Ser palmitoyltransfersase (SPT) regulation of sphingolipid homeostasis is critical for mediating plant growth and programmed cell death (PCD). As key components of this regulation, two 56–amino acid SPT-interacting proteins are shown to be essential for male gametophyte viability, and altered expression strongly affects SPT activity and sensitivity to the PCD-inducing mycotoxin fumonisin B1.
Abstract
Maintenance of sphingolipid homeostasis is critical for cell growth and programmed cell death (PCD). Serine palmitoyltransferase (SPT), composed of LCB1 and LCB2 subunits, catalyzes the primary regulatory point for sphingolipid synthesis. Small subunits of SPT (ssSPT) that strongly stimulate SPT activity have been identified in mammals, but the role of ssSPT in eukaryotic cells is unclear. Candidate Arabidopsis thaliana ssSPTs, ssSPTa and ssSPTb, were identified and characterized. Expression of these 56–amino acid polypeptides in a Saccharomyces cerevisiae SPT null mutant stimulated SPT activity from the Arabidopsis LCB1/LCB2 heterodimer by >100-fold through physical interaction with LCB1/LCB2. ssSPTa transcripts were more enriched in all organs and >400-fold more abundant in pollen than ssSPTb transcripts. Accordingly, homozygous ssSPTa T-DNA mutants were not recoverable, and 50% nonviable pollen was detected in heterozygous ssspta mutants. Pollen viability was recovered by expression of wild-type ssSPTa or ssSPTb under control of the ssSPTa promoter, indicating ssSPTa and ssSPTb functional redundancy. SPT activity and sensitivity to the PCD-inducing mycotoxin fumonisin B1 (FB1) were increased by ssSPTa overexpression. Conversely, SPT activity and FB1 sensitivity were reduced in ssSPTa RNA interference lines. These results demonstrate that ssSPTs are essential for male gametophytes, are important for FB1 sensitivity, and limit sphingolipid synthesis in planta.
INTRODUCTION
Sphingolipids are essential components of eukaryotic cells with diverse roles in membrane structure and function and mediation of basic cellular processes, such as programmed cell death (PCD) (Brodersen et al., 2002; Liang et al., 2003; Alden et al., 2011; Markham et al., 2013). In plants, sphingolipids are major lipid components of the endomembrane system, plasma membrane, and tonoplast and contribute to membrane physical properties that are important for environmental stress tolerance (Verhoek et al., 1983; Lynch and Steponkus, 1987; Sperling et al., 2005; Chao et al., 2011; Chen et al., 2012). Endomembrane-associated sphingolipids also participate in Golgi-mediated protein trafficking that affects processes such as polar auxin transport (Borner et al., 2005; Aubert et al., 2011; Markham et al., 2011; Yang et al., 2013). In addition, sphingolipids contribute to the structural integrity of raft-like domains in the plasma membrane that are important for cell surface activities, including cell wall synthesis and degradation, signaling, and trafficking (Mongrand et al., 2004; Borner et al., 2005; Melser et al., 2011). Beyond their functions in membranes, sphingolipids, acting through their ceramide and long-chain-base precursors and metabolites, are increasingly regarded as signaling molecules for regulation of a number of physiological processes (Liang et al., 2003; Coursol et al., 2005; Donahue et al., 2010). Ceramide and long-chain-base accumulation has been shown to trigger PCD, which may be important for the hypersensitive response for pathogen defense (Liang et al., 2003; Saucedo-García et al., 2011a; König et al., 2012). PCD induction by long-chain-base accumulation appears to be the mode of action for sphinganine analog mycotoxins, including fumonisin B1 (FB1) and AAL toxin (Abnet et al., 2001; Brandwagt et al., 2002). In addition, phosphorylated forms of long-chain bases (LCBs) have been implicated in abscisic acid–dependent guard cell closure and low temperature signaling in plants (Coursol et al., 2003, 2005; Chen et al., 2012; Guillas et al., 2013).
Maintenance of sphingolipid homeostasis is critical for all eukaryotic cells. Sphingolipid homeostasis, for example, is a central component of the regulation of apoptotic pathways in human cells (Rotolo et al., 2005; Chipuk et al., 2012). In plants, cell growth via expansion is dependent on sphingolipid synthesis, and elimination of sphingolipid biosynthesis results in loss of gametophytic and sporophytic cell viability (Chen et al., 2006; Dietrich et al., 2008; Teng et al., 2008). Conversely, accumulation of ceramides and LCBs triggers PCD (Liang et al., 2003; Saucedo-García et al., 2011a). Sphingolipid homeostasis is generally believed to be mediated by regulation of serine palmitoyltransferase (SPT), the first enzyme in long-chain-base biosynthesis that catalyzes the condensation of Ser with typically palmitoyl (16:0)- or stearoyl (18:0)-coenzyme A (CoA) (Hanada, 2003; see Supplemental Figure 1 online). Similar to other eukaryotes, the Arabidopsis thaliana SPT is a heterodimer consisting of LCB1 and LCB2 subunits (Chen et al., 2006; Dietrich et al., 2008; Teng et al., 2008). Together, LCB1 and LCB2 form a heterodimer with pyridoxal phosphate–dependent activity. LCB1 is encoded by a single gene in Arabidopsis designated LCB1 (Chen et al., 2006), while LCB2 is encoded by two functionally redundant genes designated LCB2a and LCB2b (Dietrich et al., 2008). How SPT activity is finely regulated in plants to support growth and modulate PCD initiation is unclear. Public microarray data, for example, suggest little transcriptional regulation of LCB1, LCB2a, or LCB2b in response to most stresses.
A major advance in understanding the regulation of SPT activity was the discovery of yeast (Saccharomyces cerevisiae) Tsc3p (Gable et al., 2000). This 80–amino acid polypeptide has no catalytic activity but stimulates SPT activity and is essential for high-temperature growth (Gable et al., 2000). Structurally unrelated but functional equivalents of Tsc3p, termed small subunits of SPT (ssSPT), have recently been identified in humans and other mammals (Han et al., 2009). In humans, two ssSPTs have been identified: ssSPTa, a 68–amino acid polypeptide, and ssSPTb, a 76–amino acid polypeptide (Han et al., 2009). Coexpression of either human ssSPTa or human ssSPTb with the human LCB1 and LCB2a or LCB2b in an lcb1Δlcb2Δ yeast mutant results in the stimulation of in vitro SPT activity by 100- to 500-fold (Han et al., 2009). In addition, it was observed that ssSPTb coexpression yielded C18 and C20 LCBs from activity with palmitoyl-CoA and stearoyl-CoA, respectively, but ssSPTa expression in this system resulted in the production of only C18 LCBs. A single amino acid difference between ssSPTa and ssSPTb was found to account for the altered substrate specificity of SPT (Harmon et al., 2013). In addition, ssSPTa and ssSPTb were found to physically interact with LCB1 and with LCB2a and LCB2b in the SPT heterodimer (Han et al., 2009). However, the mechanism by which ssSPTs enhance SPT activity and contribute to SPT acyl-CoA substrate specificity has yet to be elucidated. It is also unclear if ssSPT polypeptides are essential in eukaryotes or whether alteration in their expression can affect SPT activity in vivo and alter physiological properties of cells. A more complete understanding of ssSPT properties and functions is important for understanding the fine-tuned maintenance of sphingolipid homeostasis in eukaryotes.
Research reported here was conducted to provide insights into the regulatory system for sphingolipid synthesis in plants and to examine whether ssSPTs have critical functions in higher eukaryotic cells. Arabidopsis is particularly amenable as a eukaryotic model system for exploring ssSPT function because of the availability of insertion mutants and the relative ease of genetic manipulation and propagation. Here, we describe the identification of two 56–amino acid Arabidopsis ssSPTs, ssSPTa and ssSPTb, based on limited homology with human ssSPTs. Both polypeptides strongly stimulate Arabidopsis SPT activity in yeast SPT mutants through direct interaction with the Arabidopsis LCB1/LCB2 heterodimer. In addition, mutant and altered expression lines for the Arabidopsis ssSPT genes display strong phenotypes ranging from loss of pollen viability to altered sensitivity to FB1, which are attributable to suppression or enhancement of SPT activity.
RESULTS
Two Functional Homologs of Mammalian ssSPTs Occur in Arabidopsis
Two genes, designated ssSPTa (At1g06515) and ssSPTb(At2g30942), encoding 56–amino acid polypeptides were identified in homology searches using the human ssSPTs as query. The amino acid sequences of the Arabidopsis polypeptides share 88% identity and have predicted homologs throughout the plant kingdom (Figure 1; see Supplemental Figure 2 online). Arabidopsis ssSPTa and ssSPTb also share 25 to 30% amino acid sequence identity with human ssSPTa and 18 to 25% identity with human ssSPTb but <10% identity with the S. cerevisiae Tsc3p. Arabidopsis ssSPTa and ssSPTb have one likely transmembrane domain encompassing 19 to 23 amino acids in the central portion of the polypeptides, based on predictions from the SOSUI (Hirokawa et al., 1998), TopPred (Claros and von Heijne, 1994), and TMpred programs (Hofmann and Stoffel, 1993) (Figure 1). This is also consistent with mammalian ssSPTs, which have been shown to contain a single transmembrane domain (Han et al., 2009; Harmon et al., 2013).
Figure 1.

Amino Acid Sequence Alignment of ssSPTs from Arabidopsis and Homo sapiens.
Translated protein sequences were aligned using ClustalW. The divergent amino acid shown to influence SPT substrate specificity (M versus V) is indicated with an asterisk, and the predicted transmembrane domain is indicated with a bar. Shading represents the degree of sequence conservation.
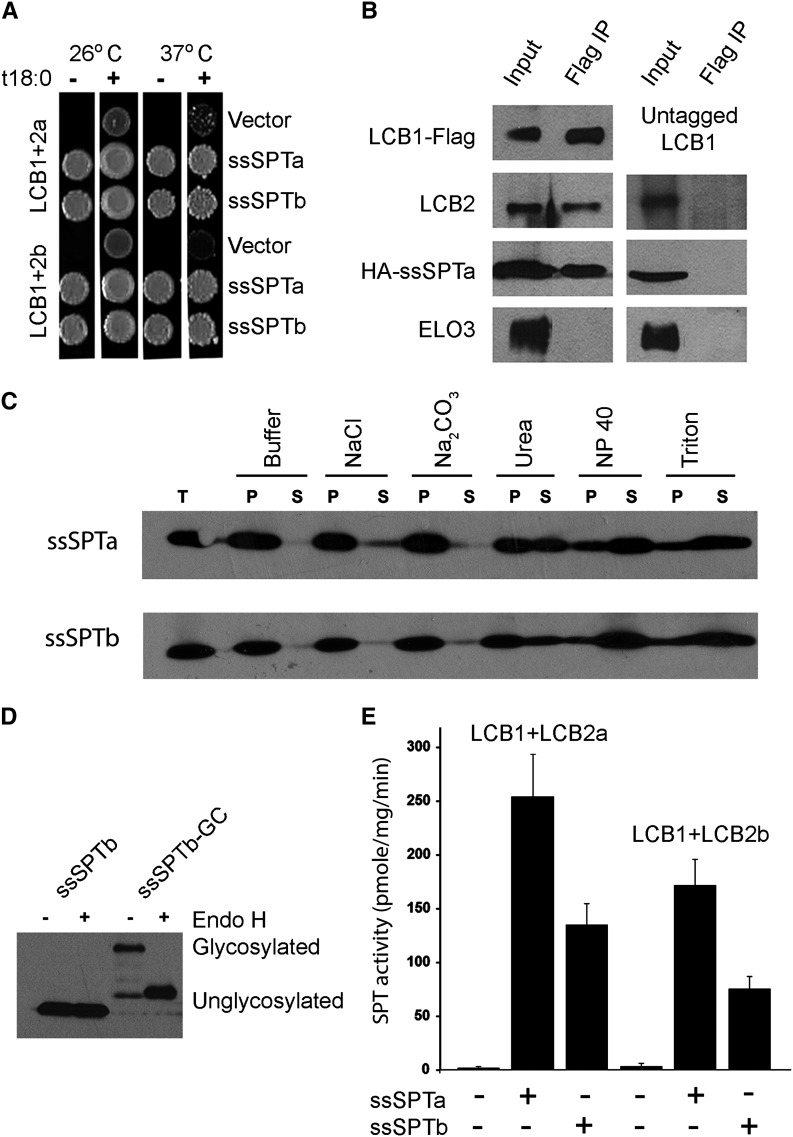
To establish their functions, Arabidopsis human influenza hemagglutinin (HA)-tagged ssSPTa and ssSPTb were expressed along with the Arabidopsis core SPT subunits LCB1 with a FLAG tag and LCB2a or LCB2b with Myc tags in a yeast mutant lacking endogenous SPT. Expression of LCB1-FLAG with the Myc-LCB2 subunits failed to complement the long-chain-base auxotrophy of the yeast mutant, but coexpression of either HA-ssSPTa or HA-ssSPTb resulted in robust growth, even at 37°C where the requirement for SPT activity is relatively high (Figure 2A). Immunoprecipitation of the LCB1-FLAG subunit resulted in copurification of the HA-ssSPTs as well as the Myc-LCB2 subunits (Figure 2B, left panel). This reflects specific binding of HA-ssSPTa with the LCB1/LCB2 heterodimer because no HA-ssSPTa bound to the beads when solubilized microsomes prepared from cells expressing untagged LCB1, Myc-LCB2, and HA-ssSPTa were used for the immunoprecipitation (Figure 2B, right panel). The ssSPTs behave like integral membrane proteins when coexpressed with LCB1 and LCB2, as the proteins are solubilized with detergents and urea, but not by salt or bicarbonate (Figure 2C). By analogy to the human ssSPTs (Harmon et al., 2013), we predicted that the Arabidopsis ssSPTs would have a single transmembrane domain and that their C termini would reside in the endoplasmic reticulum (ER) lumen. To directly address the location of the C terminus of ssSPTb, a glycosylation cassette containing three consensus AsnX(Ser/Thr) glycosylation sites was appended to the C terminus of HA-ssSPTb, and tagged protein was assessed for glycosylation based on sensitivity to Endoglycosidase H (Endo H) treatment. The increased electrophoretic mobility of HA-ssSPTb following Endo H treatment (Figure 2D) provides strong evidence that the C terminus is in the ER lumen. Consistent with the complementation data, microsomes prepared from yeast expressing the At LCB1/LCB2 heterodimers had very low SPT activity (Figure 2E), whereas coexpression of either At ssSPTa or At ssSPTb increased the microsomal SPT activities >100-fold. With all four combinations of LCB2, ssSPT, and LCB1 subunits, a high preference for palmitoyl (16:0)-CoA was observed. Activities measured with myristoyl (14:0)- or stearoyl (18:0)-CoA were <10% of the activity with palmitoyl-CoA (see Supplemental Figure 3 online). The higher SPT activities measured with ssSPTa are likely due to higher expression levels achieved with ssSPTa versus ssSPTb (Figure 2E), as indicated by protein gel blot analysis of microsomes from the recombinant yeast (see Supplemental Figure 3 online), rather than to an intrinsic functional difference between these polypeptides.
Figure 2.
Coexpression of Arabidopsis ssSPTa and ssSPTb with the Arabidopsis SPT Subunits LCB1/LCB2a or LCB1/LCB2b Complements Loss of Cell Viability in Yeast Lacking Endogenous SPT by Activating SPT through Physical Interaction with the Core SPT Heterodimer.
(A) Yeast lacking endogenous SPT activity and expressing either the Arabidopsis LCB1/LCB2a or LCB1/LCB2b are viable only when provided with the LCB phytosphingosine (t18:0). The LCB auxotrophy of these cells is rescued at 26 and 37°C when the Arabidopsis SPT heterodimers are coexpressed with either ssSPTa or ssSPTb.
(B) Solubilized microsomes from cells expressing At LCB1-FLAG, Myc-At LCB2, and HA-At ssSPTa (left panel), or untagged At LCB1, Myc-At LCB2, and HA-At ssSPTa (right panel), were incubated with anti-FLAG beads, and the beads were washed and eluted with FLAG peptide. Aliquots of the solubilized microsomes (input) and eluent (Flag IP) were resolved by SDS-PAGE, and the Arabidopsis SPT subunits were detected by immunoblotting with anti-FLAG, anti-Myc, anti-HA, and anti-Elo3p (negative control) antibodies.
(C) Microsomes from yeast expressing HA-At ssSPTa or HA-At ssSPTb, along with At LCB1and At LCB2a, were extracted on ice with an equal volume of buffer or buffer containing 1 M NaCl, 0.2 M Na2CO3, 5 M urea, 0.4% Nonidet P-40, or 2% Triton X-100 for 60 min. The samples were subjected to centrifugation at 100,000g for 30 min, and equal proportions of the supernatants and pellets were resolved by SDS-PAGE. HA-At ssSPTa and HA-At ssSPTb were detected by immunoblotting with anti-HA antibody. The ssSPTs appear to be integral membrane proteins as they are solubilized with detergents and urea but not salt or bicarbonate. T, total protein; P, pellet (insoluble protein); S, supernatant (soluble protein).
(D) HA-At ssSPTb and HA-At ssSPTb-GC were expressed in yeast along with At LCB1and At LCB2a. Microsomal proteins (20 μg, with or without Endo H treatment) were resolved by SDS-PAGE, and HA-At ssSPTb was detected by immunoblotting.
(E) SPT activity was measured in yeast microsomes expressing At LCB1+At LCB2a or At LCB1+At LCB2b with or without At ssSPTa or At ssSPTb to assess their ability to enhance SPT activity. Values shown are the average of three independent assays ± sd.
ssSPT Polypeptides Are ER Localized but ssSPTa and ssSPTb Are Differentially Expressed in Arabidopsis
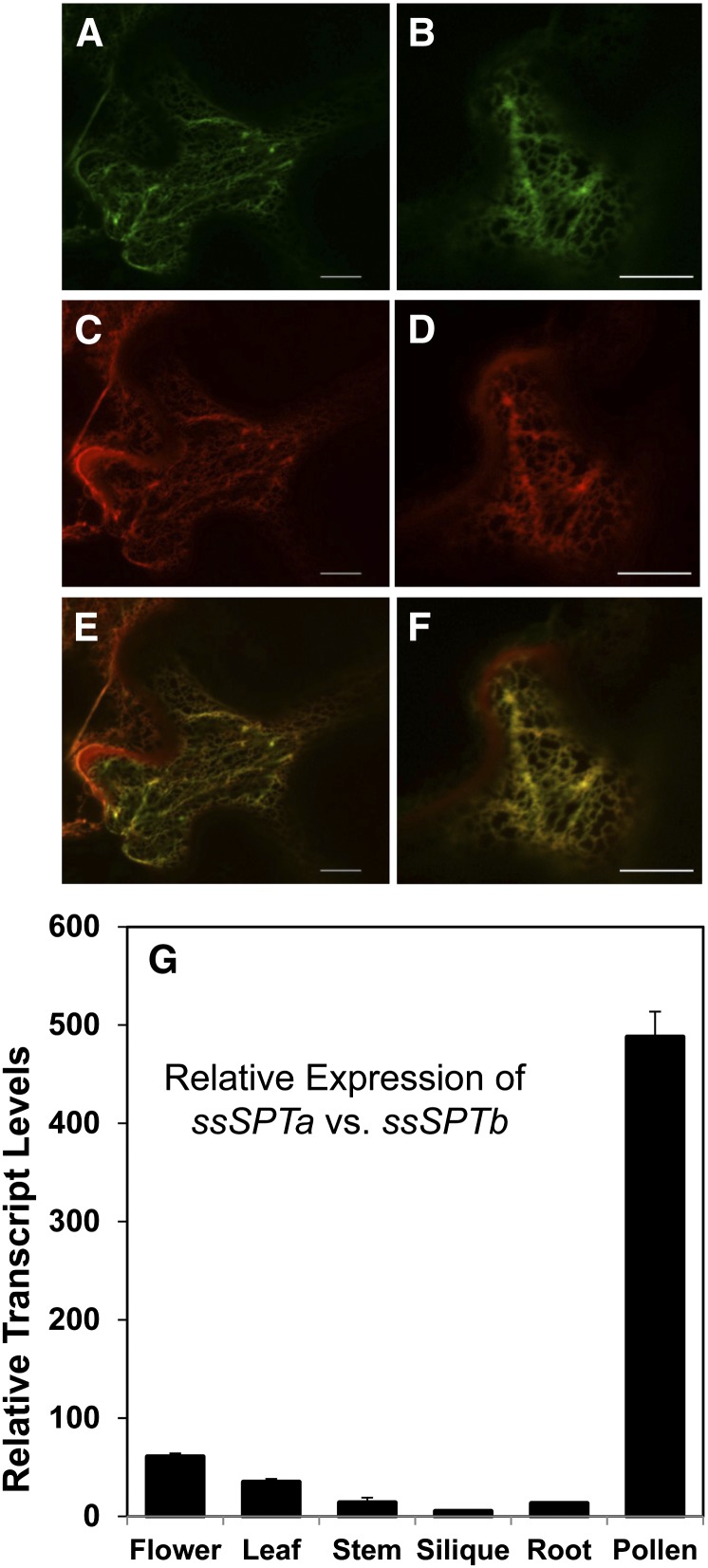
Confocal microscopy studies were conducted on transiently expressed ssSPTa and ssSPTb with N-terminal yellow fluorescent protein (YFP) tags in Nicotiana benthamiana to establish the intracellular localization of these polypeptides. Both polypeptides were found to colocalize with the ER marker mCherry-HDEL (Figures 3A to 3F). This is consistent with ER localization of LCB1, LCB2a, and LCB2b, as previously reported (Tamura et al., 2001; Chen et al., 2006).
Figure 3.
ssSPTa and ssSPTb Are ER Localized and the Corresponding Genes Are Differentially Expressed in Arabidopsis.
(A) to (F) Subcellular localization of ssSPTs was determined by transient expression in N. benthamiana. ssSPT-YFP and ER marker-mCherry fusion constructs were created in binary transformation vectors, coexpressed in tobacco by transient Agrobacterium tumefaciens–mediated transformation, and visualized by confocal microscopy. Yellow from the YFP-ssSPTa and YFP-ssSPTb fusions ([A] and [B], respectively) colocalize with red from the ER marker-mCherry fusion ([C] and [D]) as shown in the merged micrographs for YFP-ssSPTa/ER marker-mCherry (E) and YFP-ssSPTb/ER marker-mCherry (F). Bars = 5 µm.
(G) Relative expression of ssSPTa and ssSPTb. Tissues were collected from wild-type Col-0 grown under our standard conditions. qPCR was used to determine ssSPTa and ssSPTb transcript levels. Protein phosphatase 2A subunit A3 (PP2AA3) was used as a reference gene. Values shown are expression levels of ssSPTa (relative to PP2AA3) divided by expression levels of ssSPTb (relative to PP2AA3) ± sd for three independent measurements.
To assess the in planta contributions of each gene to SPT activation, transcript levels of ssSPTa and ssSPTb were measured in different organs of Arabidopsis (Figure 3G). It is notable that neither gene is represented in any of the public microarray databases. Our analyses revealed that ssSPTa is more highly expressed than ssSPTb in all organs examined (Figure 3G). Of most significance, ssSPTa transcript levels were nearly 500-fold more abundant in pollen than those for ssSPTb. ssSPTa transcripts were also 60- and 35-fold more abundant in flowers and leaves, respectively, than ssSPTb transcripts (Figure 3G).
T-DNA Disruption of ssSPTa Results in Loss of Pollen Viability
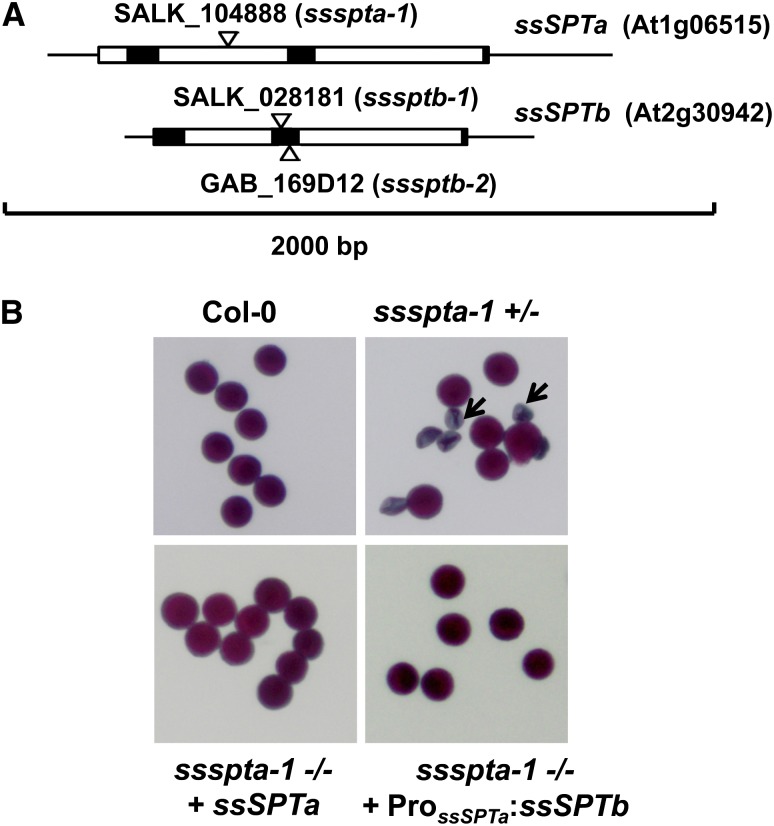
To further examine the relative in planta contributions of ssSPTa and ssSPTb, T-DNA insertion mutants for each gene were characterized. ssSPTa is on chromosome 1 and has two introns within the coding sequence, whereas ssSPTb is on chromosome 2 and has two introns within the coding sequence (Figure 4A). T-DNA insertion mutants were identified for each gene: SALK_104888 for ssSPTa (designated ssspta-1) and SALK_028181 and GAB_169D12 for ssSPTb (designated sssptb-1 and sssptb-2, respectively). In the case of sssptb-1 and sssptb-2, lines homozygous for the T-DNA insertion and lacking detectable ssSPTb transcript were identified (see Supplemental Figure 4 online). Homozygous sssptb-1 and sssptb-2 plants were indistinguishable in their growth and pollen viability from wild-type plants (see Supplemental Figure 5 online). By contrast, no homozygous ssspta-1 mutants were identified by genotyping 250 plants over three generations, suggesting that ssSPTa is an essential gene (see Supplemental Figure 6 online). Examination of pollen with viability stain revealed 50% nonviable pollen in ssspta-1 plants genotyped as heterozygous for the T-DNA insertion (Figure 4B). χ2 goodness-of-fit tests confirmed that the defect observed in pollen collected from ssspta-1 heterozygous plants segregated at the expected 1:1 ratio for a male gametophytic lethal mutation within a 90 to 95% confidence interval (χ2 = 2.627; n = 521). However, the relative numbers of aborted ovules in these plants were low and not significantly different from those in wild-type plants (see Supplemental Figure 7 online). Levels of pollen viability were restored to wild-type levels by introduction of a wild-type copy of ssSPTa (including its native promoter) into ssspta-1 plants genotyped as homozygous for the ssspta-1 allele (Figure 4B; see Supplemental Figure 8 online). Overall, these results demonstrate that ssSPTa, but not ssSPTb, is essential for pollen viability. The much higher expression of ssSPTa relative to ssSPTb in pollen (Figure 4B) likely accounts for the inability of ssSPTb to compensate for loss of ssSPTa to maintain normal male gametophytic development. To test this hypothesis, the ssSPTb cDNA was placed under control of the ssSPTa promoter and introduced into the heterozygous ssspta-1 mutant. Based on genotyping data, plants were recovered that expressed this transgene and contained the homozygous ssspta-1 allele and lacked defective pollen (Figure 4B; see Supplemental Figure 9 online). These findings indicate that the ssSPTa and ssSPTb polypeptides are functionally redundant. We also tested whether ssSPTa is essential for sporophytic (vegetative) growth by expression of an ssSPTa cDNA under control of the pollen- and ovule (meiotic cell)–specific DMC1 promoter (Klimyuk and Jones, 1997) in the ssspta-1 mutant to advance the ssspta-1 −/− allele beyond pollen development. No transgenic plants genotyped as ssspta-1 −/− and lacking the ssSPTa transcript in leaves were recovered. Instead, four independent ssspta-1 −/− lines were obtained that were severely dwarfed and had low levels of ssSPTa expression in leaves, presumably due to leaky expression from the DMC1 promoter (Figure 5). These plants had pollen with high levels of viability, and the dwarf phenotype persisted into the subsequent generation (Figure 5). This result suggests that ssSPTa is also essential for sporophytic growth.
Figure 4.
T-DNA Disruption of ssSPTa Results in Loss of Pollen Viability.
(A) Introns and exons are shown as white and black boxes, respectively, and the UTRs are shown as solid lines. ssSPTa contains an intron in the 5′ UTR. T-DNA insertion mutant alleles for ssSPTa and ssSPTb were confirmed by PCR-based genotyping (see Supplemental Figure 6 online).
(B) Mutation of ssSPTa leads to male gametophytic lethality. Pollen obtained from mature flowers of the respective genotypes were treated with Alexander stain and visualized by light microscopy. Defective pollen stains green and is folded and shriveled, whereas viable pollen is purple and round. Representative pollen collected from heterozygous plants segregating for the T-DNA insertion in the ssspta-1 mutant are shown. Pollen from segregating plants genotyped as wild type (ssspta-1 +/+) are uniformly healthy, whereas those from the heterozygous plants (ssspta-1 +/−) yield defective pollen at ∼50% frequency (black arrows). Transgenic complementation of the pollen defect was accomplished in ssspta-1 −/− plants with constructs expressing either a genomic copy of ssSPTa with native promoter (ssspta-1 −/− + ssSPTa) or the ssSPTb cDNA under control of the ssSPTa promoter (ssspta-1 −/− + ProssSPTa:ssSPTb).
Figure 5.
Complementation of ssspta-1 +/− with ssSPTa cDNA under the Control of the Gametophyte-Specific Promoter DMC1 Yields ssspta-1 −/− Plants That Have Viable Pollen and Are Dwarfed.
(A) Segregating plants from a complemented ssspta-1 line. The two larger plants are ssspta-1 +/−, while the small plant (red arrow) is ssspta-1 −/−.
(B) The progeny from the indicated ssspta-1 −/− plant in (A) are all ssspta-1 −/− and display the dwarf phenotype.
(C) Pollen from ssspta-1 −/− complemented plants appear normal, indicating successful complementation.
ssSPT Expression Levels Affect SPT Activity in Planta and Sensitivity to the Fungal Mycotoxin FB1
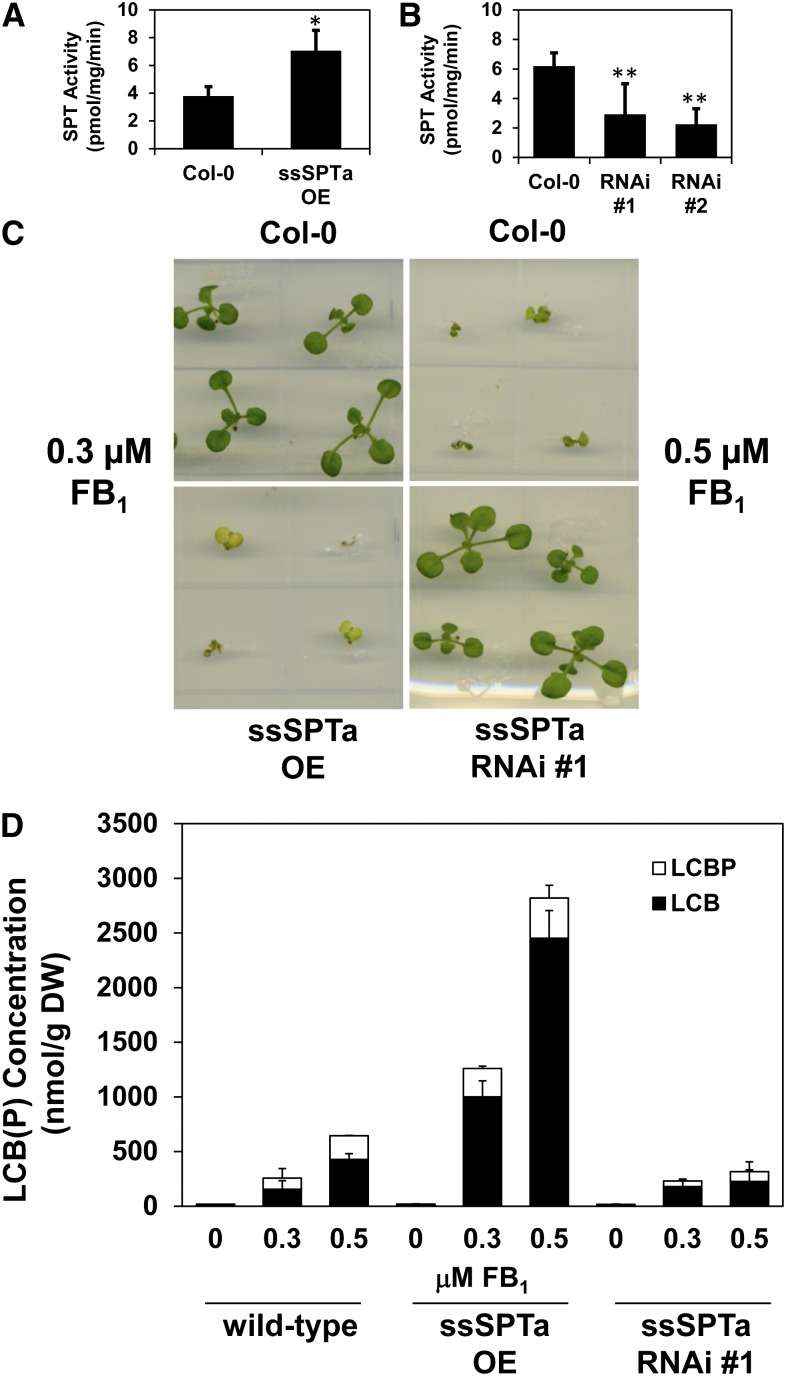
ssSPTa overexpression and suppression lines were generated in wild-type Arabidopsis to determine the impact on SPT activity in planta. Overexpression lines were prepared by placing the ssSPTa cDNA under control of the cauliflower mosaic virus 35S (CaMV35S) promoter, and RNA interference (RNAi) lines were generated using hairpin-generating constructs that targeted the ssSPTa gene under control of the CaMV35S promoter. Homozygous lines from the introduction of these transgenes in wild-type Arabidopsis were screened by quantitative PCR (qPCR) to identify those with strongly enhanced or suppressed expression of ssSPTa (see Supplemental Figure 10 online). Enzyme assays conducted with microsomes prepared from rosettes revealed an approximately twofold increase in SPT activity in a selected overexpression line and up to a twofold decrease in SPT activity in two independent suppression lines (Figures 6A and 6B). These results indicated that SPT activity in planta can be modulated by altered expression of ssSPTa, and based on the findings from the overexpression line, levels of ssSPTa expression are limiting for maximal SPT activity in planta.
Figure 6.
In Planta SPT Activity and Sensitivity to FB1 Is Modulated by Altered ssSPTa Expression.
(A) and (B) SPT activity assayed from wild-type, transgenic plants overexpressing ssSPTa, and transgenic plants with reduced expression of ssSPTa (silenced by RNAi) indicated proportional regulation of SPT activity by ssSPTa. Microsomes were prepared from the respective plant lines and SPT activity was measured with [3H]Ser and palmitoyl-CoA (C16). Overexpression of ssSPTa increased SPT activity (A), whereas suppression of ssSPTa resulted in reduced SPT activity (B). Values shown are the average of assays of three independent samples (±sd). Student’s t test was performed: *P < 0.02 and **P < 0.002.
(C) Altered expression of ssSPTa affected sensitivity to FB1, a competitive inhibitor of ceramide synthase. Seeds were sown on LS agar plates supplemented with FB1 at 0.3 and 0.5 µM as indicated, which distinguish between the FB1-resistant (fbr) and FB1-sensitive (fbs) phenotypes, respectively (Stone et al., 2000). The wild type (Col-0) is extremely sensitive to FB1 at 0.5 µM FB1 and less affected at 0.3 µM FB1. Upregulation of ssSPTa by transgenic overexpression causes an fbs phenotype (ssSPTa overexpression [OE]), whereas downregulation of ssSPTa by RNAi causes an fbr phenotype (ssSPTa RNAi). Images were taken 14 d after sowing seeds and are representative of four independent experiments.
(D) Altered ssSPTa expression affects accumulation of cytotoxic free LCB and LCB-phosphate [collectively referred to as “LCB(P)”] levels in response to FB1 treatment. Wild-type plants show increased LCB(P) levels when treated with FB1. Compared with the wild type, ssSPTa overexpression (OE) plants display strong sensitivity to FB1 and show elevated LCB(P) levels. Alternatively, ssSPTa RNAi suppression plants display resistance to FB1 and show reduced LCB(P) levels, indicating that modulation of LCB(P) levels affects FB1 sensitivity. Electrospray ionization–tandem mass spectrometry analyses were performed with a sample size of six plants per treatment with three independent biological replicates ± sd. Plants were grown on LS plates ± FB1. Plants were grown for 2 weeks before tissue collection. DW, dry weight.
Based on results from LCB1 and LCB2 Arabidopsis mutants (Shi et al., 2007; Saucedo-García et al., 2011b), altered SPT activity should affect resistance to the ceramide synthase mycotoxin inhibitor FB1 (Stone et al., 2000). Enhanced SPT activity is hypothesized to increase FB1 sensitivity by heightened production of cytotoxic LCBs, whereas reduced SPT activity is hypothesized to decrease FB1 sensitivity by lessened production of LCBs (Shi et al., 2007; Saucedo-García et al., 2011b). Consistent with these hypotheses, ssSPTa suppression lines were observed to have increased resistance to FB1: Suppression lines maintained viability at 0.5 µM FB1, which was toxic to wild-type plants (Figure 6C). Conversely, ssSPTa overexpression lines displayed strong sensitivity to 0.3 µM FB1, but wild-type plants retained viability at this FB1 concentration (Figure 6C). Consistent with this, free LCB and LCB-phosphate accumulation in the ssSPTa RNAi line was strongly reduced relative to that in wild-type plants in response to FB1, whereas free LCB and LCB-phosphate accumulation was approximately threefold higher in the ssSPTa overexpression line relative to that in wild-type plants in response to FB1 (Figure 6D). These results indicate that altered ssSPTa expression is an effective means of modulating sensitivity of plants to FB1 and likely other sphinganine analog mycotoxins by increasing or decreasing SPT activity.
Altered SPT Substrate Specificity Conferred by M19V Mutation of ssSPTs Is Conserved among Mammals and Plants
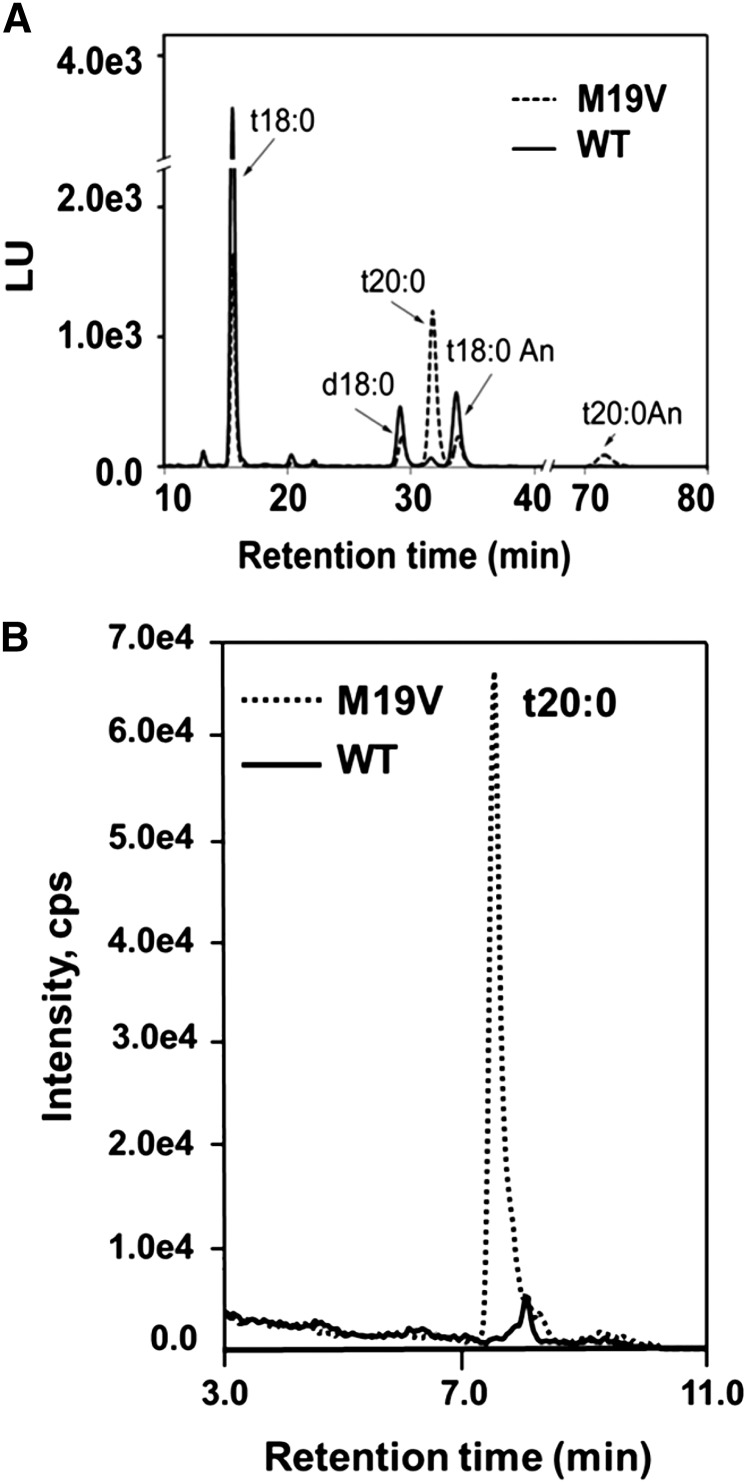
Human ssSPTa and ssSPTb confer distinct substrate specificities to human SPT: ssSPTa expression with human LCB1/LCB2 results in substrate preference for palmitoyl-CoA to generate C18 LCBs, whereas ssSPTb expression with LCB1/LCB2 results in preference for stearoyl-CoA to generate C20 LCBs (Han et al., 2009; Harmon et al., 2013). The basis for this is due to a single amino acid difference in human ssSPTa (Met-25) and ssSPTb (Val-25) (Harmon et al., 2013). Like human ssSPTa, At ssSPTa and At ssSPTb have Met at the equivalent position (Met-19), and expression of either At ssSPTa or At ssSPTb with At LCB1/At LCB2a or At LCB2b yields activity primarily with palmitoyl-CoA (Figures 1 and 2C). To test the importance of amino acid 19 of At ssSPTs for SPT substrate specificity, Met-19 in At ssSPTb was mutated to Val and expressed in a yeast SPT-null mutant along with At LCB1/At LCB2a and in wild-type Arabidopsis (Columbia-0 [Col-0]). Consistent with findings with human SPT (Han et al., 2009; Harmon et al., 2013), yeast coexpressing wild-type Arabidopsis ssSPTb and At LCB/At LCB2a accumulated exclusively C18-LCBs (produced from condensation of Ser with palmitoyl-CoA), while those expressing the Arabidopsis ssSPTb M19V mutant accumulated high levels of C20-LCBs, reflecting condensation of Ser with stearoyl-CoA (Figure 7A). Similarly, expression of At ssSPTb M19V in Arabidopsis resulted in the aberrant production of C20-LCBs that accumulated in all sphingolipid classes (Figure 7B; see Supplemental Figure 11 online). These findings indicate that a single conserved amino acid in human and Arabidopsis ssSPTs is critical for SPT substrate specificity even though these polypeptides share ≤30% amino acid sequence identity. In addition, Arabidopsis lines expressing the ssSPTb M19V mutant displayed enhanced sensitivity to FB1 and enhanced overall accumulation of free and phosphorylated LCBs in response to FB1 relative to wild-type plants (see Supplemental Figure 12 online). This effect is similar to that described above for ssSPTa overexpression.
Figure 7.
SPT Acyl-CoA Preference Is Altered with Expression of the Arabidopsis ssSPTb M19V Mutant.
(A) Yeast expressing wild-type (WT) At ssSPTb along with At LCB1 and At LCB2a accumulated exclusively C18-LCBs (solid line), while those expressing the At ssSPTb M19V mutant accumulated high levels of C20-LCBs reflecting condensation of Ser with stearoyl-CoA (broken line). LCB fluorescent derivatives were analyzed by HPLC following hydrolysis of total sphingolipids (Harmon et al., 2013). LU, Luminescence Unit (excitation at 244 nm/emission at 398 nm).
(B) Plants overexpressing the At ssSPTb M19V mutant also accumulated C20-LCBs, which were nearly undetectable in wild-type Arabidopsis. Shown is MRM for free t20:0 in electrospray ionization–tandem mass spectrometry of extracts from leaves of wild-type plants and plants expressing ssSPTb M19V. LCBs and their respective abbreviations are shown in Supplemental Figure 11 online.
DISCUSSION
Here, we show the occurrence of 56–amino acid polypeptides in Arabidopsis that share low, but significant, sequence identity with human ssSPTs. Like the human ssSPTs, the Arabidopsis ssSPT homologs physically interact with and greatly stimulate the activity of LCB1/LCB2 heterodimers. Our findings also demonstrate the importance of ssSPTs to cellular function in eukaryotes, which apart from the role of the yeast Tsc3p for high-temperature viability, has not been previously clarified in eukaryotes (Gable et al., 2000). In this regard, we show that ssSPTa is essential for male gametophyte (pollen) development and likely for sporophytic viability. In addition, our findings indicate that SPT activity can be enhanced in planta by overexpression of ssSPTa, suggesting that ssSPT levels limit SPT activity in eukaryotic cells, and, thus, that their increased expression may, in part, underlie regulation of SPT activity.
Similar to what has been reported in humans (Han et al., 2009), Arabidopsis also has two functional ssSPT genes (Figure 1), both of which are able to strongly increase SPT activity when coexpressed with the Arabidopsis LCB1 and LCB2a or 2b in a yeast SPT null mutant (Figure 2E). In addition, like the human ssSPT polypeptides, we show that Arabidopsis ssSPT polypeptides are integral membrane proteins with their C termini oriented toward the ER lumen. However, the Arabidopsis ssSPT polypeptides ssSPTa and ssSPTb share considerably higher identity with each other (88%) than do the human ssSPTa and ssSPTb polypeptides, which are 41% identical; also unlike the human ssSPTs, Arabidopsis ssSPTa and ssSPTb contain the same number of amino acids. In addition, the Arabidopsis ssSPTs both have a conserved Met at amino acid 19, whereas the analogous position of the human ssSPTs is either a Met or Val (Han et al., 2009). It has been shown that this amino acid difference is associated with defining the substrate properties of SPT (Han et al., 2009). The presence of Val at this position confers an increased ability of SPT to use the C18 substrate stearoyl-CoA (18:0-CoA), rather than the more typical C16 palmitoyl-CoA (16:0-CoA), to generate C20-LCBs in vitro and in vivo (Han et al., 2009). We show that this amino acid substitution also expands the ability of the Arabidopsis SPT to produce C20-LCBs, which is not detectable in wild-type Arabidopsis (Figure 7B).
The most striking difference between ssSPTa and ssSPTb is their expression levels in Arabidopsis plant tissues (Figure 3G). In all organs examined, ssSPTa is more highly expressed than ssSPTb. The largest difference (∼400-fold) was detected in pollen, which is consistent with the pollen lethality displayed in the ssspta-1 mutant. The higher expression levels of ssSPTa suggest that this gene is of more importance for both reproductive and vegetative growth and viability under normal growth conditions. Expression of the ssSPTb cDNA under control of the ssSPTa promoter is able to rescue the pollen lethality in ssspta-1, indicating that the ssSPTa and ssSPTb polypeptides are functionally redundant in Arabidopsis. It is unclear whether ssSPTb has a more specialized function for the response of cells to stress and altered sphingolipid homeostasis. Complicating resolution of this question is the absence of public microarray data for Arabidopsis ssSPTa and ssSPTb, likely attributable in part to the small size of their transcripts. More extensive expression profiling of Arabidopsis under a wide array of stresses as well as profiling of sphingolipid homeostatic mutants is needed to clarify differential roles of ssSPTa and ssSPTb in the in vivo regulation of SPT.
The pollen lethality observed in ssspta-1+/− is similar to that previously described in double mutants of the LCB2a and LCB2b genes (Dietrich et al., 2008; Teng et al., 2008). In the case of lcb2a-1−/−lcb2b-1+/− mutants, ∼50% of microspores did not progress to the bicellular stage of pollen development (Dietrich et al., 2008). The defective pollen lacked a well-developed endomembrane system, as indicated by vesiculated ER and the absence of Golgi stacks, and also lacked a surrounding intine layer, suggestive of disrupted Golgi trafficking (Dietrich et al., 2008). Our current findings with the ssspta-1 mutant and previous findings with lcb2a-1lcb2b-1 double mutants demonstrate the absolute requirement of sufficient SPT activity for sphingolipid synthesis to support male gametophyte development. In addition, like previous results with double mutants of LCB2a and LCB2b (Dietrich et al., 2008), ssSPTa appears to also be essential for sporophytic growth due to our failure to obtain ssspta-1 homozygous mutants devoid of ssSPTa transcript upon expression of an ssSPTa cDNA under control of the meiotic cell–specific DMC1 promoter in the ssspta-1+/−background.
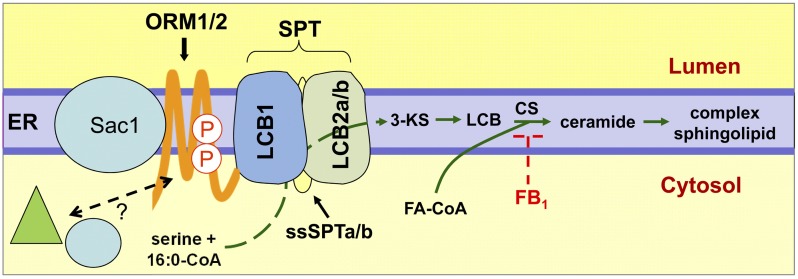
Emerging evidence, particularly from studies in yeast, points to an intricate regulation of SPT activity, which is responsive to intracellular sphingolipid levels. The current model comprises the SPT heterodimer and Tsc3p at the core of a complex that also includes the Orm1 or Orm2 polypeptide and SacI phosphatase, which is referred to as the SPOTS complex (Figure 8) (Walther, 2010). In this model, Orm1/2 polypeptides and SacI phosphatase physically interact with the SPT heterodimer to regulate its activity (Breslow et al., 2010; Han et al., 2010). Orm1 and 2 polypeptides most recently have been shown to downregulate SPT activity in response to high intracellular sphingolipid levels (Breslow et al., 2010; Roelants et al., 2011). This suppression of SPT activity is alleviated by phosphorylation of the Orm1/2 N terminus by the protein kinase Ypk1 in response to low intracellular sphingolipid levels (Roelants et al., 2011). Tsc3p in yeast and ssSPTs in other eukaryotes function in converse of Orm1/2 to stimulate SPT activity. Orm 1/2 homologs have been identified in Arabidopsis (Hjelmqvist et al., 2002), but their functions have yet to be characterized. In our studies, we show that overexpression and suppression of ssSPTa can effectively mediate SPT activity and alter mycotoxin sensitivity. Given our demonstration that the Arabidopsis ssSPT and the previous report that human ssSPT physically interact with the SPT heterodimer, it is possible that these polypeptides, which lack any known catalytic activity, facilitate or stabilize interaction of LCB1 and LCB2 to enhance SPT activity. As such, alteration in ssSPT protein levels in response to perturbations in intracellular sphingolipid levels may contribute, along with other components of the SPOTS complex, to regulation of SPT. Consistent with this, we show that SPT activity in planta can be modulated by altering ssSPTa expression. It remains to be determined whether SPT is regulated in part by mediation of ssSPT protein levels in response to perturbations in sphingolipid homeostasis.
Figure 8.
Model of SPT Regulatory Network.
The figure shown is a representation of the SPOTS complex derived from studies of yeast SPT homeostatic regulation (Gable et al., 2000; Breslow et al., 2010; Han et al., 2010; Walther, 2010; Roelants et al., 2011). The ER-associated core SPT heterodimer consisting of LCB1 and LCB2a or LCB2b in Arabidopsis (Tamura et al., 2001; Chen et al., 2006; Dietrich et al., 2008; Teng et al., 2008) catalyzes the production of the LCB precursor 3-ketosphinganine (3-KS) through condensation of Ser and palmitoyl-CoA. As shown in this article, Arabidopsis ssSPTa and ssSPTb enhance SPT activity when reconstituted in a yeast null mutant and in planta through physical interactions with LCB1/LCB2a/b. These interactions are speculated to stabilize the SPT heterodimer and/or facilitate dimerization of LCB1/LCB2a/b. Overexpression of At ssSPTa was shown to stimulate SPT activity, suggesting that ssSPTs are limiting for maximal SPT activity in Arabidopsis. Conversely, RNAi suppression of At ssSPTa reduced in planta SPT activity. Consistent with this, At ssSPTa–overexpressing plants are more sensitive to the mycotoxin FB1, which inhibits the linkage of a LCB and fatty acyl-CoA (FA-CoA) by ceramide synthase (CS), resulting in the enhanced buildup of cytotoxic LCBs. Based on the yeast SPOTs complex model, additional regulation of SPT is likely mediated through ORM1 and ORM2 polypeptides, which act as negative regulators of SPT. Orm 1 and 2 homologs have been identified in Arabidopsis (Hjelmqvist et al., 2002), but their functions have yet to be characterized. This suppression is mediated by intracellular sphingolipid levels through reversible posttranslational modification of ORM1/2, such as phosphorylation, in response to low intracellular sphingolipid levels and Sac1-mediated dephosphorylation in response to high intracellular sphingolipid levels (Breslow et al., 2010; Han et al., 2010; Walther, 2010). As indicated (green triangle, blue ball), other factors are likely involved in the fine-tuned homeostatic regulation of sphingolipid synthesis via SPT.
Finally, we show that up- or downregulation of Arabidopsis ssSPT gene expression provides a simple, single-gene approach to altering cellular functions. In the example shown, transgenic alteration of ssSPT gene expression yielded plants that are more or less tolerant to the PCD-inducing mycotoxin FB1. It has not been previously been confirmed that upregulation of LCB1 and LCB2, individually or in combination, can increase plant SPT activity in vivo, although modest increases in SPT activity have been reported in mammalian cells with the combined overexpression of LCB1 and LCB2 (Han et al., 2004). It is envisioned that ssSPT genes will be useful biotechnological or breeding targets for physiological processes and stresses that are mediated by sphingolipids, either in their structural roles in membranes and Golgi trafficking or in their signaling roles in plants.
METHODS
Yeast Cell Growth and Expression Plasmids
Yeast (Saccharomyces cerevisiae) strain TDY9113 (Mat α tsc3Δ:NAT lcb1Δ:KAN ura3 leu2 lys2 trp1Δ) lacking endogenous SPT was used for expression and characterization of the Arabidopsis thaliana ssSPT subunits. The mutant was cultured in medium containing 15 μM phytosphingosine and 0.2% tergitol. The At ssSPTa and At ssSPTb cDNA open reading frames were inserted after the 3× HA tag in pADH1 (Kohlwein et al., 2001). pAL2-TRP was constructed for divergent constitutive expression of At LCB1-FLAG and Myc-At LCB2a or Myc-At LCB2b by replacing the Gal1 and Gal10 promoters of pESC-TRP (Stratagene) with the yeast LCB2 and ADH promoters, respectively. The At ssSPTb M19V mutation was introduced by QuikChange mutagenesis (Stratagene).
SPT Assay
Yeast microsomes were prepared and SPT was assayed as described (Harmon et al., 2013) except that 75 μM myristoyl-, palmitoyl-, or stearoyl-CoA was used for the yeast microsomal SPT assays and 50 μM palmitoyl-CoA was used for the Arabidopsis microsomal SPT assays. SPT activity was measured using [3H]Ser and palmitoyl-CoA.
Protein–Protein Interactions
Immunoprecipitation was conducted as described (Breslow et al., 2010) with minor modifications. Microsomal membrane proteins were prepared from yeast cells expressing FLAG-tagged At LCB1 and Myc-tagged At LCB2a with HA-tagged At ssSPTa. Untagged At LCB1 was used to verify that At ssSPTa itself does not bind to the anti-Flag resin. Microsomal membrane proteins were resuspended at 1 mg/mL in immunoprecipitation (IP) buffer (50 mM HEPES-KOH, pH 6.8, 150 mM KOAc, 2 mM MgOAc, 1 mM CaCl2, and 15% glycerol) supplemented with 1 mM PMSF, 2 mg/mL Pepstatin A, 1 mg/mL leupeptin, and 1 mg/mL protinin and solubilized using 1% digitonin at 4°C for 2.5 h. One milliliter of solubilized microsomes was incubated with 25 μL of anti-FLAG beads (Sigma-Aldrich) at 4°C for 4 h, and the beads were washed four times with IP buffer containing 0.1% digitonin. The bound proteins were eluted in IP buffer containing 0.25% digitonin and 200 µg/mL of FLAG peptide, resolved on a 4 to 12% Bis-Tris NuPAGE gel (Invitrogen), and detected by immunoblotting with antibodies, anti-HA (Covance; 1:5000 dilution), anti-Myc (Sigma-Aldrich; 1:3000 dilution), and anti-FLAG (GenScript; 1:5000 dilution).
Membrane Association and Glycosylation Cassette Mobility Shift Assays
The assays were conducted as described (Harmon et al., 2013) with minor modifications. Microsomes prepared from yeast cells expressing HA-tagged At ssSPTa/b, At Lcb1, and At Lcb2a were incubated on ice in buffer containing 1 M NaCl, 0.2 M Na2CO3, 5 M urea, 0.4% Nonidet P-40, or 2% Triton X-100 for 60 min. The samples were subjected to centrifugation at 100,000g for 30 min, and equal proportions of the supernatants and pellets were resolved by SDS-PAGE. For topology mapping, a glycosylation cassette (GC) containing three consensus AsnX(Ser/Thr) glycosylation sites was appended to the C terminus of At ssSPTb. HA-At ssSPTb and HA-At ssSPTb-GC (along with At LCB1 and At LCB2) were expressed in yeast, and glycosylation was assessed using Endo H as previously described (Harmon et al., 2013).
Plant Material and Growth Conditions
Wild-type and mutant Arabidopsis (Col-0) were grown on soil or surface sterilized and sown on Linsmaier and Skoog (LS) agar plates. After stratification at 4°C for 4 d, plants were maintained at 22°C and 50% humidity with a 16-h-light (100 µmol/m−2/s−1)/8-h-dark cycle.
Arabidopsis Transformation and Selection
Binary vectors were introduced into Agrobacterium tumefaciens GV3101 by electroporation. Transgenic plants were created by floral dip of Arabidopsis (Col-0) or heterozygous SALK_104888 T-DNA mutant plants (Clough and Bent, 1998). Seeds were screened with a green LED light and a Red 2 camera filter to identify positive (DsRed) transformed seeds.
RNA Isolation and qPCR
For analyses of organ-specific expression of ssSPTa and ssSPTb, 6- to 8-week-old Col-0 plants were used as sources of plant material. Pollen was harvested as described previously (Johnson-Brousseau and McCormick, 2004). RNA extraction was performed using the RNeasy Plant Kit (Qiagen) according to the manufacturer’s protocol. RNA (1 µg) was treated with DNaseI (Invitrogen) according to the manufacturer’s protocol. Treated RNA was then reverse transcribed to cDNA with the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s protocol. qPCR was performed on the resulting cDNA using the Bio-Rad MyiQ iCycler qPCR instrument. Values shown are the average of three independent measurements ± sd. SYBR green was used as the fluorophore in a qPCR supermix (Qiagen). QuantiTect (Qiagen) primer sets (primers P1, P2, and P3; see Supplemental Table 1 online) were used for relative quantification. PP2AA3 (At1g13320) was used as an internal reference gene.
Subcellular Localization of ssSPTa and ssSPTb
YFP fusion proteins with ssSPTa and ssSPTb were prepared by amplification of the ssSPTa and ssSPTb open reading frames using gene-specific primers (primers P4 through P7; see Supplemental Table 1 online). PCR products were first cloned into the pENTR/D-TOPO vector (Invitrogen). The resulting plasmids were combined with the destination vector pEarlyGate 104 (Earley et al., 2006) to generate the YFP N-terminal fusion constructs. Agrobacterium-mediated infiltration of Nicotiana benthamiana leaves (English et al., 1997) was performed with ssSPTa-YFP and ssSPTb-YFP separately and in conjunction with the ER marker CD3-959 (HDEL-mCherry) (Nelson et al., 2007). Sequential imaging was performed using a Nikon A1 confocal imaging system mounted on a Nikon Eclipse 90i microscope. Excitation/emission wavelengths for YFP and mCherry were 488 nm/500 to 550 nm and 561.6 nm/570 to 620 nm, respectively.
Arabidopsis Mutant Genotyping
T-DNA insertion mutants were obtained from the ABRC and the GABI-Kat collections. Genomic DNA was extracted from plants using the REDextract-N-Amp Tissue PCR kit (Sigma-Aldrich). Genotyping was performed by PCR using gene-specific and T-DNA–specific primer sets (primers P8 through P15; see Supplemental Table 1 online).
Genetic Complementation of ssspta-1
For genomic complementation of SALK_104888 (ssspta-1) heterozygous T-DNA mutant plants, an ∼2.0-kb fragment of ssSPTa was amplified from Arabidopsis (Col-0) genomic DNA (primers P16 and P17; see Supplemental Table 1 online). The amplified product was then digested with AscI and cloned into the binary vector pB110. Heterozygous T-DNA mutants (SALK_104888) were then transformed with the pB110 construct by the floral dip method (Clough and Bent, 1998). Primary transformants were selected (DsRed) and genotyped. Selected heterozygous plants were also genotyped in the next generation (primers P18 and P19; see Supplemental Table 1 online). The ADS1 gene was used as a genotyping control (primers P20 and P21; see Supplemental Table 1 online). Pollen was analyzed from the primary and homozygous transformants.
The ssSPTa promoter:ssSPTb cDNA fusion construct was generated by amplification of ∼1.0-kb region upstream of the ssSPTa start codon (primers P22 and P23; see Supplemental Table 1 online). The product was digested with BamHI and EcoRI and cloned into the corresponding sites of pBinGlyRed2. The ssSPTb cDNA fragment was amplified from Col-0 cDNA, digested with EcoRI and XhoI, and cloned into pBinGlyRed2 (primers P24 and P25; see Supplemental Table 1 online). Transformants were selected and genotyped, and pollen was assessed for viability.
The gametophyte-specific DMC1 promoter (Klimyuk and Jones, 1997) was amplified from pPTN850 (provided by Tom Clemente, University of Nebraska–Lincoln) and cloned into the pBinGlyRed3-35S:ssSPTa cDNA construct using the BamHI and EcoRI sites, replacing the CaMV35S promoter with DMC1 promoter (primers P26 and P27; see Supplemental Table 1 online).
Microscopy
Pollen imaging was performed using an Olympus AX70 optical microscope. Anthers and siliques of mature plants were isolated using a Nikon SMZ745T dissection microscope. Anthers were smeared on a glass slide and incubated with Alexander stain (Alexander, 1969) at 4°C for 45 min before viewing. Pollen viability was assessed by shape and color.
FB1 Screening of Arabidopsis ssSPTa Overexpression and RNAi Lines
ssSPTa-overexpressing plants were generated by transforming Col-0 with the CaMV35S promoter:ssSPTa cDNA constructs. ssSPTa was cloned into the binary vector pBinGlyRed3-35S using the EcoRI-XbaI restriction sites (primers P28 and P29; see Supplemental Table 1 online). ssSPTa RNAi lines were generated by overexpressing a hairpin with the ssSPTa 3′ untranslated region (UTR). The ssSPTa 3′ UTR fragments were amplified (primers P30 and P31; see Supplemental Table 1) and cloned into the pENTR/D-TOPO vector (Invitrogen). The resulting vector was digested with ApaLI. This vector was then combined with the destination vector pFGCGW-red to generate the final ssSPTa RNAi construct. Arabidopsis (Col-0) plants were transformed with these constructs, and the resulting transformants were selected and screened by qPCR for ssSPTa transcript. Sensitivity screening relative to a wild-type control was done at 0, 0.3, and 0.5 µM FB1 (Sigma-Aldrich) concentrations in LS media. Sensitivity was determined by plant growth rate and germination at the varying FB1 concentrations.
Plant Microsomal Membrane Isolation
Microsomal membrane isolation from soil grown 3-week-old Arabidopsis rosettes was performed as described previously (Lynch and Fairfield, 1993), and protein concentration was measured using the bicinchoninic acid assay method (Smith et al., 1985).
Generation of At ssSPTb M19V Construct
The At ssSPTb M19V mutant cDNA was amplified from the corresponding yeast vector described above (primers P32 and P33; see Supplemental Table 1 online) and cloned into the pENTR/D-TOPO vector (Invitrogen). The resulting vector was linearized with ApaLI and combined with the destination vector pCD3-724-Red, resulting in the final vector pCD3-724-Red-ssSPTb M19V. The vector was transformed into Arabidopsis Col-0 as described above.
Sphingolipid Analysis
Sphingolipids were extracted from 2 to 15 mg of lyophilized seedling tissue as described (Markham and Jaworski, 2007). Sphingolipid profiling by liquid chromatography/electrospray ionization–tandem mass spectrometry was performed as described (Markham and Jaworski, 2007) with modifications, using a Shimadzu Prominence ultra-performance liquid chromatography system and a 4000 QTRAP mass spectrometer (AB SCIEX). Sphingolipids were separated on a Zorbax Eclipse Plus narrow bore RRHT C18 column, 2.1 × 100 mm, 1.8-µm particle size (Agilent) column at 40°C and a flow rate of 0.2 mL/min. Binary gradients were generated as described (Markham and Jaworski, 2007) using tetrahydrofuran/methanol/5 mM ammonium formate (3:2:5) + 0.1% formic acid (solvent A) and tetrahydrofuran/methanol/5 mM ammonium formate (7:2:1) + 0.1% formic acid (solvent B). Sphingolipid species were detected using a 4000 QTRAP mass spectrometer (AB SCIEX) with instrument settings as follows: ion spray voltage, 5000 V; source temperature, 350°C (for glycosyl inositolphosphoceramides) and 400°C; GS1, 30; GS2, 60; curtain gas, 13 p.s.i.; interface heater on at 100°C. Voltage potentials were adjusted slightly from values given by Markham and Jaworski (2007) to account for differences in column dimensions, flow rate, and source temperature. Optimized voltage potentials were determined using Arabidopsis leaf sphingolipids as a calibrant. Multiple reaction monitoring (MRM) Q1/Q3 transitions for t18:0, t18:1, d18:0, and d18:1 sphingolipid species were as described by Markham and Jaworski (2007). t20:0 free LCBs were monitored using a 346.3/328.3 mass-to-charge ratio MRM. MRM Q1/Q3 transitions for t20:0-containing sphingolipids are shown in Supplemental Table 2 online. Instrument settings for t18:0 species were used for t20:0 species. Data analysis was performed using Analyst 1.5 and Multiquant 2.1 software (AB SCIEX) as described by Markham and Jaworski (2007).
Phylogenetic Analysis
ssSPT amino acid sequence alignments were generated by ClustalW (Thompson et al., 1994) with Gonnet protein weight matrix (gap open penalty = 10, gap extension penalty = 0.1, and gap separation distance = 4). Sequence alignments are provided in Supplemental Figure 13 online. MEGA5 using the neighbor-joining method (Tamura et al., 2011) was used for phylogenetic analysis of the aligned full-length ssSPT sequences. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. All positions containing gaps and missing data were eliminated. There were a total of 40 positions in the final data set.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: ssSPTa (At1g06515), ssSPTb (At2g30942), LCB1 (At4g36480), LCB2a (At5g23670), and LCB2b (At3g48780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Long-Chain-Base Synthesis, Structures, and Phosphorylation/Dephosphorylation Reactions.
Supplemental Figure 2. Phylogenetic Analysis of ssSPTs from Diverse Eukaryotes.
Supplemental Figure 3. Comparison of Arabidopsis SPT Activity +/− ssSPTa or b with Myristoyl (14:0)-, Palmitoyl (16:0)-, and Stearoyl (18:0)-CoA with Corresponding Protein Levels Expressed in Yeast.
Supplemental Figure 4. RT-PCR Showing Complete Loss of ssSPTb Transcript in sssptb-1 −/− and sssptb-2 −/−.
Supplemental Figure 5. Pollen Isolated from sssptb-1 and ssspt-2 Homozygous Mutants Displayed High Levels of Viability.
Supplemental Figure 6. PCR-Based Genotyping of ssspta-1 Plants Identified Only Heterozygous Mutants.
Supplemental Figure 7. Siliques of Heterozygous ssspta-1 Mutants Show Normal Ovule Development.
Supplemental Figure 8. Homozygous ssspta-1 Mutants Are Recoverable by Complementation with a Wild-Type Copy of ssSPTa.
Supplemental Figure 9. RT-PCR Showing Complete Loss of ssSPTa Transcript in ssspta-1 −/− Complemented with an ssSPTb cDNA Linked to the ssSPTa Promoter.
Supplemental Figure 10. ssSPTa Expression Levels in Overexpression and RNAi Lines.
Supplemental Figure 11. Sphingolipidomic Analysis of Ceramides in Col-0 and the ssSPTb M19V Mutant Leaves.
Supplemental Figure 12. ssSPTb M19V Overexpression Lines Have Increased LCB and LCB(P) Content When Grown on FB1 and Have Enhanced FB1 Sensitivity.
Supplemental Figure 13. Amino Acid Sequence Alignment of Selected ssSPTs.
Supplemental Table 1. Oligonucleotide Primers Used in These Studies.
Supplemental Table 2. MRM Q1/Q3 Transitions for t20:0 Long-Chain-Base-Containing Sphingolipid Species Used to Monitor t20:0 Sphingolipids by LC/ESI-MS/MS.
Acknowledgments
The research was supported by funding from the U.S. National Science Foundation (MCB-1158500; to J.M.S., T.M.D., and E.B.C.).
AUTHOR CONTRIBUTIONS
A.N.K., S.M., M.C., J.M.S., T.M.D., and E.B.C. designed research. A.N.K., S.M., M.C., G.H., and R.E.C. performed research. A.N.K., S.M., M.C., G.H., T.M.D., R.E.C., and E.B.C. analyzed data. A.N.K., S.M., G.H., J.M.S., T.M.D., R.E.C., and E.B.C. wrote the article.
Glossary
- PCD
programmed cell death
- FB1
fumonisin B1
- SPT
Ser palmitoyltransfersase
- ssSPT
small subunits of SPT
- HA
human influenza hemagglutinin
- ER
endoplasmic reticulum
- YFP
yellow fluorescent protein
- CaMV35S
cauliflower mosaic virus 35S
- RNAi
RNA interference
- qPCR
quantitative PCR
- LCB
long-chain base
- Col-0
Columbia-0
- LS
Linsmaier and Skoog
- UTR
untranslated region
- MRM
multiple reaction monitoring
- IP
immunoprecipitation
References
- Abnet C.C., Borkowf C.B., Qiao Y.L., Albert P.S., Wang E., Merrill A.H., Jr, Mark S.D., Dong Z.W., Taylor P.R., Dawsey S.M. (2001). Sphingolipids as biomarkers of fumonisin exposure and risk of esophageal squamous cell carcinoma in China. Cancer Causes Control 12: 821–828 [DOI] [PubMed] [Google Scholar]
- Alden K.P., Dhondt-Cordelier S., McDonald K.L., Reape T.J., Ng C.K., McCabe P.F., Leaver C.J. (2011). Sphingolipid long chain base phosphates can regulate apoptotic-like programmed cell death in plants. Biochem. Biophys. Res. Commun. 410: 574–580 [DOI] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Aubert A., Marion J., Boulogne C., Bourge M., Abreu S., Bellec Y., Faure J.D., Satiat-Jeunemaitre B. (2011). Sphingolipids involvement in plant endomembrane differentiation: The BY2 case. Plant J. 65: 958–971 [DOI] [PubMed] [Google Scholar]
- Borner G.H., Sherrier D.J., Weimar T., Michaelson L.V., Hawkins N.D., Macaskill A., Napier J.A., Beale M.H., Lilley K.S., Dupree P. (2005). Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandwagt B.F., Kneppers T.J., Nijkamp H.J., Hille J. (2002). Overexpression of the tomato Asc-1 gene mediates high insensitivity to AAL toxins and fumonisin B1 in tomato hairy roots and confers resistance to Alternaria alternata f. sp. lycopersici in Nicotiana umbratica plants. Mol. Plant Microbe Interact. 15: 35–42 [DOI] [PubMed] [Google Scholar]
- Breslow D.K., Collins S.R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C.S., Weissman J.S. (2010). Orm family proteins mediate sphingolipid homeostasis. Nature 463: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P., Petersen M., Pike H.M., Olszak B., Skov S., Odum N., Jørgensen L.B., Brown R.E., Mundy J. (2002). Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 16: 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao D.Y., et al. (2011). Sphingolipids in the root play an important role in regulating the leaf ionome in Arabidopsis thaliana. Plant Cell 23: 1061–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Han G., Dietrich C.R., Dunn T.M., Cahoon E.B. (2006). The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18: 3576–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Markham J.E., Cahoon E.B. (2012). Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 69: 769–781 [DOI] [PubMed] [Google Scholar]
- Chipuk J.E., McStay G.P., Bharti A., Kuwana T., Clarke C.J., Siskind L.J., Obeid L.M., Green D.R. (2012). Sphingolipid metabolism cooperates with BAK and BAX to promote the mitochondrial pathway of apoptosis. Cell 148: 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claros M.G., von Heijne G. (1994). TopPred II: An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10: 685–686 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Coursol S., Fan L.M., Le Stunff H., Spiegel S., Gilroy S., Assmann S.M. (2003). Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Coursol S., Le Stunff H., Lynch D.V., Gilroy S., Assmann S.M., Spiegel S. (2005). Arabidopsis sphingosine kinase and the effects of phytosphingosine-1-phosphate on stomatal aperture. Plant Physiol. 137: 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C.R., Han G., Chen M., Berg R.H., Dunn T.M., Cahoon E.B. (2008). Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 54: 284–298 [DOI] [PubMed] [Google Scholar]
- Donahue J.L., Alford S.R., Torabinejad J., Kerwin R.E., Nourbakhsh A., Ray W.K., Hernick M., Huang X., Lyons B.M., Hein P.P., Gillaspy G.E. (2010). The Arabidopsis thaliana Myo-inositol 1-phosphate synthase1 gene is required for Myo-inositol synthesis and suppression of cell death. Plant Cell 22: 888–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K.W., Haag J.R., Pontes O., Opper K., Juehne T., Song K., Pikaard C.S. (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45: 616–629 [DOI] [PubMed] [Google Scholar]
- English J.J., Davenport G.F., Elmayan T., Vaucheret H., Baulcombe D.C. (1997). Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 12: 597–603 [Google Scholar]
- Gable K., Slife H., Bacikova D., Monaghan E., Dunn T.M. (2000). Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275: 7597–7603 [DOI] [PubMed] [Google Scholar]
- Guillas I., Guellim A., Rezé N., Baudouin E. (2013). Long chain base changes triggered by a short exposure of Arabidopsis to low temperature are altered by AHb1 non-symbiotic haemoglobin overexpression. Plant Physiol. Biochem. 63: 191–195 [DOI] [PubMed] [Google Scholar]
- Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S.D., Borovitskaya A., Harmon J.M., Dunn T.M. (2004). The topology of the Lcb1p subunit of yeast serine palmitoyltransferase. J. Biol. Chem. 279: 53707–53716 [DOI] [PubMed] [Google Scholar]
- Han G., Gupta S.D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R.H., Jr, Harmon J.M., Dunn T.M. (2009). Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA 106: 8186–8191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Lone M.A., Schneiter R., Chang A. (2010). Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA 107: 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K. (2003). Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim. Biophys. Acta 1632: 16–30 [DOI] [PubMed] [Google Scholar]
- Harmon J.M., Bacikova D., Gable K., Gupta S.D., Han G., Sengupta N., Somashekarappa N., Dunn T.M. (2013). Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem. 288: 10144–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T., Boon-Chieng S., Mitaku S. (1998). SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379 [DOI] [PubMed] [Google Scholar]
- Hjelmqvist L., Tuson M., Marfany G., Herrero E., Balcells S., Gonzàlez-Duarte R. (2002). ORMDL proteins are a conserved new family of endoplasmic reticulum membrane proteins. Genome Biol. 3: H0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K., Stoffel B. (1993). TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler 374: 166 [Google Scholar]
- Johnson-Brousseau S.A., McCormick S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Klimyuk V.I., Jones J.D. (1997). AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 11: 1–14 [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D., Eder S., Oh C.S., Martin C.E., Gable K., Bacikova D., Dunn T. (2001). Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 109–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König S., Feussner K., Schwarz M., Kaever A., Iven T., Landesfeind M., Ternes P., Karlovsky P., Lipka V., Feussner I. (2012). Arabidopsis mutants of sphingolipid fatty acid α-hydroxylases accumulate ceramides and salicylates. New Phytol. 196: 1086–1097 [DOI] [PubMed] [Google Scholar]
- Liang H., Yao N., Song J.T., Luo S., Lu H., Greenberg J.T. (2003). Ceramides modulate programmed cell death in plants. Genes Dev. 17: 2636–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D.V., Fairfield S.R. (1993). Sphingolipid long-chain base synthesis in plants. Characterization of serine palmitoyltransferase activity in squash fruit microsomes. Plant Physiol. 103: 1421–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D.V., Steponkus P.L. (1987). Plasma membrane lipid alterations associated with cold acclimation of winter rye seedlings (Secale cereale L. cv Puma). Plant Physiol. 83: 761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham J.E., Jaworski J.G. (2007). Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Markham J.E., Lynch D.V., Napier J.A., Dunn T.M., Cahoon E.B. (2013). Plant sphingolipids: Function follows form. Curr. Opin. Plant Biol. 16: 350–357 [DOI] [PubMed] [Google Scholar]
- Markham J.E., Molino D., Gissot L., Bellec Y., Hématy K., Marion J., Belcram K., Palauqui J.C., Satiat-Jeunemaître B., Faure J.D. (2011). Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23: 2362–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser S., Molino D., Batailler B., Peypelut M., Laloi M., Wattelet-Boyer V., Bellec Y., Faure J.D., Moreau P. (2011). Links between lipid homeostasis, organelle morphodynamics and protein trafficking in eukaryotic and plant secretory pathways. Plant Cell Rep. 30: 177–193 [DOI] [PubMed] [Google Scholar]
- Mongrand S., Morel J., Laroche J., Claverol S., Carde J.P., Hartmann M.A., Bonneu M., Simon-Plas F., Lessire R., Bessoule J.J. (2004). Lipid rafts in higher plant cells: Purification and characterization of Triton X-100-insoluble microdomains from tobacco plasma membrane. J. Biol. Chem. 279: 36277–36286 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Roelants F.M., Breslow D.K., Muir A., Weissman J.S., Thorner J. (2011). Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 108: 19222–19227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotolo J.A., Zhang J., Donepudi M., Lee H., Fuks Z., Kolesnick R. (2005). Caspase-dependent and -independent activation of acid sphingomyelinase signaling. J. Biol. Chem. 280: 26425–26434 [DOI] [PubMed] [Google Scholar]
- Saucedo-García M., González-Solís A., Rodríguez-Mejía P., Olivera-Flores Tde.J., Vázquez-Santana S., Cahoon E.B., Gavilanes-Ruiz M. (2011a). Reactive oxygen species as transducers of sphinganine-mediated cell death pathway. Plant Signal. Behav. 6: 1616–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-García M., Guevara-García A., González-Solís A., Cruz-García F., Vázquez-Santana S., Markham J.E., Lozano-Rosas M.G., Dietrich C.R., Ramos-Vega M., Cahoon E.B., Gavilanes-Ruíz M. (2011b). MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 191: 943–957 [DOI] [PubMed] [Google Scholar]
- Shi L., Bielawski J., Mu J., Dong H., Teng C., Zhang J., Yang X., Tomishige N., Hanada K., Hannun Y.A., Zuo J. (2007). Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 17: 1030–1040 [DOI] [PubMed] [Google Scholar]
- Smith P.K., Krohn R.I., Hermanson G.T., Mallia A.K., Gartner F.H., Provenzano M.D., Fujimoto E.K., Goeke N.M., Olson B.J., Klenk D.C. (1985). Measurement of protein using bicinchoninic acid. Anal. Biochem. 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Sperling P., Franke S., Lüthje S., Heinz E. (2005). Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 43: 1031–1038 [DOI] [PubMed] [Google Scholar]
- Stone J.M., Heard J.E., Asai T., Ausubel F.M. (2000). Simulation of fungal-mediated cell death by fumonisin B1 and selection of fumonisin B1-resistant (fbr) Arabidopsis mutants. Plant Cell 12: 1811–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Mitsuhashi N., Hara-Nishimura I., Imai H. (2001). Characterization of an Arabidopsis cDNA encoding a subunit of serine palmitoyltransferase, the initial enzyme in sphingolipid biosynthesis. Plant Cell Physiol. 42: 1274–1281 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C., Dong H., Shi L., Deng Y., Mu J., Zhang J., Yang X., Zuo J. (2008). Serine palmitoyltransferase, a key enzyme for de novo synthesis of sphingolipids, is essential for male gametophyte development in Arabidopsis. Plant Physiol. 146: 1322–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoek B., Haas R., Wrage K., Linscheid M., Heinz E. (1983). Lipids and enzymatic activities in vacuolar membranes isolated via protoplasts from oat primary leaves. Z. Naturforsch. C 38c: 770–777 [Google Scholar]
- Walther T.C. (2010). Keeping sphingolipid levels nORMal. Proc. Natl. Acad. Sci. USA 107: 5701–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Richter G.L., Wang X., Młodzińska E., Carraro N., Ma G., Jenness M., Chao D.Y., Peer W.A., Murphy A.S. (2013). Sterols and sphingolipids differentially function in trafficking of the Arabidopsis ABCB19 auxin transporter. Plant J. 74: 37–47 [DOI] [PubMed] [Google Scholar]