This work identifies the Arabidopsis thaliana mutant maigo5 (mag5), which abnormally accumulates precursor forms of storage proteins. MAG5 is a plant ortholog of yeast Sec16 that is a key component for COPII vesicle formation. MAG5/SEC16A is required for protein export from the ER and functions at ERESs that are associated with mobile Golgi stacks where it regulates COPII coat turnover.
Abstract
Plant cells face unique challenges to efficiently export cargo from the endoplasmic reticulum (ER) to mobile Golgi stacks. Coat protein complex II (COPII) components, which include two heterodimers of Secretory23/24 (Sec23/24) and Sec13/31, facilitate selective cargo export from the ER; however, little is known about the mechanisms that regulate their recruitment to the ER membrane, especially in plants. Here, we report a protein transport mutant of Arabidopsis thaliana, named maigo5 (mag5), which abnormally accumulates precursor forms of storage proteins in seeds. mag5-1 has a deletion in the putative ortholog of the Saccharomyces cerevisiae and Homo sapiens Sec16, which encodes a critical component of ER exit sites (ERESs). mag mutants developed abnormal structures (MAG bodies) within the ER and exhibited compromised ER export. A functional MAG5/SEC16A–green fluorescent protein fusion localized at Golgi-associated cup-shaped ERESs and cycled on and off these sites at a slower rate than the COPII coat. MAG5/SEC16A interacted with SEC13 and SEC31; however, in the absence of MAG5/SEC16A, recruitment of the COPII coat to ERESs was accelerated. Our results identify a key component of ER export in plants by demonstrating that MAG5/SEC16A is required for protein export at ERESs that are associated with mobile Golgi stacks, where it regulates COPII coat turnover.
INTRODUCTION
Protein export from the endoplasmic reticulum (ER) is facilitated by the coat protein complex II (COPII) machinery that selects cargo and shapes the ER membrane into specialized carriers (Barlowe et al., 1994). Assembly of the COPII coat at the ER is initiated by the GTPase Sar1 (for Secretion-associated and ras-superfamily-related1), which is activated by the ER-localized transmembrane guanine nucleotide exchange factor Sec12 (Nakano et al., 1988; Nakańo and Muramatsu, 1989; Barlowe and Schekman, 1993) and occurs sequentially (Brandizzi and Barlowe, 2013). Specifically, Sar1 recruits the Sec23/24 heterodimer through its interaction with Sec23 (Bi et al., 2002). The Sec13/31 heterodimer is then recruited through an interaction between Sec23 and Sec31 (Shaywitz et al., 1997; Bi et al., 2007; Tabata et al., 2009). Assembly of the COPII coat facilitates selective cargo recruitment through the recognition of specific cytosolic-exposed signals on transmembrane domain cargoes by Sec24. Assembly of the COPII coat is followed by deformation of the ER membrane into carriers that are destined for the Golgi. It is believed that such carriers are partially uncoated since the TRANSPORT PROTEIN PARTICLEI (TRAPPI) complexes, which function as tethering proteins on the cis-Golgi membrane, interact with the carriers through Sec23 subunits (Cai et al., 2007).
COPII coat proteins assemble at ER exit sites (ERESs) (Rossanese et al., 1999; Hammond and Glick, 2000; Ward et al., 2001; Hanton et al., 2009); however, the distribution of ERESs in relation to the Golgi apparatus varies among species (Robinson et al., 2007; Brandizzi and Barlowe, 2013). How COPII coat proteins are localized to discrete ERESs is the focus of intensive studies, and evidence in yeast (Pichia pastoris, Connerly et al., 2005; Saccharomyces cerevisiae, Shindiapina and Barlowe, 2010) and metazoans (Watson et al., 2006; Bhattacharyya and Glick, 2007; Hughes et al., 2009; Hughes and Stephens, 2010) supports a crucial role for Sec16. Sec16 depletion in yeast (P. pastoris, Connerly et al., 2005; S. cerevisiae, Castillon et al., 2009) and animal cells (Homo sapiens, Watson et al., 2006; Bhattacharyya and Glick, 2007; Drosophila melanogaster, Ivan et al., 2008) has been linked to disruption of COPII assembly at ERESs, implying that Sec16 is important in COPII-dependent ER export. Based on its ability to bind COPII components, Sec16 has been postulated to function as scaffolding in COPII coat assembly at ERESs (Supek et al., 2002). Another role of Sec16 was recently reported (Kung et al., 2012; Yorimitsu and Sato, 2012): Sec16 is involved in the regulation of COPII assembly at ERESs, most likely through an inhibitory effect on Sar1 GTP hydrolysis.
The unique mobility of the plant Golgi has posed a long-standing conundrum regarding the mechanism that enables efficient ER export to Golgi stacks (Boevink et al., 1998; Brandizzi et al., 2003; daSilva et al., 2004; Yang et al., 2005). In highly vacuolated plant cells, fluorescent fusion proteins of COPII coat components appear to be concentrated at mobile structures that are continuously associated with mobile Golgi stacks (daSilva et al., 2004; Stefano et al., 2006; Hanton et al., 2007; Faso et al., 2009). Since COPII carriers form at ERESs, this could be the result of a continuous association of ERESs at an ER/Golgi interface. However, it is also possible that the COPII coat fluorescence represents assembled coats prior to fusion with the cis-Golgi (Langhans et al., 2012). This would support the intriguing possibility that ERESs from which COPII carriers are formed are not associated with an ER domain interfacing mobile Golgi stacks. Given that Sec16 functions upstream of COPII components in D. melanogaster (Ivan et al., 2008) and H. sapiens (Hughes and Stephens, 2010), analysis of plant SEC16 would provide insight into ERES domains and the relationship between ERESs and Golgi bodies. With the exception of Sec16, COPII proteins are largely conserved at the sequence level, with plant and metazoan isoforms often outnumbering those of S. cerevisiae (Robinson et al., 2007). Although most plant COPII proteins are involved in ER export, a plant homolog of Sec16 has not been identified (Brandizzi and Barlowe, 2013).
To better understand the mechanisms that underlie protein export from the ER in plants, we performed forward genetics of Arabidopsis thaliana seeds. Maturing seed cells actively synthesize large amounts of precursors of two storage proteins (2S albumin and 12S globulin) on the ER, which are then transported into protein storage vacuoles (PSVs) (Jolliffe et al., 2005; Robinson et al., 2005; Vitale and Hinz, 2005). In PSVs, precursor proteins are converted into mature forms by the action of vacuolar processing enzymes (Shimada et al., 2003). Consequently, mutants with defects either in the transport pathway or in the conversion process should accumulate precursor proteins in dry seeds. We screened the seeds of 28,000 T-DNA–tagged lines of Arabidopsis with antibodies that specifically react with the major storage proteins and isolated maigo (mag) mutants in which the transport of storage proteins was defective (Shimada et al., 2006).
In this work, we show that one of the mag mutants, mag5, is linked to a mutation in an ortholog of Sec16. We provide functional evidence that MAG5/SEC16A is required for efficient protein export from the ER to the Golgi by regulating COPII coat dynamics at the ER. Live-cell imaging analyses with functional fusion proteins of MAG5/SEC16A showed that MAG5/SEC16A localizes at Golgi-associated cup-shaped ER areas where it functions as a scaffold and regulator of COPII coat assembly at ERESs. The study also addresses a long-standing question concerning the topological relationship of ERESs with respect to the plant Golgi and supports a model for a distribution of ERESs in continuous association with mobile Golgi stacks. These results conclusively define the plant-specific distribution of ERESs, which enables efficient protein export from the ER to mobile target membranes of the Golgi.
RESULTS
The Arabidopsis mag5-1 Mutant Has a Defect in Vacuolar Protein Transport
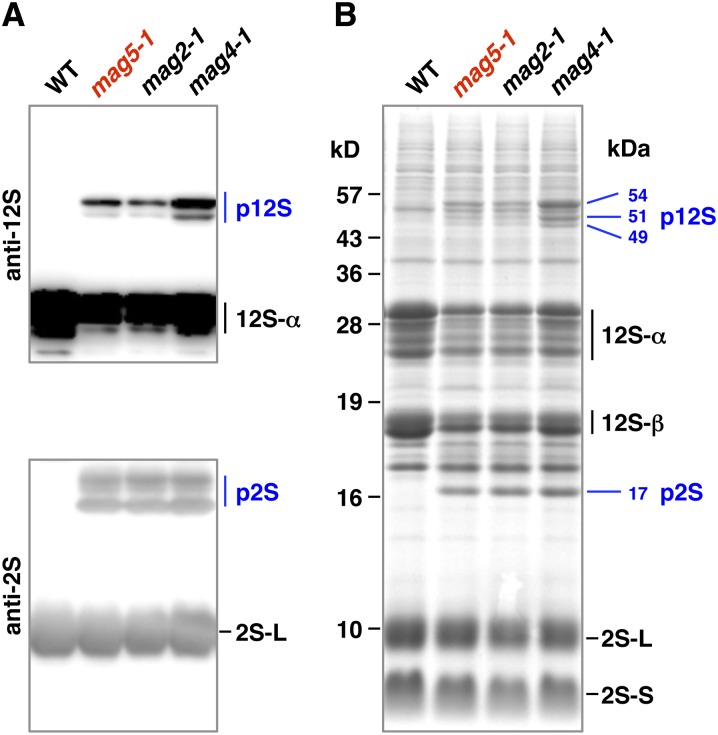
Seeds of wild-type Arabidopsis contain two major storage proteins, 12S globulin and 2S albumin, but no precursors of these proteins. Immunoblotting with a specific antibody against each of these storage proteins showed that in seeds of the mag5 mutant (designated mag5-1), precursor forms of both storage proteins were abnormally accumulated (Figure 1A, p12S and p2S). This suggested that mag5-1 has a defect either in the transport pathway from the ER to PSVs or in the conversion of precursors to mature forms. The former defect generates a 17-kD precursor of 2S albumin, whereas the latter defect generates 15- and 16-kD precursors of 2S albumin (Shimada et al., 2003). As mag5-1 accumulated a 17-kD precursor of 2S albumin, is it likely to have defective transport machinery (Figure 1B). Similar results were obtained with the other known mag mutants (Figures 1A and 1B): mag2-1 has a defect in a homolog of RINT-1/TIP20 on the ER (Li et al., 2006), whereas mag4-1 has a defect in a homolog of the Golgi tethering factor p115 (Takahashi et al., 2010). mag2-1 and mag4-1 both contained the 17-kD precursor of 2S albumin. Therefore, the mag screen successfully identified factors responsible for protein transport from the ER to vacuoles.
Figure 1.
Isolation of an Arabidopsis Mutant, mag5, That Exhibits Abnormal Accumulation of Precursors of Two Major Storage Proteins in Seeds.
(A) Immunoblot of dry seeds with anti-12S globulin and anti-2S albumin antibodies. Wild-type (WT) seeds accumulated mature forms of the major storage proteins 12S globulin (12S) and 2S albumin (2S). mag5 mutant (mag5-1) seeds accumulated precursors (p12S and p2S) of storage proteins, as did other known mag mutants (mag2-1 and mag4-1).
(B) Protein profiles of dry seeds of wild-type, mag5-1, mag2-1, and mag4-1 plants on an SDS gel stained with Coomassie blue. Molecular masses of the precursor forms (p12S and p2S) are indicated on the right in kilodaltons, and those of the markers are indicated on the left. 12S-α and 12S-β, 12S globulin subunits; 2S-L and 2S-S, 2S albumin subunits.
The mag5 Phenotype Is Linked to a Defect in the At5g47480 Gene, Which Encodes a Putative Ortholog with Low Similarity to Human Sec16A and Yeast Sec16p
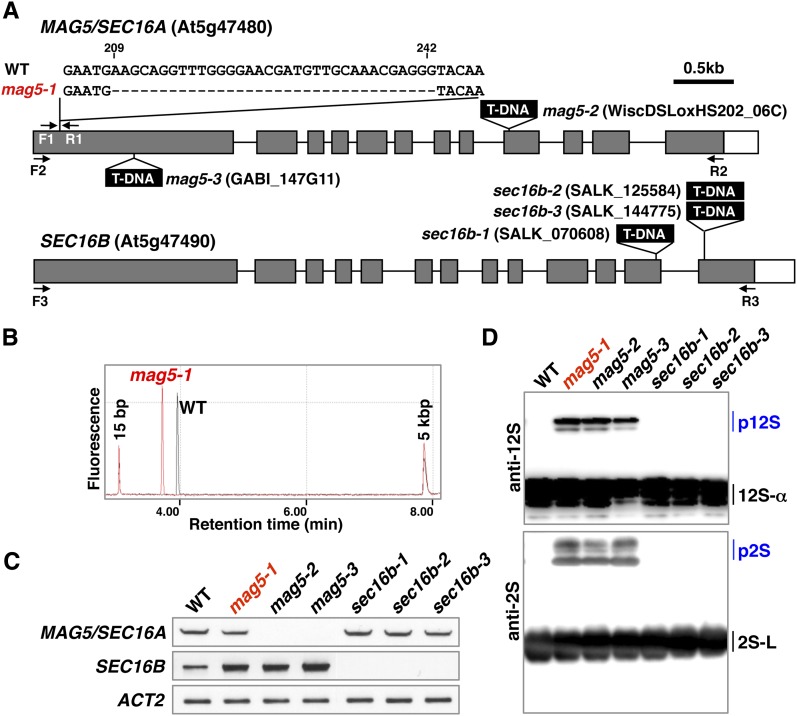
Map-based cloning and DNA sequencing revealed that mag5-1 has a 34-bp deletion in the first exon of At5g47480 (Figure 2A, top). Consistent with this, the RT-PCR product of mag5-1 generated using primers that flanked the deletion (F1 and R1 primers) was shorter than that of the wild type (Figure 2B). Sequencing analysis of MAG5 cDNA in mag5-1 revealed that the deletion caused a frameshift and a stop codon (see Supplemental Figure 1 online). Therefore, although mag5-1 produces a transcript (Figure 2C), the mutant is likely null. To further link the mag5 phenotype to disruption of the At5g47480 gene, we isolated two allelic mutants with a T-DNA insertion in the ninth or first exon of At5g47480 that we named mag5-2 and mag5-3, respectively (Figure 2A, top). Based on a lack of detectable At5g47480 transcripts, both mag5-2 and mag5-3 were RNA-null mutants (Figure 2C). Consistent with this, immunoblots clearly showed that mag5-2 and mag5-3 both accumulated abnormal levels of the precursors of storage proteins (Figure 2D), similar to mag5-1. These results, together with the complementation test results (see below), indicate that mutation of At5g47480 is responsible for the mag5 phenotype.
Figure 2.
MAG5 Encodes a Putative Ortholog with Low Sequence Similarity to Human Sec16A and Yeast Sec16p.
(A) Schematic representation of the MAG5/SEC16A gene (At5g47480) and the homologous SEC16B gene (At5g47490). Three mag5 mutants (mag5-1, mag5-2, and mag5-3) and three sec16b mutants (sec16b-1, sec16b-2, and sec16b-3) are shown, and each mutation site is indicated. F1-3 and R1-3 indicate the primers used in (B) and (C). Closed boxes indicate protein-coding regions, open boxes indicate untranslated regions, and solid lines indicate introns. WT, the wild type.
(B) RT-PCR using the F1 and R1 primers shows that a 34-bp deletion in the MAG5 gene leads to a smaller transcript in the mag5-1 mutant than in the wild type.
(C) RT-PCR of the MAG5/SEC16A and SEC16B transcripts in rosette leaves of the wild-type and mutant plants. The F2 and R2 primers were used to amplify MAG5/SEC16A, whereas the F3 and R3 primers were used to amplify SEC16B. ACT2 was included as a control.
(D) Immunoblot of dry seeds with anti-12S globulin and anti-2S albumin antibodies. Each of the mag5 mutants accumulated the storage protein precursors (p12S and p2S), whereas the sec16b mutants did not.12S-α, a 12S globulin subunit; 2S-S and 2S-L, 2S albumin subunits.
The At5g47480 protein (1350 amino acids) has low sequence similarity to human Sec16A (2357 amino acids) and yeast Sec16 (2195 amino acids;see Supplemental Figure 2 online). The central region of At5g47480 containing 223 amino acids has 31% identity to the corresponding region of human Sec16A, whereas the C-terminal region of At5g47480 containing 330 amino acids has 25% identity to the corresponding region of human Sec16A. The C-terminal region of At5g47480 containing 40 amino acids has 43% identity to the corresponding region of yeast Sec16p. These C-terminal and central regions are conserved domains among Sec16 proteins and are proposed to mediate the interactions of Sec16 proteins with other COPII components (Bhattacharyya and Glick, 2007). Thus, it seems possible that the At5g47480 protein is a homolog of Sec16. Here, we designate the At5g47480 gene as MAG5/SEC16A.
The Arabidopsis genome has another gene (At5g47490) that is similar to MAG5/SEC16A and is situated next to MAG5/SEC16A in the genome. The At5g47490 protein shares 68% identity and 75% similarity with MAG5/SEC16A and is hereafter designated as SEC16B. We established three mutants of SEC16B (sec16b-1, sec16b-2, and sec16b-3; Figure 2A, bottom), all of which were RNA-null mutants (Figure 2C). None of the sec16b mutants exhibited abnormal accumulation of storage protein precursors in seeds (Figure 2D). This is consistent with the fact that expression of SEC16B is lower than that of MAG5/SEC16A in maturing seeds (ATTED-II database).
mag5 Mutants Have Defects in Protein Export from the ER in Seed and Vegetative Cells
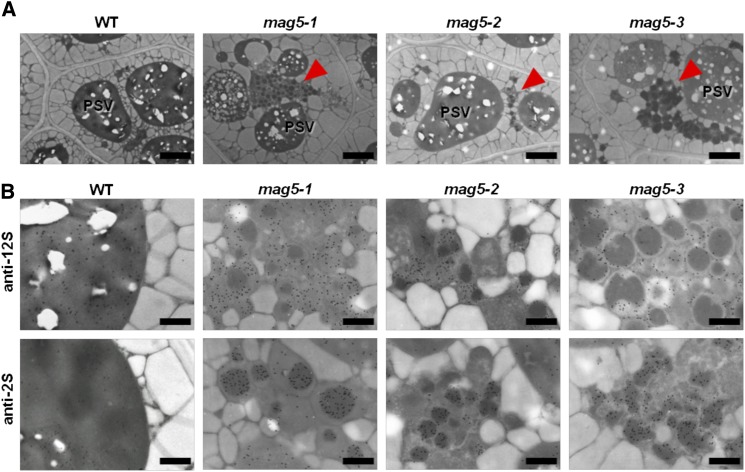
We examined whether the lack of MAG5/SEC16A induces defects at the subcellular level. Live-cell imaging is not feasible in seeds; therefore, we compared the seed cells of the three mag5 mutants with those of the wild-type plants using immunoelectron microscopy. In wild-type seed cells, the storage proteins 12S globulin and 2S albumin were localized in electron-dense PSVs (Figures 3A and 3B, WT). By contrast, all mag5 mutants (mag5-1, mag5-2, and mag5-3) developed abnormal structures that contained electron-dense cores ∼1 µm in diameter (Figure 3A, arrowheads). Immunoelectron analyses of the mag5 mutants revealed that 2S albumin was present in the cores of these structures, whereas 12S globulin was detected in the matrix region (Figure 3B). These features resemble those of MAG bodies observed in mag2 (Li et al., 2006) and mag4 (Takahashi et al., 2010) mutants, which are defective in trafficking between the ER and the Golgi. A defect in transport at the ER/Golgi interface of the mag5 mutants may lead to the generation of MAG bodies in seed cells in the mag5 mutants.
Figure 3.
mag5 Seed Cells Develop Abnormal Structures with Electron-Dense Cores Containing 2S Albumin and a Matrix Containing 12S Globulin.
(A) Electron micrographs of seed cells of wild-type (WT) and mag5 mutant (mag5-1, mag5-2, and mag5-3) plants. All mag5 mutants developed abnormal structures, designated MAG bodies, as indicated by arrowheads. Bars = 2 µm.
(B) Immunoelectron micrographs of seed cells of wild-type and mag5 mutant plants using the anti-12S globulin and anti-2S albumin antibodies. Bars = 0.5 µm.
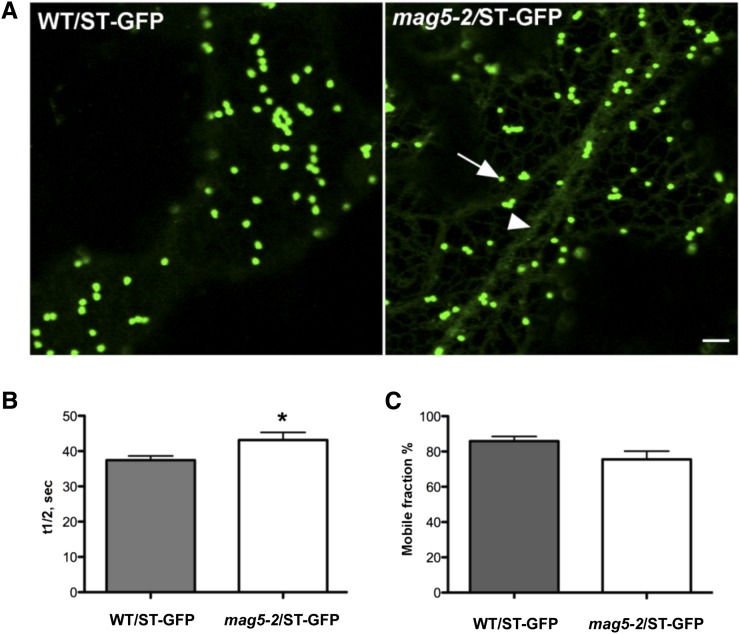
To test whether loss of MAG5/SEC16A also caused defects in Golgi membrane traffic in other tissues, the knockout line mag5-2 was stably transformed with the Golgi marker sialyl transferase (ST)-GFP (for green fluorescent protein) (Boevink et al., 1998). In epidermal cells of wild-type plants, ST-GFP clearly localized in the Golgi (Figure 4A, left panel). However, in the mag5-2 background, ST-GFP was partially distributed in the ER and Golgi (Figure 4A, right panel). To establish whether this abnormal distribution of ST-GFP was linked to defects in ER export, we performed fluorescence recovery after photobleaching (FRAP) analyses of ST-GFP (Brandizzi et al., 2002b).The fluorescence recovery rate of the marker was compared between the mutant and wild-type plants. ST-GFP cycled in and out of the Golgi significantly more slowly in the mag5-2 mutant than in the wild type (Figure 4B), although the mobile fraction of ST-GFP (i.e., the percentage of available mobile protein) was similar in the two backgrounds (Figure 4C). Consistent with these results, when ER export was indirectly inhibited using brefeldin A, which disrupts COPI assembly at the Golgi (Ritzenthaler et al., 2002), ST-GFP redistributed to the ER to a greater extent in the mag5-2 mutant compared with the wild type (see Supplemental Figure 3 online). Taken together, these data demonstrate that protein export from the ER is affected by the lack of functional MAG5/SEC16A.
Figure 4.
Lack of MAG5/SEC16A Causes an Abnormal ER Conformation and Interferes with Protein Export.
(A) Confocal images of wild-type (WT) epidermal cells of cotyledons expressing the Golgi marker ST-GFP (left panel). mag5-2 epidermal cells of cotyledons expressing the Golgi marker ST-GFP, which is partially retained in the ER (right panel). The arrow and arrowhead indicate Golgi and ER, respectively. Bar = 5 μm.
(B) and (C) FRAP analyses of ST-GFP in Golgi stacks at the cortex of cotyledon epidermal cells in wild-type and mag5-2/ST-GFP seedlings. The fluorescence recovery half-time was significantly slower in mag5-2 seedlings than in wild-type seedlings (P < 0.05) (B). The mobile fraction (%) did not significantly differ between wild-type and mag5-2 seedlings (P > 0.05) (C). Sample size in each experiment (i.e., number of bleached Golgi stacks) = 10. Error bars indicate sd.
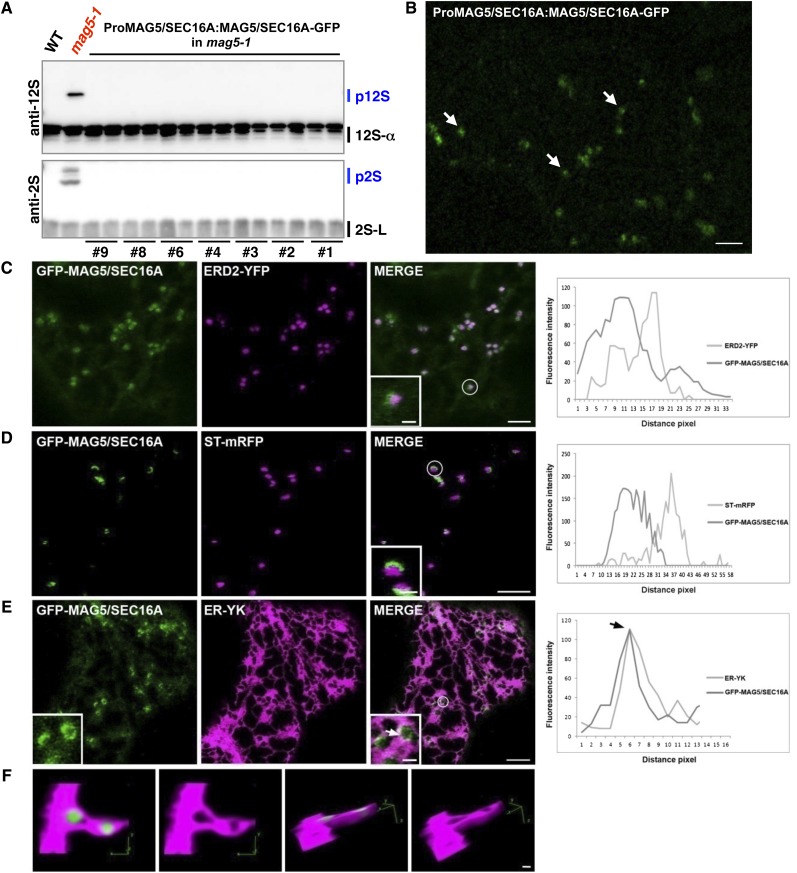
MAG5/SEC16A Is Localized in Regions of the ER That Face Associated Golgi Stacks
To determine the subcellular distribution of MAG5/SEC16A, we introduced genomic MAG5/SEC16A-GFP, whose expression was driven by the native MAG5/SEC16A promoter (ProMAG5/SEC16A:MAG5/SEC16A-GFP), into the mag5-1 mutant and obtained seven independent T1 lines. Storage protein precursors did not accumulate in T2 seeds from any of these T1 lines (Figure 5A), indicating that the expression of MAG5/SEC16A-GFP under the control of the native promoter rescued the mag5 phenotype. Hence, MAG5/SEC16A-GFP is functional in vivo. The transgenic plants expressing MAG5/SEC16A-GFP under the native MAG5/SEC16A promoter exhibited flluorescent punctate structures in epidermal cells of cotyledons (Figure 5B). Similarly, transgenic plants that expressed MAG5/SEC16A-GFP under the control of the 35S promoter displayed punctate fluorescent structures (see Supplemental Figure 4 online). The transgenic plants had a healthy phenotype and a normal morphology (see Supplemental Figure 4 online), indicating that overexpression of MAG5/SEC16A-GFP does not significantly disrupt plant growth.
Figure 5.
Punctate Structures Labeled by MAG5/SEC16A Are Surrounded by the Cup-Shaped Regions of the ER That Face Associated Golgi Stacks.
(A) Immunoblot of individual T2 seed grains from seven independent T1 lines (ProMAG5/SEC16A:MAG5/SEC16A-GFP in mag5-1) using anti-12S globulin and anti-2S albumin antibodies. The mag5-1 phenotype of abnormal accumulation of precursors was rescued in these lines. WT, the wild type.
(B) Confocal image of epidermal cells of cotyledons in T2 line number two shown in (A). MAG5/SEC16A-GFP is distributed in the cytosol and puncta (arrows). Bar = 5 µm.
(C) Confocal images of transiently transformed tobacco epidermal cells coexpressing GFP-MAG5/SEC16A and the cis-Golgi marker ERD2-YFP. The degree of colocalization between the two markers was analyzed using Zeiss software in the spot highlighted by the white circle and enlarged in the inset. The graph shows that the fluorescence signals partially overlap; however, the fluorescence peaks are shifted. In the graph, distance is presented in pixels (1 pixel = 0.09 µm)
(D) Colocalization of GFP-MAG5/SEC16A and the trans-Golgi marker ST-mRFP was analyzed using Zeiss software in the spot highlighted by the white circle and enlarged in the inset. The graph shows that the fluorescence signals do not overlap and the fluorescence peaks are even more shifted than in (C). In the graph, distance is presented in pixels (1 pixel = 0.09 µm)
(E) Confocal images of transiently transformed tobacco epidermal cells coexpressing GFP-MAG5/SEC16A and ER-YK. The degree of colocalization between the two markers was analyzed using Zeiss software in the spot highlighted by the white circle and enlarged in the inset (arrow). The graph shows a high degree of colocalization between these two markers, and their fluorescence peaks coincide (arrow). In the graph, distance is presented in pixels (1 pixel = 0.09 µm). Bars = 5 µm (main image) and 1 µm (inset).
(F) Three-dimensional projection reconstruction and rendering of images of cells expressing the same markers as shown in (E). In this projection, the ER is cup shaped around MAG5/SEC16A-positive structures, supporting the hypothesis that MAG5/SEC16A is associated with specific regions of ER. Bar = 1 µm
To determine the distribution of MAG5/SEC16A with respect to secretory organelles, GFP-MAG5/SEC16A was coexpressed with several specific markers: an inert luminal marker of the ER, ER-YK (Nelson et al., 2007); a marker of the most cis-Golgi cisternae, ERD2-YFP (for yellow fluorescent protein) (daSilva et al., 2004); a marker of trans-Golgi cisternae, ST-mRFP (for monomeric red fluorescent protein) (Saint-Jore et al., 2002); and trans-Golgi network marker YFP-SYP61 (Uemura et al., 2004). Confocal microscopy analyses showed that GFP-MAG5/SEC16A overlapped with ERD2-YFP (Figure 5C) and ST-mRFP (Figure 5D). GFP-MAG5/SEC16A was present in cup-like punctate structures within the ER-YK–labeled ER network (Figure 5E, inset). GFP fluorescence was not uniform in these structures and was more intense in some regions (Figure 5E, inset, arrow). Nonetheless, the punctate appearance of GFP-MAG5/SEC16A was maintained in cells irrespective of the abundance of the marker (i.e., Figure 5D [lower expressing cell] and Figure 5F [higher expressing cell]). We also found that GFP-MAG5/SEC16A partially overlapped with the ER-YK signal (Figure 5E) and to a lesser extent with ERD2-YFP and ST-mRFP (Figures 5C and 5D). MAG5/SEC16A did not overlap with YFP-SYP61 (see Supplemental Figure 5 online). The differences in distribution of MAG5/SEC16A and the Golgi marker with respect to a single focal imaging plane could account for an apparent lack of codistribution. To examine this, we performed time-lapse microscopy in cells coexpressing GFP-MAG5/SEC16A and ST-YFP and ensured coincident distribution of the structures labeled by MAG5/SEC16A and the Golgi markers over time (see Supplemental Figure 6 and Supplemental Movie 1 online).
To explore the distribution of GFP-MAG5/SEC16A with respect to the ER, we acquired consecutive optical slices of cells coexpressing GFP-MAG5/SEC16A and ER-YK for three-dimensional reconstruction and rendering. GFP-MAG5/SEC16A was surrounded by a cup-shaped subdomain of the ER (Figure 5F). These data indicate that MAG5/SEC16A is localized in structures that are associated with mobile Golgi stacks.
MAG5/SEC16A Interacts with SEC13 and SEC31
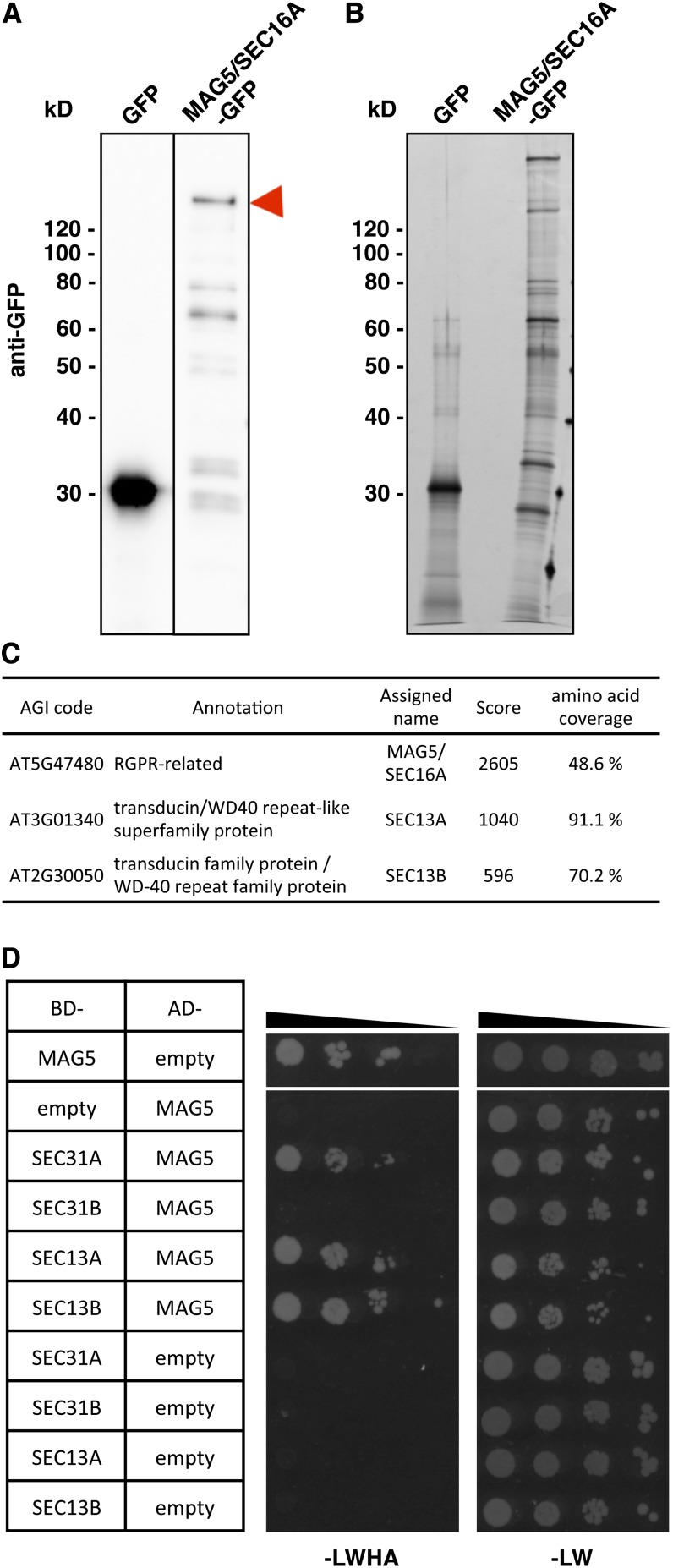
To identify MAG5/SEC16A-interacting proteins, extracts from whole seedlings of transgenic Arabidopsis plants expressing MAG5/SEC16A-GFP under the control of the 35S promoter were immunoprecipitated using an anti-GFP antibody. The immunoprecipitates were subjected to SDS-PAGE followed by immunoblotting with an anti-GFP antibody (Figure 6A) or silver staining (Figure 6B). Full-length MAG5/SEC16A-GFP and degradation products were concentrated in the precipitates along with other proteins. Linear Trap Quadrupole (LTQ)-Orbitrap mass spectrometry identified MAG5/SEC16A with the highest score (Figure 6C). SEC16B was also listed by the software (see Supplemental Table 1 online), although we could not distinguish any unique peptides of SEC16B. The Arabidopsis genome has two closely related SEC13 genes, SEC13A and SEC13B (Robinson et al., 2007). Interestingly, these SEC13 proteins were detected with the second and third highest scores with high amino acid coverage, and unique peptides were identified for both (Figure 6C; see Supplemental Figure 7 online).
Figure 6.
MAG5/SEC16A Interacts with SEC13 and SEC31.
(A) and (B) Immunoblots (A) and silver staining (B) of anti-GFP antibody immunoprecipitate samples from transgenic plants expressing GFP or MAG5/SEC16A-GFP. The arrowhead indicates full-length MAG5/SEC16A-GFP.
(C) Identification of MAG5/SEC16A-interacting proteins by mass spectrometry. The Arabidopsis Genome Initiative (AGI) codes and annotations were obtained from The Arabidopsis Information Resource database (http://www.Arabidopsis.org). Scores were calculated by Mascot (Matrix Science).
(D) Yeast two-hybrid analysis of the AH109 strain expressing a fusion protein containing the GAL4 DNA binding domain (BD) and a fusion protein containing the GAL4 activation domain (AD). Tenfold dilution series of transformants were incubated on SD/–Leu/–Trp/–His/–Ade medium (–LWHA) or SD/–Leu/–Trp medium (–LW). Negative controls (empty vector) were also included.
The interaction of MAG5/SEC16A with COPII proteins was also examined using a yeast two-hybrid assay. In agreement with the immunoprecipitation analysis, MAG5/SEC16A interacted with SEC13A and SEC13B (Figure 6D). In yeast, Sec13 forms a heterodimer with Sec31 as part of the COPII coat (Salama et al., 1993; Lederkremer et al., 2001). The Arabidopsis genome encodes two putative SEC31 orthologs (Robinson et al., 2007). We detected an interaction between MAG5/SEC16A and SEC31A (Figure 6D) and of SEC31A and SEC31B with both SEC13A and SEC13B (see Supplemental Figure 8 online). These results suggest that MAG5/SEC16A functions in concert with COPII components.
MAG5/SEC16A GFP Fusion Proteins Cycle on and off ERESs at a Much Lower Rate Than SEC24A
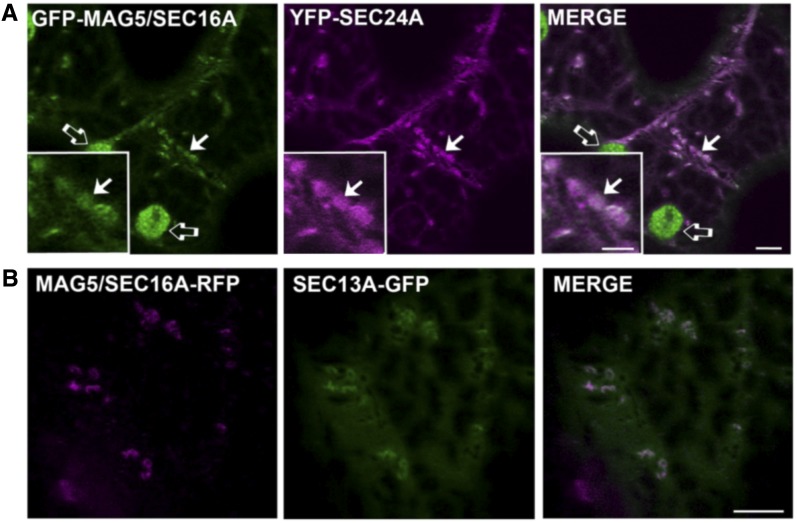
The dynamic relationship between MAG5/SEC16A and COPII components was analyzed. Plant SEC24, SEC23, SEC13, and SAR1 have all been used as COPII coat markers (Stefano et al., 2006; Hanton et al., 2007, 2008, 2009; Hawes et al., 2008; Sieben et al., 2008; Wei and Wang, 2008; Faso et al., 2009; Schoberer et al., 2009; Osterrieder et al., 2010; Ito et al., 2012; Langhans et al., 2012). Coexpression analyses showed that MAG5/SEC16A fusion proteins had a largely overlapping distribution with YFP-SEC24A and SEC13A-GFP (Figure 7), indicating that the distribution of MAG5/SEC16A within the cell coincides with that of COPII markers. This result, taken together with the association of MAG5/SEC16A with cup-shaped ER, suggests that MAG5/SEC16A could be an ideal marker of the plant ERESs.
Figure 7.
MAG5/SEC16A Fusion Proteins Colocalize with YFP-SEC24A and SEC13A-GFP.
(A) Confocal images of tobacco leaf epidermal cells coexpressing GFP-MAG5/SEC16A and YFP-SEC24A. GFP-MAG5/SEC16A is distributed in the cytosol and at puncta that colocalize with YFP-SEC24 (arrows). The inset corresponds to an enlarged region of the main image. Open arrows indicate chloroplasts, which are visible because of chlorophyll autofluorescence in the GFP channel. Bars = 5 µm (main image) and 2 µm (insets).
(B) Confocal images of tobacco leaf epidermal cells coexpressing MAG5/SEC16A-tagRFP and SEC13A-GFP. MAG5/SEC16A-tagRFP colocalizes with SEC13A-GFP. Bars = 5 µm (main image).
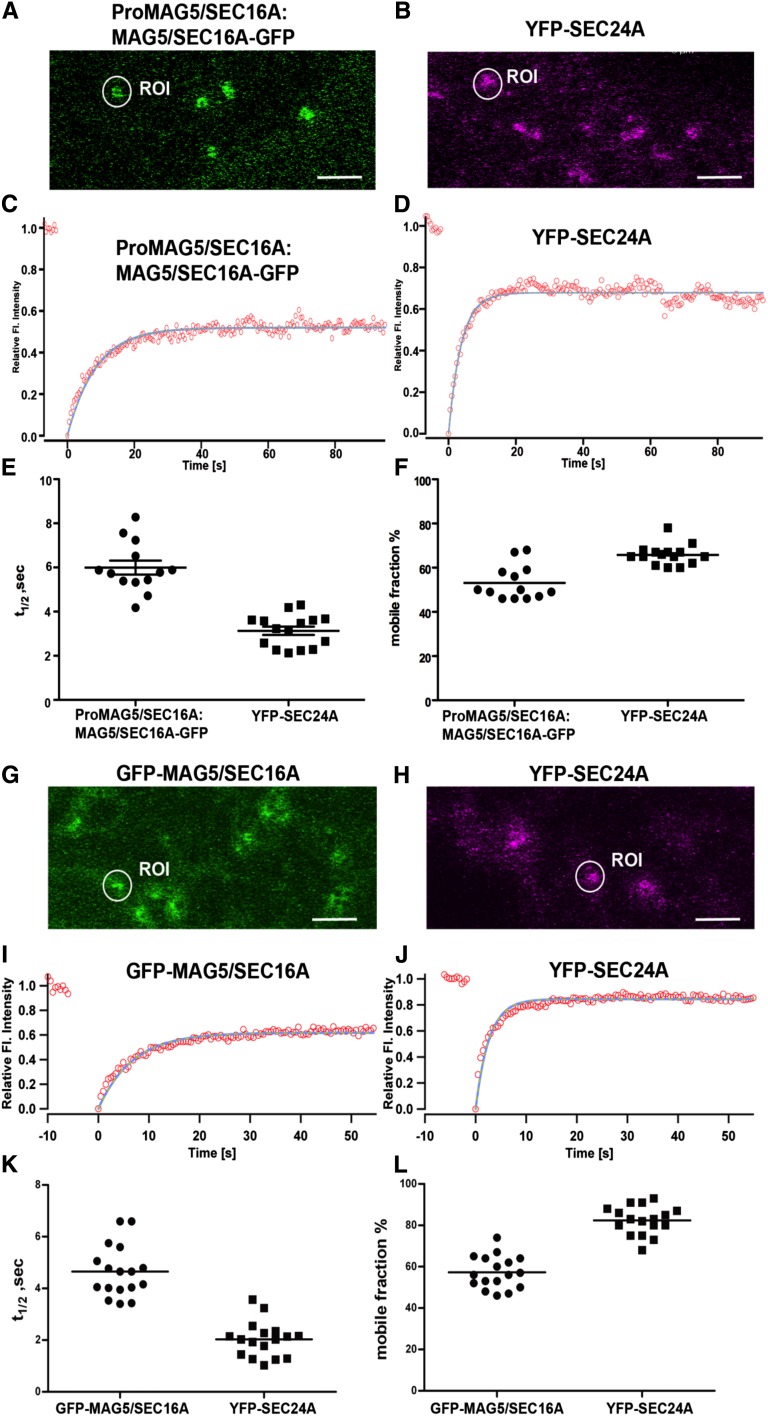
Next, we compared the dynamics of MAG5/SEC16A and SEC24A. COPII coat proteins are rapidly turned over (associate and dissociate) at ERESs (Ward et al., 2001; daSilva et al., 2004; Sato and Nakano, 2005; Hughes et al., 2009). In mammalian cells, Sec16 cycles on and off the membrane at a slower rate than other COPII components (Hughes et al., 2009). To compare the dynamics of MAG5/SEC16A with those of COPII coat proteins at plant ERESs, FRAP experiments were conducted in leaf epidermal cells of either the mag5-1 mutant rescued with Pro-MAG5/SEC16A:MAG5/SEC16A-GFP (Figure 8A) or the transgenic line expressing a functional fusion protein of SEC24A (YFP-SEC24A; Figure 8B) (Faso et al., 2009) using latrunculin B to depolymerize actin (Brandizzi et al., 2002b) and inhibit ERES movement (daSilva et al., 2004).
Figure 8.
Comparison of the Dynamics of MAG5/SEC16A with That of SEC24 at ERESs.
(A) and (B) Confocal images of leaf epidermal cells of mag5-1 rescued by expression of ProMAG5/SEC16A:MAG5/SEC16A-GFP (A) and of leaf epidermal cells of Arabidopsis plants expressing YFP-SEC24A (B). ROI, region of interest. Bar = 5 µm.
(C) and (D) Example regions of interest in (A) and (B), respectively, were used to obtain curves of FRAP events.
(E) and (F) Distributions of the individual half-time measurements (E) and mobile fractions (F) are shown. The data represent the mean of 13 to 15 independent bleaching experiments (horizontal bars). Samples were treated with latrunculin B to stop movement of ERESs, as described previously (Brandizzi et al., 2002b).
(G) and (H) Confocal images of tobacco leaf epidermal cells expressing GFP-MAG5/SEC16A (G) or YFP-SEC24 (H). Bars = 5 µm.
(I) and (J) Example ROI in (G) and (H), respectively, were used to obtain curves of FRAP events.
(K) and (L) Distributions of the individual half-time measurements (K) and mobile fractions (L) are shown. The data represent the mean of 13 to 17 independent bleaching experiments (horizontal bars). Samples were treated as described in (E) and (F).
Spot-bleach FRAP was conducted as described previously (Brandizzi et al., 2002b; daSilva et al., 2004), and the data were fitted to a single exponential (Figures 8C and 8D). MAG5/SEC16A-GFP cycled on and off ERESs with an average half-time of recovery of 5.99 ± 0.31 s (number of beached ERESs = 13) (Figure 8E). This turnover was significantly slower than that of YFP-SEC24A, which had an average half-time of recovery of 3.13 ± 0.18 s (P < 0.0001; number of bleached ERESs = 15). To compare the availabilities of MAG5/SEC16A-GFP and YFP-SEC24A for cycling on and off ERESs, the mobile fractions of the two fusion proteins were measured. A significantly smaller proportion of MAG5/SEC16A-GFP (53.15 ± 2.15%) was mobile compared with that of YFP-SEC24A (65.77% ± 1.36%) (P < 0.0001; number of bleached ERESs = 13) (Figure 8F).
Similar results were obtained from experiments using leaf epidermal cells of tobacco (Nicotiana tabacum) plants expressing GFP-MAG5/SEC16A or YFP-SEC24A (Figures 8G to 8L) and from experiments using stable Arabidopsis transformants expressing MAG5/SEC16A-GFP (see Supplemental Figure 9 online). GFP-MAG5/SEC16A had an average half-time of recovery of 4.64 ± 1.00 s, which was much slower than that of YFP-SEC24 (2.02 ± 0.46 s) (Figure 8K). The mobile fraction of GFP-MAG5/SEC16A (57.29% ± 0.62%) was lower than that of YFP-SEC24A (82.35% ± 0.47%) (Figure 8L). These data indicate that ERES protein kinetics is conserved among species (i.e., the COPII coat is turned over more rapidly than MAG5/SEC16A).
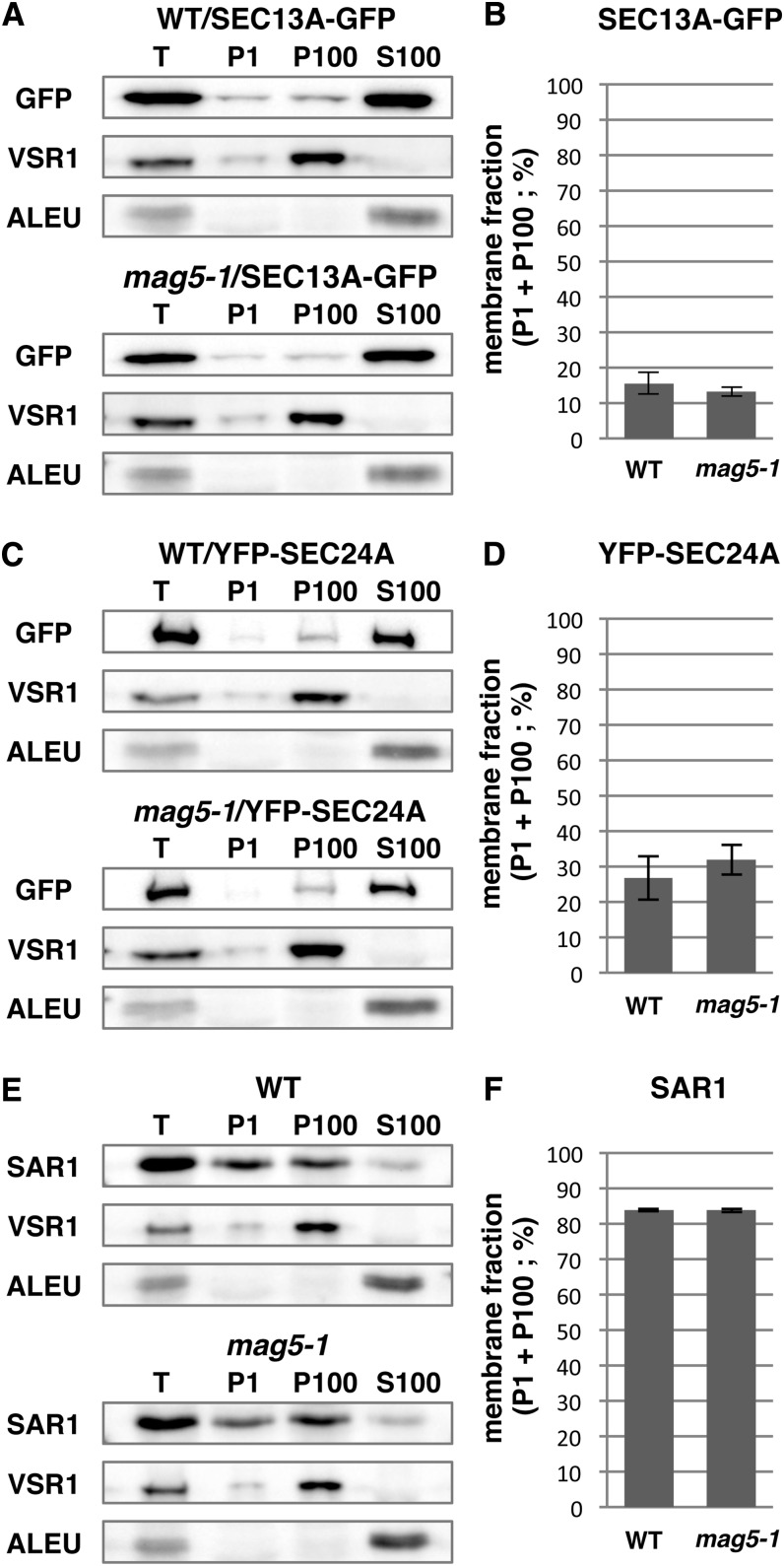
To test whether deficiency of MAG5/SEC16A affects the membrane associations of COPII components, we prepared microsomal and soluble fractions from wild-type and mag5-1 seedlings expressing SEC13A-GFP or YFP-SEC24A and then subjected these fractions to immunoblot analyses (Figure 9). The fraction of SEC13A-GFP that associated with membranes did not significantly differ between wild-type and mag5-1 seedlings. Similarly, the fraction of YFP-SEC24A that associated with membranes did not significantly differ between wild-type and mag5-1 seedlings. Furthermore, the fraction of endogenous SAR1 protein that associated with membranes did not significantly differ between wild-type and mag5-1 seedlings. Overall, these data suggest that loss of MAG5/SEC16A does not quantitatively affect the membrane association of the COPII coat components to ERESs.
Figure 9.
A Deficiency of MAG5/SEC16A Does Not Affect the Association of COPII Components with Membranes.
(A) Immunoblot of subcellular fractions generated from 5-d-old wild-type (WT) and mag5-1 seedlings showing the distribution of SEC13A-GFP. T, total extract, P1, pellet obtained following centrifugation at 1000g ; P100, pellet obtained following centrifugation at 100,000g; S100, supernatant obtained following centrifugation at 100,000g. Antibodies against GFP, VSR1 (membrane fraction marker), and ALEU (soluble fraction marker) were used.
(B) Quantitative analysis of the ratio of the SEC13A-GFP signal intensity in P1 plus P100 compared with that in S100 in the immunoblot shown in (A). The data represent the mean of three independent experiments (Student’s t test, P > 0.05, n = 3).
(C) Immunoblot of each fraction showing the distribution of YFP-SEC24A in wild-type and mag5-1 seedlings.
(D) Quantitative analysis of the ratio of YFP-SEC24A signal intensity in P1 plus P100 compared with that in S100 in the immunoblot shown in (C). The data represent the mean of three independent experiments (Student’s t test, P > 0.05, n = 3).
(E) Immunoblot of each fraction showing the distribution of SAR1 in wild-type and mag5-1 seedlings. An anti-SAR1 antibody was used.
(F) Quantitative analysis of the ratio of the SAR1 signal intensity in P1 plus P100 compared with that in S100 in the immunoblot shown in (E). The data represent the mean of three independent experiments (Student’s t test, P > 0.05, n = 3).
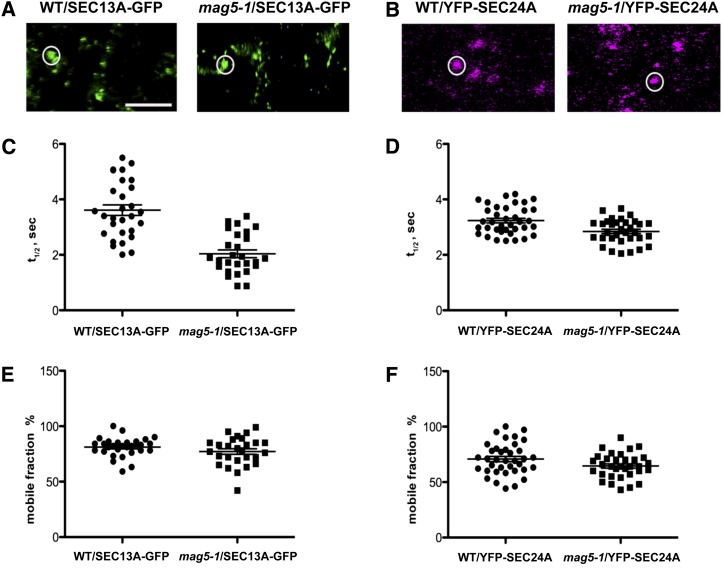
Next, we aimed to establish the effects of loss of functional MAG5/SEC16A on the dynamic assembly of COPII coat at ERESs. Therefore, we analyzed the dynamic cycling of inner and outer COPII coat components on and off ERESs in the mag5-1 background by stably expressing fluorescent fusion proteins of SEC13 and SEC24, respectively. FRAP analyses showed that cycling of these two COPII coat components on and off ERESs was significantly faster in the mag5-1 mutant than in wild-type plants (Figure 10). These data indicate that the cycling of COPII components on and off ERESs is higher in the absence of MAG5/SEC16A than in the presence of MAG5/SEC16A, which supports a regulatory role of MAG5/SEC16A in COPII coat assembly at ERESs in plants.
Figure 10.
MAG5/SEC16A Functions in the Dynamic Cycling of SEC13 and SEC24 on and off ERESs
(A) Confocal images of SEC13A-GFP in leaf epidermal cells of wild-type (WT) and mag5-1 plants. The bleached area is shown in the circle. Bar = 5 µm.
(B) Confocal images of YFP-SEC24A in leaf epidermal cells of wild-type and mag5-1 plants. The bleached area is shown in the circle. Bar = 5 µm.
(C) Distribution of the individual half-times of SEC13A-GFP in wild-type and mag5-1 plants. The data represent the mean of 27 to 29 independent bleaching experiments following treatment with latrunculin B to stop movement of ERESs. The mean half-time of SEC13A-GFP is 3.61 ± 0.19 s and 2.03 ± 0.13 s in wild-type and mag5-1 plants, respectively (P < 0.0001).
(D) Distribution of the individual half-time measurement for YFP-SEC24A in wild-type and mag5-1 plants. The data represent the mean of 33 to 37 independent bleaching experiments. The mean half-time of YFP-SEC24A is 3.23 ± 0.08 s and 2.84 ± 0.07 s in wild-type and mag5-1 plants, respectively (P < 0.005).
(E) Analysis of the mobile fraction of SEC13A-GFP in wild-type and mag5-1 plants, showing that the amount of SEC13-GFP recovered after photobleaching does not significantly differ between wild-type and mag5-1 plants. The mean mobile fraction of SEC13A-GFP is 81% ± 1.6% and 77.19% ± 2.4% in wild-type and mag5-1 plants, respectively (P > 0.05).
(F) Analysis of the mobile fraction of YFP-SEC24A in wild-type and mag5-1 plants, showing the amount of YFP-SEC24A recovered after photobleaching does not significantly differ between wild-type and mag5-1 plants. The mean mobile fraction of YFP-SEC24A is 70.70% ± 2.3% and 64.79% ± 2.05% in wild-type and mag5-1 plants, respectively (P > 0.05).
DISCUSSION
Arabidopsis MAG5/SEC16A Is the Functional Ortholog of Sec16 and Functions Redundantly with SEC16B in Plant Cells
Whereas orthologs of most COPII proteins have been characterized in plants, the plant orthologs of Sec16 had long remained unidentified. In the mag screen, identification of a deletion in MAG5 allowed us to functionally characterize endogenous Sec16. Among mag mutants, mag5 belongs to the mag mutant group that develops MAG bodies that appear as distinct domains of the ER cisternae. mag5 might form MAG bodies as a result of less efficient release of newly synthesized storage proteins from the ER. Here, we provide evidence that MAG5/SEC16A is the functional ortholog of animal and yeast Sec16. This is supported by protein sequence analyses showing that conserved functional regions of MAG5/SEC16A share partial sequence identity with human Sec16A. We also showed that functional MAG5/SEC16A is required for efficient ER export, it can bind COPII coat proteins, and its availability influences the dynamics with which COPII coat proteins are recruited to the ER (Watson et al., 2006; Bhattacharyya and Glick, 2007; Iinuma et al., 2007). The discrete steady state localization of MAG5/SEC16A at ERESs that are associated with the Golgi in concomitance with the requirement for actin integrity for the movement of the MAG5/SEC16A-labeled ERESs provides conclusive evidence that the ER/Golgi interface is uniquely organized in plants such that ERESs move together with associated Golgi stacks.
There are two SEC16 isoforms in Arabidopsis: MAG5/SEC16A and SEC16B. Notably, loss of MAG5/SEC16A partially disrupted ER export of endogenous cargo as well as that of a bulk membrane cargo in vegetative tissues and affected the dynamics of COPII coat proteins at the ER. However, the mag5 mutants are viable with no marked defects in growth or development, which contrasts the fact that Sec16 is essential in yeast (Espenshade et al., 1995). The viability of mag5, together with a high similarity between MAG5/SEC16A and SEC16B, suggests that they might redundantly function during the essential steps of ER export, although we cannot exclude the possibility that MAG5/SEC16A and SEC16B have acquired different nonessential functions. The two human Sec16 isoforms (Sec16A and Sec16B) are not functionally redundant and Sec16B has acquired distinct specialized functions (Budnik et al., 2011; Yonekawa et al., 2011). The functions of Sec16 proteins seem to have diversified so that they fulfill several functions in multicellular eukaryotes.
MAG5/SEC16A Likely Acts as a Scaffold for COPII Assembly and Influences the Turnover of COPII Coat Proteins on the ER
We demonstrated that a functional GFP fusion of MAG5/SEC16A concentrates in cup-like punctate structures on the ER. The distribution of MAG5/SEC16A at the ER mimics that of Sec16 at ERESs in mammalian cells (Hughes et al., 2009) and that of Sec13-labeled ERESs localized on saddle-shaped surfaces of the ER in yeast (Okamoto et al., 2012). Hence, the MAG5/SEC16A labels plant ERESs, which are similarly organized on the ER in different species, at least at the resolution offered by confocal microscopy.
There is continuous turnover of COPII coat proteins on and off ERESs to favor formation of COPII carriers and to recycle COPII coat proteins at the ERESs (Ward et al., 2001; daSilva et al., 2004; Hughes et al., 2009). The FRAP analyses presented here show that MAG5/SEC16A cycles on and off ERESs, although at a slower turnover rate and with a smaller mobile fraction than SEC24A. These data are consistent with the reported dynamics of Sec16 relative to the COPII coat protein at ERESs in mammalian cells (Stephens et al., 2000; Forster et al., 2006; Hughes et al., 2009). Furthermore, based on a partial distribution of the MAG5/SEC16A in the cytosol in concomitance to a distribution at ERES, we propose that the binding of SEC16A to ERESs is saturable. The large immobile population of Sec16 at ERESs has been proposed to facilitate the role of Sec16 as a template for COPII coat assembly, most likely through cooperative interactions with Sar1 (Ivan et al., 2008), Sec23 (Bhattacharyya and Glick, 2007), Sec24 (Gimeno et al., 1996), Sec13 (Hughes et al., 2009), and Sec31 (Shaywitz et al., 1997). In this study, mass spectrometry analysis detected an interaction of MAG5/SEC16A with SEC13, but not with SEC31, although yeast two-hybrid analysis detected an interaction of MAG5/SEC16A with both SEC13 and SEC31. The affinity of MAG5/SEC16A for SEC13 proteins might be higher than its affinity for other COPII components. This is supported by the proposal that the Sec16/13 heterodimer functions as a scaffold for COPII coat protein assembly in yeast (Whittle and Schwartz, 2010). It is possible that Arabidopsis MAG5/SEC16A interacts with other COPII proteins, similar to other species, but this remains to be experimentally validated.
The distribution and dynamics of plant MAG5/SEC16A at ERESs as well as the interaction of MAG5/SEC16A with SEC13 and SEC31 suggest that MAG5/SEC16A plays a conserved scaffolding role necessary for COPII assembly at ERESs. However, a deficiency of MAG5/SEC16A accelerated turnover of COPII coat proteins at ERESs. In vitro experiments recently showed that yeast Sec16 hampers the GTPase activity of the full COPII coat, supporting the hypothesis that Sec16 can regulate COPII carrier formation by affecting Sar1 activity (Kung et al., 2012; Yorimitsu and Sato, 2012). Consistent with this, the turnover of COPII coat proteins at ERESs was faster in the absence of MAG5/SEC16A than in wild-type plants, as would be expected for an accelerated formation and consumption of COPII carriers at the ERES. However, in the absence of MAG5/SEC16A, COPII coat components assembled at ERESs at similar levels as in wild-type plants. Because the steady state abundance of COPII coat proteins at ERESs is the result of two opposite events (COPII coat protein recruitment and release), our results suggest that in the absence of MAG5/SEC16A, COPII coat recruitment to the ER is increased but dynamically compensated for by an accelerated release of COPII coat proteins from the membrane compared with the wild type. This is consistent with an increase in Sar1-GTPase activity (Kung et al., 2012; Yorimitsu and Sato, 2012); however, it also implies that MAG5/SEC16A functions to limit the level of SAR1GTP-SEC23/SEC24-coated prebudding complexes at ERESs, which might ensure the fidelity with which fully competent ER-export carriers are produced or limit excessive energy consumption due to the GTPase activity of SAR1. Consistent with this model, defects in ER export concomitant with loss of MAG5/SEC16A can be explained by the faster turnover of COPII coat components at ERESs in the mag5 mutant than in the wild type. Increased turnover of the COPII coat could result in inefficient cargo loading into COPII carriers or futile cycles of binding and release of COPII coat proteins at ERESs with consequent loss of efficient ER export.
Alternatively, the ER export phenotype seen in the MAG5/SEC16A loss-of-function mutant may be linked to an imbalance between the COPII and COPI transport pathways. Accelerated production of COPII carriers from the ER could increase the ER export of components necessary for ER-Golgi transport (e.g., cargo receptors and soluble N-ethylmaleimide-sensitive factor attachment protein receptors) to the Golgi, which would not be buffered by steady state COPI-mediated protein recycling to the ER. This would indirectly cause inefficient ER membrane export. Although these possibilities require experimental validation, our in vivo results argue that MAG5/SEC16A regulates the turnover of COPII coat components at ERESs rather than merely serving as an ERES scaffolding protein necessary for the formation of COPII carriers.
MAG5/SEC16A Marks ERESs That Face the Golgi
The organization of the ER and Golgi in plant cells imparts a unique challenge to achieving efficient ER export to mobile Golgi stacks and raises a long-standing question in the field: How are proteins efficiently exported from the ER to mobile Golgi target membranes? The distribution of COPII coat markers (SEC24, SEC23, and SEC13) that associate at the interface between the ER and the Golgi (Stefano et al., 2006; Faso et al., 2009; Hanton et al., 2009; Marti et al., 2010) supports a mobile secretory unit model, in which ERESs are associated with mobile Golgi bodies and COPII coat turnover occurs during Golgi movement (daSilva et al., 2004). It is also possible that COPII carriers associate with the Golgi, as is suggested by analyses of the distribution of a YFP-SEC24 fusion protein with respect to Golgi stacks in leaf epidermal cells (Langhans et al., 2012). An association of partially uncoated COPII carriers with the Golgi raises the important question of whether structures labeled by the COPII markers represent ER subdomains or merely Golgi-associated COPII carriers that are partially uncoated. The latter would suggest the possibility that the Golgi stacks collect COPII carriers originating from unidentified regions of ER. However, our data demonstrated that a functional fusion protein of MAG5/SEC16A shows large overlap with other COPII proteins, such as SEC24A and SEC13, and that MAG5/SEC16A was located in mobile regions of the ER that faced Golgi stacks. Although the optical resolution of confocal microscopy does not allow the ERGolgi interface of highly vacuolated plant cells to be resolved, our analyses of the turnovers of MAG5/SEC16A, SEC24, and SEC13 at membranes indicate that MAG5/SEC16A is not present in partially uncoated carriers at the Golgi. If MAG5/SEC16A were included in these structures, its turnover attributable to carrier uncoating would be similar to that of COPII components such as SEC13 and SEC24. SEC13 and SEC24 cycled on and off membranes at similar rates (Figure 10). However, the cycling of MAG5/SEC16A was significantly different, both in terms of the rate of protein exchange at the bleached sites and the percentage of protein available for exchange. These data indicate that a large fraction of MAG5/SEC16A protein is excluded from the COPII coat, in agreement with the observation that a small fraction of Sec16 is associated with COPII vesicles in vivo (Espenshade et al., 1995).
Hence, although we cannot exclude the possibility that at least a portion of the foci labeled by SEC24 and SEC13 represent Golgi-associated partially uncoated COPII carriers, we propose that the structures labeled by MAG5/SEC16A are bona fide ERESs. Taken together, these observations are consistent with a previous proposal that ERESs are stably associated with Golgi stacks (daSilva et al., 2004). Laser trapping analysis revealed that Golgi bodies are physically attached to ER tubules (Sparkes et al., 2009). These reports, together with our findings, raise the possibility that, at least in highly vacuolated cells, ERESs function at an ER surface that is relatively static and interfaced with a Golgi body. The continuous association of ERESs with the Golgi might facilitate protein export from the ER to mobile Golgi stacks.
METHODS
Isolation of Arabidopsis thaliana mag Mutants
Arabidopsis thaliana ecotype Columbia-0 was used as the wild type. Arabidopsis T-DNA–tagged lines were screened as described previously (Shimada et al., 2006). Dry seeds (0.1 mg) from a mixture of one to 20 lines were subjected to SDS-PAGE and then to immunoblot analysis with anti-12S globulin and anti-2S albumin antibodies to detect storage protein precursors. Eight lines (mag mutants) were isolated that abnormally accumulated storage protein precursors. The mag5-1 line described in this article was derived from a pool of T-DNA–tagged lines that was prepared with the Ti plasmid pBI121. The T-DNA insertion mutants were obtained as follows: WiscDsLoxHS202_06C (mag5-2) from The European Arabidopsis Stock Center; SALK_070608 (sec16b-1), SALK_125584 (sec16b-2), and SALK_144775 (sec16b-3) from the ABRC at Ohio State University; and GABI_147G11 (mag5-3) from Genomanalyse im Biologischen System Pflanze (GABI-Kat).
SDS-PAGE and Immunoblotting
SDS-PAGE and immunoblot analysis were performed as described previously (Shimada et al., 2003). For immunoblotting, we newly generated antibody against 2S albumin, designated as anti-2S3M, using the synthetic peptide (C)AARAVSLQGQHGPFQSRKIY, whose sequence was derived from the mature region of Arabidopsis 2S3 (At4g27160). The peptide was conjugated to keyhole limpet hemocyanin (KLH) and then injected into a rabbit (Sigma-Aldrich Japan). Blots were probed with anti-12S globulin α-subunit (anti-12S, diluted 10,000-fold), (Shimada et al., 2003), anti-2S albumin (anti-2S3M, diluted 5000-fold), anti-GFP (JL-8, diluted 5000-fold; Clontech), anti-VSR1 (diluted 5000-fold; Yamada et al., 2005), anti-AtALEU (diluted 5000-fold; Ueda et al., 2006), or anti-SAR1 (diluted 500-fold; Agrisera) antibodies. Signals were detected with the ECL detection system (GE Healthcare).
Electron Microscopy
For electron microscopy, dry seeds were fixed, dehydrated, and embedded in LR white resin (London Resin), as described previously (Hayashi et al., 1998), with a modification in that the fixative contained 10% DMSO. Ultrathin sections were examined with a transmission electron microscope (model JEM-1015B; JEOL) at 100 kV. Immunocytochemistry was performed as described previously (Hayashi et al., 1998) with the following antibody dilutions: anti-12S, 1: 2000; anti-2S, 1:1000; and AuroProbe EM anti-rabbit IgG (H+T), 1:30 (15-nm gold; GE Healthcare).
Map-Based Cloning
The mag5-1 homozygous mutant (ecotype Columbia-0) was crossed with ecotype Landsberg erecta plants to generate a mapping population. F2 seeds were obtained by self-fertilization of the F1 plants. To isolate protein and genomic DNA from one seed grain, each F2 seed was individually homogenized in 50 μL of extraction buffer (50 mM Tris-HCl, pH 6.8, 250 mM NaCl, 25 mM EDTA, 1% [w/v] SDS, 0.5% [v/v] 2-mercaptoethanol, and 5% [v/v] glycerol). The extracts (15 µL) were subjected to SDS-PAGE and Coomassie Brilliant Blue staining to select mag5-1 mutant plants. The genomic DNA of mag5-1 mutant plants was analyzed using a combination of cleaved amplified polymorphic sequence markers and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994) with data obtained from The Arabidopsis Information Resource (http://www.Arabidopsis.org). Approximately 20 recombinant plants from the F2 progeny were screened for the mag5 phenotype for rough mapping. The position of the mag5 gene was mapped to the middle of chromosome 5. For fine-scale mapping, genomic DNA was isolated from 421 F2 plants. The nucleotide sequence was determined from both strands using the ABI Prism Big Dye Terminator cycle sequence reaction kit (Applied Biosystems) and a DNA sequencer (Prism 3100; Applied Biosystems).
RT-PCR Analyses
Total RNA was isolated from wild-type and mag5 plants using an RNeasy plant mini kit (Qiagen). RNA (1 μg) was treated with DNase I (Invitrogen) to reduce genomic DNA contamination. Reverse transcription was performed using Ready-To-Go RT-PCR beads (GE Healthcare) with an oligo(dT)12-18 primer. PCR conditions were as follows: 94°C for 2 min and 30 to 40 cycles of 98°C for 10 s, 55°C for 30 s, and 68°C for 45 s or 4 min. The primers used for PCR were provided in Supplemental Table 2 online. ACTIN2 was used as a control (Ratcliffe et al., 2003). The PCR products were analyzed by capillary electrophoresis (QIAxcel analyzer; Qiagen) or sequencing.
Fluorescent Proteins and Molecular Cloning
The fluorescent proteins used in this study were based on fusion with mGFP5 (Haseloff et al., 1997) and enhanced YFP (Clontech). MAG5/SEC16A was subcloned into the binary vector pVKH18-En6 under the control of the constitutive promoter cauliflower mosaic virus 35S (Batoko et al., 2000).
The coding region of MAG5/SEC16A was amplified by PCR with mRNA from wild-type leaves (see Supplemental Table 2 online). The amplified fragment was inserted into pENTER/D-TOPO (Invitrogen) to produce the entry clone (see Supplemental Table 3 online). We performed an L/R recombination reaction to transfer the fragment of the entry clone into the destination vector pFAST R05 (Shimada et al., 2010) for the expression of MAG5/SEC16A-GFP driven by the 35S promoter. ProMAG5/SEC16A:MAG5/SEC16A was amplified by PCR from genomic DNA of wild-type plants. The amplified fragment was inserted into pENTER/D-TOPO (Invitrogen) to produce the entry clone. Then, the stop codon and 3′ untranslated region of MAG5/SEC16A were deleted (see Supplemental Table 2 online). We performed the L/R recombination reaction to transfer the fragment of the entry clone into the destination vector pGWB4 (Nakagawa et al., 2007) for the expression of MAG5/SEC16A-GFP driven by the native promoter (see Supplemental Table 3 online). The expression vectors were introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation. Wild-type or mag5-1 mutant plants were transformed with Agrobacterium by the floral dip method (Clough and Bent, 1998). T1 seeds were selected under the fluorescence stereomicroscope (for T1 seeds carrying pFAST R05) (Shimada et al., 2010) or in a medium containing 30 mg/L of hygromycin (for T1 seeds carrying pGWB4) to establish independent transgenic lines.
Transient Expression System
Four-week-old tobacco (Nicotiana tabacum cv Petit Havana) greenhouse plants grown at 25°C were used for Agrobacterium (strain GV3101)–mediated transient expression (Batoko et al., 2000). The bacterial optical density (OD600) used for plant transformation was 0.05 for all constructs.
Confocal Laser Scanning Microscopy
Fluorescent images were captured with confocal laser scanning microscopes (LSM 510 and LSM780; Carl Zeiss) using line switching mode. For mGFP5 and YFP imaging, settings matched those of earlier studies (Brandizzi et al., 2002a; Hanton et al., 2007). FRAP analyses were conducted as described (Brandizzi et al., 2002b). Single-channel and colocalization analyses were performed using 1 Airy Unit pinhole size. FRAP analyses were performed using fully open pinhole settings. For experiments based on comparative analyses, images were acquired using the same laser intensity, line averaging, and brightness/contrast settings. Postacquisition analyses were performed with Zeiss AIM software. PaintShop Pro software was used for further image processing. For three-dimensional image acquisition and rendering, images were acquired at maximum speed using scan average = 1. Z-stacks were assembled with the Zeiss AIM software and deconvoluted using Image J software (Deconvolution and 3D plug-ins).
Brefeldin A Time-Course Treatment
Cotyledons from 10-d-old plants were treated with 100 µg/mL of brefeldin A for different times and immediately mounted on microscope slide for observation with a confocal microscope.
FRAP
Spot bleaching and quantification of recovery and mobile fraction rates were performed on individual ERESs as described earlier (Brandizzi et al., 2002b; daSilva et al., 2004). Half-time and mobile fraction values represent the average ± sd for 13 to 40 independent spot-bleach experiments per sample. The statistical significance of the experiments was determined using a Student’s two-tailed t test for two samples assuming equal variance.
Immunoprecipitation
Immunoprecipitation was performed with µMACS epitope tag protein isolation kits (Miltenyi Biotec). Whole seedlings of transgenic Arabidopsis plants expressing GFP (1.0 g) or MAG5/SEC16A-GFP (4.0 g) were homogenized on ice in 3 or 12 mL of extraction buffer containing 50 mM Tris-HCl, pH 8.0, 0.15 M NaCl, 1% (v/v) Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor (complete cocktail tablets; Roche). Homogenates were centrifuged at 9100g for 10 min to remove cellular debris. The supernatants were mixed with magnetic beads conjugated to an anti-GFP antibody (Miltenyi Biotec) and then incubated on ice for 30 min. The mixtures were applied to µColumns (Miltenyi Biotec) in a magnetic field to capture the magnetic antigen-antibody complex. After extensive washing with the buffer, immunoaffinity complexes were eluted with 50 μL of SDS sample buffer containing 50 mM Tris-HCl, pH 6.8, 2% (w/v) SDS, 10% (w/v) glycerol, and 6% (v/v) 2-mercaptoethanol.
Peptide Preparation for Tandem Mass Spectrometry Analysis
For in-gel digestion, the protein components of the immunoprecipitates were separated on a 3-cm-long SDS gel. The gel slice isolated from each lane was cut into three fractions according to molecular mass: a <40-kD fraction, a 40- to 100-kD fraction, and a >100-kD fraction. Each excised gel fraction was treated with 100% (v/v) acetonitrile for 15 min and then dried in a vacuum concentrator. The dried gel was treated with 10 mM DTT in 50 mM ammonium bicarbonate at 56°C for 45 min, followed by 55 mM 2-iodoacetamide in 50 mM ammonium bicarbonate for 30 min in darkness. The gel was washed with 25 mM ammonium bicarbonate, dehydrated with 50% (v/v) acetonitrile in 50 mM ammonium bicarbonate, and then dried in a vacuum concentrator. The dried gel was treated with 0.01 mg/mL of trypsin in 50 mM ammonium bicarbonate and incubated at 37°C for 16 h. The digested peptides were recovered with 50 μL 5% (v/v) formic acid in 50% (v/v) acetonitrile, followed by 50 μL 5% (v/v) formic acid in 70% (v/v) acetonitrile. The extracted peptides were combined and then evaporated to 10 μL in a vacuum concentrator.
Mass Spectrometry Analysis and Database Search
Liquid chromatography–tandem mass spectrometry (MS/MS) analyses were performed using the LTQ-Orbitrap XL-HTC-PAL system. Trypsin digests were loaded onto the column (100-µm internal diameter and 15-cm length; L-Column; CERI) using the Paradigm MS4 HPLC pump (Michrom BioResources) and HTC-PAL Autosampler (CTC Analytics) and were eluted by a gradient of 5 to 45% (v/v) acetonitrile in 0.1% (v/v) formic acid for 26 min. The eluted peptides were introduced directly into an LTQ-Orbitrap XL mass spectrometer (Thermo) with a flow rate of 500 nL/min and a spray voltage of 2.0 kV. The range of the mass spectrometry scan was mass-to-charge ratio 450 to 1500. The top three peaks were subjected to MS/MS analysis. MS/MS spectra were analyzed by the Mascot server (version 2.3) in house (Perkins et al., 1999) (http://www.matrixscience.com/) and compared with proteins registered in TAIR10 (The Arabidopsis Information Resource). The Mascot search parameters were set as follows: threshold of the ion score cutoff, 0.05; peptide tolerance, 10 ppm; MS/MS tolerance, ±0.8 D; and peptide charge, 2+ or 3+. The search also was set to allow one missed cleavage by trypsin, a carboxymethylation modification of Cys residues, and a variable oxidation modification of Met residues.
Yeast Two-Hybrid Assay
The coding regions of SEC13A, SEC13B, SEC31A, and SEC31B were amplified by PCR with mRNA from wild-type leaves (see Supplemental Table 2 online). The primers used for cloning of SEC13B were as described (Tamura et al., 2010). The amplified fragments were inserted into pENTER/D-TOPO (Invitrogen) to produce the entry clones. We performed an L/R recombination reaction to transfer the fragments of the entry clones into the destination vector either pDEST-GBKT7 or pDEST-GADT7 (Rossignol et al., 2007) (see Supplemental Table 3 online). The products, referred to as BD- or AD- in Figure 6D and Supplemental Figure 8 online, were transformed into Saccharomyces cerevisiae AH109 strain (Matchmaker III; Clontech). Transformed yeasts were selected on SD/-Leu/-Trp plates and the interactions were examined on SD/-Leu/-Trp/-His/-Ade plates. pGBKT7 and pGADT7 vectors were used for negative control.
Subcellular Fractionation
The wild-type plants, transgenic plants expressing SEC13A-GFP, transgenic plants expressing YFP-SEC24A (Nakano et al., 2009), mag5-1 plants, mag5-1 plants expressing SEC13A-GFP, and mag5-1 plants expressing YFP-SEC24A were used. The fractionation was performed essentially as described previously (Tamura et al., 2005). Five-day-old seedlings were homogenized in a mortar on ice in triple volume (SEC13A-GFP and SAR1) or double volume (YFP-SEC24A) of buffer (100 mM HEPES-KOH, pH 7.5, 0.3 M Suc, and protease inhibitor [complete cocktail tablets; Roche]). The homogenate was filtered through a cell strainer (70 μm) by centrifugation at 20g. An aliquot of the filtrate was used as a total fraction. The filtrate was centrifuged at 1000g and 4°C for 20 min. The supernatant was ultracentrifuged at 100,000g and 4°C for 1 h to obtain supernatant (S100) and pellet (P100) fraction. Each of total fractions, P1, P100, and S100, was subjected to immunoblot analysis. The statistical significance of the experiments was determined using a Student’s two-tailed t test for two samples assuming equal variance.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: MAG5/SEC16A (At5g47480), SEC16B (At5g47490), SEC13A (At3g01340), SEC13B (At2g30050), SEC24A (At3g07100), SEC31A (At1g18830), SEC31B (At3g63460), and ACT2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The cDNA and Protein Sequences for the N-Terminal Part of MAG5/SEC16A in the Wild Type and mag5-1 Mutant.
Supplemental Figure 2. Schematic Representation of Sec16 Proteins.
Supplemental Figure 3. Time-Course Analysis of ST-GFP in Wild-Type and mag5-2 Leaves Following Brefeldin A Treatment.
Supplemental Figure 4. Normal Growth of Transgenic Arabidopsis Plants Expressing Pro35S:MAG5/SEC16A-GFP.
Supplemental Figure 5. Localization Analyses of GFP-MAG5/SEC16A and YFP-SYP61.
Supplemental Figure 6. Images Extracted from Time-Lapse Microscopy Analysis of a Tobacco Leaf Epidermal Cell Coexpressing GFP-MAG5/SEC16A and ST-YFP.
Supplemental Figure 7. Peptides Identified by Mass Spectrometry.
Supplemental Figure 8. Yeast Two-Hybrid Assay Showing the Interaction between SEC31 and SEC13.
Supplemental Figure 9. FRAP Analysis of Pro35S:MAG5/SEC16A-GFP at ERESs.
Supplemental Table 1. Proteins Identified by Mass Spectrometry of Immunoprecipitates from MAG5/SEC16A-GFP.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Constructs Used in This Study.
Supplemental Movie 1. Time-Lapse Microscopy of Tobacco Leaf Epidermal Cells Coexpressing GFP-MAG5/SEC16A and ST-YFP.
Acknowledgments
This work was supported by Specially Promoted Research of Grant-in-Aid for Scientific Research 22000014 (to I.H.-N.) and a research fellowship (241880 to J.T.) from the Japan Society for the Promotion of Science and by Grants for Excellent Graduate Schools from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We also acknowledge support by the grants (to F.B.) from Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-91ER20021) for infrastructure and partial salary support, the National Institutes of Health (R01 GM101038-01) and the National Science Foundation (MCB 0948584 and MCB 1243792) for partial salary support. We thank Tsuyoshi Nakagawa of Shimane University for donation of the Gateway vector (pGWB4), the ABRC for providing the seeds of SALK_070608 (sec16b-1), SALK_125584 (sec16b-2), and SALK_144775 (sec16b-3), The European Arabidopsis Stock Center for providing the seeds of WiscDsLoxHS202_06C (mag5-2), and Genomanalyse im Biologischen System Pflanze for providing the seed of GABI_147G11 (mag5-3).
AUTHOR CONTRIBUTIONS
J.T., L.R., T.S., F.B., and I.H-.N. designed the research. J.T. performed forward genetics, imaging analysis, mass spectrometry, yeast two-hybrid assay, and subcellular fractionation. L.R. and G.S. performed imaging and FRAP analyses. H.T., Y.K., M.K., and M.N. performed transmission electron microscopy. H.T. performed rough mapping of mag5-1. K.T. performed the work with SEC13. Y.F. performed mass spectrometry. J.T., L.R., T.S., F.B., and I.H-.N. wrote the article. T.S., F.B., and I.H-.N. contributed equally to this work.
Glossary
- ER
endoplasmic reticulum
- ERES
ER exit site
- PSV
protein storage vacuole
- FRAP
fluorescence recovery after photobleaching
- MS/MS
tandem mass spectrometry
- Col-0
Columbia-0
References
- Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M.F., Ravazzola M., Amherdt M., Schekman R. (1994). COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77: 895–907 [DOI] [PubMed] [Google Scholar]
- Barlowe C., Schekman R. (1993). SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365: 347–349 [DOI] [PubMed] [Google Scholar]
- Batoko H., Zheng H.Q., Hawes C., Moore I. (2000). A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell C.J., Ecker J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D., Glick B.S. (2007). Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol. Biol. Cell 18: 839–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X., Corpina R.A., Goldberg J. (2002). Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419: 271–277 [DOI] [PubMed] [Google Scholar]
- Bi X., Mancias J.D., Goldberg J. (2007). Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev. Cell 13: 635–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Barlowe C. (2013). Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14: 382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F., Fricker M., Hawes C. (2002a). A greener world: The revolution in plant bioimaging. Nat. Rev. Mol. Cell Biol. 3: 520–530 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Saint-Jore C., Moore I., Hawes C. (2003). The relationship between endomembranes and the plant cytoskeleton. Cell Biol. Int. 27: 177–179 [DOI] [PubMed] [Google Scholar]
- Brandizzi F., Snapp E.L., Roberts A.G., Lippincott-Schwartz J., Hawes C. (2002b). Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: Evidence from selective photobleaching. Plant Cell 14: 1293–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budnik A., Heesom K.J., Stephens D.J. (2011). Characterization of human Sec16B: Indications of specialized, non-redundant functions. Sci. Rep. 1: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Reinisch K., Ferro-Novick S. (2007). Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12: 671–682 [DOI] [PubMed] [Google Scholar]
- Castillon G.A., Watanabe R., Taylor M., Schwabe T.M., Riezman H. (2009). Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic 10: 186–200 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Connerly P.L., Esaki M., Montegna E.A., Strongin D.E., Levi S., Soderholm J., Glick B.S. (2005). Sec16 is a determinant of transitional ER organization. Curr. Biol. 15: 1439–1447 [DOI] [PubMed] [Google Scholar]
- daSilva L.L., Snapp E.L., Denecke J., Lippincott-Schwartz J., Hawes C., Brandizzi F. (2004). Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16: 1753–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade P., Gimeno R.E., Holzmacher E., Teung P., Kaiser C.A. (1995). Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with Sec23p. J. Cell Biol. 131: 311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faso C., Chen Y.N., Tamura K., Held M., Zemelis S., Marti L., Saravanan R., Hummel E., Kung L., Miller E., Hawes C., Brandizzi F. (2009). A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21: 3655–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R., Weiss M., Zimmermann T., Reynaud E.G., Verissimo F., Stephens D.J., Pepperkok R. (2006). Secretory cargo regulates the turnover of COPII subunits at single ER exit sites. Curr. Biol. 16: 173–179 [DOI] [PubMed] [Google Scholar]
- Gimeno R.E., Espenshade P., Kaiser C.A. (1996). COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol. Biol. Cell 7: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A.T., Glick B.S. (2000). Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell 11: 3013–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton S.L., Chatre L., Matheson L.A., Rossi M., Held M.A., Brandizzi F. (2008). Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol. Biol. 67: 283–294 [DOI] [PubMed] [Google Scholar]
- Hanton S.L., Chatre L., Renna L., Matheson L.A., Brandizzi F. (2007). De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 143: 1640–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton S.L., Matheson L.A., Chatre L., Brandizzi F. (2009). Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J. 57: 963–974 [DOI] [PubMed] [Google Scholar]
- Haseloff J., Siemering K.R., Prasher D.C., Hodge S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94: 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C., Osterrieder A., Hummel E., Sparkes I. (2008). The plant ER-Golgi interface. Traffic 9: 1571–1580 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutyric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid beta-oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H., Budnik A., Schmidt K., Palmer K.J., Mantell J., Noakes C., Johnson A., Carter D.A., Verkade P., Watson P., Stephens D.J. (2009). Organisation of human ER-exit sites: Requirements for the localisation of Sec16 to transitional ER. J. Cell Sci. 122: 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H., Stephens D.J. (2010). Sec16A defines the site for vesicle budding from the endoplasmic reticulum on exit from mitosis. J. Cell Sci. 123: 4032–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iinuma T., Shiga A., Nakamoto K., O’Brien M.B., Aridor M., Arimitsu N., Tagaya M., Tani K. (2007). Mammalian Sec16/p250 plays a role in membrane traffic from the endoplasmic reticulum. J. Biol. Chem. 282: 17632–17639 [DOI] [PubMed] [Google Scholar]
- Ito Y., Uemura T., Shoda K., Fujimoto M., Ueda T., Nakano A. (2012). cis-Golgi proteins accumulate near the ER exit sites and act as the scaffold for Golgi regeneration after brefeldin A treatment in tobacco BY-2 cells. Mol. Biol. Cell 23: 3203–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan V., de Voer G., Xanthakis D., Spoorendonk K.M., Kondylis V., Rabouille C. (2008). Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol. Biol. Cell 19: 4352–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolliffe N.A., Craddock C.P., Frigerio L. (2005). Pathways for protein transport to seed storage vacuoles. Biochem. Soc. Trans. 33: 1016–1018 [DOI] [PubMed] [Google Scholar]
- Konieczny A., Ausubel F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410 [DOI] [PubMed] [Google Scholar]
- Kung L.F., Pagant S., Futai E., D’Arcangelo J.G., Buchanan R., Dittmar J.C., Reid R.J., Rothstein R., Hamamoto S., Snapp E.L., Schekman R., Miller E.A. (2012). Sec24p and Sec16p cooperate to regulate the GTP cycle of the COPII coat. EMBO J. 31: 1014–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans M., Meckel T., Kress A., Lerich A., Robinson D.G. (2012). ERES (ER exit sites) and the “secretory unit concept”. J. Microsc. 247: 48–59 [DOI] [PubMed] [Google Scholar]
- Lederkremer G.Z., Cheng Y., Petre B.M., Vogan E., Springer S., Schekman R., Walz T., Kirchhausen T. (2001). Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc. Natl. Acad. Sci. USA 98: 10704–10709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shimada T., Takahashi H., Ueda H., Fukao Y., Kondo M., Nishimura M., Hara-Nishimura I. (2006). MAIGO2 is involved in exit of seed storage proteins from the endoplasmic reticulum in Arabidopsis thaliana. Plant Cell 18: 3535–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti L., Fornaciari S., Renna L., Stefano G., Brandizzi F. (2010). COPII-mediated traffic in plants. Trends Plant Sci. 15: 522–528 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakano A., Brada D., Schekman R. (1988). A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakańo A., Muramatsu M. (1989). A novel GTP-binding protein, Sar1p, is involved in transport from the endoplasmic reticulum to the Golgi apparatus. J. Cell Biol. 109: 2677–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano R.T., Matsushima R., Ueda H., Tamura K., Shimada T., Li L., Hayashi Y., Kondo M., Nishimura M., Hara-Nishimura I. (2009). GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana. Plant Cell 21: 3672–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Okamoto M., Kurokawa K., Matsuura-Tokita K., Saito C., Hirata R., Nakano A. (2012). High-curvature domains of the ER are important for the organization of ER exit sites in Saccharomyces cerevisiae. J. Cell Sci. 125: 3412–3420 [DOI] [PubMed] [Google Scholar]
- Osterrieder A., Hummel E., Carvalho C.M., Hawes C. (2010). Golgi membrane dynamics after induction of a dominant-negative mutant Sar1 GTPase in tobacco. J. Exp. Bot. 61: 405–422 [DOI] [PubMed] [Google Scholar]
- Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. (1999). Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Ratcliffe O.J., Kumimoto R.W., Wong B.J., Riechmann J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15: 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritzenthaler C., Nebenführ A., Movafeghi A., Stussi-Garaud C., Behnia L., Pimpl P., Staehelin L.A., Robinson D.G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14: 237–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.G., Herranz M.C., Bubeck J., Pepperkok R., Ritzenthaler C. (2007). Membrane dynamics in the early secretory pathway. Crit. Rev. Plant Sci. 26: 199–225 [Google Scholar]
- Robinson D.G., Oliviusson P., Hinz G. (2005). Protein sorting to the storage vacuoles of plants: A critical appraisal. Traffic 6: 615–625 [DOI] [PubMed] [Google Scholar]
- Rossanese O.W., Soderholm J., Bevis B.J., Sears I.B., O’Connor J., Williamson E.K., Glick B.S. (1999). Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 145: 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., Doonan J.H. (2007). Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120: 3678–3687 [DOI] [PubMed] [Google Scholar]
- Saint-Jore C.M., Evins J., Batoko H., Brandizzi F., Moore I., Hawes C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29: 661–678 [DOI] [PubMed] [Google Scholar]
- Salama N.R., Yeung T., Schekman R.W. (1993). The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 12: 4073–4082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K., Nakano A. (2005). Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat. Struct. Mol. Biol. 12: 167–174 [DOI] [PubMed] [Google Scholar]
- Schoberer J., Vavra U., Stadlmann J., Hawes C., Mach L., Steinkellner H., Strasser R. (2009). Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 10: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz D.A., Espenshade P.J., Gimeno R.E., Kaiser C.A. (1997). COPII subunit interactions in the assembly of the vesicle coat. J. Biol. Chem. 272: 25413–25416 [DOI] [PubMed] [Google Scholar]
- Shimada T., Koumoto Y., Li L., Yamazaki M., Kondo M., Nishimura M., Hara-Nishimura I. (2006). AtVPS29, a putative component of a retromer complex, is required for the efficient sorting of seed storage proteins. Plant Cell Physiol. 47: 1187–1194 [DOI] [PubMed] [Google Scholar]
- Shimada T., et al. (2003). Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278: 32292–32299 [DOI] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I. (2010). A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Shindiapina P., Barlowe C. (2010). Requirements for transitional endoplasmic reticulum site structure and function in Saccharomyces cerevisiae. Mol. Biol. Cell 21: 1530–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieben C., Mikosch M., Brandizzi F., Homann U. (2008). Interaction of the K(+)-channel KAT1 with the coat protein complex II coat component Sec24 depends on a di-acidic endoplasmic reticulum export motif. Plant J. 56: 997–1006 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Ketelaar T., de Ruijter N.C., Hawes C. (2009). Grab a Golgi: Laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic 10: 567–571 [DOI] [PubMed] [Google Scholar]
- Stefano G., Renna L., Chatre L., Hanton S.L., Moreau P., Hawes C., Brandizzi F. (2006). In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 46: 95–110 [DOI] [PubMed] [Google Scholar]
- Stephens D.J., Lin-Marq N., Pagano A., Pepperkok R., Paccaud J.P. (2000). COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity to ER exit sites. J. Cell Sci. 113: 2177–2185 [DOI] [PubMed] [Google Scholar]
- Supek F., Madden D.T., Hamamoto S., Orci L., Schekman R. (2002). Sec16p potentiates the action of COPII proteins to bud transport vesicles. J. Cell Biol. 158: 1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K.V., Sato K., Ide T., Nishizaka T., Nakano A., Noji H. (2009). Visualization of cargo concentration by COPII minimal machinery in a planar lipid membrane. EMBO J. 28: 3279–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H., Tamura K., Takagi J., Koumoto Y., Hara-Nishimura I., Shimada T. (2010). MAG4/Atp115 is a Golgi-localized tethering factor that mediates efficient anterograde transport in Arabidopsis. Plant Cell Physiol. 51: 1777–1787 [DOI] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Kondo M., Nishimura M., Hara-Nishimura I. (2005). KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H., Nishiyama C., Shimada T., Koumoto Y., Hayashi Y., Kondo M., Takahashi T., Ohtomo I., Nishimura M., Hara-Nishimura I. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47: 164–175 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R.L., Nakano A., Takeyasu K., Sato M.H. (2004). Systematic analysis of SNARE molecules in Arabidopsis: Dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29: 49–65 [DOI] [PubMed] [Google Scholar]
- Vitale A., Hinz G. (2005). Sorting of proteins to storage vacuoles: How many mechanisms? Trends Plant Sci. 10: 316–323 [DOI] [PubMed] [Google Scholar]
- Ward T.H., Polishchuk R.S., Caplan S., Hirschberg K., Lippincott-Schwartz J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155: 557–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P., Townley A.K., Koka P., Palmer K.J., Stephens D.J. (2006). Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic 7: 1678–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., Wang A. (2008). Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J. Virol. 82: 12252–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle J.R., Schwartz T.U. (2010). Structure of the Sec13-Sec16 edge element, a template for assembly of the COPII vesicle coat. J. Cell Biol. 190: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Fuji K., Shimada T., Nishimura M., Hara-Nishimura I. (2005). Endosomal proteases facilitate the fusion of endosomes with vacuoles at the final step of the endocytotic pathway. Plant J. 41: 888–898 [DOI] [PubMed] [Google Scholar]
- Yang Y.D., Elamawi R., Bubeck J., Pepperkok R., Ritzenthaler C., Robinson D.G. (2005). Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 17: 1513–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekawa S., Furuno A., Baba T., Fujiki Y., Ogasawara Y., Yamamoto A., Tagaya M., Tani K. (2011). Sec16B is involved in the endoplasmic reticulum export of the peroxisomal membrane biogenesis factor peroxin 16 (Pex16) in mammalian cells. Proc. Natl. Acad. Sci. USA 108: 12746–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Sato K. (2012). Insights into structural and regulatory roles of Sec16 in COPII vesicle formation at ER exit sites. Mol. Biol. Cell 23: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]