Independent mechanisms link photosynthate availability with nitrate uptake and assimilation into amino acids.
Abstract
Mineral nutrient uptake and assimilation is closely coordinated with the production of photosynthate to supply nutrients for growth. In Arabidopsis (Arabidopsis thaliana), nitrate uptake from the soil is mediated by genes encoding high- and low-affinity transporters that are transcriptionally regulated by both nitrate and photosynthate availability. In this study, we have studied the interactions of nitrate and glucose (Glc) on gene expression, nitrate transport, and growth using glucose-insensitive2-1 (gin2-1), which is defective in sugar responses. We confirm and extend previous work by showing that HEXOKINASE1-mediated oxidative pentose phosphate pathway (OPPP) metabolism is required for Glc-mediated NITRATE TRANSPORTER2.1 (NRT2.1) expression. Treatment with pyruvate and shikimate, two products derived from intermediates of the OPPP that are destined for amino acid production, restores wild-type levels of NRT2.1 expression, suggesting that metabolites derived from OPPP metabolism can, together with Glc, directly stimulate high levels of NRT2.1 expression. Nitrate-mediated NRT2.1 expression is not influenced by gin2-1, showing that Glc does not influence NRT2.1 expression through nitrate-mediated mechanisms. We also show that Glc stimulates NRT2.1 protein levels and transport activity independently of its HEXOKINASE1-mediated stimulation of NRT2.1 expression, demonstrating another possible posttranscriptional mechanism influencing nitrate uptake. In gin2-1 plants, nitrate-responsive biomass growth was strongly reduced, showing that the supply of OPPP metabolites is essential for assimilating nitrate for growth.
The photoautotrophic metabolism of plants involves the production of carbohydrates by photosynthesis and the acquisition of minerals such as nitrogen (N) and phosphorus from the soil. Photosynthate supply is closely balanced with mineral uptake and assimilation to ensure the production of amino acids and other metabolites for plant growth. In contrast to extensive knowledge of metabolic processes involved in nitrate and carbon assimilation (Nunes-Nesi et al., 2010; Xu et al., 2012), relatively little is known about the mechanisms coupling growth to nutrient availability (Robaglia et al., 2012) and about the mechanisms integrating carbon metabolism and N uptake and assimilation. Improving our knowledge of these processes will aid the genetic and mechanistic analyses of N use efficiency in crop plants (Zhang et al., 2010).
Nitrate has profound effects on gene expression, hormone synthesis, and plant growth (Krapp et al., 2011; Krouk et al., 2011; Bouguyon et al., 2012) independent of its assimilation, demonstrating that it is sensed and signals cellular responses. Microarray analyses have shown that nitrate levels rapidly influence the expression of many classes of genes involved in their assimilation and metabolism (Wang et al., 2003; Scheible et al., 2004; Bi et al., 2007; Nero et al., 2009). These include genes of the N uptake and N assimilation pathways, including high- and low-affinity nitrate transporters, NITRATE REDUCTASE (NR), NITRITE REDUCTASE (NiR), and the genes encoding enzymes of the GOGAT ammonium assimilatory pathway (Lejay et al., 1999; Stitt, 1999; Wang et al., 2000). Nitrate influences photosynthate allocation to storage as starch, anthocyanin accumulation, and amino acid synthesis through nitrate-mediated induction of three LATERAL ORGAN BOUNDARIES (LBD) transcription factor genes (LBD37, LBD38, and LBD39; Rubin et al., 2009) that repress the expression of AtMYB75 and AtMYB90, key regulators of anthocyanin biosynthetic pathway genes (Stracke et al., 2001), as well as NR and several nitrate transporter genes. Low nitrate levels relieve LBD-mediated repression, demonstrating one transcriptional control mechanism integrating carbon and nitrate responses. Nitrate also influences the allocation of photosynthate metabolites to amino acid synthesis by repressing the expression of AGPS (encoding the regulatory subunit of AGPase), a regulatory subunit of starch synthesis, and inducing the expression of several oxidative pentose phosphate pathway (OPPP) genes (Stitt, 1999; Wang et al., 2003). Transcription factors governing the expression of genes involved in nitrate uptake and assimilation and carbon skeleton supply have been identified: DNA-BINDING ONE ZINC FINGER overexpression increases growth in low nitrate (Yanagisawa et al., 2004); the MADS transcription factor ARABIDOPSIS NITRATE REGULATED1 functions downstream of nitrate uptake to control lateral root growth (Zhang and Forde, 1998); NIN-LIKE PROTEIN7 (NLP7) controls nitrate-mediated induction of the nitrate transporter genes NITRATE TRANSPORTER2.1 (NRT2.1), NRT2.2, and NR genes (Castaings et al., 2009); and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE9 is involved in rapid nitrate-responsive gene expression (Krouk et al., 2010b). Recently, NLP7 was shown to bind to the promoters of many nitrate signaling and assimilation genes, and nitrate was shown to promote the nuclear retention of NLP7 (Marchive et al., 2013).
In Arabidopsis (Arabidopsis thaliana), the expression of genes encoding the low-affinity NRT1.1 and NRT1.2 transporters is constitutive and enhanced by high nitrate levels (Xu et al., 2012). Expression of genes encoding the high-affinity nitrate transporters NRT2.1 and NRT2.2 is repressed by high nitrate levels and activated in low-nitrate conditions to scavenge available soil nitrate (Li et al., 2006; Gojon et al., 2011; Xu et al., 2012; Wang et al., 2012b). NRT2.4 encodes an N starvation-induced high-affinity transporter (Kiba et al., 2012). NRT1.1 gene expression is also up-regulated by auxin (Guo et al., 2002), while NRT2.1 expression is down-regulated. Interestingly, in low nitrate concentrations, NRT1.1 functions as an auxin transporter, removing auxin from lateral root primordia. In high-nitrate conditions, NRT1.1-mediated auxin transport is inhibited, resulting in auxin accumulation and lateral root growth (Krouk et al., 2010a). Posttranscriptional mechanisms also influence nitrate uptake; in low-nitrate conditions, NRT1.1 is phosphorylated on its Thr-101 residue by CYSTATHIONINE BETA-LYASE-interacting protein kinase, switching it into a high-affinity transporter (Ho et al., 2009). NRT2.1 expression is repressed under high-N conditions (Lejay et al., 1999), possibly through systemic feedback repression by reduced N metabolites such as Gln (Gansel et al., 2001; Girin et al., 2010). NRT2.1 expression is also inhibited locally by NRT1.1 under high-nitrate conditions (Muños et al., 2004). Repression of NRT2.1 expression by high nitrate is associated with HIGH NITROGEN INSENSITIVE9/ARABIDOPSIS THALIANA INTERACT WITH SPT6 1-mediated increases in promoter histone methylation (Widiez et al., 2011). Constitutive expression of NRT2.1 in the nrt2.1 mutant background showed that repression of high-affinity nitrate uptake by high nitrate was not directly associated with reduced protein levels (Laugier et al., 2012), suggesting that protein modifications may be responsible.
Like nitrate, exogenous Suc and Glc influence the expression of many genes involved in metabolism and growth, including amino acid biosynthetic genes, ribosome synthesis genes, and hormone-regulated genes (Li et al., 2006). Network analyses (Gutiérrez et al., 2007) identified interdependent transcriptional responses to Glc and nitrate, including genes for auxin responses and signaling, transcription factors, and signal transduction proteins. Overexpression of the monosaccharide transporter SUGAR TRANSPORT PROTEIN13 increased growth in response to low nitrate levels, establishing a link between N use efficiency and sugar supplies (Schofield et al., 2009). Sugars also induce the expression of genes involved in the transport of nitrate, phosphate, and sulfate (Lejay et al., 2003; Li et al., 2006), revealing an intimate connection between photosynthesis, carbon metabolism, and nutrient uptake. An inhibitor of phosphogluconate dehydrogenase activity in the OPPP (Kruger and von Schaewen, 2003) reduced Glc-mediated induction of NRT2.1, NRT1.1, NRT1.5, PEPTIDE TRANSPORTER, AMMONIUM TRANSPORTER1.3(AMT1.3), SULFATE TRANSPORTER1.1 (SULTR1.1), and SULTR3.5 (Lejay et al., 2008), implicating OPPP metabolism in these sugar-mediated responses. Furthermore, reduction of HEXOKINASE1 (HXK1) gene expression by antisense RNA reduced the expression of NRT2.1 (Lejay et al., 2003), indicating that the synthesis of Glc-6-P for the OPPP is required for the sugar-mediated expression of NRT2.1. This suggested that OPPP metabolism may influence nitrate uptake by transcriptional control of nitrate transporter genes (Lejay et al., 2008). Recently, it was shown that the pgl3-1 mutant, which reduces the expression of the plastidial 6-phosphogluconolactonase gene PGL3, had reduced expression of nitrate assimilatory genes in response to Suc (Bussell et al., 2013). This established that flux through the OPPP is required for the expression of nitrate assimilation genes and growth responses to nitrate.
HXK1 has a regulatory function in addition to its catalytic activity (Moore et al., 2003) based on the Glc-insensitive responses of the glucose-insensitive2-1 (gin2-1) mutant and the maintenance of these responses in the catalytically inactive mutants HXK1G104D and HXK1S177A. We wished to further explore the influence of HXK1 on the expression of NRT2.1 and on nitrate-responsive growth in order to determine if the proposed role of the OPPP in NRT2.1 expression (Lejay et al., 2008) occurs through a novel mechanism or whether the OPPP functions through an existing NRT2.1 regulatory signaling pathway. We confirmed that Glc-induced NRT2.1 expression is highly dependent on HXK1 catalytic activity and show that OPPP intermediates and downstream metabolites rescue the impaired response of NRT2.1 to Glc in gin2-1 mutants, providing genetic and further biochemical evidence to support the conclusions of Lejay et al. (2008) and confirming the role of OPPP metabolism in nitrate assimilation and growth (Bussell et al., 2013). Nitrate-mediated changes in NRT2.1 expression were unchanged in gin2-1, indicating that nitrate and Glc influence NRT2.1 expression independently. We further show that increased Glc levels rapidly stimulate NRT2.1 protein accumulation and high-affinity nitrate uptake independently of NRT2.1 expression, possibly through posttranscriptional mechanisms.
RESULTS
Expression of N Assimilation Genes Is Reduced in the gin2-1 Hexokinase Mutant
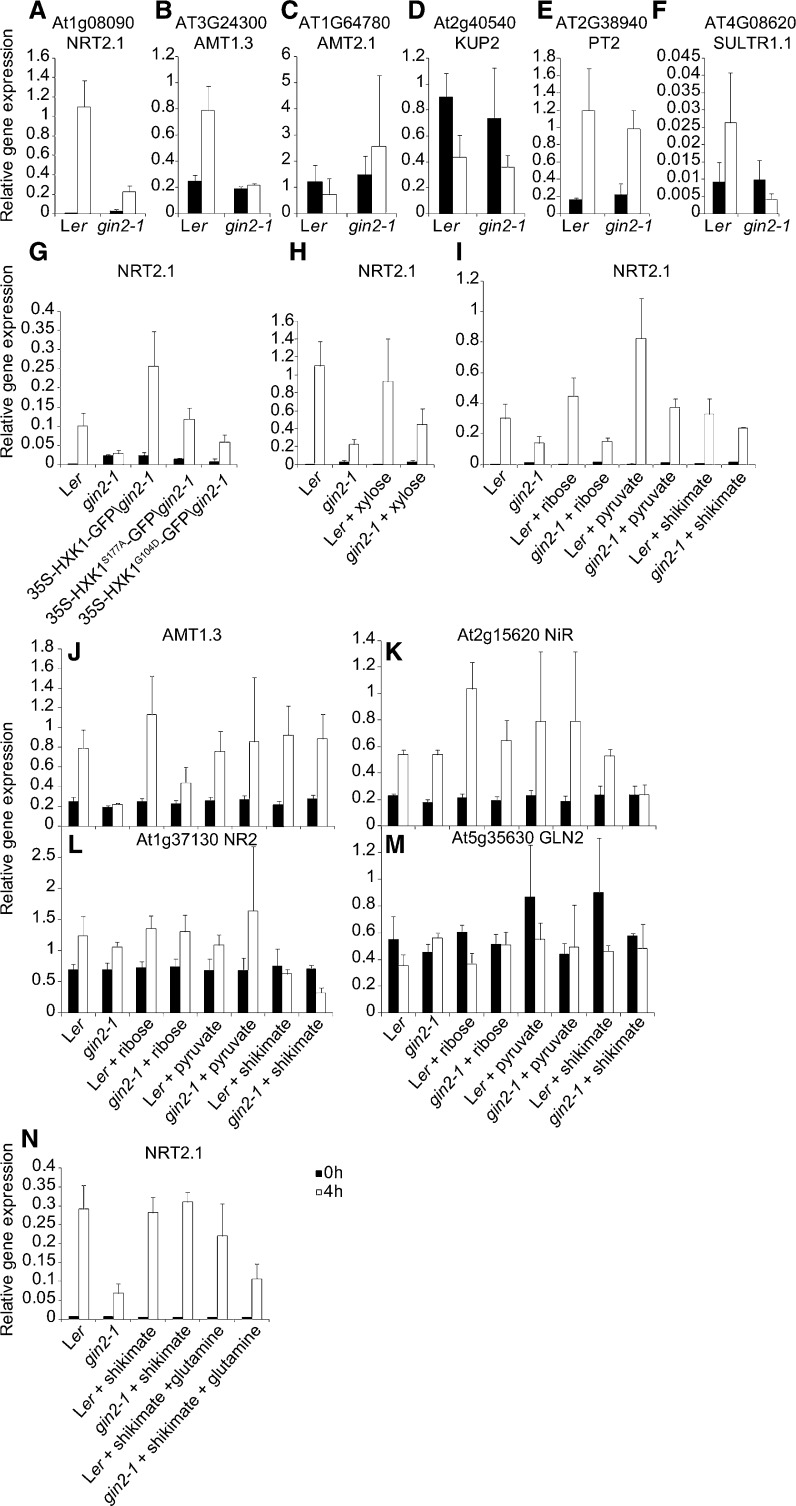
Blocking the OPPP by inhibition of phosphogluconate dehydrogenase activity reduced NRT2.1 expression in response to Suc (Lejay et al., 2008). To assess this genetically, we analyzed the expression of NRT2.1 and five other transporter genes in response to Glc in the HXK1 mutant gin2-1 (Moore et al., 2003). Landsberg erecta (Ler) and gin2-1 seedlings were grown for 7 d on Murashige and Skoog (MS) medium with 0.5% (w/v) Glc, transferred to liquid MS medium without Glc for 24 h, before placing seedlings in liquid MS medium with 3% (w/v) Glc. Roots were harvested for RNA analysis at the start of the 3% (w/v) Glc treatment and after 4 h. Figure 1A shows a large reduction in Glc-mediated induction of NRT2.1 expression in gin2-1. Of the five other transporter genes analyzed, three (AMT1.3, PHOSPHATE TRANSPORTER2, and SULTR1.1) exhibited increased expression in response to Glc, and of these, AMT1.3 and SULTR1.1 showed a significantly reduced response in gin2-1 (Fig. 1, B–F). To assess the role of HXK1 catalytic activity in NRT2.1 expression, Glc-responsive gene expression was measured in the catalytically inactive HXK1 mutants 35S-HXK1S177A and 35S-HXK1G104D (Moore et al., 2003) expressed in gin2-1. NRT2.1 expression was reduced in both 35S-HXK1S177A and 35S-HXK1G104D (Fig. 1G). The line expressing HXK1-GFP in gin2-1 rescued NRT2.1 expression. This showed that reduced Glc-responsive expression of NRT2.1 in gin2-1 resulted from reduced catalytic activity of HXK1. Analysis of the expression of seven genes encoding enzymes of the OPPP in response to Glc in Ler and gin2-1 (Supplemental Fig. S1) showed that gin2-1 did not alter their expression. This confirmed that reduced NRT2.1 expression in gin2-1 was not due to altered expression of the OPPP genes and is consistent with previous research showing that biochemical inhibition of OPPP metabolism and reduced HXK1 expression led to reduced NRT2.1 expression in response to Glc (Lejay et al., 2003, 2008) and with the genetic evidence from the pgl3-1 mutant that OPPP flux is required for sugar-induced expression of nitrate assimilation genes (Bussell et al., 2013).
Figure 1.
Glc induction of nitrate uptake and assimilation genes. Seven-day-old Ler and gin2-1 seedlings were starved of Glc for 24 h before transfer to MS medium with 3% (w/v) Glc supplemented with 30 mm Xyl, 30 mm Rib, 30 mm pyruvate, 30 mm shikimate, or 30 mm Gln. Roots were harvested either at the time of transfer (0h; black bars) or 4 h after transfer (4h; white bars). Expression levels were normalized against TUBULIN6. The values are means of three biological repeats ± se.
Reduced Glc-Responsive Expression of N Assimilation Genes in gin2-1 Is Rescued by Some Intermediates of the OPPP
To further assess the role of the OPPP in NRT2.1 expression, we tested if Xyl, which can feed into the OPPP downstream of Glc-6-P, rescued NRT2.1 Glc-responsive gene expression in gin2-1. Figure 1H shows the partial rescue of NRT2.1 expression by Xyl. To determine which of the three pathways that utilize OPPP intermediates and products (nucleotide synthesis, amino acid synthesis, and glycolysis; Kanehisa et al., 2012) might contribute to this rescue of NRT2.1 gene expression in gin2-1, several OPPP intermediates were tested. Both pyruvate, an intermediate of glycolysis, and shikimate, an intermediate of amino acid biosynthesis, fully rescued NRT2.1 responses to levels similar to those seen in Ler (Fig. 1I). Pyruvate was taken up by seedlings in these experiments (Supplemental Fig. S2H). In contrast, Rib, an intermediate of the nucleic acid biosynthesis pathway, did not rescue NRT2.1 gene expression (Fig. 1I). This supported the conclusion that reduced passage of metabolites through the OPPP in gin2-1, including those destined for amino acid biosynthesis, reduced Glc-responsive NRT2.1 expression.
We assessed the Glc-responsive expression of other high- and low-affinity nitrate transporter genes (Miller et al., 2007; Gojon et al., 2011) in gin2-1 for dependence on OPPP intermediates. The expression of NRT1.6, NRT2.2 to NRT2.5, and NRT2.7 was too low to be quantitatively analyzed by quantitative reverse transcription-PCR (data not shown; Supplemental Fig. S2). Only NRT1.1 and NRT1.7 expression was slightly Glc responsive, but it was not reduced in gin2-1 or in response to OPPP intermediates. NRT1.2 and NRT1.9 were not Glc responsive, and NRT1.2 showed only a slight influence of gin2-1. NRT1.5 and NRT1.8 expression was suppressed by Glc, and NRT1.5 was significantly lower in gin2-1, with no response to OPPP intermediates. Finally, NRT2.6 expression was not dependent on Glc but showed large responses to pyruvate and shikimate in both Ler and gin2-1 (Supplemental Fig. S2). This indicated that NRT2.1 is probably the only nitrate transporter whose Glc-responsive expression requires OPPP metabolism.
The expression of N assimilation genes is tightly coordinated (Nunes-Nesi et al., 2010); therefore, we assessed the influence of Glc on the expression of selected genes (NR2, NiR, AMT1.3, and GLUTAMINE SYNTHASE2(GLN2) in gin2-1, together with rescue by Rib, pyruvate, or shikimate. NR2, NiR, and AMT1.3 showed Glc-responsive expression in Ler that was reduced in gin2-1, and pyruvate was able to rescue their expression in gin2-1 (Fig. 1, J–L). However, shikimate only rescued the Glc response of AMT1.3 in gin2-1 (Fig. 1, J–L). GLN2 was not expressed in response to Glc in both Ler and gin2-1 (Fig. 1M). Therefore, the expression of several genes encoding enzymes of N assimilation is responsive to metabolites derived from the OPPP.
Gln, an end product of N assimilation, down-regulates NRT2.1 expression (Gansel et al., 2001; Miller et al., 2007). Therefore, we tested the effect of Gln on the rescue of NRT2.1 Glc-responsive expression by shikimate. Figure 1N shows that even though shikimate rescues the Glc response of NRT2.1 in gin2-1, Gln down-regulates NRT2.1 expression in the presence of shikimate. This suggests that OPPP metabolite-mediated increases in NRT2.1 expression can be suppressed by Gln, an end product of N assimilation.
Impairment of N Assimilation Gene Expression in gin2-1 Is Due to Reduced Responses to Glc and Not to Altered Responses to Nitrate
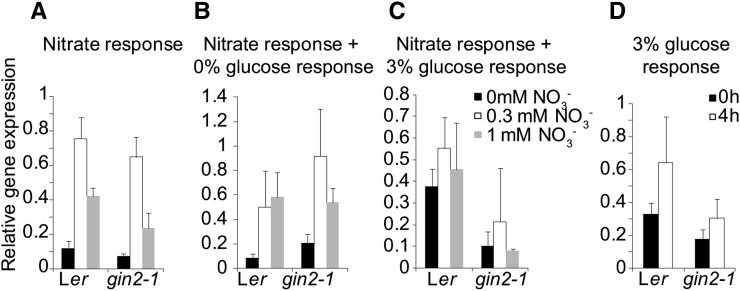
To assess the influence of nitrate on the Glc-mediated expression of NRT2.1 in gin2-1, we measured NRT2.1 expression in Ler and gin2-1 in response to different nitrate levels. Seedlings were grown for 7 d on MS medium, placed in liquid MS-N medium (MS medium without KNO3 and NH4NO3) for 24 h, and then placed in MS-N medium supplemented with 0, 0.3, or 1 mm KNO3. Glc levels were maintained at 0.5% (w/v). Seedlings were taken for RNA isolation at the start of the nitrate treatment and after 4 h. NRT2.1 was induced to similar levels in response to nitrate in both Ler and gin2-1 (Fig. 2A), showing optimal induction at low nitrate levels as reported previously (Lejay et al., 1999; Girin et al., 2010). To test if Glc- and nitrate-responsive NRT2.1 expression occurs through separate pathways, a combined analysis was performed. Seven-day-old seedlings were starved of both Glc and N for 24 h and then placed in liquid MS-N medium with either 0% or 3% (w/v) Glc combined with 0, 0.3, or 1 mm KNO3. Plants on medium with 0% (w/v) Glc showed similar nitrate-mediated induction of NRT2.1 in both Ler and gin2-1 (Fig. 2B), showing optimal expression at 0.3 mm as expected (Gojon et al., 2011). Nitrate-responsive NRT2.1 expression was significantly reduced in gin2-1 (Fig. 2C) in high Glc levels. These analyses suggested that Glc and HXK1 did not influence nitrate-mediated NRT2.1 expression significantly, apart from reduced expression in gin2-1 in response to high Glc levels at all nitrate levels tested.
Figure 2.
NRT2.1 expression in response to nitrate and Glc. Seedlings were grown on plates containing MS medium and 0.5% (w/v) Glc for 7 d before nitrate and/or Glc starvation for 24 h. Nitrate was added to a final concentration of 0, 0.3, or 1 mm, and root samples were taken for RNA analysis. A, Nitrate starvation followed by nitrate addition with constant Glc levels of 0.5% (w/v). B, Nitrate and Glc starvation followed by nitrate addition with Glc levels of 0% (w/v) after starvation. C, Nitrate and Glc starvation followed by nitrate addition with Glc levels of 3% (v/w) after starvation. D, Ler and gin2-1 seedlings (7 d old) were grown on MS-N medium containing 0.3 mm NO3− with 0.5% (w/v) Glc. Seedlings were starved of Glc for 24 h before transfer to MS-N medium containing 0.3 mm NO3− and 3% (w/v) Glc. RNA samples were taken 0 and 4 h after transfer. Gene expression levels were normalized against TUBULIN6. The values are means of three biological repeats ± se.
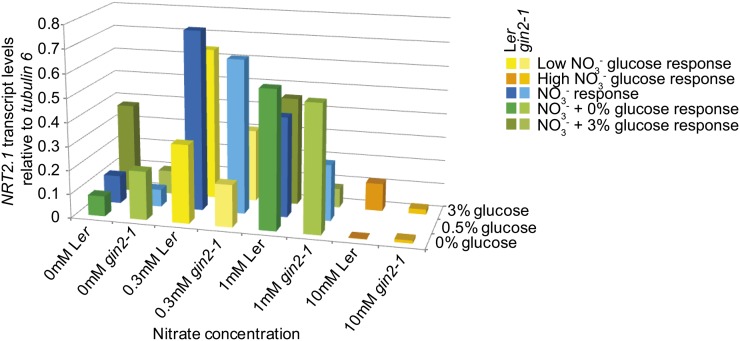
As the seedlings analyzed for Glc-responsive NRT2.1 expression in Figure 1 were all grown on MS medium containing a high level of nitrate (more than 3 mm KNO3), we analyzed Glc-induced NRT2.1 expression in gin2-1 seedlings grown on MS-N medium containing 0.3 mm KNO3, which was optimal for NRT2.1 expression (Fig. 2A). NRT2.1 expression after 4 h in response to Glc in Ler and gin2-1 (Fig. 2D) indicated that Glc had no significant effect on NRT2.1 gene expression in low-nitrate conditions, in contrast to the high induction in high nitrate (Fig. 1A). Comparison of NRT2.1 expression values across experiments showed that reduced Glc-responsive NRT2.1 expression was due to overall highly elevated expression levels in 0.3 mm nitrate (Fig. 3).
Figure 3.
Compilation of NRT2.1 gene expression levels. NRT2.1 expression levels described in Figures 1A and 2 were normalized relative to TUBULIN6 gene expression levels to permit comparisons across different nitrate and Glc conditions. To identify each data set, they are color coded according to the nitrate and Glc treatments used: low NO3− Glc response (yellow), high NO3− Glc response (orange), NO3− response (blue), NO3− + 0% (w/v) Glc response (green), and NO3− + 3% (w/v) Glc response (brown-green).
Nitrate-Promoted Growth Is Impaired in gin2-1
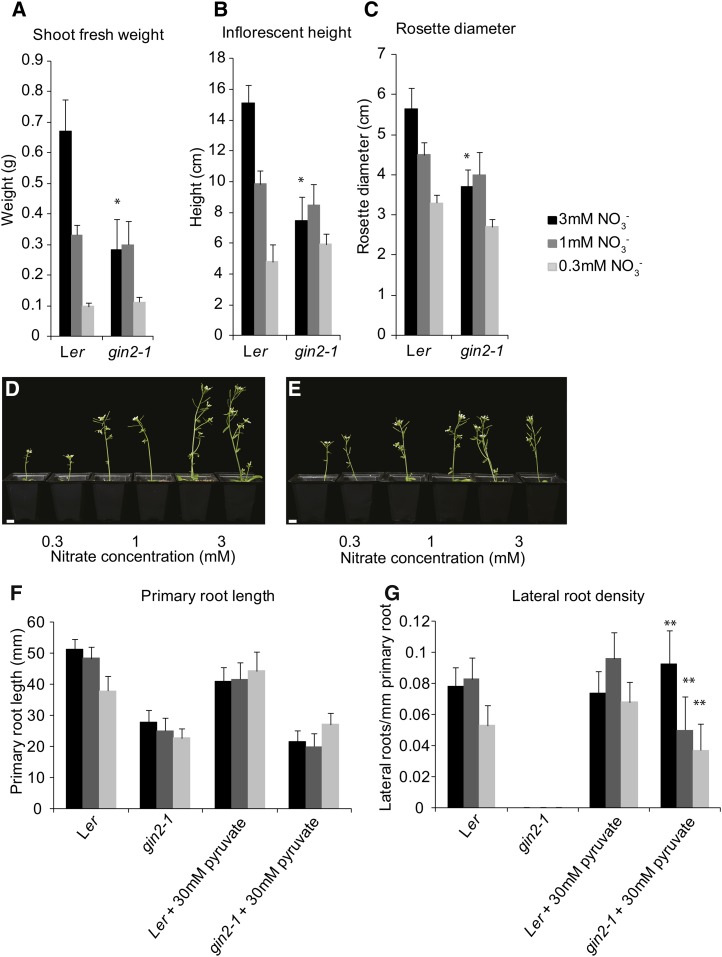
Reduced expression of several nitrate assimilatory genes in gin2-1 (Fig. 1) suggested that nitrate-responsive growth might be affected in gin2-1 plants. Growth after 5 weeks (inflorescence height, rosette diameter, and shoot fresh weight) of Ler and gin2-1 plants on perlite irrigated with 0.3, 1, and 3 mm KNO3-containing medium was measured. Figure 4, A to E, shows that Ler inflorescence height, rosette diameter, and shoot fresh weight increased progressively with increased nitrate levels. In contrast, growth in gin2-1 was not responsive to 3 mm nitrate, suggesting that nitrate assimilation and use for growth is impaired in gin2-1.
Figure 4.
Growth responses to nitrate. Ler and gin2-1 plants were grown on 0.3, 1, or 3 mm nitrate- containing growth medium (for composition, see “Materials and Methods”) for 5 weeks (A–E). A, Shoot fresh weight. B, Inflorescence height. C, Rosette diameter. The values are means of 12 plants each ± sd. *Differences between Ler and gin2-1 are significant at P < 0.05 (Student’s t test). D and E, Ler (D) and gin2-1 (E) plants photographed after 5 weeks of growth. Bars = 1 cm. F and G, Root growth after 2 weeks on agar plates containing MS-N medium and 0.1% Glc with 0.3, 1, or 3 mm nitrate and either 0 or 30 mm pyruvate. F, Primary root length. G, Lateral root density measured as the number of lateral roots per mm of primary root. The values are means of 40 plants each ± sd. **Differences between non-pyruvate- and pyruvate-treated plants are significant at P < 0.001 (Student’s t test).
We also analyzed root growth in response to nitrate levels. Figure 4F shows that primary root length is reduced and not responsive to the nitrate levels tested in gin2-1, whereas Ler primary roots were significantly longer in low and high nitrate levels. Pyruvate, which rescued NRT2.1 expression in gin2-1, had no significant influence on primary root length growth in either Ler or gin2-1 (Fig. 4F). In contrast, lateral root formation was essentially abolished in gin2-1 and was rescued to levels observed in Ler by 30 mm pyruvate (Fig. 4G). This suggested that the formation of lateral roots might be influenced by OPPP metabolites.
High-Affinity Nitrate Uptake Is Reduced in gin2-1 in High Nitrate Levels
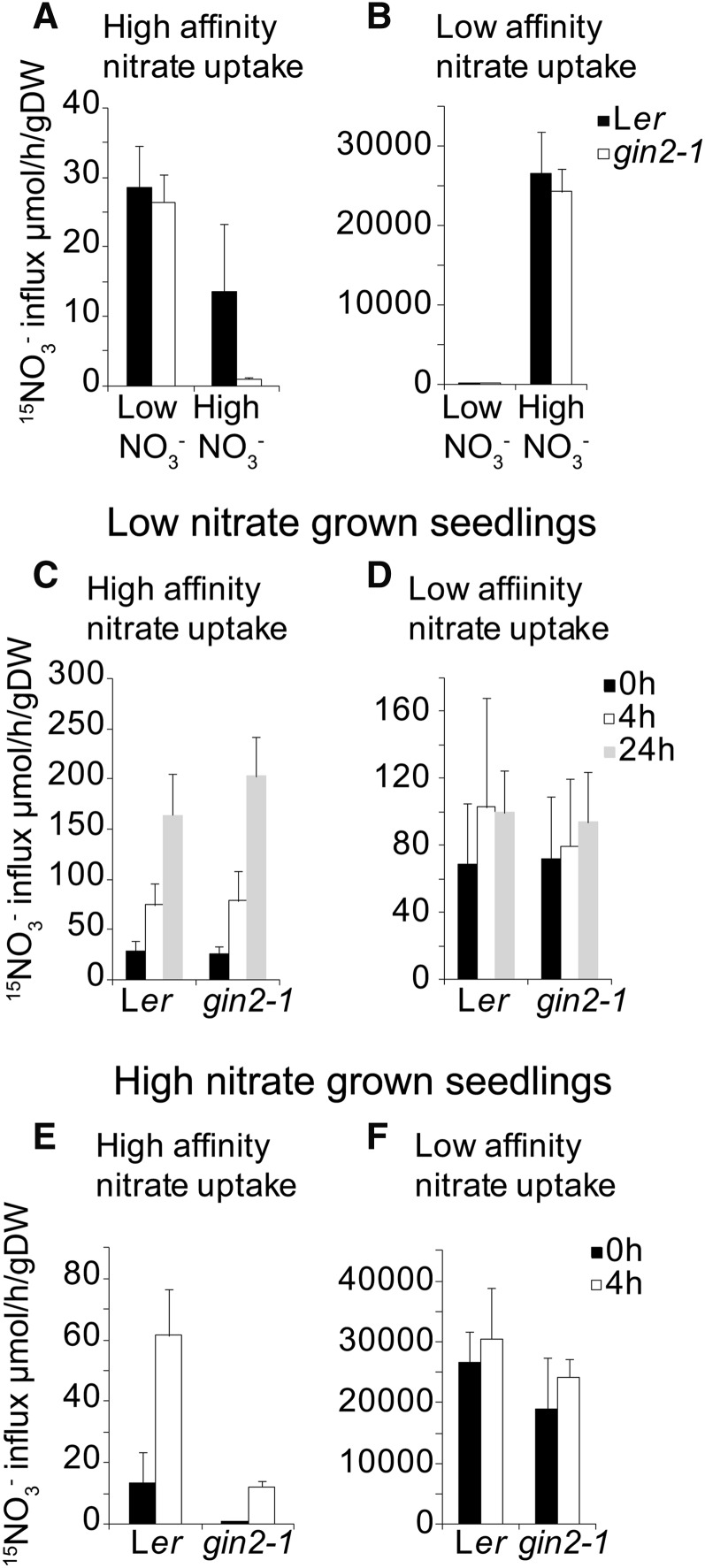
To determine the influence of reduced NRT2.1 expression in gin2-1 on nitrate uptake, both high- and low-affinity uptake were measured in response to Glc starvation and Glc treatment. The influence of Glc starvation on nitrate uptake in Ler and gin2-1 was assessed in seedlings grown for 7 d on MS medium plus 0.5% (w/v) Glc (high nitrate) or MS-N medium plus 0.5% (w/v) Glc with the addition of 0.3 mm KNO3 as the sole N source (low nitrate). Seedlings were starved of Glc for 24 h before nitrate uptake was measured. As expected (Lejay et al., 1999), Ler had reduced levels of high-affinity nitrate uptake when grown under high-nitrate conditions (Fig. 5A) and highly elevated low-affinity nitrate uptake in high-nitrate conditions (Fig. 5B). In gin2-1 seedlings grown on high nitrate, high-affinity nitrate uptake was significantly decreased compared with Ler (Fig. 5A). No significant differences were seen in low-affinity nitrate uptake in either gin2-1 or Ler in seedlings grown on high or low nitrate levels (Fig. 5B). These observations suggested that suppression of high-affinity nitrate transport by high nitrate levels is much greater in gin2-1 than in Ler, consistent with reduced expression of NRT2.1, a major high-affinity nitrate transporter, in gin2-1 (Fig. 1A). We then measured high- and low-affinity nitrate uptake activity in response to added Glc in Ler and gin2-1 seedlings grown in low-nitrate conditions. Glc induced similar rapid increases in high-affinity nitrate uptake in both Ler and gin2-1 seedlings grown on low nitrate (Fig. 5C). Glc did not influence low-affinity nitrate transport in either Ler or gin2-1 seedlings grown on low nitrate (Fig. 5D). In seedlings grown in high-nitrate conditions, Glc induced a rapid increase in high-affinity nitrate uptake in Ler (Fig. 5E), but gin2-1 seedlings had very low uptake levels. As in seedlings grown on low nitrate levels, Glc did not affect low-affinity nitrate uptake in either Ler or gin2-1 seedlings grown on high nitrate (Fig. 5F).
Figure 5.
Nitrate uptake in Ler and gin2-1 seedlings. A and B, Ler and gin2-1 seedlings (7 d old) were grown on either MS medium (high nitrate) or MS-N medium containing 0.3 mm NO3− (low nitrate) as the sole N source with 0.5% (w/v) Glc. Seedlings were starved of Glc for 24 h before nitrate uptake measurements. 15NO3− uptake was measured after 5 min in 0.2 mm 15NO3− (A; high-affinity nitrate uptake) or 6 mm 15NO3− (B; low-affinity nitrate uptake). C and D, Ler and gin2-1 seedlings (7 d old) were grown on MS-N medium containing 0.3 mm NO3− with 0.5% (w/v) Glc. Seedlings were starved of Glc for 24 h before transfer to MS-N medium containing 0.3 mm NO3− with 3% (w/v) Glc. High-affinity (C) and low-affinity (D) 15NO3− uptake was measured 0, 4, and 24 h after Glc addition as described above. E and F, Ler and gin2-1 seedlings (7 d old) were grown on MS medium with 0.5% (w/v) Glc. Seedlings were starved of Glc for 24 h before transfer to MS medium with 3% (w/v) Glc. 15NO3− uptake (high-affinity uptake [E] and low-affinity uptake [F]) was measured 0, 4, and 24 h after Glc addition as described above. All uptake measurements are means of three replicates of 10 plants each ± sd. gDW, Grams dry weight.
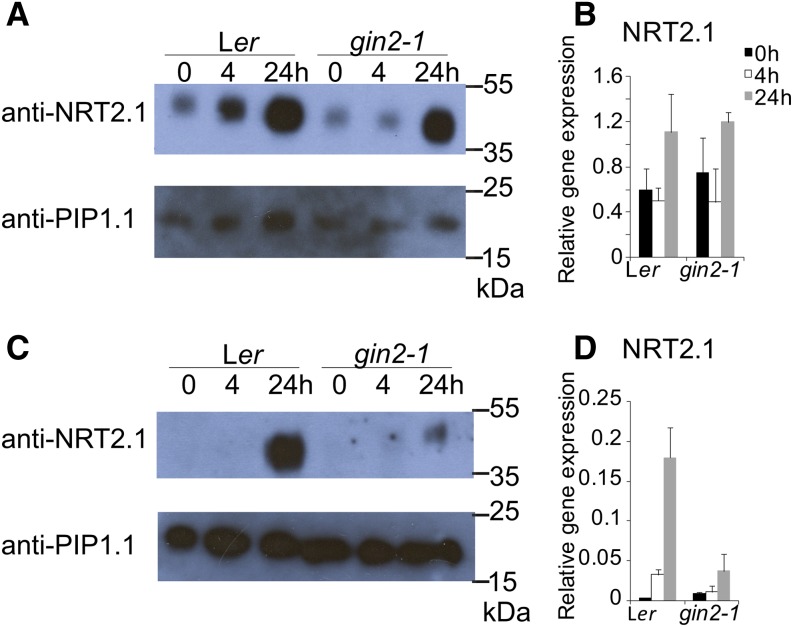
NRT2.1 protein levels were assessed in Ler and gin2-1 seedlings grown in either low- or high-nitrate conditions, together with NRT2.1 mRNA levels. Glc increased NRT2.1 protein accumulation in both Ler and gin2-1 seedlings grown in low-nitrate medium (Fig. 6A), most significantly between 4 and 24 h, consistent with increased high-affinity nitrate transport levels in response to Glc (Fig. 5C). In contrast, NRT2.1 expression was only slightly increased between 4 and 24 h (Fig. 6B). In seedlings grown in high-nitrate medium, NRT2.1 protein levels were significantly reduced in gin2-1 compared with Ler (Fig. 6C), consistent with both reduced high-affinity transport in gin2-1 in high-nitrate-grown seedlings (Fig. 5E) and reduced NRT2.1 expression (Fig. 6D). The difference between the relatively unaltered NRT2.1 expression compared with increased NRT2.1 protein accumulation in seedlings grown in low-nitrate conditions suggested that Glc may influence high-affinity nitrate uptake protein levels and transport activity posttranscriptionally, in addition to its role in stimulating NRT2.1 gene expression in high-nitrate conditions.
Figure 6.
Glc-responsive NRT2.1 protein accumulation and gene expression. Ler and gin2-1 seedlings were grown on either MS-N medium containing 0.3 mm NO3− as the sole N source (low nitrate; A and B) or MS medium (high nitrate; C and D) with 0.5% (w/v) Glc for 7 d. Seedlings were starved of Glc for 24 h before transfer to medium with 3% (w/v) Glc. Roots were harvested at the time of transfer (0h; black bars), 4 h (4h; white bars), or 24 h (24h; gray bars) after transfer. A and C show immunoblot detection of NRT2.1 and the loading control PIP1.1 in microsome extracts of 40 roots separated by 12% SDS-PAGE. B and D show NRT2.1 expression in the same root samples normalized against TUBULIN6. Values are means of three biological repeats ± se.
Accumulation of Amino-Donor Amino Acids and Ammonium in gin2-1
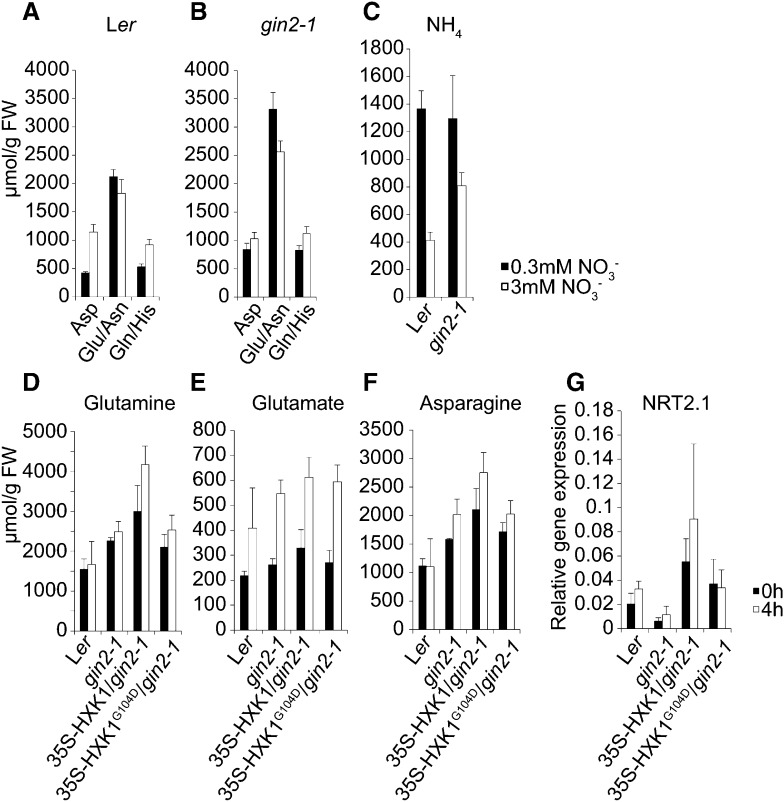
Amino acid and NH4+ levels were measured in Ler and gin2-1 seedlings grown on high- and low-nitrate medium for 7 d to assess how gin2-1 influences their accumulation. In Ler, levels of the abundant amino acids Asp and Gln/His (these amino acids were not separated by HPLC) increased in response to increased nitrate (Fig. 7A), consistent with previous studies (Nunes-Nesi et al., 2010). In gin2-1, Asp and Gln/His levels were high in low nitrate (Fig. 7B), while Glu/Asn levels were substantially higher than those observed in Ler in both low- and high-nitrate conditions. Gln/His levels remained high in gin2-1 in both high- and low-nitrate conditions, in contrast to Ler, in which levels of these amino acids were reduced in response to low nitrate (Fig. 7, A and B). The levels of other amino acids did no vary significantly in response to nitrate in Ler and gin2-1 (data not shown). NH4+ levels were twice as high in gin2-1 in plants grown on high nitrate levels (Fig. 7C) compared with Ler. Coupled to the accumulation of Asp in gin2-1 in low-nitrate conditions and increased Glu/Asn levels in gin2-1, these changes are possibly due to reduced levels of OPPP intermediates that lead to the accumulation of intermediates involved in N utilization.
Figure 7.
Amino acid and ammonium levels and NRT2.1 expression in response to nitrate and Glc. A and B, Levels of selected amino acids in 7-d-old Ler (A) and gin2-1 (B) seedlings grown on MS-N medium containing 0.3 mm NO3− (black bars) and 3 mm NO3− (white bars). C, NH3 levels in 1-week-old Ler and gin2-1 seedlings grown on MS-N medium with 0.3 mm NO3− (black bars) and 3 mm NO3− (white bars). D to G, Seven-day-old Ler and gin2-1, 35S-HXK1-GFP/gin2-1, and 35S-HXK1G104D-GFP/gin2-1 seedlings were starved of Glc for 24 h before transfer to MS medium with 3% (w/v) Glc. Seedlings were harvested either at the time of transfer (0h; black bars) or 4 h after transfer (4h; white bars). D, Gln levels. E, Glu levels. F, Asn levels. G, NRT2.1 transcript levels. The values are means of four biological repeats ± se. FW, Fresh weight.
Changes in amino acid levels were then assessed in response to Glc in gin2-1 and Ler. The increased levels of Gln and Asn seen in gin2-1 compared with Ler were not altered in response to Glc (Fig. 7, D and F). Asp and Glu levels increased in response to Glc in both Ler and gin2-1 (Fig. 7E; Supplemental Fig. S3), as did levels of Ala, Gly, and Met (Supplemental Fig. S3), while levels of Ile and Leu decreased (Supplemental Fig. S3). This showed that gin2-1 did not influence the synthesis of amino acids in response to Glc and primarily influences amino acid production in response to nitrate levels.
Pyruvate and shikimate restored Glc-mediated increases in NRT2.1 expression in gin2-1 (Fig. 1I), and Gln reversed this (Fig. 1N). This may be caused by these amino acid precursors rescuing the proposed blockage in OPPP metabolism by gin2-1. Pyruvate treatment led to its uptake (Supplemental Fig. S2H) and increases in the synthesis of Ala, a direct product of pyruvate, and Pro (Supplemental Fig. S4). Levels of Phe and Tyr, two products of shikimate, were also increased by shikimate treatment (Supplemental Fig. S4). This showed that the pyruvate and shikimate were utilized for amino acid production. However, no changes in Gln, Asn, and Glu levels in response to pyruvate or shikimate were observed (Supplemental Fig. S4). Under these conditions, the reduced levels of NRT2.1 expression in gin2-1 were fully restored (Fig. 1I; Supplemental Fig. S4). These data suggest that the rescue of Glc-mediated increases in NRT2.1 expression in gin2-1 by pyruvate and shikimate does not involve reduced levels of Gln and other amino donors, through providing intermediates for amino acid synthesis, and may involve an independent mechanism.
DISCUSSION
The mechanisms linking photosynthate availability to nitrate uptake and acquisition are centrally important for adapting growth and fitness to varying nutrient levels in the soil. Several studies (Okamoto et al., 2003; Lejay et al., 2008; Girin et al., 2010) have shown that transcriptional regulation of high-affinity nitrate transporter genes such as NRT2.1 is centrally important for optimizing plant growth in response to soil nitrate levels. In this study, we have explored relationships between carbohydrate- and nitrate-responsive gene expression, metabolism, growth, and transport using the HXK1 mutant gin2-1 that is defective in carbohydrate responses (Moore et al., 2003). We confirm and extend previous work (Lejay et al., 2003, 2008; Bussell et al., 2013) by showing that OPPP metabolism is required for Glc-mediated NRT2.1 expression and can be rescued by OPPP-derived amino acid precursor metabolites in the HXK1 mutant gin2-1. Nitrate-mediated NRT2.1 expression was not influenced by gin2-1, showing that Glc does not influence NRT2.1 expression through nitrate-mediated mechanisms. We also show that Glc stimulates NRT2.1 protein levels and transport activity independently of its HXK1-mediated stimulation of NRT2.1 expression, demonstrating two Glc-mediated mechanisms influencing nitrate uptake. The physiological significance of these mechanisms was shown by strongly reduced nitrate-responsive growth in gin2-1.
Previous studies demonstrated that inhibition of phosphogluconate dehydrogenase, the third committed step of the OPPP, reduced Suc-mediated increases of NRT2.1 expression (Lejay et al., 2008). Together with reduced NRT2.1 expression in AtHXK1 antisense lines (Lejay et al., 2003), these studies and the recent work of Bussell et al. (2013) suggested that the metabolism of sugars through the OPPP is required for NRT2.1 expression, the expression of NH4+ and sulfate transporters, and nitrate assimilatory genes NR1, NR2, and NiR. Our results using the AtHXK1 mutant gin2-1 confirmed these observations by showing large reductions in Glc-mediated increases in the expression of NRT2.1, AMT2.1, AMT2.2, and SULTR1.1 in gin2-1 (Fig. 1, A–F). The expression of seven genes encoding enzymes of the OPPP was not altered in gin2-1 (Supplemental Fig. S1), and the catalytically inactive HXK1 mutants HXK1S177A and HXK1G104D (Moore et al., 2003) did not rescue impaired Glc-responsive NRT2.1 expression in gin2-1 (Fig. 1G). However, Xyl, which feeds into the OPPP downstream of Glc-6-P (Fig. 1H), did rescue NRT2.1 expression in gin2-1. These observations suggested that disrupted OPPP metabolism due to reduced Glc-6-P production in gin2-1 leads to reduced NRT2.1 expression. The OPPP synthesizes intermediates for three major biological processes: nucleic acid biosynthesis, amino acid biosynthesis, and glycolysis (Kanehisa et al., 2012). Using intermediates destined for these pathways, respectively Rib, shikimate, and pyruvate (which is also an amino acid precursor molecule), we showed that only shikimate and pyruvate rescued NRT2.1 expression in gin2-1 (Fig. 1). This suggested that blockage of OPPP metabolites destined for amino acid synthesis contributes to reduced NRT2.1 expression and that the provision of some metabolite derived from OPPP destined for amino acid synthesis can recover reduced Glc-responsive NRT2.1 expression in gin2-1. High levels of Gln reduce NR activity and the expression of NR, NiR, and NRT2.1 genes (Miller et al., 2007; Girin et al., 2010), and we showed that exogenous Gln suppressed the restoration of NRT2.1 expression by shikimate in gin2-1 (Fig. 1N).
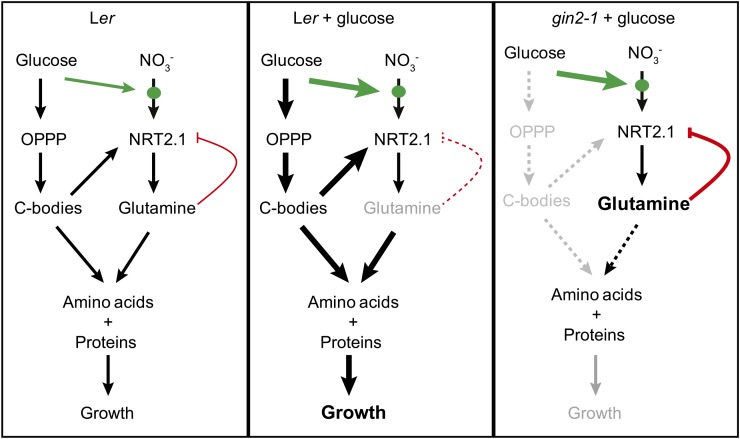
One explanation for this rescue of NRT2.1 transcription in gin2-1 by both pyruvate and shikimate involves the reduced production of amino acid precursors derived from the OPPP in gin2-1 that is rescued by providing intermediates such as pyruvate and shikimate. gin2-1 did not influence amino acid levels in response to Glc (Supplemental Fig. S4), but it did influence amino acid levels in response to nitrate. In low nitrate, Asp, Gln/His, and Glu/Asn levels were higher in gin2-1, and in high nitrate, Glu/Asn remained higher in gin2-1 in contrast to Ler (Fig. 7). In Ler, levels of these amino acids were lower in low nitrate compared with gin2-1. One possible explanation is that these N donor intermediates accumulate due to reduced amino acid synthesis and growth (Fig. 4) in gin2-1 because of the reduced supply of intermediates from the OPPP. Their buildup inhibits NRT2.1 expression, as shown by the reduction in Glc-mediated increase in NRT2.1 expression in the presence of Gln (Fig. 1N). NH4+ levels were twice as high in high nitrate in gin2-1 than in Ler, supporting this blockage hypothesis. However, although pyruvate and shikimate rescued NRT2.1 expression and increased levels of amino acids derived from them, they did not lower Asp, Gln/His, and Glu/Asn levels (Supplemental Fig. S4). The rescue of reduced Glc-responsive NRT2.1 in gin2-1 by shikimate and pyruvate, and its suppression by Gln (Fig. 1N), suggest that there may be two independent mechanisms influencing NRT2.1 expression, one involving the suppression of Glc-responsive NRT2.1 expression by Gln that may be responsible for reduced Glc-responsive NRT2.1 expression in gin2-1 and another pathway that promotes NRT2.1 expression via levels of OPPP-derived metabolites destined for amino acid synthesis. This mechanism (Figs. 3 and Fig. 8) may contribute to signaling the availability of photosynthate for nitrate uptake and assimilation for growth.
Figure 8.
Schematic summary of Glc effects on NRT2.1. Represented are, from the left, wild-type Ler in low-Glc conditions, Ler with added Glc, and gin2-1 with added Glc, all in MS medium with low nitrate levels. The black arrows describe the routes of Glc into the OPPP to produce amino acids. The route of NO3− uptake through the NRT2.1 transporter (shown by the green circle), assimilation into amino donors, and then into amino acids is also shown. The extra thickness of the arrows in the middle represents added Glc metabolism to amino acids. In gin2-1, it is proposed that this metabolism is reduced, as shown by the dotted lines. This leads to elevated levels of amino donors and increased negative feedback on NRT2.1 expression, shown by the red line. The level of this feedback is reduced in Ler due to the increased availability of carbon-bodies (C-bodies) for amino acid production. The red arrow represents the promotion of NRT2.1 expression by pyruvate, shikimate, and, possibly, other C-bodies involved in amino acid production. High levels promote NRT2.1 expression, and possible reduced levels in gin2-1 lead to reduced expression. The green arrow represents a direct influence of Glc treatment on NRT2.1 protein levels and NO3− transport activity, with higher Glc levels leading to increased protein and transport levels shown by the thicker green line.
Responses of NRT2.1 gene expression to nitrate were not affected by gin2-1, suggesting that HXK1 influences NRT2.1 expression primarily through Glc-mediated mechanisms and not through N-mediated mechanisms (Fig. 2). In contrast, Glc-mediated NRT2.1 expression was strongly influenced by external nitrate concentrations. In high nitrate concentrations, NRT2.1 expression was induced approximately 100-fold in response to Glc (Fig. 1A), compared with the 2-fold induction observed in low nitrate levels (Fig. 2D) similar to the 2-fold increase seen in gin2-1 under both high- and low-nitrate conditions. As NRT2.1 encodes a high-affinity transporter that is relatively highly expressed in response to low nitrate levels (Lejay et al., 1999), we compared absolute NRT2.1 expression levels across Glc and nitrate conditions after normalization to TUBULIN6 transcript levels (Fig. 3). This showed that, as expected, absolute NRT2.1 expression levels were much higher in low nitrate levels, peaking at 0.3 mm nitrate. Transport assays in these conditions showed that in high-nitrate conditions, Glc has no effect on low-affinity nitrate uptake but increases high-affinity uptake in Ler but not in gin2-1 (Fig. 5). This small incremental increase in nitrate uptake is unlikely to influence the reduced high-nitrate growth responses of gin2-1 measured in Figure 4. This reduced growth is consistent with recent (Bussell et al., 2013) observations that the sugar-responsive expression of several nitrate assimilation genes (NIA1, NIA2, and NiR) is reduced in the pgl3-1 mutant that also reduces OPPP flux in high-nitrate conditions. Reduced expression of AMT1.3 and SULTR1.1 transporters in gin2-1 in high-nitrate conditions (Fig. 1) may also contribute to reduced growth potential at high nitrate levels.
Our analyses provide evidence for potential posttranscriptional activation of high-affinity nitrate uptake in low-nitrate conditions by showing large, rapid (4 and 24 h), and equal increases of high-affinity nitrate uptake (Fig. 5C) and NRT2.1 protein accumulation (Fig. 6A) in Ler and gin2-1 in conditions in which NRT2.1 transcription was not significantly increased (Fig. 6B). Assuming that NRT2.1 is responsible for most high-affinity uptake, based on the very low expression levels of NRT2.2 (data not shown; Okamoto et al., 2003), these data indicate that sugar and nitrate can influence NRT2.1 expression, protein levels, and transport activity independently. Other evidence also suggests that the relationship between NRT2.1 expression and nitrate uptake is not direct (Wirth et al., 2007). Dark treatment and sugars did not influence NRT2.1 protein levels over a period of 4 h, during which NRT2.1 transcript levels were substantially reduced. Reduced NRT2.1 protein levels were only observed over a period of several days on high nitrate levels. A constitutively expressed 35S::NRT2.1 transgene in the atnrt2.1-2 null mutant has reduced high-affinity nitrate uptake in response to external ammonium nitrate, despite continued high NRT2.1 expression levels in the transgenic lines (Laugier et al., 2012). Rapid down-regulation of high-affinity nitrate uptake by dark and ammonium nitrate was not accompanied by reduced protein levels, indicating that rapidly acting posttranscriptional mechanisms control the down-regulation of NRT2.1 transport activity while protein levels remain relatively steady. Our observations are consistent with the interpretation of Laugier et al. (2012) that NRT2.1 transcriptional responses to nitrate and sugars may be long term and adaptive, while posttranslational regulation of transporter levels and activity, for example by phosphorylation (Ho et al., 2009; Wang et al., 2012a), may modulate nitrate uptake in response to rapid changes in the availability of nitrate and photosynthate.
Primary root growth in Ler was stimulated by increased nitrate levels (Fig. 4F), as shown previously (Walch-Liu et al., 2006; Walch-Liu and Forde, 2008). gin2-1 seedlings have reduced primary root length that was not significantly influenced by nitrate levels. gin2-1 seedlings also have elevated Glu/Asn and ammonium levels (Fig. 7), and primary root growth is promoted by Glu (Walch-Liu et al., 2006); therefore, the influence of gin2-1 on primary root growth is probably not through any metabolic effects on Glu levels. Lateral root formation in gin2-1 seedlings was essentially abolished at all nitrate levels (Fig. 4G), but added pyruvate fully restored gin2-1 lateral root growth to wild-type levels and had no significant effect on primary root length. Moderate levels of nitrate limitation lead to increased lateral root initiation, and nitrate starvation increases lateral root length, forming a developmental mechanism to scavenge soil nitrate (Xu et al., 2012). Loss-of-function nrt2.1 mutants display reduced lateral root initiation in response to nitrate limitation and more pronounced lateral root elongation (Remans et al., 2006). In contrast, at very high sugar concentrations (7.5% [w/v]), nrt2.1 mutants have an opposite phenotype, promoting lateral root formation, suggesting that NRT2.1 represses lateral root formation (Little et al., 2005). While our data do not reconcile these differences, which may be due to very different experimental setups and growth conditions, our experimental conditions for root analysis are more compatible with Remans et al. (2006), being conducted on relatively low Glc levels. Our data showing that gin2-1 impairs both NRT2.1 expression (Fig. 1) and lateral root formation (Fig. 4G), and the recovery of NRT2.1 expression and normal lateral root formation by pyruvate, suggest that wild-type levels of NRT2.1 expression are required for lateral root formation. This is fully consistent with observations that transfer to low nitrate levels stimulates NRT2.1 expression and lateral root formation (Remans et al., 2006).
In conclusion, we have established, with genetic, biochemical, and physiological evidence, that photosynthate availability in the form of Glc is coupled to nitrate uptake and assimilation through the metabolism of Glc by HXK1 in the OPPP, transcriptional control of NRT2.1, and posttranslational regulation of NRT2.1 protein levels and transport activities. Knowledge of these mechanisms that coordinate nitrate transport and metabolism, gene expression, and growth (Krouk et al., 2011; Bouguyon et al., 2012; Xu et al., 2012) will help improve crop nutrient acquisition using genetic analyses and transgene strategies.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seed stocks of Arabidopsis (Arabidopsis thaliana Ler) were used for all experiments. gin2-1 was obtained from Jen Sheen (Massachusetts General Hospital). Plants were stratified for 4 d at 4°C and germinated on MS medium containing 0.5% (w/v) Glc, then grown vertically for 7 d under continuous light at 22°C before placing in liquid MS medium without Glc for 24 h. To measure responses to Glc, seedlings were placed in MS medium with 3% (w/v) Glc for 4 or 24 h. To measure responses to nitrate, seedlings were placed in MS-N medium with 0, 0.3, or 1 mm nitrate for 4 h. Xyl, Rib, pyruvate, shikimate, and Gln were added when stated to a final concentration of 30 mm, and the pH of all media was adjusted to 5.8. Roots were harvested and frozen in liquid N for further analysis. For nitrate uptake and soluble amino acid analysis, seedlings were grown for 2 weeks on MS-N medium containing 0.5% (w/v) Glc supplemented with 0.3 or 1 mm KNO3 as the sole N source.
To measure root growth in response to nitrate, seedlings were grown vertically on plates containing MS-N medium with 0.1% (w/v) Glc and 0.3, 1, or 3 mm nitrate for 2 weeks with a 16-h-light/8-h-dark cycle at 20°C. Plates were then scanned at 600 dots per inch, and the root length and lateral root numbers were measured using ImageJ (Collins, 2007). The values are means ± sd for 36 to 40 plants.
Plant growth responses to nitrate were assessed in plants grown hydroponically on vermiculite (1–3 mm; Sinclair) for 5 weeks with a 16-h-light/8-h-dark cycle at 22°C. The plants were irrigated with growth medium (10 mm KH2PO4, 2 mm MgSO4, 1 mm CaCl2, 0.1 mm FeEDTA, 50 µm H3BO4, 12 µm MnSO4, 1 µm ZnCl2, 1 µm CuSO4, and 0.2 µm Na2MoO4) containing 0.3, 1, or 3 mm KNO3 as the sole N source. Inflorescence height, rosette diameter, and shoot fresh weight measurements were means ± sd for 12 to 15 plants for each condition and genotype.
Total RNA Extraction and Gene Expression Analysis
Total root RNA was extracted with the RNeasy plant mini kit from Qiagen with a DNase treatment. Gene expression was determined by quantitative real-time PCR; complementary DNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and oligo(dT)23 (Sigma) primers. PCR was performed using 40 ng of complementary DNA on a LightCycler 480 (Roche) using the LightCycler 480 SYBR Green I master kit for PCR (Roche) according to the manufacturer’s protocol. PCR conditions were as follows: 10 min at 95°C followed by 40 cycles of 10 s at 95°C, 10 s at 59°C, and 20 s at 72°C; the melting curve cycle was 5 s at 95°C, 1 min at 65°C, 0.11°C s−1 increase to 97°C, and then 10 s at 40°C. Relative transcript levels were calculated by the comparative threshold cycle method (Schmittgen and Livak, 2008). Specific primer sets were used for each tested gene (Supplemental Table S1). Expression levels were expressed as relative to AtTUB6 (At5g12250) and are means of three biological repeats each with three technical repeats ± sd.
Nitrate Uptake Measurement
Uptake of 15NO3− was assayed as described (Orsel et al., 2004). Seedlings were transferred to a 5-cm-diameter petri dish containing 0.1 mm CaSO4, with the roots in the solution and the aerial parts outside. This solution was replaced after 1 min with 0.2 or 6 mm 15NO3-−(atomic percent of 15N, 98% [w/w]) solution for 5 min. Roots were rinsed for 1 min in 0.1 mm CaSO4 before being separated, homogenized, and freeze dried overnight. Samples were analyzed using an isotope ratio mass spectrometer (model Integra CN; PDZ Europa). Influx of 15NO3− was calculated from the total 15N content of the roots (1 mg dry weight). The values are means ± sd for three replicates.
Membrane Protein Purification and Protein Immunodetection
Microsome purification was according to Laugier et al. (2012). Forty roots were harvested, snap frozen in liquid N, and ground in a Genogrinder. Tissue was then homogenized in 1 mL of buffer (50 mm Tris, pH 8.0, 500 mm Suc, 10% [v/v] glycerol, 20 mm EDTA, 20 mm EGTA, 50 mm NaF, 5 mm β-glycerophosphate, 1 mm phenanthroline, 0.6% [w/v] polyvinylpyrrolidone, 10 mm ascorbic acid adjusted to pH 8 with 1 m MES, 1 mm leupeptin, and 5 mm dithiothreitol). The homogenate was centrifuged at 2,000g at 4°C (Eppendorf 5417R) for 2 min to remove debris before the supernatant was centrifuged at 9,000g for 12 min at 4°C. The recovered supernatant was centrifuged again at 20,817g for 1 h to pellet the microsomal fraction. The pellet was resuspended in a minimal volume of conservation buffer (10 mm Tris, pH 8.0, 10 mm borate, 300 mm Suc, 9 mm KCl, and 4.2 mm leupeptin).
For western blots, proteins were separated by SDS-12% PAGE followed by electrotransfer to polyvinylidene difluoride membranes (0.45 µm; Roche). NRT2.1 was detected using the anti-NRT2.1 20 as described (Wirth et al., 2007). As a control, PLASMA MEMBRANE INTRINSIC PROTEIN1.1 was detected by using specific anti-PIP1.1 antisera (Agrisera). Immunodetection of NRT2.1 and PLASMA MEMBRANE INTRINSIC PROTEIN1.1 was done using the chemiluminescent detection system kit (SuperSignal West Femto; Pierce).
Soluble Amino Acid Analysis
Aliquots of 150 to 40 mg of seedling tissue were extracted according to Hockin et al. (2012) with modifications. Tissue was ground at −70°C in a ball mill, and 60 µL of buffer containing 20 mm HEPES (pH 7.0), 5 mm EDTA, 10 mm NaF, and 250 µL of chloroform:methanol (1.5:3.5, v/v) was added. Samples were homogenized until completely thawed and then kept on ice for 30 min. Water-soluble amino acids were extracted twice with 300 µL of water, and the aqueous phases were combined and evaporated in a rotary evaporator. The dried residue was dissolved in 100 µL of water and then filtered through an ultrafree-MC 0.22-mm filter column (Millipore). Samples were diluted 1:10, and 15 μL was derivatized using an AccQ-Tag chemistry package (Waters). For HPLC analysis, 10 μL of the final solution was injected into a Waters 2695 HPLC apparatus fitted with an AccQ-Tag column (3.9 × 150 mm) and a 474 fluorescence detector (Waters). For liquid chromatography-mass spectrometry analysis, the samples were run on an Agilent 100 liquid chromatography-mass spectrometry system equipped with a single-quad mass spectrometry detector. Amino acids were separated on a 100- × 2.1-mm, 2.6 μm Kinetex XB-C18 column (Phenomenex) using a gradient of acetonitrile in 0.1% formic acid, run at 300 μL min−1 and 25°C: 0 min, 1% acetonitrile; 2 min, 1% acetonitrile; 30 min, 25% acetonitrile; 33.5 min, 90% acetonitrile; 34.5 min, 90% acetonitrile; 35 min, 1% acetonitrile; 45 min, 1% acetonitrile. Amino acids were detected by UV A260 and by positive electrospray mass spectrometry. Full-scan data were collected from mass-to-charge ratio (m/z) 100 to 1,200, from which the amino acids were detected in extracted ion chromatograms. Spray chamber conditions were 11.5 L min−1 drying gas at 350°C, 25-pounds per square inch nebulizer pressure, and a spray voltage of 4,000 V. The instrument was set up with a ramped fragmentor voltage of 100 V between m/z 50 and 203, 130 V at m/z 273, 250 V at m/z 633, and 370 V after m/z 1,175. The values are means ± sd of four replicates.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Glc-responsive expression of OPPP genes.
Supplemental Figure S2. Nitrate transporter gene expression in response to Glc, Rib, pyruvate, and shikimate.
Supplemental Figure S3. Changes in amino acid levels in response to Glc.
Supplemental Figure S4. Amino acid levels in response to pyruvate and shikimate treatment.
Supplemental Table S1. List of genes analyzed and primer sequences for quantitative reverse transcription-PCR.
Acknowledgments
We gratefully acknowledge Dr. Tony Millar for advice on nitrate uptake measurements and Drs. Lionel Hill and Baldeep Kular for amino acid measurements.
Glossary
- OPPP
oxidative pentose phosphate pathway
- N
nitrogen
- Ler
Landsberg erecta
- MS
Murashige and Skoog
- MS-N medium
Murashige and Skoog medium without KNO3 and NH4NO3
- m/z
mass-to-charge ratio
References
- Bi YM, Wang RL, Zhu T, Rothstein SJ. (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genomics 8: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguyon E, Gojon A, Nacry P. (2012) Nitrate sensing and signaling in plants. Semin Cell Dev Biol 23: 648–654 [DOI] [PubMed] [Google Scholar]
- Bussell JD, Keech O, Fenske R, Smith SM. (2013) Requirement for the plastidial oxidative pentose phosphate pathway for nitrate assimilation in Arabidopsis. Plant J 75: 578–591 [DOI] [PubMed] [Google Scholar]
- Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutet-Mercey S, Taconnat L, Renou JP, Daniel-Vedele F, Fernandez E, et al. (2009) The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J 57: 426–435 [DOI] [PubMed] [Google Scholar]
- Collins TJ. (2007) ImageJ for microscopy. Biotechniques (Suppl) 43: 25–30 [DOI] [PubMed] [Google Scholar]
- Gansel X, Muños S, Tillard P, Gojon A. (2001) Differential regulation of the NO3− and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J 26: 143–155 [DOI] [PubMed] [Google Scholar]
- Girin T, El-Kafafi S, Widiez T, Erban A, Hubberten HM, Kopka J, Hoefgen R, Gojon A, Lepetit M. (2010) Identification of Arabidopsis mutants impaired in the systemic regulation of root nitrate uptake by the nitrogen status of the plant. Plant Physiol 153: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A, Krouk G, Perrine-Walker F, Laugier E. (2011) Nitrate transceptor(s) in plants. J Exp Bot 62: 2299–2308 [DOI] [PubMed] [Google Scholar]
- Guo FQ, Wang R, Crawford NM. (2002) The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is regulated by auxin in both shoots and roots. J Exp Bot 53: 835–844 [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. (2007) Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol 8: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. (2009) CHL1 functions as a nitrate sensor in plants. Cell 138: 1184–1194 [DOI] [PubMed] [Google Scholar]
- Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G. (2012) The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol 158: 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, Orsel M, Bréhaut V, Miller A, Daniel-Vedele F, Sakakibara H, et al. (2012) The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell 24: 245–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, Berthomé R, Orsel M, Mercey-Boutet S, Yu A, Castaings L, Elftieh S, Major H, Renou JP, Daniel-Vedele F. (2011) Arabidopsis roots and shoots show distinct temporal adaptation patterns toward nitrogen starvation. Plant Physiol 157: 1255–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, Mounier E, Hoyerova K, Tillard P, Leon S, Ljung K, et al. (2010a) Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. (2010b) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B. (2011) A framework integrating plant growth with hormones and nutrients. Trends Plant Sci 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Kruger NJ, von Schaewen A. (2003) The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 6: 236–246 [DOI] [PubMed] [Google Scholar]
- Laugier E, Bouguyon E, Mauriès A, Tillard P, Gojon A, Lejay L. (2012) Regulation of high-affinity nitrate uptake in roots of Arabidopsis depends predominantly on posttranscriptional control of the NRT2.1/NAR2.1 transport system. Plant Physiol 158: 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Müller C, Krapp A, von Wirén N, Daniel-Vedele F, Gojon A. (2003) Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15: 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L, Tillard P, Lepetit M, Olive F, Filleur S, Daniel-Vedele F, Gojon A. (1999) Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J 18: 509–519 [DOI] [PubMed] [Google Scholar]
- Lejay L, Wirth J, Pervent M, Cross JMF, Tillard P, Gojon A. (2008) Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol 146: 2036–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang Y, Okamoto M, Crawford NM, Siddiqi MY, Glass ADM. (2007) Dissection of the AtNRT2.1:AtNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol 143: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S, Smith C, Hadingham S, Sorefan K, Cawley G, Bevan MW. (2006) Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res 16: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, Malamy JE. (2005) The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci USA 102: 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A. (2013) Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun 4: 1713. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. (2007) Nitrate transport and signalling. J Exp Bot 58: 2297–2306 [DOI] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng W-H, Liu YX, Hwang I, Jones T, Sheen J. (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300: 332–336 [DOI] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. (2004) Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell 16: 2433–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nero D, Krouk G, Tranchina D, Coruzzi GM. (2009) A system biology approach highlights a hormonal enhancer effect on regulation of genes in a nitrate responsive “biomodule.” BMC Syst Biol 3: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes-Nesi A, Fernie AR, Stitt M. (2010) Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Vidmar JJ, Glass ADM. (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44: 304–317 [DOI] [PubMed] [Google Scholar]
- Orsel M, Eulenburg K, Krapp A, Daniel-Vedele FO. (2004) Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219: 714–721 [DOI] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. (2006) A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol 140: 909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robaglia C, Thomas M, Meyer C. (2012) Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr Opin Plant Biol 15: 301–307 [DOI] [PubMed] [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 21: 3567–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Schofield RA, Bi YM, Kant S, Rothstein SJ. (2009) Over-expression of STP13, a hexose transporter, improves plant growth and nitrogen use in Arabidopsis thaliana seedlings. Plant Cell Environ 32: 271–285 [DOI] [PubMed] [Google Scholar]
- Stitt M. (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Forde BG. (2008) Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises L-glutamate-induced changes in root architecture. Plant J 54: 820–828 [DOI] [PubMed] [Google Scholar]
- Walch-Liu P, Liu LH, Remans T, Tester M, Forde BG. (2006) Evidence that L-glutamate can act as an exogenous signal to modulate root growth and branching in Arabidopsis thaliana. Plant Cell Physiol 47: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Zou H, Sun SSM, He JX. (2012a) A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. J Proteome Res 11: 2301–2315 [DOI] [PubMed] [Google Scholar]
- Wang YY, Hsu PK, Tsay YF. (2012b) Uptake, allocation and signaling of nitrate. Trends Plant Sci 17: 458–467 [DOI] [PubMed] [Google Scholar]
- Widiez T, El Kafafi ES, Girin T, Berr A, Ruffel S, Krouk G, Vayssières A, Shen WH, Coruzzi GM, Gojon A, et al (2011) High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3− uptake is associated with changes in histone methylation. Proc Natl Acad Sci USA 108: 13329–13334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth J, Chopin F, Santoni V, Viennois G, Tillard P, Krapp A, Lejay L, Daniel-Vedele F, Gojon A. (2007) Regulation of root nitrate uptake at the NRT2.1 protein level in Arabidopsis thaliana. J Biol Chem 282: 23541–23552 [DOI] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63: 153–182 [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T. (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101: 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG. (1998) An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zhang N, Gibon Y, Gur A, Chen C, Lepak N, Höhne M, Zhang Z, Kroon D, Tschoep H, Stitt M, et al. (2010) Fine quantitative trait loci mapping of carbon and nitrogen metabolism enzyme activities and seedling biomass in the maize IBM mapping population. Plant Physiol 154: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]