Abstract
Mycobacterial isolates were obtained by radiometric culture from 33 different species of captive or free-ranging animals (n = 106) and environmental sources (n = 3) from six geographic zones within the United States. The identities of all 109 isolates were confirmed by using mycobactin J dependence and characterization of five well-defined molecular markers, including two integration loci of IS900 (loci L1 and L9), one Mycobacterium avium subsp. paratuberculosis (M. paratuberculosis)-specific sequence (locus 251), and one M. avium subsp. avium-specific marker (IS1245), as well as hsp65 and IS1311 restriction endonuclease analyses. Seventy-six acid-fast isolates were identified as M. paratuberculosis, 15 were identified as belonging to the M. avium-M. intracellulare complex (but not M. paratuberculosis), and the remaining 18 were identified as mycobacteria outside the M. avium-M. intracellulare complex. Fingerprinting by multiplex PCR for IS900 integration loci clustered 67 of the 76 M. paratuberculosis strains into a single clade (designated clade A18) and had a Simpson's diversity index (D) of 0.53. In contrast, sequence-based characterization of a recently identified M. paratuberculosis short sequence repeat (SSR) region enabled the differentiation of the M. paratuberculosis isolates in clade A18 into seven distinct alleles (D = 0.75). The analysis revealed eight subtypes among the 33 species of animals, suggesting the interspecies transmission of specific strains. Taken together, the results of our analyses demonstrate that SSR analysis enables the genetic characterization of M. paratuberculosis isolates from different host species and provide evidence for the host specificity of some M. paratuberculosis strains as well as sharing of strains between wild and domesticated animal species.
Paratuberculosis, or Johne's disease, is a chronic granulomatous gastroenteritis caused by Mycobacterium avium subsp. paratuberculosis (M. paratuberculosis) (46, 59). The disease occurs worldwide and is primarily a disease of domesticated ruminants, including cattle (both beef and dairy), sheep, goats, and farmed deer. Paratuberculosis has been reported to occur in wild ruminant species, including deer (10), bison (7), and elk (13, 40), as well as nonruminants, such as wild rabbits (26), their predators, including foxes and stoats (6), and primates, such as mandrills and macaques (41, 60), indicating a wide host range. In addition to the economic impact on food animal production, with losses estimated to be from $200 million to $250 million annually (47), infections in free-ranging and captive wildlife are also of great concern. Up to one-third of zoos accredited by the American Zoo and Aquarium Association have reported at least one culture-confirmed case of paratuberculosis since 1995 (39). In addition, M. paratuberculosis is of interest because of its potential association with Crohn's disease in humans (19, 28, 29, 51).
The existence and importance of wildlife reservoirs of M. paratuberculosis in the transmission cycle are still undetermined (17), and few investigations have examined the role of wildlife in the epidemiology of this important disease. There is much to learn about the dynamics of transmission of infection within animal populations and the involvement of specific subtypes in determining the characteristics of the infections and the velocity of their spread. A comprehensive analysis of the molecular diversity within M. paratuberculosis strains from various animal species will augment our understanding of the host range, distribution, and natural history of M. paratuberculosis infections and also aid in the development of a population genetic framework for this economically important bacterium. DNA-based subtyping techniques such as multiplex PCR for integration loci (MPIL), amplified fragment length polymorphism (AFLP) analyses, and IS900-based restriction fragment length polymorphism (RFLP) analyses have been used in an attempt to reveal the genetic variation in M. paratuberculosis and differentiate among strains infecting different populations (8, 15, 22, 43, 49, 56). However, these techniques have proved to have a limited ability to discriminate among strains.
The objective of this study was to analyze the distribution and molecular diversity of M. paratuberculosis strains isolated from multiple animal species (captive and free-ranging) from different geographic zones.
MATERIALS AND METHODS
Bacterial isolates.
Fecal and tissue samples from both captive and free-ranging animal species throughout the United States were submitted for radiometric mycobacterial culture (12). Samples were collected from animals suspected of infection with M. paratuberculosis on the basis of exposure to animals with confirmed infections or animals with clinical signs consistent with Johne's disease (weight loss and/or diarrhea). Water samples (n = 3) were also obtained from enclosures housing infected animals. Acid-fast isolates were tested for mycobactin J dependency (a characteristic of M. paratuberculosis) and evaluated for the presence of IS900 by PCR (24). The strains in the collection represented laboratory acquisitions from zoo specimens between 1993 and 2003 (see Table 1). The isolates were assigned a geographic zone on the basis of the locations of the host animals at the time of sample collection. Zones 1 through 6 represent the northwest, southwest, north-central, south-central, northeast, and southeast regions of United States, respectively.
TABLE 1.
Molecular characteristics of acid-fast strains by host and geographic locality
| Animal species or sample | Zonea | L1-L9b | 251c | IS1245d | hsp65e | IS1311f | DMCg | IS900h | MPIL type | G repeati | Indentification | MJ dep.j |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Transcaspian urial | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A1 | 9Gs | MAPk | 1 |
| Addax | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Angolan springbok | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Angolan springbok | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 10Gs | MAP | 1 |
| Axis deer | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Bay duiker | 4 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Bay duiker | 4 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Bay duiker | 4 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | A18 | 7Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | A18 | 7Gs | MAP | 1 |
| Blesbok | 2 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | A18 | 2GsC4Gs | MAP | 1 |
| Blesbok | 2 | 1 | 1 | 1 | 1/0 | 1 | − | 1 | A18 | 2GsC4Gs | MAP | 0 |
| British red deer | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 13Gs | MAP | 1 |
| Chinese Reeve's muntjac | 2 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | A18 | 2GsC4Gs | MAP | 1 |
| Chinese Reeve's muntjac | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Elk | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Elk | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Elk | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Elk | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Elk | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Elk | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Elk | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 13Gs | MAP | 1 |
| Elk | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 13Gs | MAP | 1 |
| Elk | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 13Gs | MAP | 1 |
| Elk | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 13Gs | MAP | 1 |
| Elk | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 15Gs | MAP | 1 |
| Ellipsen waterbuck | 2 | 1 | 1 | 1 | 1/0 | 1 | − | 1 | A18 | 9Gs | MAP | 0 |
| Environmental water | 2 | 1 | 1 | 1 | 1/0 | 1 | − | 1 | A18 | 2GsC4Gs | MAP | 1 |
| Environmental water | 2 | 1 | 1 | 1 | 1/0 | 1 | − | 1 | A18 | 2GsC4Gs | MAP | 0 |
| Gemsbok | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Goat | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Goat | 3 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | A18 | 7Gs | MAP | 1 |
| Impala | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Impala | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Impala | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Indian axis deer | 2 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | A18 | 2GsC4Gs | MAP | 0 |
| Indian axis deer | 2 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | A18 | 2GsC4Gs | MAP | 1 |
| Indian sambar | 2 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | A18 | 2GsC4Gs | MAP | 0 |
| Key deer | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 11Gs | MAP | 1 |
| Malayan sambar | 2 | 1 | 1 | 1 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Nyala | 4 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Nyala | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Oryx | 5 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Sheep | 3 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | A18 | 2GsC4Gs | MAP | 0 |
| Sika | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Thomson gazelle | 6 | 1 | 1 | 1 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Thomson gazelle | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Turkomen markhor | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Turkomen markhor | 2 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | A18 | 2GsC4Gs | MAP | 1 |
| Transcaspian urial | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Transcaspian urial | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Transcaspian urial | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Transcaspian urial | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Transcaspian urial | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Tule elk | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | − | MAP | 1 |
| Tule elk | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | A18 | 7Gs | MAP | 1 |
| Tule elk | 1 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Sitatunga | 6 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 7Gs | MAP | 1 |
| Waterbuck | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| Waterbuck | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| White-tailed gnu | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 9Gs | MAP | 1 |
| White-tailed deer | 5 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 10Gs | MAP | 1 |
| Sheep | 3 | 1 | 1 | 0 | 1 | 0 | 2 | 1 | A18 variant | 15Gs | MAP | 1 |
| Springbok | 6 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | B4 | 2GsC4Gs | MAP | 1 |
| Springbok | 3 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | B4 | 2GsC4Gs | MAP | 1 |
| Bison | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | Unique | − | MAP | 1 |
| Blesbok | 2 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | Unique | 2GsC4Gs | MAP | 1 |
| Dybowski's sika | 2 | 1 | 1 | 0 | 1 | 1 | − | 1 | Unique | − | MAP | 1 |
| Ellipsen waterbuck | 2 | 1 | 1 | 1 | 1/0 | 0 | 1 | 1 | Unique | 2GsC4Gs | MAP | 0 |
| Sika | 2 | 1 | 1 | 0 | 0 | Neg | − | 1 | Unique | − | MAP | 1 |
| Angolan springbok | 2 | 0 | 0 | 1 | 0 | 0 | − | 1 | B11 | 2GsC4Gs | MAICl | 1 |
| Bay duiker | 4 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Bighorn sheep | 1 | 0 | 0 | 1 | 0 | Neg | − | 1 | B11 | − | MAIC | 0 |
| Dama gazelle | 3 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Dom water buffalo | 2 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Eland | 6 | 0 | 0 | 1 | 0 | Neg | − | 0 | B11 | − | MAIC | 0 |
| Elk | 1 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Giraffe | 1 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Giraffe | 4 | 0 | 0 | 1 | 0 | Neg | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Impala | 6 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Munjtac | 3 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Nubian ibex | 2 | 0 | 0 | 1 | 0 | 0 | − | 1 | B11 | 2GsC4Gs | MAIC | 0 |
| Sable antelope | 2 | 0 | 0 | 1 | 1/0 | 0 | − | 1 | B11 | 2GsC4Gs | MAIC | 0 |
| Spanish ibex | 2 | 0 | 0 | 1 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | MAIC | 0 |
| Oryx | 6 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAICm | 0 |
| Armenian mouflon | 2 | 0 | 0 | 0 | 0 | 0 | − | 1 | B11 | 2GsC4Gs | Outside MAIC | 0 |
| Bison | 1 | 0 | 1 | 0 | 0 | Neg | − | 0 | B11 | 2GsC4Gs | Outside MAIC | 0 |
| Springbok | 3 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Elk | 1 | 0 | 0 | 0 | 0 | 0 | − | 0 | B11 | 2GsC4Gs | Outside MAIC | 0 |
| Environmental water | 2 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Gnu | 6 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Gnu | 6 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Goat | 3 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Impala | 6 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Indian gaur | 2 | 0 | 0 | 0 | 1/0 | Neg | − | 1 | B11 | − | Outside MAIC | 0 |
| Key deer | 6 | 0 | 0 | 0 | 1 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Macaque | 3 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Thomson gazelle | 1 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Thomson gazelle | 6 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Toucanet bird | 3 | 0 | 0 | 0 | Neg | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Unknown | 6 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | 7Gs | Outside MAIC | 0 |
| White-tailed deer | 5 | 0 | 0 | 0 | 0 | Neg | − | 0 | B11 | − | Outside MAIC | 0 |
| Transcaspian urial | 2 | 0 | 1 | 1 | 0 | 0 | − | 0 | Unique | 2GsC4Gs | MAIC | 0 |
| Control (ATCC 19698) | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 11Gs | MAP | 1 |
| Control (bovine MAP) | 3 | 1 | 1 | 0 | 1 | 1 | − | 1 | A18 | 10Gs | MAP | 1 |
Zones 1 through 6 represent the northwest, southwest, north-central, south-central, northeast, and southeast regions of United States, respectively.
L1-L9, PCR amplification and hybridization for IS900 integration loci L1 (left side integration site of IS900 into an unknown ORF) and L9 (left side integration of IS900 into alkA); 1, positive; 0, negative.
251, PCR amplification of M. paratuberculosis unique sequence locus 251; 1, positive; 0, negative.
IS1245, PCR amplification and hybridization for a region of IS1245 deleted from M. paratuberculosis; 1, positive; 0, negative.
hsp65, restriction endonuclease analysis for polymorphisms in the hsp65 gene (20, 21); 1, no restriction site; 0, one restriction site.
IS1311, restriction endonuclease analysis for polymorphisms in IS1311 gene; 0, two restriction sites; 1, three restriction sites; neg, no amplification.
DMC, a molecular marker based on a recently identified polymorphism used to differentiate cattle and sheep strains (11); —, not analyzed.
IS900, IS900 PCR amplification; 1, positive; 0, negative.
G repeat, amplification and sequencing of a locus containing variable numbers of G-residue repeats (2); −, amplification or sequencing unsuccessful.
MJ dep., mycobactin J growth dependence; 1, growth is dependent on mycobactin J supplementation; 0, growth is not dependent on mycobactin J supplementation.
MAP, M. paratuberculosis.
MAIC, M. avium-M. intracellulare complex.
Outside MAIC, mycobacteria outside M. avium-M. intracellulare complex.
Freezer stocks of the mycobacterial isolates were revived in 7H9 broth (Becton, Dickinson, Sparks, Md.) for molecular identification and diversity analysis by an independent laboratory in a blinded fashion. The identities of the strains were revealed after molecular analyses were complete. Five controls including type strain ATCC 19698, a bovine M. paratuberculosis strain, and three deliberately mixed cultures (known to contain M. paratuberculosis and M. paratuberculosis in one culture and M. paratuberculosis and M. avium subsp. avium in the other two cultures) were also analyzed.
DNA extraction.
Approximately 10 ml of turbid 7H9 broth culture was centrifuged at 2,500 rpm (Beckman GP; Beckman Institute, Urbana, Ill.) for 25 min to obtain a pellet. The pellet was resuspended in 500 μl of sterile deionized distilled water and used for DNA extraction by use of the QIAamp DNA Blood Mini kit (Qiagen Inc., Valencia, Calif.), with a few modifications, as described previously (43).
Molecular characterization of the isolates.
DNA extracted from the broth cultures was used to confirm the identities of the subspecies by characterizing all isolates with well-defined molecular markers as described previously (43): (i) PCR amplification and hybridization for two of the integration loci of insertion sequence IS900 regarded as diagnostically definitive for M. paratuberculosis (loci L1 [left side integration site of IS900 into an unknown open reading frame] and L9 [left side integration of IS900 into alkA]); (ii) PCR amplification of a recently identified unique M. paratuberculosis sequence (locus 251) (4, 43), and (iii and iv) hsp65 and IS1311 restriction endonuclease analyses for polymorphisms in the hsp65 and IS1311 genes, respectively, as described previously (43). PCR amplification of an additional DNA sequence that differed between the sheep and cattle types of M. paratuberculosis was carried out as described previously (11).
An M. avium subsp. avium-specific marker (IS1245) was used to distinguish the M. paratuberculosis and the M. avium subsp. avium isolates (27, 33). Primers were designed to amplify a 300-bp region of IS1245 in M. avium subsp. avium that is similar to but not homologous with a closely related insertion sequence (IS1311) found in M. paratuberculosis (gene information identification number [GI], 7555405 and 4210754). The DNA samples were amplified with primers 5′-biotin-GGT CGC GTG TCC GCG TGT GG-3′ and 5′-ACT TCC CGG TGG CCC ACT GGA-3′. The primers were used at final concentrations of 0.2 μM in a 50-μl PCR master mixture that also included 1.25 U of Hotstar Taq polymerase (Qiagen Inc.), 1× PCR buffer, 2.5 mM MgCl2, a 200 μM concentration of each deoxyribonucleoside triphosphate (Amersham Pharmacia Biotech Inc., Piscataway, N.J.), 5% dimethyl sulfoxide, and 5 μl (approximately 50 ng/μl) of genomic DNA. The thermal cycler parameters used for amplification included an initial incubation step at 95°C for 15 min to activate the Hotstar Taq polymerase, followed by 35 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 20 s, and extension at 72°C for 20 s, with a final extension step at 72°C for 7 min. A microtiter plate-based reverse-phase hybridization assay was carried out with the probe 5′-CTC GCT CTG CTC GAC GTC AGT GAC CAA AGT GCC GAA AC-3′ as described previously (43, 53). A negative control consisting of the PCR master mixture alone, without genomic DNA, was included in all PCR amplifications and hybridizations. The cutoff optical density was set at 0.2 on the basis of analysis of several known negative controls and M. avium subsp. avium positive controls.
IS900 PCR was performed as described previously (42). The PCR products were electrophoresed at 100 V for 2 h in a 1.5% agarose gel prestained with ethidium bromide and visualized on a UV transilluminator (Alpha Innotech Corporation, San Leandro, Calif.).
MPIL fingerprinting analysis.
Genotyping of all isolates by MPIL fingerprinting analysis was performed by a previously established method (8, 9, 43). Briefly, a total of 28 DNA fragments representing the right and left integration sites of IS900 were amplified in six sets of multiplex PCRs, referred as 9R1, 5R2, 4L1, 4L2, 3L3, and 3L4, respectively, as described previously (43). The PCR products were electrophoresed at 100 V for 4 h in a 2% agarose gel prestained with ethidium bromide and visualized on a UV transilluminator (Alpha Innotech Corporation). Cluster analysis was done with the Molecular Evolutionary Genetics Analysis (MEGA) program (version 2.1; www.megasoftware.net) by the unweighted pair-group method with arithmetic averages (36). The distance matrix for input into the MEGA program was created from the binary data by using the ETDIV and ETMEGA programs (http://foodsafe.msu.edu/whittam/programs/).
SSR analysis.
The results of the recently described multilocus short sequence repeat (MLSSR) analysis (2) for M. paratuberculosis strain differentiation indicated that the mononucleotide G-residue repeat locus within the phosphatidylethanolamine-binding domain (GI, 13881618) was the most discriminatory (Simpson's diversity index, 0.7) and was selected for use for fingerprinting in this study. Amplification of this short sequence repeat (SSR) locus was carried out with primers 5′-TCA GAC TGT GCG GTA TGG AA-3′ and 5′-GTG TTC GGC AAA GTC GTT GT-3′. The amplification parameters were as described above for IS1245 analysis. A PCR master mixture blank was included as a negative control for each batch. A total of 5 μl of the PCR product was electrophoresed at 125 V for 45 min in 1.5% agarose gels prestained with ethidium bromide and visualized on a UV transilluminator (Alpha Innotech Corporation). The PCR products were purified with a QIAquick PCR purification kit (Qiagen Inc., Valencia, Calif.) and sequenced by using standard dye terminator chemistry, and the sequences were analyzed on an automated DNA sequencer (3700 DNA Analyzer; Applied Biosystems). All chromatograms were visually inspected, and sequences were edited with the EditSeq program (DNASTAR, Madison, Wis.) to correct ambiguities and then aligned by use of the MegAlign program (DNASTAR).
A consensus phylogenetic tree was generated with the MEGA program (version 2.1) (36) by use of a maximum-parsimony model with 1,000 bootstrap replications. Numbers were assigned to each clade to reflect the number of G residues in the polymorphic region. Simpson's and Shannon-Wiener's diversity indices were calculated by use of the equations 1 − Σ(allele frequency)2 and −1 × Σ(allele frequency × ln allele frequency)2, respectively (45).
RESULTS
Molecular characterization of the isolates derived from wildlife species.
The results of the molecular target analyses are summarized in Table 1. An isolate was characterized as M. paratuberculosis if the profile was L1 and L9 positive, locus 251 positive, and IS1245 negative and carried no hsp65 polymorphism, as detected by restriction enzyme analysis (43). An isolate was characterized as belonging to the M. avium-M. intracellulare complex if the profile was L1 and L9 negative, locus 251 negative, and IS1245 positive and carried the polymorphism in hsp65, resulting in a truncated band upon restriction digestion with PstI. All other acid-fast isolates that failed to amplify any of the targets were classified as mycobacteria outside the M. avium-M. intracellulare complex. On the basis of these classification criteria, 76 isolates were identified as M. paratuberculosis, 15 were identified as belonging to the M. avium-M. intracellulare complex, and 18 were identified as mycobacteria outside the M. avium-M. intracellulare complex. All M. paratuberculosis isolates were further classified by using the IS1311 restriction patterns consistently identified in either cattle (two restriction sites) or sheep strains (one restriction site) (43, 57). Of the 76 isolates classified as M. paratuberculosis, 12 isolates from nine different animal species (Table 1) had the IS1311 restriction profile reported for sheep strains. The 12 M. paratuberculosis isolates with IS1311 profiles typical for sheep strains were further analyzed by using a recently described molecular marker to distinguish sheep and cattle isolates (11). That analysis identified 10 isolates derived from several host species with the genotype reported for cattle strains. Of the other two sheep isolates, one had a cattle strain genotype, which indicated that it was a variant. Non-M. paratuberculosis M. avium-intracellulare complex isolates (n = 12) and those outside the M. avium-M. intracellulare complex (n = 2) also had the IS1311 profile typical of sheep strains. The IS1311 region targeted failed to be amplified from 3 M. avium-M. intracellulare complex isolates and 16 mycobaterial isolates outside the M. avium-M. intracellulare complex. All the blinded controls, including the deliberately mixed cultures, were accurately identified by these molecular analyses.
All isolates classified as M. paratuberculosis by molecular analysis were positive by the IS900 PCR. Additionally, four isolates classified as belonging to the M. avium-M. intracellulare complex and two isolates classified as mycobacteria outside the M. avium-M. intracellulare complex were positive by the IS900 PCR.
Seven of the isolates classified as M. paratuberculosis by independent molecular methods as well as by IS900 PCR were apparently not mycobactin J dependent. Two isolates (one non-M. paratuberculosis M. avium-M. intracellulare complex isolate and one non-M. avium-M. intracellulare complex isolate) were positive for locus 251 by PCR. Sequence analysis of the locus 251 amplicon from the M. avium-M. intracellulare complex isolate showed 100% homology to the published sequence (GI, 20152939).
Fifteen of the M. paratuberculosis isolates were also positive for the M. avium subsp. avium-specific marker, IS1245. Since attempts were not made to isolate single colonies, it is possible that the M. paratuberculosis repository stocks contained a low level of DNA of a non-M. paratuberculosis strain whose genetic material was masked for molecular typing purposes by the M. paratuberculosis isolate in the aliquot.
MPIL fingerprinting analysis.
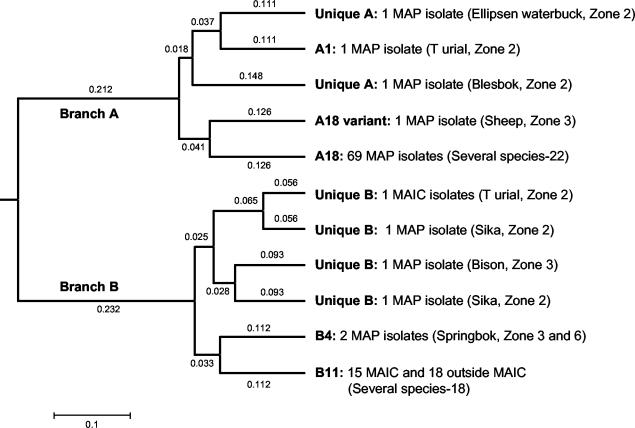
All isolates studied (n = 109) and two M. paratuberculosis control isolates (ATCC 19698 and a field isolate from a cow) were fingerprinted by MPIL analysis. Cluster analysis organized the fingerprints into 30 electrophoretic types. Electrophoretic types that were separated by distances less than 0.1 unit were assigned identical MPIL fingerprints (Fig. 1). The fingerprint nomenclature was based on a previously defined set of M. paratuberculosis MPIL fingerprints in our collection (43). Both data sets were merged for cluster analysis and fingerprint assignment. The analysis divided the isolates into two major branches. Ninety-four percent (73 of 76) of the M. paratuberculosis isolates from several species clustered in branch A and could be further divided into A18 (n = 69), A18 variant (n = 1), A1 (n = 1), and unique branch A (n = 2) fingerprints. Branch A exclusively clustered only M. paratuberculosis isolates. All non-M. paratuberculosis M. avium-M. intracellulare complex isolates (n = 15) clustered into branch B, with the fingerprints designated B11 (n = 14) and unique (n = 1). Mycobacterial isolates outside the M. avium-M. intracellulare complex (n = 18) did not amplify any target and were clustered into branch B, with the fingerprints designated B11. Five M. paratuberculosis strains derived from a bison, springbok (n = 2), and sika (n = 2) also clustered in branch B, with the fingerprints designated B4 (n = 2) and unique B (n = 3). The Simpson's and Shannon-Wiener's diversity index values for the MPIL fingerprint analysis were 0.533 and 1.067, respectively, indicating limited discriminatory capacity.
FIG. 1.
Dendrogram showing the distribution of MPIL fingerprints. The assigned fingerprints, isolate identities, and host species are shown. The proportion distance of each cluster is also indicated. MPIL analysis had a Simpson's diversity index of 0.533, indicative of a limited degree of strain discrimination capability. Shannon's diversity index was 1.067. MAP, M. paratuberculosis; MAIC, non-M. paratuberculosis M. avium-M. intracellulare complex; Outside MAIC, mycobacteria outside the M. avium-M. intracellulare complex.
SSR analysis.
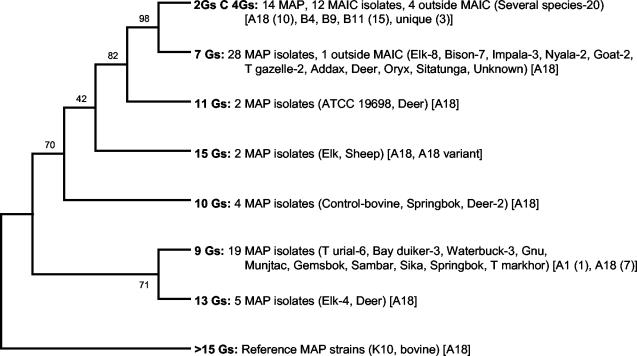
A region within the M. paratuberculosis genome that carries various numbers of G residues was amplified from all isolates. Only isolates (n = 91) with a detectable 408-bp amplification product were sequenced for further analysis. These included M. paratuberculosis (n = 72), non-M. paratuberculosis M. avium-M. intracellulare complex organisms (n = 12), organisms outside the M. avium-M. intracellulare complex (n = 5), and control M. paratuberculosis strains (ATCC 19698 and a field isolate from a cow). Of the 72 M. paratuberculosis strains analyzed, 66 had the A18 MPIL fingerprint, while 2 carried unique fingerprints. The remaining four isolates carried the A1, B4, and B11 (n = 2) fingerprints. Both of the control M. paratuberculosis isolates displayed the A18 fingerprint. Two reference strains (M. paratuberculosis K10 and a bovine M. paratuberculosis isolate [2]) were also included in the cluster analysis. Each allele was assigned a number congruent with the number of G-residue repeats. A total of eight alleles with 7 to 20 G-residue repeats (7Gs to 20Gs, respectively) were identified among 76 M. paratuberculosis isolates (including control and reference strains). Thirty-eight percent of the M. paratuberculosis isolates (n = 28) had the 7Gs repeat fingerprint. This fingerprint clustered all the bison (n = 7), impala (n = 3), nyala (n = 2), Thomson gazelle (n = 2), and goat (n = 2) M. paratuberculosis strains included in this analysis. This allele also clustered 8 of the 13 M. paratuberculosis isolates obtained from elks. The 7Gs alleles were also found in one M. paratuberculosis isolate each from an addax, axis deer, oryx, and sitatunga and in one mycobacterial isolate outside the M. avium-M. intracellulare complex from an unknown source. Alleles with 9Gs clustered all of the bay duiker (n = 3), all of the Transcaspian urial (n = 6), and three of the four waterbuck M. paratuberculosis isolates. It also included one M. paratuberculosis isolate from each of the following: springbok, munjtac, gemsbok, sambar, sika, markhor, and gnu. Alleles with 10Gs were found in M. paratuberculosis isolates from the bovine control, springbok, key deer, and white-tailed deer. Alleles with 11Gs were found in M. paratuberculosis type strain ATCC 19698 and another key deer isolate. Alleles with 13Gs were found in M. paratuberculosis strains isolated from elk (n = 4) and a British red deer (n = 1). Alleles with 15Gs were found in M. paratuberculosis isolates from an elk and a sheep. Isolates with more than 15Gs were clustered into an allele designated >15Gs. Isolates with this allele included the reference strain M. paratuberculosis K10, with 20Gs, and a bovine M. paratuberculosis isolate, with 17Gs. Allele 2GsC4Gs, in which a C residue replaced the third G residue within the repeat, was also identified. All non-M. paratuberculosis M. avium-M. intracellulare complex isolates (n = 12), four of five non-M. avium-M. intracellulare complex isolates, and some M. paratuberculosis strains (n = 13) carried this fingerprint. The species whose isolates included this allele are listed in Table 1. The phylogenetic analysis thus divided the M. paratuberculosis isolates (n = 66) with the A18 fingerprint into seven distinct alleles (Fig. 2). The Simpson's and Shannon-Wiener's diversity index values for the G-residue-repeat analysis were 0.751 and 1.593, respectively, indicating a relatively robust discriminatory capability.
FIG. 2.
Phylogenetic tree showing the distribution of strains by the numbers of G-residue repeats. The numbers of G-residue repeats, isolate identities, host species, and major fingerprints obtained by MPIL analysis are shown. Also shown at the clade origins are the bootstrap values generated by 1,000 replications in a maximum-parsimony model. SSR analysis had a Simpson's diversity index of 0.751, indicative of a high degree of strain discrimination capability. Shannon's diversity index was 1.593. MAP, M. paratuberculosis; MAIC, non-M. paratuberculosis M. avium-M. intracellulare complex; Outside MAIC, mycobacteria outside the M. avium-M. intracellulare complex.
DISCUSSION
Paratuberculosis has been well documented in a majority of domestic ruminant species. It has gained importance in the animal production industry because of the economic losses incurred from herd infections and possible human health hazards associated with M. paratuberculosis (3, 29, 51). M. paratuberculosis has also been recovered from many captive and free-ranging nondomestic animal species representing virtually all pseudoruminants and ruminants except giraffids (37, 38). The known host range of M. paratuberculosis has recently been extended to include nonruminant wildlife species, such as primates (41, 60), wild rabbits (25), and foxes and stoats (6). These reports support the contention that M. paratuberculosis has a wide host range and that disease caused by this organism may have an epidemiology more complex than was previously known.
One of the strategies for the control and eradication of paratuberculosis in an infected herd is to eliminate the transmission of M. paratuberculosis to susceptible animals. The presence of a wildlife reservoir with the potential to transmit the infection to domestic animals may affect the success of domestic agriculture control programs. Although the frequency of transmission from wildlife to domestic animals has not been documented, several reports suggest that infection may be spread from domestic animals to wildlife (13, 25). Knowledge of the extent of strain sharing across different host species is vital to understanding the dynamics of M. paratuberculosis transmission. Methods for differentiation or subtyping of bacterial strains provide important information for molecular epidemiological analyses and help provide an understanding of the population genetics of the organism. This is the first report of a comprehensive molecular analysis conducted to establish the degree of similarity or heterogeneity in M. paratuberculosis isolates from taxonomically and spatially diverse host species.
Definitive identification of M. paratuberculosis requires confirmation with multiple molecular markers.
Present methods for the diagnosis of M. paratuberculosis infection include isolation of the organism from fecal and tissue specimens, antibody detection by enzyme-linked immunosorbent assay, and IS900-based PCR (55). The culture protocol used at present relies on mycobactin J dependency as a confirmatory test. Our analysis identified seven M. paratuberculosis isolates (9%) that were not dependent on mycobactin J for growth. Mycobactin J dependency was determined by two serial subcultures on slants with and without mycobactin J supplementation for each of the seven M. paratuberculosis isolates. Although attempts were not made to isolate single colonies, all subcultures were carried out with inocula from slants without mycobactin J to rule out potential mycobactin J carryover. This discrepancy indicates that diagnostic tests that rely on mycobactin J dependency alone need to be interpreted with caution.
IS900-based PCR identification techniques have routinely been used for the detection of M. paratuberculosis (5, 30, 58). However, IS900-like elements have been found in M. avium subsp. avium isolates (44) and in some isolates outside the M. avium-M. intracellulare complex (14, 20, 34). In the present study, four isolates that were classified within the M. avium-M. intracellulare complex but that were not M. paratuberculosis and two isolates classified outside the M. avium-intracellulare complex by extensive molecular analyses were identified as M. paratuberculosis by IS900 PCR. Only one of these isolates was apparently mycobactin J dependent. This implies that the detection of M. paratuberculosis based on detection of IS900 alone needs to be evaluated with caution to avoid false-positive results.
Although a previous analysis indicated that locus 251 is unique to M. paratuberculosis (43), an M. avium-M. intracellulare complex isolate amplified a PCR product of the expected size and sequence analysis revealed 100% homology of that product to the published sequence of locus 251 (GI, 20152939). These discrepancies clearly illustrate the need for multiple molecular markers for confirmation of the identity of M. paratuberculosis as the cause of infection. Molecular characterization of acid-fast mycobacterial isolates by use of three specific markers and two polymorphic sites aided in the accurate classification of the isolates. In addition, strains that lacked mycobactin J dependency or that were positive by IS900 PCR or locus 251 PCR could be correctly identified on the basis of the presence or the absence of multiple molecular targets.
Molecular diversity analysis of M. paratuberculosis.
Several attempts have been made to identify genetic variation and host specificity in M. paratuberculosis strains isolated from different animal species. Until recently, IS900 has been the marker of choice for most fingerprinting studies that have been reported (5, 15, 50, 57). While the IS900-based RFLP analyses are fairly good at discriminating between cattle and sheep M. paratuberculosis strains, M. paratuberculosis strains from cattle and other hosts such as goats and rabbits are indistinguishable by this method (5, 25, 49). A recent study in our laboratory (43) by alternate fingerprinting techniques, MPIL and AFLP, demonstrated clustering of 73 and 56% of the M. paratuberculosis isolates, respectively, from several hosts (cattle, sheep, goats, mice, deer, and humans). These results were consistent with the hypothesis that there is a relatively small amount of genetic heterogeneity between M. paratuberculosis isolates obtained from different host species.
Many measures of diversity have been proposed, but those that are most commonly used are Simpson's and Shannon-Wiener's indices. Simpson's diversity index represents the probability that two individual isolates randomly selected from a sample will belong to different species and accounts for both the richness (diversity) and the proportion (percent) of each species. Simpson's diversity indices for RFLP analysis (n = 1,008) (50), MPIL analysis (n = 247) (43), and AFLP analysis (n = 104) (43) are 0.559, 0.597, and 0.711 respectively. The higher diversity index for AFLP analysis indicates that AFLP analysis offers a better discriminatory ability. However, the clustering by AFLP analysis was random with respect to host species and geographic location and hence was not informative (43). In addition, AFLP analysis is technically demanding, and band profiles cannot be interpreted in terms of loci and alleles.
Restricted diversity among M. paratuberculosis isolates is also revealed by MPIL analysis.
In the present study, MPIL fingerprinting analysis separated the M. paratuberculosis and M. avium-M. intracellulare complex strains into two major clusters (Fig. 1). Within the M. paratuberculosis cluster, the A18 fingerprint predominated, with 88% of the M. paratuberculosis isolates having the A18 fingerprint. The other M. paratuberculosis strains exhibited fingerprints designated A1 (n = 1), A18 variant (n = 1), B4 (n = 2), and unique (n = 5). MPIL cluster analysis had a Simpson's diversity index of 0.533 and a Shannon-Wiener's diversity index of 1.067, indicating that the MPIL technique has a limited degree of strain discrimination capability.
A previous study has shown that nine of the MPIL types correspond to a distinct PstI and BstEII RFLP types (8). This supports the idea that both MPIL and RFLP analyses address the same genetic variation and suggests that MPIL typing may be used as a substitute for RFLP typing. Hence, we predict that RFLP analysis of these isolates would result in similarly indistinguishable fingerprints. The results of MPIL analysis suggest that only a few M. paratuberculosis strains may be responsible for the widespread dissemination of infection across a variety of species. Conversely, the methods available at present lack sufficient sensitivities for the differentiation of M. paratuberculosis strains. We thus evaluated the possibility of resolving the A18 fingerprint using an alternate fingerprinting technique.
SSR sequencing enables high-resolution subtyping of M. paratuberculosis isolates from domestic and wild animal species.
Restricted allelic variation in mycobacteria is well established (54), and given the small genome size (4.83 Mbp), the expected frequency of polymorphism in M. paratuberculosis is low (18). However, there may be specific regions within the genome that have higher rates of polymorphisms. If present, these regions could be used to genotype the organisms. Recent studies (16, 52) with Bacillus anthracis, a similarly monomorphic organism, demonstrated the usefulness of single nucleotide polymorphisms, variable-number tandem repeats (VNTRs), and inserted or deleted sequences in discriminating strains within the species. VNTRs or SSRs are generated through natural events such as recombination and slipped-strand mispairing during replication (31). These have successfully been used as markers to understand the clonalities and distributions of subtypes in several bacterial species, such as Mycobacterium tuberculosis (23), Yersinia pestis (1), and B. anthracis (32, 35).
The recent completion of the whole-genome sequence of M. paratuberculosis strain K10 (L. L. Li et al., unpublished data) allowed the identification of several SSRs in the genome. Eleven of these highly polymorphic SSRs were used in a composite MLSSR analysis (2) for M. paratuberculosis strain differentiation. The results indicated that a mononucleotide G-residue-repeat locus within the phosphatidylethanolamine-binding domain (GI, 13881618) was the most discriminatory (Simpson's diversity index, 0.7) and was selected for fingerprinting in this study.
The G-residue-repeat fingerprinting analysis reported here had a Simpson's diversity index of 0.751, indicative of a comparatively higher degree of strain discrimination capability. Shannon-Wiener's diversity index was 1.593. Phylogenetic analysis of G-residue-repeat sequences divided the M. paratuberculosis isolates from the A18 cluster (n = 66) into seven different alleles (Fig. 2).
SSR analysis provides strong evidence of interspecies strain transmission and host specificity among isolates of M. paratuberculosis.
Interestingly, there appeared to be a relation between allele type and host species. For example, all the bison (n = 7), impala (n = 3), nyala (n = 2), Thomson gazelle (n = 2), and goat (n = 2) M. paratuberculosis isolates included in this analysis clustered into allele 7Gs. Similarly, the allele designated 9Gs clustered all of the bay duiker (n = 3), all of the Transcaspian urial (n = 6), and three of the four waterbuck M. paratuberculosis isolates. The elk isolates were divided between two alleles: 7Gs (n = 8) and 13Gs (n = 4). One of the elk isolates carried the 15Gs allele. This finding is significant, since all previous analyses, in which a variety of fingerprinting techniques were used, have failed to find any association between the fingerprint type and the host species (43, 48, 50). Concordance analysis of the G-residue-repeat alleles from each geographic zone showed no correlation between fingerprint types and geographic zones. Although many of the M. paratuberculosis isolates from the same host species were from the same geographic zone, kappa analysis for the concordant pairs clearly indicated that the association between allele type and host species was not due to an origin in a similar geographic region. This is not surprising, since many of the captive animals may have been born in geographic zones different from the ones in which the samples were collected. That is, they likely acquired the M. paratuberculosis infection in the first 6 months of life but had been transferred to another facility in a different zone as an adult. Animal movement among institutions is common in the zoo industry. The presence of multiple strains within a single facility suggests that more than one source for the organism (and, presumably, more than one source for the hosts) contributed to the infections in the animal collection. While the presence of multiple strains within a facility suggested that the isolates might have originated from diverse locations, the consistency of identical genotypes within host species suggests that transmission may have occurred within facilities because animals are housed in exhibits as single species or herds.
All M. paratuberculosis (n = 30) isolates from geographic zone 2 were acquired at the same time from the same animal facility during an apparent Johne's disease outbreak. Fingerprinting by MPIL analysis showed the presence of at least five distinct banding patterns: A18, B11, and unique (n = 3). Similarly, SSR analysis identified the presence of multiple strains: strains with the 9Gs, 10Gs, 13Gs, and 2GsC4Gs alleles. This clearly indicates the involvement of more than one M. paratuberculosis strain in the outbreak. This suggests the acquisition of infection from different sources. Alternately, multiple genotypes could indicate the occurrence of strains with restricted host ranges.
G-residue-repeat analysis clustered the non-M. paratuberculosis M. avium-M. intracellulare complex isolates (n = 12) and those outside the M. avium-M. intracellulare complex (n = 4) into a group with one common allele, the 2GsC4Gs allele. This group also contained 14 M. paratuberculosis isolates from several species. We noted that all 11 M. paratuberculosis isolates (Table 1) had sheep strain-like IS1311 restriction profiles, suggesting that isolates with the sheep strain genotypes are likely to be ancestral to those that are of the bovine strain genotype. The three M. paratuberculosis isolates from environmental sources (n = 2) and a blesbok that clustered into this fingerprint had IS1311 restriction profiles consistent with those of the cattle strains. An M. paratuberculosis isolate from a sheep with an A18 variant fingerprint was the only isolate that did not carry the 2GsC4Gs fingerprint. This indicates that these M. paratuberculosis isolates were phylogenetically closer to the M. avium complex strains than the cattle strain genotypes. Together the analyses suggest that the sheep strains may have emerged before the bovine host specialists.
Concluding comments.
Our analyses document a relationship between fingerprint types and host species. The results also provide strong evidence that SSR analysis with G-residue-repeat sequences can be used to study strain sharing and interspecies spread. The approach can also be applied to investigate the role of multiple or single dominant strains in the dynamics of transmission of M. paratuberculosis infections within and between domestic ruminants and wildlife reservoirs. A similar analysis could potentially be used to illustrate the presence or the lack of an association of the human and animal strains. This will allow us to bridge the gap in our understanding of the epidemiology and evolution of M. paratuberculosis, which will subsequently lead to more targeted and more robust control strategies.
Acknowledgments
This study was supported by state and federal funds appropriated to the Ohio Agricultural Research and Development Center (OARDC), including an OARDC Competitive Research Enhancement Seed Grant, awarded to S. Sreevatsan. Research in the laboratory of Vivek Kapur is supported by competitive grants from the National Institutes of Health, National Science Foundation, and U.S. Department of Agriculture's ARS, CSREES, and VS programs.
REFERENCES
- 1.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amonsin, A., L. L. Li, Q. Zhang, J. P. Bannantine, A. S. Motiwala, S. Sreevatsan, and V. Kapur. 2004. Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42:1694-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2001. CAST issue paper: Johne's disease in cattle. Council on Agriculture, Science and Technology Task Force. Council on Agriculture, Science and Technology, Washington, D.C.
- 4.Bannantine, J. P., E. Baechler, Q. Zhang, L. Li, and V. Kapur. 2002. Genome scale comparison of Mycobacterium avium subsp. paratuberculosis with Mycobacterium avium subsp. avium reveals potential diagnostic sequences. J. Clin. Microbiol. 40:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauerfeind, R., S. Benazzi, R. Weiss, T. Schliesser, H. Willems, and G. Baljer. 1996. Molecular characterization of Mycobacterium paratuberculosis isolates from sheep, goats, and cattle by hybridization with a DNA probe to insertion element IS900. J. Clin. Microbiol. 34:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beard, P. M., M. J. Daniels, D. Henderson, A. Pirie, K. Rudge, D. Buxton, S. Rhind, A. Greig, M. R. Hutchings, I. McKendrick, K. Stevenson, and J. M. Sharp. 2001. Paratuberculosis infection of nonruminant wildlife in scotland. J. Clin. Microbiol. 39:1517-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buergelt, C. D., and P. E. Ginn. 2000. The histopathologic diagnosis of subclinical Johne's disease in North American bison (Bison bison). Vet. Microbiol. 77:325-331. [DOI] [PubMed] [Google Scholar]
- 8.Bull, T. J., J. Hermon-Taylor, I. Pavlik, F. El Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146(Pt 9):2185-2197. [DOI] [PubMed] [Google Scholar]
- 9.Bull, T. J., J. Hermon-Taylor, I. I. Pavlik, F. El-Zaatari, and M. Tizard. 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing. Microbiology 146(Pt 12):3285. [DOI] [PubMed] [Google Scholar]
- 10.Chiodini, R. J., and H. J. Van Kruiningen. 1983. Eastern white-tailed deer as a reservoir of ruminant paratuberculosis. J. Am. Vet. Med. Assoc. 182:168-169. [PubMed] [Google Scholar]
- 11.Collins, D. M., M. De Zoete, and S. M. Cavaignac. 2002. Mycobacterium avium subsp. paratuberculosis strains from cattle and sheep can be distinguished by a PCR test based on a novel DNA sequence difference. J. Clin. Microbiol. 40:4760-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, M. T., K. B. Kenefick, D. C. Sockett, R. S. Lambrecht, J. McDonald, and J. B. Jorgensen. 1990. Enhanced radiometric detection of Mycobacterium paratuberculosis by using filter-concentrated bovine fecal specimens. J. Clin. Microbiol. 28:2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook, W. E., T. E. Cornish, S. Shideler, B. Lasley, and M. T. Collins. 1997. Radiometric culture of Mycobacterium avium paratuberculosis from the feces of tule elk. J. Wildl. Dis. 33:635-637. [DOI] [PubMed] [Google Scholar]
- 14.Cousins, D. V., R. Whittington, I. Marsh, A. Masters, R. J. Evans, and P. Kluver. 1999. Mycobacteria distinct from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: implications for diagnosis. Mol. Cell. Probes 13:431-442. [DOI] [PubMed] [Google Scholar]
- 15.Cousins, D. V., S. N. Williams, A. Hope, and G. J. Eamens. 2000. DNA fingerprinting of Australian isolates of Mycobacterium avium subsp. paratuberculosis using IS900 RFLP. Aust. Vet. J. 78:184-190. [DOI] [PubMed] [Google Scholar]
- 16.Cummings, C. A., and D. A. Relman. 2002. Genomics and microbiology. Microbial forensics—“cross-examining pathogens”. Science 296:1976-1979. [DOI] [PubMed] [Google Scholar]
- 17.Daniels, M. J., M. R. Hutchings, P. M. Beard, D. Henderson, A. Greig, K. Stevenson, and J. M. Sharp. 2003. Do non-ruminant wildlife pose a risk of paratuberculosis to domestic livestock and vice versa in Scotland? J. Wildl. Dis. 39:10-15. [DOI] [PubMed] [Google Scholar]
- 18.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Zaatari, F. A., M. S. Osato, and D. Y. Graham. 2001. Etiology of Crohn's disease: the role of Mycobacterium avium paratuberculosis. Trends Mol. Med. 7:247-252. [DOI] [PubMed] [Google Scholar]
- 20.Englund, S., A. Ballagi-Pordany, G. Bolske, and K. E. Johansson. 1999. Single PCR and nested PCR with a mimic molecule for detection of Mycobacterium avium subsp. paratuberculosis. Diagn. Microbiol. Infect. Dis. 33:163-171. [DOI] [PubMed] [Google Scholar]
- 21.Eriks, I. S., K. T. Munck, T. E. Besser, G. H. Cantor, and V. Kapur. 1996. Rapid differentiation of Mycobacterium avium and M. paratuberculosis by PCR and restriction enzyme analysis. J. Clin. Microbiol. 34:734-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Francois, B., R. Krishnamoorthy, and J. Elion. 1997. Comparative study of Mycobacterium paratuberculosis strains isolated from Crohn's disease and Johne's disease using restriction fragment length polymorphism and arbitrarily primed polymerase chain reaction. Epidemiol. Infect. 118:227-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gascoyne-Binzi, D. M., R. E. Barlow, R. Frothingham, G. Robinson, T. A. Collyns, R. Gelletlie, and P. M. Hawkey. 2001. Rapid identification of laboratory contamination with Mycobacterium tuberculosis using variable number tandem repeat analysis. J. Clin. Microbiol. 39:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greig, A., K. Stevenson, D. Henderson, V. Perez, V. Hughes, I. Pavlik, M. E. Hines, I. McKendrick, and J. M. Sharp. 1999. Epidemiological study of paratuberculosis in wild rabbits in Scotland. J. Clin. Microbiol. 37:1746-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greig, A., K. Stevenson, V. Perez, A. A. Pirie, J. M. Grant, and J. M. Sharp. 1997. Paratuberculosis in wild rabbits (Oryctolagus cuniculus). Vet. Rec. 140:141-143. [DOI] [PubMed] [Google Scholar]
- 27.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris, J. E., and A. M. Lammerding. 2001. Crohn's disease and Mycobacterium avium subsp. paratuberculosis: current issues. J. Food Prot. 64:2103-2110. [DOI] [PubMed] [Google Scholar]
- 29.Hermon-Taylor, J. 2001. Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:755-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hulten, K., T. J. Karttunen, H. M. El Zimaity, S. A. Naser, M. T. Collins, D. Y. Graham, and F. A. El Zaatari. 2000. Identification of cell wall deficient forms of M. avium subsp. paratuberculosis in paraffin embedded tissues from animals with Johne's disease by in situ hybridization. J. Microbiol. Methods 42:185-195. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, P. J., E. A. Walthers, A. S. Kalif, K. L. Richmond, D. M. Adair, K. K. Hill, C. R. Kuske, G. L. Andersen, K. H. Wilson, M. Hugh-Jones, and P. Keim. 1997. Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl. Environ. Microbiol. 63:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keller, A. P., M. L. Beggs, B. Amthor, F. Bruns, P. Meissner, and W. H. Haas. 2002. Evidence of the presence of IS1245 and IS1311 or closely related insertion elements in nontuberculous mycobacteria outside of the Mycobacterium avium complex. J. Clin. Microbiol. 40:1869-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, S. G., S. J. Shin, R. H. Jacobson, L. J. Miller, P. R. Harpending, S. M. Stehman, C. A. Rossiter, and D. A. Lein. 2002. Development and application of quantitative polymerase chain reaction assay based on the ABI 7700 system (TaqMan) for detection and quantification of Mycobacterium avium subsp. paratuberculosis. J. Vet. Diagn. Investig. 14:126-131. [DOI] [PubMed] [Google Scholar]
- 35.Kim, W., Y. P. Hong, J. H. Yoo, W. B. Lee, C. S. Choi, and S. I. Chung. 2002. Genetic relationships of Bacillus anthracis and closely related species based on variable-number tandem repeat analysis and BOX-PCR genomic fingerprinting. FEMS Microbiol. Lett. 207:21-27. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 37.Manning, E. J. 2001. Mycobacterium avium subspecies paratuberculosis: a review of current knowledge. J. Zoo Wildl. Med. 32:293-304. [DOI] [PubMed] [Google Scholar]
- 38.Manning, E. J., and M. T. Collins. 2001. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis and diagnosis. Rev. Sci. Tech. Off. Int. Epizoot. 20:133-150. [DOI] [PubMed] [Google Scholar]
- 39.Manning, E. J., and M. Ziccardi. 2000. Johne's disease and captive non-domestic hoofstock: prevalence and prevention, p. 432-434. In Proceedings of the American Association of Zoo Veterinarians and International Association for Aquatic Animal Medicine Conference. American Association of Zoo Veterinarians and International Association for Aquatic Animal Medicine, Media, Pa.
- 40.Manning, E. J. B., T. E. Kucera, N. B. Gates, L. M. Woods, and M. Fallon-McKnight. 2003. Testing for Mycobacterium avium subsp. paratuberculosis infection in asymptomatic free-ranging tule elk from an infected herd. J. Wildl. Dis. 39:323-328. [DOI] [PubMed] [Google Scholar]
- 41.McClure, H. M., R. J. Chiodini, D. C. Anderson, R. B. Swenson, W. R. Thayer, and J. A. Coutu. 1987. Mycobacterium paratuberculosis infection in a colony of stumptail macaques (Macaca arctoides). J. Infect. Dis. 155:1011-1019. [DOI] [PubMed] [Google Scholar]
- 42.Millar, D., J. Ford, J. Sanderson, S. Withey, M. Tizard, T. Doran, and J. Hermon-Taylor. 1996. IS900 PCR to detect Mycobacterium paratuberculosis in retail supplies of whole pasteurized cows' milk in England and Wales. Appl. Environ. Microbiol. 62:3446-3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motiwala, A. S., M. Strother, A. Amonsin, B. Byrum, S. A. Naser, J. R. Stabel, W. P. Shulaw, J. P. Bannantine, V. Kapur, and S. Sreevatsan. 2003. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: evidence for limited strain diversity, strain sharing, and identification of unique targets for diagnosis. J. Clin. Microbiol. 41:2015-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naser, S. A., J. Felix, H. Liping, C. Romero, N. Naser, A. Walsh, and W. Safranek. 1999. Occurrence of the IS900 gene in Mycobacterium avium complex derived from HIV patients. Mol. Cell. Probes 13:367-372. [DOI] [PubMed] [Google Scholar]
- 45.Nei, M. 1973. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 70:3321-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olsen, I., G. Sigurgardottir, and B. Djonne. 2002. Paratuberculosis with special reference to cattle. A review. Vet. Q. 24:12-28. [DOI] [PubMed] [Google Scholar]
- 47.Ott, S. L., S. J. Wells, and B. A. Wagner. 1999. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev. Vet. Med. 40:179-192. [DOI] [PubMed] [Google Scholar]
- 48.Pavlik, I., J. Bartl, L. Dvorska, P. Svastova, R. du Maine, M. Machackova, W. Yayo Ayele, and A. Horvathova. 2000. Epidemiology of paratuberculosis in wild ruminants studied by restriction fragment length polymorphism in the Czech Republic during the period 1995-1998. Vet. Microbiol. 77:231-251. [DOI] [PubMed] [Google Scholar]
- 49.Pavlik, I., L. Bejckova, M. Pavlas, Z. Rozsypalova, and S. Koskova. 1995. Characterization by restriction endonuclease analysis and DNA hybridization using IS900 of bovine, ovine, caprine and human dependent strains of Mycobacterium paratuberculosis isolated in various localities. Vet. Microbiol. 45:311-318. [DOI] [PubMed] [Google Scholar]
- 50.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, R. du Maine, and I. Rychlik. 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods 38:155-167. [DOI] [PubMed] [Google Scholar]
- 51.Quirke, P. 2001. Mycobacterium avium subspecies paratuberculosis is a cause of Crohn's disease. Gut 49:757-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Read, T. D., S. L. Salzberg, M. Pop, M. Shumway, L. Umayam, L. Jiang, E. Holtzapple, J. D. Busch, K. L. Smith, J. M. Schupp, D. Solomon, P. Keim, and C. M. Fraser. 2002. Comparative genome sequencing for discovery of novel polymorphisms in Bacillus anthracis. Science 296:2028-2033. [DOI] [PubMed] [Google Scholar]
- 53.Sreevatsan, S., J. B. Bookout, F. Ringpis, V. S. Perumaalla, T. A. Ficht, L. G. Adams, S. D. Hagius, P. H. Elzer, B. J. Bricker, G. K. Kumar, M. Rajasekhar, S. Isloor, and R. R. Barathur. 2000. A multiplex approach to molecular detection of Brucella abortus and/or Mycobacterium bovis infection in cattle. J. Clin. Microbiol. 38:2602-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stabel, J. R., S. J. Wells, and B. A. Wagner. 2002. Relationships between fecal culture, ELISA, and bulk tank milk test results for Johne's disease in US dairy herds. J. Dairy Sci. 85:525-531. [DOI] [PubMed] [Google Scholar]
- 56.Whittington, R., I. Marsh, E. Choy, and D. Cousins. 1998. Polymorphisms in IS1311, an insertion sequence common to Mycobacterium avium and M. avium subsp. paratuberculosis, can be used to distinguish between and within these species. Mol. Cell. Probes 12:349-358. [DOI] [PubMed] [Google Scholar]
- 57.Whittington, R. J., A. F. Hope, D. J. Marshall, C. A. Taragel, and I. Marsh. 2000. Molecular epidemiology of Mycobacterium avium subsp. paratuberculosis: IS900 restriction fragment length polymorphism and IS1311 polymorphism analyses of isolates from animals and a human in Australia. J. Clin. Microbiol. 38:3240-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whittington, R. J., L. Reddacliff, I. Marsh, and V. Saunders. 1999. Detection of Mycobacterium avium subsp. paratuberculosis in formalin-fixed paraffin-embedded intestinal tissue by IS900 polymerase chain reaction. Aust. Vet. J. 77:392-397. [DOI] [PubMed] [Google Scholar]
- 59.Whittington, R. J., and E. S. Sergeant. 2001. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust. Vet. J. 79:267-278. [DOI] [PubMed] [Google Scholar]
- 60.Zwick, L. S., T. F. Walsh, R. Barbiers, M. T. Collins, M. J. Kinsel, and R. D. Murnane. 2002. Paratuberculosis in a mandrill (Papio sphinx). J. Vet. Diagn. Investig. 14:326-328. [DOI] [PubMed] [Google Scholar]