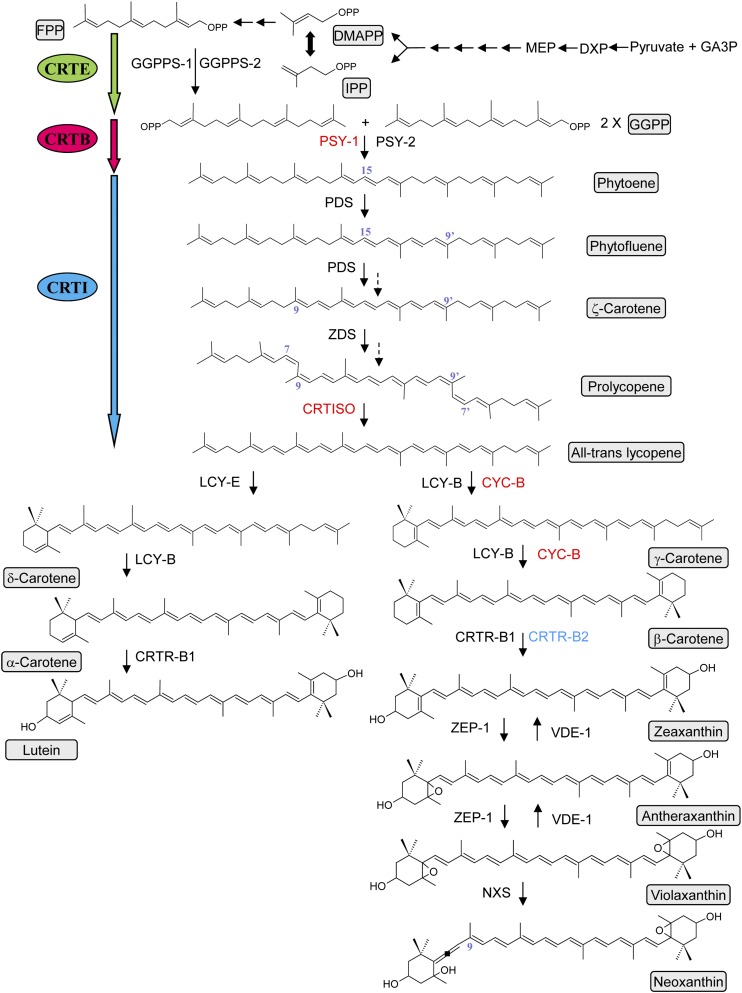
Biosynthetically, carotenoids are isoprenoids synthesized in the plastid, using isopentenyl diphosphate (C5) derived from the methylerythritol-4-phosphate pathway (Pulido et al., 2012). The conversion of two geranylgeranyl diphosphate (C20) molecules into phytoene (C40) represents the first committed step in the carotenoid pathway and is catalyzed by the enzyme phytoene synthase. Phytoene is a colorless carotene with three conjugated double bonds that can then undergo a sequential series of desaturations and isomerizations to form all-trans lycopene, which has a characteristic red coloration as a result of the 11 conjugated double bonds. In plants and algae, the optimal desaturation and isomerization of phytoene to lycopene requires four proteins, a phytoene and ζ-carotene desaturase, and two isomerases acting on ζ-carotene and poly cis-lycopene (Sandmann, 2009). Depending on the action and specificity of the cyclase enzymes, lycopene can then undergo cyclization to form β- or ε-ionone rings, yielding β-carotene and/or α-carotene. These cyclic carotenoids can then be decorated by hydroxylation and epoxidation reactions (Fraser and Bramley, 2004; Figure 1). Transcriptional regulation of the carotenoid pathway has been well documented (Fraser and Bramley, 2004; Fraser et al., 2009).
Figure 1.
Representative Scheme of the Carotenoid Biosynthetic Pathway in Higher Plants.
Enzymes in red are tomato fruit ripening specific or enhanced, and those in blue are flower specific. GA3P, glyceraldehyde-3-phosphate; DXP, 1-deoxy-d-xylulose 5-phosphate; MEP, 2-C-methyl-d-erythritol 4-phosphate; IPP, isopentenyl diphosphate; DMAPP, dimethylallyl diphosphate; FPP, farnesyl diphosphate; GGPP, geranylgeranyl diphosphate; GGPPS-1 and -2, geranylgeranyl diphosphate synthase; PSY-1, fruit-specific phytoene synthase-1; PSY-2, phytoene synthase-2; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; CRTISO, carotene isomerase; LCY-E, ε-lycopene cyclase; LCY-B, β-lycopene cyclase; CYC-B, fruit-specific β-lycopene cyclase; CRTR-B1, carotene β-hydroxylase 1; CRTR-B2, carotene β-hydroxylase 2 (flower specific); ZEP, zeaxanthin epoxidase; NXS, neoxanthin synthase; VDE, violaxanthin deepoxidase; CRTE, geranylgeranyl diphosphate synthase; CRTB, phytoene synthase; and CRTI, phytoene desaturase. The cis configurations are not shown for all molecules, but they are represented with blue numbers. The dashed arrows illustrate biochemical steps that are not represented in this scheme.
[See online article for color version of this figure.]
The heterologous expression of the Pantoea ananatis carotenoid biosynthetic genes (CrtE, CrtB, and CrtI; Misawa et al., 1990, 1991; Shimada et al., 1998; Ravanello et al., 2003) has proven to be a useful tool in the engineering of the pathway, as their low homology with respective plant genes alleviates potential silencing/cosuppression, the potential effects of endogenous allosteric regulators is reduced, and the phytoene desaturase (CRTI) from this organism is a single enzyme that will convert 15-cis phytoene to all-trans lycopene, thus overcoming the need for four proteins (Figure 1). To make further progress in carotenoid engineering of higher plants, knowledge of carotenoid pathway regulation and sequestration within the plastids are of considerable importance. Over recent years, correlations between carotenoid content and sequestration mechanisms have been highlighted (Simkin et al., 2007; Li et al., 2012; Kilambi et al., 2013). The biosynthesis and sequestration of carotenoids are two aspects that need to be considered simultaneously to optimize carotenoid accumulation through engineering of plants. Moreover, our understanding of cellular compartmentalization (Heinig et al., 2013) and plastid compartmentalization remain key in metabolic engineering.
In this article, the optimal combination of the Crt heterologous carotenoid biosynthetic genes has been ascertained in tomato (Solanum lycopersicum) fruit for increased carotenoid levels. Characterization of these events has furthered our understanding of regulation mechanisms associated with carotenoid formation, revealed subcellular adaptation to newly synthesized carotenoids, and demonstrated how perturbations to carotenoids can impact the metabolome.
RESULTS
The Combinations of Crt Genes and Their Zygosity Affect Carotenoid Content
Three homozygous tomato lines, containing the P. ananatis (1) geranylgeranyl diphosphate synthase (CrtE) and (2) phytoene synthase (CrtB), both under the control of fruit-specific promoters, and (3) phytoene desaturase (CrtI), under the control of a constitutive promoter, were used to perform genetic crosses (Figure 1). A minimum of six complementary crossing events were performed to establish the heterologous carotenoid gene combinations CrtE+B, CrtE+I, and CrtB+I. From these crosses, 10 F1 plants per cross were generated and screened by PCR for the presence of transgenes (see Supplemental Figure 1 online). Four PCR positive lines for each combination were selected for further analysis. The lines exhibiting the highest or altered carotenoid contents were then grown to maturity in the F2 generation.
To fully assess the Crt gene combinations and the effects of gene dosage, analyses of carotenoids, chlorophylls, and tocopherols were performed on hemizygous and homozygous lines over several growth cycles. Absolute levels of carotenoids did change with environmental conditions (growth season). However, the concurrent generation and analysis of wild types enable accurate evaluation of the transgenic plants. Leaf pigments revealed significant qualitative differences that could be attributed to the presence of CrtI (Table 1; see Supplemental Figure 2 online). β-carotene increased almost twofold and other β-ring–derived carotenoids, such as violaxanthin (and its isomers), displayed similar increases (Table 1). The total carotenoid content was also increased (1.3- to 1.4-fold), which contributed to a decreased chlorophyll:carotenoid ratio (Table 1). Compared with the wild type, no significant difference in carotenoid content was found in the lines expressing CrtE and CrtB alone.
Table 1. Carotenoid and Chlorophyll Contents in the Leaves of the Transgenic Lines.
| Content | AC | Homozygous |
Hemizygous |
||||
|---|---|---|---|---|---|---|---|
| CrtE | CrtB | CrtI | CrtE+B | CrtE+I | CrtB+I | ||
| β-Carotene |
219 ± 15 |
331 ± 7 |
217 ± 7 |
298 ± 6* |
228 ± 4 |
361 ± 32* |
319 ± 13* |
| Violaxanthin# |
335 ± 16 |
347 ± 10 |
322 ± 9 |
535 ± 4** |
344 ± 4 |
552 ± 22* |
549 ± 14** |
| Lutein |
303 ± 20 |
316 ± 8 |
311 ± 7 |
294 ± 9 |
310 ± 11 |
280 ± 16 |
253 ± 8 |
| Total CAR |
856 ± 51 |
894 ± 24 |
851 ± 23 |
1,127 ± 18* |
881 ± 19 |
1,193 ± 69* |
1,120 ± 26* |
| Chlorophyll a |
9,433 ± 2,210 |
10,825 ± 642 |
11,005 ± 448 |
8,107 ± 213 |
9,534 ± 1,264 |
7,501 ± 422 |
7,301 ± 507 |
| Chlorophyll b |
1,346 ± 598 |
1,817 ± 289 |
2,217.± 93 |
589 ± 190 |
1,468 ± 429 |
426 ± 274 |
519 ± 70 |
| CHL:CAR | 13 ± 3 | 15 ± 1 | 15 ± 1 | 8 ± 1 | 13 ± 2 | 7 ± 1 | 7 ± 1 |
Carotenoid and chlorophyll contents are presented as μg/g dry weight. Methods used for determinations are described in Methods. Four representative leaves from a minimum of three plants were used. The leaves were respectively pooled, and three determinations were made per sample, making a minimum of three biological and three technical replicates. The mean data are presented ± sd; # violaxanthin and isomers; CHL, chlorophyll; CAR, carotenoid. Dunnett’s test was used to determine significant differences between the wild-type background (AC) and the transgenic varieties. Values in bold indicate where significant differences have been found. P < 0.05 is designated by *.
A similar situation occurred in the tomato fruit, where the presence of CrtI in the ripe fruit conferred the greatest changes in carotenoid and tocopherol levels compared with the other genes, CrtE and CrtB (Table 2). Indeed, compared with the wild type, the hemizygous and homozygous CrtE lines showed a similar carotenoid profile, and only a modest, but significant increase of phytoene (1.2-fold) was noticeable in the hemizygous CrtB line. However, there was a significant increase of lycopene in the CrtE+B line (1.4-fold), as well as in the homozygous CrtB line (2.2-fold). The hemizygous CrtI line was characterized by a higher level of β-carotene (2.4-fold increase compared with Ailsa Craig [AC]), a greater content of γ-carotene, lutein, and α-tocopherol, and a substantial decrease of phytoene and phytofluene (0.4-fold). The presence of an early step carotenoid gene (CrtE or CrtB) in the hemizygous CrtE+I and CrtB+I lines alleviates the decrease of phytoene and phytofluene contents. Moreover, an increase in lycopene (1.4-fold) was also observed in the hemizygous CrtB+I line. The homozygous CrtI line had a similar carotenoid profile to hemizygous CrtB+I. Surprisingly, the level of lycopene in the homozygous CrtB+I line was comparable with the wild type. However, the β-carotene content of this line was the highest compared with all the CrtI-containing lines. Analysis of ripe fruit pigments from the various Crt gene combinations indicated that the CRTI enzyme solely, or in combination with the CRTE and CRTB enzymes, conferred the greatest effects on lycopene and β-carotene contents compared with the wild type, either in the hemizygous or homozygous states (Table 2).
Table 2. Carotenoid and Tocopherol Contents in the Fruits of the Transgenic Lines.
| Variety | Phytoene | Phytofluene | Lycopene | β-Carotene | γ-Carotone | Lutein | Total CAR | α-Tocopherol |
|---|---|---|---|---|---|---|---|---|
| AC | 133 ± 16 | 134 ± 24 | 1,224 ± 255 | 321 ± 32 | 62 ± 7 | 122 ± 15 | 1,995 ± 327 | 256 ± 39 |
| Hemizygous | ||||||||
| CrtE | 136 ± 4 | 141 ± 4 | 1,434 ± 55 | 322 ± 10 | 56 ± 2 | 132 ± 3** | 2,220 ± 73 | 283 ± 10 |
| CrtB | 160 ± 4* | 146 ± 11 | 1,352 ± 108 | 220 ± 19*** | 48 ± 2* | 117 ± 9 | 2,042 ± 136 | 229 ± 8 |
| CrtI | 52 ±7*** | 55 ±3*** | 1,191 ± 10 | 785 ± 11*** | 93 ± 14* | 155 ± 5* | 2,330 ± 26 | 319 ± 8** |
| CrtE+B | 143 ± 8 | 138 ± 5 | 1,681 ± 258* | 296 ± 18 | 61 ± 3 | 116 ± 2 | 2,435 ± 251 | 232 ± 21 |
| CrtE+I | 70 ± 7*** | 84 ± 2** | 1,285 ± 168 | 560 ± 39*** | 71 ± 3 | 120 ± 2 | 2,190 ± 177 | 336 ± 19** |
| CrtB+I | 56 ± 8*** | 81 ± 2** | 1,656 ± 6* | 665 ± 55*** | 74 ± 2* | 121 ± 1 | 2,649 ± 117** | 385 ± 42** |
| Homozygous | ||||||||
| CrtE | 149 ± 15 | 153 ± 11 | 1,602 ± 21 | 286 ± 8 | 66 ± 4 | 120 ± 4 | 2,376 ± 328 | 334 ± 40* |
| CrtB | 124 ± 12 | 134 ± 8 | 2,696 ± 146*** | 377 ± 40 | 71 ± 5 | 114 ± 1* | 3,517 ± 179*** | 331 ± 63 |
| CrtI | 41 ± 1*** | 79 ± 1** | 1,780 ± 74* | 680 ± 93*** | 81 ± 6** | 114 ± 2* | 2,775 ± 368* | 378 ± 24*** |
| CrtB+I | 42 ± 3*** | 40 ± 1*** | 1,109 ± 113 | 803 ± 40*** | 69 ± 3 | 117 ± 5 | 2,179 ± 140 | 343 ± 16** |
Carotenoid and tocopherol contents are presented as μg/g dry weight. Methods used for determinations are described in Methods. Three representative fruits from a minimum of three plants were used. The fruits were respectively pooled, and three determinations were made per sample, making a minimum of three biological and three technical replicates. The mean data are presented ± sd; CAR, carotenoid. Dunnett’s test was used to determine significant differences between the wild-type background (AC) and the transgenic varieties. Values in bold indicate where significant differences have been found. P < 0.05, P < 0.01, and P < 0.001 are designated by *,**, and ***, respectively.
CrtB+I Lines Showed Changes in Transcription, Plastid Ultrastructure, and Levels of Primary and Secondary Metabolites
Since analysis of carotenoids among the lines of the different Crt gene combinations revealed CrtB+I as the best line for fruit carotenoid content and showed that this combination could exist in a stable homozygous state maintaining the high β-carotene phenotype, further characterization was performed to ascertain the underlying mechanisms associated with the effects of this gene combination.
Pigment analysis of mature green, breaker, breaker + 3 d, breaker + 7 d, and breaker + 14 d, in the CrtB+I line and its wild type (AC), revealed differences in carotenoids at the breaker stage onwards (Figure 2). With the onset of ripening at the breaker stage, the phytoene content decreased, while β-carotene content increased in the CrtB+I line. These significant differences remained throughout fruit ripening. A decrease in the lycopene content occurred at the breaker + 3 d stage but was not significantly different at other stages.
Figure 2.
The Changes of Carotenoid Composition in the Fruit of CrtB+I through Ripening Compared with AC.
Lycopene, β-carotene, and phytoene contents are given as μg/g dry weight (DW). Methods used for determinations are described in Methods. Three representative fruits were used. Three determinations were made per fruit, making three biological and three technical replicates. The bars represent the mean ± sd. Student’s t test was used to determine significant differences between the wild-type background (AC) and the CrtB+I line for each ripening stage indicated. Asterisks indicate significant differences. P < 0.05, P < 0.01, and P < 0.001 are designated by *, **, and ***, respectively. The CrtB+I homozygous line was used.
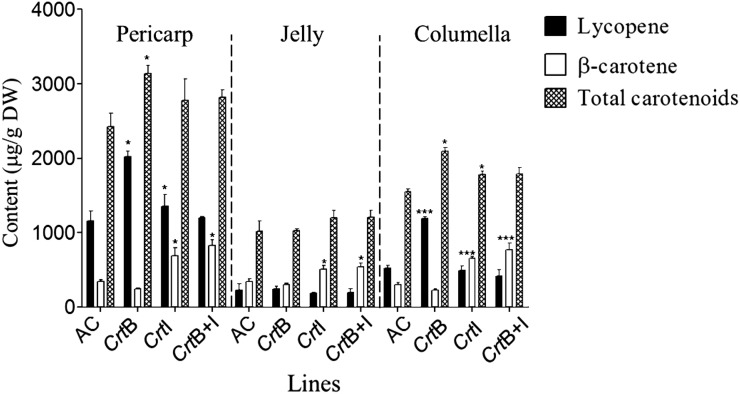
In ripe fruit, carotenoids and tocopherols were quantified in the pericarp, jelly, and columella tissues of the CrtB, CrtI, and CrtB+I lines and compared with the same material in the wild-type background (Figure 3; see Supplemental Table 1 online). Carotenoids and α-tocopherol were found in all the compartments of the fruit, but not in the same proportions. Pericarp tissue contained the most carotenoids in the control (AC), with ∼50% of total carotenoids present. The columella sequestrated 30%, while the jelly tissue contained ∼20%. In CrtB, CrtI, and CrtB+I lines, the distribution of total carotenoids within the fruit compartments was comparable to that of the control, but small increases were evident (see Supplemental Table 1 online).
Figure 3.
Lycopene and β-Carotene Content in the Pericarp, Jelly, and Columella of the Fruits of the Genetic Crosses.
Carotenoid contents are given as μg/g dry weight (DW). Methods used for determinations are described in Methods. Three representative fruits were used for a minimum of three plants. Three determinations were made per fruit, making three biological and three technical replicates. The bars represent the mean ± sd. Dunnett’s test was used to determine significant differences between the wild-type background (AC) and the transgenic lines for each compartment. Asterisks indicate significant differences. P < 0.05, P < 0.01, and P < 0.001 are designated by *,**, and ***, respectively. The CrtB+I homozygous line was used.
Lycopene was mainly found in the pericarp of all the lines, while β-carotene was the most abundant carotenoid in the jelly. However, the predominant carotenoid in the columella varied among the lines. Lycopene was the main carotenoid in the columella tissue of the wild type and the CrtB lines, whereas β-carotene predominated in CrtI and CrtB+I lines. When calculated as fold changes, although the lycopene levels increase in the pericarp and the columella of CrtB (homozygous) line, the greatest increase was in the columella (2.3-fold increase). The β-carotene levels increased in all the fruit compartments of CrtI and CrtB+I (homozygous), but the greatest change was in the columella (2.2- and 2.5-fold increase, respectively, for CrtI and CrtB+I).
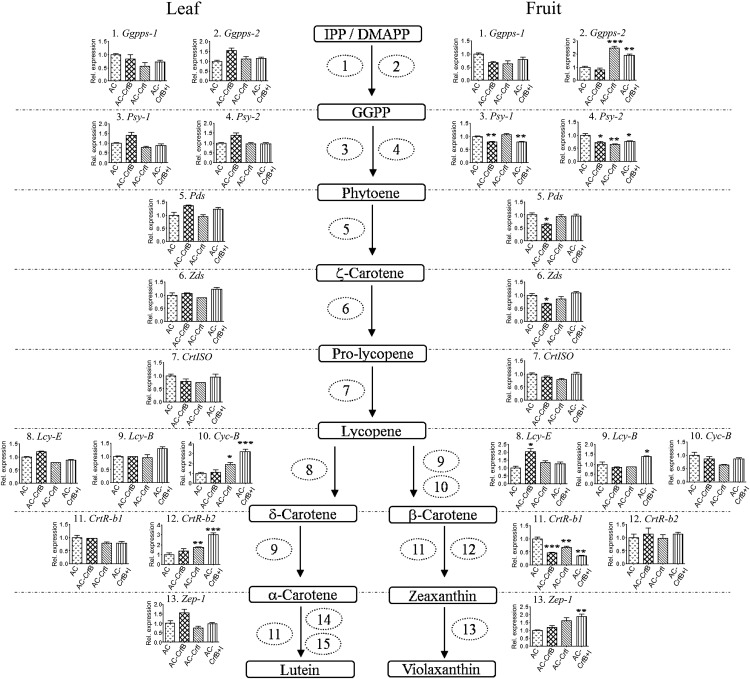
Levels of mRNAs were studied in the leaf and tomato fruit (at breaker + 3 d) of CrtB, CrtI, and CrtB+I lines and the wild type (Figure 4). In the leaf, the majority of the pathway transcripts were not significantly affected compared with the control (AC). Only the levels of the fruit-specific lycopene β-cyclase (Cyc-b) and the flower-specific carotene β-hydroxylase (CrtR-b2) transcripts were significantly different in the CrtI lines, showing a two- to threefold increase in comparison with the wild-type levels. In the transgenic fruit, several carotenogenic genes were upregulated, including geranylgeranyl pyrophosphate synthase-2 (Ggpps-2), zeaxanthin epoxidase-1 (Zep-1), ε-lycopene cyclase (Lcy-E), and the lycopene β-cyclase (Lcy-B). By contrast, the CrtB and CrtB+I lines had reduced levels of Psy-1 and Psy-2 transcripts (∼0.7-fold) compared with their control. However, CrtI only exhibited a significant reduction in Psy-2. The levels of carotene β-hydroxylase (CrtR-b1) transcripts were reduced in all transgenic lines, whereas the phytoene and ζ-carotene desaturase (Pds and Zds) transcripts were only reduced in the CrtB line (Figure 4). Several ultrastructural changes were apparent in the transgenic lines. The chromoplasts of CrtB+I were significantly larger in volume (9.1 ± 1.3 μm2) than those of the wild type (3.12 ± 0.5 μm2). In addition, the plastids contained significantly more plastoglobules in the CrtB+I line (36.3 ± 8.3) than in AC (12.9 ± 2.9). Some of these plastoglobules were larger (approximately twofold) and had different staining characteristics. Structures termed thylakoid plexus and membranous sacs were identified and were more abundant in the chromoplasts derived from this line (Figure 5). Among the different transgenic lines, similar types of chromoplasts were apparent. To verify that no heterogeneity in chromoplast structure and carotenoid content existed, separation of potential chromoplast types was performed by Suc density gradient centrifugation (see Supplemental Figure 3 online; system 3 as described in Methods). The intact chromoplasts, accumulating at different densities on the gradient, all appeared to have a similar ultrastructure and similar carotenoid profiles.
Figure 4.
Changes in the Transcript Levels of Carotenoid Biosynthetic Genes in Response to Changes in Carotenoid Content Resulting from the Expression of CrtB, CrtI, and CrtB+I Genes in Tomato.
Pooled fruit originating from three plants per genotype (AC, AC-CrtB, AC-CrtI, and AC-CrtB+I; homozygous lines) were ground in liquid nitrogen to provide a homogenous powder as described in Methods. Total RNA was then extracted from an aliquot of this material. Quantitative real-time RT-PCR was performed with gene-specific primers for (1) Ggpps-1, (2) Ggpps-2, (3) Psy-1, (4) Psy-2, (5) Pds, (6) Zds, (7) CrtISO, (8) Lcy-E, (9) Lcy-B, (10), Cyc-B, (11) CrtR-b1, (12) CrtR-b2, (13) Zep-1, (14) CYP97A, P450 β-ring hydroxylase, and (15) CYP97C, P450 hydroxylase. The expression data shown have been normalized to the expression of actin. Data are represented as relative levels found in the three varieties compared with the wild-type AC. Statistical determinations are shown as mean ± sd values, where n = 3. Dunnetts’s test illustrates statistically significant differences (*P < 0.05, **P < 0.01, and ***P < 0.001) from the wild-type levels. The first bars of the histogram indicate levels in the wild-type AC, the second bars in AC-CrtB, the third bars in AC-CrtI, and the fourth bars in AC-CrtB+I. IPP, isopentenyl pyrophosphate; DMAPP, demethylallyl diphosphate; GGPP, gerenaylgeranyl diphosphate.
Figure 5.
Electron Micrographs of Chromoplasts and Substructures of the CrtB+I Line and the AC Control at the Breaker + 5 d Stage.
Chromoplasts found in AC (control) ([A] and [B]), chromoplasts of the CrtB+I line ([C] and [D]), and substructures of the CrtB+I chromoplasts ([E] and [F]). Arrows show plastoglobules (close-up of [A]); dashed arrows show morphologically different plastoglobules (close-up of [C]). C, lycopene crystal; RC, remains of crystal; ThP, thylakoid plexus-like; MbS, membranous sac; ChE, chromoplast envelope. Methods used to obtain the electron micrographs are described in Methods. The CrtB+I homozygous line was used.
The distinct changes at the cellular level suggested perturbations beyond the isoprenoid pathway had arisen in the CrtB+I line. To assess the global effects across metabolism, metabolite profiling was performed. Over 50 metabolites were identified and quantified in a relative or absolute manner in the tomato leaf and fruit of the CrtB+I line and in the wild type. Metabolite changes relative to their control (AC) levels were determined and statistical analysis performed to assess the differences (Table 3; see Supplemental Figure 4 online). Significant changes in metabolite levels were found in all classes of compounds studied. In the fruit of CrtB+I, all amino acids and most of the sugar levels were significantly greater compared with their levels in AC, while the majority of organic acids significantly decreased in CrtB+I. Among the lipids, the level of monogalactosyldiacylglycerol (MGDG) lipids increased (3.6-fold increase) in CrtB+I. MGDG fatty acids of the CrtB+I line had increased levels of C16:0 and C16:1 cis 9 fatty acids (see Supplemental Table 2 online). No significant difference was observed in the total amount of fatty acids for all other lipids analyzed.
Table 3. Metabolite Changes Occurring in Tomato Leaves and Fruit in the CrtB+I Line Compared to the Control AC.
| Metabolite | Ratio CrtB+I to AC |
|
|---|---|---|
| Leaf | Fruit | |
| Amino acid |
||
| Ala |
1.43 ± 0.08 |
– |
| Asp |
10* |
2.10 ± 0.53 |
| β-Ala |
10* |
– |
| γ-Aminobutyric acid |
1.03 ± 0.06 |
2.44 ± 0.11 |
| Leu |
10* |
– |
| Pro |
10* |
– |
| Gln |
1.98 ± 0.2 |
1.41 ± 0.33 |
| Ser |
10* |
1.73 ± 0.29 |
| Thr |
1.32 ± 0.39 |
10* |
| Val |
1.34 ± 0.37 |
– |
| Isoprenoid |
||
| α-Tocopherol |
– |
1.34 ± 0.12 |
| Violaxanthin |
1.64 ± 0.08 |
– |
| Lutein |
0.83 ± 0.06 |
0.97 ± 0.07 |
| β-Carotene |
1.46 ± 0.21 |
2.50 ± 0.25 |
| Chlorophyll a |
0.77 ± 0.35 |
– |
| Chlorophyll b |
0.39 ± 0.32 |
– |
| γ-Carotene |
– |
1.11 ± 0.11 |
| Lutein |
– |
0.96 ± 0.07 |
| Lycopene |
– |
0.91 ± 0.19 |
| Phytoene |
– |
0.31 ± 0.04 |
| Phytofluene |
– |
0.30 ± 0.02 |
| Non-amino acid N-containing compound | ||
| Putrescine |
10* |
– |
| Lipid |
||
| DGDG |
ND |
1.05 ± 0.50 |
| MGDG |
ND |
3.61 ± 1.03 |
| PE |
ND |
1.09 ± 1.12 |
| PS/PC |
ND |
0.75 ± 0.22 |
| Triglycerides |
ND |
0.99 ± 0.10 |
| Organic acid |
||
| Aconitic acid |
– |
0.01# |
| Citric acid |
1.13 ± 0.09 |
0.98 ± 0.14 |
| Erythronic acid |
6.80 ± 1.01 |
– |
| Fumaric acid |
0.61 ± 0.09 |
0.01# |
| Glucaric acid |
8.88 ± 1.57 |
1.53 ± 0.10 |
| Gluconic acid |
0.29 ± 0.09 |
0.76 ± 0.15 |
| Glycerate |
0.01# |
– |
| Itaconic acid |
– |
0.28 ± 0.04 |
| Isocitrate |
0.01# |
– |
| Lactic acid |
10* |
– |
| Maleic acid |
0.61 ± 0.06 |
0.01# |
| Malic acid |
1.49 ± 0.17 |
1.05 ± 0.14 |
| Succinic acid |
0.96 ± 0.07 |
0.89 ± 0.07 |
| Phosphate |
||
| Glc-6-phosphate |
10* |
0.01# |
| Glycerol-3-phosphate |
10* |
– |
| Phosphate |
8.87 ± 1.91 |
0.99 ± 0.06 |
| Polyol |
||
| Glycerol |
10* |
– |
| Inositol |
0.74 ± 0.14 |
1.96 ± 0.20 |
| Sugar |
||
| Ara |
0.58 ± 0.23 |
3.45 ± 0.88 |
| Fru |
0.90 ± 0.06 |
1.10 ± 0.21 |
| Glc |
1.90 ± 0.18 |
1.15 ± 0.20 |
| Rib |
0.58 ± 0.23 |
3.45 ± 0.88 |
| Sedoheptulose |
0.18 ± 0.05 |
0.95 ± 0.17 |
| Xyl |
0.58 ± 0.23 |
3.45 ± 0.88 |
| Xylulose | – | 0.01# |
Data were compiled from multiple analytical platforms. The ratio data are presented as mean ± sd. Student’s t test analysis was carried out. Significant changes are represented in bold (P value < 0.05). 10*, theoretical value when a metabolite is unique to CrtB+I at the concentration used; 0.01#, theoretical value when a metabolite is unique to AC at the sample concentration used. – indicates metabolite not detected in both CrtB+I and AC at the sample concentration used. ND indicates metabolite not determined; PS/PC, phosphatidylserine/phosphatidylcholine; PE, phosphatidylethanolamine; DGDG; digalactosyldiacylglycerol.
Elevated Carotenoid Levels Are Sequestered in Subplastidial Compartments
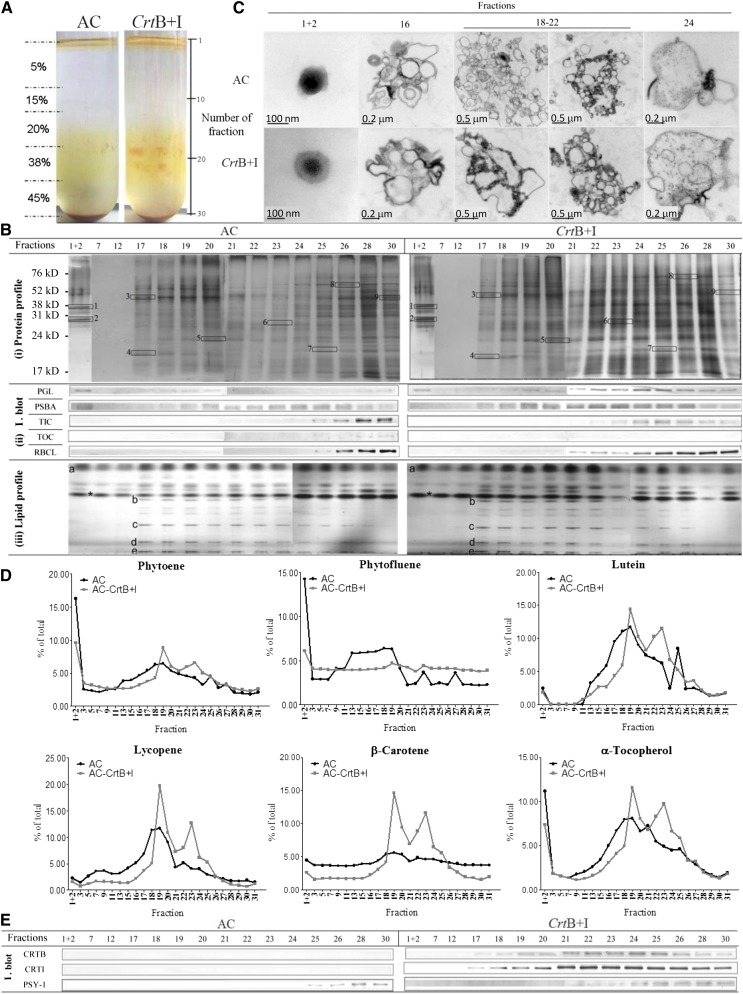
To identify the subplastidial location of carotenoids in CrtB+I and wild-type lines, chromoplasts were fractionated by Suc density gradient centrifugation (Figure 6A; see Supplemental Figure 5 online, system 1). Three distinct colored sectors in the gradient were observed, the first at the top of the gradient, in fractions 1 and 2. The red/orange color intensity of these fractions was greater in the CrtB+I preparations. In the lower part of the gradient, from fractions 16 to 24, two sectors of color intensity occurred. These two sectors were in close proximity in the gradient but separated by the less intense area arising at fractions 20 and 21. One of the distinguishing features arose from the presence of particulates, crystal-like structures, within the two sectors. Comparison between the wild-type and CrtB+I lines indicated a greater intensity of crystal-like aggregates in the upper sector derived from the wild type, with relatively few structures in the lower phase. By contrast, the CrtB+I line exhibited a greater intensity of crystalline aggregates over both sections (Figure 6A).
Figure 6.
Subplastidial Carotenoid Sequestration in Response to Elevated Carotenoid Synthesis.
(A) Fractionation of subplastidial components of chromoplasts from AC (the wild type) and CrtB+I lines. Chromoplasts were extracted from 90 g of a mix of breaker + 3 to + 5 d tomatoes and then broken with a handheld potter and separated in a discontinuous gradient of 5, 15, 20, 38, and 45% Suc (weight per volume). Fractions of 1 mL were collected for further analysis. Typically, a total of 30 fractions were collected per centrifuge tube. Fractions from six replicates were used to achieve all the experiments showed in Figure 6B. Validation of subplastidial components using antibodies to biomarker proteins and analysis of lipid species.
(B) (i) Protein profile. Proteins, extracted from each fraction, were separated and visualized using SDS-PAGE followed by silver staining. Selected proteins were identified by nano-LC-MS-MS: 1, Plastoglobulin-1; 2, plastid lipid-associated protein CHRC; 3, ATP synthase subunit b; 4, photosystem I reaction center subunit II; 5, photosystem II 22-kD protein; 6, oxygen evolving enhancer protein 1; 7, oxygen evolving enhancer protein 2; 8, heat shock cognate 70-kD protein-1; 9, ribulose-1,5-bis-phosphate carboxylase/oxygenase large subunit binding protein subunit b. Details of the identification of these proteins are shown in Supplemental Figure 6 online. (ii) Immunoblot (I. blot). Immunolocalization of biomarker proteins in the fractions were determined by immunoblotting: plastoglobulin (PGL, 35 kD), photosystem II protein D1 (PSBA, 28 kD), TIC (45 kD), TOC (75 kD), and ribulose-1,5-bis-phosphate carboxylase/oxygenase large subunit (RBCL, 52 kD). (iii) Lipid profile. Lipids derived from the fractions were separated in a thin layer chromatography silica plate with a mixture of acetone:toluene:water (91:30:7). Standards for lipid species were used for identification: a, triglycerides; b, monogalactodiacylglycerol; c, digalactodiacylglycerol; d, phosphatidylethanolamine; e, phosphatidylserine/phosphatidylcholine; asterisk, contaminant.
(C) Ultrastructure component of isolated fractions. After collection, the fractions were dialyzed against PB and then fixed in osmium tetroxide.
(D) Carotenoids and α-tocopherol contents of the isolated fractions. Metabolites were extracted from each fraction and separated by liquid chromatography using a ultrahigh performance liquid chromatograph. The carotenoids and α-tocopherol were identified and quantified using calibration curves of standards. Contents are given as a percentage in a fraction compared with the total content in the tube.
(E) Localization of the heterologous phytoene synthase (CRTB, 38 kD) and phytoene desaturase (CRTI, 56 kD) enzymes and the endogenous phytoene synthase (PSY-1, 35 kD) enzyme within the subplastidial component of the AC and CrtB+I chromoplasts. Specific antibodies were used to immunodetect these enzymes in each collected fraction. Experiments were performed with the hemizygous CrtB+I line and a concurrent control.
[See online article for color version of this figure.]
Sixteen fractions (F) throughout the gradient were analyzed to identify and designate the different subcompartments of the chromoplasts (Figure 6). The protein and lipid profiles of these fractions have been determined (Figure 6B). Their ultrastructure and carotenoid profile have been investigated (Figures 6C and 6D), and the location of heterologous enzymes (CRTB and CRTI) as well as the endogenous enzyme (the fruit-specific phytoene synthase PSY-1) have been determined (Figure 6E). The protein profile of AC and CrtB+I is similar (Figure 6B, i), with F1 and F2 characterized by two major proteins. These proteins are plastoglobule-associated plastoglobulin-1 and the Chromoplast-specifc associated protein C (CHRC). The amount of proteins present in F7 to F12 was too low for detection on SDS-PAGE stained with silver reagents. F17 to F30 contained comparatively higher protein intensity than earlier fractions, with prominent bands displaying Gaussian distribution across adjacent fractions (Figure 6B). In F17 to F24, all the proteins identified were derived from the photosynthetic systems present in the thylakoid membrane; for example, ATP synthase subunit β, photosystem I reaction center subunit II, photosystem II 22-kD protein, and the oxygen evolving enhancer proteins 1 and 2. In F24 to F30, thylakoid proteins were present, but additional proteins such as the heat shock cognate 70-kD protein were detected. This protein has been attributed to the chromoplast envelope (Ko et al., 1992). The ribulose-1,5-bisphosphate carboxylase/oxygenase protein, from the chromoplast stroma, was mainly detected in F28 and F30. Further details on the proteins identified are in Supplemental Table 3 online.
To complement the proteomic approach, immunodetection of biomarker proteins of known subplastid location was used. The immunolocalization of the plastoglobulin 35, PSBA (for photosystem II protein D1), TIC40 (for translocon at the inner envelope of chloroplasts), TOC75 (for translocon at the outer envelope of chloroplasts), and the stromal RBCL (for ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit) proteins was performed across the gradient (Figure 6B, ii). The plastoglobulin was detected in F1 and F2, but also in F18 to F30. Following comparative protein loading of wild-type and CrtB+I samples, a greater quantity of PGL was prevalent in the CrtB+I fractions. The PSBA was mainly detected in F17 to F26 fractions and at low level in control F1 and F2. TIC was found principally in F28 and F30 in AC and in F24 to F26 in CrtB+I, while TOC75, specific to the chloroplast envelope, was not detected in the fractions. The RBCL was mainly discovered at the bottom of the gradient in F26 to F30 in AC and F25 to F30 in CrtB+I.
The lipid profile of AC and CrtB+I fractions was comparable. All the fractions contained complex lipids. However, MGDG, digalactodiacylglycerol, phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine lipids were only found in F17 to F26/28 (Figure 6B, iii). These data show that F1 and F2 correspond to the free plastoglobules of the chromoplasts, F17 to F23 represent the subcompartment structure with thylakoid membrane, while F24 to F26/28 are a mix of thylakoid membrane and envelope membrane structures and the last fractions represent enriched stromal proteins (F25 to F31).
Electron microscopy was used to visualize the structures fractionated through the gradient (Figure 6C). Plastoglobules were found in F1 and F2 of both the wild type and the CrtB+I line. Membranous structures, with varying degrees of complexity and aggregation, were seen in F16 and F18 to F22, and F24, although, clear vesicle-like structures predominated in these fractions. The structures varied in the thickness of the membrane, as judged by the intensity of staining and size, with some vesicles embedded in larger membrane structures. In addition, some vesicles appeared to retain plastoglobules and/or dense staining amorphous material associated with these membranes. The different electron density levels throughout the membranes suggest a variability of the carotenoid lipoprotein structure of the membrane. The contents of fractions lower in the gradient (e.g., F24) appeared to be enriched with larger vesicles containing electron dense material.
Carotenoids and α-tocopherol were profiled in all the fractions derived from the fractionated chromoplasts (Figure 6D). The percentage of each compound studied varied through the gradient and was dependant on the line (see Supplemental Figure 6 online). Two sectors of dense pigmentation, forming peaks of metabolite intensity, were observed for lycopene, β-carotene, lutein, and α-tocopherol derived from the fractionation of the CrtB+I material. The first peak of pigmentation was located from F16 to F21 and designated as the submembrane compartment I. The second peak, located in F21 to F26, was termed submembrane compartment II. Lycopene, β-carotene, and lutein have similar profiles through the gradient. In AC, they mainly accumulate in submembrane I. In the CrtB+I line, there is an increase of lycopene, β-carotene, and lutein contents. The accumulation of these carotenoids was greater in both submembrane compartments I and II in the CrtB+I line compared with the control, especially in compartment II, which contained a higher ratio of β-carotene to lycopene, compared with compartment I. In the control, phytoene accumulated in the plastoglobules (15%) and then mainly in submembrane compartment I. In the CrtB+I line, the level of phytoene in the plastoglobules was lower (6%). The percentage of phytoene was comparable in submembrane compartments I and II.
An additional experiment was performed to ascertain if the carotenoid crystals in submembrane compartments I and II (system 1) were attached/embedded into the membrane. Another fractionation system was used (see Supplemental Figure 5 online, system 2). At the top of the gradient, a large red area was observed, while the membranes pelleted at the bottom of the gradient. The membranes were then separated using system 1. No crystals were observed in submembrane compartments I and II of the system 2 gradients (see Supplemental Figure 6A online). However, membrane-free crystals were observed by light microscopy when analyzing the red sector obtained in the first step gradient of system 2 (see Supplemental Figure 6B online). Observation of submembrane compartments I and II with a light microscope showed carotenoid crystals wrapped in a membranous structure. Therefore, carotenoid crystals are normally attached/embedded into the membrane but can be separated during fractionation. The importance of the crystalline structures in the sequestration of carotenoids in the CrtB+I line is illustrated by the comparison of carotenoid content in membranes with embedded crystals (obtained from system 1) with that in crystal-free membranes (obtained from system 2), where the ratio of carotenoid content in plastoglobules to membranes with embedded crystals is decreased in CrtB+I compared with AC (system 1) and the ratio of plastoglobules plus crystals to crystal-free membrane is increased in CrtB+I compared with AC (see Supplemental Figure 6C online). The localization of CRTB, CRTI, and PSY-1 was performed using specific antibodies (Figure 6E). The heterologous enzymes CRTB and CRTI in the CrtB+I line appeared to be strongly associated with the thylakoid-related fractions and were mainly present in submembrane compartment II. However, PSY-1 was found mainly in the stroma of both the control and CrtB+I lines.
DISCUSSION
Expression of Bacterial Carotenoid Gene Combinations Can Have a Synergistic Effect on Carotenoid Formation in Tomato Fruit
The carotenoid profiles of the homozygous CrtB and CrtI tomato lines have been studied previously by Fraser et al. (2002) and Römer et al. (2000), respectively. The results shown in this study correlate with the previous description of these lines. Moreover, the expression of CrtI in tobacco (Nicotiana tabacum) leaves was studied (Misawa et al., 1994) and also shown to result in an increase in xanthophylls. The strategy to coordinate the expression of multiple bacterial carotenoid genes to improve carotenoid formation in plants has been used previously in crops with low basal levels of carotenoids, for example, in canola (Brassica napus) seed (Ravanello et al., 2003) and potato (Solanum tuberosum) tuber (Diretto et al., 2007). The decrease of phytoene and increase in lycopene/β-carotene levels in CrtI lines was reported in both studies and is supported by the results with tomato fruit (Table 2), which are chromoplasts-containing tissues predisposed to carotenoid accumulation. The coordinate expression of two bacterial hemizygous carotenoid genes in tomato highlights synergistic effects on carotenoid formation, which were not observed in the lines expressing only one gene. For instance, there is no significant increase in lycopene in the hemizygous CrtE, CrtB, and CrtI lines compared with the control. However, there is a significant increase in the hemizygous CrtE+B and CrtB+I lines. The production of lycopene, which was not enhanced by the expression of only one bacterial carotenoid gene, is positively affected, while two bacterial carotenoid genes are expressed. This suggests that there is a functional interaction between CRTE and CRTB plus CRTB and CRTI in the fruit. By contrast, no such synergistic effect on lycopene formation was found in the CrtE+I line (Table 2), indicating that CRTE and CRTI do not interact via the endogenous phytoene synthase enzyme PSY-1. Thus, only the bacterial enzymes of consecutive steps of the carotenoid pathway have a synergistic effect on carotenoid formation. This suggests that CRTE, CRTB, and CRTI need to form an aggregate complex to be able to interact or to be sequestrated into a common microenvironment within the plastid to form a functionally complete metabolon. Previous work also suggested that a complex of the bacterial phytoene synthase, phytoene desaturase, and the lycopene cyclase enzymes allowed in vivo activity of all three proteins through substrate channeling (Ravanello et al., 2003). Although the strategy of combining expression of bacterial hemizygous carotenoid genes increased carotenoid levels, the dose of the heterologous gene(s) had a similar impact. For instance, while the level of lycopene was not significantly increased in the CrtB hemizygous and CrtI hemizygous lines compared with the wild type, it was increased in the CrtB homozygous and CrtI homozygous lines, as in the CrtE+B and CrtB+I hemizygous lines. Surprisingly, no significant increase in lycopene and total carotenoids was found in the homozygous CrtB+I line, although it did contain the highest level of β-carotene of all the lines. Consequently, the hemizygous CrtB+I line appears to be a better option than the homozygous line in regard to increasing total carotenoid levels. In summary, the choice of the heterologous carotenoid genes, their combination, and gene dosage (hemizygous or homozygous) are factors that affect carotenoid formation and that therefore need to be assessed in order to manipulate the pathway. These data also highlight why characterization of transgenic plants should not be performed solely on primary transformants. It is also feasible that the homozygous state is more prone to silencing by methylation. The involvement of epigenetic regulatory mechanisms that affect heterologous genes has not been studied intensively, but methylation has been shown to be an important mode of regulation in tomato fruit (Zhong et al., 2013). However, in this case, immunoblot analysis of CRTB and CRTI levels in the hemizygous and homozygous states indicated an increase in protein content approaching twofold in the latter. Therefore, determination of enzyme activities/flux coefficients (Fraser et al., 2002) would be an informative approach to reveal underlying mechanisms associated with the changes in carotenoid profiles between the different Crt gene combinations. This study demonstrated the potential beneficial effects of combining two Crt genes on carotenoid content. Future approaches could use the triple combination (CrtE+B+I) to potentially enhance carotenoid levels further.
The Effects of CrtB+I Go Beyond the Carotenoid Pathway
Small changes in transcription correlated, as expected, with increases in carotenoids (Tables 1 and 2). Interestingly, those transcripts most affected in the transgenic lines corresponded to genes associated with alternative tissue-specific expression (Figure 4). For example, in the leaf, transcripts of the fruit-specific lycopene cyclase (CYC-B) and the flower-specific carotene β-hydroxylase carotene (CrtR-b2) genes were altered. This suggests that transcription is tightly regulated for those carotenoid genes expressed in certain tissues, but genes not usually expressed in a given tissue are not under the same regulatory mechanisms.
The activity of CRTB and CRTI also affected the spatial accumulation of pigments over fruit development and ripening, as well as the partitioning of the carotenoids within the fruit tissues. For example, comparisons between the pericarp of the homozygous CrtB+I line with its control, at different ripening stages, showed that the timing of carotenoid formation is altered in the CrtB+I line. From the breaker stage, the CrtB+I line contained significantly more β-carotene and less phytoene compared with AC, but lycopene levels are significantly greater in the CrtB+I line at the breaker + 3 d ripening stage (Figure 2). These changes in carotenoids reflect the timing of expression of the different promoters that regulate the CrtB and CrtI genes, the CrtI gene being under constitutive control and CrtB under ripening-specific promoter control (Atkinson et al., 1998). The constitutive presence of CRTI influenced the levels of β-carotene and phytoene from the breaker stage, while synthesis of CRTB later in the ripening process affected the level of lycopene. Another phenotype resulting from CRTB and CRTI was the intrafruit partitioning of carotenoids in the pericarp, jelly, and columella. The pericarp was the fruit compartment with the greatest capacity to store carotenoids (i.e., the highest levels), but the largest changes of lycopene in the CrtB line and β-carotene level in the CrtI and CrtB+I lines, were associated with the columella (Figure 3). This suggests that the accumulation of carotenoids in the tomato fruit depends on several tissue-related factors, such as the type of cell, the composition and quantity of membranes, and the aqueous content of each tissue/cell type. Apparently, there is a saturation limit for a specific carotenoid in a given tissue and beyond that concentration, another region of the fruit must be used. In addition to the altered tissue distribution of carotenoids in the transgenic lines, structural differences of the chromoplasts were observed (Figure 5). The chromoplasts from the CrtB+I line were significantly larger (2.8-fold) and contained more membranes (previously described as thylakoid plexus in Spurr and Harris, 1968). The thylakoid plexus has also been identified in the chromoplast of ripe fruit of the high β-mutant of tomato (Harris and Spurr, 1969). The presence of the thylakoid-like membranes could allow a greater storage environment for the carotenoids. In CrtB+I, most of the plastoglobules were the same size as the control; however, some of them appeared to be larger and contained a possible crystal-like structure. Plastoglobules containing crystals have been reported in another Solanum species (Wrischer et al., 2007). Therefore, storing crystals of carotenoids in the plastoglobules of the CrtB+I chromoplasts is plausible. Membranous sacs in CrtB+I formed from the inner envelope of chromoplasts (Figure 6B). The hypothetical role of the membranous sacs is to store crystals of carotenoids (Egea et al., 2010). In conclusion, it would appear that the ultrastructure of the chromoplast adapts or responds to perturbations in carotenoid composition. This phenomenon has been observed previously (Fraser et al., 2009; Maass et al., 2009).
Metabolite profiling illustrated that, in addition to cellular changes, the expression of CrtB+I genes in the tomato fruit had effects across the metabolome (Table 3). The precise biochemical/molecular links with the increased sugars and amino acid levels, decreased organic acid levels, and altered carotenoids await further systematic analysis. However, it could be that perturbations in fruit ripening via phytohormone imbalances have arisen. Such changes in sugars, amino acids, and organic acids have the potential to alter taste, which is an important consumer attribute. It is also feasible that the changes in leaf carotenoids affect source tissue metabolism, which then manifests itself in sink tissues. Further studies under different environmental conditions would be beneficial to clarify how the modifications can alter the plant’s responses to abiotic stresses. Increased MGDG content (3.6-fold increase; Table 3) was determined in the transgenic line, especially the C16:0, C16:1 cis 9, and C18:1 fatty acid moieties of this complex lipid (see Supplemental Table 2 online). An associated increase of carotenoid and MGDG lipid (C16:0 and C18:3 fatty acids) levels was observed in a marine bacterium (Synechococcus sp) during high-light acclimation (Montero et al., 2012). It seems that the prokaryotic pool of MGDG (C16:0 plus C18:1, from plastids) is positively regulated in parallel with the carotenoid level in the CrtB+I line. MGDG is found in abundance in the inner envelope and in the thylakoid membranes of the chloroplast/chromoplast (Marechal et al., 1997). The reason for elevated MGDG in relation to carotenoid content could be explained by the fact that MGDG has a high propensity for interfacial curvature, allowing the membrane to adapt to a greater quantity of β-carotene (Szilágyi et al., 2007). In the transgenic line, it appears that this plasticity of the lipid is used to accommodate the extra carotenoid produced.
Carotenoid Sequestration Mechanisms Have Implications for Pathway Regulation
A comparison between subcompartments of the fruit chromoplasts from control and CrtB+I lines showed a number of important differences. First, an increased number of β-carotene and lycopene crystal-like structures arose in the thylakoid-like membrane fractions of the CrtB+I line (Figure 6A). Storage of endogenous carotenoids in crystal-like structures has been reported in other plant species, such as mango (Mangifera indica) (Vasquez-Caicedo et al., 2006) and red papaya (Carica papaya) (Schweiggert et al., 2011) as well as in tomato (Rosso, 1967, 1968). It seems that this sequestration mechanism has been upregulated in the transgenic lines containing increased carotenoids. This phenomenon has also been observed in Arabidopsis thaliana and carrot (Daucus carota) roots (Maass et al., 2009) and embryogenic calli from citrus (Cao et al., 2012) overexpressing the phytoene synthase gene. Secondly, the membranes of the chromoplast (envelope and thylakoid-like membrane) also appeared to play an important role. The inner envelope of the CrtB+I chromoplasts seemed to be actively producing vesicles (membranous sacs), which were visible in the electron micrographs of the CrtB+I chromoplasts (Figure 5). The thylakoid-like membranes appeared in greater quantity and electron density in the CrtB+I chromoplasts compared with those in AC. The darker and thicker membranes could be caused by a high number of lipids, proteins, and carotenoids or complexes of these components. Harris and Spurr (1969) highlighted similar characteristics of chromoplasts of the tomato high β-mutant. They described invagination of the internal membrane of the plastid envelope and swollen grana and intergrana lamellae. CRTB and CRTI are found in the same submembrane compartment, which strengthens the hypothesis that they can interact with each other and have a synergistic effect on carotenoid production. However, PSY-1 was mainly found in the stroma, whereas its product phytoene was predominantly found in the membranes and in the plastoglobules. The location of PSY-1 within the chromoplasts confirmed results found in the literature (Fraser et al., 1994, 2002). The presence of phytoene in plastoglobules means that a significant quantity of the substrate for phytoene desaturase (and carotene formation) is partitioned away from the enzyme, possibly causing it to be metabolically inert, unless it is reincorporated into the membrane enzyme complex. Collectively, the data illustrate that synthesis and sequestration are two processes that are important for engineering carotenoids in plants, with the latter requiring further investigation. Only a few proteins have been identified with a potential link to the accumulation of carotenoids in plants, including the chromoplast-specific carotenoid-associated proteins (CHRC; Kilambi et al., 2013), the plastid-encoded acetyl CoA carboxylase D (Barsan et al., 2012), and the heat shock protein 21 (Neta-Sharir et al., 2005; Carvalho et al., 2012), while the Or gene in potato enhances carotenoid accumulation and stability (Li et al., 2012).
Plant Cells Adapt to Changes in Carotenoid Content at Multiple Levels
The characterization of transgenic lines with moderate increases in carotenoid levels has demonstrated the ability of the plant to adapt to changes in the homeostatic levels of carotenoid pigments. Adaptation occurs across multiple levels of cellular regulation, including transcription, protein localization, metabolite cell/tissue type, and organelle/suborganelle structure/organization. Previous descriptions of regulatory mechanisms operating in the pathway have focused mainly on transcription and their response to developmental or environmental responses. This study also shows that the identity and level of carotenoids can trigger different regulatory mechanisms. Consequently, it implies that changes in levels of carotenoids can be sensed, causing signaling cascades that trigger adaptive mechanisms in the cell. Little is known about these sensors and signaling molecules, although cis-carotenes or cis-apocarotenoids have been proposed to act as signaling molecules (Kachanovsky et al., 2012) for the carotenoid pathway and the methylerythritol cyclodiphosphate, produced by the plastidial methylerythritol-4-phosphate pathway, has been shown to regulate the expression of nuclear stress response genes by retrograde signaling (Xiao et al., 2012). Further detailed knowledge of how the plant cell can sense, adapt, and modulate these changes is important for further exploitation of biodiversity and holistic engineering approaches.
Recently, there has been an increasing trend to create schematic representations relating to the plastid topology of carotenoid formation in the plastid. To date, these models are predominantly based on proteomic data (Barsan et al., 2012). However, valuable limitations do arise; for example, interrogating proteomic databases without implementing stringent parameters or validation procedures can lead to misleading identifications. Moreover, the expression of proteins with visualization tags, such as green fluorescent protein, could alter the protein conformation and/or the stoichiometric ratio of proteins within a complex or microenvironment of the membrane.
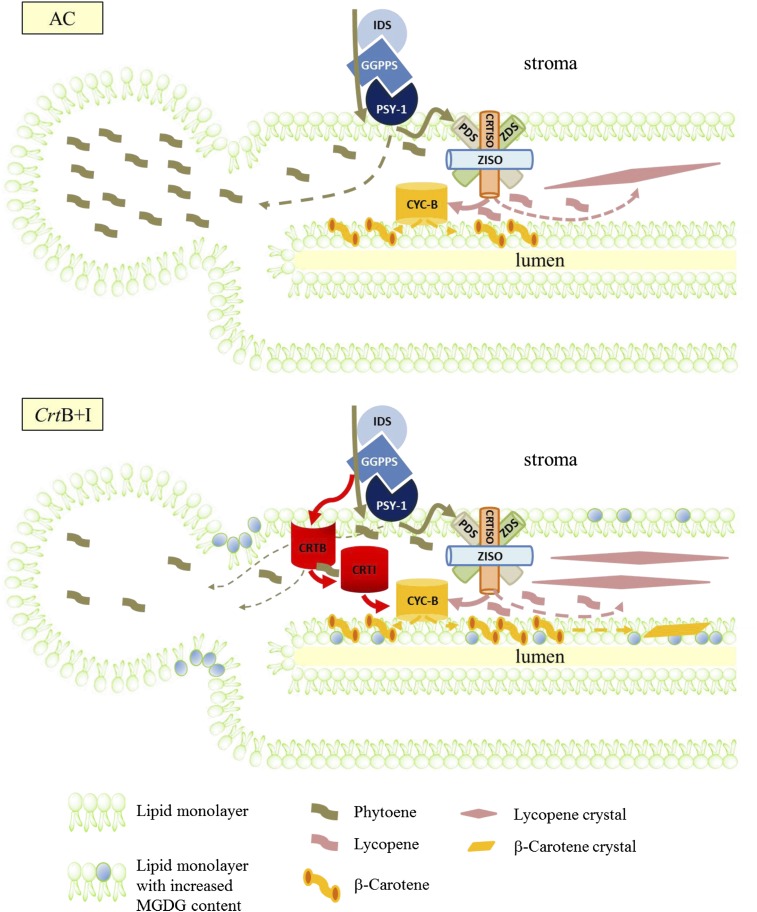
In this study, our perturbation of the system by transgenesis has enabled us to acquire valuable insight into the fundamental subplastidial organization of carotenoid synthesis and sequestration. From the experimental data, a model is presented in Figure 7. The wild-type state (AC) is illustrated with isopentenyl diphosphate isomerase, geranylgeranyl pyrophosphate synthase, and phytoene synthase(s) functioning as a metabolon for efficient channeling of precursors. This has been proven experimentally using biochemical approaches with the topology of the complex designated as membrane associated (Dogbo et al., 1988; Fraser et al., 2000). This study shows the presence of phytoene in the membrane and plastoglobule. Thus, the desaturase cannot completely use the phytoene synthesized or the delivery of the phytoene from the synthase to the desaturase is not a tethered process. Given that in green tissues no phytoene accumulates, it would appear that the desaturase is capable of converting phytoene efficiently. Thus, perhaps during the transition from chloroplast to chromoplast, the dismantling of the membrane structure affects the efficiency of the carotenoid biosynthetic pathway at the stroma–membrane juncture. The determination of phytoene in relatively high amounts in the plastoglobule suggests that the cell acts to remove excess phytoene accumulating in the membrane, presumably alleviating the potential damaging effects. In addition, the compartmentalization of enzymes is a well-documented regulatory process (Heinig et al., 2013); however, in this study, we introduce the partitioning of carotenoid precursors from their biosynthetic enzymes as another means of regulating pathway flux. The formation of β-carotene infers that the lycopene produced by the desaturase/isomerase complex can be used by the cyclase. However, the process would appear to be inefficient, with lycopene accumulating and crystalline structures arising. Concurrent analysis of the transgenic variety expressing CrtB and CrtI indicates that the bacterial phytoene synthase is membrane associated but can still synthesize and influence phytoene levels. Therefore, geranylgeranyl pyrophosphate must still be accessible. Although the procedure may not be optimal, it is still operational. The reduction in phytoene present in the membrane and plastoglobule suggests that the bacterial desaturase can act on the phytoene, providing further evidence that the phytoene is not efficiency channeled to desaturation. The interaction of the CRTI with the endogenous pathway has important fundamental implications, as it is the gene product that drives β-carotene production in Golden rice, a paradigm-changing resource of humanitarian benefit (Beyer, 2010). The increased β-carotene content in the presence of the bacterial desaturase indicates that the pool of lycopene generated by the nonendogenous enzymes is more accessible to the endogenous β-cyclase than the lycopene produced by the endogenous desaturase/isomerase complex. Adaptation of the membrane to accommodate these increases in β-carotene and-lycopene arises via the modulation of MGDG content.
Figure 7.
Schematic Representation of the Regulation of Carotenoid Production within the Thylakoid-Like Membranes of AC and CrtB+I Chromoplasts.
In the wild-type AC (top panel), the pool of phytoene, synthesized by PSY-1, can be used by PDS; however, at saturated levels, the excess phytoene appears to be sequestrated in the plastoglobules. Some lycopene and β-carotene are sequestrated in the thylakoid-like membranes and lycopene crystals are subsequently formed. In CrtB+I (bottom panel), the pool of phytoene is used by PDS and CRTI. Consequently, less phytoene is stored in the plastoglobules. A greater quantity of β-carotene and lycopene is created, leading to the formation of carotenoid crystals. Solid arrows represent a catalytic step; dashed arrows indicate movement of carotenoid. Changes in the galactolipid content of the membranes are also illustrated. IDS, isopentenyl diphosphate/dimethylallyl diphosphate isomerase; GGPPS, 1-geranylgeranyl pyrophosphate synthase; PSY-1, phytoene synthase-1; PDS, phytoene desaturase; ZDS, ζ-carotene desaturase; Z-ISO, 15-cis-ζ-carotene isomerase; CRTISO, carotene isomerase; CYC-B, β-lycopene cyclase; CRTB, phytoene synthase; CRTI, phytoene desaturase.
[See online article for color version of this figure.]
Despite modification of the membrane, it would appear that, at a certain concentration, carotenoid will crystallize. Presumably, storage of carotenoids in this manner makes them metabolically and osmotically inert and probably less prone to oxidative degradation than in solution. However, in this state, it is unlikely that carotenoids can be used by carotenoid cleavage dioxygenases. Shumskaya and Wurtzel (2013) proposed that carotenoid is resolubilized and transported to the outer envelope. This seems unlikely in tomato. It is more likely that the carotenoid cleavage dioxygenases act on the carotenoid in the membrane/or plastoglobule prior to crystallization. The molar ratios between carotenoid and derived volatiles also suggest this pattern of events, as the carotenoid content is at least two orders of magnitude more than the volatiles.
In conclusion, the use of transgenesis to modulate the carotenoid pathway has revealed important regulatory mechanisms: (1) how pathway transcription can respond to perturbations in pathway metabolites, (2) how altering the partition of precursors from the biosynthetic enzymes on a subplastid level can modulate pathway flux, and (3) how metabolite composition can direct cellular structures. The latter has a generic implication to all engineering approaches across genera, as it illustrates the need for concurrent sequestration/deposition with pathway engineering.
METHODS
Plant Material and Cultivation
The control (AC) variety of tomato (Solanum lycopersicum) had been previously transformed with the CrtB construct (Fraser et al., 2002), the CrtI construct (Römer et al., 2000), and the CrtE construct, which corresponds to the CrtB vector only with the CrtE gene from Pantoea ananatis (D90087.2) using the Agrobacterium tumefaciens strain LBA 4404. The lines were crossed by cross pollination using a small paintbrush. The plants were greenhouse grown (25°C day/15°C night) with supplementary lighting (6-h light/8-h dark). Three plants each of three to four separate cross-pollination events were grown for each transgenic line concurrently with AC.
Confirmation of Gene’s Presence by PCR
The Crt genes in the crossed lines were detected by PCR, using the set of primers shown in the Supplemental Table 4 online. PCR reactions were performed using Illustra puReTaq Ready-to-Go PCR beads (GE Healthcare), with reagents prepared to the manufacturer’s guidelines. Reactions contained 10 pmol each of the respective template’s forward (5′3′) and reverse (3′5′) primers and 50 ng of genomic DNA. Tubes were incubated at 95°C for 5 min to denature the template before 30 cycles of PCR amplification (denaturation 94°C for 30 s, annealing 50°C for 30 s, and extension 72°C for 30 s). A final incubation of 5 min at 72°C completed the reaction. Reactions were performed using a Techgene thermo cycler (Techne). PCR products were analyzed by agarose gel electrophoresis.
Determination of the CrtB+I Homozygous Line
The CrtB+I homozygous line was screened by real-time PCR. The CrtB and CrtI genes in the potential CrtB+I homozygous lines were quantified and compared to the same genes in known hemizygous and homozygous lines and normalized to Pds. The Δ-Δ cycle threshold method was used to calculate the ratios. Three young leaves (±3-cm long) from three independent plants of the same line were harvested, pooled, and frozen in liquid nitrogen. Leaf material was homogenized using a Tissue Lyser LT (Qiagen) for 1 min at 50 Hz, while keeping the samples frozen. DNA was extracted from leaf powder (100 mg) using the DNeasy reagents and protocol (Qiagen). The QuantiFast SYBR Green PCR kit (Qiagen) was used to quantify the gene of interest using a 25-ng sample of DNA. Primers were added to a final concentration of 1 μM in a reaction volume of 20 μL. Reactions were performed on a Rotor-Gene 3000 thermocycler (Qiagen). Thermocycling conditions were 95°C for 15 min followed by 40 cycles of 15 s at 94°C, 30 s at 50°C, and 15 s at 72°C. Melt curve analysis verified the reactions’ specificity. For quantification, calibration curves were run simultaneously, using actin as a housekeeping gene to normalize the data.
Measurement of Gene Expression by Real-Time Quantitative RT-PCR
Measurement of expression by real-time quantitative RT-PCR was performed as described by Enfissi et al. (2010) and using the primers reported in Supplemental Table 4 online.
Extraction and Analysis of Metabolites
Carotenoids, tocopherols, chlorophylls, and lipids were extracted from freeze-dried fruit and leaf tissue. Extractions were made from sample powder (15 mg) in 1.5-mL centrifuge tubes. Metabolites were extracted by the addition of chloroform and methanol (2:1). Samples were stored for 20 min on ice. Subsequently, water (1 volume) was added. Samples were centrifuged for 5 min at top speed in a Heraeus Pico21 centrifuge (Thermo Scientific). The organic phase, containing the pigment extract, was placed in a fresh centrifuge tube, and the aqueous phase was reextracted with chloroform (×2 by volume). Organic phases were pooled and dried using the Genevac EZ.27. Dried samples were stored at −20°C and resuspended in ethyl acetate prior to spectrophotometric and chromatographic analysis.
Total carotenoids and chlorophyll a and b were determined spectrophotometrically, as described by Wellburn (1994).
Chromatographic Analysis
Carotenoids were separated and identified by ultrahigh performance liquid chromatography with photodiode array detection. An Acquity ultrahigh performance liquid chromatography system (Waters) was used with an Ethylene Bridged Hybrid (BEH C18) column (2.1 × 100 mm, 1.7 μm) with a BEH C18 VanGuard precolumn (2.1 × 50 mm, 1.7 μm). The mobile phase used was A, methanol/water (50/50), and B, acetonitrile (ACN)/ethyl acetate (75:25). All solvents used were HPLC grade and filtered prior to use through a 0.2-μm filter. The gradient was 30% A:70% B for 0.5 min and then stepped to 0.1% A:99.9% B for 5.5 min and then to 30% A:70% B for the last 2 min. Column temperature was maintained at 30°C and the temperature of samples at 8°C. Online scanning across the UV/visible range was performed in a continuous manner from 250 to 600 nm, using an extended wavelength photo diode array detector (Waters). Carotenoids were quantified from dose–response curves. The HPLC separation, detection, and quantification of carotenoids, tocopherols, and chlorophylls have been described in detail previously (Fraser et al., 2000).
Lipid Analysis
Lipids were analyzed on high-performance thin layer chromatography silica gel 60 F254 plates (Merck) using a solvent system of acetone/toluene/water (91:30:7). They were visualized with iodine vapor and identified by cochromatography with lipids of known composition. For quantitative analysis, individual lipids were isolated from thin layer chromatography plates and extracted in chloroform/methanol (1:1). Then, 50 μg of internal standard (myristic acid D27) was added to the extract prior to drying. To transmethylate the lipids, the samples were resuspended in hexane (2 mL) and methanol (4 mL) plus 1% sulfuric acid and incubated at 85°C for 2 h. Two milliliters of hexane and 1 mL of water plus 5% KCl were then added. The hexane phase was dried and resuspended in 20 μL of methanol for quantification by gas chromatography–mass spectrometry, as described below.
Extraction, Derivatization, and Gas Chromatography–Mass Spectrometry Analysis
Extraction and analysis of polar metabolites was performed as described previously (Enfissi et al., 2010; Jones et al., 2012), with slight modifications. Freeze-dried powder (10 mg) was extracted in methanol (400 μL)/water (400 μL)/chloroform (800 μL). An aliquot (20 μL) was removed from the extract, the internal standard (ribitol, 10 μg) was added to the aliquot, and the samples were dried. Six extractions were performed on each biological replicate. Derivatization was performed by the addition of methoxyamine hydrochloride (30 μL; Sigma-Aldrich) at 20 mg/mL, in pyridine. Samples were incubated at 40°C for 1 h, after which N-methyl-N-trimethylsilyltrifluoroacetamide (Sigma-Aldrich; 70 μL) was added and the samples incubated for 2 h at 40°C before analysis. Gas chromatography–mass spectrometry was performed as described previously (Enfissi et al., 2010), using a 20:1 split injector. To identify chromatogram components found in the tomato profiles, a mass spectral (MS) library was constructed from in-house standards, as well as the NIST08 MS library. Retention time calibration was performed on all standards to facilitate the determination of retention indices. Using the retention indices and MS, identification was performed by comparison with the MS library. Relative quantification to the internal standard was performed.
Subcellular Fractionation and Cellular Analysis
Electron Microscopy of Intact Chromoplasts
Tomato fruit was cut into 2-mm cubes using a new sharp razor blade in a drop of cold fixative (2.5% glutaraldehyde in 100 mM sodium cacodylate buffer [CAB], pH 7.2) on dental wax. Pieces of tissue were transferred into a glass vial (with a cocktail stick) containing cold fixative (∼2 mL). Lids were placed on the vials and tissue was fixed in the fridge (4°C) overnight. Tissue was washed in CAB (2 × 10 min) and then postfixed in 1% osmium tetroxide in CAB for 1 h at room temp (20°C). Tissue was washed (2 × 10) min in milliQ water. Tissue was then dehydrated in increasing concentrations of ethanol as follows: 50, 70, and 90% (10 min) and 3 × 100% (15 min). Tissue was then washed (3×10 min) in the transition solvent propylene oxide. Tissue was then transferred into 50% propylene oxide and 50% agar low-viscosity resin (Agar Scientific) for 30 min. Tissue was then placed in 100% agar low-viscosity resin (2 × 1.5 h) with a vacuum applied four times during the incubation. Tissue pieces were then placed in labeled silicone molds and polymerized in the oven (60°C) for 24 h. Polymerized blocks were sectioned (70 nm) on a RMC MTXL ultramicrotome, and sections were collected on 400 mesh copper grids. Sections were counterstained with 4.5% uranyl acetate in 1% acetic acid for 45 min and Reynolds lead citrate for 7 min. Sections were viewed in a Jeol 1230 transmission electron microscope with an accelerating voltage of 80 kV. Images were recorded with a Gatan digital camera. The images shown in Figure 5 are representative of three biological replicates for each line, from which 12 sections where taken per biological replicate. Volumes of chromoplasts were measured using ImageJ software.
Electron Microscopy of Subplastidial Components of Chromoplasts
Two previous methods were combined (De Camilli et al., 1983; Angaman et al., 2012). Subchromoplast fractions were pelleted in a microfuge tube and fixed in 2.5% glutaraldehyde in 100 mM phosphate buffer (PB), pH 7.4, for 1 h at room temperature. Fixed subchromoplast components were pelleted in a microfuge and washed (2 × 10 min) in PB. After the final wash, components were resuspended in PB (100 μL). Aliquots (100 μL) of fixed component were added to tubes immersed in a 54°C water bath. After a 15-s interval, which allowed the suspension to warm up, 100 μL of a solution (at 54°C) containing 3% agarose in 5 mM PB was added to the subchromoplast suspension. The suspension obtained was quickly mixed, while still immersed in the warm water bath, by forcing it up and down through a Pasteur pipette prewarmed in a 60°C oven. Care was taken to prevent foaming. Immediately afterwards, the agarose-subchromoplast suspension was transferred by pipetting into a frame made from two glass slides separated by a shaped acetate gasket and held together by bull dog clips. The frame had also been prewarmed in a 60°C oven. The agarose mixture was then allowed to cool and solidify. At this point, the two glass slides were separated, and the agarose gel, attached to one of the glass slides, was cut into 2-mm squares with a razorblade. The gel squares were then washed off the glass slide into a Petri dish by a stream of 0.1% Alcian blue in 1% acetic acid from a Pasteur pipette. These agarose squares were then transferred to glass vials and washed (2 × 10 min) in PB to remove stain. The Alcian blue stain made the agarose squares visible and aided subsequent processing and resin embedding. Samples were postfixed in 1% OsO4 containing potassium ferricyanide (0.8%) in the same PB for 1 h at room temperature. Then, after three washes in milliQ water, transmission electron microscopy was performed as described. Fractions from three technical replicates, which correspond to the mix of ∼10 tomatoes, were fixed and photographed. The procedures were repeated independently at least three times. With each CrtB+I preparation, an AC control preparation was also performed concurrently and comparative cellular analysis performed.
Subcellular Fractionation of Chromoplasts
System 1
All procedures were performed at 4°C. Approximately 10 fresh tomatoes (Breaker + 3 to 5 d) were harvested and the pericarp was cut into 1-cm2 pieces (80 to 150 g) and stored at 4°C overnight. Tomato tissue was added to extraction buffer (0.4 M Suc, 50 mM Tris, pH 7.8, 1 mM EDTA, and 1 mM DTT) and homogenized for 2 × 3 s in a Waring blender. The homogenate was filtered through four layers of muslin. Extraction buffer was added to the filtrate in a 500-mL centrifuge tube. Tubes were centrifuged for 10 min at 5000g in a Sorvall RC5C centrifuge (Thermo Scientific) with a GSA-3 rotor. The supernatant was discarded, and the pellet was resuspended in extraction buffer and transferred into 50-mL centrifuge tubes. The tubes were centrifuged for 10 min at 9000g with a GSA-5 rotor. The supernatant was discarded. Pellets were resuspended in 45% Suc buffer (45% [w/v] Suc, 50 mM Tricine, 2 mM EDTA, 2 mM DTT, and 5 mM sodium bisulphite, pH 7.9; 3 mL). The chromoplasts were physically broken using a handheld potter homogenizer (10 times). The solution was then resuspended in 45% Suc buffer (5 mL), and the 8 mL was placed in a 38.5-mL Ultra-Clear centrifuge tube (Beckmann Coulter). Subsequently, other layers of discontinuous Suc gradient were overlaid, consisting of 38% Suc buffer (6 mL), then 20% Suc buffer (6 mL), 15% Suc buffer (4 mL), and 5% Suc buffer (8 mL). Gradients were centrifuged for 17 h at 100,000g and 4°C, using an L7 ultracentrifuge with an SW28 swing out rotor (Beckman Coulter). Fractions (1 mL) were collected, from the top of the gradients using a Minipuls 3 peristaltic pump and FC203B fraction collector (Gilson). Samples were dialyzed overnight against PB (50 mM, pH 7.5) for electron microscopy analysis.
System 2
Chromoplasts were isolated as in system 1. However, instead of resuspending the pellets obtained after the centrifugation step at 9000g, they were resuspended in 0.6 M Suc buffer (0.6 M Suc, 50 mM Tris, 1 mM DTT, and 1 mM EDTA, pH 7.8; 3 mL) and homogenized using a handheld potter homogenizer to break the chromoplasts. The solution was then resuspended in 0.6 M Suc buffer to a final volume of 35 mL in 38.5-mL Ultra-Clear centrifuge tubes (Beckmann Coulter). Tubes were centrifuged for 1 h at 100,000g and 4°C, using an L7 ultracentrifuge with an SW28 swing-out rotor (Beckman Coulter). The red supernatants, which correspond to the chromoplast crystals and plastoglobules, were transferred into Eppendorf tubes and stored at −20°C. The pellets were resuspended in 45% Suc buffer (8 mL) and placed in a 38.5-mL Ultra-Clear centrifuge tube (Beckmann Coulter). The same gradient as that used in system 1 was created; subsequently, the same protocol followed as for the system 1. The two systems are represented in Supplemental Figure 5 online.
Isolation of Different Types of Chromoplasts
System 3
Chromoplast extraction was undertaken in a cold room at 4°C. Fresh tomato fruits (200 g) were harvested from selected plants, cut into pieces of ∼1 cm2, covered in foil, and stored at 4°C overnight to reduce starch content. Tomato tissue was homogenized in prechilled chromoplast buffer A (100 mM Tris-HCl, pH 8.2, 0.33 M sorbitol, 2 mM MgCl2, 10 mM KCl, 8 mM EDTA, 10 mM ascorbic acid, 5 mM l-Cys, 1 mM phenylmethylsulfonyl fluoride, 1% polyvinylpyrrolidone, and 1 mM DTT), twice for 3 s in a small laboratory blender (Waring Products). The resulting slurry was filtered through four layers of muslin cloth and the liquid decanted into a 500-mL screw cap centrifuge tube. Subsequently, tubes were centrifuged for 15 min at 5000g and 4°C in a Sorval RC5C centrifuge (Thermo Scientific) with a GSA-3 rotor. The supernatants were discarded. The pellets were resuspended in buffer B (buffer A without polyvinylpyrrolidone) and transferred into 50-mL centrifuge tubes. The tubes were centrifuged for 15 min at 5000g and 4°C with the GSA-5 rotor. The supernatants were discarded. The pellets were resuspended in 4 mL of buffer B. In 38.5-mL Ultra-Clear centrifuge tubes, a discontinuous Suc gradient (Suc [w/v] in 50 mM Tris-HCl, pH 7.4, supplemented with 1 mM DTT) with the following steps was constituted: 50% (9 mL), 40% (7 mL), 30% (7 mL), and 15% (7 mL) of Suc. The chromoplast solutions (4 mL) were placed on top of the gradients. Gradients were centrifuged for 1 h at 100,000g and 4°C, using an L7 ultracentrifuge with an SW28 swing-out rotor (Beckman Coulter). The chromoplast fractions were recovered with a Pasteur pipette in an Eppendorf tube, washed in 1 volume of buffer B followed by centrifugation at 6000g for 10 min. The supernatants were discarded and the pellets of chromoplasts kept at −20°C.
Protein Analysis
Extraction, separation, immunodetection, and liquid chromatography–mass spectrometry (LC-MS) analysis of proteins were performed following the protocols of Robertson et al. (2012) and Mora et al. (2013).
Extraction, Separation (SDS-PAGE), and LC-MS Analysis of Proteins
Proteins contained in the subplastidial fractions were precipitated with methanol and resuspended in sample buffer solution (0.5 M Tris-HCl, pH 6.8, 10% [v/v] glycerol, 10% [w/v] SDS, 1.4% [v/v] β-mercaptoethanol, and 0.05% [w/v] bromophenol blue). After a heat denaturation step at 95°C for 4 min, a volume of 5 μL of this solution was loaded into a 12.5% acrylamide gel (Laemmli, 1970). The ProteoSilver Silver Stain kit (Sigma-Aldrich) was used to develop the gel after running at 80 V. High-range rainbow molecular weight marker (RPN756E) from GE Healthcare Life Sciences was used.
In-Gel Digestion of Protein Band
Stained bands corresponding to the protein of interest were excised using a scalpel and cut into 1 mm3 and then introduced in centrifuge tubes (0.5 mL) and washed three times for 10 min with 50 mM ammonium bicarbonate, pH 8.0 (50 μL). After that, gel pieces were dried three times with ACN (50 μL) for 10 min. Once the gel pieces shrank and turned opaque, 12.5 ng/μL of trypsin (10 μL) dissolved in 50 mM ammonium bicarbonate, pH 8.0, was added. An additional 15 μL of 50 mM ammonium bicarbonate was added to each tube in order to cover the gel pieces. The tubes were incubated at 37°C overnight, and the supernatant was transferred to a clean Eppendorf tube. Tryptic peptides were sequentially extracted with 25 μL of ACN/water (50:50, v/v) with 0.1% (v/v) trifluoroacetic acid, while sonicating for 10 min (two times). The peptide extracts were combined and dried in a GeneVac Ez-2 Plus rotary evaporator and reconstituted in 5 μL of 0.1% (v/v) trifluoroacetic acid. The peptide samples were cleaned with ZipTip C18 (Millipore) prior to the nano–liquid chromatography–tandem mass spectrometry (nano-LC-MS-MS) analysis, and peptides were eluted with 10 μL of water:ACN (50:50, v/v) with 0.1% (v/v) formic acid.
Nano-LC-MS-MS Conditions
The nano-LC-MS-MS analysis was performed in an Ultimate 3000 rapid separation liquid chromatography nanosystem from Dionex (Thermo Fisher Scientific) coupled to an AmaZon ETD ion-trap mass spectrometer equipped with a nanoelectrospray ionization source (Bruker Daltonik).
Twenty microliters of loading buffer (water:ACN [98:2, v/v] with 0.1% [v/v] formic acid) was added to the sample, and 2 μL was injected into the LC-MS system using the autosampler. Sample was preconcentrated on a Dionex Acclaim PepMap 100 column (100 μm × 2 cm, C18, 5 μm, 100Å) (Dionex, LC Packings) at a flow rate of 4 μL/min and using 0.1% of trifluoroacetic acid as mobile phase. After 3 min of preconcentration, the trap column was automatically switched in-line with a Dionex Acclaim PepMap rapid separation liquid chromatography nanocolumn (75 μm × 15 cm, C18, 2 μm, 100Å) (Dionex, LC Packings). Mobile phases consisted of solvent A, containing 0.1% (v/v) formic acid in water, and solvent B, containing 0.1% (v/v) formic acid in 100% ACN. Chromatographic conditions were a linear gradient from 95 to 60% (v/v) solvent A in 45 min at a flow rate of 0.250 μL/min at 30°C.
The column outlet was directly coupled to a nanoelectrospray ion source. The positive mass spectrum was recorded for a mass-to-charge ratio range of 300 to 2000 followed by MS-MS scans of the three most intense peaks. Typical ion spray voltage was in the range of 2.5 to 3.0 kV, and nitrogen was used as the collision gas. The ion trap was used in ultrascan mode with a maximum accumulation time of 200 ms and an average of 5. Other source parameters and spray positions were optimized with a tryptic digest of BSA protein.
Database Search
Automated spectral processing and peak list generation were performed using Mascot Distiller v2.4.2.0 software (Matrix Science). The database search was done through Mascot Daemon software in combination with the Mascot interface 2.2 (Matrix Science). Regarding searching parameters, Mascot searches were done with no enzymatic specificity and a peptide tolerance on the mass measurement of 100 ppm and 0.6 Da for MS-MS ions. Carbamidomethylation and oxidation of Met were used as variable modifications. Identification of the protein origin of the identified peptides was done using the UniProt protein database. BLAST was used as a basic local alignment search tool to find regions of local similarity between the identified proteins and the protein sequences of tomato (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Immunodetection by Immunoblot Analysis
Proteins were separated by SDS-PAGE (12.5%) for 3 h at a constant current of 80 mA. Proteins were transferred onto polyvinylidene difluoride membranes, and immunodetection was performed as described by Fraser et al. (1994).
Statistical Analysis
A minimum of three biological and three technical replicates were analyzed for every experiment unless stated otherwise. Metabolite levels from the different technology platforms were combined. Principal component analysis was performed on these data matrices. SIMCA-P+ software v. 13.0.2 (Umetrics) was used to carry out and display clusters derived from the principal component analysis. GraphPadPrism software v.5 (GraphPad Software) or Excel (Microsoft) embedded algorithms were used to perform Student’s t tests or Dunnett’s test to determine significant differences between the transgenic lines and the control AC. Where appropriate, P < 0.05, P < 0.01, and P < 0.001 are indicated by one asterisk, two asterisks, and three asterisks, respectively.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Ggpps-1: DQ267902; Ggpps-2: SGNU223568; Psy-1: Y00521; Psy-2: L23424; Pds, X59948; ζ-carotene desaturase, AF195507; carotene isomerase, AF416727; ε-lycopene cyclase, Y14387; Lcy-B, AF254793; fruit-specific β-lycopene cyclase, Y18297; CrtR-b1, Y14809; CrtR-b2, DQ864755; Zep-1: Z83835.1; CrtE, CrtB, and CrtI, D90087.2.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. PCR Confirmation of the Presence of CrtE and CrtI Genes in the CrtE+I Lines.
Supplemental Figure 2. Chromatographic Profiles of Carotenoids, Chlorophylls, and α-Tocopherol of AC and CrtB+I Tomatoes.
Supplemental Figure 3. Separation of Chromoplast Types on a Sucrose Gradient.
Supplemental Figure 4. Principal Component Analysis of All Metabolites Detected in AC and CrtB+I Tomato Lines.
Supplemental Figure 5. Schematic Representation of the Systems (1 and 2) Used to Fractionate Chromoplasts and Isolate Their Respective Submembrane Compartments.
Supplemental Figure 6. Evidence and Importance of the Carotenoid Crystals Embedded in the Chromoplast Membranes.
Supplemental Table 1. Carotenoid Content in the Pericarp, Jelly, and Columella Tissues of Ripe Fruit Derived from the Genetic Crosses Containing Different Gene Combinations.
Supplemental Table 2. Changes in the Composition (in Percentage) of Fatty Acids Present in the Lipids Species Found in CrtB+I and Control AC.
Supplemental Table 3. Identification of Proteins from the Isolated Fractions by nESI-LC-MS-MS.
Supplemental Table 4. Sequences of Primers Used in Real-Time PCR and PCR.
Acknowledgments
This work was supported through the European Union Framework Program 7 METAPRO and DISCO project grants (244348 and 613513, respectively) funded under the Knowledge-Based Bio-Economy program, coordinated by P.D.F. L.M. received a Marie Curie International Fellowship (FOOSAF Project) under European Union Framework Program 7. We thank Ian Brown (University of Kent, UK) for the work provided in electron microscopy analysis and expertise. We also thank Chris Gerrish for excellent technical assistance.
AUTHOR CONTRIBUTIONS
M.N., L.M., P.D.F., and E.M.A.E. performed the experiments. P.D.F., M.N., E.M.A.E., and P.M.B. designed the research program. M.N., P.D.F., P.M.B., and L.M. wrote the article. P.D.F. acquired the funding.
Glossary
- CRTI
phytoene desaturase
- AC
Ailsa Craig
- MGDG
monogalactosyldiacylglycerol
- CRTB
phytoene desaturase
- CRTE
geranylgeranyl diphosphate synthase
- ACN
acetonitrile
- MS
mass spectral
- CAB
cacodylate buffer
- PB
phosphate buffer
- LC-MS
liquid chromatography–mass spectrometry
- nano-LC-MS-MS
nano–liquid chromatography–tandem mass spectrometry
References
- Angaman D.M., Petrizzo R., Hernández-Gras F., Romero-Segura C., Pateraki I., Busquets M., Boronat A. (2012). Precursor uptake assays and metabolic analyses in isolated tomato fruit chromoplasts. Plant Methods 8: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson R.G., Bolitho K.M., Wright M.A., Iturriagagoitia-Bueno T., Reid S.J., Ross G.S. (1998). Apple ACC-oxidase and polygalacturonase: Ripening-specific gene expression and promoter analysis in transgenic tomato. Plant Mol. Biol. 38: 449–460 [DOI] [PubMed] [Google Scholar]
- Auldridge M.E., McCarty D.R., Klee H.J. (2006). Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 9: 315–321 [DOI] [PubMed] [Google Scholar]
- Barsan C., Zouine M., Maza E., Bian W.P., Egea I., Rossignol M., Bouyssie D., Pichereaux C., Purgatto E., Bouzayen M., Latché A., Pech J.C. (2012). Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol. 160: 708–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer P. (2010). Golden Rice and 'Golden' crops for human nutrition. New Biotechnol. 27: 478–481 [DOI] [PubMed] [Google Scholar]
- Cao H., Zhang J., Xu J., Ye J., Yun Z., Xu Q., Xu J., Deng X. (2012). Comprehending crystalline β-carotene accumulation by comparing engineered cell models and the natural carotenoid-rich system of citrus. J. Exp. Bot. 63: 4403–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho L.J.C.B., Lippolis J., Chen S., Batista de Souza C.R., Vieira E.A., Anderson J.V. (2012). Characterization of carotenoid-protein complexes and gene expression analysis associated with carotenoid sequestration in pigmented cassava (Manihot esculenta Crantz) storage root. The Open Biochem. J. 6: 116–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Osto L., Fiore A., Cazzaniga S., Giuliano G., Bassi R. (2007). Different roles of alpha- and beta-branch xanthophylls in photosystem assembly and photoprotection. J. Biol. Chem. 282: 35056–35068 [DOI] [PubMed] [Google Scholar]
- De Camilli P., Harris S.M., Jr, Huttner W.B., Greengard P. (1983). Synapsin I (Protein I), a nerve terminal-specific phosphoprotein. II. Its specific association with synaptic vesicles demonstrated by immunocytochemistry in agarose-embedded synaptosomes. J. Cell Biol. 96: 1355–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B., Adams W.W., III (2002). Antioxidants in photosynthesis and human nutrition. Science 298: 2149–2153 [DOI] [PubMed] [Google Scholar]
- Diretto G., Al-Babili S., Tavazza R., Papacchioli V., Beyer P., Giuliano G. (2007). Metabolic engineering of potato carotenoid content through tuber-specific overexpression of a bacterial mini-pathway. PLoS ONE 2: e350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogbo O., Laferriére A., D’Harlingue A., Camara B. (1988). Carotenoid biosynthesis: Isolation and characterization of a bifunctional enzyme catalyzing the synthesis of phytoene. Proc. Natl. Acad. Sci. USA 85: 7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I., Barsan C., Bian W.P., Purgatto E., Latché A., Chervin C., Bouzayen M., Pech J.C. (2010). Chromoplast differentiation: Current status and perspectives. Plant Cell Physiol. 51: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Enfissi E., et al. (2010). Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P.D., Bramley P.M. (2004). The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 43: 228–265 [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Enfissi E.M., Bramley P.M. (2009). Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch. Biochem. Biophys. 483: 196–204 [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Romer S., Shipton C.A., Mills P.B., Kiano J.W., Misawa N., Drake R.G., Schuch W., Bramley P.M. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. USA 99: 1092–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P.D., Schuch W., Bramley P.M. (2000). Phytoene synthase from tomato (Lycopersicon esculentum) chloroplasts—Partial purification and biochemical properties. Planta 211: 361–369 [DOI] [PubMed] [Google Scholar]
- Fraser P.D., Truesdale M.R., Bird C.R., Schuch W., Bramley P.M. (1994). Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol. 105: 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris W.M., Spurr A.R. (1969). Chromoplasts of tomato fruits. II. The red tomato. Am. J. Bot. 56: 380–389 [Google Scholar]
- Heinig U., Gutensohn M., Dudareva N., Aharoni A. (2013). The challenges of cellular compartmentalization in plant metabolic engineering. Curr. Opin. Biotechnol. 24: 239–246 [DOI] [PubMed] [Google Scholar]
- Hirschberg J. (2001). Carotenoid biosynthesis in flowering plants. Curr. Opin. Plant Biol. 4: 210–218 [DOI] [PubMed] [Google Scholar]
- Jones M.O., Piron-Prunier F., Marcel F., Piednoir-Barbeau E., Alsadon A.A., Wahb-Allah M.A., Al-Doss A.A., Bowler C., Bramley P.M., Fraser P.D., Bendahmane A. (2012). Characterisation of alleles of tomato light signalling genes generated by TILLING. Phytochemistry 79: 78–86 [DOI] [PubMed] [Google Scholar]
- Kachanovsky D.E., Filler S., Isaacson T., Hirschberg J. (2012). Epistasis in tomato color mutations involves regulation of phytoene synthase 1 expression by cis-carotenoids. Proc. Natl. Acad. Sci. USA 109: 19021–19026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilambi H.V., Kumar R., Sharma R., Sreelakshmi Y. (2013). Chromoplast-specific carotenoid-associated protein appears to be important for enhanced accumulation of carotenoids in hp1 tomato fruits. Plant Physiol. 161: 2085–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko K., Bornemisza O., Kourtz L., Ko Z.W., Plaxton W.C., Cashmore A.R. (1992). Isolation and characterization of a cDNA clone encoding a cognate 70-kDa heat shock protein of the chloroplast envelope. J. Biol. Chem. 267: 2986–2993 [PubMed] [Google Scholar]
- Krinsky N.I., Johnson E.J. (2005). Carotenoid actions and their relation to health and disease. Mol. Aspects Med. 26: 459–516 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Li L., Yang Y., Xu Q., Owsiany K., Welsch R., Chitchumroonchokchai C., Lu S., Van Eck J., Deng X.X., Failla M., Thannhauser T.W. (2012). The Or gene enhances carotenoid accumulation and stability during post-harvest storage of potato tubers. Mol. Plant 5: 339–352 [DOI] [PubMed] [Google Scholar]
- Maass D., Arango J., Wüst F., Beyer P., Welsch R. (2009). Carotenoid crystal formation in Arabidopsis and carrot roots caused by increased phytoene synthase protein levels. PLoS ONE 4: e6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal E., Block M.A., Dorne A.J., Douce R., Joyard J. (1997). Lipid synthesis and metabolism in the plastid envelope. Physiol. Plant. 100: 65–77 [Google Scholar]
- Misawa N., Masamoto K., Hori T., Ohtani T., Boger P., Sandmann G. (1994). Expression of an erwinia phytoene desaturase gene not only confers multiple resistance to herbicides interfering with carotenoid biosynthesis but also alters xanthophyll metabolism in transgenic plants. Plant J. 6: 481–489 [Google Scholar]
- Misawa N., Nakagawa M., Kobayashi K., Yamano S., Izawa Y., Nakamura K., Harashima K. (1990). Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 172: 6704–6712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa N., Yamano S., Ikenaga H. (1991). Production of beta-carotene in Zymomonas mobilis and Agrobacterium tumefaciens by introduction of the biosynthesis genes from Erwinia uredovora. Appl. Environ. Microbiol. 57: 1847–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero O., Sánchez-Guijo A., Lubián L.M., Martínez-Rodríguez G. (2012). Changes in membrane lipids and carotenoids during light acclimation in a marine cyanobacterium Synechococcus sp. J. Biosci. 37: 635–645 [DOI] [PubMed] [Google Scholar]
- Mora L., Bramley P.M., Fraser P.D. (2013). Development and optimisation of a label-free quantitative proteomic procedure and its application in the assessment of genetically modified tomato fruit. Proteomics 13: 2016–2030 [DOI] [PubMed] [Google Scholar]
- Neta-Sharir I., Isaacson T., Lurie S., Weiss D. (2005). Dual role for tomato heat shock protein 21: Protecting photosystem II from oxidative stress and promoting color changes during fruit maturation. Plant Cell 17: 1829–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P., Perello C., Rodriguez-Concepcion M. (2012). New insights into plant isoprenoid metabolism. Mol. Plant 5: 964–967 [DOI] [PubMed] [Google Scholar]
- Ravanello M.P., Ke D., Alvarez J., Huang B., Shewmaker C.K. (2003). Coordinate expression of multiple bacterial carotenoid genes in canola leading to altered carotenoid production. Metab. Eng. 5: 255–263 [DOI] [PubMed] [Google Scholar]
- Robertson F.P., Koistinen P.K., Gerrish C., Halket J.M., Patel R.K.P., Fraser P.D., Bramley P.M. (2012). Proteome changes in tomato lines transformed with phytoene synthase-1 in the sense and antisense orientations. J. Exp. Bot. 63: 6035–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer S., Fraser P.D., Kiano J.W., Shipton C.A., Misawa N., Schuch W., Bramley P.M. (2000). Elevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 18: 666–669 [DOI] [PubMed] [Google Scholar]
- Rosso S.W. (1967). An ultrastructural study of the mature chromoplasts of the tangerine tomato (Lycopersicon esculentum var. “golden jubilee”). J. Ultrastruct. Res. 20: 179–189 [DOI] [PubMed] [Google Scholar]
- Rosso S.W. (1968). The ultrastructure of chromoplast development in red tomatoes. J. Ultrastruct. Res. 25: 307–322 [DOI] [PubMed] [Google Scholar]
- Sandmann G. (2009). Evolution of carotene desaturation: The complication of a simple pathway. Arch. Biochem. Biophys. 483: 169–174 [DOI] [PubMed] [Google Scholar]
- Schweiggert R.M., Steingass C.B., Heller A., Esquivel P., Carle R. (2011). Characterization of chromoplasts and carotenoids of red- and yellow-fleshed papaya (Carica papaya L.). Planta 234: 1031–1044 [DOI] [PubMed] [Google Scholar]
- Shimada H., Kondo K., Fraser P.D., Miura Y., Saito T., Misawa N. (1998). Increased carotenoid production by the food yeast Candida utilis through metabolic engineering of the isoprenoid pathway. Appl. Environ. Microbiol. 64: 2676–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumskaya M., Wurtzel E.T. (2013). The carotenoid biosynthetic pathway: Thinking in all dimensions. Plant Sci. 208: 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin A.J., Gaffé J., Alcaraz J.P., Carde J.P., Bramley P.M., Fraser P.D., Kuntz M. (2007). Fibrillin influence on plastid ultrastructure and pigment content in tomato fruit. Phytochemistry 68: 1545–1556 [DOI] [PubMed] [Google Scholar]
- Spurr A.R., Harris M. (1968). Ultrastructure of chloroplasts and chromoplasts in Capsicum annuum l. Thylakoid membrane changes during fruit ripening. Am. J. Bot. 55: 1210–1224 [Google Scholar]
- Szilágyi A., Sommarin M., Akerlund H.E. (2007). Membrane curvature stress controls the maximal conversion of violaxanthin to zeaxanthin in the violaxanthin cycle—Influence of alpha-tocopherol, cetylethers, linolenic acid, and temperature. Biochim. Biophys. Acta 1768: 2310–2318 [DOI] [PubMed] [Google Scholar]
- Vasquez-Caicedo A.L., Heller A., Neidhart S., Carle R. (2006). Chromoplast morphology and beta-carotene accumulation during postharvest ripening of Mango Cv. ‘Tommy Atkins’. J. Agric. Food Chem. 54: 5769–5776 [DOI] [PubMed] [Google Scholar]
- Wellburn A. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Plant Physiol. 144: 307–313 [Google Scholar]
- Wrischer M., Prebeg T., Magnus V., Ljubesic N. (2007). Crystals and fibrils in chromoplast plastoglobules of Solanum capsicastrum fruit. Acta Bot. Croat. 66: 81–87 [Google Scholar]
- Xiao Y.M., Savchenko T., Baidoo E.E.K., Chehab W.E., Hayden D.M., Tolstikov V., Corwin J.A., Kliebenstein D.J., Keasling J.D., Dehesh K. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Chen Y.R., Zheng Y., Huang M., Vrebalov J., McQuinn R., Gapper N., Liu B., Xiang J., Shao Y., Giovannoni J.J. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31: 154–159 [DOI] [PubMed] [Google Scholar]