This work examines the role of the NAC transcription factor ANAC096, finding that ANAC096 interacts with specific bZIP transcription factors to globally affect abscisic acid–responsive transcription during osmotic and drought stresses.
Abstract
Multiple transcription factors (TFs) play essential roles in plants under abiotic stress, but how these multiple TFs cooperate in abiotic stress responses remains largely unknown. In this study, we provide evidence that the NAC (for NAM, ATAF1/2, and CUC2) TF ANAC096 cooperates with the bZIP-type TFs ABRE binding factor and ABRE binding protein (ABF/AREB) to help plants survive under dehydration and osmotic stress conditions. ANAC096 directly interacts with ABF2 and ABF4, but not with ABF3, both in vitro and in vivo. ANAC096 and ABF2 synergistically activate RD29A transcription. Our genome-wide gene expression analysis revealed that a major proportion of abscisic acid (ABA)–responsive genes are under the transcriptional regulation of ANAC096. We found that the Arabidopsis thaliana anac096 mutant is hyposensitive to exogenous ABA and shows impaired ABA-induced stomatal closure and increased water loss under dehydration stress conditions. Furthermore, we found the anac096 abf2 abf4 triple mutant is much more sensitive to dehydration and osmotic stresses than the anac096 single mutant or the abf2 abf4 double mutant. Based on these results, we propose that ANAC096 is involved in a synergistic relationship with a subset of ABFs for the transcriptional activation of ABA-inducible genes in response to dehydration and osmotic stresses.
INTRODUCTION
Abiotic stresses such as water deficit and high soil salinity can cause severe damage to plant growth and development. To cope with these stresses, plants have evolved adaptive molecular mechanisms that result in the expression of a large number of genes that alter plant physiology depending on the specific type of environmental stresses. One of the pivotal events in these responses is the perception of stress, which activates the expression of stress-response genes via signal transduction (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2001; Bray, 2002). The core signaling components involved in the regulation of the expression of stress-responsive genes during abiotic stress responses include kinases, phosphatases, and transcription factors (TFs) (Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000; Zhu, 2001; Bray, 2002).
Many different types of TFs are involved in the induction of stress-responsive genes during abiotic stress responses. One type of TFs includes members of the ABRE binding factor/ABRE binding protein (ABF/AREB) family proteins, which contain a bZIP structure conserved in plants and animals (Choi et al., 2000; Newman and Keating, 2003; Kim et al., 2004; Corrêa et al., 2008). The expression of these ABFs/AREBs is regulated by signaling pathways under the control of abscisic acid (ABA), a phytohormone that plays an essential role in adaptation to abiotic stress. The genes transcribed by these ABFs/AREBs contain an ABA-responsive cis-acting element in their promoter regions referred to as the ABA-responsive element (ABRE; PyACGTGGC) (Yamaguchi-Shinozaki et al., 1990).
NAC (for NAM, ATAF1/2, and CUC2) TFs are plant specific and contain the highly conserved NAC DNA binding domain and variable C-terminal domains. They play pivotal roles in plant development, senescence, auxin responses, and biotic stress responses (Aida et al., 1997; Olsen et al., 2005a, 2005b; Lu et al., 2007; Kim et al., 2009; Jensen et al., 2010; Hao et al., 2011; Nakashima et al., 2012; Puranik et al., 2012; Sun et al., 2012). The Arabidopsis thaliana genome encodes more than 100 NAC domain–containing genes, which can be divided into 10 major groups based on phylogenetic analysis (Jensen et al., 2010). Group III NAC TFs, such as ANAC019, ANAC055, and ANAC072, play important roles in abiotic stress responses (Duval et al., 2002; Tran et al., 2004; Lu et al., 2007; Jensen et al., 2010). For instance, ectopic expression of Arabidopsis RD26/ANAC072 and Arachis hypogaea NAC2 resulted in ABA-hypersensitive phenotypes and increased resistance to dehydration stress (Fujita et al., 2004; Liu et al., 2011). Thus, they are considered to act as positive regulators of abiotic stress or ABA signaling. Besides this clade, it still remains unclear whether other NAC protein subfamilies play similar roles in abiotic stress and ABA signaling. ANAC096, which belongs to the IV-1 subfamily with unknown functions, physically interacts with TOPLESS (TPL). TPL also interacts with other stress-responsive TFs, although the biological consequence of this interaction remains elusive (Causier et al., 2012). Moreover, other NAC TFs, such as soybean (Glycine max) NAC20 and Setaria italica NAC, regulate an ABA-independent pathway involved in plant responses to dehydration (Hao et al., 2011; Puranik et al., 2011, 2012). Although extensive efforts have been made to identify different NAC TFs in response to abiotic stress responses, the specific and precise transcriptional and physiological outputs of each NAC TF remain obscure.
NAC TFs recognize the consensus cis-acting elements CGT(G/A) and CACG (Simpson et al., 2003; Ernst et al., 2004; Tran et al., 2004), and different NAC TFs adopt different binding modes for these NAC recognition sequences. For instance, Arabidopsis vascular plant one zinc-finger protein (VOZ) recognizes only a pair of palindromic binding sequences separated by nearly 10 bp, whereas NAC019 bind to both single and palindromic recognition sequences, suggesting the evolutionary diversification of the DNA recognition mode of NACs (Mitsuda et al., 2004; Tran et al., 2004). The highly conserved N-terminal region has the DNA binding activity as well as motifs for dimerization (Ernst et al., 2004; Welner et al., 2012). Biophysical and crystal structural analyses revealed that NAC proteins contain a TF fold with a twisted β-sheet surrounded by a few helical elements (Ernst et al., 2004). In addition, the dimerization is crucial for the DNA binding (Olsen et al., 2005a, 2005b).
During abiotic stress responses, the expression of the different families of TFs was originally thought to be regulated by specific and separate signaling pathways with unique sets of components and a specific set of target genes (Shinozaki and Yamaguchi-Shinozaki, 2000). However, recent studies revealed that the signaling pathways inducing the different types of TFs are more complex (Lee et al., 2010; Fujita et al., 2011; Martinez and Rao, 2012). For example, instead of a linear relationship between the signaling pathway and the type of TF, recent studies raised the intriguing possibility of crosstalk between signaling pathways and between different types of TFs (Lee et al., 2010). Indeed, DREB1A/CBF3, DREB2A, and DREB2C TFs physically interact with ABFs/AREBs (Lee et al., 2010). However, how extensive the crosstalk is between the different types of TFs during abiotic stress remains unclear.
To further elucidate the molecular mechanisms underlying ABA-mediated dehydration stress responses, we screened for mutants that showed an ABA-hyposensitive phenotype from a pool of activation-tagged mutants (Weigel et al., 2000). One loss-of-function mutant carried a T-DNA insertion in a gene encoding a NAC domain–containing protein, ANAC096. In this study, we show that ANAC096 functions as a positive regulator of ABA-responsive signaling under conditions of dehydration and osmotic stress. ANAC096 acted synergistically with ABF2/4 to activate the transcription of a subset of ABA- and/or dehydration stress–inducible genes, resulting in increased resistance to dehydration stress.
RESULTS
Exogenous ABA Affects Germination and Postgermination Growth of anac096 Mutants and ANAC096-Overexpressing Plants
To gain insight into the mechanism underlying dehydration stress responses in plants, we screened for ABA-insensitive mutants from a population of activation-tagging mutants using cotyledon greening in the presence of 1 μM exogenous ABA (Weigel et al., 2000). We isolated 10 putative mutants and focused on one that carried a T-DNA insertion in exon 3 of At5g46590, which encodes a NAC domain protein called ANAC096 (see Supplemental Figures 1A and 1B online). This mutant, which we named anac096-1 (see Supplemental Figure 1B online), carries a T-DNA insertion in the downstream region of the highly conserved NAC domain of ANAC096. NAC domain proteins harbor a highly conserved, plant-specific DNA binding domain and play important roles in various cellular processes, including osmotic stress responses in plants (Duval et al., 2002; Tran et al., 2004; Lu et al., 2007; Jensen et al., 2010; Hao et al., 2011). As observed during screening, anac096-1 plants were less sensitive to exogenous ABA during germination (Figures 1A and 1C). Despite the fact that the mutant was isolated from the activation-tagging mutant population, RT-PCR analysis showed that ANAC096 (At5g46590) transcripts were not detectable (see Supplemental Figure 1C online). This indicated that the reduced ABA sensitivity of anac096-1 was due to loss of function rather than overexpression of ANAC096. To confirm this, we obtained another independent mutant carrying a T-DNA insertion in ANAC096 (SALK_078797C) from the ABRC stock center and named it anac096-2 (see Supplemental Figure 1B online). RT-PCR showed that neither mutant expressed the ANAC096 transcript (see Supplemental Figure 1C online). Similar to anac096-1, anac096-2 showed reduced sensitivity to exogenous ABA during germination (Figures 1A and 1C). Additionally, postgermination growth of the mutant plants (anac096-1 and anac096-2), measured as root growth and plant fresh weight, was less sensitive to exogenous ABA (Figures 1D, 1F, and 1G), suggesting that ANAC096 acts as a positive factor for the mediation of ABA responses. To further confirm that these phenotypes were caused by a mutation in ANAC096, we examined the ABA sensitivity of the F1 progeny generated by crossing anac096-1 and anac096-2 plants. The F1 progeny of the cross also showed reduced sensitivity to exogenous ABA (Figures 1A and 1C), confirming that the mutant phenotype is caused by the mutation in ANAC096.
Figure 1.
Phenotype of anac096 Mutants and ANAC096OX Transgenic Plants.
(A) and (B) Effect of exogenous ABA on seed germination. Wild-type (WT) plants, two independent alleles of anac096 mutants, F1 progeny from a cross between anac096-1 and anac096-2 mutants, and two independent ANAC096OX lines were planted on one-half-strength MS plates supplemented with DMSO or ABA, and the germination rates were examined. Plant images were obtained at two different magnifications at 4 d (A) and 5 d (B) after planting on half-strength MS plates supplemented with DMSO or 1 μM ABA.
(C) The germination rates were scored 5 d after planting on the plates supplemented with the indicated ABA concentrations. Three independent experiments were performed with 100 plants or seeds per experiment. Error bars indicate sd (n = 3). Statistical analysis was performed between the wild type and anac096 mutants or between the wild type and ANAC096OXs. *P value < 0.01 (Student’s t test).
(D) to (G) Effect of exogenous ABA on vegetative growth. Plants grown on half-strength MS plates for 3 d were transferred vertically to half-strength MS plates supplemented with DMSO or 10 μM ABA.
(D) and (E) Images of the indicated plants were taken at 12 d after transfer. White underbars indicate the root tip region.
(F) and (G) To assess vegetative growth quantitatively, the primary root length (F) and the fresh weight of whole plants (G) were measured at 12 d after transfer. Three independent experiments were performed using 20 plants per experiment. Error bars indicate sd (n = 3). Vertical bars = 2 cm. Statistical analysis was performed between the wild type and anac096 mutants or between the wild type and ANAC096OX. *P value < 0.01 (Student’s t test).
To obtain independent evidence that ANAC096 is involved in ABA-mediated processes, we generated transgenic plants that overexpressed ANAC096 under the control of the strong cassava vein mosaic virus promoter. The transgenic ANAC096-overexpressing (ANAC096OX) plants showing a dramatic increase in ANAC096 transcript levels (see Supplemental Figure 1D online) were examined for sensitivity to exogenous ABA. In contrast with the mutant plants, the germination (Figures 1B and 1C) and postgermination growth (Figures 1E to 1G) of two independent lines of ANAC096OX plants showed increased sensitivity to exogenous ABA. These results confirm that ANAC096 is a positive factor mediating ABA responses in plants.
anac096 Mutant Plants and ANAC096OX Transgenic Plants Have Altered Resistance to Dehydration Stress
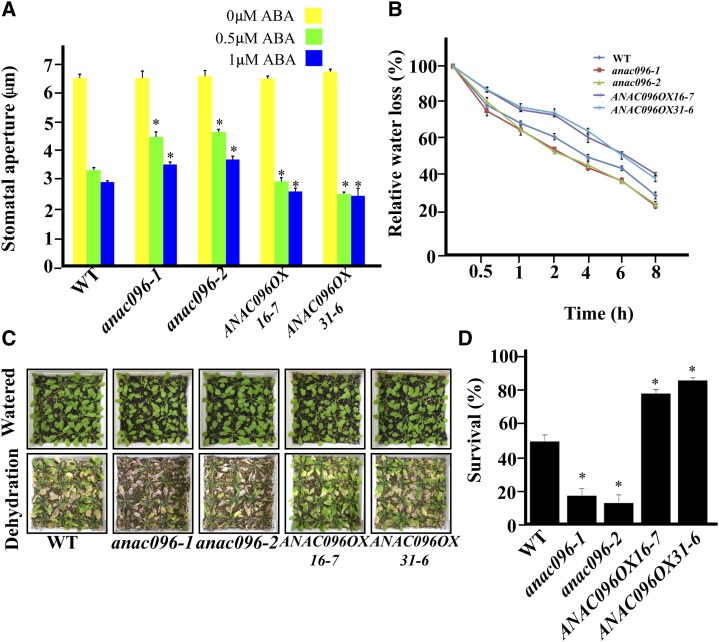
To understand the physiological role of ANAC096, we examined the effect of the mutation or ectopic expression on various ABA-related physiological responses and dehydration stress responses. High levels of ABA induce stomatal closure (Jeon et al., 2008). Thus, we compared ABA-mediated stomatal closure of anac096 mutants and ANAC096OX plants with that of wild-type plants. Stomatal closure was examined in the guard cells of epidermal peels of wild-type, anac096 mutant, and ANAC096OX plants after treating with 0.5 or 1 μM ABA for 1 h. The stomatal aperture in anac096 mutant plants was larger than that in wild-type plants (Figure 2A). However, ANAC096OX plants showed an opposite phenotype, more closed stomata (Figure 2A). These results indicate that ANAC096 positively regulates ABA-mediated stomatal closure.
Figure 2.
ANAC096 Plays a Positive Role in Both Stomatal Closure and Water Loss under Dehydration Stress Conditions.
(A) Role of ANAC096 in ABA-induced stomata closure. Epidermal peels from wild-type (WT), anac096, and ANAC096OX leaf tissues were exposed to high-light conditions for 3 h to induce full opening of the stomata and subsequently treated with 0.5 or 1 μM ABA. To quantify the stomatal closure, the width of stomatal opening was measured in a triplicate experiment with 120 pairs of guard cells per experiment. Error bars indicate sd (n = 3). Statistical analysis was performed by comparing with the wild type. *P < 0.01 (Student’s t test).
(B) Water loss from wild-type, anac096, and ANAC096OX plants. The aerial parts of Columbia-0 (WT), anac096 mutants, and ANAC096OX transgenic plants grown on plates for 10 d were excised and exposed to 30% relative humidity condition. To quantify water loss, fresh weight of the excised plant tissues was measured at the indicated time points. Error bars indicate sd (n = 3).
(C) and (D) Dehydration stress sensitivity of wild-type, anac096 mutants, and ANAC096OX plants.
(C) Wild-type, anac096, and ANAC096OX plants grown for 9 d in soil under normal growth conditions (watered) were kept in a greenhouse without watering for 14 d (dehydration), and the images of surviving plants were taken at 3 d after rewatering (rehydration).
(D) To quantify the survival rate, 16 plants of each plant type were used in each experiment, and five independent experiments were performed. Error bars indicate sd (n = 5). Statistical analysis was performed between the wild type and anac096 mutants or between the wild type and ANAC096OXs. *P value < 0.01 (Student’s t test).
The function of ANAC096 in ABA-mediated stomatal closure prompted us to test whether it plays the similar regulatory roles in plant water conservation under dehydration stress response. To test this, the aerial parts of the anac096 mutant, ANAC096OX, and wild-type plants were excised, and water loss was examined over time. The results showed that anac096 plants lost water more quickly than wild-type plants (Figure 2B). To obtain further evidence for the role of ANAC096 in regulating water loss, we examined water loss in ANAC096OX plants. Under the same conditions, ANAC096OX plants lost water more slowly than wild-type plants, confirming that ANAC096 is involved in regulating water loss (Figure 2B). To examine the long-term effect of ANAC096 on dehydration stress responses, anac096 mutant and ANAC096OX plants grown for 9 d under normal growth conditions were exposed to dehydration stress by withholding water for 14 d. Survival rates were examined 3 d after rewatering. Under these conditions, the survival rates of the anac096 mutant, wild-type plants, and ANAC096OX plants were ∼18, 52, and 82%, respectively, of that of the hydrated plants (Figures 2C and 2D). These results are consistent with a positive role of ANAC096 in dehydration stress responses.
anac096 Mutants Show a Delay in the Induction of ABA-Responsive Genes
The phenotype of the anac096 mutants raised the possibility that dehydration stress and ABA regulate the expression of ANAC096. To determine the expression patterns of ANAC096 under different abiotic stresses as well as ABA responses, wild-type plants grown on plates were treated with different conditions such as ABA, dehydration, mannitol, and cold stress. As a positive control, we included RD29A, which is activated transcriptionally under these conditions (Xu et al., 2012). The transcript levels of ANAC096 gradually increased after 1 and 2 h treatment of exogenous ABA as well as dehydration and osmotic stresses (Figure 3A). Under these conditions, the transcript levels of RD29A also increased rapidly (Figure 3B). However, ANAC096 expression altered a little under cold shock, despite the fact that the transcript levels of RD29A showed a dramatic increase under this condition. These results indicate that the ANAC096 expression is not responsive to cold stress (Figures 3A and 3B). To examine the spatial and temporal expression patterns of ANAC096, transgenic plants were generated using an ANAC096pro:GUS construct containing a 3.0-kb fragment from the region upstream of the ANAC096 start codon and the β-glucuronidase (GUS) coding region. At the seedling stage, GUS was detected in the veins of the cotyledons, hypocotyls, and roots (see Supplemental Figure 2A online). At later stages of plant development, the GUS expression was detected in true leaves and in primary and secondary roots (see Supplemental Figure 2A online). In addition, GUS was expressed at higher levels in young leaves than in older leaves. The ANAC096 transcript levels detected by quantitative RT-PCR (qRT-PCR) were in agreement with the GUS expression patterns (see Supplemental Figure 2B online). The transgenic plants harboring ANAC096pro:GUS showed an increase in GUS staining when treated with exogenous ABA and dehydration stress, confirming that the ANAC096 expression is regulated by ABA and dehydration stress (see Supplemental Figure 2C online).
Figure 3.
ANAC096 Induced by ABA and Dehydration Stress Plays a Positive Role in the Expression of ABA-Inducible Genes.
(A) and (B) Induction of the ANAC096 expression by exogenous ABA and different abiotic stresses. Total RNA from wild-type plants treated with exogenous ABA (10 μM), dehydration stress (30% relative humidity), mannitol (300 mM), or cold stress (4°C) at the indicated time points were used for qRT-PCR analysis of ANAC096 (A) and RD29A (B). RD29A was used as a positive control for the treatments. ACT2 was used as an internal control for qRT-PCR. Error bars indicate sd (n = 3).
(C) Defect in induction of ABA-responsive genes in anac096 mutants. Total RNA from two alleles of anac096 mutant plants as well as wild-type (WT) plants that had been treated with exogenous ABA (100 μM) for 0.5 or 1 h was used for qRT-PCR analysis of RD29A, RD29B, and COR47 transcript levels. ACT2 was used as an internal control for qRT-PCR. Error bars indicate sd (n = 3).
Previous studies showed that the NAC TFs ANAC019, ANAC055, and ANAC072 were involved in the induction of various osmotic stress–inducible genes (Tran et al., 2004; Jensen et al., 2010). To examine whether loss of function of ANAC096 causes the defect in the expression of early ABA-responsible genes, the anac096-1 and anac096-2 mutant plants and wild-type plants were treated with 100 μM ABA, and the transcript levels of three ABA-inducible genes (RD29A, RD29B, and COR47) (Xiong et al., 2001) were examined by qRT-PCR at 0.5- and 1-h time points. As shown in Figure 3C, the ABA-mediated induction dynamics of these genes were significantly impaired in mutants compared with the wild type, suggestive of the partial positive contribution of ANAC096 to the early ABA-responsive gene expression.
ANAC096 Regulates the Expression of a Major Proportion of ABA-Responsive Genes
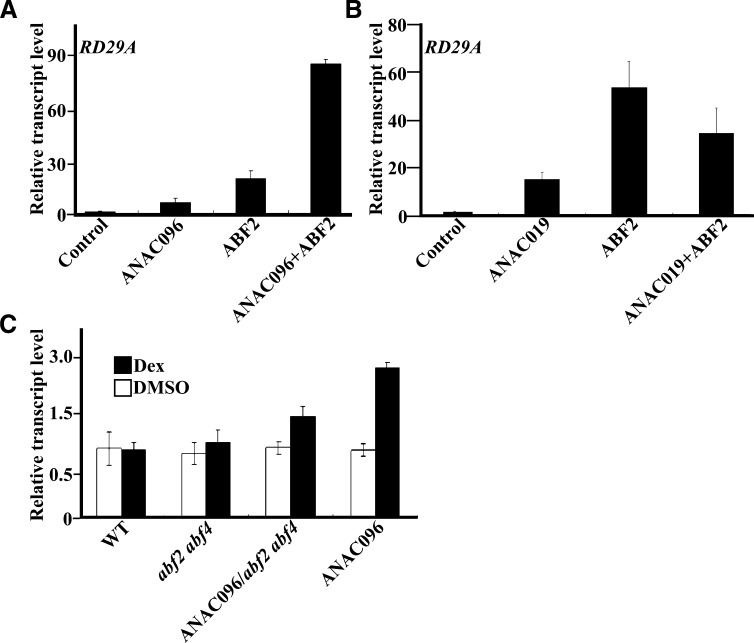
To investigate the transcriptional regulation governed by ANAC096, we identified the genes under the transcriptional control of ANAC096. To minimize any secondary effects caused by ectopic expression of ANAC096, we generated transgenic plants expressing ANAC096 under the control of a dexamethasone (Dex)–inducible promoter (Aoyama and Chua, 1997). First, we confirmed ANAC096-mediated gene expression using two independent lines of pTA7002-ANAC096 transgenic plants to avoid any positional effects of the transgene expressing ANAC096. In both lines, the expression of ANAC096 was rapidly induced by 30 μM Dex (see Supplemental Figure 3A online). At this condition, the transcript levels of RD29A rapidly increased in both alleles (see Supplemental Figure 3B online). Moreover, the increase of the RD29A transcript levels correlated with that of ANAC096, raising the possibility that ANAC096 is involved in the expression of RD29A.
To examine the extent of transcriptional regulation governed by ANAC096 and its role in ABA-mediated transcriptional regulation, we performed genome-wide gene expression profiling of pTA-ANAC096 plants treated with Dex (30 μM) for 1 h and wild-type plants treated with ABA (2 μM) for 0.5 h using Agilent-031025 Arabidopsis 8x60k arrays. ANAC096- and ABA-responsive genes (P value < 0.05 and fold change ≥ 2) were identified by an integrative statistical method (Chae et al., 2013; see Methods). We identified 1351 and 451 genes that were up- and downregulated by ANAC096, respectively, and 924 and 279 genes that were up- and downregulated by exogenous ABA, respectively (Figure 4A; see Supplemental Figure 4A online). Interestingly, more than 60% of the genes upregulated by exogenous ABA overlapped with those upregulated by ANAC096 (Figure 4A; P < 1×10−10 by Fisher’s exact test; see Supplemental Figure 5 online). Moreover, the set of genes upregulated by ABA closely overlaps with those affected by ANAC096, further supporting our notion that ANAC096 is involved in the expression of ABA-responsive genes (Figure 4A; see Supplemental Figure 5 online). In the case of downregulated genes, a relatively smaller portion (35%) of ABA-responsive genes, compared with the upregulated genes, overlapped with those of ANAC096 (see Supplemental Figure 4A online), although this overlap is still significant (P < 1×10−10 by Fisher’s exact test; see Supplemental Figure 5 online). Considering the role of ANAC096 as a transcriptional activator (see Supplemental Figure 3B online), it is possible that the ANAC096-repressed genes are secondary targets of ANAC096-induced genes. To systematically explore the differential expression of the genes regulated by ANAC096, the ANAC096-induced or -repressed genes were divided in two clusters, that is, up- or downregulated by both ANAC096 and ABA conditions (C1) and up- or downregulated only by ANAC096 (C2; Figure 4B; see Supplemental Figure 4B online). Remarkably, the genes in the C1 cluster are mainly involved in ABA stimulus, water deprivation, and osmotic stress responses (Figure 4C), indicating that ANAC096 is involved in abiotic stress and ABA-mediated responses. Also, it is likely that ANAC096 acts as a transcriptional regulator for a large number of genes encoding TFs, phosphatases, kinases, transporters, and a myriad of components in theABA signaling circuit, thereby further suggesting the importance of ANAC096 in ABA response (see Supplemental Figure 6 online). By contrast, the genes that were suppressed by ANAC096 are involved in auxin stimulus and shoot development (see Supplemental Figure 4C online). The suppression of auxin- and shoot development–related genes is consistent with the fact that dehydration stress and high levels of ABA inhibit plant growth.
Figure 4.
ANAC096 Regulates Positively a Major Proportion of ABA-Responsive Genes.
(A) Venn diagram of ANAC096 and ABA upregulated genes (P = 2.49 × 10−321 by Fisher’s exact test; see Supplemental Figure 5 online).
(B) Hierarchical clustering of ANAC096-induced genes in ANAC096-induced (ANAC096) and ABA-treated (ABA) conditions. E1 and E2 represent each sample in the duplicates. C1 and C2 represent the genes upregulated in both conditions and only ANAC096-induced condition, respectively. Heat color gradation in red denotes the increase in log2 fold change.
(C) GOBPs enriched by the genes in C1 and C2 (P < 0.05). The color bar represents the gradient of −log10(P), where P is the enrichment of a P value computed by DAVID.
(D) qRT-PCR analysis of ABA-responsive genes. Total RNA from pTA, pTA-ANAC096, and anac096-1 plants that had been treated with 30 μM Dex only for 1 h (Dex) or with 30 μM Dex only for 0.5 h followed by additional 0.5-h incubation with both 2 μM ABA and 30 μM Dex (Dex + ABA) were used for qRT-PCR analyses. ACT2 was used as an internal control for qRT-PCR. Error bars indicate sd (n = 3).
To confirm the data from the microarray analysis, we examined the expression of representative ABA-responsive genes in anac096 mutant plants by qRT-PCR. The ABA-induced expression of the eight genes whose expression was upregulated by ANAC096 was greatly impaired in the anac096 mutant plants (Figure 4D), supporting the notion that ANAC096 plays a role in ABA-mediated signaling.
ANAC096 Localizes to the Nucleus and Recognizes Consensus Core cis-Elements within the RD29A Promoter
Members of the NAC TF family harbor a highly conserved N-terminal DNA binding domain and a C-terminal variable domain that functions as either a transcriptional activator or repressor depending on the specific isoform (Olsen et al., 2005a; Hao et al., 2010; Nakashima et al., 2012). The activation of the transcription of a subset of osmotic stress–inducible genes by ANAC096 raised the possibility that the C-terminal region of ANAC096 contains a transcriptional activation domain. To test this, we used the yeast one-hybrid approach (Ouwerkerk and Meijer, 2011). The GAL4 DNA binding domain was fused to full-length ANAC096, the N-terminal domain, or the C-terminal domain to generate GAL4BD-ANAC096F, GAL4BD-ANAC096N, or GAL4BD-ANAC096C, respectively (see Supplemental Figure 7A online). These constructs were then transformed into yeast together with the reporter construct GAL4p:His, and the growth of the transformants was examined on His-deficient medium. Transformants harboring GAL4BD-ANAC096F and GAL4BD-ANAC096C displayed normal growth on this medium, indicating that the C-terminal region of ANAC096 contains a transcriptional activation domain (see Supplemental Figure 7B online).
NAC TFs recognize the consensus cis-acting elements, CGT(G/A) and CACG, or NAC-recognition sequences (NACRSs) (Simpson et al., 2003; Tran et al., 2004). Sequence analysis revealed that RD29A, an ANAC096-inducible gene, contained two NACRSs in the −99- to −50-bp region of its promoter (Figure 5A) (Kasuga et al., 2004). To examine whether ANAC096 binds to these elements, we performed an electrophoresis mobility shift assay (EMSA) using a 50-bp DNA fragment containing the −99- to −50-bp region of the RD29A promoter (Figures 5A and 5B). The N-terminal domain of ANAC096 (ANAC096N) was expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein, and purified recombinant GST-ANAC096N or GST alone was incubated with 32P-labeled RD29A promoter probes. A slowly migrating band was detected for GST-ANAC096N, but not for GST alone, indicating that ANAC096N binds to the RD29A promoter fragment containing the NACRSs (Figure 5B). To further test the binding specificity, the two NAC domain protein recognition sites (CGTA [inverted sequence] and CACG) were mutated to AAAA (either one by one or simultaneously). GST-ANAC096N still bound to the DNA fragments containing a single mutation; however, GST-ANAC096N failed to bind to fragments harboring mutations at both sites, indicating that ANAC096 specifically recognized the two NACRSs within the RD29A promoter (Figure 5B).
Figure 5.
ANAC096 Activates Transcription by Binding to the Consensus Core cis-Acting Elements at the RD29A Promoter.
(A) Nucleotide sequence containing the NACRS at the promoter region of RD29A from −99 to −50 bp. The letters in bold indicate the consensus sequences of the NACRS at the promoter region of RD29A. The two consensus binding sites were mutated either individually (1A, 2A) or simultaneously (1A/2A). The bases in lowercase letters indicate the mutated sequences in the RD29A promoter. WT, wild-type sequence.
(B) Binding of ANAC096 to NACRSs at the RD29A promoter. Purified GST-ANAC096N (1 μg) was incubated with 0.025 pmol 32P-labeled wild-type or mutant fragments. GST alone was included as a negative control. Samples were analyzed by EMSA on 4 to 16% gradient gels, and the gels were exposed to x-ray film.
(C) Dimer formation of ANAC096. GST-ANAC096 or GST alone (immobilized on glutathione-agarose beads) was incubated with His-ANAC096N, His-ANAC096N(R11A/E18A), or His-ANAC096N(d2-10), and proteins bound to glutathione-agarose beads were precipitated. The precipitates were analyzed by immunoblotting using an anti-His antibody. Subsequently, the membrane was stained with Coomassie blue.
(D) The role of dimerization in the DNA binding. Purified His-ANAC096N, His-ANAC096N(R11A/E18A), or His-ANAC096N(d2-10) (1 μg) was incubated with 0.025 pmol 32P-labeled 1A probe. Probe alone was used as a negative control. Samples were analyzed by EMSA on 4 to 16% gradient gels, and the gels were exposed to x-ray film.
(E) Schematic representation of the constructs used for transient expression. Intact and mutated RD29A promoter fragments were inserted into the upstream region of the LUC reporter gene. The GUS gene, which was used to normalize transformation efficiency, was placed under the control of the UBQ10 promoter. ANAC096 was placed under the Dex-inducible promoter in the pTA vector. NOS-T, Nos terminator.
(F) Transcriptional activation of the RD29A promoter by ANAC096 through recognition of the consensus NACRSs. Protoplasts from transgenic plants harboring pTA-ANAC096 or empty vector were cotransformed with reporter and normalizing plasmids, incubated for 23 h, and then incubated an additional 1 h with 30 μM Dex. Total RNA from transformed protoplasts was used for qRT-PCR. The transcript level of LUC was normalized using that of GUS. Error bars indicate sd (n = 3). Statistical analysis was performed. *P value < 0.01 (Student’s t test).
To further gain insight into the mode of transcriptional activation mediated by ANAC096, we examined whether dimer formation is required for the activation of transcription. Dimerization of DNA binding domains is the common feature of NAC TFs (Ernst et al., 2004). The N-terminal DNA binding domain of ANAC096 showed a high degree of sequence homology to that of ANAC019, including the residues responsible for the salt bridge at the dimerization interface (see Supplemental Figure 8 online), raising the possibility that ANAC096 also forms a dimer. To support this idea at the biochemical level, we generated deletion [ANAC096N(d2-10)] or substitution [ANAC096N(R11A/E18A)] mutants of the ANAC096 N-terminal domain (ANAC096N). The deletion mutant was generated by deleting an N-terminal nine–amino acid region that is crucial for the antiparallel β-sheet, whereas the substitution mutant was generated by substituting Arg-11 and Glu-18 with Ala residues. Arg-11 and Glu-18 are responsible for the formation of the two prominent salt bridges at the dimer interface (Ernst et al., 2004; Olsen et al., 2005b). Recombinant wild-type and mutant forms of ANAC096N tagged with His epitope were incubated with recombinant GST or GST-ANAC096 bound to beads. Wild-type ANAC096N was detected in the precipitates (Figure 5C). By contrast, the deletion and substitution mutant proteins were not detected in the precipitates. These results indicate that ANAC096N forms a dimer similar to ANAC019 (Olsen et al., 2005b), but these mutants cannot make the homomeric interaction. To examine whether the dimerization of ANAC096 is crucial for binding to NACRS, we performed EMSA using oligonucleotide A1, which has a single binding site. Compared with ANAC096N, which showed slow-migrating bands, two mutant forms of ANAC096N that cannot make the homomeric interaction failed to bind to the single NACRS, indicating that the dimerization of ANAC096 through the conserved dimerization interface is necessary for DNA binding (Figure 5D).
Next, we examined whether these NACRSs were required for ANAC096-mediated transcriptional activation. We generated a reporter chimeric construct, RD29A50pro:luciferase (LUC), by fusing the −99- to −50-bp promoter region of RD29A (RD29A50pro) to the LUC gene (Figure 5E). The three mutant forms of RD29A50pro used in the EMSA experiments also were included. The wild-type or mutant forms of RD29A50pro:LUC were cotransformed into protoplasts from ANAC096-inducible transgenic plants along with UBQ10pro:GUS, a chimeric construct of GUS under the control of the UBQ10 promoter, using the polyethylene glycol–mediated transformation method (Hyunjong et al., 2006). UBQ10pro:GUS was used to monitor transformation efficiency (Figure 5E). After transformation, protoplasts were incubated for 23 h and then were incubated for an additional hour in the presence of 30 μM Dex. The expression of ANAC096 was induced for only 1 h to minimize any secondary effects on the transcription of the reporter gene through the activation of dehydration stress signaling. The transcript levels of LUC were then determined by qRT-PCR. Total RNA isolated from the protoplasts was used for qRT-PCR analysis, and the transcript levels of LUC were normalized against those of GUS. The LUC transcript levels expressed from the wild-type RD29A50pro in ANAC096 protoplasts increased ∼2.5-fold compared with those in the empty vector controls (Figure 5F). By contrast, the two single mutant constructs showed only slightly increased LUC expression, and the construct harboring mutations at both sites failed to show any increase. This confirms that ANAC096 activates transcription via the NACRSs within the RD29A promoter. Of note, the substitution of the most upstream located CGT to AAA did not affect RD29A promoter activity, indicating that this sequence is not required for ANAC096 binding (see Supplemental Figures 9A and 9B online). Moreover, ANAC096 with the C-terminal deletion failed to induce the expression of LUC from intact RD29A50pro, confirming that the C-terminal domain was involved in transcriptional activation (see Supplemental Figure 10 online). Taken together, these results demonstrate that ANAC096 activates transcription via recognition of the NACRSs within the RD29A promoter.
ANAC096 Directly Binds ABF2/4 and Has a Synergistic Effect on Transcriptional Activation
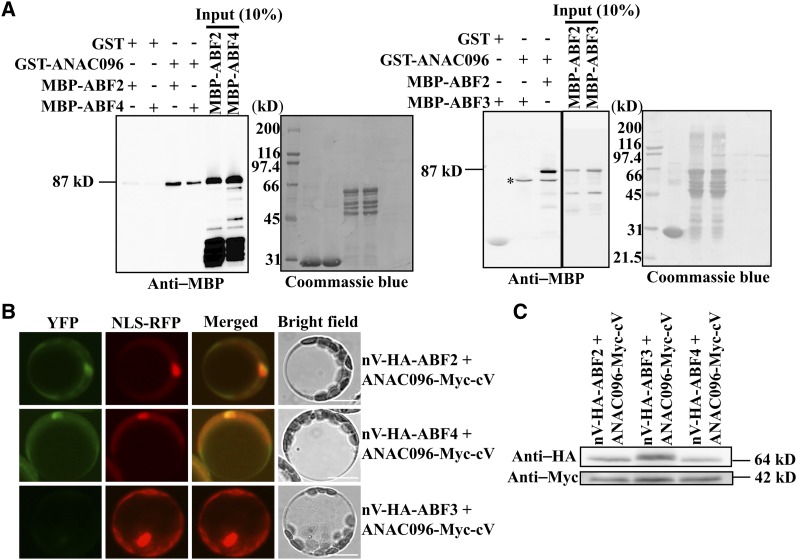
Many ANAC096-inducible genes were also targets of ABFs (Choi et al., 2000). The cis-acting ABRE element of ABFs overlapped with or was located close to NACRSs in the −99- to −50-bp region of the RD29A promoter (Figure 5A). The results showed that ABF2 and ABF4 directly bind to the RD29A promoter (see Supplemental Figure 11 online). Together with the fact that ANAC096 was responsible for only part of the total ABA-mediated activation of RD29A, this result raised the possibility that ANAC096 and ABFs cooperate for the full activation of RD29A expression. To test this idea, we examined whether ANAC096 and ABFs directly bind to each other by performing a protein pull-down experiment using recombinant ABFs and ANAC096. Of the four ABFs (ABF1 to 4), ABF1 was induced only by cold stress, whereas the other three ABFs were induced by osmotic stress (Fujita et al., 2011). Thus, we examined whether any of these three osmotic stress–induced ABFs binds to ANAC096. We expressed ABF2, ABF3, and ABF4 as maltose binding protein (MBP) fusion proteins and ANAC096 as a GST fusion protein in E. coli. Purified GST-ANAC096 was incubated with MBP-ABF2, MBP-ABF3, or MBP-ABF4, and the proteins were precipitated with glutathione-agarose beads. GST alone was included as a negative control. The precipitates were analyzed by immunoblotting using anti-GST and anti-MBP antibodies. MBP-ABF2 and MBP-ABF4, but not MBP-ABF3, were detected in the precipitates of GST-ANAC096 (Figure 6A). This indicated that ANAC096 binds to ABF2 and ABF4, but not to ABF3, in vitro. To confirm this result in vivo, the interaction between ABFs and ANAC096 was examined using a bimolecular fluorescence complementation (BiFC) approach (Müller-Taubenberger and Anderson, 2007). Full-length ANAC096 was fused to the C-terminal half of Myc-tagged Venus (Myc-cV), and ABF2, ABF3, or ABF4 was fused to the N-terminal half of HA-tagged Venus (nV-HA). These constructs were transformed into plant protoplasts. In addition, NLS-RFP, a chimeric red fluorescent protein (RFP) construct containing a nuclear localization signal (Lee et al., 2001), was included in the transformation. When ANAC096-Myc-cV was cotransformed with nV-HA-ABF2 or nV-HA-ABF4, strong BiFC signals were observed in both the cytosol and nucleus of protoplasts (Figure 6B). The localization of these proteins in the nucleus was confirmed by colocalization with NLS-RFP (Figures 6B and 6C; see Supplemental Figure 12 online). By contrast, when ANAC096-Myc-cV was cotransformed with nV-HA-ABF3, no BiFC signals were detected (Figures 6B and 6C). These results indicate that ANAC096 interacts directly with ABF2 and ABF4, but not with ABF3, in vivo.
Figure 6.
Interaction of ANAC096 with ABF2 and ABF4 in Vitro and in Vivo.
(A) Interaction of ANAC096 with ABF2 and ABF4, but not with ABF3, in vitro. GST-ANAC096 or GST alone (immobilized on glutathione-agarose beads) was incubated with MBP-ABF2, MBP-ABF3, and MBP-ABF4, and the proteins were precipitated. The precipitates were analyzed by immunoblotting with an anti-MBP antibody. Subsequently, the membrane was stained with Coomassie blue. Asterisk indicates a nonspecific band that was often copurified with GST-ANAC096 depending on the growth conditions of the E. coli.
(B) Interaction of ANAC096 with ABF2 and ABF4, but not with ABF3, in vivo. Protoplasts were transformed with the indicated constructs and BiFC signals were observed under a fluorescence microscope. NLS-RFP was included as a marker for the nucleus. YFP, yellow fluorescent protein. Bars = 20 μm.
(C) Expression of proteins from the BiFC constructs. Protein extracts from protoplasts transformed with the indicated BiFC constructs were analyzed by protein gel blotting using anti-HA and anti-Myc antibodies.
To further test the role played by the interaction between ANAC096 and ABFs in increasing transcriptional activation during dehydration or osmotic stress, we again used the transient expression system in protoplasts using RD29Apro:LUC as a reporter construct. When ANAC096 and ABF2 were introduced separately into protoplasts, both induced the expression of LUC, although ABF2 was more effective than ANAC096 (Figure 7A). When both ANAC096 and ABF2 were cotransformed into protoplasts, RD29A expression was more strongly induced; the transcript levels of RD29A were much higher than the combined values observed with ABF alone or ANAC096 alone (Figure 7A). This suggests that the two TFs act synergistically, rather than additively, in the induction of RD29A expression. However, when ANAC019, another NAC protein involved in dehydration stress responses, was cotransformed with ABF2, it suppressed the ABF2-mediated expression of RD29A, indicating that the interaction between different NACs and their partners can give a different outcome in the expression of target genes (Figure 7B). To further confirm this, we examined the ability of ANAC096 to induce the RD29Apro:LUC expression in the abf2 abf4 double mutant background. In abf2 abf4 double mutant protoplasts, ANAC096 increased the transcript levels of LUC only slightly, indicating that ANAC096 functions cooperatively with ABF2 and ABF4 for the full induction of RD29A expression (Figure 7C).
Figure 7.
Synergistic Activation of RD29A Expression in Protoplasts by ANAC096 and ABF2.
(A) and (B) The effect of coexpressing ANAC096 or ANAC019 with ABF2 on the induction of RD29A. Protoplasts from wild-type plants were transformed with ANAC096 and ABF2 (A) or ANAC019 and ABF2 (B), and total RNA from the transformed protoplasts was used to qRT-PCR to measure the transcript levels of RD29A. ACT2 was used as an internal control for qRT-PCR. Error bars indicate sd (n = 3).
(C) Effect of the abf2 abf4 mutations on ANAC096-mediated transcriptional activation of RD29A. Protoplasts from wild-type (WT), abf2 abf4, ANAC096/abf2 abf4 (pTA-ANAC096 crossed with abf2 abf4), and ANAC096 (pTA-ANAC096) plants were cotransformed with RD29Apro:LUC and UBQ10pro:GUS and incubated in the presence of DMSO or Dex (30 μM) for 1 h. Total RNA from the protoplasts was subjected to qRT-PCR analysis to measure the transcript levels of LUC. The expression level was normalized using the transcript level of GUS. Error bars indicate sd (n = 3).
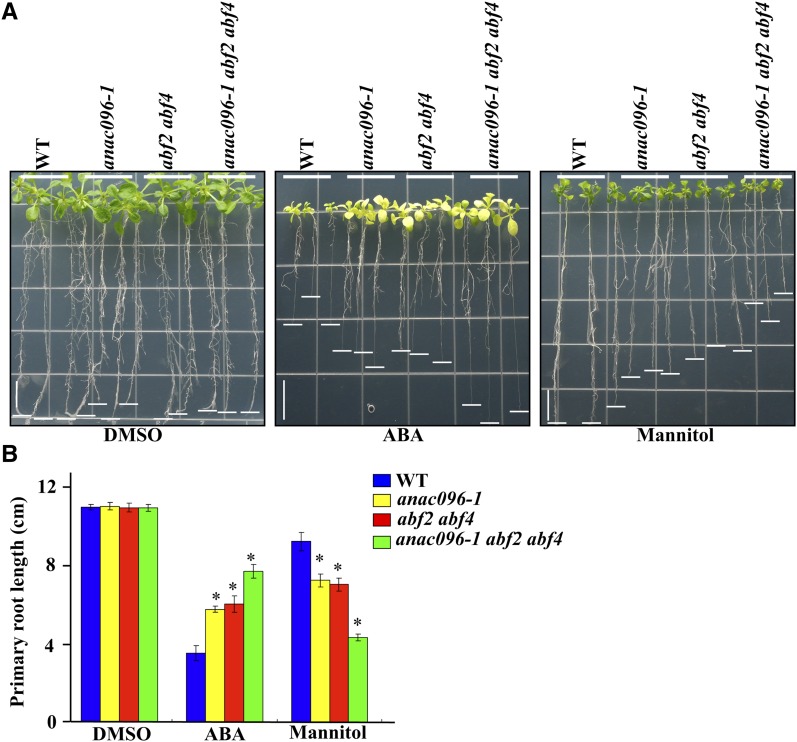
To find genetic evidence for this interaction, we generated anac096 abf2 abf4 triple knockout plants. Previous studies showed that abf2 abf4 double mutants were less sensitive to exogenously applied ABA and more sensitive to dehydration and osmotic stresses (Yoshida et al., 2010). When anac096 abf2 abf4 mutants were grown on plates supplemented with 10 μM ABA, they displayed even greater hyposensitivity to exogenous ABA compared with anac096 single mutant or abf2 abf4 double mutant plants (Figures 8A and 8B). Moreover, anac096 abf2 abf4 plants showed higher sensitivity to mannitol (130 mM) than did anac096 single mutant and abf2 abf4 double mutant plants (Figures 8A and 8B). These results indicate that the two different types of TFs act together to mediate responses to ABA and osmotic stress.
Figure 8.
The anac096 Mutation Enhances the Phenotype of the abf2 abf4 Double Mutant under Conditions of Exogenous ABA Application and Osmotic Stress.
(A) Phenotype of the indicated mutant plants. The seedlings grown on half-strength MS plates for 3 d were transferred onto MS plates supplemented with DMSO, 10 μM ABA, or 130 mM mannitol and grown vertically for 12 d. White underbars indicate the root tip region. Vertical bars = 2 cm. WT, the wild type.
(B) Quantification of the mutant phenotypes. To quantify the phenotypes, root length was measured in a triplicate experiment with 20 plants per experiment. Error bar indicate sd (n = 3). Statistical analysis was performed between wild-type and anac096 plants, between wild-type and abf2 abf4 plants, or between wild-type and anac096-1 abf2 abf4 triple mutant plants. *P value < 0.01 (Student’s t test).
DISCUSSION
The Arabidopsis genome encodes a large number of NAC TFs, which can be classified into 10 groups, with some groups being divided further into subgroups (Olsen et al., 2005a; Jensen et al., 2010; Nakashima et al., 2012). NAC TFs act as either transcriptional activators or repressors in various physiological processes, including abiotic and biotic stress responses, hormone signaling, and development (Olsen et al., 2005a; Lu et al., 2007; Hao et al., 2010). Previous studies demonstrated that ectopic expression of ANAC019, ANAC055, ANAC072, A. hypogaea NAC2, and soybean NAC20 led to the induction of the dehydration-responsive genes COR47, RD29B, ERD11, RD29A, RAB18, and ADH1 (Tran et al., 2004; Jensen et al., 2010; Hao et al., 2011; Liu et al., 2011). However, sequence homology alone is insufficient for determining their physiological roles and transcriptional specificities. NAC TFs regulate the expression of both ABA-dependent and ABA-independent genes even during abiotic stress responses (Tran et al., 2004; Hao et al., 2010; Jensen et al., 2010; Puranik et al., 2011, 2012). In this study, we demonstrated that the NAC TF ANAC096 is a positive component of the ABA-dependent signaling pathway involved in adaptation to dehydration and osmotic stress. This conclusion was based on the results showing that anac096 loss-of-function mutants exhibited hyposensitivity to exogenous ABA and increased sensitivity to dehydration stress during germination and postgermination growth, whereas ANAC096OX plants exhibited hypersensitivity to exogenous ABA and enhanced resistance to dehydration stress. Because ANAC096 is a TF, it likely plays a role in osmotic stress responses by regulating gene expression. Indeed, a major proportion of ABA-induced genes is also upregulated by ANAC096, suggesting that ANAC096 acts as a transcriptional activator in the ABA-dependent signaling pathway under dehydration stress conditions. There were also genes that were upregulated by ANAC096 but not by ABA. They are involved in cell organization, oxidative stress, photosynthesis, and defense responses. However, some of them are known to be regulated by ABA (Ton et al., 2009; Kar, 2011; Sreenivasulu et al., 2012; Tseng et al., 2013). It is possible that many of these genes in this group may be transcriptionally activated by ABA at a later time point or higher ABA concentration than we used. Of course, some of these genes show truly ABA-independent expression. Further studies are necessary to clarify this. In addition to these genes upregulated by ANAC096, a large number of genes were suppressed by induction of ANAC096. Genes involved in auxin stimulus and shoot development belong to this category. Considering that ANAC096 functions as a transcriptional activator, the genes downregulated by ANAC096 may belong to the secondary targets of ANAC096-induced genes. However, we cannot rule out the possibility that ANAC096 directly suppresses these genes, which should be clarified in the future.
NAC TFs recognize a conserved consensus sequence known as the NACRS (Simpson et al., 2003; Tran et al., 2004). ANAC096 bound to two NACRSs at the promoter region of RD29A. However, ANAC096 may not recognize all NACRS sequences because it does not induce the expression of ERD1, which harbors the conserved NACRS in its promoter region. The ERD1 expression is induced by ANAC019, another NAC TF (Tran et al., 2004; Jensen et al., 2010). According to phylogenetic tree analysis, ANAC096 belongs to NAC group IV-1 TFs, whereas ANAC019, ANAC055, and ANAC072 belong to NAC group III-3 TFs (Jensen et al., 2010). This raises the possibility that individual proteins belonging to different NAC groups may have differential binding affinity for NACRS. Indeed, VOZ TFs require a pair of palindromic DNA sequences for appropriate DNA binding; however, in the case of ANAC019, a single NACRS is sufficient for DNA recognition (Mitsuda et al., 2004; Tran et al., 2004; Olsen et al., 2005b). Similarly, ANAC096 binds to a single NACRS. However, it is not fully understood how this would occur at the molecular level. One possibility is that this could be determined by sequence variation within NACRSs. Alternatively, cofactors may contribute to the recognition of the cis-acting elements. Another characteristic feature of NAC proteins in the DNA binding is that they bind to DNA as a dimer. The N-terminal region contains the highly conserved dimer interface composed of two short antiparallel β-sheets and two prominent salt bridges formed by the conserved Arg and Glu residues (Ernst et al., 2004; Olsen et al., 2005b). ANAC096 also forms a dimer in vitro. Moreover, the dimer formation is crucial for its binding to DNA. Consistent with this observation, the amino acid residues critical for dimerization are conserved in ANAC096 and show a high degree of sequence similarity with that of ANAC019, ANAC055, and ANAC072 belonging to NAC group III-3 TFs.
Often, many different types of TFs regulate the expression of a particular gene. Therefore, fine-tuning among these different types of TFs is necessary; this is thought to occur through the formation of enhanceosome complexes that stabilize protein–protein and protein–DNA contacts (Martinez and Rao, 2012). Previous studies suggested that NAC TFs form heterodimers with other TFs (Duval et al., 2002; Lee et al., 2010). ANAC096 interacts with ABF2 and ABF4, which belong to the bZIP-type TF family and play a crucial role in the transcription of many ABA- and dehydration stress–inducible genes (Choi et al., 2000; Kim et al., 2004; Fujita et al., 2005). However, ANAC096 did not interact with ABF3, suggesting that ANAC096 interacts specifically with other TFs. Similarly, a previous study showed that DREB1A and DREB2A bind to ABF2 and ABF4 but not ABF3 (Lee et al., 2010). Currently it is not clearly understood how this differential interaction occurs among the members of ABFs. Similarly, ABFs also form homodimers and heterodimers to regulate transcriptional activation (Lee et al., 2010). The interaction of ANAC096 with ABFs resulted in the synergistic activation of the common target gene RD29A. However, the fact that ANAC019 has an antagonistic relationship with ABF2 suggests differential cooperation among different NACs and other types of TFs. This conclusion is also supported by a recent publication showing that ANAC019 physically interacts with a zinc finger homeodomain, ZFHD1, thereby synergistically activating the expression of ERD1 (Tran et al., 2007). ANAC096 also functions independently of ABFs. This conclusion is based on the fact that overexpression of ANAC096 caused ABA-mediated growth suppression in abf2 abf4 double mutant plants, although this suppression was less severe in abf2 abf4 double mutant plants than that in wild-type plants. The interaction among these highly conserved TFs raises an intriguing question of how they exert specificity toward their binding partners. Despite the fact that ANAC096 and ANAC019 share the highly conserved NAC DNA binding domain, they interact with different partners (Tran et al., 2007), which suggests that the C-terminal variable domain of ANAC096 is responsible for the specific binding to its partners. This idea was also supported by a previous study showing that the ANAC096-interacting protein TPL can recognize the repression domains of other TFs but not their DNA binding domains (Causier et al., 2012). Similarly, although the bZIP DNA binding domains of ABFs are highly conserved, different ABFs have different transactivation activities and different binding partners (Lee et al., 2010; Yoshida et al., 2010), which also suggests that the transactivation domain of ABFs confers specificity to their interactions with their partners. The interaction of ANAC096 with ABF2/4 raises an interesting question of why plants use such interactions in abiotic stress responses. Under abiotic stress, plants need to rapidly adjust their gene expression profile to optimize their physiological fitness for survival (Zhu, 2001). For robust responses to these abiotic stresses, it is clearly advantageous to employ multiple pathways that employ different TFs (Choi et al., 2000; Chen et al., 2012; Mizoi et al., 2012; Nakashima et al., 2012; Puranik et al., 2012). Multiple synergistic interactions between different pathways could be critically important for eliciting the diverse outputs in terms of specificity and amplitude. Thus, the interplay between different types of TFs, as observed in the case of ANAC096 and ABFs, may be an essential part of the mechanism for adapting to abiotic stress. Further work, including the identification of the full range of target genes regulated by ANAC096, will be necessary to understand the exact mechanism via which ANAC096 intervenes in dehydration stress responses mediated by ABA signaling in plants.
METHODS
Plant Growth
Arabidopsis thaliana plants (ecotype Columbia) were grown either on Murashige and Skoog (MS) plates at 23°C in a culture room or in a greenhouse with 70% relative humidity and a 16-h/8-h light/dark cycle. Plants were harvested and frozen immediately in liquid nitrogen for RNA preparation. For ABA treatment, plants grown in MS liquid medium for 1 week were treated with 100 μM ABA, as described previously (Jang et al., 1998; Piao et al., 1999). To determine the germination rate, sterilized seeds were planted on one-half-strength MS medium containing 1% Suc and different concentrations of ABA. Germination rates were scored based on radicle protrusion and the greening of expanded cotyledons. For growth measurements, 3-d-old seedlings grown on one-half-strength MS agar plates were transferred to one-half-strength MS agar plates supplemented with 10 μM ABA. Root growth was measured 12 d after transplantation. Similarly, growth measurements of wild-type, anac096, abf2 abf4, and anac096 abf2 abf4 mutant plants were conducted in one-half-strength MS agar plates supplemented with DMSO, 10 μM ABA, or 130 mM mannitol. To determine the expression patterns of ANAC096 in different conditions, plants grown on agar plates were treated with 10 μM ABA, incubated for 1 and 2 h in a chamber with 30% relative humidity, 300 mM mannitol, and a cold chamber (4°C). To examine the expression of ABA-responsive genes, wild-type and anac096 mutant plants grown in liquid medium were treated with 100 μM ABA for 1 h, and the expression levels of RD29A, RD29B, and COR47 were determined at varying time points. For statistical analysis, at least three independent experiments were performed. P values were calculated using the Student’s t test.
Screening of Mutants
Transgenic Arabidopsis plants showing green cotyledons on MS plates supplemented with 1 μM ABA were isolated from a pool of activation-tagging transgenic plants (Weigel et al., 2000). To determine the T-DNA insertion site, the T-DNA (together with the flanking region) was isolated from genomic DNA of mutant plants using the plasmid rescue method (Weigel et al., 2000). The flanking regions of the T-DNA insertion sites were sequenced to identify the target genes.
Construction of Plasmids
ANAC096 cDNA was isolated from a cDNA library by PCR using the gene-specific primers ANAC096F (XbaI) and ANAC096F (BamHI). PCR products were inserted into a plant binary pCsV1300 vector (Invitrogen), which harbors the cassava vein mosaic virus promoter, via the XbaI and BamHI sites. To generate pTA-ANAC096, PCR products were inserted into the pTA7002 binary vector via the XhoI and SpeI sites. To generate the ANAC096-GFP construct, ANAC096 cDNA without a termination codon was ligated in frame to the N terminus of green fluorescent protein (GFP) in the plant expression vector 326-GFP (Jin et al., 2001; Kim et al., 2001) via the XbaI and BamHI sites. To generate GAL4-ANAC096F, GAL4-ANAC096N, and GAL4-ANAC096C, the full-length, N-terminal region and C-terminal region of ANAC096 were generated by PCR using the primers GAL4F and GAL4R, GAL4NF and GAL4NR, and GAL4CF and GAL4CR, respectively, and were inserted into the pGBKT7 vector via the NdeI and PstI sites. The DNA fragment containing 100 bp (−1 to −99 nucleotide positions) of the RD29A promoter region was generated by PCR using primers RD29ApF and RD29ApR. RD29A-50 (a fragment containing from −50 to −99 nucleotide positions of the RD29A promoter) and its three substitution mutants RD29A-50[1A] (1A), RD29A-50[2A] (2A), and RD29A-50[1A/2A] (1A/2A), carrying the AAAA substitution at the first, second, or both of NACRS sites, respectively, were amplified by PCR. The primers used were RD29A-50[1A]F and RD29A-50[1A]R for RD29A-50[1A]; RD29A-50[2A]F and RD29A-50[2A]R for RD29A-50[2A]; and RD29A-50[1A]F and RD29A-50[1A]R, and RD29A-50[2A]F and RD29A-50[2A]R for RD29A-50[1A/2A]. For RD29A[CGT/AAA]pro, fragments were amplified using specific primers RD29A[CGT/AAA]pF and RD29A[CGT/AAA]pR. To construct GST-ANAC096, the ANAC096 fragment was produced by PCR using primers GST-ANAC096F and GST-ANAC096R, and the PCR product was ligated into pGEX-5X via the BamHI and SalI sites. To generate MBP-ABF2, MBP-ABF3, and MBP-ABF4, the fragments for ABF2, ABF3, and ABF4 were generated by PCR using the specific primers MBP-ABF2F and MBP-ABF2R, MBP-ABF3F and MBP-ABF3R, and MBP-ABF4F and MBP-ABF4R, respectively. The fragments were ligated into pMAL-c2X via the BamHI and SalI sites for ABF3 and via the EcoRI and SalI sites for ABF2 and ABF4. For His-ANAC096N, His-ANAC096N(R11A/E18A), and His-ANAC096N(d2-10), the corresponding DNA fragments were amplified by the pair of primers His-ANAC096NF(BamHI) and His-ANAC096NR(EcoRI), His-ANAC096N(R11A/E18A)F and His-ANAC096N(R11A/E18A)R, His-ANAC096N(d2-10)F and His-ANAC096N(d2-10)R, respectively. The PCR fragments were ligated to pRSET digested with BamHI and EcoRI. For nV-HA-ABF2, nV-HA-ABF3, and nV-HA-ABF4, ABF2, ABF3, and ABF4 were amplified by PCR using primers nVenus-ABF2F and nVenus-ABF2R, nVenus-ABF3F and nVenus-ABF3R, and nVenus-ABF4F and nVenus-ABF4R, respectively, and the PCR products were ligated into nVenus via the BamHI site for ABF3 and via the BamHI and EcoRI sites for ABF2 and ABF4. For ANAC096-Myc-cV, ANAC096 was amplified using primers cVenus-ANAC096F and cVenus-ANAC096R, and the PCR product was ligated into cVenus via the BamHI and EcoRI sites. To generate the ANAC096pro:GUS construct, a 3.0-kb fragment containing the promoter region of ANAC096 was amplified by PCR using primers ANAC096pF and ANAC096pR, and the PCR product was inserted into the binary vector pCAMBIA3301 (Invitrogen) via the BamHI and NcoI sites. All the PCR products were sequenced to confirm the nucleotide sequence. All primer sequences are listed in Supplemental Table 1 online.
Generation of Transgenic Plants
The binary constructs pCAMBIA-ANAC096pro:GUS, pTA-ANAC096, and pCSV1300-ANAC096 were transformed into wild-type plants (Clough and Bent, 1998). Transgenic plants were screened on MS plates supplemented with hygromycin (25 mg/L) or phosphinothricin (25 mg/L). To generate anac096-1 abf2 abf4 triple mutants, anac096 was crossed with abf2 abf4 double mutants, and triple mutants were screened by PCR-mediated genotyping using primers abf2LP and abf2RP, abf4LP and abf4RP, and anac096F and anac096R for abf2, abf4, and anac096 loci, respectively.
Stomatal Aperture, Water Loss, and Survival Rate Measurements
Epidermal peels from rosette leaves were incubated in a solution containing 10 mM MES-Tris, pH 6.15, 50 μM CaCl2, and 10 mM KCl and exposed to light (150 μmol m−2 s−1) for 3 h. Subsequently, 0.5 or 1 μM ABA was added to the solution to induce stomatal closure, and stomatal apertures were measured 1 h after the treatment (Jeon et al., 2008). In each experiment, measurements were performed on 120 pairs of guard cells in the epidermal peels from three different plants. To measure water loss, the aerial parts of plants grown on plates for 10 d were excised and placed on a bench (30% relative humidity), and the fresh weight of plants was measured (Xu et al., 2012). Plants grown for 9 d in the soil under normal growth conditions (watered) were kept in a greenhouse without watering for 14 d (dehydration), and the survival rates of plants were determined 3 d after rewatering (rehydration) (Xu et al., 2012). Five independent experiments were performed with 16 plants per experiment.
qRT-PCR Analysis of Transcript Levels
Total RNA was prepared from plant tissues using an RNeasy plant mini kit (Qiagen) and treated with TURBO DNase (Ambion). Total RNA was reverse-transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems). qRT-PCR was performed using a SYBR Green kit to detect transcript levels. ACT2 was used as an internal control. Primer sequences are listed in Supplemental Table 1 online. PCR was performed in a 20-μL reaction volume containing 10 ng of cDNA, 0.5 μM of each primer, and 1 unit of Taq polymerase. Amplification conditions were as follows: denaturation at 94°C for 4 min, followed by 30 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 60 s
Yeast Transformation and Growth
The full-length, N-terminal, or C-terminal regions of ANAC096 were ligated to the vector pGBKT7 containing the GAL4 DNA binding domain. The yeast strain AH109 harboring the LacZ and His reporter genes was transformed with GAL4BD-ANAC096F, GAL4BD-ANAC096N, or GAL4BD-ANAC096C. Serial dilutions of the transformed yeast culture were dropped onto SD plates with or without His, and growth was monitored (Ouwerkerk and Meijer, 2011).
Expression of GUS under the ANAC096 Promoter
To detect the expression of GUS from ANAC096pro:GUS in transgenic plants, F2 plants from 10 independent transgenic lines were stained with X-GLUC as previously described (Xu et al., 2012). To observe the tissue-specific expression of ANAC096, whole plants were soaked in X-GLUC solution at 37°C for 3 h. To examine the ABA- and dehydration stress–induced expression, the ANAC096pro:GUS plants were treated with 10 μM ABA and 30% relative humidity conditions for 1 h and were subsequently soaked in X-GLUC solution at 37°C for 1.5 h.
Purification of Recombinant Protein and EMSA
The fusion constructs GST, GST-ANAC096N, GST-ABF2, GST-ABF4, His-ANAC096N, His-ANAC096N(R11A/E18), and His-ANAC096(d2-10) were transformed into Escherichia coli BL21(DE3), and the transformed E. coli cells were cultured overnight at 16°C in the presence of 1 mM isopropyl β-d-1-thiogalactopyranoside. Recombinant fusion proteins were purified using glutathione-agarose (Thermo Scientific) beads or nickel-nitrilotriacetic acid agarose beads (Qiagen). EMSA was performed essentially as described previously (Ren et al., 2010). Briefly, purified recombinant proteins GST, GST-ANAC096N, GST-ABF2, GST-ABF4, His-ANAC096N, His-ANAC096N(R11A/E18), or His-ANAC096(d2-10) (1 μg) were incubated with 0.025 pmol of 32P-labeled probes RD29A-50 (wild type), RD29A-50[1A] (1A), RD29A-50[2A] (2A), or RD29A-50[1A/2A] (1A/2A) in a total volume of 20 μL. The reaction mixtures were analyzed on 4 to 16% gradient gels (Native PAGE Novex 4 to 16% Bis-Tris gel; Invitrogen), and gels and blots were exposed to x-ray film.
Transient Expression in Protoplasts
Plasmid DNAs were prepared using Qiagen columns. Protoplasts were prepared from leaf tissues from 1- to 2-week-old plants grown on MS plates (Jin et al., 2001; Hyunjong et al., 2006). After transformation, GFP, RFP, or BiFC signals were observed directly under a fluorescence microscope (Kim et al., 2001; Bae et al., 2008). To detect protein expression, transformed protoplasts were pelleted by centrifugation (500 rpm). Protein extracts were prepared by gentle lysis of transformed protoplasts using sonication and analyzed by immunoblotting using appropriate antibodies. To examine reporter gene expression, protoplasts from transgenic plants harboring pTA-ANAC096 or empty vector were transformed with the appropriate reporter constructs and incubated for 23 h followed by 1-h treatment with 30 μM Dex. Transformed protoplasts were pelleted by low-speed centrifugation (500 rpm). Total RNA was prepared using an RNA extraction kit (Ambion) and used for qRT-PCR analysis by the comparative cycle threshold method, in which LUC transcript levels were normalized using the GUS transcript (Yuichi et al., 2000).
Protein Pull-Down Experiments
For protein pull-down experiments, GST alone or GST-ANAC096 (3 μg) was immobilized onto glutathione beads and incubated with MBP-ABF2, MBP-ABF3, MBP-ABF4, His-ANAC096N, His-ANAC096N(R11A/E18A), or His-ANAC096N(d2-10) in binding buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 3 mM MgCl2, 1 mM DTT, and 0.1% Triton X-100) at 4°C for 3 h. The beads were then washed three times with binding buffer. Subsequently, SDS-PAGE sample buffer was added to the beads. The samples were boiled, separated by SDS-PAGE, and analyzed by immunoblotting with an anti-MBP antibody.
Microarray Experiments
pTA plants (12 d old) cultured in liquid medium were treated with 30 μM Dex alone for 1 h (Control) or 30 μM Dex alone for 30 min followed by an additional treatment of 30 min with both 30 μM Dex and 2 μM ABA. For pTA-ANAC096 plants, 12-d-old seedlings were treated with 30 μM Dex for 1 h (ANAC096). Total RNAs were isolated from two biological replicates at each condition. RNA integrity was evaluated using the Bioanalyzer 2100 (Agilent), and the RNA integrity numbers were larger than 9.5 for all samples, which is sufficient for gene expression analysis. RNA was reverse-transcribed and amplified using the standard Agilent protocols and then hybridized to the array (Agilent-031025 Arabidopsis 8x60k), which includes 62,976 probes corresponding to 28,949 genes (TAIR10). The gene expression data set was deposited at the Gene Expression Omnibus database (GSE51218).
Statistical Analysis of Gene Expression Data
The probe intensities measured from the microarray experiments were first converted into log2 intensities, which were then normalized using quantile normalization (Bolstad et al., 2003). To identify present probes, a Gaussian mixture model (one for present probes and the other for absent probes) was fitted to distribution of the normalized log2 intensities in each sample. The probes with intensities larger than a cutoff in which two Gaussian distributions meet were determined to be present. To identify differentially expressed genes (DEGs), an integrative statistical method previously reported was applied (Chae et al., 2013). In brief, Student’s t test and log2 median ratio test were performed to calculate T values and log2 median ratios for all of the probes. Empirical null distributions of T values and log2 median ratio were estimated by random permutation experiments of the six samples (i.e., 90 possible permutations). P values for T values and log2 median ratios were calculated by two-tailed tests with their corresponding empirical distributions and then combined using the Stouffer’s method (Hwang et al., 2005). The present genes with the combined P ≤ 0.05 and absolute log2 fold changes larger than a cutoff (1.00; twofold change) were identified as DEGs.
Enrichment Analysis of Gene Ontology Biological Processes
Before the enrichment analysis of Gene Ontology biological processes (GOBPs), the DEGs under the ANAC096-induced condition were categorized into four groups based on differential expression patterns under ANAC096-induced and ABA-treated conditions: 1 and 2, the DEGs whose expression increased or decreased commonly at both conditions; 3 and 4, the DEGs whose expression increased or decreased only in the ANAC096-induced condition (Figure 4B; see Supplemental Figure 4B online). For each group of DEGs, enrichment analysis of GOBPs was performed to compute P values that indicate the significance of each GOBP being represented by the genes using DAVID software (Huang et al., 2009). GOBPs with P < 0.05 were identified as enriched processes by the genes in each cluster.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome initiative or GenBank/EMBL databases under the following accession numbers: ANAC096, At5g46590; RD29A, At5g52310; RD29B, At5g52300; COR47, At1g20440; HVA22D, At4g24960; RAB18, At1g43890; NCED3, At3g14440; HAI2, At1g07430; ABF2, At1g45249; ABF3, At4g34000; and ABF4, At3g19290.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Screening of anac096 Mutants Showing the ABA-Hyposensitive Phenotype during Germination.
Supplemental Figure 2. The Tissue-Specific Expression Pattern of ANAC096 Examined by GUS Staining and qRT-PCR Analysis.
Supplemental Figure 3. The Expression of ANAC096 and RD29A Was Rapidly Induced in pTA-ANAC096 Plants by Dex.
Supplemental Figure 4. ANAC096 Negatively Regulates a Large Number of Genes, Including a Subset of ABA Downregulated Genes.
Supplemental Figure 5. Comparison of Fold Changes between ANAC096-Induced and ABA-Treated Conditions.
Supplemental Figure 6. Genes Identified from the Genome-Wide Expression Profiling Show Complicated Networks with Respect to the ABA-Related Cellular Processes.
Supplemental Figure 7. ANAC096 Functions as a Transcriptional Activator in Yeast Cells.
Supplemental Figure 8. Amino Acid Sequence Alignment of ANAC096 with ANAC019, ANAC055, and ANAC072.
Supplemental Figure 9. The Most 5′ CGT at RD29A Promoter Is Not Required for Transcriptional Activation by ANAC096.
Supplemental Figure 10. The ANAC096 C-Terminal Region Is Required for Transcriptional Activation.
Supplemental Figure 11. ABF2 and ABF4 Bind to the RD29A Promoter.
Supplemental Figure 12. ANAC096 Localizes to the Nucleus.
Supplemental Table 1. Primers Used for qRT-PCR Analysis.
Acknowledgments
This work was supported in part by grants from the National Research Foundation, the Ministry of Science, Information Computer Technology and Future Planning, the Ministry of Agriculture, Food and Rural Affairs (Korea), and the Functional Genomics Center, Pohang University of Science and Technology. D.H. was supported by the Institute for Basic Science (CA1308), Korea.
AUTHOR CONTRIBUTIONS
Z.-Y.X., S.Y.K., and I.H. conceived the project and designed the research strategies. Z.-Y.X. and S.Y.K. isolated anac096 mutants, generated most of the transgenic plants, and performed the majority of genetic, biochemical, and cell biological assays. Z.-Y.X. conducted most of the genetic, biochemical, and physiological assays. S.Y.K. conducted imaging-related experiments and genetic and physiological assays. D.H.K. performed the EMSA assays. D.Y.H. and D.H. analyzed the microarray data. T.D. and Y.P. performed some qRT-PCR analyses. S.-H.J and S.-K.K. examined ABA-related phenotypes. J.B.J. conducted dehydration stress–related work. J.C.H. provided abf2 and abf4 mutants. Z.-Y.X. and I.H. wrote the article.
Glossary
- ABA
abscisic acid
- qRT-PCR
quantitative RT-PCR
- Dex
dexamethasone
- NACRS
NAC-recognition sequence
- EMSA
electrophoresis mobility shift assay
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
- DEG
differentially expressed gene
- GOBP
Gene Ontology biological process
References
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bae W., Lee Y.J., Kim D.H., Lee J., Kim S., Sohn E.J., Hwang I. (2008). AKR2A-mediated import of chloroplast outer membrane proteins is essential for chloroplast biogenesis. Nat. Cell Biol. 10: 220–227 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Bray E.A. (2002). Abscisic acid regulation of gene expression during water-deficit stress in the era of the Arabidopsis genome. Plant Cell Environ. 25: 153–161 [DOI] [PubMed] [Google Scholar]
- Causier B., Ashworth M., Guo W., Davies B. (2012). The TOPLESS interactome: A framework for gene repression in Arabidopsis. Plant Physiol. 158: 423–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S., Ahn B.Y., Byun K., Cho Y.M., Yu M.H., Lee B., Hwang D., Park K.S. (2013). A systems approach for decoding mitochondrial retrograde signaling pathways. Sci. Signal. 6: rs4. [DOI] [PubMed] [Google Scholar]
- Chen L., Song Y., Li S., Zhang L., Zou C., Yu D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819: 120–128 [DOI] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J., Kim S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corrêa L.G., Riaño-Pachón D.M., Schrago C.G., dos Santos R.V., Mueller-Roeber B., Vincentz M. (2008). The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS ONE 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M., Hsieh T.F., Kim S.Y., Thomas T.L. (2002). Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50: 237–248 [DOI] [PubMed] [Google Scholar]
- Ernst H.A., Olsen A.N., Larsen S., Lo Leggio L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5: 297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Maruyama K., Seki M., Hiratsu K., Ohme-Takagi M., Tran L.S., Yamaguchi-Shinozaki K., Shinozaki K. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Satoh R., Maruyama K., Parvez M.M., Seki M., Hiratsu K., Ohme-Takagi M., Shinozaki K., Yamaguchi-Shinozaki K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124: 509–525 [DOI] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y.-J., Song Q.-X., Chen H.-W., Zou H.-F., Wei W., Kang X.-S., Ma B., Zhang W.-K., Zhang J.-S., Chen S.-Y. (2010). Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232: 1033–1043 [DOI] [PubMed] [Google Scholar]
- Hao Y.-J., et al. (2011). Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68: 302–313 [DOI] [PubMed] [Google Scholar]
- Huang W., Sherman B.T., Lempicki R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Hwang D., Rust A.G., Ramsey S., Smith J.J., Leslie D.M., Weston A.D., de Atauri P., Aitchison J.D., Hood L., Siegel A.F., Bolouri H. (2005). A data integration methodology for systems biology. Proc. Natl. Acad. Sci. USA 102: 17296–17301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyunjong B., Lee D.S., Hwang I. (2006). Dual targeting of xylanase to chloroplasts and peroxisomes as a means to increase protein accumulation in plant cells. J. Exp. Bot. 57: 161–169 [DOI] [PubMed] [Google Scholar]
- Jang H.J., Pih K.T., Kang S.G., Lim J.H., Jin J.B., Piao H.L., Hwang I. (1998). Molecular cloning of a novel Ca2+-binding protein that is induced by NaCl stress. Plant Mol. Biol. 37: 839–847 [DOI] [PubMed] [Google Scholar]
- Jensen M.K., Kjaersgaard T., Nielsen M.M., Galberg P., Petersen K., O’Shea C., Skriver K. (2010). The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Jeon B.W., Hwang J.-U., Hwang Y., Song W.-Y., Fu Y., Gu Y., Bao F., Cho D., Kwak J.M., Yang Z., Lee Y. (2008). The Arabidopsis small G protein ROP2 is activated by light in guard cells and inhibits light-induced stomatal opening. Plant Cell 20: 75–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J.B., Kim Y.A., Kim S.J., Lee S.H., Kim D.H., Cheong G.-W., Hwang I. (2001). A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 13: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar R.K. (2011). Plant responses to water stress: Role of reactive oxygen species. Plant Signal. Behav. 6: 1741–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Miura S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45: 346–350 [DOI] [PubMed] [Google Scholar]
- Kim D.H., Eu Y.J., Yoo C.M., Kim Y.W., Pih K.T., Jin J.B., Kim S.J., Stenmark H., Hwang I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13: 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.H., Woo H.R., Kim J., Lim P.O., Lee I.C., Choi S.H., Hwang D., Nam H.G. (2009). Trifurcate feed-forward regulation of age-dependent cell death involving miR164 in Arabidopsis. Science 323: 1053–1057 [DOI] [PubMed] [Google Scholar]
- Kim S., Kang J.Y., Cho D.-I., Park J.H., Kim S.Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40: 75–87 [DOI] [PubMed] [Google Scholar]
- Lee S.-J., Kang J.-Y., Park H.-J., Kim M.D., Bae M.S., Choi H.-I., Kim S.Y. (2010). DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 153: 716–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Kim D.H., Kim Y.-W., Hwang I. (2001). Identification of a signal that distinguishes between the chloroplast outer envelope membrane and the endomembrane system in vivo. Plant Cell 13: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Hong L., Li X.-Y., Yao Y., Hu B., Li L. (2011). Improved drought and salt tolerance in transgenic Arabidopsis overexpressing a NAC transcriptional factor from Arachis hypogaea. Biosci. Biotechnol. Biochem. 75: 443–450 [DOI] [PubMed] [Google Scholar]
- Lu P.-L., Chen N.-Z., An R., Su Z., Qi B.-S., Ren F., Chen J., Wang X.-C. (2007). A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 63: 289–305 [DOI] [PubMed] [Google Scholar]
- Martinez G.J., Rao A. (2012). Immunology. Cooperative transcription factor complexes in control. Science 338: 891–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Hisabori T., Takeyasu K., Sato M.H. (2004). VOZ; isolation and characterization of novel vascular plant transcription factors with a one-zinc finger from Arabidopsis thaliana. Plant Cell Physiol. 45: 845–854 [DOI] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Müller-Taubenberger A., Anderson K.I. (2007). Recent advances using green and red fluorescent protein variants. Appl. Microbiol. Biotechnol. 77: 1–12 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819: 97–103 [DOI] [PubMed] [Google Scholar]
- Newman J.R., Keating A.E. (2003). Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300: 2097–2101 [DOI] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. (2005a). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Olsen A.N., Ernst H.A., Leggio L.L., Skriver K. (2005b). DNA binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169: 785–797 [Google Scholar]
- Ouwerkerk P.B., Meijer A.H. (2011). Yeast one-hybrid screens for detection of transcription factor DNA interactions. Methods Mol. Biol. 678: 211–227 [DOI] [PubMed] [Google Scholar]
- Piao H.L., Pih K.T., Lim J.H., Kang S.G., Jin J.B., Kim S.H., Hwang I. (1999). An Arabidopsis GSK3/shaggy-like gene that complements yeast salt stress-sensitive mutants is induced by NaCl and abscisic acid. Plant Physiol. 119: 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Bahadur R.P., Srivastava P.S., Prasad M. (2011). Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet [Setaria italica (L.) P. Beauv]. Mol. Biotechnol. 49: 138–150 [DOI] [PubMed] [Google Scholar]
- Puranik S., Sahu P.P., Srivastava P.S., Prasad M. (2012). NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 17: 369–381 [DOI] [PubMed] [Google Scholar]
- Ren X., Chen Z., Liu Y., Zhang H., Zhang M., Liu Q., Hong X., Zhu J.-K., Gong Z. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3: 217–223 [PubMed] [Google Scholar]
- Simpson S.D., Nakashima K., Narusaka Y., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 33: 259–270 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N., Harshavardhan V.T., Govind G., Seiler C., Kohli A. (2012). Contrapuntal role of ABA: Does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506: 265–273 [DOI] [PubMed] [Google Scholar]
- Sun L.J., Li D.Y., Zhang H.J., Song F.M. (2012). Functions of NAC transcription factors in biotic and abiotic stress responses in plants. Yi Chuan 34: 993–1002 [DOI] [PubMed] [Google Scholar]
- Thomashow M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Ton J., Flors V., Mauch-Mani B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14: 310–317 [DOI] [PubMed] [Google Scholar]
- Tran L.-S., Nakashima K., Sakuma Y., Osakabe Y., Qin F., Simpson S.D., Maruyama K., Fujita Y., Shinozaki K., Yamaguchi-Shinozaki K. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 49: 46–63 [DOI] [PubMed] [Google Scholar]
- Tran L.-S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng I.C., Hong C.Y., Yu S.M., Ho T.H. (2013). Abscisic acid- and stress-induced highly proline-rich glycoproteins regulate root growth in rice. Plant Physiol. 163: 118–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner D.H., Lindemose S., Grossmann J.G., Møllegaard N.E., Olsen A.N., Helgstrand C., Skriver K., Lo Leggio L. (2012). DNA binding by the plant-specific NAC transcription factors in crystal and solution: A firm link to WRKY and GCM transcription factors. Biochem. J. 444: 395–404 [DOI] [PubMed] [Google Scholar]
- Xiong L., Ishitani M., Lee H., Zhu J.-K. (2001). The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13: 2063–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z.-Y., Lee K.H., Dong T., Jeong J.C., Jin J.B., Kanno Y., Kim D.H., Kim S.Y., Seo M., Bressan R.A., Yun D.J., Hwang I. (2012). A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24: 2184–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Mino M., Mundy J., Chua N.H. (1990). Analysis of an ABA-responsive rice gene promoter in transgenic tobacco. Plant Mol. Biol. 15: 905–912 [DOI] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zhu J.-K. (2001). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4: 401–406 [DOI] [PubMed] [Google Scholar]