An apparently simple trade-off in plant defense against bacterial pathogens and insect herbivores shows unexpected mechanistic complexity.
Abstract
Multicellular eukaryotic organisms are attacked by numerous parasites from diverse phyla, often simultaneously or sequentially. An outstanding question in these interactions is how hosts integrate signals induced by the attack of different parasites. We used a model system comprised of the plant host Arabidopsis thaliana, the hemibiotrophic bacterial phytopathogen Pseudomonas syringae, and herbivorous larvae of the moth Trichoplusia ni (cabbage looper) to characterize mechanisms involved in systemic-induced susceptibility (SIS) to T. ni herbivory caused by prior infection by virulent P. syringae. We uncovered a complex multilayered induction mechanism for SIS to herbivory. In this mechanism, antiherbivore defenses that depend on signaling via (1) the jasmonic acid–isoleucine conjugate (JA-Ile) and (2) other octadecanoids are suppressed by microbe-associated molecular pattern–triggered salicylic acid (SA) signaling and infection-triggered ethylene signaling, respectively. SIS to herbivory is, in turn, counteracted by a combination of the bacterial JA-Ile mimic coronatine and type III virulence-associated effectors. Our results show that SIS to herbivory involves more than antagonistic signaling between SA and JA-Ile and provide insight into the unexpectedly complex mechanisms behind a seemingly simple trade-off in plant defense against multiple enemies.
INTRODUCTION
In both natural and agricultural environments, plants are challenged by numerous pathogens and herbivores, making multiway interactions between plants, pathogens, and herbivorous insects the rule rather than the exception. Bacterial phytopathogens of the genus Pseudomonas can spread efficiently across landscapes via the water cycle or insect herbivores (Hirano and Upper, 2000; Morris et al., 2008; Stavrinides et al., 2009; Chung et al., 2013), and plants may therefore encounter them early in the growing season. Such early Pseudomonas infections affect subsequent plant–pathogen or plant–herbivore interactions, either compromising or aiding the response to a second attacker of similar or different identity (Cui et al., 2002, 2005; Fu and Dong, 2013). With only a few exceptions, most studies have focused on crosstalk that takes place locally in a pathogen-infected or partially consumed leaf, leaving questions about systemic crosstalk between defense response pathways largely unaddressed. However, systemic crosstalk is likely to be more important ecologically than local crosstalk because the entire plant rather than a single leaf is potentially involved in a change in susceptibility to attack. To study the underlying molecular mechanisms mediating the outcome of such complex interactions, we previously analyzed a three-way interaction system consisting of the model genetic plant Arabidopsis thaliana, the plant pathogen Pseudomonas syringae, and leaf-chewing caterpillars, Trichoplusia ni (Lepidoptera: Noctuidae) (Cui et al., 2002, 2005).
Plant defenses are regulated by a multilayered system that is shaped through coevolution of plants and their many enemies (Jones and Dangl, 2006). Plants recognize pathogens that have breached a first layer of constitutive barriers, such as the leaf cuticle, by the binding of conserved microbe-associated molecular pattern (MAMP) epitopes to pattern recognition receptors, which elicits pattern-triggered immunity (PTI). Examples of such epitopes are the synthetic peptides flg22 and elf26, which are synthetic 22- and 26-amino acid peptides derived from bacterial flagellin and elongation factor Tu, respectively (Zipfel et al., 2004, 2006). Pathogens have evolved arrays of effector proteins to suppress PTI at various stages of the process (Lindeberg et al., 2012). Bacteria such as P. syringae inject these effectors into host cells via the type III secretion system. In a subsequent layer of the plant immune system, intracellular surveillance proteins of the nucleotide-binding site-leucine-rich repeat family directly recognize pathogen-encoded effectors or their effects on host factors to elicit effector-triggered immunity (ETI) (Jones and Dangl, 2006).
Downstream of pathogen recognition, the plant immune system is regulated by a hormonal signaling network that for simplicity can be considered to consist of four main signaling sectors that interact to provide specificity to the defense response: (1) salicylic acid (SA)-, (2) jasmonic acid (JA)-, (3) ethylene (ET)-, and (4) PHYTOALEXIN DEFICIENT4 (PAD4)-mediated signaling (Tsuda et al., 2009). Dependent on the particular mode of attack and the activity of MAMPs, effectors, or other compounds that affect defense signaling, each pathogen generates a specific signal signature (de Vos et al., 2005). In the case of P. syringae, this signature is complex because all four main signaling sectors are induced, making the underlying mechanisms mediating crosstalk between them difficult to analyze (de Vos et al., 2005).
Defense against hemibiotrophic pathogens such as P. syringae is mediated primarily via SA signaling. SA production via the enzyme isochorismate synthase (ICS1; also referred to as SALICYLIC ACID INDUCTION DEFICIENT2 [SID2]) is stimulated by the lipase-like protein PAD4 (Wildermuth et al., 2001). Downstream SA responses are largely controlled by the transcriptional regulator NON-EXPRESSER OF PATHOGENESIS-RELATED GENES1 (NPR1) (Cao et al., 1994). SA-related signals also move systemically to immunize distal tissue in a process described as systemic acquired resistance (SAR) (Fu and Dong, 2013). Although SAR is generally triggered by effector recognition, it may also be induced by MAMPs (Mishina and Zeier, 2007).
Whereas the SA-signaling sector appears to be the main sector activated by pathogen attack, JA signaling is triggered by both herbivorous insects and necrotrophic fungal pathogens (Glazebrook, 2005). Even though JA is mainly active in an isoleucine-conjugated state (JA-Ile), other octadecanoid JA precursors and derivatives act as signaling molecules in their own right and have both overlapping and distinct roles (Stintzi et al., 2001; Taki et al., 2005; Bruce et al., 2008; Wang et al., 2008a). When triggered simultaneously, SA and JA-Ile signaling mostly counteract each other in a process referred to as SA/JA antagonism (Thaler et al., 2012). Some P. syringae strains exploit this antagonism by producing the JA-Ile–mimicking compound coronatine (COR), which weakens both local and systemic SA-mediated defenses (Zheng et al., 2012).
ET signaling functions in concert with JA-Ile or SA to activate particular branches of the defense network and repress others. When signaling in conjunction with JA-Ile, ET-mediated signaling results in the activation of ETHYLENE RESPONSE FACTOR family transcription factors, whereas JA-Ile signaling without the influence of ET leads to activation of MYC family transcription factors. The ETHYLENE RESPONSE FACTOR and MYC branches are mutually antagonistic, primarily activating antifungal and antiherbivore defenses, respectively (Lorenzo et al., 2004; Fernández-Calvo et al., 2011; Verhage et al., 2011). ET can also have antagonistic effects on SA signaling. For example, ET blocks SA production by repressing transcription of ICS1/SID2 (Chen et al., 2009). Indeed, during a virulent P. syringae infection, at least two type III effectors, AvrPto and AvrPtoB, induce ET signaling in tomato (Solanum lycopersicum), which enhances susceptibility to the bacterial pathogen (Lund et al., 1998; Cohn and Martin, 2005). By contrast, during ETI, when plant immunity is triggered by recognition of pathogen-encoded effectors, the ET-signaling sector functions synergistically with the SA and other signaling sectors to robustly activate antimicrobial responses (Tsuda et al., 2009).
PAD4 is involved in regulating at least three important aspects of the plant immune signaling network. First, in association with its signaling partner ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1), it regulates hormone-dependent and -independent signaling during PTI and ETI (Feys et al., 2001; Rustérucci et al., 2001; Rietz et al., 2011). Second, PAD4 stimulates SA production as part of a positive feedback loop that also includes ICS1/SID2 (Zhou et al., 1998; Jirage et al., 1999). Third, full production of ET after a plant is subjected to pathogen infection or other stresses relies on PAD4 (Heck et al., 2003; Mühlenbock et al., 2008). Regulation of these three processes is essential for the biosynthesis of sufficient antimicrobials during infection (Glazebrook et al., 1997).
In addition to the four main signaling sectors mediated by SA, JA-Ile, ET, and PAD4, the plant immune signaling network also receives inputs from other hormones, such as abscisic acid, gibberellic acid, brassinosteroid, cytokinin, and auxin, which have traditionally been studied in the context of plant growth and development and the response to abiotic stresses (Robert-Seilaniantz et al., 2011; Pieterse et al., 2012). Examples of P. syringae effectors that modify hormone signaling to suppress SA-mediated defenses are AvrPtoB, which modulates abscisic acid signaling (de Torres-Zabala et al., 2007), and AvrRpt2, which activates auxin signaling (Chen et al., 2007).
Here, we use the Arabidopsis–P. syringae–T. ni model system to investigate mechanisms behind systemic crosstalk in three-way plant pathogen–herbivore interactions. Previously, we found that infection of Arabidopsis lower rosette leaves with P. syringae pv maculicola (Psm) ES4326 triggers systemic-induced susceptibility (SIS) to herbivory by T. ni in the upper rosette leaves. Surprisingly, we found that SIS to herbivory is independent of SA signaling because SIS was still present in the SA-signaling mutant npr1-1 and in SA-deficient transgenic nahG plants (Cui et al., 2002). In addition, we found that elicitation of SIS to herbivory was counteracted by COR produced by P. syringae (Cui et al., 2005) or by ETI triggered by P. syringae expressing the type III effectors AvrRpt2 or AvrB (Cui et al., 2002). Because SIS to herbivory is counteracted by COR and does not rely on SA signaling, we concluded that P. syringae–elicited susceptibility to T. ni occurs independently of SA/JA antagonism. This was a surprising conclusion since the limited number of previous studies in this area pointed at an important role for SA/JA antagonism in regulating the outcomes of crosstalk in three-way plant pathogen–herbivore interactions (Felton et al., 1999; Thaler et al., 2010).
In this study, we show that plant infection with COR-deficient Psm ES4326 cfa6 leads to systemic transcriptional changes in the ET-, PAD4-, and EDS1-signaling sectors. Genetic analysis showed that ET signaling is both necessary and sufficient for pathogen-triggered SIS to herbivory. This is congruent with our previous results showing that SIS depends on PAD4 (Cui et al., 2002), as PAD4 is essential for boosting ET production after pathogen infection and other stresses (Heck et al., 2003; Mühlenbock et al., 2008). We propose a multilayered induction mechanism underlying SIS to herbivory. In the first layer, MAMPs trigger SIS to herbivory dependent on SA/JA antagonism, which can be neutralized by COR and the action of certain type III bacterial effectors that stimulate signaling through the JA-Ile receptor complex containing CORONATINE INSENSITIVE1 (COI1) (He et al., 2004). In a subsequent layer, other type III effectors trigger SIS to herbivory by a mechanism that relies on ET and interferes with octadecanoid-dependent but not COI1-dependent defenses. Together, the results provide insight into the unexpectedly complex mechanisms behind a seemingly simple trade-off in plant defense against multiple attackers.
RESULTS
Pathogen-Triggered SIS to T. ni Requires PAD4 but not SA or JA Signaling
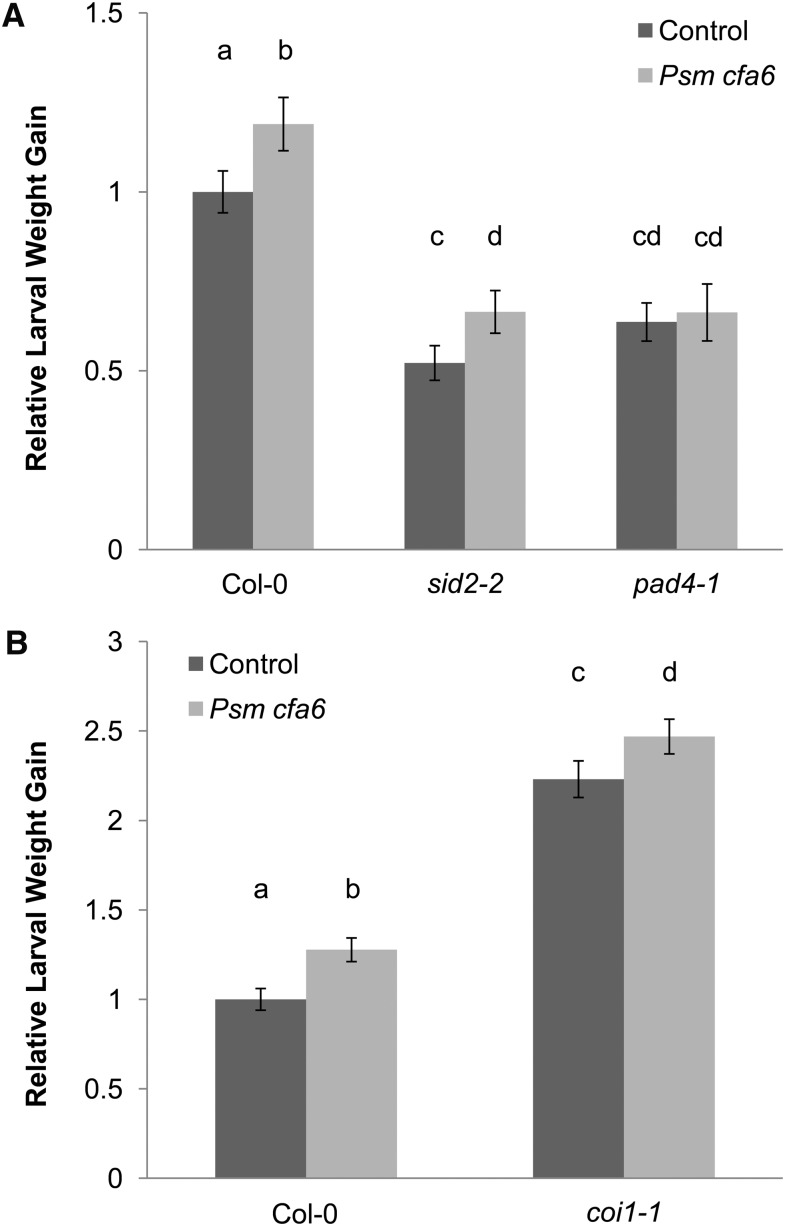
Even though their baseline resistance is greater than wild-type plants, pathogen-triggered SIS to T. ni herbivory is present in the nahG and npr1-1 plants, suggesting that SIS is independent of SA signaling (Cui et al., 2002). To collect additional data to test this hypothesis more thoroughly, we tested the sid2-2 mutant, which is deficient in SA biosynthesis. Since COR counteracts SIS to herbivory (Cui et al., 2005), we infiltrated sid2-2 plants with COR-deficient Psm ES4326 cfa6 to enhance the magnitude of the SIS phenotype. We found that infiltration of Psm ES4326 cfa6 in the sid2-2 mutant induced SIS to T. ni herbivory similar to wild-type plants (Figure 1A). By contrast, SIS was absent in a pad4-1 mutant, confirming our earlier finding that SIS to herbivory relies on signaling through PAD4, but not SA (Figure 1A).
Figure 1.
PAD4 Is Involved in Pathogen-Triggered SIS to Herbivory Independent of SA Production and the COI1 JA-Signaling Module.
Lower Arabidopsis rosette leaves were inoculated with a sterile 10 mM MgSO4 control solution (dark-gray bars) or Psm ES4326 cfa6 suspension (OD600 = 0.2; light-gray bars). Four days later, the inoculated leaves were removed and newly hatched T. ni larvae were placed on the upper rosette leaves. Larval weight gain was measured after 7 d of feeding. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on mock-inoculated wild-type (Col-0) plants. The bars represent the means (±se) of relative weight gain from two independent experiments with n = 36 larvae per treatment per experiment.
(A) Relative larval weight gain in wild-type (Col-0), sid2-2, and pad4-1 plants (ANOVA; treatment, P < 0.05; genotype, P < 0.05; treatment × genotype, P < 0.05).
(B) Relative larval weight gain in wild-type (Col-0) and coi1-1 plants (ANOVA; treatment, P < 0.05; genotype, P < 0.05, treatment × genotype, not significant). Letters above the bars signify statistically significant differences among groups (Tukey, P < 0.05).
The fact that pathogen-triggered SIS to herbivory is independent of SA signaling suggests that it does not rely on SA/JA antagonism, which involves JA-Ile that is perceived via COI1. Indeed, although the coi1-1 mutant was more susceptible to T. ni because JA-mediated defenses are severely compromised, we found that Psm ES4326 cfa6 nevertheless triggered SIS to herbivory in the coi1-1 mutant (Figure 1B). These data strongly suggest that pathogen-triggered SIS to herbivory is not a consequence of SA/JA antagonism, as the latter relies on functional SA and COI1-mediated JA signaling.
Psm ES4326 cfa6 Induces Transcriptional Changes Associated with ET Signaling in Systemic Tissue
We performed whole-genome transcriptional profiling on systemic leaves of Arabidopsis plants infected with Psm ES4326 cfa6 versus mock-inoculated plants to identify candidate genes that could underlie SIS to herbivory. The plants used in the expression profiling analysis exhibited SIS to herbivory when infiltrated with Psm ES4326 cfa6 (similar to that shown for Columbia-0 [Col-0] in Figure 1A). We profiled the transcriptome at three time points representing early (3 h), intermediate (48 h), and late (96 h) stages of bacterial infection in our experimental setup. The 48-h time point has previously been identified as a stage where significant changes in gene expression and plant defense phenotype can be observed in systemic rosette leaves after infection of lower rosette leaves with Psm ES4326 (Mishina and Zeier, 2007).
Three independent biological RNA replicates per time point were individually hybridized to Affymetrix ATH1 GeneChips. For each probe set, signal intensities, mean expression fold change, and P values were calculated after data normalization. Because changes in gene expression levels in the systemic rosette leaves were relatively modest following Psm ES4326 cfa6 infiltration of lower rosette leaves (see Supplemental Table 1 online), we searched for differentially expressed genes without using the false discovery rate (FDR) method (Benjamini and Hochberg, 1995) or a fold change cutoff. Lists of differentially expressed genes at the 3-, 48-, and 96-h time points are given in Supplemental Data Set 1 online. The maximum number of differentially expressed genes at P < 0.05 identified in this way in Psm ES4326 cfa6-infected plants compared with mock-inoculated controls was observed at the 48-h time point (see Supplemental Table 1 online), and we focused on this time point for further analysis. A primary objective of the transcriptional profiling was to determine whether any of the canonical plant hormonal and defensive signaling sectors could be involved in the elicitation of SIS to T. ni herbivory. To this end, we conducted a more detailed analysis of the data for the 48-h time point in which we only considered candidate genes involved in plant hormone biosynthesis or hormonal regulation of immune signaling based on The Arabidopsis Information Resource annotation (www.arabidopsis.org; see Supplemental Data Set 2 online). We found that many genes of interest (see below) were in the top 5% of those differentially regulated or were among the lowest 5% in terms of uncorrected P values (see Supplemental Data Set 2 online).
At the 48-h time point, several genes regulated by JA-Ile were repressed, including PDF1.2b and CHI-B (not listed in Supplemental Data Set 2 online as these are marker genes and not actively involved in hormonal regulation of immune signaling), and the DELLA protein-encoding RGL3 (see Supplemental Table 2 online). The repressor of gibberellic acid–signaling RGL3 is necessary for full JA responses (Wild et al., 2012). By contrast, the ET precursor 1-aminocyclopropane-1-carboxylate (ACC) synthase-encoding gene ACS2, which mediates a rate-limiting step in ET production (Liu and Zhang, 2004) and whose transcriptional regulation correlates with enhanced pathogen-induced ET biosynthesis (Li et al., 2012), and EOL2, which encodes a negative regulator of ACS5, were induced 48 h after inoculation (see Supplemental Table 2 online). This could represent a switch from ACS5 to ACS2 as the main ACS involved in ET production, which has been previously observed after wounding (Tsuchisaka and Theologis, 2004). Two additional genes that were repressed at the 48-h time point were NUDT7, which encodes a hydrolase that negatively regulates SA-independent EDS1- and PAD4-mediated signaling (Bartsch et al., 2006), and the mitogen-activated protein kinase kinase kinase–encoding gene MEKK1 (see Supplemental Table 2 online). MEKK1 is involved in the activation of MPK4, a negative regulator of EDS1 and PAD4 (Ichimura et al., 2006; Suarez-Rodriguez et al., 2007).
The transcriptional profiling data showed that in plants that exhibited SIS to herbivory, very few differentially expressed genes could be identified in the systemic leaves, at least using Affymetrix GeneChip technology. Although the data were not sufficiently robust to correlate specific hormone signaling pathways with SIS, they suggested that ET, PAD4, and EDS1 signaling might be systemically affected following infection of lower rosette leaves with Psm ES4326 cfa6. Because (1) ET, PAD4, and EDS1 are associated with susceptibility to tissue-chewing insect herbivores (Kahl et al., 2000; Stotz et al., 2000; Winz and Baldwin, 2001; Cui et al., 2002), (2) ET production and transcription of PAD4 and EDS1 are induced by P. syringae and/or associated with leaf senescence in local, infected tissue (Bent et al., 1992; Magalhaes et al., 2000; Huang et al., 2005; Wang et al., 2008b; Mur et al., 2009), and (3) pathogen-triggered SIS to herbivory relies on PAD4 (Figure 1A), we hypothesized that SIS to herbivory may depend on ET signaling, and this prompted us to test a variety of ET-related signaling mutants for their ability to display the SIS to herbivory phenotype.
Pathogen-Triggered SIS to Herbivory Depends on ET Signaling
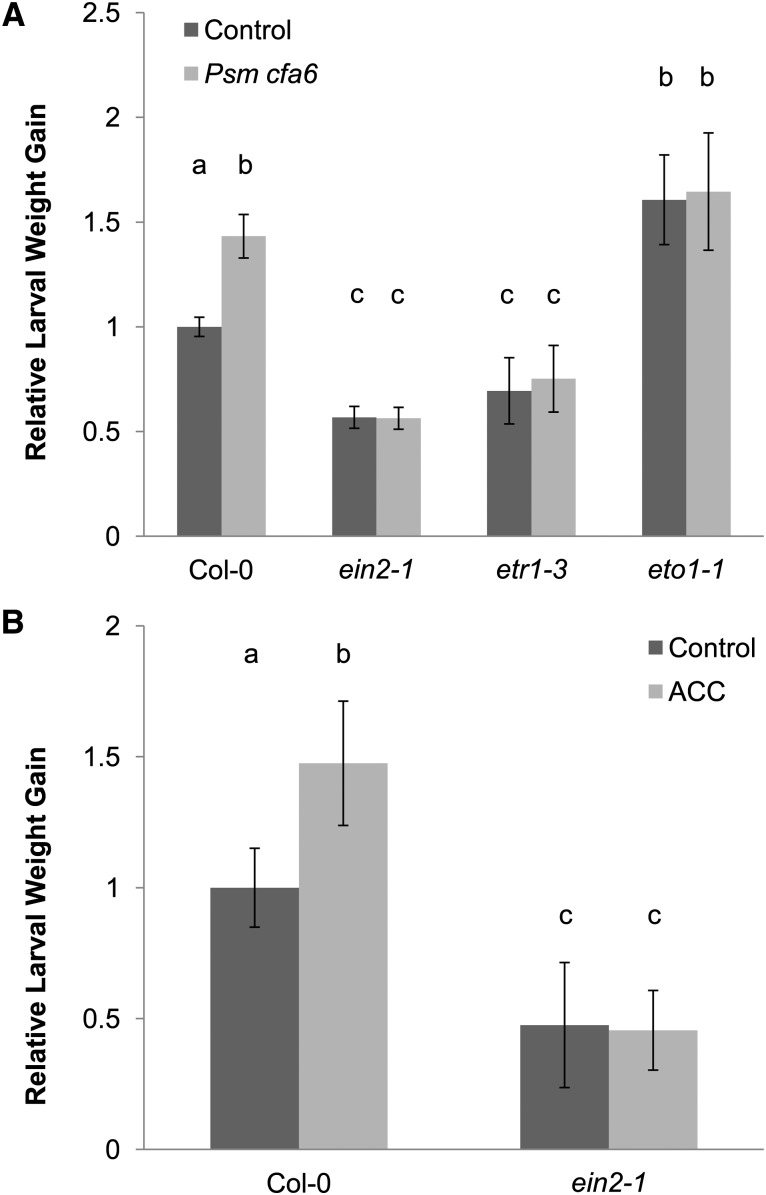
To test the hypothesis that pathogen-triggered SIS to herbivory depends on ET signaling, we phenotyped mutants with reduced and enhanced ET signaling. Indeed, although the ein2-1 and etr1-3 mutants, which are disrupted in ET signaling, were more resistant to T. ni herbivory than wild-type plants, we found no SIS to herbivory in these mutants (Figure 2A). Given that the ein2-1 and etr1-3 mutants were more resistant to T. ni, in agreement with the published data demonstrating that ET suppresses antiherbivore defense (Kahl et al., 2000; Stotz et al., 2000; Winz and Baldwin, 2001), we reasoned that the eto1-1 mutant, which overproduces ET, would be more susceptible to T. ni, which was indeed the case (Figure 2A). Moreover, infiltration of eto1-1 with Psm ES4326 cfa6 did not induce systemic susceptibility, again showing that ET represses systemic antiherbivore defense in response to bacterial infection (Figure 2A). To confirm that ET signaling has systemic effects and can elicit susceptibility to T. ni in distal leaves, we treated three lower rosette leaves of wild-type or ein2-1 mutant plants with the ET precursor ACC. As expected, ACC elicited SIS to herbivory in wild-type plants but not in the ET-insensitive ein2-1 mutant (Figure 2B), showing that local induction of ET production, which has been observed previously after virulent P. syringae infection and associated senescence (Bent et al., 1992; Magalhaes et al., 2000; Huang et al., 2005; Mur et al., 2009), is sufficient to block antiherbivore defenses systemically.
Figure 2.
Pathogen-Triggered SIS to Herbivory Is Mediated via ET Signaling.
Lower Arabidopsis rosette leaves were inoculated with a control solution (dark-gray bars), Psm ES4326 cfa6 suspension (OD600 = 0.2; light-gray bars; [A]), or a 100 μM solution of the ET precursor ACC (light-gray bars; [B]), respectively. Four days later, the inoculated or treated leaves were removed and newly hatched T. ni larvae were placed on the upper rosette leaves. Larval weight gain was measured after 7 d of feeding. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on mock-inoculated (A) or control-treated (B) wild-type (Col-0) plants. The bars represent the means (±se) of relative weight gain from two independent experiments with n = 36 larvae per treatment per experiment.
(A) Relative larval weight gain in wild-type (Col-0), ein2-1, etr1-3, and eto1-1 plants (ANOVA; treatment, P < 0.05; genotype, P < 0.05; treatment × genotype, P < 0.05).
(B) Relative larval weight gain in wild-type (Col-0) and ein2-1 plants (ANOVA; treatment, P < 0.05; genotype, P < 0.05; treatment × genotype, P < 0.05). Letters above the bars signify statistically significant differences among groups (Tukey, P < 0.05).
ET Regulates Octadecanoid-Dependent Defenses Independently of COI1
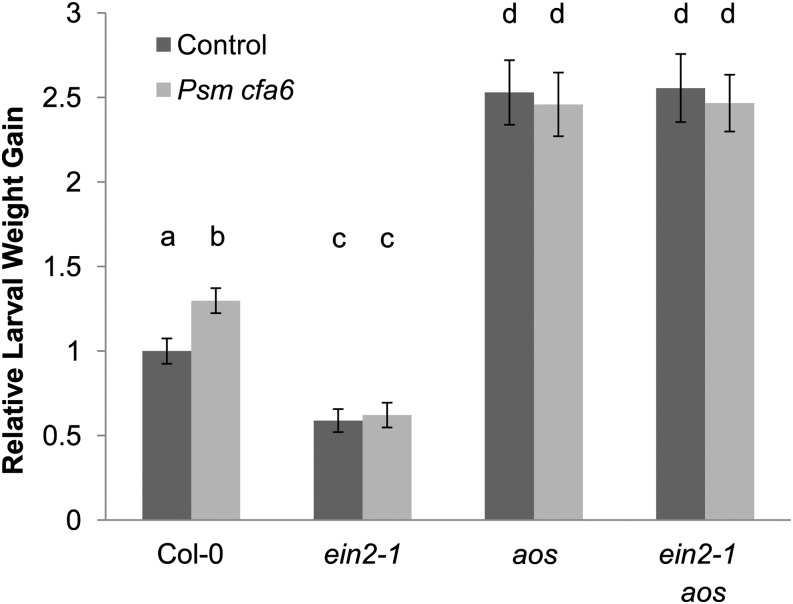
SIS to herbivory does not rely on the core COI1 JA-signaling component (Figure 1). However, other octadecanoids (including JA unconjugated from Ile and other JA precursors or derivatives) also regulate antiherbivore defenses independently of COI1 (Stintzi et al., 2001; Schweizer et al., 2013). Production of JA and its precursor 12-oxo-phytodienoic acid (OPDA) is disrupted in the aos mutant, which is defective in the enzyme ALLENE OXIDE SYNTHASE (AOS; CYP74A) (Park et al., 2002). As expected, the aos mutant and the coi1-1 mutant were highly susceptible to T. ni (Figure 3). However, in contrast with the coi1-1 mutant (Figure 1B), Psm ES4326 cfa6 did not induce further susceptibility in aos plants (Figure 3), suggesting that repression of the activity of JA precursors or derivatives other than JA-Ile contribute to SIS to T. ni.
Figure 3.
During Pathogen-Triggered SIS to Herbivory, ET Regulates Octadecanoid-Dependent, JA-Ile–Independent Defenses.
Lower Arabidopsis rosette leaves were inoculated with a sterile 10 mM MgSO4 control solution (dark-gray bars) or Psm ES4326 cfa6 suspension (OD600 = 0.2; light-gray bars). Four days later, the inoculated leaves were removed and newly hatched T. ni larvae were placed on the upper rosette leaves. Larval weight gain was measured after 7 d of feeding. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on mock-inoculated wild-type (Col-0) plants. The bars represent the means (±se) of relative weight gain from two independent experiments with n = 36 larvae per treatment per experiment. Relative larval weight gain in wild-type (Col-0), ein2-1, aos, and ein2-1 aos plants (ANOVA; treatment, P < 0.05; genotype, P < 0.05; treatment × genotype, P < 0.05). Letters above the bars signify statistically significant differences among groups (Tukey, P < 0.05).
ET signaling is required for SIS to T. ni and ET-signaling mutants are much more resistant to herbivory (Figure 2), presumably because they have a high level of JA-mediated signaling (Adie et al., 2007). If this interpretation is correct, we reasoned that the enhanced susceptibility of the aos mutant would be epistatic to the enhanced resistance of the ein2-1 mutant. Indeed, disruption of AOS (as in the aos mutant allele) in an ein2-1 background completely rescued the enhanced resistance to herbivory of the ein2-1 mutant and even elevated susceptibility to T. ni herbivory to a level indistinguishable from that seen in the aos single mutant (Figure 3). Moreover, as in the ein2-1 and aos single mutants, pathogen-triggered SIS to herbivory was absent in an aos ein2-1 double mutant (Figure 3), strengthening the conclusion that ET is a negative regulator of octadecanoid-dependent, but COI1-independent, antiherbivore defense signaling.
MAMP-Triggered SIS to Herbivory Depends on SA/JA Antagonism, Not on ET Signaling
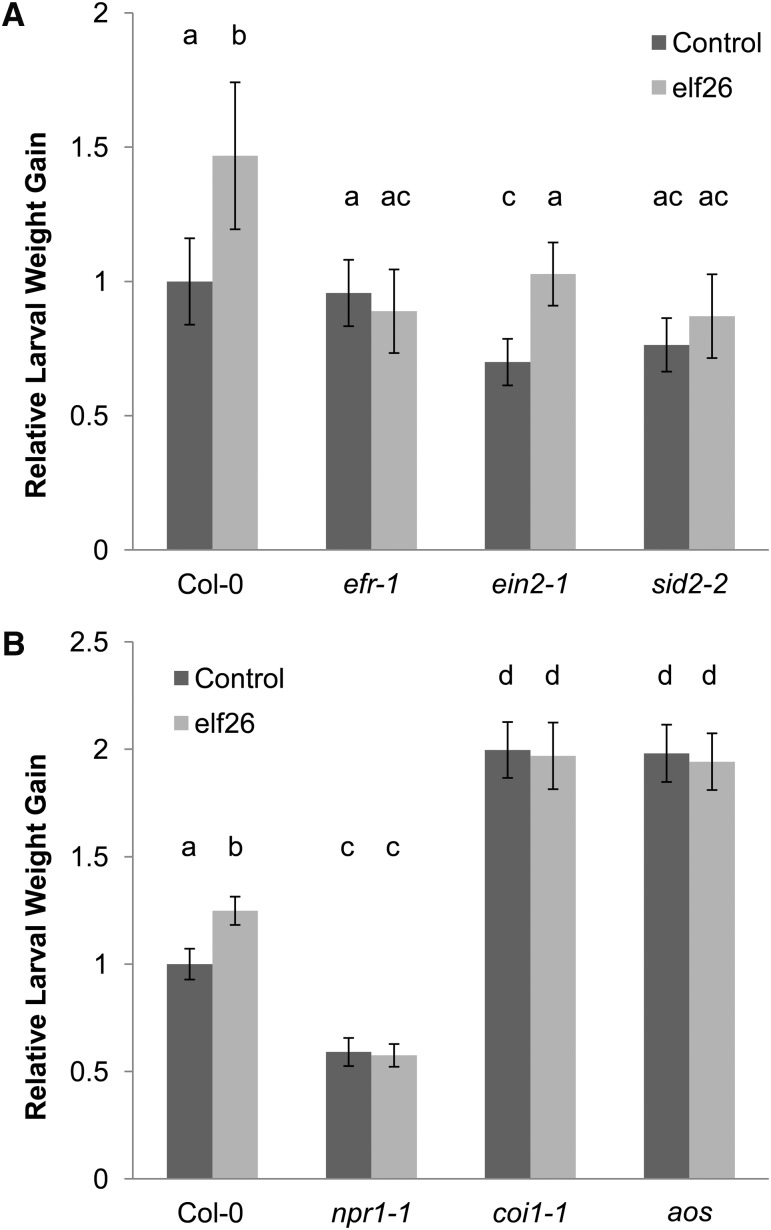
MAMPs have previously been shown to induce both ET production and SAR to subsequent infection with virulent P. syringae (Navarro et al., 2004; Mishina and Zeier, 2007). We hypothesized that MAMPs might induce an ET response strong enough to elicit SIS to herbivory. Because expression of the flagellin (flg22) receptor FLS2 depends on ET signaling (Boutrot et al., 2010; Mersmann et al., 2010), we primarily used treatment with the MAMP peptide elf26 in these experiments. Expression of the elf26 receptor EFR does not depend on ET signaling (Tintor et al., 2013). Instead of being infiltrated with bacteria, three lower rosette leaves were treated with 10 µM elf26 or flg22, and 4 d later T. ni larvae were added to systemic rosette leaves of the plants, after clipping off the infiltrated (local) leaves. Surprisingly, SIS to herbivory was observed in ein2-1 but not in sid2-2 after treatment with elf26 (Figure 4A). Likewise, SIS to herbivory did not occur in sid2-2 mutants after treatment with flg22 (see Supplemental Figure 1 online). In agreement with the dependency of FLS2 accumulation on functional ET signaling, SIS to herbivory did not occur in flg22-treated ein2-1 mutants either (see Supplemental Figure 1 online).
Figure 4.
MAMP-Triggered SIS to Herbivory Depends on SA/JA Antagonism, not ET Signaling.
Lower Arabidopsis rosette leaves were inoculated with a control solution (dark-gray bars) or suspensions of 10 μM elf26 (light-gray bars). Four days later, the treated leaves were removed and newly hatched T. ni larvae were placed on the upper rosette leaves. Larval weight gain was measured after 7 d of feeding. For each experiment, weight gain data were normalized to the weight gain of larvae feeding on control-treated wild-type (Col-0) plants. The bars represent the means (±se) of relative weight gain from two independent experiments with n = 36 larvae per treatment per experiment.
(A) Relative larval weight gain in wild-type (Col-0), efr-1, ein2-1, and sid2-2 plants (ANOVA; treatment, P < 0.05; genotype, P < 0.05; treatment × genotype, P < 0.05). Letters above the bars signify statistically significant differences among groups (Tukey, P < 0.05).
(B) Relative larval weight gain in wild-type (Col-0), npr1-1, coi1-1, and aos plants (ANOVA; treatment, P < 0.05; genotype, P < 0.05; treatment × genotype, P < 0.05). Letters above the bars signify statistically significant differences among groups (Tukey, P < 0.05).
The finding that MAMP-triggered SIS to herbivory (in contrast with pathogen-elicited SIS to herbivory) depends on SA but not ET signaling suggests that it may depend on SA/JA antagonism. SA/JA antagonism depends on functional COI1 and NPR1. When aos, coi1-1, and npr1-1 mutants were treated with 10 µM elf26, MAMP-induced SIS to herbivory was absent in aos, coi1-1, and npr1-1, whereas it was present in wild-type plants (Figure 4B). Because pathogen-triggered SIS to herbivory does not rely on functional COI1 (Figure 1B) and NPR1 (Cui et al., 2002), these results indicate that pathogen-triggered SIS to herbivory and MAMP-triggered SIS to herbivory rely on different mechanisms. Pathogen-triggered SIS to herbivory relies on ET and octadecanoid signaling but is independent of COI1, SA signaling, and NPR1. By contrast, MAMP-triggered SIS to herbivory relies on SA/JA antagonism and depends on functional COI1 and NPR1.
Pathogen-Triggered SIS to Herbivory Is Mediated by Pathogen Effectors
Having established that pathogen-triggered SIS to herbivory is not mediated solely by MAMPs, we hypothesized that bacterial type III effectors might elicit SIS to herbivory. Type III effectors are possible candidates because they enhance pathogen virulence (if they or their actions are not recognized by a corresponding nucleotide-binding site-leucine-rich repeat resistance receptor) by interfering with PTI-mediated signaling, in some cases suppressing SA or stimulating ET signaling (Cohn and Martin, 2005; de Torres-Zabala et al., 2007; Jelenska et al., 2007). This hypothesis is consistent with our results showing that pathogen-triggered SIS to herbivory relies on ET but not on SA signaling. Moreover, the finding that hemibiotrophic pathogens activate ET signaling, which may enhance pathogen proliferation and disease symptoms, suggests some manipulation of the interaction by the pathogen, which could be achieved through the action of pathogen-encoded effectors (Lund et al., 1998; Magalhaes et al., 2000; Cohn and Martin, 2005; Huang et al., 2005; Chen et al., 2009; Mur et al., 2009; Wi et al., 2012).
To study the role of pathogen-encoded effectors in SIS to T. ni herbivory, we inoculated wild-type, ein2-1, and npr1-1 plants with P. syringae strains that were unable to produce COR (cfa6 cmaA mutants) or unable to inject type III effectors into plant cells in addition to being COR deficient (hrcC cfa6 cmaA mutants) or with the isogenic wild-type P. syringae pv tomato (Pto) DC3000 parent. In these experiments, we used Pto DC3000 instead of Psm ES4326 so we could further test if pathogen-triggered SIS to herbivory is elicited by more than one virulent P. syringae strain. We found that infiltrating COR-deficient Pto cfa6 cmaA into wild-type plants elicits SIS to herbivory (see Supplemental Figure 2 online), similar to Psm ES4326 cfa6 (Cui et al., 2005). Compared with the COR-deficient mutant Pto cfa6 cmaA, wild-type Pto DC3000 elicited lower levels of SIS in wild-type plants (see Supplemental Figure 2 online), again similar to our previous observations on plants infected with wild-type Psm ES4326 compared with Psm ES4326 cfa6 (Cui et al., 2005) and consistent with the fact that COR induces JA signaling, which activates antiherbivore defenses (Cui et al., 2005).
By testing these Pto mutants on wild-type, ein2-1, and npr1-1 Arabidopsis, we were able to separate the effects of MAMPs and effectors. COR-deficient Pto cfa6 cmaA, which is still able to employ type III effectors, elicited SIS in npr1-1 but not ein2-1 plants (see Supplemental Figure 2 online). By contrast, the Pto hcC cfa6 cmaA mutant, which also has a disrupted type III secretion system due to the hrcC null mutation, triggered slightly enhanced susceptibility in ein2-1 but not in npr1-1 plants (see Supplemental Figure 2 online). These results suggest that type III effectors suppress SA signaling, which interferes with COI1-dependent antiherbivore defenses via SA/JA antagonism, and trigger ET signaling, which interferes with COI1-independent antiherbivore defenses. In line with the result in wild-type plants, when COR was added to the interaction by infiltrating plants with wild-type Pto DC3000, SIS to herbivory was diminished in npr1-1 plants and weak resistance to herbivory was induced in ein2-1 plants (see Supplemental Figure 2 online).
In ein2-1, the weak additional resistance to T. ni on Pto DC3000–infiltrated plants and the slightly induced susceptibility to T. ni on Pto hcC cfa6 cmaA resulted in a significant difference in susceptibility to herbivory between these two treatment groups (see Supplemental Figure 2 online), in line with results of previous experiments showing a resistance-enhancing role for COR (Cui et al., 2005) and a susceptibility-enhancing role for MAMPs (Figure 4). The result that Pto hcC cfa6 cmaA did not induce significant SIS on wild-type and ein2-1 plants (see Supplemental Figure 2 online), but treatment of plants with the MAMP elf26 did (Figure 4), may be explained by the fact that this P. syringae mutant does not reach as high a titer as wild-type Pto DC3000 and the signaling it induces is simply not strong enough. Taken together, our data support a role for type III effectors suppressing MAMP-triggered SIS to herbivory by inhibiting SA signaling and simultaneously eliciting pathogen-triggered SIS to herbivory by suppressing COI1-independent antiherbivore defenses via ET signaling.
DISCUSSION
A Multilayered Induction Mechanism behind SIS to Herbivory
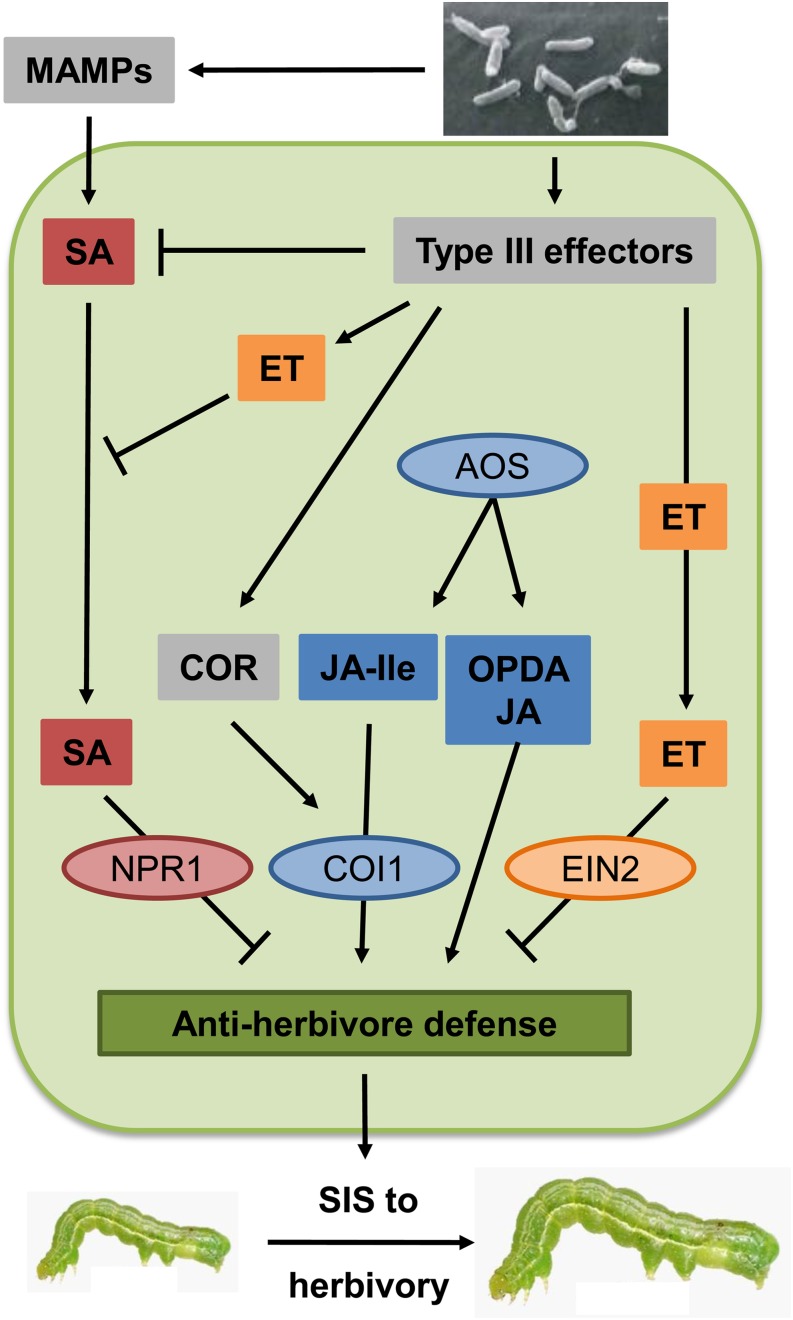
Using a combination of Arabidopsis and P. syringae mutants in sensitive T. ni feeding assays, we uncovered a multilayered mechanism behind pathogen-triggered SIS to herbivory. Although not yet biochemically validated, our genetic studies lead to a new model for SIS to herbivory described in Figure 5. This model resembles the zigzag model illustrating the quantitative output of plant immune signaling in response to microbial attack (Jones and Dangl, 2006). In one layer, MAMP-triggered systemic SA signaling antagonizes COI1-dependent JA signaling via SA/JA antagonism that relies on NPR1, thereby weakening antiherbivore defenses and causing SIS to herbivory. This is neutralized in a second layer where SA signaling is suppressed by COR and some type III effectors that interfere with MAMP-triggered signaling. Additionally, enhanced ET signaling, also mediated by effectors, interferes with antiherbivore defenses that rely on octadecanoids produced via AOS but that function independent of COI1 (Stintzi et al., 2001; Schweizer et al., 2013), leading again to SIS to herbivory. When one of the type III effectors elicits ETI, the induced susceptibility is nullified and effector-triggered signaling synergizes with COR to induce systemic resistance to herbivory (Cui et al., 2002, 2005).
Figure 5.
Simplified Model of Pathogen-Triggered SIS to Herbivory.
In the first layer, MAMP-triggered SA signaling (red) antagonizes COI1-dependent JA signaling (blue) via SA/JA antagonism that relies on NPR1, thereby weakening antiherbivore defenses and causing SIS to herbivory. This is neutralized in the next layer where SA signaling is suppressed by COR and some type III effectors that interfere with MAMP-triggered signaling. At the same time, enhanced ET signaling (orange), also mediated by type III effectors, interferes with antiherbivore defenses that rely on octadecanoids produced via AOS, but that function independent of COI1, leading again to SIS to herbivory. When one of the type III effectors elicits ETI, the induced susceptibility is nullified and effector-triggered signaling synergizes with COR to induce systemic resistance to herbivory. P. syringae–associated factors are indicated in gray squares, Arabidopsis hormones in colored squares, and Arabidopsis proteins in colored ellipses. (T. ni photo courtesy of Nestor Bautista Martínez.)
The dominant role of effector-mediated ET signaling over MAMP-triggered SA signaling in SIS to herbivory is consistent with previous studies, which found that ET suppresses transcription of the SA biosynthesis enzyme ICS1 (SID2) (Chen et al., 2009) and overrides NPR1-mediated SA-JA antagonism (Leon-Reyes et al., 2009, 2010). The involvement of ET signaling in SIS to herbivory may also explain our earlier observation that variability exists in the strength of the SIS to herbivory phenotype dependent on environmental circumstances (Cui et al., 2005). ET levels respond to environmental cues such as light, water, and temperature. However, while ET signaling is both necessary and sufficient for pathogen-triggered SIS to herbivory, signaling by other hormones in addition to ET could potentially be involved in SIS. Type III effectors, such as HopI1, which actively perturbs SA biogenesis (Jelenska et al., 2007), AvrRpt2, which activates auxin signaling (Chen et al., 2007), and AvrPtoB, which modulates abscisic acid signaling (de Torres-Zabala et al., 2007), all interfere with SA-regulated pathogen defenses and could also influence antiherbivore defenses. The contributions of individual type III effectors and plant hormones other than ET should be investigated in future studies.
Virulent Hemibiotrophic Pathogens May Generally Induce ET Production
Our findings contribute to a growing body of literature showing that virulent hemibiotrophic pathogens actively suppress plant immunity by enhancing ET signaling. In tobacco (Nicotiana tabacum), both of the virulent hemibiotrophic pathogens P. syringae pv tabaci and Phytophthora parasitica var nicotianae induce ET production starting around 24 to 48 h and peaking at 72 h after inoculation (Huang et al., 2005; Wi et al., 2012). Despite the fact that we have not measured ET levels directly, we found evidence of an ET response in our transcriptional profiling data of the systemic response to Psm ES4326 cfa6 in Arabidopsis as well, which corroborates previous observations of ET production induced by virulent P. syringae infection and/or associated tissue senescence in local, infected Arabidopsis leaves (Bent et al., 1992; Magalhaes et al., 2000; Mur et al., 2009).
By inducing ET signaling, virulent hemibiotrophic pathogens may subvert successful plant immune responses. ET-defective or -insensitive mutants of Arabidopsis, tobacco, and tomato show reduced pathogen proliferation and/or disease symptoms after infection (Bent et al., 1992; Lund et al., 1998; Wi et al., 2012). The hypothesis that hemibiotrophic pathogens actively induce ET signaling to enhance plant disease susceptibility is supported by the findings that ET production is induced by several pathogen effectors (Cohn and Martin, 2005) and that ET suppresses production of SA that is necessary for full immune signaling (Chen et al., 2009). From this perspective, SIS to herbivory may be a by-product of pathogens subverting the plant immune system to their own benefit.
Pathogen-Induced Susceptibility to Insects Could Have Epidemiological Consequences
Our results suggest that inducing susceptibility to insect herbivores could conceivably have epidemiological consequences and facilitate the dispersal of both insect and pathogen, especially when the insect can act as vector to transmit the pathogen to new hosts. Many examples of this phenomenon exist in the plant virus literature (Mauck et al., 2012), including one provided by the begomovirus tomato yellow leaf curl China virus. This virus makes tomato more susceptible to the invasive silverleaf whitefly Bemisia tabaci B, which also transmits the virus, promoting the spread of both (Jiu et al., 2007). The suppression of JA-mediated signaling by the viral virulence factors C2 and βC1 likely contributes to the facilitation of the herbivore (Yang et al., 2008; Lozano-Durán et al., 2011).
Pseudomonas spp can be transported by leaf-chewing and phloem-feeding insects (Lilley et al., 1997; Hirano and Upper, 2000; Stavrinides et al., 2009; Chung et al., 2013), and Pseudomonas spp facilitate feeding by the chewing herbivore Leptinotarsa decemlineata or Colorado potato beetle (Chung et al., 2013). Interestingly, a positive association between endophytic Pseudomonas spp abundance and damage by specialist, dipteran herbivores from the genus Scaptomyza (Whiteman et al., 2011) has been observed in the wild mustard species Cardamine cordifolia in a relatively intact ecosystem (Parris T.H. Humphrey and Noah K. Whiteman, unpublished data). Moreover, the suppression of antiherbivore defenses in JA-signaling mutants of the wild tobacco Nicotiana attenuata led to increased vulnerability of plants to insect herbivores in field experiments and natural populations (Kessler et al., 2004; Kallenbach et al., 2012), as well as the attraction of herbivores that did not normally feed on these plants (Kessler et al., 2004). Future field studies with these and other pathosystems may shed further light on the role that SIS to herbivory might play in shaping plant–herbivore interactions in nature.
METHODS
Plant Lines and Growth Conditions
Seeds of Arabidopsis thaliana accession Col-0 (wild-type) plants and mutants (all Col-0 background) were cold-stratified in 0.1% agarose for 3 d at 4°C. Subsequently, plants were grown in 36-cell trays under a 12-h-day:12-h-night cycle on Fafard #2 soil mix at 23/20°C day/night receiving water twice per week until 5 weeks old. The Arabidopsis mutants used were as follows: efr-1 (SALK_044334, At5g20480), fls2-c (SAIL_691C04, At5g46330), sid2-2 (At1g74710), pad4-1 (At3g52430), ein2-1 (At5g03280), etr1-3 (At1g66340, formerly ein1), eto1-1 (At3g51770), aos/cyp74a (SALK_017756, At5g42650), coi1-1 (At2g39940,), and an aos ein2-1 double mutant. Homozygous coi1-1 mutants were selected by spraying seedling offspring of selfing coi1-1/COI1 heterozygotes with 1 mM methyl jasmonate in a solution containing 5% ethanol. After 7 d, heterozygotes and COI1/COI1 homozygotes were removed. Wild-type control plants and other plants accompanying coi1-1 mutants in the same experiments were treated with 5% ethanol in parallel.
Bacterial Strains and Cultivation
The COR-deficient bacterial strain Pseudomonas syringae pv maculicola ES4326 cfa6::Kmr (Psm ES4326 cfa6) carrying the empty vector pLAFR3 (Tetr) has been described previously (Cui et al., 2005). Pto CUCPB5532 (Pto hcC cfa6 cmaA; originally created by Brian Kvitko, Cornell University, Ithaca, NY) and Pto DB29 (Pto cfa6 cmaA) are derived from wild-type Pto DC3000. Pto DB29 has been described previously (Millet et al., 2010). P. syringae bacterial strains were cultured on King’s B media plates supplemented with appropriate antibiotics: 50 mg mL−1 rifampicin for Pto DC3000 and Pto CUCPB5532; 50 mg mL−1 streptomycin for Psm ES4326; 50 mg mL−1 kanamycin for Pto DB29, and Psm ES4326 cfa6. On the third day, single colonies were inoculated into King’s B media liquid cultures containing the same antibiotics. Overnight cultures were grown in a shaking incubator at 28°C to OD600 = 1.0. The tubes harboring the cultures were then spun down at 5000 rpm for 5 min. The King’s B media was removed, and the bacterial cells were washed by resuspending in sterile 10 mM MgSO4. This was repeated twice, and prior to inoculation in Arabidopsis leaves, bacteria were resuspended at a final concentration of OD600 = 0.2.
Trichoplusia ni Weight Gain Assay
Trichoplusia ni eggs (Benzon Research) were incubated in a box containing a moist paper towel at 30°C for 2 d to synchronize hatching. One neonate caterpillar was transferred to each experimental plant using a fine paint brush. The plants with caterpillars were kept individually in Magenta GA-7 boxes (Sigma-Aldrich) with insect-proof mesh lids or in pots wrapped in mesh cloth (DC May), and caterpillars were weighed to the nearest 0.01 mg after 7 d of feeding on the plants using a micro-balance (Mettler-Toledo). The weight of the caterpillars reflects weight gain because the weight of the neonates, including relative differences between individuals at the start of the feeding trial, is negligible. Each feeding trial was performed at least twice independently with comparable results. All weight gain assays were run blind in that student volunteers did not know which plant accessions were fed to which caterpillars and had no prior expectation regarding experimental outcome.
T. ni SIS to Herbivory Weight Gain Assay
Three lower rosette leaves (true leaves three, four, and five) of 5-week-old plants were inoculated with a suspension of Psm ES4326 cfa6 or Pto DC3000 (and mutants thereof) at a concentration OD600 = 0.2 (or ∼108 colony-forming units mL−1; described in Cui et al., 2005). As a control, a solution of sterile 10 mM MgSO4 was used. After 4 d, the leaves infected with bacteria were cut off and a neonate T. ni caterpillar was added to each plant. This ensured minimal direct contact between insect and pathogen, thereby allowing the interpretation that the observed phenotypes are plant-mediated through systemic leaves. The latter part of the assay is as described above for the T. ni weight gain assay.
T. ni Weight Gain Assay after Preinoculation with ACC, elf26, or flg22
The herbivore weight gain assay was executed as described above for the T. ni SIS to herbivory weight gain assay with the exception that instead of live bacteria, a solution of 100 μM ACC (Sigma-Aldrich) or solution of 10 μM flg22 or elf26 in sterile 10 mM MgSO4 was used to pretreat plants (Navarro et al., 2004).
Statistical Analysis of Weight Gain Assays
All statistical analyses were performed using Minitab. Weight gain data from experiments on Arabidopsis mutants alongside wild-type plants subjected to different treatments were analyzed with analysis of variance (ANOVA) including plant line and treatment as factors.
Harvesting Leaf Tissue for the Microarray Experiment
Plants were grown and infiltrated as described above for the T. ni SIS to herbivory weight gain assay. For subsets of plants at the three time points (3, 48, and 96 h) after infection, leaves 6 and 7 were cut off at the petiole with scissors, flash-frozen in liquid nitrogen in paper envelopes, and stored at −80°C until RNA was extracted. Leaves 6 and 7 were stored separately; leaf 6 was used for gene expression profiling unless the yield of RNA had a concentration below 500 ng μL−1: in that case, leaf 7 was used. Leaves from eight replicate plants were pooled to form each RNA sample.
RNA Extraction for the Microarray Experiments
For the microarray experiments, total RNA was extracted from ∼200 mg of leaf tissue using the Qiagen RNeasy plant mini kit with the on-column DNase digestion (Qiagen). Pooled leaves from each treatment group were briefly ground in liquid nitrogen using a mortar and pestle. Two aliquots of ∼100 mg of tissue were processed following the manufacturer’s protocol. RNA yield and quality were assessed using fluorimetry in an Agilent bioanalyzer. For labeling and hybridization, standard Affymetrix protocols were used (Affymetrix). Five micrograms of RNA was taken to generate cDNA for each sample. After subsequent synthesis of complementary strand cDNA and purification, samples were transcribed into biotinylated cRNA, cut into fragments, and hybridized to Affymetrix ATH1 microarrays.
Microarray Experimental Design and Data Analysis
Eighteen Affymetrix ATH1 whole-genome arrays were divided evenly among a combination of the following factors: treatment group (inoculation with sterile 10 mM MgSO4 as a mock control and Psm ES4326 cfa6); time point (3, 48, and 96 h after inoculation); and trial (three independent trials). Successful establishment of infection was confirmed visually by following the progress of disease symptoms on the inoculated leaves. Leaves inoculated with Psm ES4326 cfa6 started showing symptoms after 2 to 3 d when the leaves began to turn yellow compared with mock-inoculated leaves. Furthermore, to ascertain that infection with Psm ES4326 cfa6 was sufficient to cause the expected phenotype, a T. ni weight gain assay was conducted. The assays were performed as described above. Raw data for each experiment are available at NASCArrays (experiment reference: NASCARRAYS-705 “systemic response of Arabidopsis to Pseudomonas syringae pv maculicola ES4326 cfa6”).
Data from the microarray experiment were analyzed using Bioconductor (Gentleman et al., 2004). Affymetrix (.cel) files were loaded into the program. Background correction, normalization, and expression summaries were performed using the robust multiarray average approach (Irizarry et al., 2003) and then combined into experiment definitions to compare the different time points. Linear modeling of microarray data and identification of differentially expressed genes were performed with the limma package (Smyth, 2005). P values were adjusted to control the FDR using the method of Benjamini and Hochberg (1995), which was performed with the statistical package implemented in R 2.10.1 (Ihaka and Gentleman, 1996). Because this proved to be too stringent to detect significant changes in gene expression levels in the systemic leaves following Psm ES4326 cfa6 infiltration of lower leaves, no FDR method or fold change cutoffs were used to generate the lists of differentially expressed genes in Supplemental Data Set 1 online.
However, subsequently, a more detailed analysis of 750 candidate genes involved in plant hormone biosynthesis or hormonal regulation of immune signaling, which were selected based on The Arabidopsis Information Resource annotation (www.arabidopsis.org), was conducted (see Supplemental Data Set 2 online). Although applying the FDR method on the resulting group of genes revealed that none of the genes were below the new significance threshold, genes in the top 5% of those differentially regulated (based on absolute fold change) or among the lowest 5% in terms of P values without FDR correction were considered for generating hypotheses to test in the T. ni weight gain experiments.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Flg22 Triggers SIS to Herbivory, which Requires SA and ET Signaling.
Supplemental Figure 2. Bacterial Type III Effectors Mediate Pathogen-Triggered SIS to Herbivory via ET Signaling Suppressing NPR1-Mediated MAMP-Triggered SIS to Herbivory.
Supplemental Table 1. Number of Differentially Expressed Genes in Systemic Leaves of Psm ES4326 cfa6- versus 10 mM MgSO4-Inoculated Plants.
Supplemental Table 2. Selected Differentially Expressed Genes in Systemic Leaves of Psm ES4326 cfa6- versus 10 mM MgSO4-Inoculated Plants.
Supplemental Data Set 1. Genes Differentially Expressed in Systemic Leaves of Psm ES4326 cfa6- versus 10 mM MgSO4-Inoculated Plants.
Supplemental Data Set 2. Response of Genes Involved in Hormone Biosynthesis and Hormonal Regulation of Immune Signaling in Systemic Leaves of Psm ES4326 cfa6- versus 10 mM MgSO4-Inoculated Plants at 48 h after Inoculation.
Acknowledgments
N.K.W. was supported by a Kirschstein National Research Service Award from the National Institutes of Health (F32AI069732), a grant from the National Science Foundation (DEB-1256758), and a grant from the University of Arizona’s Seed Grant for Faculty Research Program. F.M.A. was supported by a grant from the National Science Foundation (MCB-0519898) and two grants from the National Institutes of Health (R37 GM48707 and P30 DK040561), and N.E.P. was supported by a grant from the National Science Foundation (SES-0750480). A.K.B. was supported by a Harvard University Microbial Sciences Initiative Postdoctoral Fellowship. N.K.W., S.C.G., F.M.A., and N.E.P. were supported by a grant from Harvard University’s Provost Fund for Interfaculty Collaboration and a grant from the Harvard University Science and Engineering Committee Seed Fund for Interdisciplinary Science. We thank Suresh Gopalan (ReSurfX) for help with the analysis of microarray data. We thank the following members of the Pierce laboratory and others who volunteered to help with the caterpillar feeding assays (in alphabetical order): Deborah Anderson, Lina Arcila, Ashley Bae, Chris Baker, Katie Berry, Megan Berry, Leonora Bittleston, John Boyle, Andrew Brownjohn, Lian Bruno, Alanna Callendrello, Norma-Rashid Che Yusoff, Mark Cornwall, James Crall, Seth Donoghue, Rodney Eastwood, Marianne Espeland, Abby Finkelstein, Joshua Fries, Masaru Hojo, Jay Iwasaki, Zofia Kaliszewska, Sarah Kocher, Petra Kubikova, Bonnie Lei, Patrick McCormack, Bruno de Medeiros, John Mewherter, Gabriel Miller, Julianne Pelaez, Jon Sanders, Mariah Slone, Jared Squires, Lauren Tomkinson, and Brian Trippe. We thank the following people for their help and advice during the experiments and data analyses: Reddy Gali, Jennifer Couget, Claire Reardon (Harvard University Faculty of Arts and Sciences Systems Biology Core), Julia Dewdney, and Nicole Clay. We also thank Fumiaki Katagiri for nonglabrous coi1-1 seeds, Carol Bender for providing the COR-deficient mutants of Pto DC3000, and Alan Collmer and Brian Kvitko for the nonpolar hrc cfa6 cmaA mutant of Pto DC3000 Pto CUCPB5532. The manuscript was improved by comments from the editorial team at The Plant Cell and two anonymous reviewers.
AUTHOR CONTRIBUTIONS
S.C.G., N.K.W., A.K.B., J.C., and A.C.-J. performed experiments and data analysis. A.K.B., A.M.W., and J.A.R. designed and conducted microarray experiments. I.A.B., J.D.R., and G.-H.H. organized and carried out feeding trials. J.B. grew plants and provided advice on the project. S.C.G., N.K.W., F.M.A., and N.E.P. formulated the project, were responsible for the coordination and supervision of research, and wrote the article. F.M.A. and N.E.P. are joint senior authors. All authors contributed to and approved the final article.
Glossary
- MAMP
microbe-associated molecular pattern
- PTI
pattern-triggered immunity
- ETI
effector-triggered immunity
- SA
salicylic acid
- JA
jasmonic acid
- ET
ethylene
- SAR
systemic acquired resistance
- COR
coronatine
- SIS
systemic induced susceptibility
- Psm
P. syringae pv maculicola
- Col-0
Columbia-0
- FDR
false discovery rate
- ACC
1-aminocyclopropane-1-carboxylate
- Pto
P. syringae pv tomato
- ANOVA
analysis of variance
References
- Adie B.A.T., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.-J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M., Gobbato E., Bednarek P., Debey S., Schultze J.L., Bautor J., Parker J.E. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57: 289–300 [Google Scholar]
- Bent A.F., Innes R.W., Ecker J.R., Staskawicz B.J. (1992). Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant Microbe Interact. 5: 372–378 [DOI] [PubMed] [Google Scholar]
- Boutrot F., Segonzac C., Chang K.N., Qiao H., Ecker J.R., Zipfel C., Rathjen J.P. (2010). Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proc. Natl. Acad. Sci. USA 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce T.J.A., Matthes M.C., Chamberlain K., Woodcock C.M., Mohib A., Webster B., Smart L.E., Birkett M.A., Pickett J.A., Napier J.A. (2008). cis-Jasmone induces Arabidopsis genes that affect the chemical ecology of multitrophic interactions with aphids and their parasitoids. Proc. Natl. Acad. Sci. USA 105: 4553–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.M., Xue L., Chintamanani S., Germain H., Lin H.Q., Cui H.T., Cai R., Zuo J.R., Tang X.Y., Li X., Guo H.W., Zhou J.M. (2009). ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 21: 2527–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.Y., Agnew J.L., Cohen J.D., He P., Shan L.B., Sheen J., Kunkel B.N. (2007). Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc. Natl. Acad. Sci. USA 104: 20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.H., Rosa C., Scully E.D., Peiffer M., Tooker J.F., Hoover K., Luthe D.S., Felton G.W. (2013). Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc. Natl. Acad. Sci. USA 110: 15728–15733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn J.R., Martin G.B. (2005). Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44: 139–154 [DOI] [PubMed] [Google Scholar]
- Cui J., Bahrami A.K., Pringle E.G., Hernandez-Guzman G., Bender C.L., Pierce N.E., Ausubel F.M. (2005). Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc. Natl. Acad. Sci. USA 102: 1791–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Jander G., Racki L.R., Kim P.D., Pierce N.E., Ausubel F.M. (2002). Signals involved in Arabidopsis resistance to Trichoplusia ni caterpillars induced by virulent and avirulent strains of the phytopathogen Pseudomonas syringae. Plant Physiol. 129: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres-Zabala M., Truman W.M., Bennett M.H., Lafforgue G., Mansfield J.W., Rodriguez Egea P., Bögre L., Grant M.R. (2007). Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 26: 1434–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos M., van Oosten V.R., van Poecke R.M.P., van Pelt J.A., Pozo M.J., Mueller M.J., Buchala A.J., Métraux J.P., van Loon L.C., Dicke M., Pieterse C.M.J. (2005). Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Felton G.W., Korth K.L., Bi J.L., Wesley S.V., Huhman D.V., Mathews M.C., Murphy J.B., Lamb C.J., Dixon R.A. (1999). Inverse relationship between systemic resistance of plants to microorganisms and to insect herbivory. Curr. Biol. 9: 317–320 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P., et al. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Moisan L.J., Newman M.A., Parker J.E. (2001). Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO J. 20: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X. (2013). Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Gentleman R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Zook M., Mert F., Kagan I., Rogers E.E., Crute I.R., Holub E.B., Hammerschmidt R., Ausubel F.M. (1997). Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P., Chintamanani S., Chen Z.Y., Zhu L.H., Kunkel B.N., Alfano J.R., Tang X.Y., Zhou J.M. (2004). Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 37: 589–602 [DOI] [PubMed] [Google Scholar]
- Heck S., Grau T., Buchala A., Métraux J.P., Nawrath C. (2003). Genetic evidence that expression of nahG modifies defence pathways independent of salicylic acid biosynthesis in the Arabidopsis-Pseudomonas syringae pv. tomato interaction. Plant J. 36: 342–352 [DOI] [PubMed] [Google Scholar]
- Hirano S.S., Upper C.D. (2000). Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae- A pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64: 624–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Schmelz E.A., Alborn H., Engelberth J., Tumlinson J.H. (2005). Phytohormones mediate volatile emissions during the interaction of compatible and incompatible pathogens: The role of ethylene in Pseudomonas syringae infected tobacco. J. Chem. Ecol. 31: 439–459 [DOI] [PubMed] [Google Scholar]
- Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281: 36969–36976 [DOI] [PubMed] [Google Scholar]
- Ihaka R., Gentleman R.C. (1996). R: A language for data analysis and graphics. J. Comput. Graph. Statist. 5: 299–314 [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J., Yao N., Vinatzer B.A., Wright C.M., Brodsky J.L., Greenberg J.T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17: 499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D., Tootle T.L., Reuber T.L., Frost L.N., Feys B.J., Parker J.E., Ausubel F.M., Glazebrook J. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiu M., Zhou X.P., Tong L., Xu J., Yang X., Wan F.H., Liu S.S. (2007). Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS ONE 2: e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kahl J., Siemens D.H., Aerts R.J., Gäbler R., Kühnemann F., Preston C.A., Baldwin I.T. (2000). Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta 210: 336–342 [DOI] [PubMed] [Google Scholar]
- Kallenbach M., Bonaventure G., Gilardoni P.A., Wissgott A., Baldwin I.T. (2012). Empoasca leafhoppers attack wild tobacco plants in a jasmonate-dependent manner and identify jasmonate mutants in natural populations. Proc. Natl. Acad. Sci. USA 109: E1548–E1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A., Halitschke R., Baldwin I.T. (2004). Silencing the jasmonate cascade: Induced plant defenses and insect populations. Science 305: 665–668 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A., Du Y., Koornneef A., Proietti S., Körbes A.P., Memelink J., Pieterse C.M.J., Ritsema T. (2010). Ethylene signaling renders the jasmonate response of Arabidopsis insensitive to future suppression by salicylic acid. Mol. Plant Microbe Interact. 23: 187–197 [DOI] [PubMed] [Google Scholar]
- Leon-Reyes A., Spoel S.H., de Lange E.S., Abe H., Kobayashi M., Tsuda S., Millenaar F.F., Welschen R.A.M., Ritsema T., Pieterse C.M.J. (2009). Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149: 1797–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Meng X., Wang R., Mao G., Han L., Liu Y., Zhang S. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8: e1002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley A.K., Hails R.S., Cory J.S., Bailey M.J. (1997). The dispersal and establishment of pseudomonad populations in the phyllosphere of sugar beet by phytophagous caterpillars. FEMS Microbiol. Ecol. 24: 151–157 [Google Scholar]
- Lindeberg M., Cunnac S., Collmer A. (2012). Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends Microbiol. 20: 199–208 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16: 3386–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O., Chico J.M., Sánchez-Serrano J.J., Solano R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R., Rosas-Díaz T., Gusmaroli G., Luna A.P., Taconnat L., Deng X.W., Bejarano E.R. (2011). Geminiviruses subvert ubiquitination by altering CSN-mediated derubylation of SCF E3 ligase complexes and inhibit jasmonate signaling in Arabidopsis thaliana. Plant Cell 23: 1014–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund S.T., Stall R.E., Klee H.J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10: 371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes J.R., Monte D.C., Durzan D. (2000). Nitric oxide and ethylene emission in Arabidopsis thaliana. Physiol. Mol. Biol. Plants 6: 117–127 [Google Scholar]
- Mauck K.E., Bosque-Perez N.A., Eigenbrode S.D., De Moraes C.M., Mescher M.C. (2012). Transmission mechanisms shape pathogen effects on host-vector interactions: Evidence from plant viruses. Funct. Ecol. 26: 1162–1175 [Google Scholar]
- Mersmann S., Bourdais G., Rietz S., Robatzek S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet Y.A., Danna C.H., Clay N.K., Songnuan W., Simon M.D., Werck-Reichhart D., Ausubel F.M. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22: 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina T.E., Zeier J. (2007). Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Morris C.E., Sands D.C., Vinatzer B.A., Glaux C., Guilbaud C., Buffière A., Yan S.C., Dominguez H., Thompson B.M. (2008). The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2: 321–334 [DOI] [PubMed] [Google Scholar]
- Mühlenbock P., Szechynska-Hebda M., Plaszczyca M., Baudo M., Mateo A., Mullineaux P.M., Parker J.E., Karpinska B., Karpinski S. (2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356 Erratum. Plant Cell 20: 3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur L.A., Lloyd A.J., Cristescu S.M., Harren F.J., Hall M.A., Smith A.R. (2009). Biphasic ethylene production during the hypersensitive response in Arabidopsis: A window into defense priming mechanisms? Plant Signal. Behav. 4: 610–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L., Zipfel C., Rowland O., Keller I., Robatzek S., Boller T., Jones J.D.G. (2004). The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol. 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Halitschke R., Kim H.B., Baldwin I.T., Feldmann K.A., Feyereisen R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pieterse C.M.J., van der Does D., Zamioudis C., Leon-Reyes A., van Wees S.C.M. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Rietz S., Stamm A., Malonek S., Wagner S., Becker D., Medina-Escobar N., Vlot A.C., Feys B.J., Niefind K., Parker J.E. (2011). Different roles of ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) bound to and dissociated from PHYTOALEXIN DEFICIENT4 (PAD4) in Arabidopsis immunity. New Phytol. 191: 107–119 [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D.G. (2011). Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Rustérucci C., Aviv D.H., Holt B.F., III, Dangl J.L., Parker J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F., Bodenhausen N., Lassueur S., Masclaux F.G., Reymond P. (2013). Differential contribution of transcription factors to Arabidopsis thaliana defence against Spodoptera littoralis. Front. Plant Sci. 4: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth, G.K.(2005). Limma Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, R.C. Gentleman, V.J. Carey, S. Dut, R. Irizarry, and W. Huber, eds (New York Springer), pp. 397–420. [Google Scholar]
- Stavrinides J., McCloskey J.K., Ochman H. (2009). Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl. Environ. Microbiol. 75: 2230–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A., Weber H., Reymond P., Browse J., Farmer E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz H.U., Pittendrigh B.R., Kroymann J., Weniger K., Fritsche J., Bauke A., Mitchell-Olds T. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol. 124: 1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez-Rodriguez M.C., Adams-Phillips L., Liu Y., Wang H., Su S.H., Jester P.J., Zhang S., Bent A.F., Krysan P.J. (2007). MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 143: 661–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N., et al. (2005). 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 139: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler J.S., Agrawal A.A., Halitschke R. (2010). Salicylate-mediated interactions between pathogens and herbivores. Ecology 91: 1075–1082 [DOI] [PubMed] [Google Scholar]
- Thaler J.S., Humphrey P.T.H., Whiteman N.K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17: 260–270 [DOI] [PubMed] [Google Scholar]
- Tintor N., Ross A., Kanehara K., Yamada K., Fan L., Kemmerling B., Nürnberger T., Tsuda K., Saijo Y. (2013). Layered pattern receptor signaling via ethylene and endogenous elicitor peptides during Arabidopsis immunity to bacterial infection. Proc. Natl. Acad. Sci. USA 110: 6211–6216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A., Theologis A. (2004). Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 136: 2982–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Sato M., Stoddard T., Glazebrook J., Katagiri F. (2009). Network properties of robust immunity in plants. PLoS Genet. 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage A., Vlaardingerbroek I., Raaymakers C., van Dam N.M., Dicke M., van Wees S.C.M., Pieterse C.M.J. (2011). Rewiring of the jasmonate signaling pathway in Arabidopsis during insect herbivory. Front. Plant Sci. 2: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Allmann S., Wu J.S., Baldwin I.T. (2008a). Comparisons of LIPOXYGENASE3- and JASMONATE-RESISTANT4/6-silenced plants reveal that jasmonic acid and jasmonic acid-amino acid conjugates play different roles in herbivore resistance of Nicotiana attenuata. Plant Physiol. 146: 904–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Mitra R.M., Hasselmann K.D., Sato M., Lenarz-Wyatt L., Cohen J.D., Katagiri F., Glazebrook J. (2008b). The genetic network controlling the Arabidopsis transcriptional response to Pseudomonas syringae pv. maculicola: Roles of major regulators and the phytotoxin coronatine. Mol. Plant Microbe Interact. 21: 1408–1420 [DOI] [PubMed] [Google Scholar]
- Whiteman N.K., Groen S.C., Chevasco D., Bear A., Beckwith N., Gregory T.R., Denoux C., Mammarella N., Ausubel F.M., Pierce N.E. (2011). Mining the plant-herbivore interface with a leafmining Drosophila of Arabidopsis. Mol. Ecol. 20: 995–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wi S.J., Ji N.R., Park K.Y. (2012). Synergistic biosynthesis of biphasic ethylene and reactive oxygen species in response to hemibiotrophic Phytophthora parasitica in tobacco plants. Plant Physiol. 159: 251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M., Davière J.M., Cheminant S., Regnault T., Baumberger N., Heintz D., Baltz R., Genschik P., Achard P. (2012). The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Winz R.A., Baldwin I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-Induced ethylene reduces jasmonate-induced nicotine accumulation by regulating putrescine N-methyltransferase transcripts. Plant Physiol. 125: 2189–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.Y., Iwasaki M., Machida C., Machida Y., Zhou X., Chua N.H. (2008). betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes Dev. 22: 2564–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X.Y., Spivey N.W., Zeng W., Liu P.P., Fu Z.Q., Klessig D.F., He S.Y., Dong X. (2012). Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N., Tootle T.L., Tsui F., Klessig D.F., Glazebrook J. (1998). PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. Plant Cell 10: 1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C., Kunze G., Chinchilla D., Caniard A., Jones J.D.G., Boller T., Felix G. (2006). Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D.G., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]