Leaf oil bodies are the source of a novel phytoalexin produced in response to fungal infection and senescence.
Abstract
Oil bodies are intracellular structures present in the seed and leaf cells of many land plants. Seed oil bodies are known to function as storage compartments for lipids. However, the physiological function of leaf oil bodies is unknown. Here, we show that leaf oil bodies function as subcellular factories for the production of a stable phytoalexin in response to fungal infection and senescence. Proteomic analysis of oil bodies prepared from Arabidopsis (Arabidopsis thaliana) leaves identified caleosin (CLO3) and α-dioxygenase (α-DOX1). Both CLO3 and α-DOX1 were localized on the surface of oil bodies. Infection with the pathogenic fungus Colletotrichum higginsianum promoted the formation of CLO3- and α-DOX1-positive oil bodies in perilesional areas surrounding the site of infection. α-DOX1 catalyzes the reaction from α-linolenic acid (a major fatty acid component of oil bodies) to an unstable compound, 2-hydroperoxy-octadecatrienoic acid (2-HPOT). Intriguingly, a combination of α-DOX1 and CLO3 produced a stable compound, 2-hydroxy-octadecatrienoic acid (2-HOT), from α-linolenic acid. This suggests that the colocalization of α-DOX1 and CLO3 on oil bodies might prevent the degradation of unstable 2-HPOT by efficiently converting 2-HPOT into the stable compound 2-HOT. We found that 2-HOT had antifungal activity against members of the genus Colletotrichum and that infection with C. higginsianum induced 2-HOT production. These results defined 2-HOT as an Arabidopsis phytoalexin. This study provides, to our knowledge, the first evidence that leaf oil bodies produce a phytoalexin under a pathological condition, which suggests a new mechanism of plant defense.
Lipids have broad and important functions for living organisms. Oil bodies (lipid droplets) are known to be universal cellular organelles that act as lipid storage compartments such as triacylglycerols (Farese and Walther, 2009; Krahmer et al., 2009). Oil bodies are usually surrounded with phospholipids and various oil body proteins (Murphy and Vance, 1999; Murphy, 2012). Plant cells develop a number of oil bodies (Murphy, 2001). The lipids stored in seeds supply energy for seed germination and have drawn attention as a bioenergy resource. However, vegetative organs, including leaves, which do not require storage lipids for growth, also develop oil bodies (Lersten et al., 2006), suggesting that these structures may have unknown function(s) in addition to acting as a source of energy. The physiological roles of oil bodies are poorly understood, especially in vegetative tissues.
Plants produce a variety of secondary metabolites, including phytoalexins (antimicrobial compounds that are synthesized after stresses) and bioactive lipids. Bioactive lipids, such as jasmonate (Li et al., 2002) and green leaf volatiles (Matsui, 2006), have activities to defend against pathogens and herbivores. Some oxygenated lipids, which are referred to as oxylipins, are synthesized from linoleic acid by lipoxygenases within chloroplasts (Porta and Rocha-Sosa, 2002). It is known that oxylipins play an important role as signaling compounds and defense compounds related to biotic stresses and insect infestation (Blée, 2002; Prost et al., 2005; Nalam et al., 2012). However, there are few reports describing the synthesis of bioactive oxylipins in oil bodies (Yadav and Bhatla, 2011). Furthermore, it is not clear whether oil bodies, which consist mainly of triacylglycerols, supply the source materials for the production of bioactive oxylipins.
Fungal infection can dramatically affect crop production. The genus Colletotrichum is a common infectious and pathogenic fungus in a variety of crops, and many species of this fungus use a hemibiotrophic infectious strategy to invade host plants (Perfect et al., 1999). Adapted Colletotrichum species form appressoria and later primary hyphae when they infect plants (Shimada et al., 2006). Experimental systems have been established using Colletotrichum species that consist of an infection assay, a conidia germination assay, and stable transformation procedures (Kimura et al., 2001; Takano et al., 2001; Hiruma et al., 2010). Recently, the genus Colletotrichum genome and transcriptome were analyzed (O’Connell et al., 2012). An isolate of Colletotrichum higginsianum has been reported to infect Arabidopsis (Arabidopsis thaliana; Narusaka et al., 2004; O’Connell et al., 2004). The use of model plants enabled the characterization of the interaction between the fungus and Arabidopsis plants in the infected area (Narusaka et al., 2009; Hiruma et al., 2010, 2011). However, little is known about the defense responses in the perilesional area surrounding the infected area.
Plants produce antifungal compounds in response to biotic stresses. These biotic stress-induced antifungal compounds are defined as phytoalexins (Ahuja et al., 2012). Phytoalexins have a crucial role in plant defense against pathogens. Camalexin (Tsuji et al., 1992; Glazebrook and Ausubel, 1994) and rapalexin A (Pedras and Adio, 2008; Ahuja et al., 2012) are known to be phytoalexins of Arabidopsis. The biosynthesis pathway and the related enzymes of camalexin have been well characterized (Ahuja et al., 2012). PHYTOALEXIN DEFICIENT3, which encodes an enzyme catalyzing the final step of camalexin biosynthesis, is required for defense against the necrotrophic fungus Alternaria brassicicola (Thomma et al., 1999). Arabidopsis plants are supposed to have more phytoalexins unidentified.
Seeds have oil bodies surrounded by oleosins, caleosins, and steroleosins (Shimada and Hara-Nishimura, 2010). Seed oleosins have a central hydrophobic domain and a Pro knot motif responsible for targeting the protein to oil bodies (Abell et al., 1997; Chen et al., 1999; Takahashi et al., 2000; Chen and Tzen, 2001; Aubert et al., 2010), and they confer freezing tolerance to seedlings by maintaining proper oil body size within seed cells (Siloto et al., 2006; Shimada et al., 2008). Caleosins have a Ca2+-binding motif (an EF hand motif) in addition to the oil body-targeting domains (Abell et al., 1997; Chen et al., 1999; Takahashi et al., 2000; Chen and Tzen, 2001; Aubert et al., 2010). Seed caleosin has been shown to possess peroxygenase activity (Hanano et al., 2006). Recent studies show that caleosin homologs are expressed in vegetative tissues (Aubert et al., 2010; Kim et al., 2011). However, the other proteins on leaf oil bodies are not known. In this study, α-dioxygenase (α-DOX1) was identified as an oil body protein and shown to function together with caleosin in dying leaf cells to produce a novel phytoalexin with antifungal activity against Colletotrichum species.
RESULTS
Identification of α-DOX1 as an Oil Body-Localized Protein
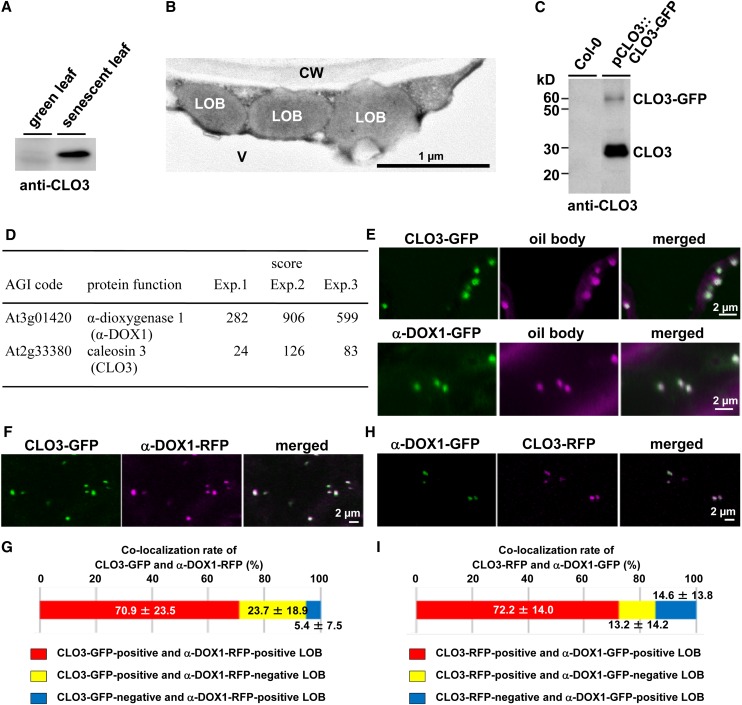
To investigate the role of leaf oil bodies, we first focused on the oil body protein caleosin3/responsive to dessication20/peroxygenase3 (CLO3/RD20/PXG3) of Arabidopsis (Takahashi et al., 2000; Hanano et al., 2006; Aubert et al., 2010). CLO3 was present in senescent leaves but undetectable in green leaves (Fig. 1A), suggesting that dying cells synthesize CLO3. Spherical oil bodies are observed in cytoplasm of the senescent leaf cells but not in chloroplasts (Fig. 1B), suggesting that the oil bodies we focused on in this study were not related to plastoglobules of chloroplasts. To identify the molecular components of leaf oil bodies, transgenic plants expressing CLO3-GFP under the control of the native CLO3 promoter (pCLO3::CLO3-GFP) were generated. Immunoprecipitation of CLO3-GFP-tagged oil bodies from senescent leaves of transgenic plants using anti-GFP antibodies yielded two bands corresponding to CLO3-GFP and endogenous CLO3, which were identified with an anti-CLO3 antibody (Fig. 1C). Mass spectrometric analysis of CLO3-GFP-tagged oil bodies was performed in triplicate on the LTQ-Orbitrap (Supplemental Data S1). In this study, we especially focused on α-DOX1, with a high score (Fig. 1D). α-DOX1 was remarkably up-regulated in senescent leaves, as was CLO3 (Supplemental Fig. S1). As expected, CLO3-GFP was localized to the leaf oil bodies (Fig. 1E). Although α-DOX1 is thought to be a soluble cytosolic protein (Arabidopsis database ATTEDII; Obayashi et al., 2009), α-DOX1-GFP was also detected on the leaf oil bodies (Fig. 1E). Colocalization of α-DOX1 and CLO3 on leaf oil bodies was further demonstrated by the transient expression of α-DOX1-RFP (for red fluorescent protein) and CLO3-GFP (Fig. 1, F and G) and by the transient expression of α-DOX1-GFP and CLO3-RFP (Fig. 1, H and I). Importantly, these analyses identified α-DOX1 as a new oil body-localized protein.
Figure 1.
Colocalization of CLO3 and α-DOX1 on the leaf oil bodies. A, Immunoblots showing the induction of CLO3 by senescence. B, Electron micrographs of senescent leaves showing spherical leaf oil bodies in cytoplasm. CW, Cell wall; LOB, leaf oil body; V, vacuole. C, Immunoblots showing that the pull-down CLO3-GFP-tagged oil bodies contained both CLO3-GFP and endogenous CLO3. D, LTQ-Orbitrap MS of CLO3-GFP-tagged oil bodies identified CLO3 and α-DOX1. Arabidopsis Genome Initiative (AGI) codes and annotations are from The Arabidopsis Information Resource database (http://www.arabidopsis.org). Scores were calculated using the Mascot program (Matrix Science). Raw data are given in Supplemental Data S1. E, Localization of CLO3 and α-DOX1 to oil bodies stained with Nile red. Either CLO3-GFP or α-DOX1-GFP was transiently expressed in N. benthamiana leaves. F, Colocalization of CLO3-GFP and α-DOX1-RFP by transient expression assay in N. benthamiana leaves. G, Quantitative analysis of the colocalization rate of CLO3-GFP and α-DOX1-RFP in seven fluorescence images. A total of 229 oil bodies were examined, most of which (yellow) were CLO3-GFP- and α-DOX1-RFP-positive oil bodies. Data represent average values ± sd. H, Colocalization of CLO3-RFP and α-DOX1-GFP by transient expression assay in N. benthamiana leaves. I, Quantitative analysis of the colocalization rate of CLO3-RFP and α-DOX1-GFP in nine fluorescence images. A total of 188 oil bodies were examined, most of which (yellow) were CLO3-RFP- and α-DOX1-GFP-positive oil bodies. Data represent average values ± sd.
Development of CLO3- and α-DOX1-Positive Oil Bodies in Perilesional Areas of Leaves Infected with C. higginsianum
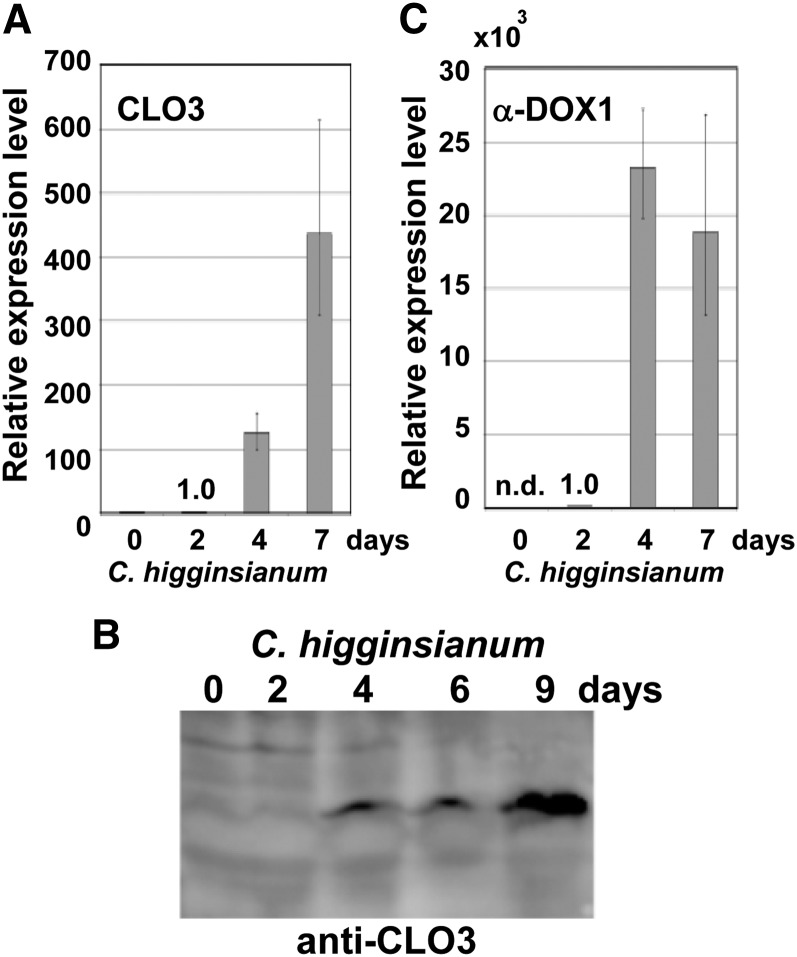
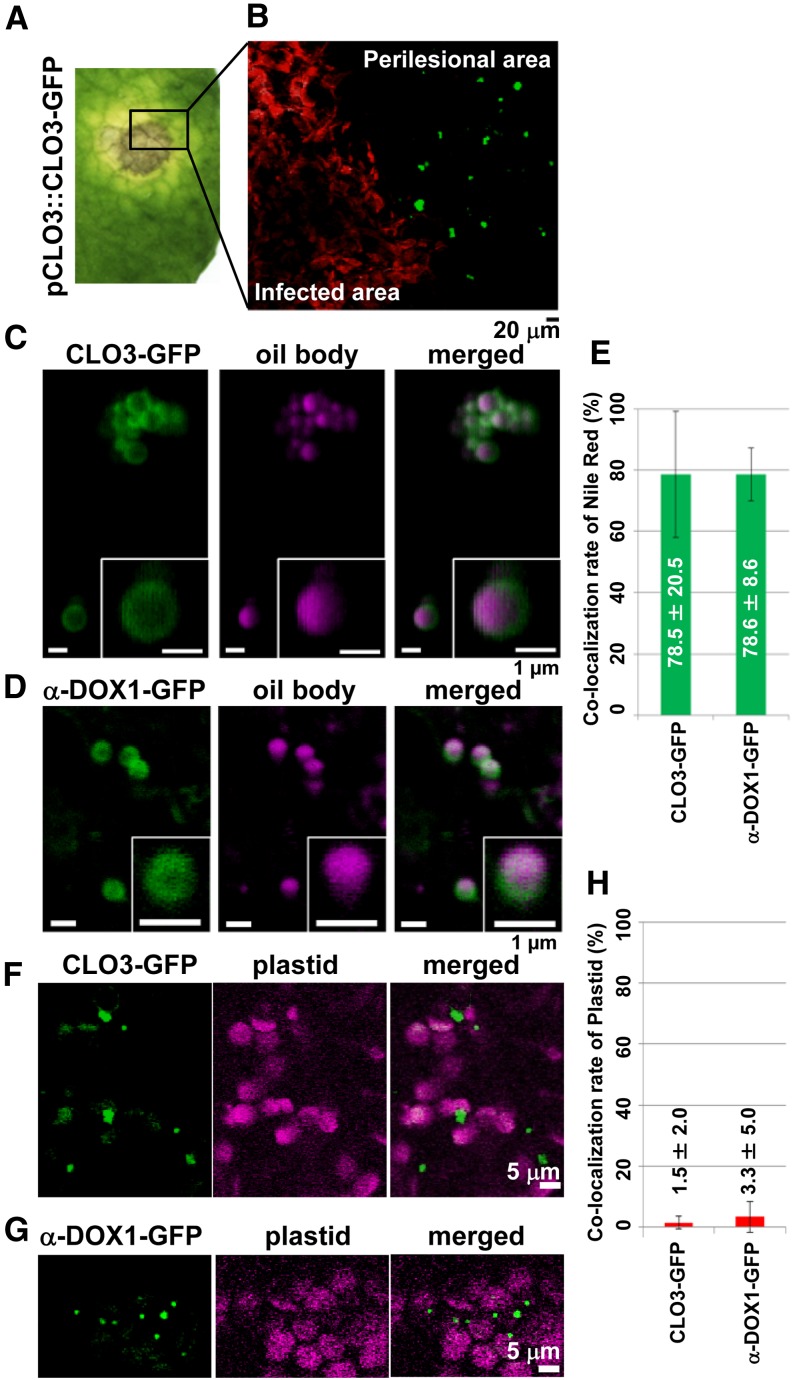
Infection of C. higginsianum, which is pathogenic to Arabidopsis (Narusaka et al., 2004; O’Connell et al., 2004), resulted in the formation of lesions on leaves 4 d after infection. CLO3 mRNA began to accumulate 4 d after infection with C. higginsianum, and the levels increased significantly within 7 d (Fig. 2A), similar to the increase in CLO3 protein (Fig. 2B). α-DOX1 mRNA also began to accumulate 4 d after infection (Fig. 2C). These results indicated that CLO3 and α-DOX1 are inducible by fungal infection. We next examined the emergence of CLO3-positive and α-DOX1-positive oil bodies in greater detail after infection of C. higginsianum. Leaves showed yellowing (chlorosis) of the tissues surrounding the infected area 4 d after infection (Fig. 3A). Interestingly, we found that CLO3-GFP expression in the pCLO3::CLO3-GFP transgenic plants was clearly limited to the yellowing perilesional areas (an area free of fungi) but was not detected in areas where C. higginsianum proliferated (Fig. 3B). On the other hand, transgenic plants expressing α-DOX1-GFP driven by the native α-DOX1 promoter (pα-DOX1::α-DOX1-GFP) were infected with C. higginsianum. α-DOX1-GFP was detected both in the yellowing perilesional areas and in areas where C. higginsianum proliferated (see below). By observation at higher resolution, CLO3-GFP and α-DOX1-GFP were detected on the surface of oil bodies of the perilesional cells (Fig. 3, C and D). Most of the Nile red-stained oil bodies had CLO3-GFP and α-DOX1-GFP (Fig. 3E), showing the colocalization of both proteins on the leaf oil bodies of the transgenic plants. The stainability of oil bodies with Nile red shows that the oil bodies surrounded by CLO3 and α-DOX1 contained neutral lipids including triacylglycerol. Therefore, the oil bodies are defined as normal leaf oil bodies but not lipid droplets derived from damaged membranes. The fluorescence signals of CLO3-GFP or α-DOX1-GFP were not merged with those of plastids (Fig. 3, F–H), indicating that the leaf oil bodies are not related to plastoglobules of plastids. These results suggest that perilesional dying cells are involved in the active induction of both CLO3 and α-DOX1 on leaf oil bodies.
Figure 2.
CLO3 and α-DOX1 are induced remarkably after infection with C. higginsianum. A, mRNA levels of CLO3 in whole leaves after fungal infection. The mRNA level at 2 d was defined as 1.0. Vertical bars indicate the 95% confidence intervals for three experiments. The ACTIN2 gene was included as a control. B, Changes in the levels of the CLO3 protein in response to infection with C. higginsianum. Extracts from leaves infected with C. higginsianum were subjected to immunoblotting with a specific antibody against CLO3. C, mRNA levels of α-DOX1 in whole leaves after fungal infection. The mRNA level at 2 d was defined as 1.0. Vertical bars indicate the 95% confidence intervals for three experiments. The ACTIN2 gene was included as a control. n.d., Not detected.
Figure 3.
CLO3 and α-DOX1 localize to leaf oil bodies of the perilesional area after infection with C. higginsianum. A, Leaves of transgenic plants expressing CLO3-GFP under the control of the endogenous CLO3 promoter were infected with C. higginsianum. The yellow perilesional area surrounds the infected area 4 d after infection. B, Fluorescence image of a section of the infected leaf in A showing that the detection of CLO3-GFP was limited to the perilesional area (an area free of fungi) and was not observed in areas where RFP-labeled fungi proliferated. C, Localization of CLO3-GFP on the surface of oil bodies stained with Nile red in the perilesional area of a 4-d-infected leaf. Insets show magnified views. D, Localization of α-DOX1-GFP on the surface of oil bodies stained with Nile red in the perilesional area of a 6-d-infected leaf of a transgenic plant expressing α-DOX1-GFP under the control of the endogenous α-DOX1 promoter. Insets show magnified views. E, Quantitative analysis of the rate of oil bodies labeled with CLO3-GFP (C) or α-DOX1-GFP (D). A total of 73 oil bodies in three fluorescence images of CLO3-GFP and a total of 101 oil bodies in five fluorescence images of α-DOX1-GFP were examined in 4-d-infected leaves. Data represent average values ± sd (error bars). F and G, Autofluorescence images of plastids and GFP fluorescence images of CLO3-GFP (F) and α-DOX1-GFP (G) in the perilesional area of 4-d-infected leaves. H, Quantitative analysis of the rate of CLO3-GFP-positive dots (F) or α-DOX1-GFP-positive dots (G) overlapped with autofluorescence images of plastids. A total of 135 CLO3-GFP-positive dots in five fluorescence images and a total of 210 α-DOX1-GFP-positive dots in five fluorescence images were examined. Data represent average values ± sd (error bars).
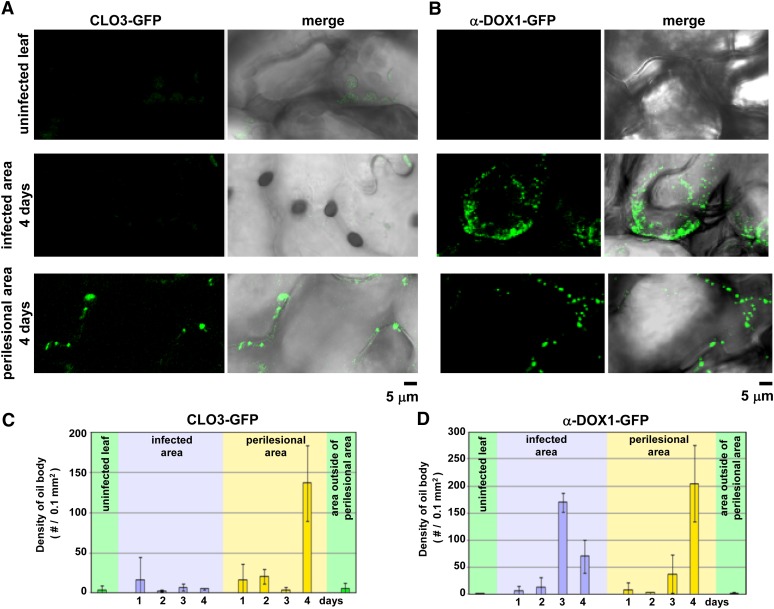
The numbers of CLO3-GFP-positive oil bodies (Fig. 4A; Supplemental Fig. S2) and α-DOX1-GFP-positive oil bodies (Fig. 4B; Supplemental Fig. S3) were measured in a 0.1-mm2 tissue section, and the results are summarized in Figure 4, C and D, respectively. Few CLO3- or α-DOX1-GFP-positive oil bodies were detected in uninfected leaves or in the green areas outside of the yellowing perilesional areas. In the perilesional areas, CLO3- and α-DOX1-GFP-positive oil bodies increased in density 4 d after infection. On the other hand, in the infected area, although CLO3-GFP-positive oil bodies did not develop, α-DOX1-GFP-positive oil bodies increased in density 3 d after infection (Fig. 4). These results indicated that the perilesional cells actively synthesized both CLO3 and α-DOX1 and deposited them on leaf oil bodies.
Figure 4.
Changes in the number of CLO3-GFP-positive and α-DOX1-GFP-positive oil bodies in the infected area and the yellow perilesional area. A, Punctate fluorescent structures in the leaves of pCLO3::CLO3-GFP transgenic plants 0, 2, and 4 d after infection with C. higginsianum. Uninfected leaves, the infected area, and the perilesional area were observed by confocal laser scanning microscopy (CLO3-GFP) and differential interference contrast (DIC). Right panels show merged images of CLO3-GFP and DIC. B, Punctate fluorescent structures in the leaves of pα-DOX1::α-DOX1-GFP transgenic plants 0, 2, and 4 d after infection with C. higginsianum. Uninfected leaves, the infected area, and the perilesional area were observed by confocal laser scanning microscopy (α-DOX1-GFP) and DIC. Right panels show merged images of α-DOX1-GFP and DIC. C and D, Changes in the density of CLO3-GFP-positive (C) and α-DOX1-positive (D) oil bodies (number of oil bodies per 0.1 mm2 of tissue) in the respective leaf area of the transgenic plants after infection. Vertical bars indicate sd of three individual leaves.
Coordinated Activity of α-DOX1 and CLO3 in Oxylipin Synthesis
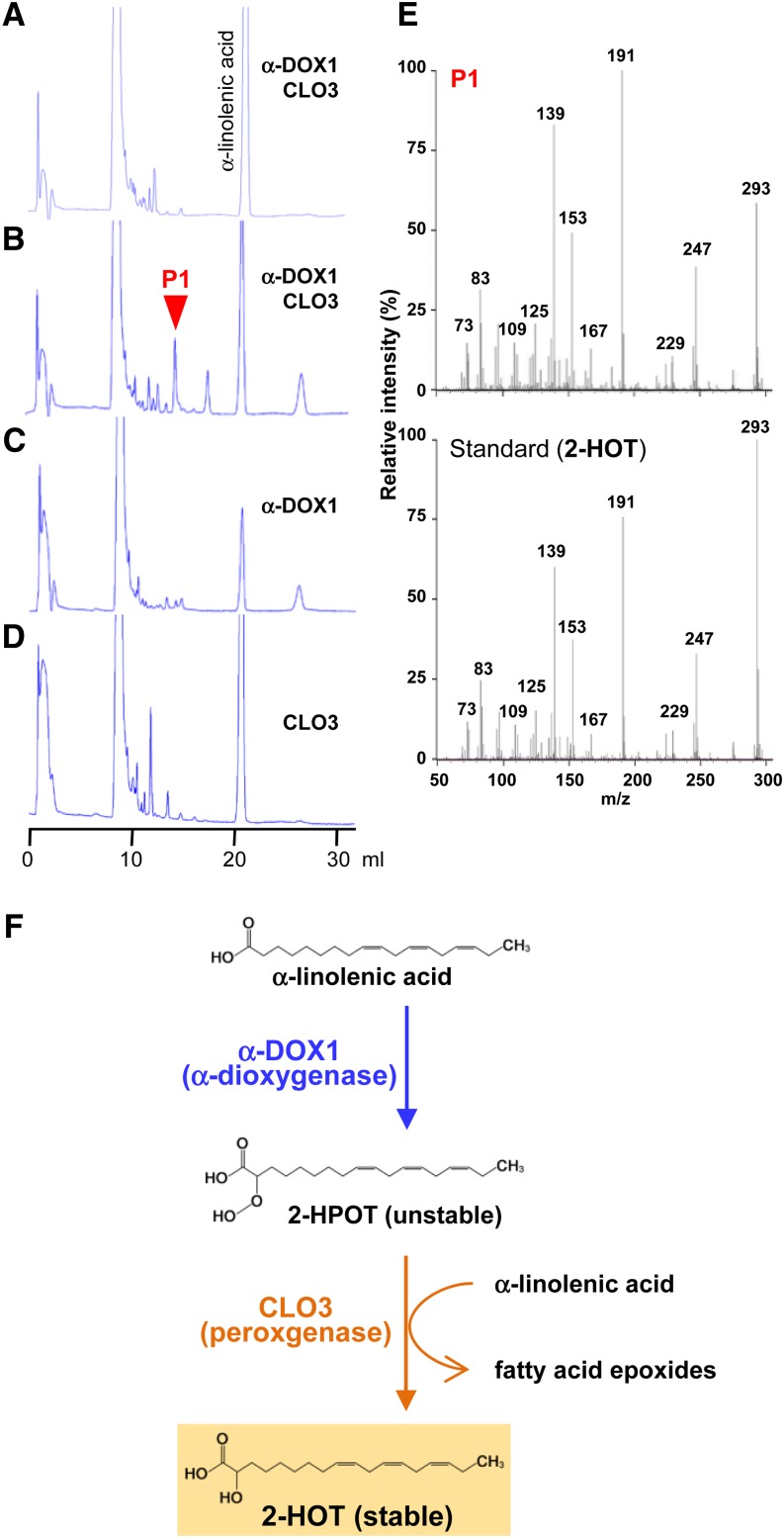
The subcellular colocalization of CLO3 and α-DOX1 suggested that they may function together on the leaf oil bodies. To clarify the roles of α-DOX1 and CLO3, recombinant proteins were synthesized (Supplemental Fig. S4) and incubated with α-linolenic acid (a major fatty acid component of oil bodies) in vitro. The reaction products of α-DOX1 and CLO3 were examined by HPLC. Prior to the reaction, α-linolenic acid produced a single peak (Fig. 5A); however, after incubation with α-DOX1 and CLO3 for 24 h, the α-linolenic acid peak decreased and a new peak (P1) was detected (Fig. 5B). P1 was not present after the incubation of α-linolenic acid with α-DOX1 (Fig. 5C) or CLO3 (Fig. 5D) alone. P1 was identified as 2-hydroxy-octadecatrienoic acid (2-HOT) by tandem mass spectrometry (MS/MS; Fig. 5E). These results indicated that the coordinated activity of α-DOX1 and CLO3 resulted in the production of 2-HOT from α-linolenic acid. Figure 5F illustrates a possible pathway (see below). The oxygenated fatty acid metabolite 2-HOT is one of the oxylipins. Considering the fact that typical plant oxylipins are synthesized within chloroplasts (Porta and Rocha-Sosa, 2002), the oil body might be a novel subcellular site of oxylipin biosynthesis in plants.
Figure 5.
Recombinant proteins of CLO3 and α-DOX1 act cooperatively to catalyze the conversion of α-linolenic acid into 2-HOT. A to D, HPLC patterns of the products of the reaction of α-linolenic acid with α-DOX1 and CLO3 for 0 h (A) and 24 h (B) and with either α-DOX1 (C) or CLO3 (D) for 24 h. α-DOX1 and CLO3 together generate a new peak (P1) from α-linolenic acid. E, MS/MS profiles of the P1 fraction (top) and a standard of 2-HOT (bottom) identifying the product of the P1 fraction as 2-HOT. F, Possible α-dioxygenase pathway for the production of 2-HOT from α-linolenic acid involving the cooperative action of α-DOX1 and CLO3.
The Oxylipin 2-HOT Has Antifungal Activity against Members of the Genus Colletotrichum
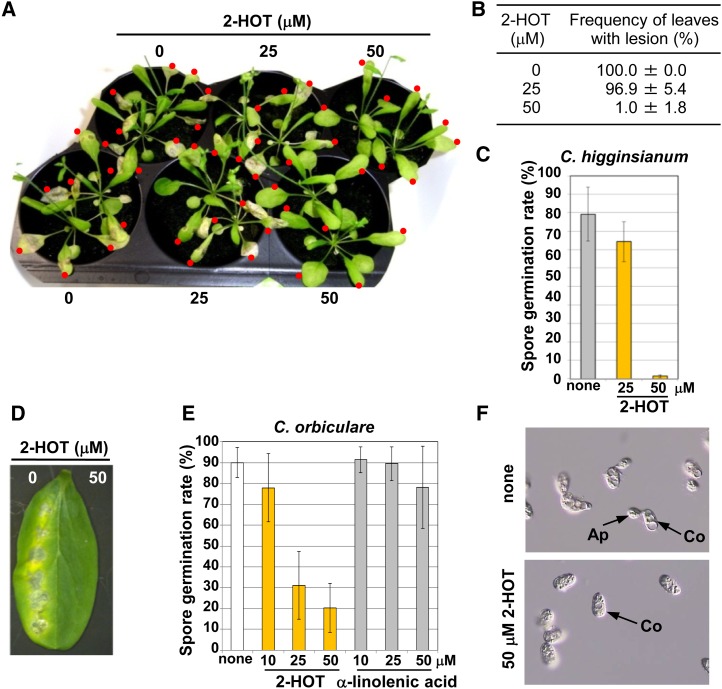
Plant oxylipins are known to have a variety of biological activities and are involved in the defense against pathogens (Namai et al., 1993; Prost et al., 2005; Walters et al., 2006). We examined the potential antifungal activity of 2-HOT against the genus Colletotrichum. Infection of C. higginsianum resulted in the formation of lesions on leaves 7 d after infection (Fig. 6A). Treatment with 50 µm 2-HOT strongly suppressed lesion formation by C. higginsianum (Fig. 6A; Supplemental Fig. S5) and caused a 99% decrease in the number of leaves forming lesions (Fig. 6B). Additionally, 2-HOT at 50 µm clearly inhibited C. higginsianum spore germination (Fig. 6C). The activity of another species of Colletotrichum that infects cucumber (Cucumis sativus) leaves, Colletotrichum orbiculare (Takano et al., 2009), was also suppressed by 2-HOT at 50 µm (Fig. 6D), suggesting that 2-HOT has antifungal activity against the genus Colletotrichum. Furthermore, 2-HOT reduced C. orbiculare spore germination in a dose-dependent manner but α-linolenic acid was ineffective (Fig. 6, E and F). The effective concentration of 2-HOT was comparable with that reported for other antifungal oxylipins (Namai et al., 1993; Prost et al., 2005; Walters et al., 2006). Taken together, these findings suggested that oil bodies may play a role in converting a nonantifungal compound (α-linolenic acid) into a stable antifungal compound (2-HOT). Thus, plants might have evolved an oil body-mediated toxin production system to protect healthy tissues and young plants from fungal infection.
Figure 6.
Antifungal activity of 2-HOT against C. higginsianum and C. orbiculare. A, Arabidopsis leaves that were drop inoculated with spore suspensions of C. higginsianum containing 2-HOT (0, 25, and 50 µm). Images were obtained 7 d after inoculation. Red dots indicate drop-inoculated leaves. B, Number of leaves forming lesions after infection with C. higginsianum. Statistical analyses were performed on three independent infection experiments. Photographs of the infected plants are shown in A and Supplemental Figure S5. C, Effect of 2-HOT on the germination rate of C. higginsianum spores on Arabidopsis leaves. Drops containing spores and the indicated compounds were placed on the leaves and viewed after 1 d. none, Solvent only (0.3% ethanol). Vertical bars indicate sd of three repeated experiments. D, Cucumber cotyledons were drop inoculated with spore suspensions of another species of Colletotrichum, C. orbiculare, containing 2-HOT (0, 25, and 50 µm). Images were obtained 7 d after the inoculation. E, Effect of 2-HOT on the germination rate of C. orbiculare spores. Drops containing spores and the indicated compounds were placed on a microscope slide and viewed after 3 h. none, Solvent only (0.3% ethanol). Vertical bars indicate sd of three repeated experiments. F, Photographs of the germination rate of C. orbiculare spores. Spore suspensions with or without 2-HOT-containing spores and the indicated compounds were placed on a microscope slide and viewed after 3 h. Ap, Appressorium; Co, conidia; none, spore suspension (0.3% ethanol).
The Oxylipin 2-HOT Is Defined as a Novel Phytoalexin and Is Synthesized in Response to Fungal Infection and Senescence
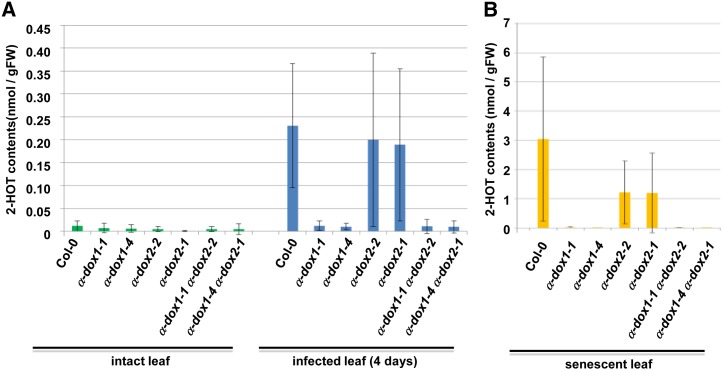
To explore if 2-HOT is a phytoalexin, which is synthesized de novo after the perception of stresses, including pathogen infection (Ahuja et al., 2012), we examined the induction of 2-HOT accumulation in response to C. higginsianum infection by using liquid chromatography (LC)-mass spectrometry (MS). LC-MS analysis showed that the 2-HOT level in infected leaves was approximately 19-fold higher than that in uninfected leaves (Fig. 7A; Supplemental Fig. S6A). This result indicates that the stable oxylipin 2-HOT is a novel phytoalexin.
Figure 7.
The accumulation of 2-HOT in infected and senescent leaves is dependent on α-dioxygenase. 2-HOT contents in the wild type and the α-dioxygenase mutants (α-dox1-1, α-dox1-4, α-dox2-2, α-dox2-1, α-dox1-1 α-dox2-2, and α-dox1-4 α-dox2-1) were measured by LC-MS. Leaves at 4 d after C. higginsianum infection (A; infected leaf), senescent leaves (B), and intact leaves were used. Data represent average values ± sd (error bars). Details are provided in Supplemental Figure S6A. FW, Fresh weight.
To determine whether α-DOX1 and α-DOX2 are involved in the production of 2-HOT in vivo, we generated α-dioxygenase mutants, including two α-dox1 single mutant alleles (α-dox1-1 and α-dox1-4), two α-dox2 single mutant alleles (α-dox2-2 and α-dox2-1), and two double mutant alleles (α-dox1-1 α-dox2-2 and α-dox1-4 α-dox2-1). The 2-HOT levels in these mutants were determined in the leaves before and after C. higginsianum infection. Intriguingly, a deficiency of α-DOX1 caused a significant reduction in the 2-HOT levels, whereas a deficiency of α-DOX2 did not (Fig. 7A; Supplemental Fig. S6A). This result indicates that α-DOX1, but not α-DOX2, contributes to the phytoalexin 2-HOT synthesis.
We next examined the induction of 2-HOT synthesis during senescence. As expected from the up-regulation of CLO3 and α-DOX1 in response to senescence (Fig. 1A; Supplemental Fig. S7), the 2-HOT level in senescent leaves was much higher than that in intact leaves (Fig. 7; Supplemental Fig. S6A). The results identified a 2-HOT level of approximately 2 µg g−1 fresh weight (Supplemental Fig. S6B). The 2-HOT level in senescent leaves of the α-dox1-1 α-dox2-2 double mutant was 3 orders of magnitude lower than that in senescent wild-type leaves (Supplemental Fig. S6B). The α-dox1 mutants expressed no α-DOX1 mRNA, whereas the α-dox2 mutants expressed only a very small amount of α-DOX2 mRNA in senescent leaves (Supplemental Fig. S6C). A deficiency in α-DOX1 caused a significant reduction of the 2-HOT levels in senescent leaves, whereas a deficiency in α-DOX2 did not (Fig. 7B; Supplemental Fig. S6A). These results indicate that senescence and fungal infection both induce α-DOX1-dependent 2-HOT production. The in vitro data (Fig. 5) and these results suggest that α-DOX1 functions coordinately with CLO3 in the synthesis of the novel phytoalexin 2-HOT in response to fungal infection and senescence.
We also generated the clo3 mutant (Supplemental Fig. S8A) and determined the 2-HOT content in this mutant. In contrast to α-dox1 mutants, the clo3 mutant exhibited no reduction of 2-HOT levels (Supplemental Fig. S8B). Arabidopsis has eight caleosin homologs, including CLO3 (Hanano et al., 2006; Partridge and Murphy, 2009). In addition, one of the closely related homologs, CLO4, is localized at oil bodies in Arabidopsis leaves (Aubert et al., 2010; Kim et al., 2011). Hence, the other homologs might compensate for the lack of CLO3, resulting in no reduction of the 2-HOT content in the clo3 mutant. Whereas caleosins could be related to 2-HOT production, we cannot eliminate the possibility that some other oil body proteins play the same role.
DISCUSSION
This study shows that leaf oil bodies play other roles in addition to oil storage. The results indicate that oil bodies function as subcellular factories for the production of a novel phytoalexin. To our knowledge, this is the first evidence of an active function of leaf oil bodies. The production of 2-HOT from α-linolenic acid required the coordinated action of two enzymes, α-dioxygenase and peroxygenase. CLO3 may act as a peroxygenase (Fig. 5F), which is consistent with the fact that the seed-type caleosin CLO1/PXG1 has peroxygenase activity (Hanano et al., 2006). α-DOX1 catalyzes the conversion of α-linolenic acid to 2-hydroperoxy-octadecatrienoic acid (2-HPOT; Fig. 5F), which is unstable and easily degraded to heptadecatrienal (Hamberg et al., 1999). Without 2-HPOT degradation, the peroxygenase CLO3 is hypothesized to catalyze a coupling reaction in which 2-HPOT and α-linolenic acid are converted into 2-HOT and a fatty acid epoxide (Fig. 5F). The colocalization of α-DOX1 and CLO3 on oil bodies would prevent the degradation of unstable 2-HPOT, resulting in the efficient production of the more stable 2-HOT. This might explain the slow in vitro rate of production of 2-HOT in the presence of both α-DOX1 and CLO3 (Fig. 5B), because of a diffusion-limited time lag for the two enzymes and the substrate to combine. The colocalization of α-DOX1 and CLO3 together with α-linolenic acid might be necessary for efficient 2-HOT production.
The phytoalexin 2-HOT might be mainly produced in senescent leaves and in the perilesional area of the infected leaves, where both α-DOX1 and CLO3 genes are up-regulated (Fig. 2; Supplemental Fig. S1). On the other hand, no 2-HOT might be produced in the infected area, where only the α-DOX1 gene was up-regulated (Fig. 4), although we cannot exclude the possibility that α-DOX1, together with other homologs of caleosins, functions in the production of 2-HOT in the infected area.
Although α-DOX1-GFP was detected to be localized on the leaf oil bodies (Fig. 1E), it cannot be excluded that α-DOX1 is also distributed in the cytosol. However, it appears that such non-oil body α-DOX1, if any, is not involved in 2-HOT production, for three reasons as follows. (1) Although α-DOX2 has a similar dioxygenase activity toward α-linolenic acid to α-DOX1 (Bannenberg et al., 2009), α-DOX2 did not contribute to 2-HOT production (Fig. 7). Interestingly, we found that α-DOX2 was not localized on oil bodies but on endoplasmic reticulum-like structures (Supplemental Fig. S9). These findings suggest that the 2-HOT production by α-dioxygenases occurs on oil bodies. Consequently, oil body-localized α-DOX1 might mainly contribute to produce 2-HOT. (2) CLO3 (a caleosin isoform) is localized on oil bodies (Aubert et al., 2010). For the efficient production of 2-HOT via the unstable intermediate 2-HPOT, α-DOX1 should be close to CLO3. Hence, non-oil body α-DOX1 hardly contributes to the production of 2-HOT. (3) The substrate α-linolenic acid might be concentrated in oil bodies. The probability of collision between non-oil body α-DOX1 and the substrates is considered to be low.
Several phytoalexins have been reported; however, the organelle(s) involved in the synthesis of phytoalexins were not fully known. Therefore, this study provides a valuable insight into the production of the novel Arabidopsis phytoalexin 2-HOT. Previous reports showed that 2-HOT suppressed the cell death that resulted from infection with pathogenic bacteria (Hamberg et al., 2003; Vicente et al., 2012). A recent report suggested that 2-HOT correlated with defense activation during insect feeding in Nicotiana attenuata (Gaquerel et al., 2012). These reports imply that 2-HOT is a multifunctional metabolite against fungi, bacteria, and insects and that the oil body-mediated 2-HOT production system functions in defense against these invaders. The novel phytoalexin 2-HOT also was found in N. attenuata (Steppuhn et al., 2010). Genes with homology to CLO3 and α-DOX1 were found in rice (Oryza sativa), grape (Vitis vinifera), fern (Selaginella moellendorffii), and moss (Physcomitrella patens; SALAD database [http://salad.dna.affrc.go.jp/salad/]; Mihara et al., 2010). Consequently, the oil body-mediated 2-HOT production system could be widespread in land plants.
The bioactive compound-producing oil bodies develop in dying cells of the perilesional tissues and senescent leaves. They are responsible for the accumulation of stable phytoalexins in dead tissues and fallen leaves, which are unable to generate active defenses. Little is known about immunity in the perilesional area in plants, although immunity in the infected regions is well characterized (Collins et al., 2003; Hatsugai et al., 2004; Lipka et al., 2005; Jones and Dangl, 2006; Stein et al., 2006; Hatsugai et al., 2009; Narusaka et al., 2009; Shirasu, 2009; Hiruma et al., 2010; Hara-Nishimura and Hatsugai, 2011). This study provides a conceptual advance to our understanding of oil body function by revealing a form of perilesional defense in leaf oil bodies that is designed to block the spread of fungi from dead tissues to healthy tissues.
Plant defense responses against pathogen infection involve the activation of cell-autonomous immunity in the infected region (Hatsugai et al., 2004, 2009; Hara-Nishimura and Hatsugai, 2011). Infection-site defense strategies have been well characterized and work primarily to stop pathogen proliferation (Collins et al., 2003; Lipka et al., 2005; Jones and Dangl, 2006; Stein et al., 2006; Narusaka et al., 2009; Shirasu, 2009; Hiruma et al., 2010). This study shows that the perilesional tissues may also be involved in the prevention of fungal propagation through oil body-mediated toxin production. This perilesional system might function as a secondary defense against fungal infection. The expression of CLO3 and α-DOX1 was markedly enhanced by senescence and the plant hormone abscisic acid (ABA; Supplemental Fig. S7). Therefore, ABA might be an inducer of the oil body system in perilesional cells. ABA, which is an essential signal for plant resistance to pathogens (Adie et al., 2007), might be translocated from fungus-infected areas to the perilesional area to induce the expression of CLO3 and α-DOX1. The plant hormone methyl jasmonate does not significantly induce the expression of α-DOX1 and CLO3 (Supplemental Fig. S7), suggesting that the senescence and infection conditions to induce these two genes are not linked to the wounding or damage condition.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) Columbia (Col-0) and Landsberg erecta (Ler) were used as wild-type plants. Arabidopsis seeds were surface sterilized with 70% ethanol, sown on Murashige and Skoog medium (Wako) with 1% (w/v) Suc and 0.8% (w/v) agar, and incubated at 4°C for 1 to 3 d to break seed dormancy. The seeds were germinated and grown at 22°C under continuous light (100 µE s−1 m−2) for 3 weeks and then transferred to vermiculite for growth at 22°C using a 16-h-light (100 µE s−1 m−2)/8-h-dark photoperiod. Four transfer DNA mutants (Col-0 background) were obtained from the Arabidopsis Biological Resource Center at Ohio State University: SALK_005633 (α-dox1-1), SM_3_27646 (α-dox1-4), SALK_029547 (α-dox2-1), and SALK_089649 (α-dox2-2); α-dox1-1, α-dox2-1, and α-dox2-2 have been described by Bannenberg et al. (2009). The double mutants α-dox1-1 α-dox2-2 and α-dox1-4 α-dox2-1 were generated by crossing. A transfer DNA mutant (Ler background) was obtained from the Cold Spring Harbor Laboratory: GT15023 (clo3 KO).
Fungal Cultures
Colletotrichum higginsianum (MAFF305635) and Colletotrichum orbiculare (syn. Colletotrichum lagenarium) strain 104-T (MAFF240422) were obtained from the MAFF GenBank. A transgenic C. higginsianum expressing RFP has been described previously (Hiruma et al., 2010). The fungal cultures were maintained on potato dextrose agar medium (Difco Laboratories) at 24°C in the dark. Conidia were obtained by gentle scraping of cultures incubated for 7 to 14 d, followed by filtration through two layers of sterile cheesecloth.
Chemicals
Two optical isomers of 2-HOT were used: 2(R)-hydroxy-9(Z),12(Z),15(Z)-octadecatrienoic acid [2(R)-HOT] and 2(R,S)-hydroxy-9(Z),12(Z),15(Z)-octadecatrienoic acid [2(R,S)-HOT]. Both 2(R)-HOT and 2(R,S)-HOT were obtained from Larodan Fine Chemicals, although 2(R)-HOT was not available in May 2011. 2(R)-HOT was used in the experiments of Figures 5E and 6, D to F, and Supplemental Figure S6B, while 2(R,S)-HOT was used in the experiments of Figures 6, A to C, and 7 and Supplemental Figures S5 and S6A. α-Linolenic acid was obtained from Cayman Chemical.
Plasmid DNA Constructs
The plasmid constructs were produced using Gateway Technology (Invitrogen) with the destination vectors pGWB405 (Nakagawa et al., 2007), pGWB560 (AB543177; Nakagawa et al., 2007; Nakamura et al., 2010), pBGWF7 (Karimi et al., 2002; Shimada et al., 2010; Plant System Biology), and FAST-R7 (Shimada et al., 2010; Plant System Biology). The CLO3 complementary DNA (cDNA) fragment from nucleotide 1, corresponding to A of the start codon ATG, to nucleotide 708 was amplified by PCR using the primer set 5′-CACCATGGCAGGAGAGGCAGAGGCTT-3′ and 5′-GTCTTGTTTGCGAGAATTGGCCC-3′. The α-DOX1 cDNA fragment (from nucleotide 1 to 1,917) was amplified by PCR using the primer set 5′-CACCATGAAAGTAATTACTTCCCTAAT-3′ and 5′-AGAGGGAATTCGGAGATAGAGAG-3′. The α-DOX2 cDNA fragment (from nucleotide 1 to 1,893) was amplified by PCR using the primer set 5′-CACCATGGGTTTCTCTCCATCCTCTTC-3′ and 5′-TGGAGCGGATCGGAGGTACAAAG-3′. PCR was performed with KOD Plus DNA polymerase (Toyobo) and PrimeSTAR HS DNA polymerase (Takara). The cDNA fragments of CLO3, α-DOX1, and α-DOX2 were inserted into pENTER/D-TOPO (Invitrogen) by a TOPO reaction to produce an entry clone. Clones were then transferred into the destination vector pGWB405 or pGWB560 by a recombination reaction with LR Clonase in Gateway system (LR recombination reaction; Invitrogen), creating five vectors: pGWB405-CLO3-GFP, pGWB405-α-DOX1-GFP, pGWB405-α-DOX2-GFP, pGWB560-CLO3-RFP, and pGWB560-α-DOX1-RFP.
The CLO3 genomic DNA fragment (from nucleotide −1,127 to 1,388) was amplified by PCR using the primer set 5′-CACCGTTAATGAATATCTTGACAACA-3′ and 5′-GGTCTTGTTTGCGAGAATTGGC-3′. PCR was performed with Arabidopsis genomic DNA and KOD Plus (Toyobo). The CLO3 genomic fragment was inserted into pENTER/D-TOPO (Invitrogen) by a TOPO reaction to produce an entry clone. The clone was then transferred into the destination vector pBGWF7 by an LR recombination reaction (Invitrogen) to create vector pBGWF7-pCLO3-CLO3. The α-DOX1 genomic DNA fragment (from nucleotide −2,181 to 3,036) was amplified by PCR using the primer set 5′-CACCGACTCTTTATTTCTTTGTCAAAC-3′ and 5′-GAGAGGGAATTCGGAGATAGAGAG-3′. PCR was performed with Arabidopsis genomic DNA and PrimeSTAR HS DNA polymerase. The α-DOX1 genomic fragment was inserted into pENTER/D-TOPO (Invitrogen) by a TOPO reaction to produce an entry clone. The clone was then transferred into the destination vector pFAST-R7 by an LR recombination reaction (Invitrogen) to create the pFAST-R-pα-DOX1-α-DOX1 vector.
Stable Transformation of Arabidopsis Plants
Three vectors of pBGWF7-pCLO3-CLO3 or pFAST-R-pα-DOX1-α-DOX1 were each introduced into Arabidopsis (Col-0) plants via Agrobacterium tumefaciens (strain GV3101) using the floral dip method (Bechtold and Pelletier, 1998) to generate pCLO3::CLO3-GFP or pα-DOX1::α-DOX1-GFP, respectively.
Immunoprecipitation
Extracts from senescent leaves of the wild type (Col-0) and pCLO3::CLO3-GFP (0.8 g fresh weight) were ground with 1.5 mL of extraction buffer containing 20 mm Tris-HCl, 150 mm NaCl, 1 mm CaCl2, 1 mm MgCl2, and protein inhibitor (complete, EDTA free; Roche). The extracts were centrifuged at 500g for 1 min at 4°C; the supernatants were centrifuged at 1,000g for 5 min at 4°C followed by 8,000g for 10 min at 4°C. The supernatants (1 mL each) were subjected to immunoprecipitation with 50 μL of anti-GFP microbeads (μMACS GFP tag protein isolation kit; Milteny Biotech) and then were applied to a column (μ column; Milteny Biotech). Pure immunoprecipitates were eluted with 50 μL of elution buffer (2% [w/v] SDS).
Immunoblotting
Immunoblot procedures followed those of previous studies (Shimada et al., 2008). Arabidopsis leaves were homogenized in 2- to 10-fold SDS sample buffer (100 mm Tris-HCl, pH 6.8, 4% [w/v] SDS, 20% [v/v] glycerol, and 10% [v/v] 2-mercaptoethanol) under the same concentrations of samples. The homogenates were subjected to SDS-PAGE on 15% (w/v) acrylamide gels. Immunoprecipitated samples (25 μL) were added to 25 μL of SDS sample buffer, and the mixtures were subjected to SDS-PAGE on 7.5% (w/v) to 15% (w/v) acrylamide gradient gels (Bio Craft). The membranes were treated with blocking solution (5% [w/v] skim milk, 100 mM Tris-HCl, pH 7.5, and 0.05% [v/v] Tween 20) and then incubated for 1 h in anti-CLO3 antibody (diluted 1:2,000; Shimada et al., 2010).
Proteomic Analysis
The procedures for proteomic analysis have been described previously (Fujiwara et al., 2009). Immunoprecipitated samples (25 μL) were added to 25 μL of SDS sample buffer and then subjected to SDS-PAGE on 7.5% (w/v) to 15% (w/v) acrylamide gradient gels (Bio Craft). For shotgun analysis, gel lanes were divided into three blocks. Each block was cut into small pieces, washed with 100% (v/v) acetonitrile (Kanto Chemical), and dried in a vacuum concentrator. The dried gel pieces were reduced with 10 mm dithiothreitol and 50 mm NH4HCO3 for 45 min at 56°C and alkylated with 55 mm iodoacetamide and 50 mm NH4HCO3 in the dark for 30 min at room temperature. After dilution with 25 mm NH4HCO3, gel pieces were washed twice with HPLC-grade water containing 50% (v/v) acetonitrile. The dried gel pieces were treated with 10 µg mL−1 trypsin (sequence grade; Promega) and 50 mm NH4HCO3 and incubated at 37°C for 16 h. Digested peptides in the gel pieces were recovered with 30 µL of 5% (v/v) formic acid and 50% (v/v) acetonitrile and then with 30 µL of 5% (v/v) formic acid and 70% (v/v) acetonitrile. Combined extracts were dried in a vacuum concentrator and dissolved in 5% (v/v) acetonitrile and 0.1% (v/v) acetic acid. Finally, the peptide extracts were filtered with an Ultra free-MC 0.45-µm Filter Unit (Millipore).
Mass spectrometric analyses were performed using an LTQ-Orbitrap XL-HTC-PAL-Paradigm MS4 system. Trypsin-digested peptides were loaded on the column (75-µm internal diameter, 15 cm; L-Column; CERI) using a Paradigm MS4 HPLC pump (Michrom BioResources) and an HTC-PAL autosampler (CTC Analytics). Two solutions were used: 0.1% (v/v) acetic acid and 2% (v/v) acetonitrile in water (solvent A) and 0.1% (v/v) acetic acid and 90% (v/v) acetonitrile in water (solvent B). A linear gradient from 5% to 45% (solvent B) was applied for 25 min, and peptides eluted from the column were introduced directly into an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific) at a flow rate of 300 nL min–1 and a spray voltage of 2.0 kV. The resulting spectra were compared with a protein database (20080607) from the National Center for Biotechnology Information using the Mascot server (version 2.1; Matrix Science).
Electron Microscopy
2The aerial parts of 6-week-old plants (Col-0) were transferred to the dark condition at 24°C and incubated for 7 d to obtain senescent leaves. The senescent leaves were cut into small pieces and fixed with 2% (w/v) paraformaldehyde and 2% (v/v) glutaraldehyde in 0.05 m cacodylate buffer, pH 7.4, at 4°C overnight. Then, the samples were rinsed three times with 0.05 m cacodylate buffer for 30 min each followed by post fixation with 2% (v/v) osmium tetroxide in 0.05 m cacodylate buffer at 4°C for 3 h. The samples were dehydrated through an ethanol series (50%, 70%, 90%, and 100%). The schedule was as follows: 50% and 70% for 30 min each at 4°C, 90% for 30 min at room temperature, and four changes of 100% for 30 min each at room temperature. After these dehydration processes, the samples were continuously dehydrated with 100% ethanol at room temperature overnight. The samples were infiltrated with propylene oxide two times for 30 min each and put into a 70:30 mixture of propylene oxide and resin (Quetol-651; Nisshin EM) for 1 h; then, the cap of the tube was kept open and propylene oxide was volatilized overnight. The samples were transferred to fresh 100% resin and polymerized at 60°C for 48 h. The blocks were ultrathin sectioned at 70 nm with a diamond knife using an ultramicrotome (Ultracut UCT; Leica), and sections were placed on copper grids. They were stained with 2% (w/v) uranyl acetate at room temperature for 15 min, rinsed with distilled water, and then secondarily stained with lead stain solution (Sigma-Aldrich) at room temperature for 3 min. The grids were observed by transmission electron microscopy (JEM-1400Plus; JEOL) at an acceleration voltage of 80 kV. Digital images (2,048 × 2,048 pixels) were taken with a CCD camera (VELETA; Olympus Soft Imaging Solutions).
Confocal Laser Scanning Microscopy
GFP images of transgenic plants were obtained with a confocal laser scanning microscope (LSM510 META; Carl Zeiss) using the 488-nm line of a 40-mV argon/krypton laser or the 544-nm line of a 1-mW helium/neon laser. Differential interference contrast images were obtained with the same microscope. Procedures followed those described previously (Shimada et al., 2008). The 488-nm laser was used to excite GFP, whereas the 543-nm laser was used to excite RFP and for autofluorescence of plastids. The leaves of transgenic plants (pCLO3::CLO3-GFP and pα-DOX1::α-DOX1-GFP) infected with C. higginsianum were observed using a confocal laser scanning microscope to determine the numbers of punctate fluorescent structures in primary lesion areas, secondary lesion areas, and areas outside of secondary lesions.
Nile Red Staining
Arabidopsis leaves were vacuum infiltrated for 5 min with 2.5 µg mL−1 Nile red stain in water. The fluorescence of GFP and Nile red was inspected using the confocal laser scanning microscope. Their fluorescence spectra were separated using the LSM510 META device.
Transient Expression Assay in Nicotiana benthamiana
Five vectors (pGWB405-CLO3-GFP, pGWB405-α-DOX1-GFP, pGWB405-α-DOX2-GFP, pGWB560-CLO3-RFP, and pGWB560-α-DOX1-RFP) were developed with transient expression assays as described previously (Sparkes et al., 2006). The vectors were independently transformed into A. tumefaciens (strain GV3101). Transformed A. tumefaciens strains were inoculated (approximately 1.0 optical density) into the leaves of 5- to 7-week-old N. benthamiana plants. One day after A. tumefaciens inoculation, 100 µm ABA was infiltrated into the inoculated leaves to induce leaf oil bodies. Two days after A. tumefaciens inoculation, the inoculated leaves were observed with a confocal laser-scanning microscope (LSM510 META; Carl Zeiss).
Measurement of Antifungal Activity
To measure antifungal activity on cucumber (Cucumis sativus), conidial suspensions of C. orbiculare (5 × 105 conidia mL−1) with 0.3% ethanol (control), 25 µm 2(R)-HOT, and 50 µm 2(R)-HOT were drop inoculated onto detached cotyledons of cucumber, as described previously (Asakura et al., 2009). The cucumber cotyledons were incubated for 7 d at 24°C. This experiment was replicated three times.
Enzyme Reactions of CLO3 and α-DOX1
Purified CLO3 and α-DOX1 solutions were used in three forms: 60-μL CLO3 solution and 60-μL α-DOX1 solution (CLO3 and α-DOX1), 120-μL CLO3 solution (only CLO3), and 120-μL α-DOX1 solution (only α-DOX1). The enzyme solutions were made with 2 μL of 100 mm α-linolenic acid. A 55-μL portion of each enzyme solution was removed (0 h). The remaining enzyme solutions were incubated at 25°C for 24 h. The enzyme solutions (0 and 24 h) were added to 1 mL of acetonitrile:water:trifluoroacetic acid (5:95:0.1, v/v), filtered with an Ultra free-MC 0.45-μm Filter Unit, and then analyzed by HPLC with AKTApurifier 10 (GE Healthcare).
C. higginsianum Inoculation
Arabidopsis plants for fungal inoculation were grown at 22°C for 3 weeks under continuous light (100 µE s−1 m−2) and then transferred to vermiculite at 22°C for 4 weeks under a 16-h-light (100 µE s−1 m−2)/8-h-dark photoperiod. One drop (2 µL each) of conidial suspension (5 × 105 conidia mL−1) was spotted on each leaf of the plants, as described previously (Hiruma et al., 2010). Inoculated plants were placed at 22°C under continuous light (100 µE s−1 m−2) and 100% relative humidity.
To measure antifungal activity, conidial suspensions of C. higginsianum (5 × 105 conidia mL−1) with 1.5% ethanol (control), 25 µm 2(R,S)-HOT, and 50 µm 2(R,S)-HOT were prepared. Four independent 4-week-old Arabidopsis plants were used for each conidial suspension. We made eight spots of each suspension on four leaves of a single plant. The plants were incubated for 7 d at 24°C using a 16-h-light (100 µE s−1 m−2)/8-h-dark photoperiod under 100% relative humidity. This experiment was replicated three times. We determined the frequency rates of leaves that formed lesions after infection with C. higginsianum.
C. higginsianum Spore Germination Assay on Arabidopsis Leaves
Conidial suspensions of C. higginsianum (5 × 105 conidia mL−1) with 1.5% ethanol, 25 µm 2(R,S)-HOT, and 50 µm 2(R,S)-HOT were prepared. The conidial suspensions were spotted on 4-week-old Arabidopsis leaves. The plants were incubated for 1 d at 24°C using a 16-h-light (100 µE s−1 m−2)/8-h-dark photoperiod under 100% relative humidity. The number of germinated conidia, which was defined as conidia with appressorium or hyphae, was counted. This experiment was repeated three times.
C. orbiculare Spore Germination Assay on Glass Slides
Conidia were suspended in 0.1% (w/v) yeast extract (Nacalai Tesque) with each solution of 0.3% ethanol (control), 2(R)-HOT (10, 25, or 50 µm), and α-linolenic acid (10, 25, or 50 µm). The conidial suspensions of C. orbiculare (5 × 105 conidia mL−1) were incubated on multiwell glass microscope slides (ICN Biomedicals) at 24°C for 3 h, as described previously (Takano et al., 2001). The number of germinated conidia, which was defined as conidia with appressorium or hyphae, was counted. This experiment was repeated three times.
Expression and Purification of Recombinant CLO3 and α-DOX1
Recombinant CLO3 and α-DOX1 were expressed in tobacco BY2 cells using the tomato mosaic virus infection system, according to previous studies (Dohi et al., 2006, 2008). At 2 d after subculturing the cells, β-estradiol was added at a final concentration of 10 μm. After a 3-d incubation period, the cells were freeze thawed, sonicated, and then centrifuged for 10 min at 30,000 rpm and 4°C. The supernatant was applied to an Ni2+ affinity column to purify the recombinant protein using a His tag (6×His). The eluted fraction was dialyzed in 0.1 m HEPES (pH 7.2) to remove imidazole. The purified proteins were subjected to SDS-PAGE followed by either staining with Coomassie blue or immunoblotting with anti-CLO3 antibody.
HPLC Analysis and MS/MS Analysis
HPLC was performed with a μRPC C2/C18 ST 4.6/100 column (GE Healthcare). The solvent systems consisted of mixtures of acetonitrile, water, and trifluoroacetic acid in volume proportions of 5:95:0.1 (solvent A) and 95:5:0.1 (solvent B). The absorbance (210 nm) was assessed using an ultraviolet detector (AKTApurifier 10). Enzyme solutions were subjected to HPLC using a solvent system with 100% solvent A at 6.6 mL, followed by a linear increase in solvent B up to 60% solvent B at 0.8 mL, and finally, by an isocratic postrun with 60% solvent B at 22.6 mL. The flow rate was 0.75 mL min−1. The P1 peak appeared with an elution volume of 14.6 mL. The fractions of the enzyme solutions of CLO3 and α-DOX1 for 24 h with elution volumes of 14.5 to 15.5 mL were gathered.
For qualitative analysis of metabolism, 14.5- to 15.5-mL (P1) fractions (CLO3 and α-DOX1 for 24 h) were analyzed by using 2(R)-HOT as a standard with a Waters Q-TOF Premier high-resolution mass spectrometer. Ion detection was achieved in negative mode using a source capillary voltage of 3.5 kV, a sampling cone voltage of 25 V, a source temperature of 80°C, and a cone gas flow of 40 L h−1 (N2).
Quantification of 2-HOT in the Wild Type and α-Dioxygenase Mutants by LC-MS
Arabidopsis plants (the wild type [Col-0 and Ler], α-dox1-1, α-dox1-4, α-dox2-2, α-dox2-1, α-dox1-1 α-dox2-2, α-dox1-4 α-dox2-1, and clo3 KO) were sown on rockwool, incubated at 4°C for 1 to 3 d, and grown at 24°C. Leaves of 4- to 6-week-old plants were used as uninfected leaves. Four-week-old plants were infected with C. higginsianum, and leaves at 4 d after the fungal inoculation were used as infected leaves. The aerial parts of 4- to 6-week-old plants were transferred to the dark condition at 24°C and incubated for 7 d to obtain the senescent leaves.
To measure the 2-HOT contents by LC-MS, samples were prepared from the leaves as reported previously (Okazaki et al., 2009, 2013), except that the samples were finally dissolved in ethanol. High-resolution electrospray MS was performed in negative ion mode using a Xevo G2 QTof mass spectrometer combined with an ACQUITY UPLC system (Waters). Conditions for LC were as follows: column, ACQUITY UPLC HSS T3 1.8 μm (1.0 mm i.d., 50 mm long; Waters); solvent A, acetonitrile:water:1 m ammonium acetate:formic acid, 200:800:10:1 (v/v/v/v); solvent B, acetonitrile:2-propanol:1 m ammonium acetate:formic acid, 100:900:10:1 (v/v/v/v); gradient, 35% to 70% B (0–3 min), 70% to 85% B (3–7 min), 85% to 90% B (7–10 min), 90% B (10–12 min), 90% to 35% B (12–12.5 min), 35% B (12.5–15 min); flow rate, 0.15 mL min–1; column temperature, 55°C. Conditions for MS were as follows: scan range, mass-to-charge ratio (m/z) 100 to 2,000; capillary voltage, 2.5 kV; cone voltage, 40 V; source temperature, 120°C; desolvation temperature, 450°C; cone gas flow, 50 L h−1; desolvation gas flow, 600 L h−1; nebulizer gas, N2. The level of 2-HOT was analyzed using QuanLynx software (Waters) on the basis of the intensity of [M–H]–.
Quantitative Measurement of 2-HOT by LC-Fourier Transform-Mass Spectroscopy
Senescent leaves (approximately 200 mg fresh weight) of the wild type (Col-0) and α-dox1-1 α-dox2-2 were frozen by liquid nitrogen and freeze dried. The leaf sample was homogenized in 100× its weight of 100% ethanol with an internal standard (12.5 µm 7-hydroxy-5-methylflavone; Extrasynthese) and was centrifuged at 20,400g for 15 min at 4°C. The pellet was homogenized and centrifuged again. The pooled supernatant solution (1 mL) was dried with a vacuum evaporator at 40°C, redissolved in 200 µL of 100% ethanol, and then centrifuged at 20,400g for 15 min at 4°C. The supernatants were filtered through a 0.2-µm hydrophilic PTFE filter (Millipore) and subjected to LC-Fourier transform-MS analysis.
An Agilent 1100 system (http://www.home.agilent.com/agilent/home.jspx?lc=eng&cc=US) was used for HPLC. The filtered extract was applied to an Ascentis Express C18 column (4.6 × 150 mm, 2.7 µm; SPELCO) at 40°C. Solvent A of 0.1% (v/v) formic acid and solvent B of acetonitrile with 0.1% (v/v) formic acid served as a gradient with the following program: 30% to 95% solvent B from 0 to 40.0 min, 95% solvent B from 40.0 to 45.0 min, followed by 30% solvent B from 45.1 to 55.0 min, at a flow rate of 0.5 mL min−1.
Fourier transform-ion cyclotron resonance-mass spectrometry was performed with Finnigan LTQ-FT (Themo Electron) under the following conditions: detection mode, electrospray ionization negative; sheath gas, nitrogen; auxiliary gas, nitrogen; spray voltage, 4.0 kV; capillary temperature, 300°C; mass range, m/z = 150 to 800; resolution, 100,000 at m/z = 400. Authentic 2(R)-HOT (m/z = 293.21 [M–H]–) was used.
Reverse Transcription-PCR Analysis
Total RNA was isolated from senescent leaves of the wild type and α-dox1 α-dox2 using the RNeasy Plant Mini Kit (Qiagen). RNA was then treated with DNase I. cDNA prepared from total RNA with Ready-To-Go RT-PCR beads (GE Healthcare) and with the oligo(dT) primer was subjected to PCR for 40 cycles with ExTaq polymerase (Takara) and the following primer sets: 5′-TTGCTTAAATTCATCCACAAAG-3′ and 5′-TGACGATCAATCACATCCTTGAG-3′ for α-DOX1, 5′-GGATCCAGTTCATGATCCATG-3′ and 5′-ATCGCATCCATTGATCGGTTAAC-3′ for α-DOX2, and 5′-AGAGATTCAGATGCCCAGAAGTCTTGTTCC-3′ and 5′-AACGATTCCTGGACCTGCCTCATCATACTC-3′ for ACTIN2.
Quantitative Reverse Transcription-PCR
Arabidopsis leaves were used 0, 2, 4, or 7 d after inoculation with C. higginsianum. Total RNA was isolated from the leaves, and cDNA was prepared as described above. The cDNA was quantified with gene-specific probes (At02162482_g1 for α-DOX1 and At02335270_gH for ACTIN2; Applied Biosystems) and the TaqMan Gene Expression Assay Kit of the 7500 Real-Time PCR System (Applied Biosystems). The relative quantity of target mRNA was calculated using ACTIN2 as a control (Shirakawa et al., 2010).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: CLO1 (At4g26740), CLO2 (At5g55240), CLO3 (At2g33380), CLO4 (At1g70670), α-DOX1 (At3g01420), α-DOX2 (At1g73680), and ACTIN2 (At3g18780).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene expression patterns of CLO3 and α-DOX1 in developing and senescing leaves.
Supplemental Figure S2. Punctate fluorescent structures in leaves of pCLO3::CLO3-GFP transgenic plants 0, 2, and 4 d after infection with C. higginsianum.
Supplemental Figure S3. Punctate fluorescent structures in leaves of pα-DOX1::α-DOX1-GFP transgenic plants 0, 2, and 4 d after infection with C. higginsianum.
Supplemental Figure S4. Purified recombinant proteins CLO3 and α-DOX1.
Supplemental Figure S5. Antifungal activity of 2-HOT against C. higginsianum.
Supplemental Figure S6. 2-HOT contents in uninfected leaves, infected leaves, and senescent leaves of the wild type or α-dioxygenase mutants.
Supplemental Figure S7. Gene expression patterns of four caleosins (CLO1, CLO2, CLO3, and CLO4) and two α-dioxygenases (α-DOX1 and α-DOX2).
Supplemental Figure S8. Accumulation of 2-HOT in senescent leaves of the wild type and the clo3 mutant.
Supplemental Figure S9. Distinct subcellular localization of α-DOX2 from that of α-DOX1.
Supplemental Data S1. LTQ-Orbitrap raw data of the identified proteins.
Acknowledgments
We are grateful to Tetsuro Mimura and Aya Anegawa of Kobe University and Naoharu Watanabe of Shizuoka University for the analysis of oxylipins, to Tsuyoshi Nakagawa of Shimane University for his kind donation of Gateway vectors (pGWB560 and pGWB405), to Tomoko Mori and Yumiko Makino of the National Institute for Basic Biology for their technical support in the MS/MS analysis, to Kyoko Koumoto of Kyoto University for her technical support in the HPLC analysis, to the Arabidopsis Biological Resource Center for providing the seeds of SALK_005633 (α-dox1-1), SM_3_27646 (α-dox1-4), SALK_029547 (α-dox2-1), and SALK_089649 (α-dox2-2), and to the Cold Spring Harbor Laboratory for providing the seeds of KO GT15023 (clo3).
Glossary
- RFP
red fluorescent protein
- 2-HOT
2-hydroxy-octadecatrienoic acid
- LC
liquid chromatography
- MS
mass spectrometry
- 2-HPOT
2-hydroperoxy-octadecatrienoic acid
- ABA
abscisic acid
- Col-0
Columbia
- Ler
Landsberg erecta
- 2(R)-HOT
2(R)-hydroxy-9(Z),12(Z),15(Z)-octadecatrienoic acid
- 2(R,S)-HOT
2(R,S)-hydroxy-9(Z),12(Z),15(Z)-octadecatrienoic acid
- cDNA
complementary DNA
- m/z
mass-to-charge ratio
- MS/MS
tandem mass spectrometry
References
- Abell BM, Holbrook LA, Abenes M, Murphy DJ, Hills MJ, Moloney MM. (1997) Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9: 1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R. (2007) ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja I, Kissen R, Bones AM. (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Asakura M, Ninomiya S, Sugimoto M, Oku M, Yamashita S, Okuno T, Sakai Y, Takano Y. (2009) Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 21: 1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Vavasseur A, Galaud JP. (2010) RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 51: 1975–1987 [DOI] [PubMed] [Google Scholar]
- Bannenberg G, Martínez M, Rodríguez MJ, López MA, Ponce de León I, Hamberg M, Castresana C. (2009) Functional analysis of α-DOX2, an active α-dioxygenase critical for normal development in tomato plants. Plant Physiol 151: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. (1998) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol 82: 259–266 [DOI] [PubMed] [Google Scholar]
- Blée E. (2002) Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7: 315–322 [DOI] [PubMed] [Google Scholar]
- Chen JC, Tsai CC, Tzen JT. (1999) Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol 40: 1079–1086 [DOI] [PubMed] [Google Scholar]
- Chen JC, Tzen JT. (2001) An in vitro system to examine the effective phospholipids and structural domain for protein targeting to seed oil bodies. Plant Cell Physiol 42: 1245–1252 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Dohi K, Nishikiori M, Tamai A, Ishikawa M, Meshi T, Mori M. (2006) Inducible virus-mediated expression of a foreign protein in suspension-cultured plant cells. Arch Virol 151: 1075–1084 [DOI] [PubMed] [Google Scholar]
- Dohi K, Tamai A, Mori M. (2008) Insertion in the coding region of the movement protein improves stability of the plasmid encoding a tomato mosaic virus-based expression vector. Arch Virol 153: 1667–1675 [DOI] [PubMed] [Google Scholar]
- Farese RV, Jr, Walther TC. (2009) Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139: 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Hamada S, Hiratsuka M, Fukao Y, Kawasaki T, Shimamoto K. (2009) Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol 50: 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaquerel E, Steppuhn A, Baldwin IT. (2012) Nicotiana attenuata α-DIOXYGENASE1 through its production of 2-hydroxylinolenic acid is required for intact plant defense expression against attack from Manduca sexta larvae. New Phytol 196: 574–585 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Ausubel FM. (1994) Isolation of phytoalexin-deficient mutants of Arabidopsis thaliana and characterization of their interactions with bacterial pathogens. Proc Natl Acad Sci USA 91: 8955–8959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castresana C. (1999) Alpha-oxidation of fatty acids in higher plants: identification of a pathogen-inducible oxygenase (piox) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem 274: 24503–24513 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Rodriguez MJ, Calvo AP, Castresana C. (2003) Activation of the fatty acid alpha-dioxygenase pathway during bacterial infection of tobacco leaves: formation of oxylipins protecting against cell death. J Biol Chem 278: 51796–51805 [DOI] [PubMed] [Google Scholar]
- Hanano A, Burcklen M, Flenet M, Ivancich A, Louwagie M, Garin J, Blée E. (2006) Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J Biol Chem 281: 33140–33151 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I, Hatsugai N. (2011) The role of vacuole in plant cell death. Cell Death Differ 18: 1298–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, Nishimura M, Hara-Nishimura I. (2009) A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev 23: 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. (2011) Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. Plant J 67: 980–992 [DOI] [PubMed] [Google Scholar]
- Hiruma K, Onozawa-Komori M, Takahashi F, Asakura M, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. (2010) Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell 22: 2429–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim YY, Jung KW, Yoo KS, Jeung JU, Shin JS. (2011) A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol 52: 874–884 [DOI] [PubMed] [Google Scholar]
- Kimura A, Takano Y, Furusawa I, Okuno T. (2001) Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Farese RV, Jr, Walther TC. (2009) SnapShot: lipid droplets. Cell 139: 1024, e1 [DOI] [PubMed] [Google Scholar]
- Lersten NR, Czlapinski AR, Curtis JD, Freckmann R, Horner HT. (2006) Oil bodies in leaf mesophyll cells of angiosperms: overview and a selected survey. Am J Bot 93: 1731–1739 [DOI] [PubMed] [Google Scholar]
- Li L, Li C, Lee GI, Howe GA. (2002) Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc Natl Acad Sci USA 99: 6416–6421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Matsui K. (2006) Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr Opin Plant Biol 9: 274–280 [DOI] [PubMed] [Google Scholar]
- Mihara M, Itoh T, Izawa T. (2010) SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res 38: D835–D842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ. (2001) The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog Lipid Res 40: 325–438 [DOI] [PubMed] [Google Scholar]
- Murphy DJ. (2012) The dynamic roles of intracellular lipid droplets: from archaea to mammals. Protoplasma 249: 541–585 [DOI] [PubMed] [Google Scholar]
- Murphy DJ, Vance J. (1999) Mechanisms of lipid-body formation. Trends Biochem Sci 24: 109–115 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nakamura S, Mano S, Tanaka Y, Ohnishi M, Nakamori C, Araki M, Niwa T, Nishimura M, Kaminaka H, Nakagawa T, et al. (2010) Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci Biotechnol Biochem 74: 1315–1319 [DOI] [PubMed] [Google Scholar]
- Nalam VJ, Keeretaweep J, Sarowar S, Shah J. (2012) Root-derived oxylipins promote green peach aphid performance on Arabidopsis foliage. Plant Cell 24: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namai T, Kato T, Yamaguchi Y, Hirukawa T. (1993) Anti-rice blast activity and resistance induction of C-18 oxygenated fatty acids. Biosci Biotechnol Biochem 57: 611–613 [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. (2009) RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J 60: 218–226 [DOI] [PubMed] [Google Scholar]
- Narusaka Y, Narusaka M, Park P, Kubo Y, Hirayama T, Seki M, Shiraishi T, Ishida J, Nakashima M, Enju A, et al. (2004) RCH1, a locus in Arabidopsis that confers resistance to the hemibiotrophic fungal pathogen Colletotrichum higginsianum. Mol Plant Microbe Interact 17: 749–762 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell R, Herbert C, Sreenivasaprasad S, Khatib M, Esquerré-Tugayé MT, Dumas B. (2004) A novel Arabidopsis-Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Mol Plant Microbe Interact 17: 272–282 [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Thon MR, Hacquard S, Amyotte SG, Kleemann J, Torres MF, Damm U, Buiate EA, Epstein L, Alkan N, et al. (2012) Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet 44: 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Kamide Y, Hirai MY, Saito K. (2013) Plant lipidomics based on hydrophilic interaction chromatography coupled to ion trap time-of-flight mass spectrometry. Metabolomics (Suppl 1) 9: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Shimojima M, Sawada Y, Toyooka K, Narisawa T, Mochida K, Tanaka H, Matsuda F, Hirai A, Hirai MY, et al. (2009) A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21: 892–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge M, Murphy DJ. (2009) Roles of a membrane-bound caleosin and putative peroxygenase in biotic and abiotic stress responses in Arabidopsis. Plant Physiol Biochem 47: 796–806 [DOI] [PubMed] [Google Scholar]
- Pedras MS, Adio AM. (2008) Phytoalexins and phytoanticipins from the wild crucifers Thellungiella halophila and Arabidopsis thaliana: rapalexin A, wasalexins and camalexin. Phytochemistry 69: 889–893 [DOI] [PubMed] [Google Scholar]
- Perfect SE, Hughes HB, O’Connell RJ, Green JR. (1999) Colletotrichum: a model genus for studies on pathology and fungal-plant interactions. Fungal Genet Biol 27: 186–198 [DOI] [PubMed] [Google Scholar]
- Porta H, Rocha-Sosa M. (2002) Plant lipoxygenases: physiological and molecular features. Plant Physiol 130: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerré-Tugayé MT, Rosahl S, et al. (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139: 1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada C, Lipka V, O’Connell R, Okuno T, Schulze-Lefert P, Takano Y. (2006) Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol Plant Microbe Interact 19: 270–279 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Hara-Nishimura I. (2010) Oil-body-membrane proteins and their physiological functions in plants. Biol Pharm Bull 33: 360–363 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Hara-Nishimura I. (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J 61: 519–528 [DOI] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Shirakawa M, Ueda H, Shimada T, Koumoto Y, Shimada TL, Kondo M, Takahashi T, Okuyama Y, Nishimura M, Hara-Nishimura I. (2010) Arabidopsis Qa-SNARE SYP2 proteins localized to different subcellular regions function redundantly in vacuolar protein sorting and plant development. Plant J 64: 924–935 [DOI] [PubMed] [Google Scholar]
- Shirasu K. (2009) The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol 60: 139–164 [DOI] [PubMed] [Google Scholar]
- Siloto RM, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18: 731–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steppuhn A, Gaquerel E, Baldwin IT. (2010) The two alpha-dox genes of Nicotiana attenuata: overlapping but distinct functions in development and stress responses. BMC Plant Biol 10: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Katagiri T, Yamaguchi-Shinozaki K, Shinozaki K. (2000) An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol 41: 898–903 [DOI] [PubMed] [Google Scholar]
- Takano Y, Asakura M, Sakai Y. (2009) Atg26-mediated pexophagy and fungal phytopathogenicity. Autophagy 5: 1041–1042 [DOI] [PubMed] [Google Scholar]
- Takano Y, Oshiro E, Okuno T. (2001) Microtubule dynamics during infection-related morphogenesis of Colletotrichum lagenarium. Fungal Genet Biol 34: 107–121 [DOI] [PubMed] [Google Scholar]
- Thomma BP, Nelissen I, Eggermont K, Broekaert WF. (1999) Deficiency in phytoalexin production causes enhanced susceptibility of Arabidopsis thaliana to the fungus Alternaria brassicicola. Plant J 19: 163–171 [DOI] [PubMed] [Google Scholar]
- Tsuji J, Jackson EP, Gage DA, Hammerschmidt R, Somerville SC. (1992) Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv syringae. Plant Physiol 98: 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J, Cascón T, Vicedo B, García-Agustín P, Hamberg M, Castresana C. (2012) Role of 9-lipoxygenase and α-dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol Plant 5: 914–928 [DOI] [PubMed] [Google Scholar]
- Walters DR, Cowley T, Weber H. (2006) Rapid accumulation of trihydroxy oxylipins and resistance to the bean rust pathogen Uromyces fabae following wounding in Vicia faba. Ann Bot (Lond) 97: 779–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav MK, Bhatla SC. (2011) Localization of lipoxygenase activity on the oil bodies and in protoplasts using a novel fluorescence imaging method. Plant Physiol Biochem 49: 230–234 [DOI] [PubMed] [Google Scholar]