Aldoximes are precursors of several classes of direct plant defense compounds. This study characterizes two cytochrome P450 monooxygenase enzymes, CYP79D6/7, which are involved in gypsy moth–induced aldoxime formation in western balsam poplar. Volatile formation mediated by CYP79s may be a general phenomenon in the plant kingdom.
Abstract
Aldoximes are known as floral and vegetative plant volatiles but also as biosynthetic intermediates for other plant defense compounds. While the cytochrome P450 monooxygenases (CYP) from the CYP79 family forming aldoximes as biosynthetic intermediates have been intensively studied, little is known about the enzymology of volatile aldoxime formation. We characterized two P450 enzymes, CYP79D6v3 and CYP79D7v2, which are involved in herbivore-induced aldoxime formation in western balsam poplar (Populus trichocarpa). Heterologous expression in Saccharomyces cerevisiae revealed that both enzymes produce a mixture of different aldoximes. Knockdown lines of CYP79D6/7 in gray poplar (Populus × canescens) exhibited a decreased emission of aldoximes, nitriles, and alcohols, emphasizing that the CYP79s catalyze the first step in the formation of a complex volatile blend. Aldoxime emission was found to be restricted to herbivore-damaged leaves and is closely correlated with CYP79D6 and CYP79D7 gene expression. The semi-volatile phenylacetaldoxime decreased survival and weight gain of gypsy moth (Lymantria dispar) caterpillars, suggesting that aldoximes may be involved in direct defense. The wide distribution of volatile aldoximes throughout the plant kingdom and the presence of CYP79 genes in all sequenced genomes of angiosperms suggest that volatile formation mediated by CYP79s is a general phenomenon in the plant kingdom.
INTRODUCTION
Plants have developed a complex arsenal of chemical defense strategies. They produce toxins and feeding deterrents that serve as direct defenses to herbivores and release volatile compounds that repel the herbivore or even attract herbivore enemies as indirect defenses. Among the many classes of defense chemicals, some may participate in direct and indirect defense (Fürstenberg-Hägg et al., 2013).
Aldoximes are well-known precursors of several classes of direct-defense compounds. They are produced from their corresponding amino acids through the action of multifunctional cytochrome P450 monooxygenases (CYP) of the CYP79 family (recently reviewed in Hamberger and Bak, 2013). The first described CYP79 enzyme, CYP79A1, isolated and characterized from sorghum (Sorghum bicolor) (Sibbesen et al., 1995), catalyzes the conversion of Tyr to p-hydroxyphenylacetaldoxime, the rate-limiting step in cyanogenic glucoside biosynthesis. This reaction involves two successive N-hydroxylations, a dehydration, a decarboxylation, and a final isomerization reaction (Halkier et al., 1995; Sibbesen et al., 1995). p-hydroxyphenylacetaldoxime is further converted to p-hydroxymandelonitrile, and a subsequent glycosylation results in the formation of dhurrin, the major cyanogenic glucoside in sorghum (Møller and Conn, 1980). Aliphatic or aromatic aldoximes produced by CYP79 enzymes also participate in the biosynthesis of other cyanogenic glycosides and noncyanogenic hydroxynitriles like lotaustralin or the rhodiocyanosides from lotus (Lotus japonicus), linamarin from cassava (Manihot esculenta), and taxiphyllin from sea arrowgrass (Triglochin maritima) (Andersen et al., 2000; Nielsen and Møller, 2000; Forslund et al., 2004; Saito et al., 2012). Moreover, the biosynthesis of glucosinolates, the most important defense compounds in the Brassicaceae, involves a CYP79-catalyzed conversion of amino acids to aldoximes (Wittstock and Halkier, 2000). One of these aldoxime enzyme products, indole-3-acetaldoxime, produced by CYP79B2 and CYP79B3 in Arabidopsis thaliana, is not only metabolized to indole glucosinolates but can be further converted to camalexin, a major phytoalexin occurring in several Brassicaceae (Rauhut and Glawischnig, 2009). Indole-3-acetaldoxime is also thought to be a precursor of auxin biosynthesis (Pollmann et al., 2006). As aldoxime intermediates were not found to accumulate in the plant, it was proposed that they are channeled through a large protein complex called a metabolon (Møller and Conn, 1980; Møller, 2010). This mechanism prevents the release of putative toxic and reactive intermediates (Grootwassink et al., 1990; Møller, 2010)
Besides their role as intermediates in the formation of direct defenses, aldoximes have also been reported to be released as volatiles from a large number of different plant species (Knudsen et al., 2006). The flowers of many moth-pollinated, night-blooming plants like evening primroses (Oenothera spp), wild tobacco (Nicotiana spp), and orchids are reported to emit aliphatic and aromatic volatile aldoximes, which are considered to function as specific attractants for pollinators (Kaiser, 1993; Raguso, 2008; Vergara et al., 2011). Volatile aldoximes have also been detected from vegetative organs in the headspace of herbivore-damaged or pathogen-infested plants. For example, spider mite infestation was shown to induce the emission of aliphatic and aromatic aldoximes from golden chain (Laburnum anagyroides), black locust (Robinia pseudoacacia), tobacco (Nicotiana tabacum), eggplant (Solanum melongena), apple (Malus domestica), and cucumber (Cucumis sativus) (Takabayashi et al., 1991, 1994; van den Boom et al., 2004), while caterpillar damage resulted in the release of volatile aldoximes from maize (Zea mays) seedlings (Takabayashi et al., 1995) and lima bean (Phaseolus lunatus) (Wei et al., 2006), and pathogen infection induced the production of aldoximes in grapefruit plants (Citrus × paradisi) (Zhang and Hartung, 2005).
In general, herbivore-induced volatile blends are often dominated by terpenes and green leaf volatiles, while aldoximes are released as minor components (Takabayashi et al., 1991, 1994; van den Boom et al., 2004). However, in a recent study, it was demonstrated that aldoximes released from black poplar (Populus nigra) upon feeding by gypsy moth (Lymantria dispar) caterpillars attract braconid wasps (Glyptapantheles liparidis) that parasitize the caterpillars (A.C. McCormick, S. Irmisch, A. Reinecke, G.A. Boeckler, D. Veit, M. Reichelt, B. Hansson, J. Gershenzon, T.G. Köllner, and S.B. Unsicker, unpublished data). The two tested aliphatic aldoxime mixtures, 2-methylbutyraldoxime (E:Z, 3:1) and 3-methylbutyraldoxime (E:Z, 2:1), elicited stronger physiological responses by the parasitic wasp antennae than the major blend constituents like green leaf volatiles or terpenoids. Despite the widespread distribution of volatile aldoximes throughout angiosperms, little is known about the biochemistry of their formation. Most likely they are produced from amino acids in a similar manner as described for nonvolatile aldoxime intermediates in cyanogenic glucoside and glucosinolate formation (Hamberger and Bak, 2013). However, the enzymes involved are so far unclear. The aim of this study was to investigate the herbivore-induced formation of volatile aldoximes in western balsam poplar (Populus trichocarpa). As fast-growing trees, poplars play an important role in forestry and as energy crops (Beringer et al., 2011; Aust et al., 2013). During recent years, an extensive arsenal of resources has been developed for functional genomics and genetic studies of poplars. This and the availability of the full genome sequence of P. trichocarpa (Tuskan et al., 2006) make poplar species ideal model organisms for studying woody plant–herbivore interactions on a molecular level. Here, we report the identification and characterization of two P450 enzymes, CYP79D6v3 and CYP79D7v2, which have broad substrate specificity and catalyze the conversion of at least five different amino acids to their corresponding aldoximes. Overexpression of the CYP79 genes in N. benthamiana and the RNA interference (RNAi)–mediated knockdown of the two genes in Populus × canescens revealed that CYP79D7v3 and CYP79D7v2 are key enzymes for the production of a complex volatile blend comprising not only aldoximes but also their corresponding nitriles, nitro compounds, and alcohols.
RESULTS
The Herbivore-Induced Volatile Blend of Poplar Comprises Aldoximes and Related Nitrogenous Compounds
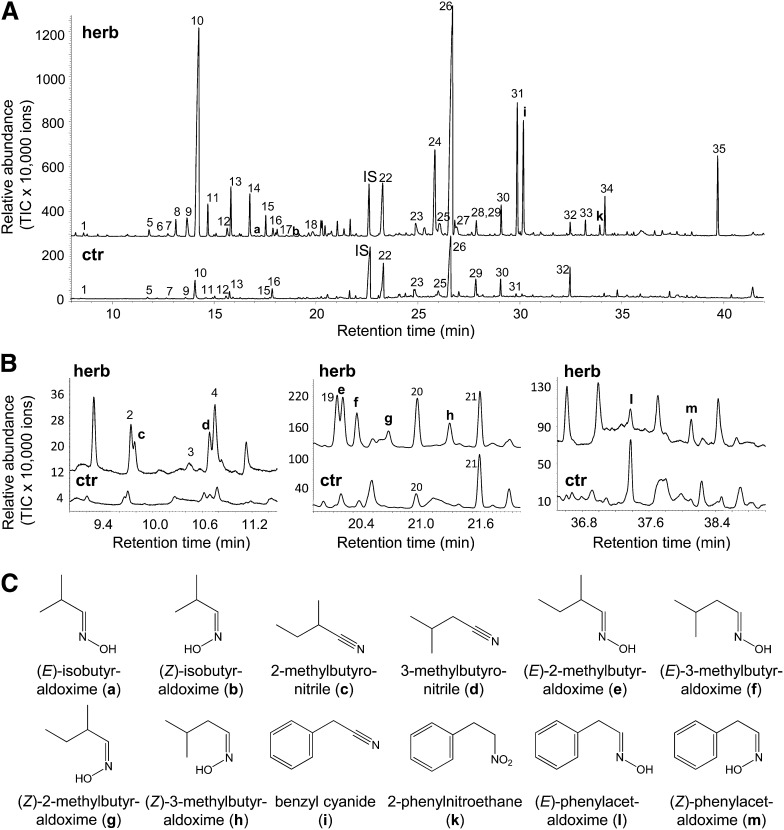
In a previous study, we documented the volatiles released by P. trichocarpa upon infestation by gypsy moth caterpillars (Danner et al., 2011). Besides terpenes, green leaf volatiles, aromatic aldehydes, and alcohols, P. trichocarpa releases significant amounts of the nitrile benzyl cyanide. Since other nitriles and related aldoximes have been reported for P. simonii × pyramibalis (Zeng-hui et al., 2004) and P. nigra (A.C. McCormick, S. Irmisch, A. Reinecke, G.A. Boeckler, D. Veit, M. Reichelt, B. Hansson, J. Gershenzon, T.G. Köllner, and S.B. Unsicker, unpublished data), we performed another volatile analysis on P. trichocarpa with the focus on nitrogenous volatiles. To ensure a good chromatographic separation of the different compounds, we used a polyethylene glycol column with a length of 60 m for gas chromatography–mass spectrometry (GC-MS) analysis. Comparison of the volatile bouquet released from gypsy moth–damaged trees with that from undamaged control trees showed many substances whose emission was induced by feeding (Figure 1A). The monoterpene (E)-β-ocimene and the sesquiterpene (E,E)-α-farnesene were the most abundant compounds in the caterpillar-induced volatile blend as previously described (Danner et al., 2011). However, herbivore-damaged trees also released a complex mixture of nitrogenous volatiles (Figures 1A and 1B). In addition to benzyl cyanide, two other nitriles, 2-methylbutyronitrile and 3-methylbutyronitrile, and the nitro compound 2-phenylnitroethane as well as eight different aldoximes, isobutyraldoxime (E:Z, 2:1), 2-methylbutyraldoxime (E:Z, 2.9:1), 3-methylbutyraldoxime (E:Z, 1.7:1), and phenylacetaldoxime (E:Z, 1:1.7), could be identified (Figures 1A to 1C).
Figure 1.
Volatiles Emitted from Gypsy Moth–Damaged and Undamaged P. trichocarpa Trees.
(A) and (B) The volatile profiles of herbivore-treated (herb) and undamaged control (ctr) P. trichocarpa trees were measured and analyzed using GC-MS. The panels in (B) are enlargements of three regions of the full chromatogram depicted in (A). 1, α-Pinene; 2, hexanal; 3, β-pinene; 4, isoamyl acetate 5, myrcene; 6, 3/2-methyl-1-butanol; 7, limonene; 8, (E)-2-hexenal; 9, (Z)-β-ocimene; 10, (E)-β-ocimene; 11, hexyl acetate; 12, (3E)-4,8-dimethyl-1,3,7-nonatriene; 13, (Z)-hexenyl acetate; 14, 1-hexanol; 15, (Z)-3-hexenol; 16, nonanal; 17, 2-hexen-1-ol; 18, α-cubebene; 19, (Z)-epoxy-ocimene; 20, benzaldehyde; 21, linalool; IS, nonyl acetate (internal standard); 22, (E)-β-caryophyllene; 23, salicyl aldehyde; 24, germacrene D; 25, (Z,E)-α-farnesene; 26, (E,E)-α-farnesene; 27, δ-cadinene; 28, 2-phenylethyl ester; 29, (3E,7E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene; 30, benzyl alcohol; 31, 2-phenylethanol; 32, nerolidol; 33, hexyl benzoate; 34, (Z)-hexenyl benzoate; 35, indole. TIC, total ion chromatogram.
(C) Structures of aldoximes and related nitrogen-containing volatiles; a, (E)-isobutyraldoxime; b, (Z)-isobutyraldoxime; c, 2-methylbutyronitrile; d, 3-methylbutyronitrile; e, (E)-2-methylbutyraldoxime; f, (E)-3-methylbutyraldoxime; g, (Z)-2-methylbutyraldoxime; h, (Z)-3-methylbutyraldoxime; i, benzyl cyanide; k, 2-phenylnitroethane; l, (E)-phenylacetaldoxime; m, (Z)-phenylacetaldoxime.
l-Phe Is the Precursor of Aromatic Nitrogenous Poplar Volatiles
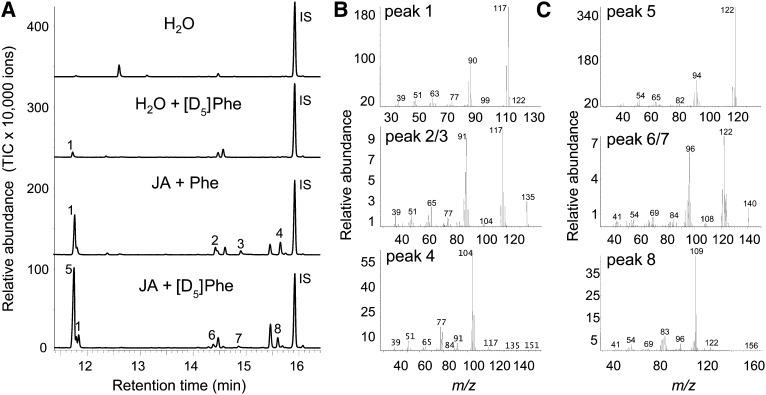
Aldoximes are known to be produced from their corresponding amino acid precursors and can be further converted to nitriles (Møller and Conn, 1980; Du and Halkier, 1996). To investigate whether a similar biosynthetic route occurs in P. trichocarpa, excised leaves induced with jasmonic acid were fed with deuterium-labeled l-Phe, the supposed substrate for (E/Z)-phenylacetaldoxime and benzyl cyanide formation, labeled with deuterium on the ring, and a volatile collection was conducted. The label incorporation into the nitrogenous volatiles was analyzed by mass spectrometry and could be observed as a mass shift of five atomic mass units for the molecular ions of (E)-phenylacetaldoxime (mass-to-charge ratio [m/z] 135 → m/z 140), (Z)-phenylacetaldoxime (m/z 135 → m/z 140), benzyl cyanide (m/z 117 → m/z 122), and 2-phenylnitroethane (m/z 104 → m/z 109) (Figure 2). These results indicate that l-Phe acts as the precursor for the formation of aromatic nitrogenous volatiles in P. trichocarpa.
Figure 2.
Deuterium-Labeled Nitrogenous Volatiles Emitted from Excised P. trichocarpa Leaves Incubated with [ring-D5]l-Phe.
(A) Volatiles were collected from single leaves kept in a solution of water, water with [ring-D5]l-Phe ([D5]Phe) (H2O + [D5]Phe), water with jasmonic acid and unlabeled l-Phe (JA + Phe), or water with jasmonic acid and [D5]Phe (JA + [D5]Phe). Collections were analyzed by GC-MS. IS, internal standard (nonyl acetate). TIC, total ion chromatogram.
(B) Mass spectra of [ring-D5]–labeled nitrogenous compounds.
(C) Mass spectra of unlabeled nitrogenous compounds. 1, Benzyl cyanide; 2, (E)-phenylacetaldoxime; 3, (Z)-phenylacetaldoxime; 4, 2-phenylnitroethane; 5, [ring-D5]-benzyl cyanide; 6, [ring-D5]-(E)-phenylacetaldoxime; 7, [ring-D5]-(Z)-phenylacetaldoxime; 8, [ring-D5]-2-phenylnitroethane. The spectra of compounds 2 and 3, on the one hand, and 6 and 7, on the other hand, were nearly identical.
The P. trichocarpa Genome Contains Four CYP79 Genes
To identify the enzymes involved in aldoxime biosynthesis in poplar, we conducted a BLAST search of the P. trichocarpa genome (http://www.phytozome.net/poplar) with CYP79A1 from sorghum (GenBank Q43135) as template. This analysis revealed four putative CYP79 genes in the P. trichocarpa genome, which were designated as CYP79D5 (Potri.013G157400), CYP79D6 (Potri.013G157200), CYP79D7 (Potri.013G157300), and CYP79D8 (Potri.004G055200) according to the general P450 nomenclature (D.R. Nelson, P450 Nomenclature Committee). CYP79D6 and CYP79D7 were found to cluster within 8 kb of each other in the same orientation on chromosome 13 (see Supplemental Figure 1A online). They are 98% identical at the nucleotide level and, according to the database, share 98.9% identity of their single 277(6)-bp introns and 96.8% identity within the first 300 nucleotides of their promoter regions (see Supplemental Figure 1B online), suggesting a recent tandem gene duplication. CYP79D5 was also located on chromosome 13 ∼33.5 kb downstream of CYP79D7. It shares 97.8 and 98.3% nucleotide identity with CYP79D6 and CYP79D7, respectively. However, the promoter region contains a 55-nucleotide deletion within the first 300 bases in comparison to CYP79D6 and CYP79D7 (see Supplemental Figure 1B online). The fourth CYP79 gene, CYP79D8, was found to be located on chromosome 4 and shared 75.7, 75.9, and 75.6% nucleotide sequence identity to CYP79D6, CYP79D7, and CYP79D5, respectively.
The P. trichocarpa CYP79 Proteins Are Typical Members of the CYP79 Family
While the complete open reading frames of CYP79D6 and CYP79D7 could be isolated from cDNA attained from herbivore-damaged leaves of P. trichocarpa, the amplification of CYP79D5 and CYP79D8 from the same cDNA failed, suggesting that the latter two genes were not transcribed in herbivore-damaged leaves. The two obtained genes were designated as CYP79D6v3 and CYP79D7v2 to distinguish them from their corresponding alleles in the database derived from the P. trichocarpa genotype ‘Nisqually-1’ (D.R. Nelson, P450 Nomenclature Committee). The proteins encoded by CYP79D6v3 and CYP79D7v2 showed 96.8% amino acid identity to each other and shared 60.6% identity with CYP79D4 from L. japonicus (Forslund et al., 2004), the closest CYP79 characterized so far (see Supplemental Figure 2 online). A phylogenetic analysis of all characterized CYP79 enzymes and the four P. trichocarpa CYP79 sequences showed that the poplar CYP79 proteins group together with other members of the CYP79D subfamily (see Supplemental Figure 2 online), which were described to be involved in the biosynthesis of cyanogenic glycosides (Andersen et al., 2000; Forslund et al., 2004). Motifs reported to be conserved in nearly all P450 enzymes, such as the ProProxxPro motif right before the N-terminal membrane anchor, the heme binding site including the conserved Cys residue, and the ProGluArgPhe motif (Durst and Nelson, 1995), were also found in the P. trichocarpa sequences (see Supplemental Figure 3 online). Moreover, the heme binding site as well as the ProGluArgPhe motif showed substitutions unique for the CYP79 family. In comparison to the consensus sequence ProPheGlyxGlyArgArgxCysxGly described for A-type P450s (Durst and Nelson, 1995), the heme binding motifs of both CYP79D6v3 and CYP79D7v2 (457-SerPheSerThrGlyLysArgGlyCysIleGly-467; see Supplemental Figure 3 online) contained three amino acid substitutions which have been reported for other members of the CYP79 family (Bak et al., 1998a; Andersen et al., 2000; Nielsen and Møller, 2000). The ProGluArgPhe motif (residues 434 to 440; see Supplemental Figure 3 online) was altered to ProGluArgHis with a His substitution being unique for the CYP79 family (Bak et al., 1998a). The replacement of two conserved Thr-(Thr/Ser) residues in a proposed substrate binding pocket (Durst and Nelson, 1995) with Asn-Pro (see Supplemental Figure 3 online; residues 328/329 for CYP79D6) is also a typical feature for enzymes of the CYP79 family.
CYP79D6v3 and CYP79D7v2 Catalyze the Formation of Aromatic and Aliphatic Aldoximes
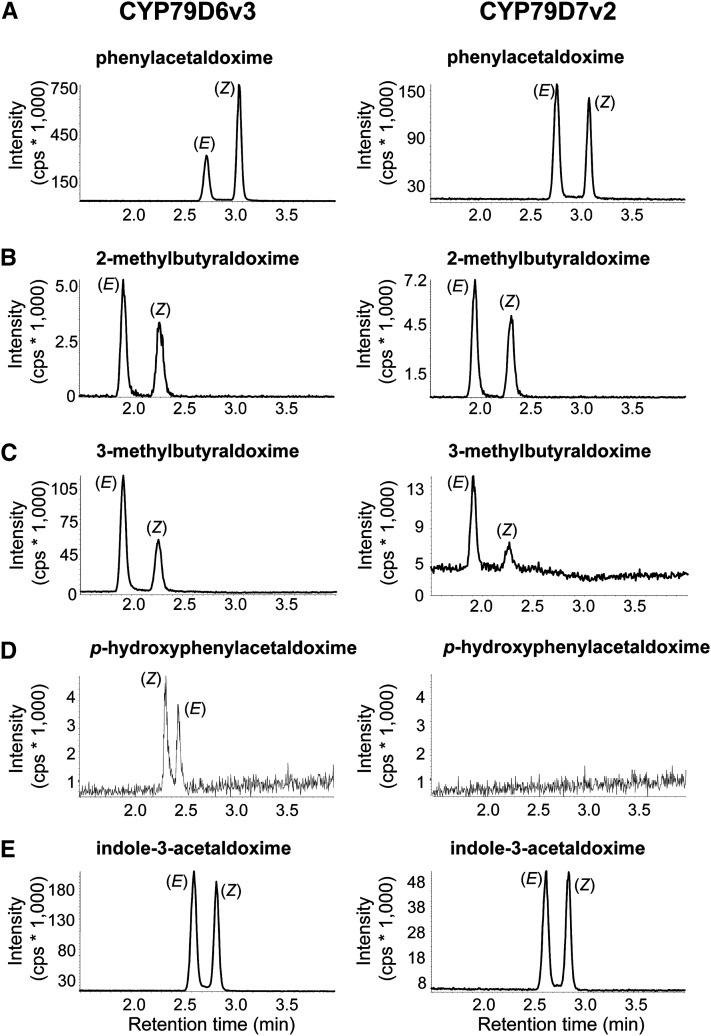
To investigate the biochemical properties of CYP79D6v3 and CYP79D7v2, the enzymes were heterologously expressed in yeast (Saccharomyces cerevisiae) coexpressing the Arabidopsis cytochrome P450 reductase 1 (S. cerevisiae strain WAT11; Pompon et al., 1996). Prepared microsomes were incubated with the cosubstrate NADPH and the potential amino acid substrates l-Phe, l-Leu, l-Ile, l-Trp, l-Val, and l-Tyr. Products were detected using liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis and verified by the use of authentic standards obtained as described in Methods. CYP79D6v3 and CYP79D7v2 were able to accept l-Phe, l-Leu, l-Ile, and l-Trp as substrates and converted them to the (E)- and (Z)-isomers of phenylacetaldoxime, 3-methylbutyraldoxime, 2-methylbutyraldoxime, and indole-3-acetaldoxime, respectively (Figure 3). l-Tyr was only accepted by CYP79D6v3 and was converted to p-hydroxyphenylacetaldoxime. No enzyme activity could be detected when l-Val was used as substrate for both CYP79D6v3 and CYP79D7v2. Assays containing microsomes of an empty vector expression as well as assays performed without NADPH showed no activity (see Supplemental Figure 4 online). Although both enzymes mostly accepted the same substrates, they differed in their substrate affinity (Table 1). The Km values for l-Leu and l-Ile were similar for both enzymes, but CYP79D6v3 showed a better affinity for l-Phe (Km = 744 µM) than for l-Trp (Km = 1427 µM), whereas CYP79D7v2 clearly preferred l-Trp (Km = 285 µM) over the other amino acids. Since measurements of carbon monoxide difference spectra were not successful, we could not determine the protein concentration in the microsomes and were therefore not able to calculate kcat values. Instead, the maximal velocity (Vmax) after separate assays with each amino acid at 1 mM was measured. With two exceptions, the results showed that the Vmax values of aldoximes formed by the enzymes reflected their Km values, indicating similar turnover numbers for most of the different amino acid substrates (see Supplemental Table 1 online). The efficient conversion of l-Phe into phenylacetaldoxime by CYP79D6v3 suggested a higher turnover number for l-Phe than for the aliphatic amino acids. For CYP79D7v2, the less efficient formation of indole-3-acetaldoxime indicated a lower Vmax for l-Trp than for the other amino acid substrates despite the low Km value for this substrate. Another striking difference between CYP79D6v3 and CYP79D7v2 was observed in their isomeric specificity. While both enzymes converted l-Leu and l-Ile mainly to the (E)-isomers of the respective aldoximes and l-Trp to an equal mixture of (E)- and (Z)-indole-3-acetaldoxime (Figures 3B, 3C, and 3E), CYP79D6v3 produced mainly (Z)-phenylacetaldoxime, whereas CYP79D7v2 formed both E and Z isomers in equal amounts (Figure 3A).
Figure 3.
Catalytic Activity of CYP79D6v3 and CYP79D7v2.
Yeast microsomes containing heterologously expressed enzymes were prepared and incubated with the substrates l-Phe (A), l-Ile (B), l-Leu (C), l-Tyr (D), and l-Trp (E). Products were detected using LC-MS/MS analysis with MRM in the positive mode. Diagnostic reactions for each product: phenylacetaldoxime m/z 136.0/119.0 (A); 2-methylbutyraldoxime m/z 102.0/69.0 (B); 3-methylbutyraldoxime m/z 102.0/46.0 (C); p-hydroxyphenylacetaldoxime m/z 152.0/107.0 (D); indole-3-acetaldoxime m/z 175.0/158.0 (E). cps, counts per second.
Table 1. Substrate Affinity (Km Values) of CYP79D6v3 and CYP79D7v2.
| Amino Acid Substrate | CYP79D6v3 (µM) | CYP79D7v2 (µM) |
|---|---|---|
| l-Phe | 744 ± 83 | 2901 ± 300 |
| l-Ile | 526 ± 52 | 633 ± 39 |
| l-Leu | 447 ± 29 | 851 ± 115 |
| l-Trp | 1427 ± 134 | 285 ± 39 |
| l-Tyr | 1828 ± 181 | – |
Enzymes were heterologously expressed in S. cerevisiae, and prepared microsomes were incubated with different concentrations of l-Phe, l-Ile, l-Leu, l-Trp, and l-Tyr. Products were detected using LC-MS/MS analysis, and Km values were calculated. Means and se are shown (n = 3). –, No product detected.
N. benthamiana Plants Expressing CYP79 Genes from P. trichocarpa Produce Complex Mixtures of Nitrogenous and Non-Nitrogenous Volatiles
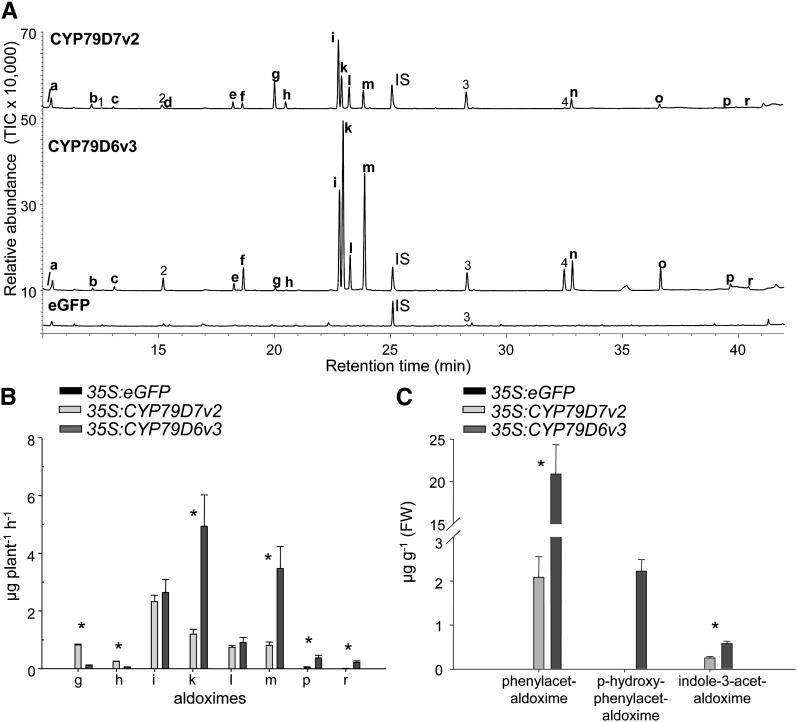
To verify the biochemical properties of CYP79D6v3 and CYP79D7v2 in vivo, the genes were transiently expressed in N. benthamiana. Agrobacterium tumefaciens transformed with CYP79D6v3, CYP79D7v2, or the gene for the enhanced green fluorescent protein (eGFP) were used to infiltrate N. benthamiana plants. In all transformations, a construct encoding the silencing suppressor protein p19 (Voinnet et al., 2003) was coinfiltrated to increase expression. Control plants expressing eGFP showed bright fluorescence 3 d after Agrobacterium infiltration. Thus, a volatile collection of control plants (35S:eGFP) and CYP79-overexpressing plants (35S:CYP79D6v3 and 35S:CYP79D7v2) was conducted on the third day after infiltration, and the volatile blends were analyzed using gas chromatography-mass spectrometry (GC-MS) and gas chromatography-flame ionization detector (GC-FID). Plants overexpressing CYP79D6v3 and CYP79D7v2 emitted complex mixtures of nitrogenous volatiles with (E/Z)-3-methylbutyraldoxime and (E/Z)-2-methylbutyraldoxime, respectively, as main components (Figures 4A and 4B; see Supplemental Table 2 online). Interestingly both volatile blends also contained (E)- and (Z)-isobutyraldoxime, suggesting that l-Val was accepted as substrate in vivo (Figures 4A and 4B). In addition to the aldoximes, CYP79-overexpressing plants also released different nitriles, nitro compounds, and related alcohols (Figure 4A; see Supplemental Table 2 online), indicating a further conversion of the aldoximes in planta. In contrast with the CYP79-overexpressing plants, control plants expressing eGFP produced none of the above-mentioned volatiles (Figures 4A and 4B; see Supplemental Table 2 online).
Figure 4.
Volatiles, Volatile Aldoximes, and Nonvolatile Aldoximes Produced by Transgenic N. benthamiana Plants Overexpressing P. trichocarpa CYP79 Genes.
Plants were infiltrated with Agrobacterium containing 35S:eGFP; 35S:CYP79D6v3, or 35S:CYP79D7v2.
(A) and (B) Volatiles were collected on the third day after infiltration. Identification of compounds was done with GC-MS, and total ion chromatograms are depicted in (A). Quantification was done with GC-FID, and results are depicted in (B). IS, internal standard; TIC, total ion chromatogram.
(C) Nonvolatile aldoximes were extracted with methanol and analyzed using LC-MS/MS. Nitrogen-containing volatiles: a, isobutyronitrile; b, 2-methylbutyronitrile; c, 3-methylbutyronitrile; d, 2-methyl-1-nitropropane; e, 1-nitro-2-methylbutane; f, 1-nitro-3-methylbutane; g, (E)-isobutyraldoxime; h, (Z)-isobutyraldoxime; i, (E)-2-methylbutyraldoxime; k, (E)-3-methylbutyraldoxime; l, (Z)-2-methylbutyraldoxime; m, (Z)-3-methylbutyraldoxime; n, benzyl cyanide; o, 2-phenylnitroethane; p, (E)-phenylacetaldoxime; r, (Z)-phenylacetaldoxime. Other volatiles: 1, 2-methylpropanol; 2, 3/2-methyl-1-butanol; 3, 5-epi-aristolochene; 4, 2-phenylethanol. Means and se are shown (n = 5). The Student’s t test was used to analyze the data for significant differences between the two expressed enzymes, and asterisks indicate P < 0.05. FW, fresh weight.
To measure nonvolatile CYP79 enzyme products, methanol extracts of infiltrated N. benthamiana leaves were prepared and analyzed using LC-MS/MS. As expected, large amounts of the less volatile compound phenylacetaldoxime but also small amounts of the nonvolatile indole-3-acetaldoxime were found in the CYP79D6v3- and CYP79D7v2-overexpressing lines, while p-hydroxyphenylacetaldoxime could only be detected in the CYP79D6v3-overexpressing line (Figure 4C). Control plants expressing eGFP showed no accumulation of aldoximes (Figure 4C; see Supplemental Table 3 online). To test for significant differences between 35S:CYP79D6v3 and 35S:CYP79D7v2, a Student´s t test was used. The t and P values are given in Supplemental Tables 2 and 3 online.
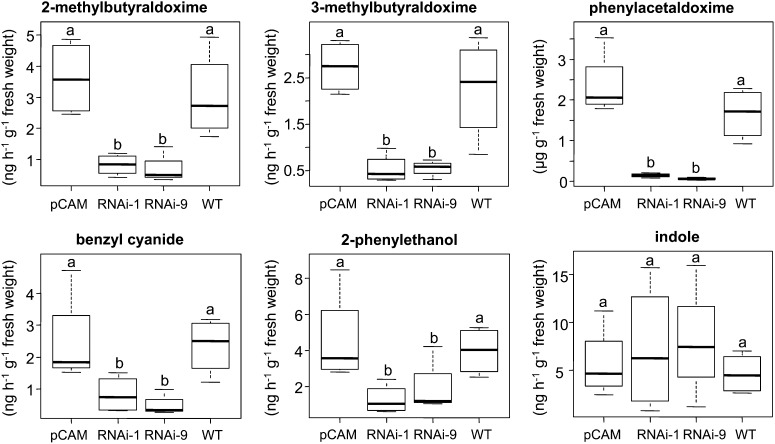
RNAi-Mediated Knockdown of CYP79D6 and CYP79D7 in P. × canescens Resulted in a Decreased Production of Aldoximes, Nitriles, and Alcohols
The biochemical characterization of CYP79D6v3 and CYP79D7v2 revealed broad substrate specificity, which can explain the occurrence of all aldoximes present in the herbivore-induced volatile blend of P. trichocarpa. Furthermore, overexpression of CYP79D6 and D7 in N. benthamiana suggested a further conversion of the produced aldoximes to other nitrogenous compounds as well as non-nitrogenous alcohols in vivo (see Supplemental Table 2 online). To test the role of CYP79D6 and D7 genes in planta, we created RNAi-mediated knockdown lines with reduced levels of CYP79D6v3 and CYP79D7v2 transcripts after wounding (see Supplemental Figure 5 online). Since there is no established transformation method for P. trichocarpa, we produced these lines in a P. × canescens background, the hybrid usually used for poplar transformation (Leplé et al., 1992). A volatile collection made from herbivore-damaged trees of the two independent CYP79D6 and D7 knockdown lines, a line transformed with the empty vector, and the P. × canescens wild type showed a significant reduction of aldoximes, benzyl cyanide, and 2-phenylethanol emission in the knockdown lines in comparison to the controls, while the emission of indole, a nitrogenous volatile not related to the above-mentioned compounds, was not affected (Figure 5; see Supplemental Table 4 online). In addition the amount of accumulated phenylacetaldoxime was significantly reduced in CYP79-RNAi lines (Figure 5; see Supplemental Table 4 online). Generally, fewer nitrogenous compounds could be detected in P. × canescens trees compared with P. trichocarpa. To test for significance, data were analyzed by one-way analysis of variance (ANOVA). Since there was no significant difference between the controls and between the two RNAi lines, a factor level reduction was performed. F-values and P values are shown in Supplemental Table 4 online.
Figure 5.
Volatile Emission and Phenylacetaldoxime Accumulation of CYP79 Knockdown and Control Lines in the P. × canescens Background.
Transgenic trees resulting from two different CYP79-RNAi transformation events (RNAi-1 and RNAi-9), trees transformed with an empty vector (pCAM), and nontransgenic trees (the wild type [WT]) were subjected to gypsy moth feeding, and volatiles were analyzed using GC-MS/FID. The less volatile phenylacetaldoxime was extracted with methanol and analyzed using LC-MS/MS. Means and se are shown (n = 4). Different letters above each box indicate significant differences (P < 0.05).
Gene Expression, Product Formation, and Amino Acid Substrate Availability for CYP79D6v3 and CYP79D7v2 Are Upregulated in Herbivore-Damaged P. trichocarpa Leaves
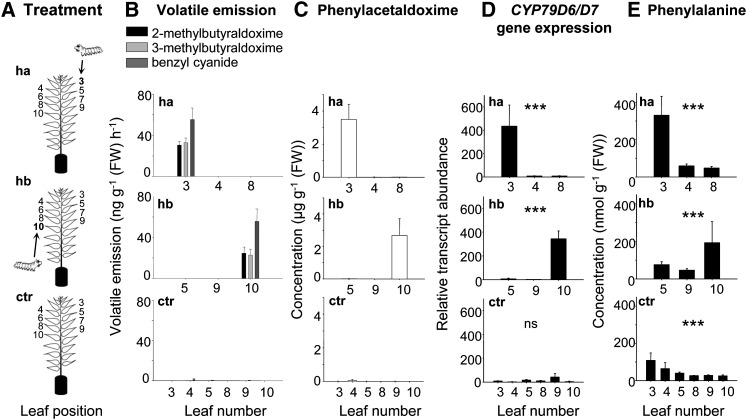
To investigate the spatial regulation of nitrogenous volatile emission within the tree, we collected volatiles from single leaves of undamaged control trees and trees that received herbivory on either one young apical leaf (leaf plastochron index [LPI] 3; Frost et al., 2007) or on a single older basal leaf (LPI 10; Figure 6A). Nitrogenous volatiles could only be detected in the headspace of the gypsy moth–damaged leaves independent from their apical or basal location in the tree (Figure 6B; see Supplemental Table 5 online). Neither neighboring leaves nor leaves with possible vascular connections to the damaged leaf (LPI 5 and 8, respectively; Frost et al., 2007) emitted nitrogenous compounds. Furthermore, LC-MS/MS analysis of methanol extracts made from single leaves revealed an accumulation of phenylacetaldoxime (E:Z, 1:1.8) in the herbivore-damaged leaves (Figure 6C). Trace amounts of indole-3-acetaldoxime, 2-methylbutyraldoxime, and 3-methylbutyraldoxime were also found in these leaf extracts.
Figure 6.
The Response of Single P. trichocarpa Leaves to Herbivory Restricted to an Apical or Basal Leaf.
(A) Single leaves were measured from either control trees (ctr), trees that received herbivory apically on leaf LPI 3 (ha, herbivory apical) or basally on leaf LPI 10 (hb, herbivory basal).
(B) to (E) The release of 2- and 3-methylbutyraldoxime and benzyl cyanide (B), the accumulation of phenylacetaldoxime (C), the gene expression of CYP79D6/7 (D), and the concentration of Phe (E) are shown for the treated leaf (LPI 3 or 10), its nearest neighbor (LPI 4 and 9, respectively), possible vascularly connected leaves (leaf LPI 8 and 5, respectively), and the corresponding controls. Leaves were numbered according to the LPI described by Frost et al. (2007). Volatiles were collected from single leaves and analyzed using GC-MS and GC-FID. Phenylacetaldoxime and Phe were extracted with methanol and analyzed by LC-MS/MS. Gene expression was determined by qRT-PCR. Means and se are shown (n = 5). Asterisks in (D) and (E) indicate statistical significance in likelihood ratio tests performed after mixed effect models with “leaf number” as an explanatory term. ns, non significant; FW, fresh weight. Gene expression: herbivory apical (P < 0.0001, F = 23.617), herbivory basal (P < 0.0001, F = 28.966), and control (P = 0.25, F = 6.607); Phe: herbivory apical (P = 0.0007, F = 14.477), herbivory basal (P = 0.082, F = 4.999), and control (P < 0.0001, F = 29.285).
In addition to the analysis of nitrogenous compounds, the expression patterns of CYP79D6v3 and CYP79D7v2 were analyzed in the same tissues. Both genes were strongly upregulated in damaged leaves (quantification cycle (Cq), 23.3 ± 0.5) in comparison to undamaged control leaves (Cq, 30.1 ± 0.4) (gene expression herbivory apical, P < 0.001, t = −6.8; herbivory basal, P = 0.001, t = −5.028; Figure 6D). Moreover, the high Cq values for control leaves indicated only trace expression in undamaged leaves. As CYP79D6v3 and CYP79D7v2 share 98% nucleotide identity, it was not possible to distinguish between the two genes in the quantitative RT-PCR (qRT-PCR) analysis. However, repeated sequencing of qRT-PCR fragments revealed that 79% ± 6% of the amplified fragments were assignable to CYP79D6v3, indicating higher transcript abundance for CYP79D6v3 in comparison to CYP79D7v2.
As l-Phe, l-Ile, l-Leu, l-Val, l-Trp, and l-Tyr serve as substrates for the poplar CYP79s, we additionally measured the concentrations of these amino acids in single leaves (Figure 6E; see Supplemental Tables 6 and 7 online). For all of these amino acids, a significant increase or a trend for elevated levels was found in the caterpillar-damaged LPI 3 leaf and/or LPI 10 leaf (Figure 6E; see Supplemental Table 7 online) in comparison to the corresponding undamaged leaf of undamaged control or apical/basal damaged trees. In general, there was even a trend for elevated levels of l-Phe, l-Trp, l-Tyr, l-Leu, l-Ile, and l-Val in the herbivore-treated leaves compared with noninfested leaves on the same tree (see Supplemental Tables 6 and 7 online).
Phenylacetaldoxime Reduces Herbivore Growth and Survival
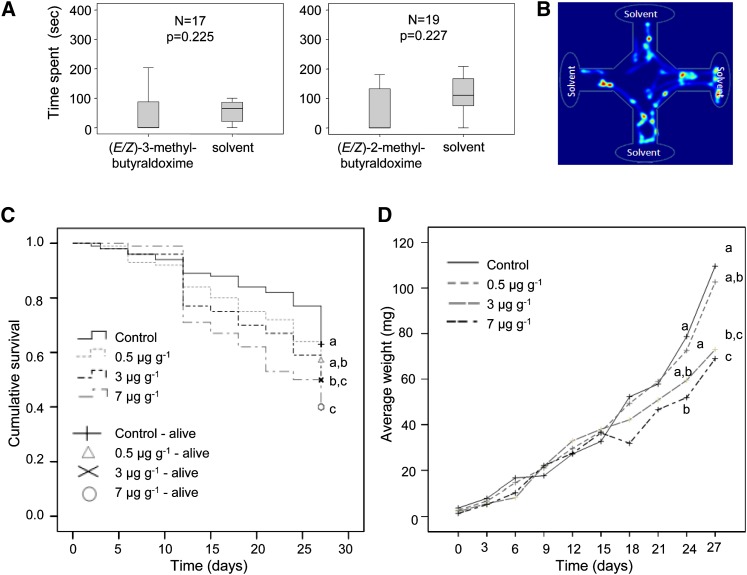
To investigate the biological activity of the herbivore-induced aldoximes against a poplar herbivore, the more volatile methylbutyraldoximes were tested in an olfactometer with gypsy moth caterpillars, while the less volatile phenylacetaldoxime was added to artificial diet upon which caterpillars were raised. Isomeric mixtures of 2-methylbutyraldoxime (E:Z, 3:1) and 3-methylbutyraldoxime (E:Z, 2:1) offered in a four field olfactometer had no repellent or attractive effects on early instar gypsy moth caterpillars (Friedman test; Figure 7A). On the other hand, phenylacetaldoxime (E:Z, 1:1.5) offered in artificial wheat germ diet had negative effects on caterpillar performance. For concentrations of 3 and 7 μg g−1 fresh weight (natural concentrations in poplar), caterpillar survival was significantly lower than in the control treatment ([Kaplan Meier survival analysis followed by Breslow pairwise comparisons, χ2 = 13.607 and 5.020 and P = 0.000 and 0.025 for 7 and 3 µg g−1 (E/Z)-phenylacetaldoxime, respectively; Figure 7B]. Larval weight showed a progressive decrease when larvae were fed with diet containing 3 and 7 µg g−1 phenylacetaldoxime and was significantly different from the control at the last two measurement points (24 and 27 d) for the 7 µg g−1 concentration and the last time point 27 d) for the 3 µg g−1 concentration (one-way ANOVA followed by Tukey test; at 24 d, P = 0.029 for 7 µg g−1 versus control, and at 27 d, P = 0.01 and 0.02 for 3 and 7 µg g−1 versus control, respectively; Figure 7C). Furthermore, the time until pupation was significantly delayed in caterpillars exposed to the 7 µg g−1 concentration of phenylacetaldoxime (Kruskall-Wallis ANOVA followed by Dunn’s multiple comparisons Q = 2.778, P < 0.05) (Table 2). No significant differences were found between male and female pupal weights (one-way ANOVA for males; Kruskal-Wallis for females), and there was no effect of phenylacetaldoxime on the sex distribution (Pearson χ2 test of independence), although there was a trend toward more male-biased ratios for the 3 and 7 µg g−1 treatments (Table 2).
Figure 7.
Effect of Poplar Aldoximes on Gypsy Moth Larvae.
(A) Box whisker plots showing average time spent by second instar gypsy moth caterpillars in the fields of a four-arm olfactometer containing 2-methylbutyraldoxime (E:Z, 3:1) or 3-methylbutyraldoxime (E:Z, 2:1) dissolved in dichloromethane versus a dichloromethane control during 5 min of observation. Values for the control are an average of three arms. n = number of responding larvae out of 20 tested; P = asymptotic significance after a nonparametric Friedman Test. The error bars represent se.
(B) The heat map shows the movement of the larvae during a 5-min period when all arms contain dichloromethane. In this case, there is no behavioral preference/repellence.
(C) Cumulative survival of gypsy moth larvae for three different concentrations of phenylacetaldoxime (E:Z, 1:1.5) (0.5, 3, and 7 µg g−1 diet) and control for 10 measurements (3 d apart) during a 1-month period. Treatments with the same letter are not significantly different (Kaplan Meier survival analysis followed by Breslow-Generalized Wilcoxson test).
(D) Average larval weights (mg) for the same treatments and time points as in (B). Treatments with the same letter at the same time point are not significantly different (one-way ANOVA-Tukey test).
[See online article for color version of this figure.]
Table 2. Effect of Dietary (E/Z)-Phenylacetaldoxime on Adult Emergence, Pupal Weight, Days to Pupation, and Sex Ratios of Gypsy Moth.
| Parameters | Control |
0.5 µg g−1 |
3 µg g−1 |
7 µg g−1 |
||||
|---|---|---|---|---|---|---|---|---|
| Males | Females | Males | Females | Males | Females | Males | Females | |
| N | 14 | 13 | 17 | 11 | 12 | 3 | 11 | 4 |
| Pupal wt | 324.2 ± 30.5a | 1313.8 ± 69.2a | 332.3 ± 38.1a | 1101.6 ± 113.8a | 255.0 ± 24.7a | 1231.9 ± 150.6a | 304.6 ± 36.8a | 1373.1 ± 64.2a |
| Time to pupation | 39.6 ± 1.3a | 42.2 ± 0.8a | 44.5 ± 1.0a | 45.0 ± 1.1b | ||||
| Sex ratio (males/N) | 0.52a | 0.61a | 0.8a | 0.73a | ||||
Number of adults (N) per gender emerging from pupae for three different (E/Z)-phenylacetaldoxime concentrations (0.5, 3, and 7 µg g−1 diet) and control. Pupal weights in mg ± se. Time to pupation in days ± se. Statistical tests performed for each set were as follows: pupal weights were compared for each gender by one-way ANOVA for males and Kruskal-Wallis ANOVA for females; time until pupation was compared among treatments by Kruskal-Wallis followed by Dunn’s test; effect of the treatments on sex ratio was compared by Pearson-χ2 test of independence. Values with the same letter (superscript a or b) on the same row do not differ significantly.
DISCUSSION
We are exploring the hypothesis that aldoximes from the herbivore-induced volatile blend of black poplar function as important attractants for herbivore enemies that might help protect plants from further damage. Therefore, we have begun to investigate the biosynthesis of these volatile aldoximes in the model poplar species P. trichocarpa. Here, we have shown that P450 enzymes of the CYP79 family, previously demonstrated to convert amino acids to aldoximes as intermediates in cyanogenic glucoside and glucosinolate formation (Andersen et al., 2000; Wittstock and Halkier, 2000), are also responsible for volatile aldoxime biosynthesis in poplar and most likely in many other plant species.
CYP79D6v3 and CYP79D7v2 Have Broad Substrate Specificity and Produce a Mixture of Aldoximes
The CYP79s functioning in cyanogenic glucoside and glucosinolate biosynthesis are entry-level enzymes that usually possess high specificities for their amino acid substrates and so determine the specificity of the entire pathway (Koch et al., 1992; Andersen et al., 2000; Wittstock and Halkier, 2000; Forslund et al., 2004). By contrast, recombinant CYP79D6v3 and CYP79D7v2 from P. trichocarpa each accepted a variety of different amino acids as substrates, converting them into the corresponding aldoximes (Figure 3). Although both enzymes possessed high Km values ranging from ∼0.3 to 3 mM (Table 1), they were apparently able to accept all substrates in planta also since the various aldoxime products were detected both in herbivore-induced P. trichocarpa (Figures 1 and 6; see Supplemental Table 5 online) as well as in N. benthamiana overexpressing the two CYP79 genes (Figure 4; see Supplemental Tables 2 and 3 online). High Km values for amino acid substrates have been reported for other CYP79 enzymes (Koch et al., 1992; Halkier et al., 1995; Andersen et al., 2000; Forslund et al., 2004), and it has been suggested that low substrate affinity has evolved to avoid possible depletion of the free amino acid pool in plants (Andersen et al., 2000).
Like CYP79D6v3 and CYP79D7v2, other enzymes involved in volatile formation often show broad substrate or product specificities. Terpene synthases, for example, frequently produce many different products from a single substrate (e.g., Degenhardt et al., 2009), while methyltransferases and acyltransferases involved in the formation of volatile esters often accept several alcohols as substrates (Effmert et al., 2005; D’Auria, 2006). Thus, using a limited number of different enzymes, plants are able to produce complex volatile blends comprising dozens of compounds. This complexity might provide an evolutionary advantage for defense compounds since it is likely harder for herbivores or pathogens to adapt to a complex mixture than to a single compound (Gershenzon et al., 2012). Furthermore, the uniqueness of a volatile blend increases with its complexity, allowing complex blends to serve as more specific signals in plant–insect or plant–plant communication. Therefore, one can speculate that the broad substrate specificity of CYP79D6v3 and CYP79D7v2 evolved from a substrate-specific ancestor as a result of its recruitment for the biosynthesis of herbivore-induced volatiles.
CYP79D6v3- and CYP79D7v2-Produced Aldoximes Are Released Directly and Are Further Converted to Other Volatile Compounds
Beside aldoximes, herbivore-damaged P. trichocarpa was shown to release related nitrogenous volatiles, namely, 2-methylbutyronitrile, 3-methylbutyronitrile, benzyl cyanide, and 2-phenylnitroethane (Figure 1; see Supplemental Table 5 online). Since these nitriles and nitro compounds were also emitted from transgenic N. benthamiana plants overexpressing P. trichocarpa CYP79 genes (Figure 4; see Supplemental Tables 2 and 3 online) and the knockdown of CYP79D6v3 and CYP79D6v2 in P. × canescens resulted in a significantly reduced emission of aldoximes and benzyl cyanide (Figure 5; see Supplemental Table 4 online), these other nitrogenous volatiles are most likely derived from the aldoximes. This assumption is further supported by the similar ratios between the aldoximes and their respective conversion products emitted from N. benthamiana expressing the P. trichocarpa CYP79 genes (Figure 4; see Supplemental Tables 2 and 3 online). However, the mechanism of volatile nitrile and 2-phenylnitroethane formation in poplar and N. benthamiana is still unclear. In general, nitriles can be generated from their aldoxime precursors in three different ways. First, a spontaneous dehydration of aldoximes can occur, but this is rather unlikely in planta since harsh reaction conditions are usually required (Kato et al., 1999; Li et al., 2005). Second, aldoxime dehydratases widely distributed in microorganisms catalyze nitrile formation (Kato et al., 2000). However, so far, no aldoxime dehydratases are known in plants and a BLAST search revealed no homologous genes in the P. trichocarpa genome. The third and most likely mode of volatile nitrile formation in planta is the oxidation of aldoximes by P450 enzymes of the CYP71 or CYP736 family as described for the biosynthesis of cyanogenic glycosides and camalexin (Bak et al., 1998b; Nafisi et al., 2007; Takos et al., 2011). The P. trichocarpa genome contains 25 CYP71 genes and six CYP736 genes (Nelson et al., 2008), and microarray analysis revealed that several of these genes were upregulated after herbivore feeding (Ralph et al., 2006). Therefore, they might play a role in herbivore-induced volatile nitrile formation in poplar. Nitro compounds have been reported as intermediates and possible side products of glucosinolate and cyanogenic glycoside biosynthesis (Matsuo et al., 1972; Sibbesen et al., 1995). Starting from an oxime, nitro compounds are proposed to arise via a reactive aci-nitro intermediate and a prototropic shift (Matsuo et al., 1972). Sibbesen et al. (1995) reported that 1-nitro-2-p-hydroxyphenylethane can occur as a side product of the CYP79A1-catalyzed conversion of l-Tyr to (Z)-p-hydroxyphenylacetaldoxime. However, whether poplar nitro compounds are produced from CYP79s or other enzymes is still unknown.
In addition to nitrogenous volatiles, both herbivore-damaged P. trichocarpa trees as well as transgenic N. benthamiana plants overexpressing P. trichocarpa CYP79 genes released alcohols like 2-methylbutanol, 3-methylbutanol, and 2-phenylethanol (Figures 1 and 4; see Supplemental Tables 2 and 5 online). Moreover, decreased CYP79 gene expression in transgenic P. × canescens resulted in a significantly reduced emission of 2-phenylethanol (Figure 5; see Supplemental Table 4 online), suggesting that 2-phenylethanol and these other alcohols may be derived from their corresponding aldoximes or nitriles. Conversion of an aldoxime to an alcohol has been shown in the biosynthesis of tyrosol from p-hydroxyphenylacetaldoxime in white mustard (Sinapis alba) (Kindl and Schieffe, 1971). Furthermore, overexpression of the p-hydroxyphenylacetaldoxime–producing CYP79A1 from sorghum in tobacco and Arabidopsis plants resulted in the accumulation of the corresponding alcohol (Bak et al., 2000). Thus, in addition to being important herbivore-inducible volatiles themselves, aldoximes may be precursors of a variety of other volatile compounds.
Nitrogenous Volatiles Are Emitted Exclusively from Damaged Leaves and Their Formation Is Closely Regulated by CYP79 Gene Expression
In response to local herbivore feeding, plants frequently show a systemic induction of defense reactions in adjacent organs, such as the production of defensive proteins or the formation of volatiles (Heil and Ton, 2008; Howe and Jander, 2008). For example, feeding of forest tent caterpillars (Malacosoma disstria) on P. trichocarpa × deltoides resulted in local and systemic emission of a complex volatile blend comprising mainly mono- and sesquiterpenes, but also benzyl cyanide (Arimura et al., 2004). Here, we demonstrated that aldoximes and related nitrogen-containing volatiles in P. trichocarpa were only produced and released from herbivore-damaged leaves and not from undamaged neighboring leaves (Figures 6B and 6C; see Supplemental Table 5 online). In addition, expression of CYP79D6v3 and CYP79D7v2 was strongly upregulated in caterpillar-damaged leaves in comparison to neighboring leaves and undamaged controls (Figure 6D). This suggests that the formation of nitrogenous volatiles is closely regulated by the transcription of CYP79 genes. Transcriptional regulation of induced volatile biosynthesis has been described for terpenes and aromatic esters in other plant species (Schnee et al., 2006; Köllner et al., 2010). However, the elevated levels of amino acid concentrations in herbivore-damaged P. trichocarpa leaves (Figure 6E; see Supplemental Tables 6 and 7 online) might also contribute to induced formation of aldoximes by CYP79D6v3 and CYP79D7v2.
Poplar Aldoximes May Have Important Roles in Direct Defense against Herbivorous Caterpillars
Plant volatiles can be involved in direct defense against insect herbivores (for references, see Unsicker et al., 2009). For example, maize volatiles emitted from herbivore-damaged seedlings were shown to repel the corn leaf aphid (Rhopalosiphum maidis) (Bernasconi et al., 1998). However, in this study, we did not observe any repellence of early instar gypsy moth caterpillars by the major poplar volatile aldoximes (E/Z)-2-methylbutyraldoxime and (E/Z)-3-methylbutyraldoxime in the course of our olfactometer experiments (Figure 7A).
We also tested the effects on herbivores of (E/Z)-phenylacetaldoxime, which is emitted (Figure 1B) and accumulates in P. trichocarpa leaves after gypsy moth herbivory (Figure 6C). This less volatile aldoxime was tested in artificial diet for effects on gypsy moth caterpillar performance. At natural concentrations, (E/Z)-phenylacetaldoxime had pronounced negative effects on gypsy moth caterpillar survival, growth, and time until pupation (Figures 7B and 7C, Table 2). Aldoximes have been reported to be toxic to mammals, fungi, and microorganisms (Drumm et al., 1995; Bartosova et al., 2006), but their effect on insects was previously unclear. Our results suggest that aldoximes can now be added to the list of plant compounds that defend plants against herbivores.
Aldoximes have also been suggested to have a role in plant defense against pathogens (Møller, 2010). As (E/Z)-phenylacetaldoxime accumulates at the site of herbivory in P. trichocarpa (Figure 6C), it is conceivable that this metabolite also serves to protect damaged tissue against pathogen attack following insect herbivore damage. Further conversion of the accumulated aldoxime into other nonvolatile compounds could provide additional direct defenses against pathogens or herbivores in poplar.
The Production of Volatile Aldoximes by CYP79s Is Widespread in Angiosperms
The P. trichocarpa CYP79 enzymes represent CYP79s that can mediate volatile formation in plants, and many other enzymes of this family may also participate in the production of volatiles. A comprehensive survey of the plant volatile literature revealed that volatile aldoximes are emitted from a large number of plant species belonging to at least 15 different families of diverse plant orders (see Supplemental Table 8 online). Moreover, all angiosperm genomes sequenced so far possess putative CYP79 genes independent of their ability to produce glucosinolates, cyanogenic glycosides, hydroxynitrile glycosides, or camalexin (see Supplemental Table 9 online). Thus, floral and vegetative plant volatile aldoximes in general are likely to be produced by CYP79 enzymes in many plant taxa, and the study of these enzymes can help to understand the biosynthesis, regulation, and biological function of volatile aldoximes in various plant–insect or plant–pathogen interactions.
METHODS
Plant and Insect Material
Western balsam poplar (Populus trichocarpa) trees were propagated from monoclonal stem cuttings (clone 625, Nordwestdeutsche Forstliche Versuchsanstalt) and grown under summer conditions in a greenhouse (24°C, 60% relative humidity, 16-h/8-h light/dark cycle) in a 1:1 mixture of sand and soil (Klasmann potting substrate; Klasmann-Deilmann), until they reached ∼1 m in height. Wild-type and transgenic gray poplar (Populus × canescens) plants were amplified by micropropagation as described by Behnke et al. (2007). Saplings of ∼10-cm high were repotted to soil (Klasmann potting substrate) and propagated in a controlled environment chamber for 1 month (day, 22°C; night, 18°C; 65% relative humidity; 16-h/8-h light/dark cycle) before they were transferred to the greenhouse (day, 23 to 25°C; night, 19 to 23°C; 45 to 55% relative humidity; 16-h/8-h light/dark cycle).
Gypsy moth (Lymantria dispar) egg batches were kindly provided by Hannah Nadel, United States Department of Agriculture- Animal and Plant Health Inspection Service. After hatching, the caterpillars were reared on an artificial diet (gypsy moth diet; MP Biomedicals). Nicotiana benthamiana Domin plants were grown in soil (Klasmann potting substrate) and maintained in the greenhouse under the same conditions as described above.
Poplar Volatile Collection and Analysis
The caterpillar treatment and the volatile collections of whole trees were performed as previously described by Danner et al. (2011). To investigate whether induced volatile emission is restricted to the damaged leaf only, we conducted single-leaf volatile collections. Trees were infested with gypsy moth larvae on either an apical leaf (LPI 3; Frost et al., 2007) or a basal leaf (LPI 10), and volatiles were separately collected from leaves LPI 3 to LPI10. Five replicates each of LPI 3–treated trees, LPI 10–treated trees, and undamaged control trees were measured. Single leaves were enclosed with a polyethylene terephthalate bag (Bratschlauch; Toppits) by fixing the ends of the bags with cable binders. Five gypsy moth caterpillars in third to fourth instar starved for 12 h were released on the leaves. The caterpillars were fed with poplar leaves for 1 week prior to the onset of the experiment. Caterpillars were allowed to feed for 24 h (16.00 to 16.00 h). Thereafter, volatiles were collected for 6 h during the middle of the next light period (10.00 to 16.00 h) using a push-pull system and a filter packed with 30 mg of SuperQ (Analytical Research Systems) as described by Danner et al. (2011). After collection, the volatiles were desorbed by eluting the filter twice with 100 μL of dichloromethane containing nonyl acetate as an internal standard (10 ng µL−1).
Qualitative and quantitative volatile analysis was conducted using an Agilent 6890 Series gas chromatograph (Agilent Technologies) coupled to an Agilent 5973 quadrupole mass selective detector (interface temperature, 270°C; quadrupole temperature, 150°C; source temperature, 230°C; electron energy, 70 eV) or a flame ionization detector operated at 300°C. The constituents of the volatile bouquet were separated using a polyethylene glycol (ZB-WAX) column (Phenomenex; 60 m × 0.25 mm × 0.15 µm) and He (mass spectrometer) or H2 (flame ionization detector) as carrier gas. The sample (1 µL) was injected without split at an initial oven temperature of 40°C. The temperature was held for 2 min and then increased to 225°C with a gradient of 5°C min−1, held for another 2 min, and then further increased to 250°C with 100°C min−1 and a hold for 1 min.
Compounds were identified by comparison of retention times and mass spectra to those of authentic standards obtained from Fluka, Roth, Sigma-Aldrich, Bedoukian, and PIN Chemicals or by reference spectra in the Wiley and National Institute of Standards and Technology libraries. Aldoxime standards were synthesized as described below. The absolute amount of all compounds was determined based on their flame ionization detection peak area in relation to the area of the internal standard.
Plant Tissue Sampling, RNA Extraction, and Reverse Transcription
Poplar leaf material was harvested immediately after the volatile collection, flash-frozen with liquid nitrogen, and stored at −80°C until further processing. After grinding the frozen leaf material in liquid nitrogen to a fine powder, total RNA was isolated using an Invisorb Spin Plant RNA mini kit (Invitek) according to the manufacturer’s instructions. RNA concentration, purity, and quality were assessed using a spectrophotometer (NanoDrop 2000c; Thermo Scientific) and an Agilent 2100 bioanalyzer. Prior to cDNA synthesis, 0.75 µg of RNA was DNase treated using 1 μL of DNase (Fermentas). Single-stranded cDNA was prepared from the DNase-treated RNA using SuperScript III reverse transcriptase and oligo(dT12-18) primers (Invitrogen).
Identification and Isolation of CYP79 Genes
To identify putative poplar CYP79 genes, a BLAST search against the poplar genome database (http://www.phytozome.net/poplar) was conducted using the amino acid sequence of CYP79A1 from sorghum (Sorghum bicolor) (GenBank Q43135) as input sequence. Four sequences representing putative P450 enzymes within the CYP79 family were identified. Two of these sequences could be amplified from cDNA attained from herbivore-induced leaves of poplar. Primer sequence information is available in Supplemental Table 9 online. PCR products were cloned into the sequencing vector pCR-Blunt II-TOPO (Invitrogen), and both strands were fully sequenced using the Sanger method.
Heterologous Expression of CYP79D6v3 and CYP79D7v2 in Saccharomyces cerevisiae
The complete open reading frames of CYP79D6v3 and CYP79D7v2 were cloned into the pESC-Leu2d vector (Ro et al., 2008) as NotI-SacI fragments. For expression, the resulting constructs were transferred into the S. cerevisiae strain WAT11 (Pompon et al., 1996). For gene expression, a single yeast colony was picked to inoculate a starting culture that contained 30 mL of synthetic complete minimal medium lacking Leu (6.7 g L−1 yeast nitrogen base without amino acids, but with ammonium sulfate). Other components included 100 mg L−1 of l-adenine, l-Arg, l-Cys, l-Lys, l-Thr, l-Trp, and uracil; 50 mg L−1 of the amino acids l-Asp, l-His, l-Ile, l-Met, l-Phe, l-Pro, l-Ser, l-Tyr, l-Val; 20 g L−1 d-Glc. The culture was grown overnight at 28°C and 180 rpm. One OD of this culture (∼2 × 107 cells mL−1) was used to inoculate 100 mL of yeast-peptone-Glc-agar full medium (10 g L−1 yeast extract, 20 g L−1 bactopeptone, 74 mg L−1 adenine hemisulfate, and 20 g L−1 d-Glc), which was grown for 32 to 35 h (until OD ∼5), induced by the addition of Gal, and cultured for another 15 to 18 h. Cells were harvested and yeast microsomes were isolated according to the procedures described by Pompon et al. (1996) and Urban et al. (1994) with minor modifications. Briefly, the cultures were centrifuged (7500g, 10 min, 4°C), the supernatant was decanted, and the pellet was resuspended in 30 mL of TEK buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 100 mM KCl) and centrifuged again. Then, the cell pellet was carefully resuspended in 2 mL of TES buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 600 mM sorbitol, 10 g L−1 bovine serum fraction V protein, and 1.5 mM β-mercaptoethanol) and transferred to a 50-mL conical tube. Glass beads (0.45- to 0.50-mm diameter; Sigma-Aldrich Chemicals) were added so that they filled the full volume of the cell suspension. Yeast cell walls were disrupted by five cycles of 1 min of shaking by hand and subsequent cooling down on ice for 1 min. The crude extract was recovered by washing the glass beads four times with 5 mL of TES. The combined washing fractions were centrifuged (7500g, 10 min, 4°C), and the supernatant was transferred in another tube and centrifuged again (100,000g, 60 min, 4°C). The resulting microsomal protein fraction was homogenized in 2 mL of TEG buffer (50 mM Tris-HCl, 1 mM EDTA, and 30% [w/v] glycerol) using a glass homogenizer (Potter-Elvehjem, Fisher Scientific). Aliquots were stored at −20°C and were used for protein assays.
Vector Construction and Transformation of P. × canescens
The construction of the binary vector was described by Levée et al. (2009). The transformation of the P. × canescens clone INRA 7171-B4 followed a protocol published by Meilan and Ma (2006). To target only CYP79 mRNA, a fragment between position 310 and 534 of the coding sequence was selected. Transgenic RNAi poplar plants were amplified by micropropagation as described by Behnke et al. (2007). To test the level of transgenicity, qRT-PCR analysis was done on wild-type, vector control (pCAM), and RNAi plants (line 1 and line 9) before and after wounding. Plants were wounded with a razor blade in the evening and again on the following morning (day 2). Plants were harvested in the late afternoon of day 2.
Analysis of Recombinant CYP79
To determine the substrate specificity of CYP79D6v3 and CYP79D7v2, yeast microsomes harboring recombinant protein were incubated for 30 min at 25°C and 300 rpm individually with the potential substrates l-Phe, l-Val, l-Leu, l-Ile, l-Tyr, and l-Trp in glass vials containing 300 μL of the reaction mixture (75 mM sodium phosphate buffer, pH 7.0, 1 mM substrate [concentration was variable for Km determination], 1 mM NADPH, and 10 μL of the prepared microsomes). Reaction mixtures containing boiled microsomes, no NADPH, or microsomes prepared from WAT11 transformed with the empty vector served as negative controls. Reaction products where analyzed using LC-MS/MS as described below.
For the determination of the Km values, assays were performed as triplicates and stopped by placing on ice after 300 μL of methanol were added. Enzyme concentrations and incubation times were chosen so that the reaction velocity was linear during the incubation time period.
qRT-PCR Analysis of CYP79D6 and D7 Expression
cDNA was prepared as described above and diluted 1:3 with water. For the amplification of the CYP79D6 gene fragments with a length of ∼150 bp, a primer pair was designed having a Tm ≥ 58°C, a GC content between 35 and 55%, and a primer length in the range of 20 to 25 nucleotides (see Supplemental Table 9 online for primer information). The primer recognized CYP79D7 as well as CYP79D6 (98% nucleotide identity), but no other genes. Primer specificity was confirmed by agarose gel electrophoresis, melting curve analysis, and standard curve analysis and by sequence verification of cloned PCR amplicons. Ubiquitin was used as a reference gene (Ramírez-Carvajal et al., 2008). Samples were run in triplicate using Brilliant III SYBR Green QPCR Master Mix (Stratagene) with ROX as reference dye. The following PCR conditions were applied for all reactions: initial incubation at 95°C for 3 min followed by 40 cycles of amplification (95°C for 20 s and 60°C for 20 s). Plate reads were taken during the annealing and the extension steps of each cycle. Data for the melting curves were recorded at the end of cycling from 55 to 95°C.
All samples were run on the same PCR machine (Mx3000P; Agilent Technologies) in an optical 96-well plate. Five biological replicates were analyzed as triplicates in the qRT-PCR for each of the three treatments. Data for the relative quantity to calibrator average (dRn) were exported from the MXPro software. Sequencing of PCR products obtained from four biological replicates revealed the average percentage of transcripts of CYP79D6v3 versus CYP79D7v2.
Transient Expression of CYP79D6 and D7 Genes in N. benthamiana
For gene expression in N. benthamiana, the coding regions of CYP79D6v3 and CYP79D7v2 were cloned into the pCAMBiA2300U vector. After verification of the sequence integrity, pCAMBiA vectors carrying the CYP79D6v3, CYP79D7v2, or eGFP construct and the construct pBIN:p19 were separately transferred into Agrobacterium tumefaciens strain LBA4404. The pCAMBiA 2300U vector and vectors carrying eGFP and p19 were kindly provided by the group of D. Werck-Reichhart, Strasbourg, France. The transformation was confirmed by PCR. Five milliliters of an overnight culture (220 rpm, 28°C) was used to inoculate 50 mL of Luria-Bertani media (50 µg mL−1 kanamycin, 25 µg mL−1 rifampicin, and 25 µg mL−1 gentamicin) for overnight growth. The following day, the cultures were centrifuged (4000g, 5 min), and the cells were resuspended in infiltration buffer (10 mM MES, 10 mM MgCl2, and 100 µM acetosyringone, pH 5.6) to reach a final OD of 0.5. After shaking for at least 1 h at room temperature, the cultures carrying CYP79 or eGFP were mixed with an equal volume of cultures carrying pBIN:p19. Since p19 functions as a suppressor of silencing, it enhances the expression of the desired coexpressed protein in planta (Voinnet et al., 2003).
For transformation, 3- to 4-week-old N. benthamiana plants were dipped upside down in an Agrobacterium solution and vacuum was applied to infiltrate the leaves. Infiltrated plants were shaded with cotton tissue to protect them from direct irradiation. Volatiles were measured on the third day after transformation. For volatile collection, plants were separately placed in gas-tight 3-liter glass desiccators. Charcoal-filtered air was pumped into the desiccators at a flow rate of 2 L min−1, and the air left the desiccators through a filter packed with 30 mg of Porapaq Q (ARS). Volatiles were collected for 5 h (10 am to 3 pm). The volatile compounds were desorbed from the filters and analyzed by GC-MS and gas chromatography–flame ionization detection (GC-FID) as described above. Transfected leaves were labeled after infiltration, harvested after the volatile collection, ground in liquid nitrogen, and stored at −80°C until further analysis.
LC-MS/MS Analysis of Aldoximes and Amino Acids
For determining amino acid and aldoxime concentration, 100 mg of plant powder was extracted with 1 mL methanol. For the measurement of amino acids, the methanol extract was diluted 1:10 with water and spiked with 13C,15N-labeled amino acids (algal amino acids 13C,15N; Isotec) at a concentration of 10 µg of the mix per milliliter. The concentration of the individual, labeled amino acids in the mix had been previously determined by classical HPLC-fluorescence detection analysis after precolumn derivatization with o-phthaldialdehyde-mercaptoethanol using external standard curves made from standard mixtures (amino acid standard mix and Gln, Asn, and Trp; Fluka). Amino acids in the diluted methanol extract were directly analyzed by LC-MS/MS. The method described by Jander et al. (2004) was used with some modifications. Briefly, chromatography was performed on an Agilent 1200 HPLC system. Separation was achieved on a Zorbax Eclipse XDB-C18 column (50 × 4.6 mm, 1.8 µm; Agilent Technologies) with aqueous formic acid (0.05%) and acetonitrile employed as mobile phases A and B, respectively. The elution profile was 0 to 1 min, 97% A; 1 to 2.7 min, 3 to 100% B in A; 2.7 to 3 min 100% B; and 3.1 to 6 min 97% A. The mobile phase flow rate was 1.1 mL min−1, and the column temperature was maintained at 25°C. The liquid chromatography was coupled to an API 5000 tandem mass spectrometer (Applied Biosystems) equipped with a Turbospray ion source operated in positive ionization mode (ion spray voltage, 5500 eV; turbo gas temperature, 700°C; nebulizing gas, 70 p.s.i.; curtain gas, 35 p.s.i.; heating gas, 70 p.s.i.; collision gas, 2 p.s.i.). Multiple reaction monitoring (MRM) was used to monitor a parent ion → product ion reaction for each analyte. MRM was chosen as described by Jander et al. (2004), except for Arg (m/z 175 → 70) and Lys (m/z 147 → 84). Both Q1 and Q3 quadrupoles were maintained at unit resolution. Analyst 1.5 software (Applied Biosystems) was used for data acquisition and processing. Individual amino acids in the sample were quantified from corresponding peaks in the 13C,15N-labeled amino acid internal standard, except for Trp, which was quantified using 13C,15N-Phe applying a response factor of 0.42.
Aldoximes were measured from methanol extracts using the same LC-MS/MS system as described for amino acid analysis. Formic acid (0.2%) in water and acetonitrile were employed as mobile phases A and B, respectively, on a Zorbax Eclipse XDB-C18 column (50 × 4.6 mm, 1.8 µm; Agilent Technologies). The elution profile (gradient 1) was 0 to 0.5 min, 30% B; 0.5 to 3 min, 30 to 66% B; 3 to 3.1 min, 66 to 100% B; 3.1 to 4 min 100% B; and 4.1 to 6 min 30% B at a flow rate of 0.8 mL min−1 at 25°C. For the analysis of the more polar p-hydroxyphenylacetaldoxime, the gradient was modified as follows (gradient 2): 0 to 4 min, 10 to 70% B; 4 to 4.1 min, 70 to 100% B; 4.1 to 5 min 100% B; and 5.1 to 7 min, 10% B at a flow rate of 1.1 mL min−1. The API 5000 tandem mass spectrometer was operated in positive ionization mode (ion spray voltage, 5500 eV; turbo gas temperature, 700°C; nebulizing gas, 60 p.s.i.; curtain gas, 30 p.s.i.; heating gas, 50 p.s.i.; collision gas, 6 p.s.i.). MRM was used to monitor parent ion → product ion reactions for each analyte as follows: m/z 136.0 → 119.0 (collision energy [CE], 17 V; declustering potential [DP], 56 V) for phenylacetaldoxime; m/z 102.0 → 69.0 (CE, 13 V; DP, 31 V) for 2-methylbutyraldoxime; m/z 102.0 → 46.0 (CE, 15 V; DP, 31 V) for 3-methylbutyraldoxime; m/z 175.0 → 158.0 (CE, 17 V; DP, 56 V) for indole-3-acetaldoxime; and m/z 152.0 → 107.0 (CE, 27 V; DP, 100 V) for p-hydroxyphenylacetaldoxime. The concentration of aldoximes was determined using external standard curves made with authentic standards synthesized as described below.
Synthesis of Aldoximes
Aldoximes were synthesized by condensation of the respective aldehyde with hydroxylamine. Briefly, 10 mL of 50% NaOH was added drop-wise to a mixture of 50 mmol aldehyde, 55 mmol hydroxylamine hydrochloride, 12.5 mL of water, 2.5 mL of ethanol, and ∼20 g of ice. After stirring for 1 h, the mixture was extracted with 20 mL of diethylether, and the separated aqueous phase was adjusted to pH 6.0 with hydrochloric acid and extracted twice with 20 mL of diethylether. The two latter ether extracts were combined and dried over CaCl2, and the solvent was evaporated. Aldoximes were yielded from the residue by recrystallization with diluted ethanol (phenylacetaldoxime) or vacuum distillation (2-methylbutyraldoxime, 15 mbar, 73 to 74°C; 3-methylbutyraldoxime, 96 mbar, 99 to 102°C).
Indole-3-acetaldoxime was prepared in a two-step synthesis from Trp via indoleacetaldehyde. Following a protocol from Glawischnig et al. (2004), 50 mg of Trp was dissolved in 12 mL of a 1 M sodium carbonate solution that was saturated with mannitol and sodium chloride and adjusted to pH 10. Toluol (100 mL) was added and under constant stirring, 200 μL of a freshly prepared 0.5% NaOCl solution, saturated with sodium chloride, was pipetted to the reaction mixture every 2 min until a total of 6 mL was added. Toluol was replaced every 12 min, and the combined toluol extracts were dried on sodium sulfate overnight. Afterwards, 25 mL of a 100 mM hydroxylamine hydrochloride solution at pH 7.0 was added, and the mixture was stirred at room temperature for 1 h. The separated toluol phase was again dried on sodium sulfate and filtered, and the solvent was evaporated to yield solid indole-3-acetaldoxime. The product identity was verified by LC-MS/MS, GC-MS, and 1H-NMR. p-Hydroxyphenylacetaldoxime was kindly provided by B.L. Møller (University of Copenhagen).
NMR Analysis of Aldoximes
The samples were dissolved in 150 μL of acetone-d6 (99.96%; Deutero) and transferred into a 2.5-mm-diameter NMR tube for analysis. 1H-NMR spectra were recorded at 297K using an Avance NMR spectrometer operating at a proton resonance frequency of 500.13 MHz (Bruker-Biospin). The spectrometer was equipped with a Bruker TCI cryoprobe (5 mm). The residual signal of acetone-d6 at δH 2.04 was used as a chemical shift reference. Ratios of E/Z-isomers were determined by integrating signals of H(a), which shows characteristic chemical shifts depending on the configuration (see Supplemental Figure 6 online) (Karabatsos et al., 1963).
Sequence Analysis and Phylogenetic Tree Construction
An alignment of the poplar CYP79 enzymes and other characterized CYP79 enzymes was constructed and visualized using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html) and the ClustalW algorithm. For the estimation of a phylogenetic tree, we used the MUSCLE algorithm (gap open, −2.9; gap extend, 0; hydrophobicity multiplier, 1.5; clustering method, upgmb) implemented in MEGA5 (Tamura et al., 2011) to compute an amino acid alignment (see Supplemental Data Set 1 online). Based on the MUSCLE alignment, the tree was reconstructed with MEGA5 using a neighbor-joining algorithm (Poisson model). A bootstrap resampling analysis with 1000 replicates was performed to evaluate the tree topology. A second tree was computed based on the same MUSCLE alignment with a model-based phylogenetic analysis using Bayesian Markov chain Monte Carlo inference, consisting of four Markov chains, using MrBayes 3.1.2. The analysis was run for 106 generations, with sampling from the trees every 100 generations. The first 1000 generations were discarded as burn-in, and remaining trees were combined into a single summary tree. Posterior probability values given by the Bayesian phylogeny were included into the neighbor-joining tree created with MEGA5. Treeview (www.taxonomy.zoology.gla.ac.uk/rod/treeview.html) was used to visualize the tree.
Four-Armed Olfactometer Bioassays with Gypsy Moth Caterpillars
To test the behavior of gypsy moth in response to the volatile aldoximes (E/Z)-2-methylbutyraldoxime and (E/Z)-3-methylbutyraldoxime, we used a four-arm olfactometer (see Supplemental Figure 7 online), which was located inside a white box to avoid contact of the larvae with visual stimuli. A digital video camera was placed on top to record larval behavior. Incoming airflow of 0.2 L min−1 entered each of the four arms of the arena, and air was pumped out at the center of the arena at a flow of 0.8 L min−1. The odor samples were prepared using a concentration of 0.5 µg µL−1 of each compound diluted in dichloromethane, which after a GC-MS measurement corresponded approximately to the amounts observed in headspace collection from damaged plants (∼50 ng g−1 dry weight of leaf material). Five microliter of the odor sample was tested in each trial. The samples were dispensed onto filter paper disks and then placed inside tightly sealed 100-mL glass containers connected through Teflon tubing at the end of each arm, through which the air was blown in. An initial test was performed using solvent only (5 μL of dichloromethane) in each of the arms to exclude bias in the arena (Figure 7A). In all behavioral trials with caterpillars, three arms of the arena had a filter paper disk with 5 μL of solvent, and the fourth arm had a filter paper with the compound to be tested. All video clips were analyzed using the software EthoVision XT (Noldus Information Technology). Twenty second-instar caterpillars previously fed on commercially available wheat germ diet (gypsy moth diet; MP Biomedicals) were placed individually in the center of the arena and observed for 5 min. The time spent in each of the odor permeated arms of the arena was recorded. When no choice was made by the caterpillars within the observation period (5 min), the trial was not considered in data analysis.
Feeding Experiments with (E/Z)-Phenylacetaldoxime and Gypsy Moth Caterpillars
To test the impact of (E/Z)-phenylacetaldoxime on the performance of a generalist caterpillar, gypsy moth caterpillars were fed on artificial diet containing three different concentrations of (E/Z)-phenylacetaldoxime (0.5 µg g−1, 3 µg g−1, and 7 µg g−1 fresh weight wheat germ diet), and artificial diet without (E/Z)-phenylacetaldoxime was used as a control. Neonate caterpillars were transferred to Petri dishes (9-cm diameter) in groups of 10 with the respective diet. Each treatment was replicated 10 times resulting in an overall number of 400 caterpillars in the experiment. The concentrations of (E/Z)-phenylacetaldoxime offered in artificial diet resembled natural concentrations of this compound in planta. Caterpillars were allowed to feed ad libitum, and the diet was replaced every third day. During every diet change, the caterpillars were weighed with a fine balance and survival was checked. Both parameters were measured 10 times over a period of 1 month before the first pupation events occurred. At pupation, the pupae were separated and weighed, and the time until pupation was recorded. Adults were sexed right after emergence from the pupae.
Statistical Analysis
Whenever necessary, the data were log or arcsine transformed to meet statistical assumptions, such as normality and homogeneity of variances. Throughout the article, data are presented as means ± se. To compare N. benthamiana plants expressing 35S:CYP79D6v3 and 35S:CYP79D7v2 and the gene expression of CYP79 in herbivore-treated and control leaves, Student’s t tests were performed with SigmaPlot 11.0 for Windows (Systat Software). To compare amino acid concentrations and gene expression in single leaves of poplar, mixed-effects models (nlme package) with leaf number as fixed effect and plant identity as random effect were used, followed by a maximum likelihood ratio test (L and P values are shown in Supplemental Table 10 online). One-way ANOVAs were used to analyze the differences of RNAi and non-RNAi trees. To test whether amino acid concentrations changed due to direct herbivory (treatment), ANOVAs were applied for leaf LPI 3 and 10 separately. All were simplified by factor-level reduction (Crawley, 2002) and were done with R2.15.2 (R Development Core Team; http://www.R-project.org). To test the effects of volatile aldoximes on caterpillar behavior, a Friedman rank test was used comparing the average time spent on the three control fields versus the time spent in the field with the test odor. A Kaplan Meier survivorship analysis followed by Breslow-Generalized Wilcoxon test for treatment comparisons was applied to test for differences in caterpillar performance in response to different (E/Z)-phenylacetaldoxime concentrations. Differences in larval weight at individual time points were tested by ANOVA followed by a Tukey posthoc comparison. Pupal weights were compared for each gender through ANOVA (for males) and the Kruskal-Wallis test (for females). To compare time until pupation in the caterpillar feeding experiment, a Dunn’s test was performed following Kruskal-Wallis ANOVA. The effect of the treatments on the sex distribution was tested using a Pearson χ2 test of independence. For caterpillar behavior, survival and performance as well as pupation weights and sex ratio data were analyzed using SigmaPlot 11.0 for Windows (Systat Software).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CYP79D6v3 (KF562515), CYP79D7v2 (KF562516), CYP79D3 (AAT11920), CYP79D4 (AAT11921), CYP79D1 (AAF27289), CYP79D2 (AAF27290), CYP79A1 (Q43135), CYP79B3 (AEC07294), CYP79B1 (AAD03415), CYP79B2 (AEE87143), CYP79A2 (AAF70255), CYP79E1 (AF140609), CYP79E2 (AF140610), CYP79F2 (AAG24796), CYP79F1 (AEE29448), and CYP71E1 (AAC39318).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Chromosomal Location of the Three Poplar CYP79 Genes and Sequence Comparison of the CYP79 Promoter Regions.
Supplemental Figure 2. Phylogenetic Tree of CYP79 Enzymes from Poplar and Other Plant Species.
Supplemental Figure 3. Amino Acid Sequence Alignment of CYP79D6v3 and CYP79D7v2 with Representative CYP79 Sequences from Other Plants and CYP71E1 from Sorghum bicolor.
Supplemental Figure 4. Empty Vector Control and Assay without NADPH.
Supplemental Figure 5. Relative Abundance of mRNA Transcripts in Wild-Type, Vector Control, and RNAi Plants.
Supplemental Figure 6. Partial 1H-NMR Spectra of Aldoxime E/Z-Isomer Mixtures.
Supplemental Figure 7. Schematic Representation of the Four-Armed Olfactometer.
Supplemental Table 1. Product Accumulation of CYP79 Enzymes (ng Assay−1 h−1) after in Vitro Assay with Various Amino Acid Substrates.
Supplemental Table 2. Amounts of Volatiles Emitted from Transfected N. benthamiana Plants (ng Plant−1 h−1) Transformed with CYP79D6 and D7 Compared with Controls Transformed with eGFP.
Supplemental Table 3. Amounts of Aldoximes (ng g−1 Fresh Weight−1) in Transfected N. benthamiana Leaves Transformed with CYP79D6 and D7 Compared with Controls Transformed with eGFP.
Supplemental Table 4. Volatile Emission and Phenylacetaldoxime Accumulation of P. × canescens Trees with RNAi-Mediated Knockdown of CYP79D6 and D7 Genes.
Supplemental Table 5. Nitrogenous Volatiles Emitted from Single Leaves of Intact P. trichocarpa Trees (ng g−1 Fresh Weight h−1).
Supplemental Table 6. Amino Acid Concentrations in Single Leaves of Intact P. trichocarpa Trees (nmol g−1 Fresh Weight).
Supplemental Table 7. Statistical Analysis of Amino Acid Concentrations in Single Leaves of P. trichocarpa in Response to L. dispar Herbivory Restricted to a Single Leaf.
Supplemental Table 8. Plant Families Reported to Produce Volatile Aldoximes.
Supplemental Table 9. Number of CYP79 Genes in Sequenced Genomes of Angiosperms.
Supplemental Table 10. Oligonucleotides Used for Isolation, qRT-PCR Analysis of CYP79 Genes, and Construction of the Binary RNAi Vector.
Supplemental Data Set 1. Amino Acid Alignment Data.
Acknowledgments
We thank Grit Kunert for help with the statistics, Tamara Krügel and all the Max Planck Institute for Chemical Ecology gardeners for help with rearing the poplar trees, Marion Stäger for maintenance of transgenic poplars cultures, and Horst Stockhecke and Bernd Pogodda from Nordwestdeutsche Forstliche Versuchsanstalt (Münden, Germany) for kindly providing poplar stem cuttings. The research was funded by the Max Planck Society.
AUTHOR CONTRIBUTIONS
S.I., A.C.M., J.-P.S., J.G., S.B.U., and T.G.K. designed research. S.I., A.C.M., G.A.B., A.S., M.R., B.S., and K.B. carried out the experimental work. S.I., A.C.M., A.S., B.S., and S.B.U. analyzed data. S.I. and T.G.K. wrote the article. All authors read and approved the final article.
Glossary
- RNAi
RNA interference
- GC-MS
gas chromatography–mass spectrometry
- m/z
mass-to-charge ratio
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- ANOVA
analysis of variance
- LPI
leaf plastochron index
- qRT-PCR
quantitative RT-PCR
- GC-FID
gas chromatography–flame ionization detection
- MRM
multiple reaction monitoring
- CE
collision energy
- DP
declustering potential
References
- Andersen M.D., Busk P.K., Svendsen I., Møller B.L. (2000). Cytochromes P-450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. Cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J. Biol. Chem. 275: 1966–1975 [DOI] [PubMed] [Google Scholar]
- Arimura G., Huber D.P.W., Bohlmann J. (2004). Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa x deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (-)-germacrene D synthase, PtdTPS1. Plant J. 37: 603–616 [DOI] [PubMed] [Google Scholar]
- Aust C., Schweier J., Brodbeck F., Sauter U., Becker G., Schnitzler J.-P. (2013). Land availability and potential biomass production with poplar and willow short rotation coppices in Germany. GCB Bioenergy, 10.1111/gcbb.12083. [Google Scholar]
- Bak S., Kahn R.A., Nielsen H.L., Møller B.L., Halkier B.A. (1998b). Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol. Biol. 36: 393–405 [DOI] [PubMed] [Google Scholar]
- Bak S., Nielsen H.L., Halkier B.A. (1998a). The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acid to aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol. Biol. 38: 725–734 [DOI] [PubMed] [Google Scholar]
- Bak S., Olsen C.E., Halkier B.A., Møller B.L. (2000). Transgenic tobacco and Arabidopsis plants expressing the two multifunctional sorghum cytochrome P450 enzymes, CYP79A1 and CYP71E1, are cyanogenic and accumulate metabolites derived from intermediates in Dhurrin biosynthesis. Plant Physiol. 123: 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartosova L., Kuca K., Kunesova G., Jun D. (2006). The acute toxicity of acetylcholinesterase reactivators in mice in relation to their structure. Neurotox. Res. 9: 291–296 [DOI] [PubMed] [Google Scholar]
- Behnke K., Ehlting B., Teuber M., Bauerfeind M., Louis S., Hänsch R., Polle A., Bohlmann J., Schnitzler J.P. (2007). Transgenic, non-isoprene emitting poplars don’t like it hot. Plant J. 51: 485–499 [DOI] [PubMed] [Google Scholar]
- Beringer T., Lucht W., Schaphoff S. (2011). Bioenergy production potential of global biomass plantations under environmental and agricultural constraints. GCB Bioenergy 3: 299–312 [Google Scholar]
- Bernasconi M.L., Turlings T.C.J., Ambrosetti L., Bassetti P., Dorn S. (1998). Herbivore-induced emissions of maize volatiles repel the corn leaf aphid, Rhopalosiphum maidis. Entomol. Exp. Appl. 87: 133–142 [Google Scholar]
- Crawley, M.J. (2002). Statistical Computing. An Introduction to Data Analysis Using S-Plus. (Chichester, West Sussex, UK: John Wiley & Sons, Ltd.). [Google Scholar]
- Danner H., Boeckler G.A., Irmisch S., Yuan J.S., Chen F., Gershenzon J., Unsicker S.B., Köllner T.G. (2011). Four terpene synthases produce major compounds of the gypsy moth feeding-induced volatile blend of Populus trichocarpa. Phytochemistry 72: 897–908 [DOI] [PubMed] [Google Scholar]
- D’Auria J.C. (2006). Acyltransferases in plants: A good time to be BAHD. Curr. Opin. Plant Biol. 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Degenhardt J., Köllner T.G., Gershenzon J. (2009). Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637 [DOI] [PubMed] [Google Scholar]
- Drumm J.E., Adams J.B., Brown R.J., Campbell C.L., Erbes D.L., Hall W.T., Hartzell S.L., Holliday M.J., Kleier D.A., Martin M.J., Pember S.O., Ramsey G.R. (1995). Oxime fungicides: Highly-active broad-spectrum protectants. Synth. Chem. Agrochem. IV 584: 396–405 [Google Scholar]
- Du L.C., Halkier B.A. (1996). Isolation of a microsomal enzyme system involved in glucosinolate biosynthesis from seedlings of Tropaeolum majus L. Plant Physiol. 111: 831–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst F., Nelson D.R. (1995). Diversity and evolution of plant P450 and P450-reductases. Drug Metabol. Drug Interact. 12: 189–206 [DOI] [PubMed] [Google Scholar]
- Effmert U., Saschenbrecker S., Ross J., Negre F., Fraser C.M., Noel J.P., Dudareva N., Piechulla B. (2005). Floral benzenoid carboxyl methyltransferases: From in vitro to in planta function. Phytochemistry 66: 1211–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Morant M., Jørgensen B., Olsen C.E., Asamizu E., Sato S., Tabata S., Bak S. (2004). Biosynthesis of the nitrile glucosides rhodiocyanoside A and D and the cyanogenic glucosides lotaustralin and linamarin in Lotus japonicus. Plant Physiol. 135: 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost C.J., Appel H.M., Carlson J.E., De Moraes C.M., Mescher M.C., Schultz J.C. (2007). Within-plant signalling via volatiles overcomes vascular constraints on systemic signalling and primes responses against herbivores. Ecol. Lett. 10: 490–498 [DOI] [PubMed] [Google Scholar]
- Fürstenberg-Hägg J., Zagrobelny M., Bak S. (2013). Plant defense against insect herbivores. Int. J. Mol. Sci. 14: 10242–10297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon, J., Fontana, A., Burow, M., Wittstock, U., and Degenhardt, J.(2012). Mixtures of plant secondary metabolites: Metabolic origins and ecological benefits. In Ecology of Plant Secondary Metabolites: From Genes to Landscapes, G.R. Iason, M. Dicke and S.E. Hartley, eds (Cambridge, UK: Cambridge University Press), pp. 56–77. [Google Scholar]
- Glawischnig E., Hansen B.G., Olsen C.E., Halkier B.A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grootwassink J.W.D., Balsevich J.J., Kolenovsky A.D. (1990). Formation of sulfatoglucosides from exogenous aldoximes in plant-cell cultures and organs. Plant Sci. 66: 11–20 [Google Scholar]
- Halkier B.A., Nielsen H.L., Koch B., Møller B.L. (1995). Purification and characterization of recombinant cytochrome P450TYR expressed at high levels in Escherichia coli. Arch. Biochem. Biophys. 322: 369–377 [DOI] [PubMed] [Google Scholar]
- Hamberger B., Bak S. (2013). Plant P450s as versatile drivers for evolution of species-specific chemical diversity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M., Ton J. (2008). Long-distance signalling in plant defence. Trends Plant Sci. 13: 264–272 [DOI] [PubMed] [Google Scholar]
- Howe G.A., Jander G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59: 41–66 [DOI] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Joshi V., Fraga M., Rugg A., Yu S., Li L., Last R.L. (2004). Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J. 39: 465–475 [DOI] [PubMed] [Google Scholar]
- Kaiser R.A.J. (1993). On the scent of orchids. Bioactive volatile compounds from plants. Acs. Sym. Ser. 525: 240–268 [Google Scholar]
- Karabatsos G.J., Taller R.A., Vane F.M. (1963). Structural studies by nuclear magnetic resonance. 3. Syn-anti assignments from solvent effects. J. Am. Chem. Soc. 85: 2327–2328 [Google Scholar]
- Kato Y., Ooi R., Asano Y. (1999). A new enzymatic method of nitrile synthesis by Rhodococcus sp. strain YH3-3. J. Mol. Cat. B Enzymatic 6: 249–256 [Google Scholar]
- Kato Y., Ooi R., Asano Y. (2000). Distribution of aldoxime dehydratase in microorganisms. Appl. Environ. Microbiol. 66: 2290–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindl H., Schieffe S. (1971). Aldoximes as intermediates in biosynthesis of tyrosol and tyrosol derivatives. Phytochemistry 10: 1795–1802 [Google Scholar]
- Knudsen J.T., Eriksson R., Gershenzon J., Stahl B. (2006). Diversity and distribution of floral scent. Bot. Rev. 72: 1–120 [Google Scholar]
- Koch B., Nielsen V.S., Halkier B.A., Olsen C.E., Møller B.L. (1992). The biosynthesis of cyanogenic glucosides in seedlings of cassava (Manihot esculenta Crantz). Arch. Biochem. Biophys. 292: 141–150 [DOI] [PubMed] [Google Scholar]
- Köllner T.G., Lenk C., Zhao N., Seidl-Adams I., Gershenzon J., Chen F., Degenhardt J. (2010). Herbivore-induced SABATH methyltransferases of maize that methylate anthranilic acid using s-adenosyl-L-methionine. Plant Physiol. 153: 1795–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplé J.C., Brasileiro A.C.M., Michel M.F., Delmotte F., Jouanin L. (1992). Transgenic poplars: Expression of chimeric genes using four different constructs. Plant Cell Rep. 11: 137–141 [DOI] [PubMed] [Google Scholar]
- Levée V., Major I., Levasseur C., Tremblay L., MacKay J., Séguin A. (2009). Expression profiling and functional analysis of Populus WRKY23 reveals a regulatory role in defense. New Phytol. 184: 48–70 [DOI] [PubMed] [Google Scholar]
- Li D.M., Shi F., Guo S., Deng Y.Q. (2005). Highly efficient Beckmann rearrangement and dehydration of oximes. Tetrahedron Lett. 46: 671–674 [Google Scholar]
- Matsuo M., Kirkland D.F., Underhill E.W. (1972). 1-Nitro-2-phenylethane, a possible intermediate in the biosynthesis of benzylglucosinolate. Phytochemistry 11: 697–701 [Google Scholar]
- Meilan, R., and Ma, C. (2006). Populus. In Methods in Molecular Biology, Vol. 344: Agrobacterium Protocols, 2nd ed., K. Wang, ed (Totowa, NJ: Humana Press), pp. 143–152. [Google Scholar]
- Møller B.L., Conn E.E. (1980). The biosynthesis of cyanogenic glucosides in higher plants. Channeling of intermediates in dhurrin biosynthesis by a microsomal system from Sorghum bicolor (linn) Moench. J. Biol. Chem. 255: 3049–3056 [PubMed] [Google Scholar]
- Møller B.L. (2010). Plant science. Dynamic metabolons. Science 330: 1328–1329 [DOI] [PubMed] [Google Scholar]
- Nafisi M., Goregaoker S., Botanga C.J., Glawischnig E., Olsen C.E., Halkier B.A., Glazebrook J. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D., Ming R., Alam M., Schuler M. (2008). Comparison of cytochrome P450 genes from six plant genomes. Trop. Plant Biol. 1: 216–235 [Google Scholar]
- Nielsen J.S., Møller B.L. (2000). Cloning and expression of cytochrome P450 enzymes catalyzing the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of cyanogenic glucosides in Triglochin maritima. Plant Physiol. 122: 1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S., Müller A., Weiler E.W. (2006). Many roads lead to “auxin”: Of nitrilases, synthases, and amidases. Plant Biol. (Stuttg.) 8: 326–333 [DOI] [PubMed] [Google Scholar]
- Pompon D., Louerat B., Bronine A., Urban P. (1996). Yeast expression of animal and plant P450s in optimized redox environments. Method Enzymol. 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Raguso R.A. (2008). Wake up and smell the roses: The ecology and evolution of floral scent. Annu. Rev. Ecol. Evol. Syst. 39: 549–569 [Google Scholar]
- Ralph S., et al. (2006). Genomics of hybrid poplar (Populus trichocarpax deltoides) interacting with forest tent caterpillars (Malacosoma disstria): Normalized and full-length cDNA libraries, expressed sequence tags, and a cDNA microarray for the study of insect-induced defences in poplar. Mol. Ecol. 15: 1275–1297 [DOI] [PubMed] [Google Scholar]
- Ramírez-Carvajal G.A., Morse A.M., Davis J.M. (2008). Transcript profiles of the cytokinin response regulator gene family in Populus imply diverse roles in plant development. New Phytol. 177: 77–89 [DOI] [PubMed] [Google Scholar]
- Rauhut T., Glawischnig E. (2009). Evolution of camalexin and structurally related indolic compounds. Phytochemistry 70: 1638–1644 [DOI] [PubMed] [Google Scholar]
- Ro D.K., Ouellet M., Paradise E.M., Burd H., Eng D., Paddon C.J., Newman J.D., Keasling J.D. (2008). Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 8: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Motawia M.S., Olsen C.E., Møller B.L., Bak S. (2012). Biosynthesis of rhodiocyanosides in Lotus japonicus: Rhodiocyanoside A is synthesized from (Z)-2-methylbutanaloxime via 2-methyl-2-butenenitrile. Phytochemistry 77: 260–267 [DOI] [PubMed] [Google Scholar]
- Schnee C., Köllner T.G., Held M., Turlings T.C.J., Gershenzon J., Degenhardt J. (2006). The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc. Natl. Acad. Sci. USA 103: 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbesen O., Koch B., Halkier B.A., Møller B.L. (1995). Cytochrome P-450TYR is a multifunctional heme-thiolate enzyme catalyzing the conversion of L-tyrosine to p-hydroxyphenylacetaldehyde oxime in the biosynthesis of the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench. J. Biol. Chem. 270: 3506–3511 [DOI] [PubMed] [Google Scholar]
- Takabayashi J., Dicke M., Posthumus M. (1991). Variation in composition of predator-attracting allelochemicals emitted by herbivore-infested plants: Relative influence of plant and herbivore. Chemoecology 2: 1–6 [Google Scholar]
- Takabayashi J., Dicke M., Takahashi S., Posthumus M.A., Vanbeek T.A. (1994). Leaf age affects composition of herbivore-induced synomones and attraction of predatory mites. J. Chem. Ecol. 20: 373–386 [DOI] [PubMed] [Google Scholar]
- Takabayashi J., Takahashi S., Dicke M., Posthumus M.A. (1995). Developmental stage of herbivore Pseudaletia separata affects production of herbivore-induced synomone by corn plants. J. Chem. Ecol. 21: 273–287 [DOI] [PubMed] [Google Scholar]
- Takos A.M., Knudsen C., Lai D., Kannangara R., Mikkelsen L., Motawia M.S., Olsen C.E., Sato S., Tabata S., Jørgensen K., Møller B.L., Rook F. (2011). Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant J. 68: 273–286 [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G.A., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Unsicker S.B., Kunert G., Gershenzon J. (2009). Protective perfumes: The role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 12: 479–485 [DOI] [PubMed] [Google Scholar]
- Urban P., Werck-Reichhart D., Teutsch H.G., Durst F., Regnier S., Kazmaier M., Pompon D. (1994). Characterization of recombinant plant cinnamate 4-hydroxylase produced in yeast. Kinetic and spectral properties of the major plant P450 of the phenylpropanoid pathway. Eur. J. Biochem. 222: 843–850 [DOI] [PubMed] [Google Scholar]
- van den Boom C.E.M., van Beek T.A., Posthumus M.A., de Groot A., Dicke M. (2004). Qualitative and quantitative variation among volatile profiles induced by Tetranychus urticae feeding on plants from various families. J. Chem. Ecol. 30: 69–89 [DOI] [PubMed] [Google Scholar]
- Vergara R.C., Torres-Araneda A., Villagra D.A., Raguso R.A., Arroyo M.T.K., Villagra C.A. (2011). Are eavesdroppers multimodal? Sensory exploitation of floral signals by a non-native cockroach Blatta orientalis. Curr. Zool. 57: 162–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wei J.N., Zhu J.W., Kang L. (2006). Volatiles released from bean plants in response to agromyzid flies. Planta 224: 279–287 [DOI] [PubMed] [Google Scholar]
- Wittstock U., Halkier B.A. (2000). Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 275: 14659–14666 [DOI] [PubMed] [Google Scholar]
- Zeng-hui H., Di Y., Ying-bai S. (2004). Difference in volatiles of poplar induced by various damages. J. For. Res. 15: 280–282 [Google Scholar]
- Zhang A.J., Hartung J.S. (2005). Phenylacetaldehyde O-methyloxime: A volatile compound produced by grapefruit leaves infected with the citrus canker pathogen, Xanthomonas axonopodis pv. citri. J. Agric. Food Chem. 53: 5134–5137 [DOI] [PubMed] [Google Scholar]