A MYB domain protein maintains petiole identity by repressing expression of a motor organ identity gene and promoting petiole and stipule development in Medicago truncatula.
Abstract
Plant leaves, simple or compound, initiate as peg-like structures from the peripheral zone of the shoot apical meristem, which requires class I KNOTTED-LIKE HOMEOBOXI (KNOXI) transcription factors to maintain its activity. The MYB domain protein encoded by the ASYMMETRIC LEAVES1/ROUGH SHEATH2/PHANTASTICA (ARP) gene, together with other factors, excludes KNOXI gene expression from incipient leaf primordia to initiate leaves and specify leaf adaxial identity. However, the regulatory relationship between ARP and KNOXI is more complex in compound-leafed species. Here, we investigated the role of ARP and KNOXI genes in compound leaf development in Medicago truncatula. We show that the M. truncatula phantastica mutant exhibited severe compound leaf defects, including curling and deep serration of leaf margins, shortened petioles, increased rachises, petioles acquiring motor organ characteristics, and ectopic development of petiolules. On the other hand, the M. truncatula brevipedicellus mutant did not exhibit visible compound leaf defects. Our analyses show that the altered petiole development requires ectopic expression of ELONGATED PETIOLULE1, which encodes a lateral organ boundary domain protein, and that the distal margin serration requires the auxin efflux protein M. truncatula PIN-FORMED10 in the M. truncatula phantastica mutant.
Plant leaves are the primary photosynthetic organs and play a key role in plant growth and biomass production. Leaves are derived from leaf founder cells developed at the periphery of the shoot apical meristem (SAM), a pluripotent structure capable of self-renewal. The meristematic activity of SAM is maintained by class I KNOTTED-LIKE HOMEOBOX genes (KNOXIs; Clark et al., 1996; Long et al., 1996). Early events marking the recruitment of leaf founder cells to the incipient leaf primordia at the peripheral zone of the SAM involve the down-regulation of KNOXI gene expression, the expression of the MYB domain transcription factor gene, ARP, for ASYMMETRIC LEAVES1 (AS1) in Arabidopsis (Arabidopsis thaliana; Byrne et al., 2000), ROUGH SHEATH2 (RS2) in maize (Zea mays; Timmermans et al., 1999; Tsiantis et al., 1999), and PHANTASTICA (PHAN) in Antirrhinum species (Waites and Hudson, 1995; Waites et al., 1998), and the formation of auxin activity maxima (Reinhardt et al., 2000; Vernoux et al., 2000). It has been shown that AS1 acts together with the lateral organ boundary (LOB) domain transcription factor, AS2, to exclude the expression of KNOXI genes in incipient leaf primordia and that SHOOT MERISTEMLESS (STM; a KNOXI protein) acts to exclude AS1 expression in the SAM, and these regulatory relationships are not only important for the maintenance of the meristematic activity of SAM but also for the development of leaf primordia in Arabidopsis (Byrne et al., 2000, 2002; Semiarti et al., 2001; Guo et al., 2008). Leaf primordia initiate and expand along the proximodistal, mediolateral, and adaxial-abaxial axes. ARP and the class III HOMEODOMAIN-LEUCINE ZIPPER genes are known to specify leaf adaxial identity (McConnell and Barton, 1998; McConnell et al., 2001; Emery et al., 2003). On the other hand, YABBY (Siegfried et al., 1999; Golz et al., 2004) and KANADI (Kerstetter et al., 2001; Emery et al., 2003) specify leaf abaxial identity.
Plant leaves can be categorized as either simple or compound. A simple leaf is composed of a single undivided blade, and a compound leaf is composed of multiple blades known as leaflets. In compound-leafed species, specific regions of the leaf margin acquire a transient meristematic activity and initiate leaflet primordia in a species-specific pattern. The KNOXI genes are initially down-regulated in incipient leaf primordia and subsequently reactivated to promote the leaf marginal meristematic activity to initiate leaflet primordia in compound-leafed species such as tomato (Solanum lycopersicum) and Cardamine hirsuta (Kim et al., 2003a; Hay and Tsiantis, 2006; Barkoulas et al., 2008). In C. hirsuta as1 mutants, STM is ectopically expressed in leaf primordia, leading to an increased compoundness of leaves (Hay and Tsiantis, 2006; Barkoulas et al., 2008). However, the regulatory relationship between the ARP and KNOXI genes is more complex in tomato (Zoulias et al., 2012). It has been shown that down-regulation of the tomato ARP gene, SlPHAN, results in radialized or peltately palmate compound leaves and a loss of the typical wild-type pinnate compound leaves (Kim et al., 2003b).
In C. hirsuta and tomato, the down-regulation of KNOXI genes results in compound leaves with reduced leaflets (Hay and Tsiantis, 2006; Shani et al., 2009; Burko et al., 2013). In tomato, overexpression of the KNOXI gene, LeT6, results in supercompound leaves (Hareven et al., 1996). Intriguingly, in the inverted repeat-lacking clade of legumes, including garden pea (Pisum sativum) and Medicago species, the KNOXI genes are not expressed in leaf primordia and therefore are not likely involved in compound leaf development, although the genetic evidence is lacking (Hofer et al., 2001; Champagne et al., 2007; Peng et al., 2011). Instead, the FLO/LFY orthologs, UNIFOLIATA (UNI) and SINGLE LEAFLET1 (SGL1), play a key role in compound leaf development in pea and Medicago truncatula, respectively (Hofer et al., 1997; Wang et al., 2008). Pea crispa (cri) mutants, in which the pea PHAN gene is mutated, exhibit leaf adaxial-abaxial and proximodistal polarity defects and ectopic stipules on the adaxial lamina surface, but the typical pinnate compound leaf pattern is not altered (Tattersall et al., 2005). RNA in situ hybridization shows patchy expression of the pea KNOXI gene, BREVIPEDICELLUS (BP), in leaves of cri mutants (Tattersall et al., 2005). This is thought to be associated with the development of ectopic stipules on the adaxial lamina surface in cri mutants (Tattersall et al., 2005).
Auxin convergent points or activity maxima mediated by auxin efflux transporter PIN proteins mark and precede the initiation of leaf primordia in both simple- and compound-leafed species (Barkoulas et al., 2008; Koenig et al., 2009). In compound-leafed species, auxin activity maxima also mark and precede leaflet and lobe initiation (Koenig et al., 2009; Ben-Gera et al., 2012). In tomato entire (e) mutants that affect the auxin response inhibitor, SlIAA9 (Zhang et al., 2007; Koenig et al., 2009), the auxin signal monitored by the auxin response sensor, DR5, expands to include the complete leaf margin (Ben-Gera et al., 2012). Similar to e mutants, tomato goblet (gob) mutants affecting the CUP-SHAPED COTYLEDON transcription factor gene develop only primary leaflets (Berger et al., 2009). Inhibition of auxin transport or activity suppresses the GOB overexpression phenotype (Ben-Gera et al., 2012), consistent with the notion that auxin mediates GOB-regulated leaf patterning. Down-regulation of both E and GOB results in complete loss of leaflet initiation and strong auxin signals throughout the leaf margin (Ben-Gera et al., 2012). These observations support the hypothesis that proper leaflet initiation and separation requires distinct boundaries between regions of lamina growth and adjacent regions of growth suppression (Ben-Gera et al., 2012).
In M. truncatula, mutations in the PIN10 gene encoding an auxin efflux transporter result in complete loss of serrations at the distal leaf margin and a variable number of leaflets likely due to the fusion of leaf primordia during leaf development (Peng and Chen, 2011; Zhou et al., 2011). Interestingly, in Arabidopsis, AS1 and auxin converge to suppress the expression of KNAT1/BP and promote the leaf fate, and the interactions between auxin, AS1, and KNOX activities control both leaf initiation and leaf form (Hay et al., 2006).
To address the role of ARP and KNOXI genes in compound leaf development in M. truncatula, we isolated and characterized Tnt1 retrotransposon insertion mutants of M. truncatula PHAN and BP genes. Our results show that the mtphan mutant exhibits multiple defects in compound leaf development, including curling and deep serration of leaf margins, shortened petioles, increased rachises, petioles acquiring motor organ characteristics, and ectopic development of petiolules. On the other hand, the mtbp mutant did not exhibit visible defects in compound leaf development. We show that the altered petiole development requires ectopic expression of ELONGATED PETIOLULE1 (ELP1) and that the leaf margin serration requires the auxin efflux protein M. truncatula PIN-FORMED10 (MtPIN10) in the mtphan mutant. However, the development of ectopic tissues on the leaf adaxial surface does not require MtBP activity.
RESULTS
Isolation of the M. truncatulatphan Mutant
Genome analysis identified a single ARP gene, MtPHAN (Medtr7g061550.1), in M. truncatula and four copies of ARP genes in soybean (Glycine max). In Lotus japonicus, a tandem repeat of ARP genes, LjPHAN1 and LjPHAN2, has been reported (Luo et al., 2005). Amino acid sequence alignments of ARP genes from legume and nonlegume species revealed extensive amino acid sequence similarities in the NH2-terminal MYB domain and the COOH-terminal domain among the sequences (Supplemental Fig. S1). Phylogenetic analysis grouped the legume ARP genes into a single clade, in which the pea PHAN ortholog, CRI, was most closely related to MtPHAN (Supplemental Fig. S2A). The ARP genes examined also exhibited a conserved intron-exon structure, one intron and two exons (Supplemental Fig. S2B).
Using a reverse genetic screen (Cheng et al., 2011), we isolated an insertion mutant of the MtPHAN gene with the tobacco (Nicotiana tabacum) Tnt1 retrotransposon inserted at the 3′ end of the coding region (Fig. 1A; Supplemental Fig. S1). Reverse transcription (RT)-PCR analysis indicates that the isolated mutant lacked the full-length transcripts of the corresponding gene (Fig. 1B). Quantitative RT-PCR analysis using primers located upstream of the Tnt1 insertion site revealed that the transcript level was extremely low compared with wild-type (R108) plants (Fig. 1C), suggesting that mtphan is a loss-of-function or partially reduced function mutant.
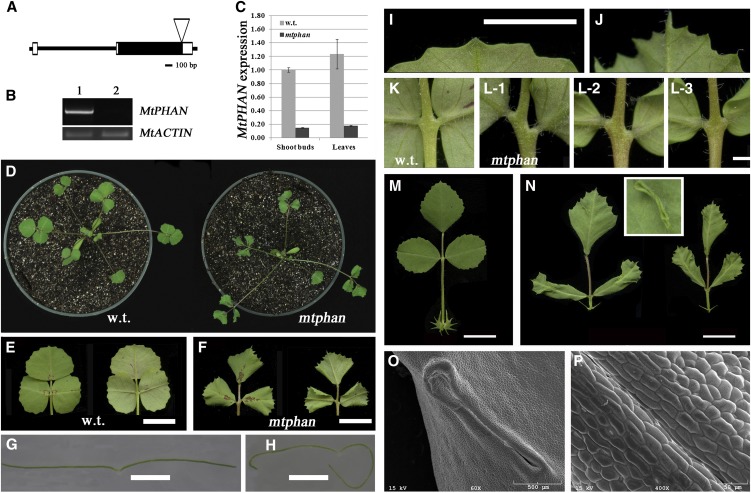
Figure 1.
Compound leaf phenotypes of the mtphan mutant. A, MtPHAN gene structure and Tnt1 insertion site. Black boxes, Exons; white boxes, 5′ and 3′ untranslated regions; horizontal lines, introns. B, RT-PCR analysis of MtPHAN gene expression in 21-d-old shoot buds. Lane 1, The wild type (R108); lane 2, mtphan. An MtACTIN gene was used as a loading control. PCR cycles used were 32 and 28 for MtPHAN and MtACTIN, respectively. C, Quantitative RT-PCR analysis of MtPHAN gene expression in 21-d-old plants. MtACTIN was used as the reference gene. D, Images of 21-d-old wild type (w.t.; left) and mtphan mutant (right) plants. E and F, Closeup views of adaxial (left) and abaxial (right) sides of wild-type (E) and mtphan (F) compound leaves. G and H, Cross sections of leaflets of the wild type (G) and the mtphan mutant (H). I and J, Closeup views of distal leaf margins of the wild type (I) and the mtphan mutant (J). K and L, Closeup views of the typical symmetric lateral leaflet placement in the wild type (K) and various lateral leaflet placements in the mtphan mutant (L-1 to L-3). M and N, Compound leaves of 70-d-old wild-type (M) and mtphan (N) plants. The inset shows ectopic tissues developed on the adaxial leaf surface of the mutant. O and P, SEM images show ectopic tissues and epidermal cell morphologies on the adaxial leaf surface of the mtphan mutant. Bars = 1 cm (E, F, M, and N), 2 mm (G, H, K, and L), and 5 mm (I and J).
MtPHAN Regulates Leaf Margin Development, Lateral Leaflet Placement, and Leaf Adaxial-Abaxial Polarity
In wild-type plants, adult leaves were trifoliate, with one terminal leaflet and two lateral ones attached to a rachis and a petiole, respectively (Fig. 1, D and E). Leaflets were folded upward along the central axis during early stages of development and subsequently unfolded to expose the adaxial surface (Fig. 1D; Supplemental Fig. S3C). The distal margin of leaflets was slightly serrated (Fig. 1, E and I).
In contrast to the flat wild-type leaflets (Fig. 1, E and G), the proximal margin of leaflets of compound leaves and the juvenile leaf of the mtphan mutant curled downward (Fig. 1, D, F, and H). This phenotype was observed in young leaflets when they were still folded along the central axis (Supplemental Fig. S3, C and D); however, microscopic dissection of shoot buds revealed that curling of the proximal leaf margin did not occur when laminae were only a few millimeters in length. Leaf primordia development was not different between mtphan and wild-type plants (Supplemental Fig. S3, A and B). The distal margin of mtphan mutant leaflets did not curl. Instead, it exhibited deeper serrations than the wild-type counterpart (Fig. 1, E, F, I, and J). These results indicate that MtPHAN plays roles in distal and proximal leaf margin development.
In a wild-type compound leaf, a pair of lateral leaflets always developed symmetrically on the petiole (Fig. 1, D, E, and K). Inspection of a large number of plants revealed that 107 out of 131 (82%) mtphan mutant plants developed asymmetric lateral leaflets on petioles (Fig. 1, K and L), although the number of compound leaves with asymmetric lateral leaflets was variable among plants, ranging from one to all compound leaves in 3-week-old plants. This phenotype, however, was much less pronounced in 70-d-old plants.
Next, we compared leaf epidermal cell morphologies between wild-type and mtphan plants at different developmental stages, using scanning electron microscopy (SEM). In 21-d-old plants, epidermal cell size and the shape of both terminal and lateral leaflets were not different between the wild type and mtphan. However, in 70-d-old plants, epidermal cells of the adaxial surface of fully expanded leaves (leaves on the fourth node from the top) of the mtphan mutant were smooth and appeared to be less differentiated, in contrast to the jigsaw puzzle-like leaf pavement cells of corresponding wild-type plants, while the leaf abaxial epidermal cells were similar between mtphan and wild-type plants, suggesting that leaf adaxial differentiation or identity was affected in mature mtphan mutant plants (Supplemental Fig. S4, A–H). In addition, some ectopic tissues frequently formed on the adaxial surface of leaflets in 70-d-old mtphan plants, in contrast to wild-type plants (Fig. 1, M and N, inset). SEM images show that these ectopic tissues appeared as ring-like extrusions with distinct boundaries and consisted of rod-shaped smooth cells (Fig. 1, O and P). Based on the morphological changes, we conclude that MtPHAN regulates lamina adaxial-abaxial polarity, and this regulatory role appears to be strongly dependent on the developmental stage.
MtPHAN Regulates Petiole, Rachis, and Petiolule Development
We next examined the role of MtPHAN in petiole and rachis development. In 4-week-old plants, rachises and petioles that developed on apical nodes 1 to 3 were not different between the wild type and the mtphan mutant (Supplemental Fig. S5, A–D). However, rachises and petioles on the fourth and fifth nodes from the top were longer and shorter, respectively, in the mtphan mutant than in wild-type plants (Supplemental Fig. S5, A–D). Measurements further showed that epidermal cells of rachises and petioles on the apical fourth and fifth nodes were longer and shorter, respectively, in the mtphan mutant than in the wild type (Supplemental Fig. S5E), suggesting that MtPHAN regulates petiole and rachis development primarily through the modulation of cell elongation.
Differences in rachis and petiole development between the wild type and mtphan were much pronounced in 70-d-old plants (Fig. 2, A and B; Supplemental Fig. S6). While petioles and rachises developed coordinately along the primary stem in wild-type plants, resulting in a narrow range of rachis-petiole ratios (0.3–0.5; Supplemental Fig. S6), rachises and petioles of leaves on apical nodes 1 to 4 were much longer and shorter, respectively, in mtphan than in wild-type plants (Supplemental Fig. S6, C and D). The rachis length on apical nodes 5 to 7 was not much different between the wild type and the mtphan mutant; however, the petiole length was still largely reduced. As a result, high rachis-petiole ratios were observed in mtphan plants compared with wild-type plants (Supplemental Fig. S6, C–E).
Figure 2.
Development of ectopic petiolules and altered petioles in mtphan plants. A and B, Representative 70-d-old wild-type (A) and mtphan mutant (B) compound leaves. Closeup images are shown at the bottom. Horizontal lines indicate the regions where SEM (E–H) and cross sections (I and J) were made. Bars = 1 cm. C and D, SEM images of wild-type (C) and mtphan mutant (D) compound leaves. Asterisks indicate pulvini, and arrows indicate ectopic petiolules. E to H, SEM images of petioles of 70-d-old wild-type (E) and mtphan mutant (F–H) plants. I and J, Cross sections of petioles of 70-d-old wild-type (I) and mtphan mutant (J) plants. Bar = 100 μm.
In a wild-type compound leaf, lateral leaflets were attached to the petiole through pulvini (Fig. 2, A and C, asterisks). In an mtphan compound leaf, lateral leaflets were attached to the petiole through pulvini and petiolules (Fig. 2, B and D). This phenotype was variable among plants but was much pronounced in 70-d-old plants. These results suggest that MtPHAN is also involved in the suppression of petiolule development in wild-type plants.
SEM analysis shows that petiole epidermal cells were greatly reduced in length and altered in morphology in the 70-d-old mtphan mutant compared with wild-type counterparts (Fig. 2, E–H). Petiole epidermal cells of the mtphan plants developed longitudinal and transverse folds on their surface (Fig. 2, F–H). These morphological modifications resembled those of pulvini (Fig. 2, A–H). Cross-section images show that wild-type petioles exhibited the characteristic adaxial-abaxial polarity, with one large vascular bundle and two small ones at the abaxial and adaxial sides, respectively (Fig. 2I). By contrast, petioles of the mtphan plants contained an enlarged central vascular bundle (Fig. 2J), resembling that of pulvini. On the other hand, rachis epidermal cells were elongated and smooth in both wild-type and mtphan mutant plants (Supplemental Fig. S7, A and B). Taken together, these results reveal a novel role of MtPHAN in the maintenance of petiole identity through preventing ectopic acquisition of the motor organ characteristics in petioles.
MtPHAN Regulates Stipule Development
In a wild-type leaf, a pair of stipules with three to five digitations developed at the base of the petiole (Couzigou et al., 2012; Supplemental Fig. S8A). Both the size of stipule lamina and the number of digitations were significantly reduced in the mtphan mutant compared with wild-type plants (Supplemental Fig. S8, A and B). SEM analysis shows that epidermal cells of both stipule lamina and digitation were less curving in mtphan than in wild-type plants, suggesting altered stipule epidermal cell differentiation or stipule identity in the mtphan mutant (Supplemental Fig. S8, C–F).
Functional Rescue of mtphan Mutant Defects by MtPHAN
To confirm that the observed mtphan mutant phenotypes were indeed caused by the mutation of the MtPHAN gene, we introduced the MtPHAN genomic sequence including its promoter and the coding sequence fused to the GFP into the mtphan mutant by stable transformation. Supplemental Figure S9 shows that the mtphan mutant phenotypes were rescued in stable transgenic plants. Therefore, the mutation in the MtPHAN gene was responsible for the mtphan mutant phenotypes.
MtPHAN Functionally Rescued Arabidopsis as1 Mutant Phenotypes
To test whether M. truncatula and Arabidopsis PHAN genes are functional orthologs, we introduced the same MtPHAN genomic sequence-GFP fusion construct into the Arabidopsis as1-1 mutant by stable transformation. Supplemental Figure S10, A and B, shows that both leaf and inflorescence defects of the as1-1 mutant were rescued in transgenic lines by MtPHAN. Laser confocal microscopic analysis reveals that high MtPHAN-GFP signals were localized in nuclei of leaf epidermal cells at both adaxial and abaxial surfaces in transgenic as1-1 plants (Supplemental Fig. S11, A–C).
Next, we tested whether the MtPHAN sequence can rescue the expression of AS1 downstream target genes. It has been shown that AS1 negatively regulates KNAT1/BP but not STM in Arabidopsis (Byrne et al., 2000). In transgenic as1-1 plants expressing MtPHAN, the ectopic expression of KNAT1/BP was suppressed in leaves, similar to that of wild-type plants (Supplemental Fig. S10C). The expression of STM, however, was not affected in leaves of transgenic plants as expected (Supplemental Fig. S10C). These results indicate that (1) MtPHAN and AS1 are functional orthologs and (2) the MtPHAN promoter sequence is correctly recognized in Arabidopsis.
MtPHAN Tissue-Specific Expression
RNA in situ hybridization shows that MtPHAN transcripts were strongly expressed in incipient and developing leaf primordia and in lamina tissues (Fig. 3A). MtPHAN transcripts were also detectable in SAM, albeit at a lower level (Fig. 3A). As a negative control, an MtPHAN sense probe did not result in any detectable signals (Fig. 3B).
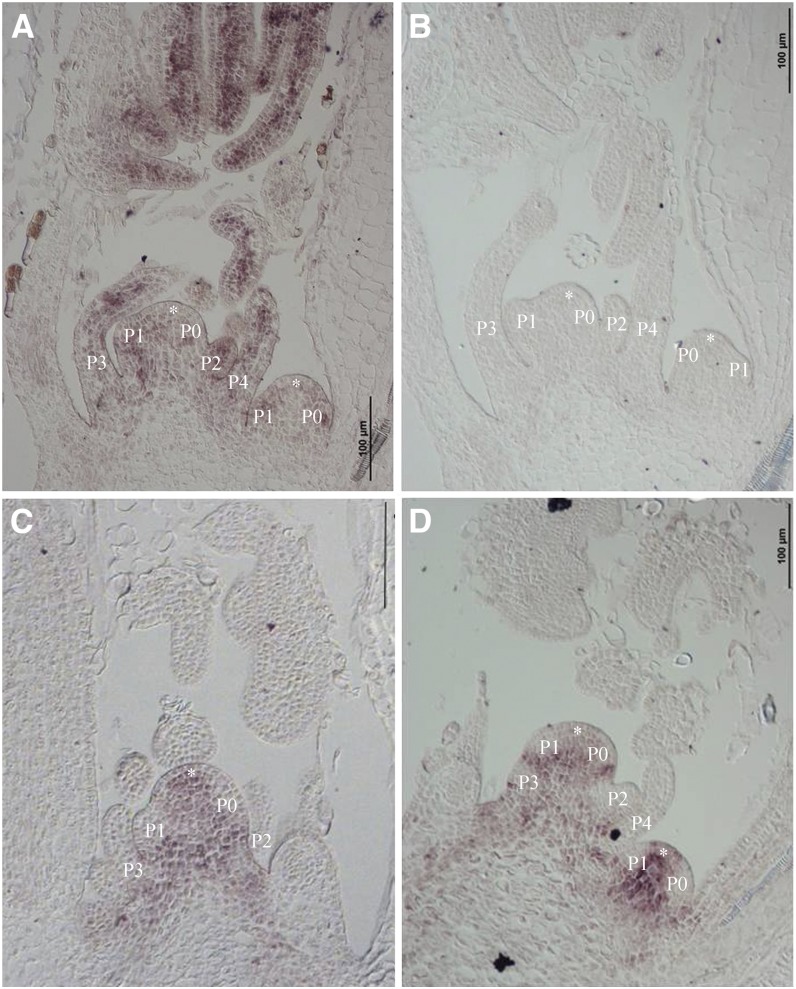
Figure 3.
RNA in situ hybridization of MtPHAN and MtBP. A, Longitudinal section of a vegetative shoot bud of wild-type plants showing MtPHAN transcripts in SAM (asterisks), leaf primordia at various stages, and young leaves. B, An adjacent section hybridized with a sense probe did not yield any signals, serving as a negative control. C, MtBP transcripts were detected in SAM (asterisk) but not in leaf primordia at various stages and young leaves of wild-type plants. D, Similarly, MtBP transcripts were detected in SAM (asterisks) but not in leaf primordia at various stages and young leaves of mtphan mutant plants. Bars = 100 µm.
Interactions between MtPHAN and ELP1
ELP1, encoding a LOB domain protein, is required to maintain the motor organ identity in M. truncatula (Chen et al., 2012; Zhou et al., 2012). Ectopic expression of ELP1 results in both petioles and rachises acquiring motor organ characteristics (i.e. reduced cell size and altered cell surface morphology; Chen et al., 2012). Since the altered petiole morphologies in mature mtphan plants (Fig. 2) resemble those of petioles of ELP1 ectopic expression lines (Chen et al., 2012), we hypothesized that ELP1 is possibly involved in this process. To test this hypothesis, we first examined ELP1 gene expression in rachises and petioles of mature mtphan mutant plants. RT-PCR results show that ELP1 was ectopically expressed in petioles but not in rachises of mtphan plants, in contrast to wild-type plants (Fig. 4A). On the other hand, ELP1 was similarly expressed in pulvini of both mtphan and wild-type plants (Fig. 4A). Figure 4A also shows ectopic expression of ELP1 in petioles of an ELP1 overexpression line, as reported previously (Chen et al., 2012).
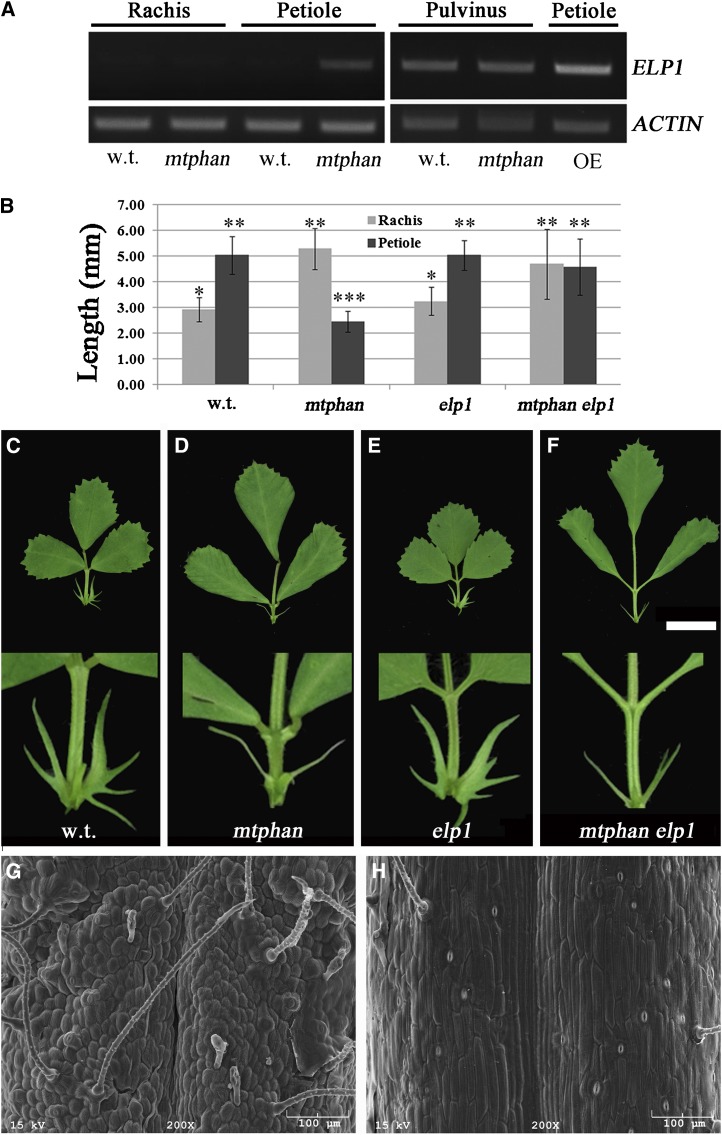
Figure 4.
M. truncatula ELP1 is required for the altered petiole identity in mature mtphan mutant plants. A, RT-PCR analysis of ELP1 gene expression in rachises, petioles, and pulvini of 70-d-old wild-type (w.t.), mtphan, and 35S::ELP1 plants. MtACTIN was used as a loading control. PCR cycles were 32 and 28 for ELP1 and MtACTIN, respectively. B, Measurements of the petiole length of compound leaves on the apical third node of 70-d-old wild-type, mtphan, elp1, and mtphan elp1 double mutant plants. Values shown are means ± sd; n = 12, and statistical significance was calculated by Tukey’s honestly significant difference test (P < 0.05). C to F, Images of compound leaves on the apical third node of 70-d-old wild-type (C), mtphan (D), elp1 (E), and mtphan elp1 (F) plants. Closeup views are shown at the bottom. G and H, SEM images of petioles of leaves on the third node of mature mtphan (G) and mtphan elp1 mutant (H) plants. Bars = 1 cm (C–F) and 100 µm (G and H). [See online article for color version of this figure.]
We next generated the mtphan elp1 double mutant (Fig. 4F). Similar to the elp1 mutant, the mtphan elp1 double mutant developed petiolules in place of pulvini (Fig. 4, E and F; Supplemental Fig. S12, arrows and asterisks). Measurements show that the reduced petiole length in 70-d-old mtphan plants was restored in the mtphan elp1 double mutant to the level of wild-type and elp1 mutant plants, whereas the elongated rachis phenotype of the mtphan mutant plants remained unchanged in the double mutant (Fig. 4, B–F). SEM analysis shows that the reduced cell size and altered cell morphology of petioles of mature mtphan plants were restored in the double mutant to that of the wild type and the elp1 mutant (Fig. 4, G and H). These results indicate that the altered development of petioles but not that of rachises of mature mtphan plants is a result of ectopic expression of ELP1 in petioles.
Distal Leaf Margin Serration Requires the Auxin Efflux Protein, MtPIN10
The leaf distal margin serration was much deeper in the mtphan plants than in wild-type plants (Fig. 1, I and J). Since the auxin efflux protein MtPIN10 is required for leaf distal margin development in M. truncatula (Peng and Chen, 2011; Zhou et al., 2011), we tested whether MtPIN10 is involved in this process in the mtphan mutant. For this, we generated mtphan mtpin10 double mutants. Figure 5 shows that mtphan mtpin10 double mutants exhibited compound leaves with smooth leaf margins, resembling those of the mtpin10 single mutant. This genetic interaction result suggests that the development of the deeply serrated leaf margin of the mtphan mutant is dependent on auxin activity maxima mediated by the auxin efflux protein MtPIN10. The proximal leaf margin of the mtphan mtpin10 double mutant remained curled like that in the mtphan single mutant, indicating that MtPIN10 is not involved in MtPHAN-dependent proximal leaf margin development (Fig. 5).
Figure 5.
Compound leaf phenotypes of the mtphan mtpin10 double mutant. Adaxial (A) and abaxial (B) views show compound leaves of 70-d-old wild-type (w.t.), mtphan, mtpin10, and mtphan mtpin10 double mutant (from left to right) plants. The inset shows a closeup view of the distal leaflet margin of the mtphan mtpin10 double mutant. Bar = 1 cm. [See online article for color version of this figure.]
Genetic Analysis of MtBP in Compound Leaf Development
The role of the class I KNOX genes in compound leaf development in M. truncatula has not been tested genetically. To investigate the involvement of MtBP in compound leaf development, we isolated an M. truncatula mutant with Tnt1 retrotransposon inserted in the first exon of MtBP (Supplemental Fig. S13A). RT-PCR analysis shows that MtBP transcripts were not detectable in vegetative shoot buds in the mtbp mutant, suggesting that mtbp is a null allele (Supplemental Fig. S13B). As a control, we show that FUSED COMPOUND LEAF1 (FCL1; Peng et al., 2011) was similarly expressed in mtbp and wild-type plants (Supplemental Fig. S13B). Phenotypic analyses reveal that the mtbp mutant did not show obvious defects in compound leaf development (Supplemental Fig. S13, C, D, H, and I). We generated mtbp mtphan double mutants (Supplemental Fig. S13, C and D). In mature mtbp mtphan double mutant plants, extra tissues developed similarly to the mtphan single mutant on the adaxial leaf surface (Supplemental Fig. S13, C and D, inset), suggesting that MtBP is not responsible for the development of ectopic tissues in the mtphan mutant.
RNA in situ hybridization shows that MtBP transcripts were strongly detected in SAM, but they were excluded from incipient and subsequent leaf primordia (Fig. 3C). In the mtphan mutant, although MtBP transcripts were detected in SAM, they were similarly excluded from leaf primordia as in wild-type plants (Fig. 3D). Real-time RT-PCR results show that MtBP gene expression was up-regulated in shoot buds in 21- and 70-d-old mtphan mutants compared with wild-type plants (Supplemental Fig. S13E). However, its expression was extremely low in leaves of both mtphan mutant and wild-type plants at different developmental stages (Supplemental Fig. S13E). Similarly, transcripts of two other KNOXI genes, MtSTM and MtKNOX6, were also up-regulated in shoot buds in the mtphan mutant but remained extremely low in leaves of both mtphan mutant and wild-type plants (Supplemental Fig. S13, F and G).
Genetic Interactions between MtPHAN and SGL1
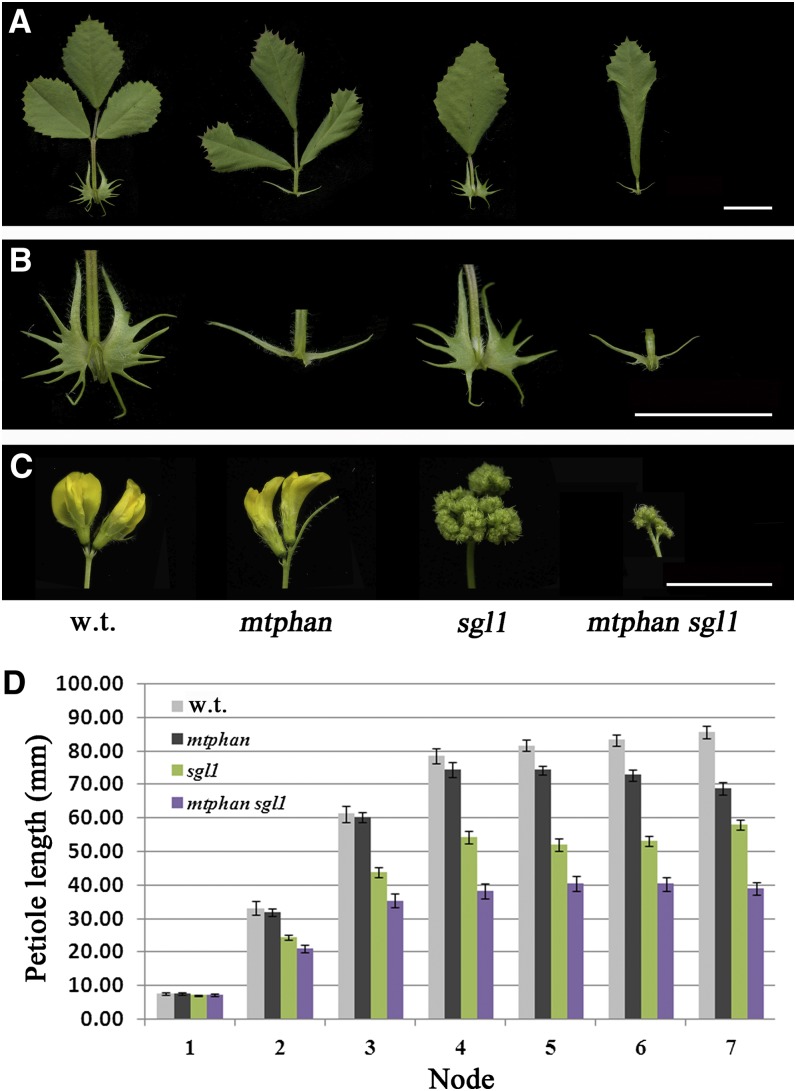
SGL1 has been shown to act as an indeterminate factor in the control of lateral leaflet initiation in M. truncatula, a similar role played by KNOXI in tomato and C. hirsuta (Wang et al., 2008). Given the known KNOXI-ARP interactions, we investigated genetic interactions between MtPHAN and SGL1. For this, we generated mtphan sgl1 double mutants (Fig. 6; Supplemental Fig. S14). mtphan sgl1 double mutants exhibited simple leaves with downwardly curled and deeply serrated leaf margins and simplified stipules (Fig. 6, A and B; Supplemental Fig. S14, A–D). These results suggest that MtPHAN and SGL1 act independently in compound leaf patterning and polarity development. sgl1 mutants produced cauliflower-like inflorescences due to defects in flower meristem development (Fig. 6C). mtphan sgl1 double mutants produced small cauliflower-like inflorescences, resembling that of the sgl1 single mutants (Fig. 6C). On the other hand, the petiole length was further reduced in the mtphan sgl1 double mutant compared with single mutants, suggesting additive interactions between MtPHAN and SGL1 in leaf petiole development (Fig. 6D).
Figure 6.
Compound leaf phenotypes of the sgl1 mtphan double mutant. A, Compound leaves of 70-d-old wild-type (w.t.), mtphan, sgl1, and mtphan sgl1 double mutant (from left to right) plants. B, Closeup images of stipules. C, Flowers and inflorescences of wild-type, mtphan, sgl1, and mtphan sgl1 double mutant (from left to right) plants. Bars = 1 cm. D, Measurements of petiole lengths in compound leaves on apical nodes 1 to 7 of 4-week-old wild-type, mtphan, sgl1, and mtphan sgl1 double mutant plants. Values shown are means ± se; n = 11.
DISCUSSION
In this study, we demonstrate novel roles of M. truncatula PHAN in trifoliate leaf development, in addition to its conserved role in regulating leaf adaxial-abaxial polarity. In particular, our work shows that (1) MtPHAN is required to maintain petiole identity by repressing the ectopic expression of ELP1 in petioles; (2) MtPHAN represses rachis and petiolule but promotes petiole development; (3) MtPHAN maintains proper leaf margin development and lateral leaflet placement; and (4) MtPHAN promotes stipule development.
MtPHAN Is Required to Maintain Petiole Identity by Repressing the Ectopic Expression of ELP1
The petiole identity was altered in mature mtphan mutant plants, as indicated by the facts that (1) petioles exhibited a nearly radial symmetry in contrast to wild-type petioles with an adaxial-abaxial polarity and (2) petiole epidermal cells were small and developed longitudinal and transverse folds on their surfaces. This structural feature, absent in wild-type petioles, resembles but is not identical to that of pulvini (Chen et al., 2012). It has been shown that the motor organ identity is controlled by the ELP1 gene. ELP1 is expressed in the motor organ precursor cells, and its expression is restricted to the motor organ (Chen et al., 2012; Zhou et al., 2012). Ectopic expression of ELP1 results in petioles and rachises acquiring motor organ characteristics (Chen et al., 2012). Our gene expression analysis shows that in mature mtphan mutant plants, ELP1 was ectopically expressed in petioles but not in rachises, indicating a specific effect of the mtphan mutation on ELP1 gene expression. Our mtphan elp1 double mutant analysis shows that the altered petiole development in mature mtphan mutant plants was rescued when ELP1 was mutated, confirming the involvement of ectopic ELP1 gene expression in the acquisition of motor organ characteristics in petioles of mature mtphan plants.
In Arabidopsis, AS1 forms a complex with AS2, a LOB domain protein, and the AS1-AS2 nuclear complex interacts with promoter elements and represses the transcription of KNOX genes and other downstream targets such as ETTIN/AUXIN RESPONSE FACTOR3 (Guo et al., 2008; Iwasaki et al., 2013). It is not clear whether the same AS1-AS2 complex functions in compound leaf development in M. truncatula. Several AS2-like sequences are present in the M. truncatula genome. Future work is needed to examine the role of M. truncatula AS2 ortholog(s) in compound leaf development. It is intriguing that ELP1 also encodes a LOB domain protein, although it is most closely related to LOB, the founding member of the LBD family, but not AS2.
MtPHAN Represses Rachis and Petiolule But Promotes Petiole Development
PHAN has been shown to play a role in leaf adaxial-abaxial polarity and proximodistal axis development in simple- and compound-leafed species. In compound-leafed species such as C. hirsuta, tomato, and garden pea, PHAN plays a role in leaf patterning and development, albeit in a species-specific manner. For example, in pea cri mutants, compound leaves remain pinnate, although multiple polarity defects are observed in mutant leaves. By contrast, down-regulation of the tomato PHAN gene, SlPHAN, in the leaf proximal region leads to an absence of the adaxial domain in the proximal region, resulting in cup-shaped simple leaves or peltately palmate compound leaves (Kim et al., 2003b;; Zoulias et al., 2012).
In 3-week-old mtphan mutant plants, leaves that developed on the fourth and fifth nodes had slightly increased rachis length and reduced petiole length compared with wild-type leaves. In 70-d-old mtphan mutant plants, however, young leaves had extremely increased and reduced rachises and petioles, respectively, compared with those of wild-type plants. This results in significantly large rachis-petiole ratios. In older leaves, the petiole length was reduced but the rachis length was not increased, still resulting in large rachis-petiole ratios for mtphan mutant leaves. Cell size measurement results show that epidermal cells of rachises and petioles were increased and reduced, respectively, in the mtphan mutant plants compared with wild-type plants, suggesting that MtPHAN primarily regulates rachis and petiole cell differentiation.
In mature mtphan mutant plants, we observed that a large number of compound leaves developed petiolules at the proximal ends of pulvini of lateral leaflets. By contrast, lateral leaflets were attached to petioles by pulvini in wild-type plants. This abnormality was present, but less prevalent, in 3-week-old mtphan mutant plants. This phenotype, together with the effect of the mtphan mutation on rachis elongation, supports that MtPHAN normally represses the development of rachises and petiolules in wild-type compound leaves.
MtPHAN Maintains Proper Leaf Margin Development and Lateral Leaflet Placement
Deep serration and curling of distal and proximal leaf margins in the mtphan mutant indicate that MtPHAN plays key roles in leaf margin development. Previous studies have shown that auxin convergent points mediated by auxin efflux transporter PIN proteins are prerequisites for the initiation of leaf primordia and leaf serrations in simple- and compound-leafed species and for the initiation of leaflet primordia in compound-leafed species. In M. truncatula, MtPIN10 plays an essential role in distal leaf margin development, as indicated by the smooth leaf margin phenotype of mtpin10 mutants (Peng and Chen, 2011; Zhou et al., 2011). Because the deeply serrated distal leaf margin of the mtphan mutant was changed to the smooth margin in mtphan mtpin10 double mutants, similar to the mtpin10 mutant, MtPIN10-mediated auxin maxima are required for the MtPHAN-regulated distal leaf margin development in M. truncatula.
In mature mtphan mutant plants, leaf epidermal cells on the adaxial surface appeared to be less differentiated, compared with the jigsaw puzzle-like epidermal cells of wild-type plants. In addition, ectopic tissues developed on the adaxial surface of mature mtphan mutant plants. These phenotypes suggest that the adaxial-abaxial polarity was altered in the mtphan mutant. AS1/RS2/PHAN is known to negatively regulate the expression of KNOX genes in Arabidopsis and other species. This is shown by the ectopic expression of KNOX genes in leaves and the formation of ectopic tissues or meristems on the leaf adaxial surface or sinuses in arp mutants.
Depending on the timing and extent of spatiotemporal expression of the KNOX genes and species studied, overexpression of KNOX genes may lead to different developmental consequences. Analysis of tomato mutants suggests that weak LeT6 (a tomato KNOX gene) overexpression and SlPHAN down-regulation lead to LeT6 overexpression phenotypes and leaf lobing and compoundness, whereas strong LeT6 overexpression and SlPHAN down-regulation lead to SlPHAN down-regulation phenotypes, such as cup-shaped and wire-like leaves (Janssen et al., 1998; Kim et al., 2003a;). In maize rs2 mutants, KNOX genes may not be overexpressed to a high enough level to exhibit leaf lobing or PHAN down-regulation phenotypes (Timmermans et al., 1999; Tsiantis et al., 1999). In pea cri mutants, some ectopic stipules are formed on the adaxial leaf surface, and this is attributed to patches of BP ectopic expression in leaflets (Tattersall et al., 2005).
We isolated a knockout mutant of the M. truncatula BP gene. Phenotypic analysis of mtphan mtbp double mutant plants shows that ectopic tissues developed on the adaxial leaf surface in mature mtphan mtbp double mutant as in the mtphan single mutant, suggesting that MtBP is not involved in this process. Consistent with this conclusion, RNA in situ hybridization shows that MtBP transcripts are detected in SAM but not in leaf primordia and leaflets in both wild-type and mtphan mutant plants. Interestingly, quantitative RT-PCR shows that the expression level of MtBP, as well as MtSTM and MtKNOX6, two other KNOXI genes in M. truncatula, was elevated in vegetative shoot buds in the mtphan mutant compared with wild-type plants, supporting a negative regulation of MtKNOXI genes by MtPHAN in the shoot apex. In M. truncatula, three KNOXI genes have been identified thus far, and it is possible that redundant gene functions among the class I KNOX genes exist to mask the phenotypes of single mutants. Future experiments are required to further test this hypothesis.
In tomato plants with PHAN down-regulation, the characteristic pinnate compound leaves change to peltately palmate compound leaves (Kim et al., 2003b;; Zoulias et al., 2012). Interestingly, we observed altered leaflet placement in young mtphan mutant plants. However, the number of compound leaves with asymmetric lateral leaflets and the degree of displacement were variable. It is not yet clear how MtPHAN regulates leaflet placement. In the mtpin10 mutant, we also observed asymmetric leaflets in compound leaves (Peng and Chen, 2011). Since mtpin10 mtphan double mutants resemble the mtpin10 single mutant, we reasoned that PHAN may be involved in the synchronized lateral leaflet initiation through MtPIN10-mediated auxin activity maxima.
MtPHAN Promotes Stipule Development
In the mtphan mutant, stipules were greatly reduced in size and digitations and altered in epidermal cell identity. Recently, it has been shown that M. truncatula NODULE ROOT (NOOT) and pea COCHLEATA (COCH) genes, orthologs of the Arabidopsis BLADE-ON-PETIOLE (BOP) gene, play a role in stipule and nodule development (Couzigou et al., 2012). The stipule phenotypes of noot and coch mutants resemble that of the mtphan mutant, raising the possibility that PHAN and BOP may interact to regulate stipule development in M. truncatula.
Our observation that mtphan mutant phenotypes are strongly dependent on developmental stages appears to be consistent with previous studies, which show that AS1-AS2 has multiple downstream targets (Iwasaki et al., 2013), and multiple pathways, including chromatin modification, cell proliferation, ribosomal proteins, and trans-acting small, interfering RNA biogenesis, are involved in regulating leaf development in the as1 or as2 background (Horiguchi et al., 2011; Kojima et al., 2011; Ishibashi et al., 2012; Nakagawa et al., 2012; Xu et al., 2012). Alternatively, mtphan represents a partially reduced function mutant, and the residual level of gene expression in the mutant may be sufficient to mask some phenotypes. However, some mtphan mutant phenotypes, such as serration and curling of distal and proximal leaf margins and asymmetric placement of lateral leaflets, appear early and thus may not be in support of the latter hypothesis.
There are reports that leaf adaxial-abaxial polarity and expansion in some phan mutants are conditional. For example, in Arabidopsis as1 mutants, a high rate of radialized leaves is only observed in the Landsberg erecta genetic background (Xu et al., 2003). The Antirrhinum phan mutants exhibit completely abaxialized, needle-like leaves at a late developmental stage or at a low temperature (17°C). The first leaves are usually broader and heart shaped in Antirrhinum species phan mutants than in wild-type plants (Waites and Hudson, 1995; Waites et al., 1998). In maize rs2 mutants, about 90% of the plants display narrow, bladeless leaves in the B73 genetic background, whereas this phenotype is rare in the Mo17 background (Schneeberger et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999). The conditional and stage-dependent phenotypes observed in phan mutants may be explained by the existence of redundant genes or complex mechanisms (Tattersall et al., 2005).
Genetic Interactions between MtPHAN and SGL1
SGL1, the M. truncatula FLO/LFY/UNI ortholog, is required to maintain a transient indeterminacy for the initiation of lateral leaflets in M. truncatula, a role played by the class I KNOX genes in compound-leafed species outside the inverted repeat-lacking clade legumes (Wang et al., 2008). Consistent with this, sgl1 mutants have simple leaves with shortened petioles (Wang et al., 2008; Peng et al., 2011). Interestingly, phenotypes of the sgl1 mtphan double mutant suggest independent functions of MtPHAN and SGL1 in compound leaf patterning and leaf margin development. On the other hand, the further reduced petiole phenotype in sgl1 mtphan double mutants suggests additive interactions between SGL1 and MtPHAN in petiole development. MtPHAN and SGL1 functions appear to be consistent with their partially overlapping expression patterns. SGL1 is highly expressed in SAM and leaf primordia, but its expression is rapidly decreased in expanding leaves (Wang et al., 2008), whereas MtPHAN is weakly expressed in SAM but strongly expressed in leaf primordia and leaflets.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Medicago truncatula phan (mtphan) and bp (mtbp) mutants were obtained from reverse screens of the Tnt1 retrotransposon insertion pool in the R108 background (Cheng et al., 2011). Mutants were backcrossed to R108. The BC1 mutant and its descendants were used for phenotypic characterization. sgl1, mtpin10, and elp1 mutants, all in the R108 background, were as described previously (Wang et al., 2008; Peng and Chen, 2011; Chen et al., 2012). Arabidopsis (Arabidopsis thaliana) as1-1 was from the Arabidopsis Stock Center. Arabidopsis and M. truncatula plants were grown in growth chambers and glasshouses (16-h/8-h day/night light cycle; 22°C/20°C day/night temperature), respectively.
Sequence Alignment and Phylogenetic Analysis
ARP amino acid sequences were aligned using ClustalX 1.83. The aligned sequences were edited using BioEdit (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The maximum parsimony phylogenetic tree was reconstructed by PAUP 4.0 beta 10 in Geneious Pro 5.6.3, with 1,000 bootstrap replications.
SEM
SEM was performed as described previously (Wang et al., 2008). Briefly, plant tissues were fixed with 2.5% glutaraldehyde in phosphate saline solution (pH 7.0) overnight, followed by fixation with 1% OsO4 at 4°C for 1 h. The fixed samples were dehydrated with an ethanol series, critical point dried, and mounted for imaging.
Genetic Complementation
M. truncatula PHAN genomic DNA, including promoter and coding sequences (without the stop codon), was cloned into a binary vector in frame with GFP. The complementation construct was introduced into the mtphan mutant plants by Agrobacterium tumefaciens EHA105-mediated stable transformation, as reported previously (Wang et al., 2008), and into the Arabidopsis as1-1 mutant by A. tumefaciens GV3101-mediated stable transformation.
RNA Isolation, RT-PCR, and Quantitative PCR
Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) and quantified with a Nanodrop spectrophotometer. RT was performed using the Qiagen SuperScript II Kit. Quantitative PCR was conducted on the 7900HT Fast Real-Time PCR system (Applied Biosystems). An M. truncatula ACTIN gene was used as the control. LinRegPCR (Ramakers et al., 2003) and SDS2.2.1 (Applied Biosystems) were used for data analysis. Statistical analysis was carried out using Student’s t test. Primer sequences are listed in Supplemental Table S1.
RNA in Situ Hybridization
RNA in situ hybridization was performed as described previously (Ferrándiz et al., 2000) with minor modifications. The MtPHAN probes correspond to a 445-bp sequence of the MtPHAN coding sequence. The MtBP probes correspond to a 613-bp sequence of the MtBP coding sequence. Eight-micrometer sections from shoot apices of 2- to 4-week-old seedlings were processed and hybridized with digoxigenin-labeled sense and antisense probes.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank under the following accession numbers: MtPHAN (DQ468322), GmPHAN1 (NP_001236839.1), GmPHAN2 (NP_001235251), GmPHAN3 (KC737842), GmPHAN4 (KC737843), LjPHAN1 (AAX21343.1), LjPHAN2 (AAX21344.1), ELP1 (JQ653161.1), MtSTM (EF128056.1), MtBP (EF128057.1), MtKNOX6 (EF128061.1), SGL1/UNI (AY928184.1), FCL1 (HQ695002), AS1 (AT2G37630), STM (AT1G62360), and BP/KNAT1 (AT4G08150).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid sequence alignment of MtPHAN and its related sequences.
Supplemental Figure S2. Phylogenetic analysis and gene structures of MtPHAN and its related sequences.
Supplemental Figure S3. Leaf primordia development of the mtphan mutant.
Supplemental Figure S4. Leaf epidermal cell morphologies of the mtphan mutant.
Supplemental Figure S5. Compound leaf phenotypes of mtphan mutant seedlings.
Supplemental Figure S6. Compound leaf phenotypes of 70-d-old mtphan mutant plants.
Supplemental Figure S7. SEM analysis of rachis epidermal cells of compound leaves of 70-d-old wild-type and mtphan mutant plants.
Supplemental Figure S8. Altered stipule development of the mtphan mutant.
Supplemental Figure S9. Functional rescue of the mtphan mutant by MtPHAN.
Supplemental Figure S10. Functional rescue of the Arabidopsis as1 mutant by MtPHAN.
Supplemental Figure S11. Nuclear localization of MtPHAN-GFP fusion proteins in transgenic Arabidopsis plants.
Supplemental Figure S12. Compound leaf phenotypes of the mtphan elp1 double mutant.
Supplemental Figure S13. Characterization of mtbp and mtphan mtbp mutant plants.
Supplemental Figure S14. Compound leaf phenotypes of the sgl1 mtphan double mutant.
Supplemental Table S1. Primer sequences used in this study.
Acknowledgments
We thank members of the Chen laboratory for helpful comments on the manuscript; Guangming Li (Noble Foundation) for technical support; Colleen Elles (Noble Foundation) for greenhouse assistance; Preston Larson (University of Oklahoma) for assistance with SEM; Guifen Li and Yuhong Tang (Noble Foundation) for assistance in tissue sectioning; Xiaofei Cheng and Jiangqi Wen (Noble Foundation) for reverse genetic screening; and Kirankumar Mysore (Noble Foundation) for providing Tnt1 lines.
Glossary
- SAM
shoot apical meristem
- LOB
lateral organ boundary
- RT
reverse transcription
- SEM
scanning electron microscopy
References
- Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. (2008) A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet 40: 1136–1141 [DOI] [PubMed] [Google Scholar]
- Ben-Gera H, Shwartz I, Shao MR, Shani E, Estelle M, Ori N. (2012) ENTIRE and GOBLET promote leaflet development in tomato by modulating auxin response. Plant J 70: 903–915 [DOI] [PubMed] [Google Scholar]
- Berger Y, Harpaz-Saad S, Brand A, Melnik H, Sirding N, Alvarez JP, Zinder M, Samach A, Eshed Y, Ori N. (2009) The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136: 823–832 [DOI] [PubMed] [Google Scholar]
- Burko Y, Shleizer-Burko S, Yanai O, Shwartz I, Zelnik ID, Jacob-Hirsch J, Kela I, Eshed-Williams L, Ori N. (2013) A role for APETALA1/fruitfull transcription factors in tomato leaf development. Plant Cell 25: 2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. (2000) Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. (2002) ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965 [DOI] [PubMed] [Google Scholar]
- Champagne CE, Goliber TE, Wojciechowski MF, Mei RW, Townsley BT, Wang K, Paz MM, Geeta R, Sinha NR. (2007) Compound leaf development and evolution in the legumes. Plant Cell 19: 3369–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Moreau C, Liu Y, Kawaguchi M, Hofer J, Ellis N, Chen R. (2012) Conserved genetic determinant of motor organ identity in Medicago truncatula and related legumes. Proc Natl Acad Sci USA 109: 11723–11728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Wen J, Tadege M, Ratet P, Mysore KS. (2011) Reverse genetics in Medicago truncatula using Tnt1 insertion mutants. Methods Mol Biol 678: 179–190 [DOI] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. (1996) The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Couzigou JM, Zhukov V, Mondy S, Abu el Heba G, Cosson V, Ellis TH, Ambrose M, Wen J, Tadege M, Tikhonovich I, et al. (2012) NODULE ROOT and COCHLEATA maintain nodule development and are legume orthologs of Arabidopsis BLADE-ON-PETIOLE genes. Plant Cell 24: 4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. (2003) Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol 13: 1768–1774 [DOI] [PubMed] [Google Scholar]
- Ferrándiz C, Gu Q, Martienssen R, Yanofsky MF. (2000) Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127: 725–734 [DOI] [PubMed] [Google Scholar]
- Golz JF, Roccaro M, Kuzoff R, Hudson A. (2004) GRAMINIFOLIA promotes growth and polarity of Antirrhinum leaves. Development 131: 3661–3670 [DOI] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MC. (2008) Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20: 48–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. (1996) The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell 84: 735–744 [DOI] [PubMed] [Google Scholar]
- Hay A, Barkoulas M, Tsiantis M. (2006) ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 133: 3955–3961 [DOI] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. (2006) The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38: 942–947 [DOI] [PubMed] [Google Scholar]
- Hofer J, Gourlay C, Michael A, Ellis TH. (2001) Expression of a class 1 knotted1-like homeobox gene is down-regulated in pea compound leaf primordia. Plant Mol Biol 45: 387–398 [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Hellens R, Ambrose M, Matthews P, Michael A, Ellis N. (1997) UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr Biol 7: 581–587 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Nakayama H, Ishikawa N, Kubo M, Demura T, Fukuda H, Tsukaya H. (2011) ANGUSTIFOLIA3 plays roles in adaxial/abaxial patterning and growth in leaf morphogenesis. Plant Cell Physiol 52: 112–124 [DOI] [PubMed] [Google Scholar]
- Ishibashi N, Kanamaru K, Ueno Y, Kojima S, Kobayashi T, Machida C, Machida Y. (2012) ASYMMETRIC-LEAVES2 and an ortholog of eukaryotic NudC domain proteins repress expression of AUXIN-RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol Open 1: 197–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki M, Takahashi H, Iwakawa H, Nakagawa A, Ishikawa T, Tanaka H. (2013) Dual regulation of AUXIN-RESPONSE-FACTOR3 gene expression by AS1-AS2, which maintains the status of DNA methylation, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 40: 1958–1969 [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Lund L, Sinha N. (1998) Overexpression of a homeobox gene, LeT6, reveals indeterminate features in the tomato compound leaf. Plant Physiol 117: 771–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Bollman K, Taylor RA, Bomblies K, Poethig RS. (2001) KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kim M, McCormick S, Timmermans M, Sinha N. (2003b) The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424: 438–443 [DOI] [PubMed] [Google Scholar]
- Kim M, Pham T, Hamidi A, McCormick S, Kuzoff RK, Sinha N. (2003a) Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130: 4405–4415 [DOI] [PubMed] [Google Scholar]
- Koenig D, Bayer E, Kang J, Kuhlemeier C, Sinha N. (2009) Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136: 2997–3006 [DOI] [PubMed] [Google Scholar]
- Kojima S, Iwasaki M, Takahashi H, Imai T, Matsumura Y, Fleury D, Van Lijsebettens M, Machida Y, Machida C. (2011) Asymmetric leaves2 and Elongator, a histone acetyltransferase complex, mediate the establishment of polarity in leaves of Arabidopsis thaliana. Plant Cell Physiol 52: 1259–1273 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. (1996) A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Luo JH, Yan J, Weng L, Yang J, Zhao Z, Chen JH, Hu XH, Luo D. (2005) Different expression patterns of duplicated PHANTASTICA-like genes in Lotus japonicus suggest their divergent functions during compound leaf development. Cell Res 15: 665–677 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. (2001) Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Takahashi H, Kojima S, Sato N, Ohga K, Cha BY, Woo JT, Nagai K, Horiguchi G, Tsukaya H, et al. (2012) Berberine enhances defects in the establishment of leaf polarity in asymmetric leaves1 and asymmetric leaves2 of Arabidopsis thaliana. Plant Mol Biol 79: 569–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Chen R. (2011) Auxin efflux transporter MtPIN10 regulates compound leaf and flower development in Medicago truncatula. Plant Signal Behav 6: 1537–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Yu J, Wang H, Guo Y, Li G, Bai G, Chen R. (2011) Regulation of compound leaf development in Medicago truncatula by fused compound leaf1, a class M KNOX gene. Plant Cell 23: 3929–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM. (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeberger R, Tsiantis M, Freeling M, Langdale JA. (1998) The rough sheath2 gene negatively regulates homeobox gene expression during maize leaf development. Development 125: 2857–2865 [DOI] [PubMed] [Google Scholar]
- Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y. (2001) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128: 1771–1783 [DOI] [PubMed] [Google Scholar]
- Shani E, Burko Y, Ben-Yaakov L, Berger Y, Amsellem Z, Goldshmidt A, Sharon E, Ori N. (2009) Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 21: 3078–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried KR, Eshed Y, Baum SF, Otsuga D, Drews GN, Bowman JL. (1999) Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Tattersall AD, Turner L, Knox MR, Ambrose MJ, Ellis TH, Hofer JM. (2005) The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. Plant Cell 17: 1046–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans MC, Hudson A, Becraft PW, Nelson T. (1999) ROUGH SHEATH2: a Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Tsiantis M, Schneeberger R, Golz JF, Freeling M, Langdale JA. (1999) The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156 [DOI] [PubMed] [Google Scholar]
- Vernoux T, Kronenberger J, Grandjean O, Laufs P, Traas J. (2000) PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127: 5157–5165 [DOI] [PubMed] [Google Scholar]
- Waites R, Hudson A. (1995) phantastica: a gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Waites R, Selvadurai HR, Oliver IR, Hudson A. (1998) The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789 [DOI] [PubMed] [Google Scholar]
- Wang H, Chen J, Wen J, Tadege M, Li G, Liu Y, Mysore KS, Ratet P, Chen R. (2008) Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula. Plant Physiol 146: 1759–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Huang W, Li Y, Wang H, Huang H, Cui X. (2012) Elongator complex is critical for cell cycle progression and leaf patterning in Arabidopsis. Plant J 69: 792–808 [DOI] [PubMed] [Google Scholar]
- Xu L, Xu Y, Dong A, Sun Y, Pi L, Xu Y, Huang H. (2003) Novel as1 and as2 defects in leaf adaxial-abaxial polarity reveal the requirement for ASYMMETRIC LEAVES1 and 2 and ERECTA functions in specifying leaf adaxial identity. Development 130: 4097–4107 [DOI] [PubMed] [Google Scholar]
- Zhang J, Chen R, Xiao J, Qian C, Wang T, Li H, Ouyang B, Ye Z. (2007) A single-base deletion mutation in SlIAA9 gene causes tomato (Solanum lycopersicum) entire mutant. J Plant Res 120: 671–678 [DOI] [PubMed] [Google Scholar]
- Zhou C, Han L, Fu C, Chai M, Zhang W, Li G, Tang Y, Wang ZY. (2012) Identification and characterization of petiolule-like pulvinus mutants with abolished nyctinastic leaf movement in the model legume Medicago truncatula. New Phytol 196: 92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Hou C, Metelli A, Qi L, Tadege M, Mysore KS, Wang ZY. (2011) Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant Cell 23: 2106–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulias N, Koenig D, Hamidi A, McCormick S, Kim M. (2012) A role for PHANTASTICA in medio-lateral regulation of adaxial domain development in tomato and tobacco leaves. Ann Bot (Lond) 109: 407–418 [DOI] [PMC free article] [PubMed] [Google Scholar]