Fungal pectin-degrading enzymes act as microbe-associated molecular patterns that are recognized by a pattern recognition receptor from Arabidopsis.
Abstract
Plants perceive microbial invaders using pattern recognition receptors that recognize microbe-associated molecular patterns. In this study, we identified RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1 (RBPG1), an Arabidopsis (Arabidopsis thaliana) leucine-rich repeat receptor-like protein, AtRLP42, that recognizes fungal endopolygalacturonases (PGs) and acts as a novel microbe-associated molecular pattern receptor. RBPG1 recognizes several PGs from the plant pathogen Botrytis cinerea as well as one from the saprotroph Aspergillus niger. Infiltration of B. cinerea PGs into Arabidopsis accession Columbia induced a necrotic response, whereas accession Brno (Br-0) showed no symptoms. A map-based cloning strategy, combined with comparative and functional genomics, led to the identification of the Columbia RBPG1 gene and showed that this gene is essential for the responsiveness of Arabidopsis to the PGs. Transformation of RBPG1 into accession Br-0 resulted in a gain of PG responsiveness. Transgenic Br-0 plants expressing RBPG1 were equally susceptible as the recipient Br-0 to the necrotroph B. cinerea and to the biotroph Hyaloperonospora arabidopsidis. Pretreating leaves of the transgenic plants with a PG resulted in increased resistance to H. arabidopsidis. Coimmunoprecipitation experiments demonstrated that RBPG1 and PG form a complex in Nicotiana benthamiana, which also involves the Arabidopsis leucine-rich repeat receptor-like protein SOBIR1 (for SUPPRESSOR OF BIR1). sobir1 mutant plants did not induce necrosis in response to PGs and were compromised in PG-induced resistance to H. arabidopsidis.
Microbe-associated molecular patterns (MAMPs) are molecular signatures of entire groups of microbes and have key roles in activation of the defense response in plants (Jones and Dangl, 2006; Boller and Felix, 2009). Well-characterized proteinaceous MAMPs are bacterial flagellin, Elongation Factor Tu (EF-Tu), and Ax21, fungal xylanase, and oomycete pep13, an epitope of a secreted transglutaminase (Boller and Felix, 2009; Monaghan and Zipfel, 2012). Plants recognize MAMPs by means of pattern recognition receptors (PRRs), comprising a group of leucine-rich repeat (LRR) receptor-like kinases (RLKs) or LRR receptor-like proteins (RLPs) located in the plasma membrane (Greeff et al., 2012; Monaghan and Zipfel, 2012). The LRR-RLKs FLS2 and EFR recognize flg22 (the 22-amino acid eliciting epitope from the conserved flagellin domain) and elf18/elf26 (peptides derived from the N terminus of translation elongation factor EF-Tu), respectively (Gómez-Gómez and Boller, 2000; Kunze et al., 2004; Chinchilla et al., 2006; Zipfel et al., 2006). The fungal protein ethylene-inducing xylanase (EIX) is recognized by the tomato (Solanum lycopersicum) LRR-RLPs LeEIX1 and LeEIX2, of which only the latter mediates a necrotic response (Ron and Avni, 2004).
BRI1-ASSOCIATED KINASE1/SOMATIC EMBRYOGENESIS RECEPTOR KINASE3 (BAK1/SERK3) is an LRR-RLK acting as a common component in many RLK signaling complexes (Monaghan and Zipfel, 2012). Although it was originally identified as a protein that interacts with the brassinosteroid receptor BRI1 (Li et al., 2002; Nam and Li, 2002), BAK1 also forms ligand-induced complexes with FLS2 and EFR and contributes to disease resistance against the pathogens Pseudomonas syringae, Hyaloperonospora arabidopsidis (Hpa), and Phytophthora infestans (Chinchilla et al., 2007; Heese et al., 2007; Chaparro-Garcia et al., 2011; Roux et al., 2011). Tomato BAK1 interacts in a ligand-independent manner with LeEIX1 but not with LeEIX2, and the BAK1-LeEIX1 interaction is required for the ability of LeEIX1 to attenuate the signaling of LeEIX2 (Bar et al., 2010). BAK1 has also been shown to interact with another LRR-RLK, BAK1-INTERACTING RECEPTOR-LIKE KINASE1 (BIR1). A bir1 mutant showed extensive cell death, activation of constitutive defense responses, and impairment in the activation of the mitogen-activated protein kinase MPK4 (Gao et al., 2009). sobir1 (for suppressor of BIR1) mutants suppress BIR1 phenotypes, and overexpression of SOBIR1 triggers cell death and defense responses (Gao et al., 2009). SOBIR1 does not physically interact with BIR1, suggesting that SOBIR1 mediates an alternative signal transduction route. A recent study showed that the tomato SOBIR1 interacts with RLPs and is required for RLP-mediated disease resistance (Liebrand et al., 2013).
Endopolygalacturonases (PGs) are a class of pectinases that hydrolyze the homogalacturonan domain of pectic polysaccharides (van den Brink and de Vries, 2011). Secreted PGs are able to cause cell wall decomposition and tissue maceration and thereby act as virulence factors in several fungal pathogens, such as Aspergillus flavus, Claviceps purpurea, and Alternaria citri (Shieh et al., 1997; Isshiki et al., 2001; Oeser et al., 2002). The most extensively studied PGs from fungal plant pathogens are those of Botrytis cinerea (for review, see Zhang and van Kan, 2013b), a necrotrophic broad-host-range pathogen that contains six PG genes (designated Bcpg1–Bcpg6) in its genome (Wubben et al., 1999). Deletion of either Bcpg1 or Bcpg2 resulted in a strong reduction in virulence on tomato and broad bean (Vicia faba) leaves (ten Have et al., 1998; Kars et al., 2005), presumably because the enzymes have a detrimental effect on the integrity of host cell walls and tissues. Indeed, infiltrating BcPG2 into broad bean leaves or transient expression of BcPG2 in Nicotiana benthamiana led to tissue collapse and necrosis, and the necrotic response was abolished when the catalytic domain of the PG was mutated (Kars et al., 2005; Joubert et al., 2007). By contrast, Poinssot et al. (2003) reported that BcPG1 can activate plant defense responses in grapevine (Vitis vinifera) cell suspensions independently of its enzymatic activity, suggesting that the protein itself might be recognized by plant cells as an elicitor. These studies with seemingly opposing conclusions were conducted with different isozymes on distinct cell types of different plant species. Thus, it remained inconclusive whether plant responses observed after exposure to PGs are due to the structural damage resulting from pectin hydrolysis or to recognition of the protein as a MAMP (followed by downstream signaling responses).
The degradation of pectin by PGs leads to the release of oligogalacturonides (OGAs), which may activate a variety of defense responses (Prade et al., 1999; D’Ovidio et al., 2004). OGAs act as damage-associated molecular patterns (DAMPs) via their perception by the cell wall-associated receptor Wall-Associated Kinase1 (WAK1; Brutus et al., 2010). Overexpression of WAK1 in Arabidopsis (Arabidopsis thaliana) enhances resistance to B. cinerea (Brutus et al., 2010).
Here, we describe the occurrence of natural variation among Arabidopsis accessions in responsiveness to fungal PGs. Two accessions that strongly differed in their response to PGs were selected for further analysis. Cloning and functional characterization demonstrated that the gene RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1 (RBPG1) encodes an LRR-RLP. Finally, we demonstrate that the LRR-RLK SUPPRESSOR OF BIR1 (SOBIR1) interacts with RBPG1 and is essential for responsiveness to fungal PGs.
RESULTS
Map-Based Cloning of RBPG1
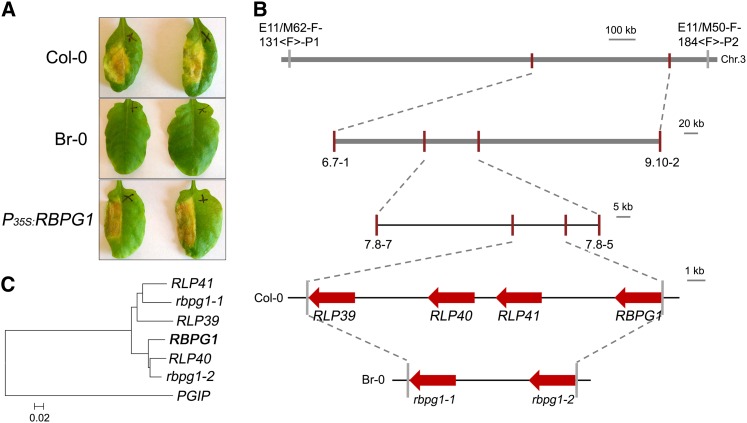
In order to study the response of Arabidopsis to B. cinerea PGs, leaves of 47 accessions (Supplemental Table S1) were infiltrated with pure BcPG2, BcPG3, BcPG4, and BcPG6 (Kars et al., 2005). Considerable variation in responses was observed among these accessions, ranging from no visible symptoms to full necrosis of the infiltrated area. Eleven of the 47 accessions, representing the spectrum of phenotypic variation, were selected for further study (Supplemental Table S1). The responses were scored in five classes ranging from 0 (no visible symptom) to 4 (full necrosis of the infiltrated area; Supplemental Fig. S1A). The accessions Columbia (Col-0), Kashmir-1 (Kas-1), and Kas-2 showed the most severe symptoms in response to BcPGs, whereas the accessions Brno (Br-0) and Estland (Est-0) showed no symptoms (Fig. 1A; Supplemental Fig. S1B). Plants from accessions Col-0 and Br-0 were crossed, and F1 progeny were all responsive to BcPGs, indicating that responsiveness is a dominant trait. F1 plants were selfed to obtain a segregating F2 population (n > 350). A total of 183 F2 progeny were tested for their responsiveness to BcPGs in a quantitative manner and used for amplified fragment-length polymorphism (AFLP) analysis. Quantitative trait locus (QTL) mapping identified a single locus governing the responsiveness to all BcPGs tested (Supplemental Fig. S2). The QTL is designated RBPG1 and is positioned on chromosome 3 at a distance of approximately 10 centimorgan (cM) from the GLABROUS1 (gl1) locus (Hauser et al., 2001), which also segregated in this cross. The primary mapping showed that the RBPG1 locus is in a 1.6-Mb region between AFLP markers E11/M62-F-131<F>-P1 and E11/M50-F-184-<F>-P2 (Fig. 1B; Supplemental Fig. S2A). Additional single-nucleotide polymorphism (SNP) markers were designed in this region based on sequence polymorphisms between Col-0 and Br-0 (http://signal.salk.edu/atg1001/3.0/gebrowser.php). Further mapping was performed with F8 recombinant inbred lines (RILs), obtained by single-seed descent of the F2 population (n = 310), and placed the RBPG1 locus in a smaller region of approximately 500 kb between SNP markers 6.7-1 and 9.10-2 (Fig. 1B). To further narrow the interval, fine-mapping of the RBPG1 locus was performed on a backcross F2 population of Phytoalexin deficient3 (pad3, a camalexin biosynthesis-deficient mutant in Col-0; Zhou et al., 1999) × BC41 (an F8 RIL containing rbpg1 as well as the gl1 mutation). The RBPG1 locus could be pinpointed to a region of approximately 85 kb delimited by SNP markers 7.8-7 and 7.8-5 (Fig. 1B), which contains 21 candidate genes (Supplemental Table S2).
Figure 1.
Map-based cloning of RBPG1. A, Response to BcPG3 of Arabidopsis accessions Col-0 and Br-0 and Br-0 transgenic plants expressing P35S:RBPG1 (1.5 µm). Leaves were photographed 7 d after infiltration with BcPG3. B, Map of the RBPG1 locus. C, Phylogenetic tree of RBPG1 homologs in Col-0 and Br-0. The amino acid sequences were aligned by ClustalX 1.83, and the phylogenetic tree was generated by using MEGA4 by the neighbor-joining method with 1,000 bootstrap replicates.
As responsiveness to BcPGs is a dominant trait, a homozygous RBPG1 transfer DNA (T-DNA) mutant in the Col-0 background would be expected to be unresponsive to BcPGs. To find out which of the 21 candidate genes is RBPG1, the available T-DNA insertion mutants of 17 of these candidate genes (in a Col-0 background) were investigated (Supplemental Table S2). Homozygosity of the T-DNA mutants was confirmed by PCR, and the mutants were phenotyped by infiltrating with BcPG3. All of the T-DNA mutants tested were equally responsive to BcPG3 as Col-0. According to these results, the 17 genes of which the homozygous T-DNA mutants were responsive to BcPG3 could be eliminated as candidates. No T-DNA mutants were available for the genes At3g24770, At3g24890, and At3g25020, whereas the T-DNA mutant in the At3g24800 gene could not be made homozygous, possibly due to lethality. Therefore, these four genes remained candidates.
The genome sequences of several Arabidopsis accessions were released by the Arabidopsis 1001 Genomes project (Cao et al., 2011), including Br-0, Eden-2, Est-1, and Gy-0. Among these accessions, Eden-2 was equally unresponsive to BcPG3 as Br-0, while Est-1 and Gy-0 were equally responsive to BcPG3 as Col-0. The different responsiveness of Arabidopsis accessions to BcPGs could be due to amino acid substitutions in RBPG1. Thus, SNPs in the 21 candidate genes were compared between accessions in order to identify SNPs that are associated with the responsive phenotype. In the 21 genes in the region, there were SNPs in four genes (from At3g24770 to At3g24860) that result in amino acid substitutions between Col-0 and Br-0. However, none of these substitutions was specifically associated with the phenotype in accessions Eden-2, Est-1, and Gy-0 (http://signal.salk.edu/atg1001/3.0/gebrowser.php). Surprisingly, the sequences in the RBPG1 locus downstream of At3g24890 were very poorly represented in accessions Br-0, Eden-2, and Gy-0 and also in many other accessions (http://signal.salk.edu/atg1001/3.0/gebrowser.php); especially, the region that comprises four RLP paralogs in Col-0 is barely covered by mappable sequence reads in many other accessions. In combination with the results of T-DNA mutant analysis, At3g24890 and At3g25020, therefore, were considered to be the remaining candidates for being the RBPG1 gene (Supplemental Table S2).
To determine the function of At3g24890 and At3g25020, both genes (under the control of the 35S promoter) were transformed into the BcPG-unresponsive accession Br-0. Transgenic plants expressing At3g25020 displayed the BcPG-responsive phenotype (Fig. 1A), whereas transgenic plants containing At3g24890 constructs remained equally unresponsive to BcPG3 as the recipient accession Br-0 (Supplemental Fig. S3). The At3g25020 gene encodes an LRR-RLP and is one of a family of four RLP-encoding genes that occur in tandem in a cluster within this region of the Col-0 genome (Fig. 1B). The four genes are At3g24900, At3g24982, At3g25010, and At3g25020 and correspond to the genes RLP39 to RLP42 according to the classification of Wang et al. (2008). Genome reassembly of raw Br-0 sequence data and resequencing of amplicons spanning the assembled region showed that this region in accession Br-0 contains only two RLP-encoding genes, which we designated rbpg1-1 and rbpg1-2 (Fig. 1B), with 90% nucleotide identity and 84% amino acid identity to each other (Supplemental Fig. S4; the sequence of the region covering the Br-0 rbpg1 locus is deposited in GenBank with accession no. KF684938). Phylogenetic analysis shows that RLP39 and RLP41 cluster with rbpg1-1, whereas RLP40 and RLP42 cluster with rbpg1-2 (Fig. 1C). To determine whether the three Col-0 paralogs, RLP39, RLP40, and RLP41, can also confer responsiveness to BcPGs, these genes (under the control of the 35S promoter) were transformed into BC41 (an F8 RIL of Col-0 × Br-0, unresponsive to BcPG3). None of the transgenic plants carrying RLP39, RLP40, or RLP41 showed a BcPG-responsive phenotype (Supplemental Fig. S5). These data demonstrate that RLP42 is the only gene in this region that confers responsiveness to BcPG3; hereafter, it is referred to as the RBPG1 gene.
The C-Terminal Intracellular Domain of RBPG1 Is Not Required for Responsiveness
One striking difference between the protein sequences of RBPG1, on the one hand, and the three homologs from accession Col-0 (RLP39–RLP41) and two homologs from accession Br-0 (rbpg1-1 and rbpg1-2), on the other hand, is the presence of an extended intracellular, C-terminal domain of nine amino acids in RBPG1 (Supplemental Figs. S4 and S6A). To investigate whether this domain is required for responsiveness, two constructs were generated in which the C-terminal tail of RBPG1 is truncated: RBPG1_Trunc1 lacking the C-terminal 18 amino acids and RBPG1_Trunc2 lacking the C-terminal 10 amino acids. Also, a construct was generated in which the C-terminal residue of rbpg1-2 is replaced with the C-terminal 10 residues of RBPG1 (rbpg1-2_Swap1). These three gene constructs (under the control of the 35S promoter) were transformed into BC41 (an F8 RIL of Col-0 × Br-0, unresponsive to BcPG3). RBPG1_Trunc1 and RBPG1_Trunc2 conferred responsiveness to BcPG3, similar to that of the full-length RBPG1 construct, whereas recipient line BC41 and the plants that were transformed with rbpg1-2_Swap1 constructs were unresponsive to BcPG3 (Supplemental Fig. S6B). These results show that the C-terminal intracellular domain of RBPG1 is not required for the responsiveness.
RBPG1 Recognizes Multiple Active and Inactive PGs But Not a Conserved Peptide
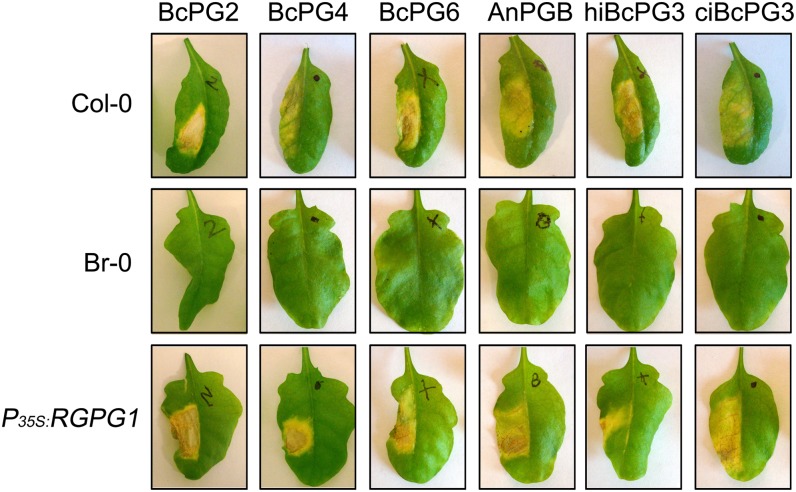
Plants that are responsive to BcPG3 (Col-0 and transgenic plants expressing RBPG1) also showed necrotic symptoms upon infiltration with B. cinerea proteins BcPG2, BcPG4, and BcPG6 as well as AnPGB from the saprotrophic fungus Aspergillus niger (Fig. 2). The minimal concentration required to consistently induce necrosis on responsive plants ranged from 0.1 µm (BcPG2) to 3 µm (BcPG4). In screening for the presence of the RBPG1 gene, BcPG3 was routinely used at a concentration of 1.5 µm. At higher protein concentrations, BcPG2 and AnPGB caused water-soaked symptoms, reflecting the occurrence of pectin degradation and maceration (as opposed to necrosis), because of the high specific enzyme activity of these two proteins, as compared with BcPG3, BcPG4, and BcPG6 (data not shown).
Figure 2.
Response to fungal PGs of Arabidopsis accessions Col-0, Br-0 and Br-0 transgenic plants expressing RBPG1. Col-0 and P35S:RBPG1 transgenic plants are responsive to BcPG2 (0.1 µm), BcPG4 (3 µm), BcPG6 (1 µm), AnPGB (0.3 µm), hiBcPG3 (1.5 µm), and ciBcPG3 (1.5 µm). Leaves were photographed 7 d after infiltration.
It is known that cell wall-derived OGAs may serve as DAMPs (D’Ovidio et al., 2004; Boller and Felix, 2009; Hématy et al., 2009; Rasul et al., 2012). To distinguish whether the RBPG1-dependent responsiveness to BcPG3 is through the recognition of the protein itself (acting as a MAMP) or through the recognition of OGAs released in planta (acting as DAMPs), heat-inactivated BcPG3 (hiBcPG3) and catalytically inactive BcPG3D353E/D354N (ciBcPG3) were infiltrated into leaves of Col-0, Br-0, and Br-0 transgenic plants expressing RBPG1. Both hiBcPG3 and ciBcPG3 proteins induced necrosis in Col-0 and in P35S:RBPG1 transgenic plants but not in Br-0 (Fig. 2). Since multiple PGs were able to induce necrosis, we considered the possibility that a peptide motif that is conserved among fungal PGs might act as an epitope that is recognized by RBPG1. In the case of flagellin and EF-Tu, conserved short peptide motifs are sufficient to trigger similar responses as the proteins that they are derived from (Felix et al., 1999; Brunner et al., 2002; Kunze et al., 2004). Amino acid sequence alignment of several fungal PGs and three Arabidopsis PGs (Ogawa et al., 2009) showed that an 11-amino acid stretch, just downstream of the catalytic site, is highly conserved among fungal PGs (Supplemental Fig. S7); the homologous region in Arabidopsis PGs contains a Gly-to-Pro substitution. Other conserved peptide stretches in fungal PGs were seven residues (the catalytic site) or shorter than four residues in length (data not shown). A synthetic 22-amino acid peptide corresponding to BcPG3 residues 367 to 388 (with the 11-amino acid peptide stretch in the middle; Supplemental Fig. S7) was infiltrated in leaves of Col-0 and Br-0. In concentrations ranging from 10 µm to 1 mm (100-fold higher than normally used for the BcPG3 protein), the peptide did not induce any symptoms until 7 d after infiltration.
In order to examine whether the Gly-to-Pro substitution at the beginning of the conserved peptide stretch (as present in Arabidopsis PGs) would affect the necrosis-inducing activity of fungal PGs, due to its presumed impact on protein folding, we produced a mutant BcPG3G373P protein in Pichia pastoris. The mutant BcPG3G373P protein induced necrosis in Col-0 and in P35S:RBPG1 transgenic plants (Supplemental Fig. S8).
RBPG1 Does Not Confer Responsiveness to OGAs Acting as DAMPs
In order to further explore whether the response to PGs was due to recognition of the protein as a MAMP or to the recognition of pectin breakdown products as a DAMP, the alcohol-insoluble residue (AIR) fraction, consisting mainly of cell wall polysaccharides, was extracted from leaves of Col-0 and Br-0 and hydrolyzed with active BcPG3. The solubilized cell wall fragments released from the AIR did not induce any visible symptom in either Col-0 or Br-0. In addition, a set of distinct, partially purified OGAs with a degree of polymerization ranging from 3 to 20, at concentrations ranging from 10 µm up to 1 mm, did not induce any visible symptom upon infiltration into leaves of Col-0 or Br-0 (data not shown). Collectively, the above data lead to the conclusion that the RBPG1-mediated necrotic response to PG is not due to DAMP recognition but results from the recognition by RBGP1 of PG proteins as MAMPs.
The Role of RBPG1 in Immunity in Response to BcPG3
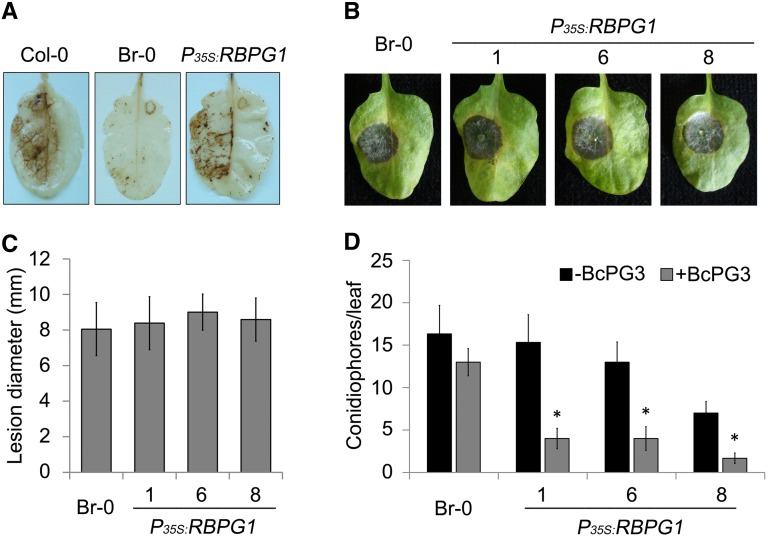
MAMP recognition leads to a set of signaling events and activates basal defense in plants (Jones and Dangl, 2006). RBPG1-mediated responsiveness to BcPG3 was accompanied by the induction of the formation of reactive oxygen species (ROS) 3 d after infiltration (Fig. 3A). To examine whether RBPG1 contributes, either negatively or positively, to B. cinerea disease development, P35S:RBPG1 T3 transgenic Br-0 lines (homozygous T2 lines with single-copy integration) and recipient accession Br-0 were inoculated with B. cinerea. Lesion development on transgenic and recipient lines was indistinguishable over the entire course of the experiment (Fig. 3, B and C). To study whether RBPG1 modulates resistance to other pathogens, unrelated to B. cinerea, three independent P35S:RBPG1 transgenic Br-0 lines and recipient Br-0 were inoculated with Hpa isolate Maks9. No consistent difference was observed between transgenic and recipient plants in susceptibility to Hpa (Fig. 3D). When transgenic and recipient plants were pretreated with hiBcPG3 and 1 d later inoculated with Hpa isolate Maks9, all three hiBcPG3-treated transgenic lines displayed a significant increase in resistance to Hpa as compared with the mock-treated transgenic plants, while the hiBcPG3-treated recipient Br-0 was equally susceptible to Hpa (Fig. 3D).
Figure 3.
Role of RBPG1 in immunity in response to BcPG3. A, ROS accumulation in Arabidopsis leaves 4 d after infiltration of BcPG3 (1.5 µm). B, Disease symptoms of B. cinerea on Arabidopsis leaves at 3 d dpi. One representative leaf is shown. C, Lesion diameter caused by B. cinerea at 3 dpi on Arabidopsis leaves. Data represent means ± sd (n ≥ 15). D, Br-0 and P35S:RBPG1 transgenic plants were infiltrated with water or BcPG3 and 24 h later inoculated with Hpa isolate Maks9. Conidiophores were counted at 10 dpi. Results are averages ± se (n = 15). *P < 0.01 by Student’s t test. All experiments were repeated at least three times with similar results.
RBPG1 Interacts with BcPG3
All previous results pointed to the recognition of PG proteins as MAMPs by the RLP RBPG1. In order to study whether recognition was due to direct interaction between both proteins, immunoprecipitation analysis was performed upon transient coexpression of 10xMyc-tagged BcPG3 and RBPG1-GFP in N. benthamiana. Two days after agroinfiltration, RBPG1-GFP proteins were immunoprecipitated using GFP affinity beads, and purified proteins were analyzed by western blot using antibodies against GFP and Myc (Fig. 4). BcPG3 coimmunoprecipitated with RBPG1-GFP but not with the GFP beads alone. GFP-tagged EFR was used as a negative control to exclude that BcPG3 interacts nonspecifically with extracellular LRR domain-containing proteins. To further confirm the interaction between BcPG3 and RBPG1, we swapped the epitope tags and coexpressed 10xMyc-tagged RBPG1 and GFP-tagged BcPG3 in N. benthamiana. BcPG3-GFP was equally capable of immunoprecipitating RBPG1-myc (Fig. 4), corroborating that BcPG3 directly interacts with RBPG1.
Figure 4.
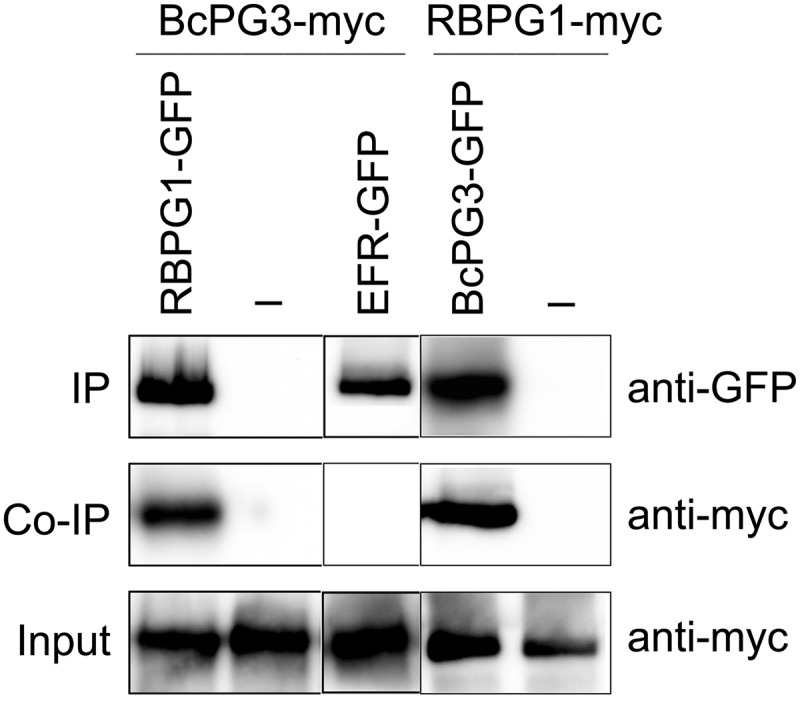
BcPG3 forms a complex with RBPG1 and its homologs in N. benthamiana. Coimmunoprecipitation (Co-IP) of BcPG3 and RBPG1 or EFR is shown. Total proteins expressed in N. benthamiana leaves were subjected to immunoprecipitation (IP) with GFP Trap beads followed by immunoblot analysis with anti-myc antibodies to detect BcPG3-myc and RBPG1-myc as well as anti-GFP antibodies to detect RBPG1-GFP, EFR-GFP, and BcPG3-GFP. All experiments were repeated at least three times with similar results.
SOBIR1 Is Required for the RBPG1-Dependent Response to BcPG3
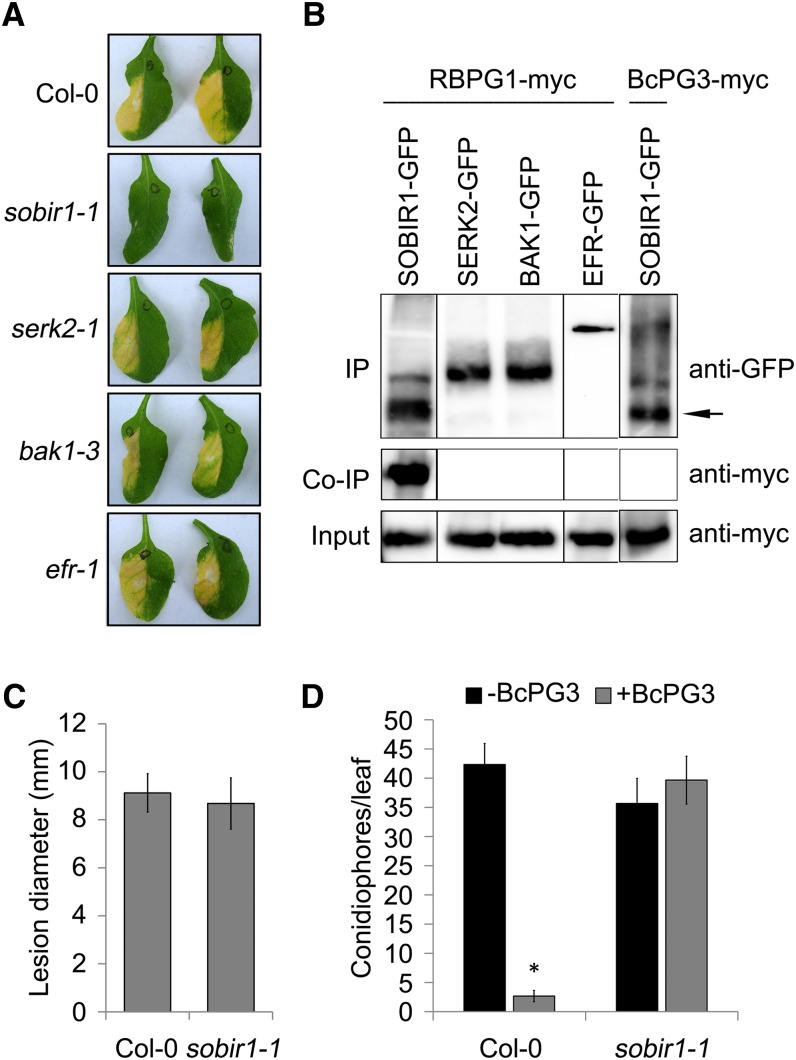
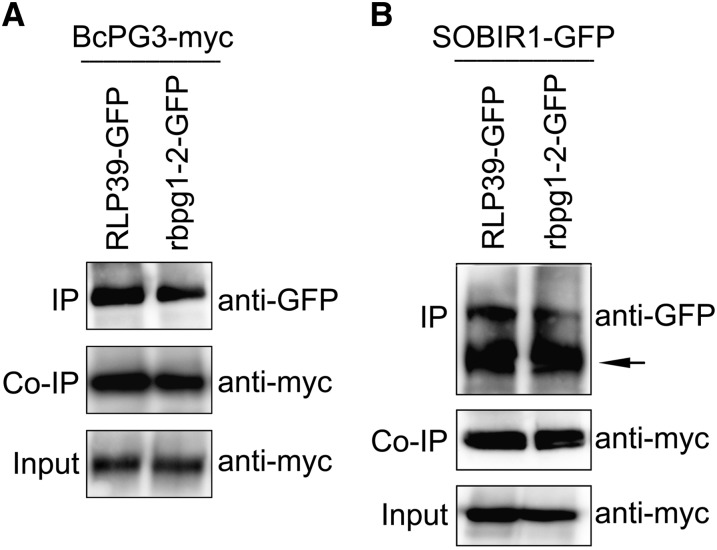
BAK1/SERK3 and other SERK family members play important roles in plant defense responses by forming complexes with a number of LRR-RLKs such as FLS2 and EFR (Chinchilla et al., 2009; Roux et al., 2011). Recently, the tomato LRR-RLK SOBIR1 was shown to interact specifically with a number of LRR-RLPs (Liebrand et al., 2013). To test whether LRR-RLKs are involved in the response of RBPG1 to BcPG3, Arabidopsis plants carrying mutations in different LRR-RLK genes were analyzed (Fig. 5A). Two serk mutants and an efr mutant were unaltered in their responsiveness to BcPG3, whereas the sobir1-1 mutant did not show any symptoms following infiltration with BcPG3. Coimmunoprecipitation analysis showed that RBPG1 interacts with SOBIR1 but not with SERK2, BAK1/SERK3, or EFR (Fig. 5B) and also not upon the addition of ligand (data not shown). GFP-tagged SOBIR1 was incapable to immunoprecipitate 10xMyc-tagged BcPG3 (Fig. 5B).
Figure 5.
SOBIR1 is required for RBPG1-dependent responsiveness. A, Response of Arabidopsis mutants to BcPG3. Leaves were photographed 7 d after infiltration with BcPG3 (1.5 µm). B, Coimmunoprecipitation (Co-IP) of RBPG1 and SOBIR1, SERK2, BAK1, or EFR and of BcPG3 and SOBIR1 is shown. Total proteins expressed in N. benthamiana leaves were subjected to immunoprecipitation (IP) with GFP Trap beads, followed by immunoblot analysis with anti-myc antibodies to detect RBPG1-myc and BcPG3-myc as well as with anti-GFP antibodies to detect SOBIR1-GFP, SERK2-GFP, BAK1-GFP, and EFR-GFP. The black arrow indicates the full-length SOBIR1-GFP protein, and the other bands are unspecific products. C, Lesion diameter caused by B. cinerea at 3 dpi on Arabidopsis leaves. Data represent means ± sd (n ≥ 15). D, Col-0 and sobir1-1 mutant plants were infiltrated with water or BcPG3 and 24 h later inoculated with Hpa isolate Noco2. Conidiophores were counted at 6 dpi. Results are averages ± se (n = 15). *P < 0.01 by Student’s t test. All experiments were repeated at least three times with similar results.
As mentioned above, a T-DNA insertion mutant in the RBPG1 gene was not available. The requirement of SOBIR1 for the RBPG1-mediated response to PGs enabled us to use sobir1-1 mutant plants as (indirect) functional RBPG1 knockouts in pathogen infection experiments. Inoculation of Col-0 and the sobir1-1 mutant with B. cinerea resulted in lesions of equal sizes in the wild type and the mutant (Fig. 5C). Furthermore, Col-0 and the sobir1-1 mutant were inoculated with Hpa isolate Noco2, either with or without infiltration of the heat-inactivated hiBcPG3 protein, 1 d before inoculation. The untreated Col-0 and sobir1-1 plants were equally susceptible to Hpa (Fig. 5D). hiBcPG3 pretreatment induced increased resistance to Hpa in Col-0 but not in the sobir1-1 mutant (Fig. 5D).
RBPG1 Homologs Physically Interact with BcPG3 and SOBIR1
Among the six RLP homologs encoded within the RBPG1 locus from Col-0 and Br-0, RBPG1 is the only receptor that confers responsiveness to BcPG3. To test whether specificity is determined by the interaction of the ligand (BcPG3) with the receptor, coimmunoprecipitation was also performed with RBPG1 homologs by transient coexpression of 10xMyc-tagged BcPG3 with GFP-tagged RLP39, RLP40, RLP41, and rbpg1-2 in N. benthamiana (cloning of the rbpg1-1 construct was unsuccessful). RLP39 and rbpg1-2 were able to coimmunoprecipitate BcPG3 (Fig. 6A). Moreover, GFP-tagged SOBIR1 was able to coimmunoprecipitate 10xMyc-tagged RLP39 and rbpg1-2 (Fig. 6B). Due to low expression of either GFP-tagged or 10xMyc-tagged RLP40 and RLP41 proteins, their interaction with BcPG3 or SOBIR1 could not be studied.
Figure 6.
RLP39 and rbpg1-2 interact with BcPG3 and SOBIR1 in N. benthamiana. Coimmunoprecipitation (Co-IP) of RLP39 or rbpg1-2 with BcPG3 (A) and with SOBIR1 (B) is shown. Total proteins expressed in N. benthamiana leaves were subjected to immunoprecipitation (IP) with GFP Trap beads, followed by immunoblot analysis with anti-myc antibodies to detect BcPG3-myc, RLP39-myc, and rbpg1-2-myc as well as with anti-GFP antibodies to detect SOBIR1-GFP, RLP39-GFP, and rbpg1-2-GFP. The black arrow indicates the full-length SOBIR1-GFP protein, and the other bands are unspecific products. All experiments were repeated at least three times with similar results.
DISCUSSION
This study started off with the observation that Arabidopsis accessions show natural variation in response to PGs, with symptoms ranging from no detectable damage to full necrosis of the infiltrated area. Accessions Col-0 and Br-0 were selected for detailed studies, as representatives of the extremes in the spectrum of responses. We initially considered the possibility that the lack of symptoms in accession Br-0 might be related to differences in pectin architecture or modification, rendering the pectin a poor substrate for PGs and thereby preventing damage inflicted upon PG infiltration. Cell wall preparations of both ecotypes, however, were highly similar in their monosaccharide composition, degree of methylation, and degree of acetylation (L. Zhang, unpublished data). This result hinted at the possibility that tissue necrosis observed upon PG infiltration might not be a direct consequence of pectin decomposition and loss of cell wall integrity.
A QTL mapping approach on the F2 progeny of Col-0 × Br-0 identified a single recessive locus determining the response to PGs, designated as RBPG1. The RBPG1 gene in the locus was isolated by map-based cloning and a combination of functional and comparative genomics analysis and appeared to encode an LRR-RLP (RLP42). In Arabidopsis, the family of LRR-RLPs comprises 57 members (Wang et al., 2008), of which the few that have been functionally characterized are involved in developmental processes (CLAVATA2 [RLP10] and TOO MANY MOUTHS [RLP17]; Jeong et al., 1999; Nadeau and Sack, 2002) or hormone-induced senescence (RLP41; Wang et al., 2008). A recent study showed that ReMAX (RLP1) functions as a PRR sensing the MAMP eMax from Xanthomonas spp. (Jehle et al., 2013). Here, we demonstrate that RBPG1 (RLP42) functions as a novel PRR recognizing fungal PGs, not only from B. cinerea but also one from the saprotroph A. niger.
There is at present no evidence that recognition of PGs by LRR-RLPs and the subsequent necrotic response occurs outside Arabidopsis. We previously showed that PGs from B. cinerea are able to induce necrosis in tomato, broad bean, and N. benthamiana (Kars et al., 2005; Joubert et al., 2007). This response required enzyme activity, and there were marked differences in necrosis-inducing activities between individual PGs, with BcPG2 causing massive and rapid tissue collapse (less than 30 min in broad bean) and BcPG3 causing no detectable symptoms in any of the three plants mentioned (Kars et al., 2005; Joubert et al., 2007). The response in Arabidopsis to PGs, therefore, appears to be fundamentally different from that in tomato, broad bean, and N. benthamiana.
Several lines of evidence demonstrated that PGs act as true MAMPs. First, the necrosis-inducing activity was independent of enzymatic activity, since heat-inactivated BcPG3 and catalytically inactive BcPG3D353E/D354N proteins triggered necrotic responses like the active enzyme. This result is analogous to observations reported for the Trichoderma spp. EIX and B. cinerea xylanase xyn11A, proteins that are both able to induce a necrotic response independent of their catalytic activity (Enkerli et al., 1999; Furman-Matarasso et al., 1999; Noda et al., 2010). Second, the protein concentrations required for necrosis-inducing activity were quite similar for all five PGs tested in this study (approximately 10 µm), in spite of the fact that the enzymes differ markedly in their specific activities (ranging from 20 to 900 units mg−1 for B. cinerea PGs; Kars et al., 2005). Furthermore, crude OGA fragments released from Arabidopsis cell walls upon incubation with BcPG3, as well as a set of OGAs of defined size ranges, failed to trigger any visible symptoms in BcPG3-responsive plants.
A conserved amino acid stretch that serves as an epitope is sufficient for the MAMP activity of flagellin, EF-Tu, or EIX (Rotblat et al., 2002; Kunze et al., 2004; Chinchilla et al., 2006). Amino acid sequence alignment identified an 11-amino acid peptide as the longest highly conserved sequence stretch among fungal PGs. This sequence is less conserved in PGs of bacteria, oomycetes, and plants. A synthetic peptide encompassing the 11-amino acid stretch was incapable of inducing necrosis. Either the tertiary structure of the stretch could differ between the full-length protein and the synthetic peptide or the peptide was unstable after infiltration in the plant. Notably, the corresponding region in the sequence of Arabidopsis PGs starts with a Pro residue instead of Gly (corresponding to BcPG3 residue 373). This Pro likely alters the tertiary structure in this region and might enable Arabidopsis PGs to escape self-recognition by the RBPG1 receptor. However, a mutated BcPG3G373P protein remained able to induce necrosis in RBPG1-expressing plants. This observation questions the involvement of the 11-residue peptide stretch in PG recognition by RBPG1. It remains to be established which epitope(s) of fungal PGs are essential for MAMP activity.
The B. cinerea genome contains in total six PG genes (Wubben et al., 1999), and the genes display a complex temporal and spatial expression pattern during plant infection, depending on the host species, tissue type, and incubation temperature (ten Have et al., 2001). Expression of Bcpg1, Bcpg4, and Bcpg6 during infection of Arabidopsis was demonstrated (Zhang and van Kan, 2013a). Thus, Arabidopsis genotypes expressing the RBPG1 gene should be able to recognize the protein(s) and respond with the induction of necrosis. One might consider that B. cinerea, as a necrotroph, would possibly benefit from the recognition of PGs, leading to the induction of necrosis. However, the necrotic response to PGs was too slow to have an impact on disease development. Typically, infiltration of PGs into responsive plants caused the first symptoms 3 to 4 d after infiltration. Under our experimental conditions, B. cinerea produced expanding lesions on Arabidopsis leaves from 48 h post inoculation onward, and leaves were entirely colonized and dead within 5 d. This explains that the presence of the RBPG1 gene did not promote B. cinerea disease development to any detectable degree, since the RBPG1-mediated response to PGs is too slow to make a difference. We could not directly examine the susceptibility of RBPG1 mutants to B. cinerea, because there were no T-DNA insertion lines in public mutant collections. However, the observation that SOBIR1 is required for the RBPG1-mediated necrotic response to PGs enabled us to indirectly test the effect of RBPG1 on susceptibility to B. cinerea. We did not observe any difference in B. cinerea disease development between Col-0 and the sobir1 mutant (Fig. 5C), suggesting that RBPG1 indeed has no impact on susceptibility to B. cinerea, as was also concluded from the observation that transgenic Br-0 plants expressing RBPG1 were equally susceptible to B. cinerea (Fig. 3B).
If not against B. cinerea, could RBPG1 be involved in immunity to other pathogens, such as biotrophs? Inoculations were performed with Hpa, but we did not observe any difference in susceptibility to Hpa between Br-0 (lacking RBPG1) and transgenic lines expressing RBPG1 (Fig. 3D), nor between Col-0 and the sobir1 mutant (nonresponsive to PGs; Fig. 5D). The genome of Hpa (an oomycete) contains three PG genes, which are divergent in sequence from the PGs of fungi (data not shown). Thus, the observation that the RBPG1 gene by itself does not influence susceptibility to Hpa was to be expected. However, infiltration of BcPG3 into RBPG1-expressing plants prior to inoculation with Hpa led to a marked reduction in disease development, as evidenced by strongly reduced Hpa conidiophore production (Fig. 3D). The inoculation with Hpa was performed 1 d after protein application, which is at least 2 d before the PG-induced onset of ROS production (Fig. 3A) and the appearance of necrotic symptoms (Fig. 1A). Yet, pretreatment of RBPG1-expressing plants with BcPG3 triggered defense responses that led to increased resistance to Hpa (Fig. 3D). The increased resistance to Hpa following BcPG3 application was abolished in the sobir1 mutant (Fig. 5D), corroborating the crucial role of the RLK SOBIR1 in the function of the RLP RBPG1.
The current paradigm for signaling via RLPs is heterodimerization with RLKs (Bleckmann et al., 2010; Liebrand et al., 2013). The RLK SOBIR1 was initially identified as a suppressor of BIR1 that regulates cell death (Gao et al., 2009). More recent studies show that tomato SOBIR1 particularly interacts with LRR-RLPs that play a role in defense against fungal pathogens (Liebrand et al., 2013). Experiments presented here show that Arabidopsis SOBIR1 interacts with RBPG1 and is essential for RBPG1-mediated responsiveness to PGs. The presence of the BcPG3 ligand did not in any detectable way affect the interaction between RBPG1 and SOBIR1, showing that the interaction of SOBIR1 and RBPG1 is ligand independent, in agreement with the study by Liebrand et al. (2013) on the tomato SOBIR1 homolog.
RBPG1 is one of a family of four LRR-RLP-encoding genes (RLP39, RLP40, RLP41, and RLP42) that occur in a cluster (Wang et al., 2008) within the RBPG1 locus of the Col-0 genome. By contrast, accession Br-0 contains only two LRR-RLP-encoding genes (rbpg1-1 and rbpg1-2) in this region. Phylogenetic analysis showed that RLP39 and RLP41 cluster with rbpg1-2, whereas RLP40 and RBPG1 cluster with rbpg1-1. Among these six paralogs, RBPG1 is the only gene that confers responsiveness to BcPG3. It is likely that a duplication of a gene cluster with two paralogs occurred in the lineage to Col-0 and, subsequently, RBPG1 gained the responsiveness to BcPG3. This region in the Arabidopsis genome shows great diversity, which cannot easily be unraveled. Accession Col-0 contains four paralogs with high nucleotide identity. In many other accessions, the region is barely covered by mappable sequence reads (http://signal.salk.edu/atg1001/3.0/gebrowser.php; Cao et al., 2011). In order to determine the structure of this region in the Br-0 genome, it appeared essential to perform a de novo assembly of Br-0 sequence reads and manually validate the assembled contigs. For many other Arabidopsis accessions, it was equally unclear from the 1001 Genomes browser how many RLP paralogs are present in this region because of the lack of mappable reads. Likewise, genome assembly is probably required to study the diversity between these LRR-RLPs in different Arabidopsis accessions in more detail.
Not only RBPG1 physically interacts with BcPG3; RLP39 and rbpg1-2 (from accession Br-0) also were able to coimmunoprecipitate BcPG3, even though they do not confer responsiveness to BcPG3. Thus, the specificity of the RLP-mediated response to PGs seems to be unrelated to the ligand-binding capacity. This situation is analogous to the tomato LRR-RLPs LeEIX1 and LeEIX2, encoded by paralogous genes in a cluster, that act as receptors for fungal EIX. Both receptors bind EIX, but only LeEIX2 mediates defense responses (Ron and Avni, 2004). We considered the possibility that differences in the cytoplasmic domain between RBPG1 and its paralogs might account for the ability of RBPG1 to transduce a signal. Genetic complementation studies, however, demonstrated that the intracellular C-terminal domain of RBPG1 is not required for inducing necrosis in response to PGs. Several studies suggest that MAMP receptors must heterodimerize with RLKs for proper signal transduction to occur (Chinchilla et al., 2007; Roux et al., 2011; Monaghan and Zipfel, 2012). An alternative reason for the inability of RLP39, RLP40, and RLP41 to respond to BcPG3 could be the lack of interaction with other RLKs. We showed that the LRR-RLK SOBIR1 interacts with RBPG1 and is required for the responsiveness to BcPG3. RLP39 as well as rbpg1-2 were equally able to interact with SOBIR1. Therefore, it remains to be studied whether the recognition of BcPG3 by RBPG1 changes the phosphorylation status of SOBIR1, thereby allowing additional components to associate with the complex.
A recent study by Zhang et al. (2013) has demonstrated that a different Arabidopsis LRR-RLP, RLP30, can recognize an as yet unidentified MAMP from culture medium of Sclerotinia sclerotiorum, and the recognition results in the induction of ethylene production. T-DNA insertion mutants in the RLP30 gene failed to produce ethylene in response to the MAMP, and they were more susceptible to S. sclerotiorum and to B. cinerea. The RLP30-mediated response to the unidentified MAMP requires BAK1 and SOBIR1, and Zhang et al. (2013) reported that bak1 and sobir1 mutants were more susceptible to S. sclerotiorum and B. cinerea.
Among the 57 RLP genes in the Arabidopsis genome, described by Wang et al. (2008), the functions of only six have been reported (Jeong et al., 1999; Nadeau and Sack, 2002; Wang et al., 2008; Zhang et al., 2010, 2013; Yang et al., 2012; Jehle et al., 2013). The results presented here have revealed a novel aspect of fungal PGs and have unraveled the function of the RLP42 gene (RBPG1) as a receptor for fungal PGs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) plants used in this study were grown in a greenhouse at 20°C or in a growth chamber at 20°C and 70% relative humidity under a 12-h light/dark cycle (short-day conditions of 8 h of light/16 h of dark). Arabidopsis accessions were kindly provided by Maarten Koornneef, Corry Hanhart, and Joost Keurentjes. Transgenic seeds were grown on one-half-strength Murashige and Skoog medium plates with 1% (w/v) Suc and 1% (w/v) plant agar containing 20 µg mL−1 hygromycin or 50 µg mL−1 kanamycin for approximately 2 weeks. Antibiotic-resistant seedlings were transferred into soil, and the copy number of the introduced gene was determined by quantitative PCR using the actin gene as an internal standard. T-DNA insertion mutants were obtained from the Arabidopsis Biological Resource Center or the Nottingham Arabidopsis Stock Centre. The homozygosity of the T-DNA mutants was checked by PCR using the primers listed in Supplemental Table S3.
Phenotypic Scoring
Six- to 8-week-old plants were infiltrated with BcPGs purified from culture filtrates of Pichia pastoris, as described previously (Kars et al., 2005). In the initial screening, 47 Arabidopsis accessions (Supplemental Table S1) were infiltrated with BcPG2, BcPG3, BcPG4, and BcPG6 at 3 units mL−1 (diluted in 10 mm sodium acetate buffer, pH 4.2) in duplicate. Multiple rosette leaves per plant were infiltrated with one BcPG on either side of the midvein. Eleven selected accessions (Supplemental Table S1) were subsequently infiltrated with BcPG2, BcPG3, and BcPG6 in duplicate with 3 units mL−1. Plants of the F2 population of Col-0 × Br-0 were infiltrated with BcPG2 in triplicate and with BcPG3, BcPG4, and BcPG6 in duplicate. Plants of the F2 and F3 population of BC41 × pad3 were infiltrated with BcPG3 in duplicate.
The response to each infiltration was visually scored on a scale ranging from 0 to 4, as follows: 0, no symptoms; 1, chlorotic spots within the infiltrated zone; 2, chlorosis covering the infiltration zone; 3, abundant chlorosis with necrotic spots; 4, complete necrosis (Supplemental Fig. S1A).
Genotyping of the F2 Population of Col-0 × Br-0
Genotypic data on the F2 population of Col-0 × Br-0 were generated (Keygene) using AFLP markers (Vos et al., 1995) with the restriction enzymes EcoRI and MseI. AFLP markers were amplified using adapter-specific primers containing two (E+2) or three (M+3) selective nucleotides. Five different E+2/M+3 primer combinations were used (Supplemental Table S4). AFLP amplification reactions were performed in a Perkin-Elmer 9600 thermocycler. The amplified DNA products were separated on a MegaBACE 1000 capillary electrophoresis system (Amersham BioSciences). Proprietary AFLP marker analysis software (Keygene) was used to score the markers codominantly on the basis of peak intensities. Data that could not be scored codominantly unambiguously were scored dominantly. The genetic linkage map of the F2 population was constructed using the JoinMap 3.0 program (Stam, 1993; van Ooijen and Voorrips, 2001) applying the Kosambi mapping function.
QTL Analysis
QTL mapping was performed using the software packages MapQTL version 4.0 (van Ooijen et al., 2002) and WinQTLcart version 2.5 (Wang et al., 2006). For each BcPG, the QTL analysis was performed on the individual replicates and on the averages of the replicates. The data were analyzed using the interval-mapping method, calculating the log-likelihood (LOD) values every 1 cM along the chromosome. QTLs were significant when the LOD score exceeded the significance threshold (P = 0.05), which represents 95% confidence intervals for normally distributed data. Empirical thresholds for interval mapping were obtained by permutations (10,000), as implemented in the package (Churchill and Doerge, 1994). The resulting genome-wide LOD threshold for all traits in the F2 population was 3.3 (Supplemental Fig. S2B).
Further Mapping with the F8 RILs of Col-0 × Br-0
Individuals of the F2 population of Col-0 × Br-0 were propagated by single-seed descent to generate an F8 RIL population comprising 310 RILs. Three recombinants between markers E11/M62-F-131<F>-P1 and E11/M50-F-184<F>-P2 (Supplemental Fig. S2A) were identified by PCR with SNP markers (Supplemental Table S4), which were designed based on the SNPs between the sequences of Col-0 and Br-0 (http://signal.salk.edu/atg1001/3.0/gebrowser.php).
Fine-Mapping with the F2 Population of BC41 × pad3
To fine-map RBPG1, BC41 (an F8 RIL of Col-0 × Br-0, which is homozygous for the Br-0 allele in the RBPG1 locus and unresponsive to BcPGs) was crossed with the pad3 mutant in the Col-0 background (Zhou et al., 1999). BC41, alike accession Br-0, is glabrous and has the mutated gl1 gene (GL1 is involved in trichome synthesis; Hauser et al., 2001). The RBPG1 locus and the GL1 locus are both located on chromosome 3. The genetic distance between RBPG1 and GL1 is estimated to be 6 to 10 cM, based on the RIL population. F2 plants that had trichomes and were unresponsive to BcPG3 were putative recombinants and were used for linkage analysis with SNP markers (Supplemental Table S5). In total, over 4,000 F2 plants were grown and phenotyped; 400 of those plants were genotyped and 10 new recombinants were identified, reducing the RBPG1 locus to a region delimited by SNP markers 7.8-7 and 7.8-5 (Fig. 1B).
Genome Sequence Reassembly of the RBPG1 Locus in Accession Br-0
The raw sequencing data of Br-0 were kindly provided by Dr. R. Schmitz (SALK Institute). The reads were mapped on the genome of accession Col-0 (The Arabidopsis Information Resource 10), subtracting the sequences between coordinates 9,090,000 and 9,250,000 on chromosome 3, which spans the region from about 8 kb upstream of At3g24890 to 100 kb downstream of At3g25060, using Bowtie 2 version 2.0.0-beta4 (Langmead and Salzberg, 2012). Unmapped Br-0 reads were sorted and filtered accordingly using SAMtools version 0.1.18 (Li et al., 2009) and FastQC (http://www.bioinformatics.babraham.ac.uk./projects/fastqc/; version 0.10.0). Filtered reads were assembled with Velvet version 1.1.07 (Zerbino and Birney, 2008) using the K-mer setting as 57. Six assembled nodes were mapped to the RBPG1 locus, and 14 pairs of primers (Supplemental Table S6) were used to fill gaps between the nodes, enabling us to assemble them into one contig. The entire contig was finally confirmed by sequencing.
AIR Digestion with BcPG3 and OGA Preparation for Infiltration in Plants
Leaves of 5- to 6-week-old plants were freeze dried and milled. AIR was extracted with 70% ethanol at 50°C as described by Hilz et al. (2005). Twenty milligrams of AIR was suspended in 10 mm sodium acetate, pH 4.2, and incubated with 200 units of BcPG3 at room temperature. After 42 h of hydrolysis, the supernatant was obtained by centrifugation at 13,000 rpm for 10 min and filtrated through an Amicon ultra 0.5-mL centrifugal filter with a 3 K membrane (Millipore) that eliminates OGAs with a degree of polymerization greater than 15. The collected filtrate containing hydrolyzed cell wall fragments was infiltrated into Arabidopsis leaves directly. The BcPG3 proteins that remained in the filter were also collected and diluted to 100 units mL−1 for leaf infiltration.
OGAs of defined length were prepared and quantified according to Kester and Visser (1990). Samples containing pure compounds (GalpA)3 and (GalpA)4 as well as mixtures of (GalpA)4-5, (GalpA)2-6, (GalpA)4-10, and (GalpA)7-9 were each infiltrated in leaves at final concentrations of 0.1, 0.3, and 1 mm. OGAs of various lengths were generated by incubating 200 mL of 1% (w/v) sodium polygalacturonate in 10 mm sodium acetate buffer, pH 4.2, with 25 units of either Aspergillus niger PGII or BcPG2 for 10 min at 30°C. The partial digests were boiled for 5 min to stop digestion and treated three times with 3 g of Dowex 50W-X8 (H+) (Bio-Rad) to convert all pectic fragments into the acidic form. The sodium polygalacturonate/AnPGII digest was separated by gel filtration using a 60- × 2.6-cm Superdex 200 column (Amersham Biosciences). Elution was carried out with water, and fractions containing reducing sugars were collected in six pools. Pool 1 contained (GalpA)6-20 (approximately 10 µm each); pool 2 contained mainly (GalpA)4-12 (approximately 20 µm each); pool 3 comprised primarily (GalpA)1-7 (approximately 10 µm each); pool 4 consisted especially of GA monomers (70 µm) and (GalpA)2-5 (approximately 10 µm each); pools 5 and 6 primarily contained (GalpA)1-4 (approximately 5 µm each).
Plasmid Construction and Transformation
For complementation analysis in the Br-0 background, the genomic DNA sequence of At3g24890 was amplified from Col-0 with primers AT174/AT176 and cloned into pDONR207 (Invitrogen) by Gateway cloning to obtain the entry vector pENTR-At3g24890. Due to the high sequence identity in the coding regions, we first amplified approximately 3.9-kb genomic DNA fragments encompassing RLP39, RLP40, RLP41, and RBPG1 from Col-0 with primers AT188/AT189, AT170/AT171, AT190/AT191, and AT172/AT173, which are respectively specific for each gene fragment; an approximately 3.9-kb genomic DNA fragment containing rbpg1-2 was amplified from Br-0 with primers AT219/AT204. The resulting fragments were used as templates to amplify RLP39, RLP40, RLP41, RBPG1, and rbpg1-2 with primers AT192/AT193, AT194/AT195, AT196/AT198, AT177/AT178, and AT196/AT250, respectively, which were subsequently cloned into pDONR207 to generate the entry vectors. Two C-terminally truncated RBPG1 fragments (RBPG1_Trunc1 and RBPG1_Trunc2) were amplified from pENTR-RBPG1 with primers AT177/AT251 and AT177/AT252, respectively, and the swapped rbpg1-2_Swap1 fragment was amplified from pENTR-rbpg1-2 with primers AT196/AT254. The resulting fragments were cloned into pDONR207 to obtain the entry vectors. All the genes in entry vectors were confirmed by sequencing. Expression vectors (containing the 35S promoter) were created by Gateway cloning of pMDC32 (Curtis and Grossniklaus, 2003) with the corresponding entry vectors; binary constructs were transformed into Agrobacterium tumefaciens GV3101 and subsequently into Arabidopsis plants by floral dipping (Clough and Bent, 1998).
For transient expression in Nicotiana benthamiana, Bcpg3 with a plant PR1 signal peptide was amplified from pAT2-3 (Joubert et al., 2007) with primers AT265/AT268 and cloned into donor vector pDONR207 to obtain the entry vector, which was checked by sequencing. Expression vectors were created by Gateway cloning into pSOL2095 (Liebrand et al., 2012) or pGWB20 (Nakagawa et al., 2007) with the corresponding entry vectors and transformed into A. tumefaciens C58C1, carrying helper plasmid pCH32 (Liebrand et al., 2012), or into A. tumefaciens GV3101 (only for SERK2 and BAK1 constructs).
The substitutions chosen for generating the catalytically inactive Bcpg3D353E/D354N protein were designed based on studies of Armand et al. (2000) on the catalytic residues of Aspergillus niger pgB. For mutant protein production in P. pastoris, site-directed mutagenesis was carried out by amplifying two fragments of mutant allele Bcpg3D353E/D354N from pPIC3.5-Bcpg3 (Kars et al., 2005) with primers AT261/AT269 and AT270/AT262, which were linked by overlap PCR with primers AT261/AT262. The resulting mutant allele Bcpg3D353E/D354N was cloned into pGEM-T easy vector (Promega) and checked by sequencing. Mutant allele Bcpg3G373P was cloned by the same method with primers AT261/AT272 and AT273/AT262 followed by AT261/AT262. To generate N-terminal Myc-tagged BcPG3, wild-type and mutant alleles of Bcpg3 were amplified from pPIC3.5-Bcpg3, pGEMT-Bcpg3D353E/D354N, and pGEMT-Bcpg3G373P, respectively, with primers AT261/AT262 and linked with 10xMyc (derived from pGWB20 with primers AT263/AT264) by overlap PCR with primers AT261/AT264 and subsequently cloned into pGEM-T easy vector and checked by sequencing. The EcoRI/NotI-digested fragments of Bcpg3-myc, Bcpg3D353E/D354N-myc, and Bcpg3G373P-myc derived from pGEM-T constructs were subcloned into the corresponding sites of pPIC3.5K (Invitrogen), yielding the P. pastoris expression vectors. The SalI-linearized pPIC3.5K-Bcpg3, pPIC3.5K-Bcpg3D353E/D354N, and pPIC3.5K-Bcpg3D373P were used to transform P. pastoris GS115 by electroporation (Kars et al., 2005). Protein production in P. pastoris was performed according to the expression manual (Invitrogen) with minor modifications. The constructs generated are listed in Supplemental Table S7, and primers used for cloning are shown in Supplemental Table S8.
ROS Assay
ROS accumulation in planta was visualized by diaminobenzidine staining. Arabidopsis leaves were vacuum infiltrated for 2 min with diaminobenzidine solution (1 mg mL−1, pH 3.8), incubated on a shaker for 1 h, and subsequently destained with 96% (v/v) ethanol.
Infection Assay
Three- to 4-week-old Arabidopsis plants were infiltrated with heat-inactive BcPG3 (0.75 µm). After 1 d of infiltration, the same leaves were infected with Hyaloperonospora arabidopsidis.
Hpa isolate Maks9 (kindly provided by Dr. Eric Holub) and Noco2 were maintained on Arabidopsis accession Br-0. Hpa maintenance and infection were performed as described previously (Van Damme et al., 2005) with minor modifications. Plants were inoculated with spores (5 × 104 spores mL−1) by using a spray gun (Holub et al., 1994), air dried for approximately 30 min, and incubated under a sealed lid at 100% relative humidity in a growth chamber at 16°C with 9 h of light per day. Sporulation levels were quantified at 6 or 10 d post inoculation (dpi) by counting the number of sporangiophores per leaf.
Transient Expression in N. benthamiana
A. tumefaciens (strain C58C1, except for SERK2 and BAK1, which were in strain GV3101) was grown in Luria-Bertani medium with appropriate antibiotics at 28°C overnight. Cultures were harvested and resuspended in 2.0% (w/v) Suc, 0.5% (w/v) Murashige and Skoog salts without vitamins, 0.2% (w/v) MES, and 0.2 mm acetosyringone, pH 5.6, to optical density at 600 nm (OD600) = 2.0. Cultures carrying pGWB20-Bcpg3 were resuspended at OD600 = 0.2, and cultures carrying pZP-SERK2 or pZP-BAK1 were resuspended at OD600 = 0.5. For coexpression, two cultures carrying appropriate constructs were mixed in a 1:1 ratio, incubated for 2 h, and infiltrated into 6-week-old N. benthamiana leaves. Samples were collected 2 d after agroinfiltration for coimmunoprecipitation analysis.
Immunoprecipitation and Immunoblotting
Immunoprecipitation was performed as described (Liebrand et al., 2012). N. benthamiana membrane fractions were extracted in buffer (150 mm NaCl, 1.0% [v/v] IGEPAL CA-630 [Nonidet P-40], 0.5% [w/v] sodium deoxycholate, 0.1% [w/v] SDS, and 50 mm Tris, pH 8.0). After centrifugation at 14,000 rpm for 15 min, 15 µL of GFPTrap_A beads (Chromotek) was added to the supernatant and incubated for 1 h at 4°C. After washing the beads five times with extraction buffer, immunoprecipitated proteins were separated by 8% (w/v) SDS-PAGE and electroblotted onto Immun-Blot polyvinylidene difluoride membranes (Bio-Rad) at 22 V overnight. Membranes were rinsed in Tris-buffered saline and blocked for 1 h in 5% (w/v) skim milk in Tris-buffered saline plus Tween (0.1% [v/v]). GFP-tagged proteins were detected with 1:5,000 diluted anti-GFP-horseradish peroxidase (MACS antibodies), whereas myc-tagged proteins were detected with 1:2,000 diluted anti-myc (cMyc 9E10, sc-40; Santa Cruz) and subsequently with 1:2,000 diluted anti-mouse immunoglobulin-horseradish peroxidase (Amersham). SuperSignal West Femto chemiluminescent substrate (Thermo) was applied for signal development.
Sequence data from Col-0 can be found in the Arabidopsis Genome Initiative databases under the following accession numbers: RLP39, At3g24900; RLP40, At3g24982; RLP41, At3g25010; RBPG1, At3g25020; SOBIR1, At2g31880; EFR, At5g20480; SERK2, At1g34210; BAK1/SERK3, At4g33430. Sequence data from the syntenic region of the RBPG1 locus in accession Br-0 (14.3 kb) containing the RLP genes rbpg1-1 and rbpg1-2 were deposited in the National Center for Biotechnology Information under accession number KF684938.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Responsiveness of Arabidopsis accessions to B. cinerea PGs.
Supplemental Figure S2. QTL mapping of the RBPG1 locus from the F2 population of Col-0 × Br-0.
Supplemental Figure S3. Responsiveness of Arabidopsis accessions Col-0 and Br-0, and Br-0 transgenic plants expressing P35S:At3g24890, to BcPG3.
Supplemental Figure S4. Amino acid alignment of RLP39, RLP40, RLP41, RBPG1, rbpg1-1, and rbpg1-2.
Supplemental Figure S5. Responsiveness of Arabidopsis F8 RIL line BC41 and BC41 transgenic plants expressing P35S:RBPG1, P35S:RLP39, P35S:RLP40, and P35S:RLP41 to BcPG3.
Supplemental Figure S6. The C-terminal domain of RBPG1 is not required for the responsiveness to BcPG3.
Supplemental Figure S7. Amino acid sequence alignment of fungal PGs and Arabidopsis PGs.
Supplemental Figure S8. Responsiveness of Arabidopsis accessions Col-0 and Br-0, and Br-0 transgenic plants expressing P35S:At3g24890, to BcPG3G373P.
Supplemental Table S1. Arabidopsis accessions used in this study.
Supplemental Table S2. Candidate genes in the RBPG1 locus.
Supplemental Table S3. Primers used in the analysis of Arabidopsis homozygous T-DNA mutants.
Supplemental Table S4. Number of polymorphic AFLP markers found between the Arabidopsis accessions Col-0 and Br-0 using five primer combinations.
Supplemental Table S5. Primers used in genetic mapping.
Supplemental Table S6. Primers used for Br-0 sequence assembly and confirmation.
Supplemental Table S7. Plasmids used in this study.
Supplemental Table S8. Primers used for plasmid construction.
Acknowledgments
We thank Maarten Koornneef, Corry Hanhart, and Joost Keurentjens (Wageningen University) for providing Arabidopsis accessions; Hanneke Witsenboer (Keygene) for facilitating the initial AFLP mapping; Rik van Wijk (Nickerson-Zwaan Group) for assistance in QTL mapping analysis; Bob Schmitz and Joe Ecker (Salk Institute) for providing the Br-0 sequence reads; our colleagues Ronnie de Jonge and Luigi Faino (Wageningen University) for assistance in genome sequence reassembly of accession Br-0; Catherine Albrecht and Sacco de Vries (Wageningen University) for providing SERK-GFP constructs; Silke Robatzek and Cyril Zipfel (Sainsbury Laboratory) for providing the EFR entry vector and efr mutant; Yuelin Zhang (University of British Columbia) for providing the sobir1-1 mutant; Peter Schaap (Wageningen University) for providing advice on the targeted mutagenesis of BcPG3; and our colleagues Bart Thomma and Matthieu Joosten (Wageningen University) for critical reading of the manuscript.
Glossary
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- LRR
leucine-rich repeat
- RLK
receptor-like kinase
- RLP
receptor-like protein
- PG
endopolygalacturonase
- OGA
oligogalacturonide
- DAMP
damage-associated molecular pattern
- AFLP
amplified fragment-length polymorphism
- QTL
quantitative trait locus
- cM
centimorgan
- SNP
single-nucleotide polymorphism
- RIL
recombinant inbred line
- T-DNA
transfer DNA
- AIR
alcohol-insoluble residue
- ROS
reactive oxygen species
- Hpa
Hyaloperonospora arabidopsidis
- LOD
log-likelihood
- dpi
days post inoculation
- OD600
optical density at 600 nm
References
- Armand S, Wagemaker MJ, Sánchez-Torres P, Kester HC, van Santen Y, Dijkstra BW, Visser J, Benen JA. (2000) The active site topology of Aspergillus niger endopolygalacturonase II as studied by site-directed mutagenesis. J Biol Chem 275: 691–696 [DOI] [PubMed] [Google Scholar]
- Bar M, Sharfman M, Ron M, Avni A. (2010) BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J 63: 791–800 [DOI] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Brunner F, Rosahl S, Lee J, Rudd JJ, Geiler C, Kauppinen S, Rasmussen G, Scheel D, Nürnberger T. (2002) Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J 21: 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc Natl Acad Sci USA 107: 9452–9457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Schneeberger K, Ossowski S, Günther T, Bender S, Fitz J, Koenig D, Lanz C, Stegle O, Lippert C, et al. (2011) Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43: 956–963 [DOI] [PubMed] [Google Scholar]
- Chaparro-Garcia A, Wilkinson RC, Gimenez-Ibanez S, Findlay K, Coffey MD, Zipfel C, Rathjen JP, Kamoun S, Schornack S. (2011) The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS ONE 6: e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Shan L, He P, de Vries S, Kemmerling B. (2009) One for all: the receptor-associated kinase BAK1. Trends Plant Sci 14: 535–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ovidio R, Mattei B, Roberti S, Bellincampi D. (2004) Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim Biophys Acta 1696: 237–244 [DOI] [PubMed] [Google Scholar]
- Enkerli J, Felix G, Boller T. (1999) The enzymatic activity of fungal xylanase is not necessary for its elicitor activity. Plant Physiol 121: 391–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Furman-Matarasso N, Cohen E, Du Q, Chejanovsky N, Hanania U, Avni A. (1999) A point mutation in the ethylene-inducing xylanase elicitor inhibits the beta-1-4-endoxylanase activity but not the elicitation activity. Plant Physiol 121: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, Bi D, Cheng YT, Chen S, Li X, et al (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6: 34–44 [DOI] [PubMed] [Google Scholar]
- Gómez-Gómez L, Boller T. (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Greeff C, Roux M, Mundy J, Petersen M. (2012) Receptor-like kinase complexes in plant innate immunity. Front Plant Sci 3: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser MT, Harr B, Schlötterer C. (2001) Trichome distribution in Arabidopsis thaliana and its close relative Arabidopsis lyrata: molecular analysis of the candidate gene GLABROUS1. Mol Biol Evol 18: 1754–1763 [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. (2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K, Cherk C, Somerville S. (2009) Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol 12: 406–413 [DOI] [PubMed] [Google Scholar]
- Hilz H, Bakx EJ, Schols HA, Voragen AGJ. (2005) Cell wall polysaccharides in black currants and bilberries: characterisation in berries, juice, and press cake. Carbohydr Polym 59: 477–488 [Google Scholar]
- Holub EB, Beynon LJ, Crute IR. (1994) Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol Plant Microbe Interact 7: 223–239 [Google Scholar]
- Isshiki A, Akimitsu K, Yamamoto M, Yamamoto H. (2001) Endopolygalacturonase is essential for citrus black rot caused by Alternaria citri but not brown spot caused by Alternaria alternata. Mol Plant Microbe Interact 14: 749–757 [DOI] [PubMed] [Google Scholar]
- Jehle AK, Lipschis M, Albert M, Fallahzadeh-Mamaghani V, Fürst U, Mueller K, Felix G. (2013) The receptor-like protein ReMAX of Arabidopsis detects the microbe-associated molecular pattern eMax from Xanthomonas. Plant Cell 25: 2330–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE. (1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Joubert DA, Kars I, Wagemakers L, Bergmann C, Kemp G, Vivier MA, van Kan JA. (2007) A polygalacturonase-inhibiting protein from grapevine reduces the symptoms of the endopolygalacturonase BcPG2 from Botrytis cinerea in Nicotiana benthamiana leaves without any evidence for in vitro interaction. Mol Plant Microbe Interact 20: 392–402 [DOI] [PubMed] [Google Scholar]
- Kars I, Krooshof GH, Wagemakers L, Joosten R, Benen JA, van Kan JAL. (2005) Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J 43: 213–225 [DOI] [PubMed] [Google Scholar]
- Kester HC, Visser J. (1990) Purification and characterization of polygalacturonases produced by the hyphal fungus Aspergillus niger. Biotechnol Appl Biochem 12: 150–160 [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110: 213–222 [DOI] [PubMed] [Google Scholar]
- Liebrand TW, Smit P, Abd-El-Haliem A, de Jonge R, Cordewener JH, America AH, Sklenar J, Jones AM, Robatzek S, Thomma BP, et al (2012) Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiol 159: 1819–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TW, van den Berg GC, Zhang Z, Smit P, Cordewener JH, America AH, Sklenar J, Jones AM, Tameling WI, Robatzek S, et al (2013) Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc Natl Acad Sci USA 110: 10010–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD. (2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nam KH, Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110: 203–212 [DOI] [PubMed] [Google Scholar]
- Noda J, Brito N, González C. (2010) The Botrytis cinerea xylanase Xyn11A contributes to virulence with its necrotizing activity, not with its catalytic activity. BMC Plant Biol 10: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeser B, Heidrich PM, Müller U, Tudzynski P, Tenberge KB. (2002) Polygalacturonase is a pathogenicity factor in the Claviceps purpurea/rye interaction. Fungal Genet Biol 36: 176–186 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Kay P, Wilson S, Swain SM. (2009) ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinssot B, Vandelle E, Bentéjac M, Adrian M, Levis C, Brygoo Y, Garin J, Sicilia F, Coutos-Thévenot P, Pugin A. (2003) The endopolygalacturonase 1 from Botrytis cinerea activates grapevine defense reactions unrelated to its enzymatic activity. Mol Plant Microbe Interact 16: 553–564 [DOI] [PubMed] [Google Scholar]
- Prade RA, Zhan DF, Ayoubi P, Mort AJ. (1999) Pectins, pectinases and plant-microbe interactions. Biotechnol Genet Eng Rev 16: 361–391 [DOI] [PubMed] [Google Scholar]
- Rasul S, Dubreuil-Maurizi C, Lamotte O, Koen E, Poinssot B, Alcaraz G, Wendehenne D, Jeandroz S. (2012) Nitric oxide production mediates oligogalacturonide-triggered immunity and resistance to Botrytis cinerea in Arabidopsis thaliana. Plant Cell Environ 35: 1483–1499 [DOI] [PubMed] [Google Scholar]
- Ron M, Avni A. (2004) The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotblat B, Enshell-Seijffers D, Gershoni JM, Schuster S, Avni A. (2002) Identification of an essential component of the elicitation active site of the EIX protein elicitor. Plant J 32: 1049–1055 [DOI] [PubMed] [Google Scholar]
- Roux M, Schwessinger B, Albrecht C, Chinchilla D, Jones A, Holton N, Malinovsky FG, Tör M, de Vries S, Zipfel C. (2011) The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 23: 2440–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh MT, Brown RL, Whitehead MP, Cary JW, Cotty PJ, Cleveland TE, Dean RA. (1997) Molecular genetic evidence for the involvement of a specific polygalacturonase, P2c, in the invasion and spread of Aspergillus flavus in cotton bolls. Appl Environ Microbiol 63: 3548–3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam P. (1993) Construction of integrated genetic-linkage maps by means of a new computer package: Joinmap. Plant J 3: 739–744 [Google Scholar]
- ten Have A, Breuil WO, Wubben JP, Visser J, van Kan JAL. (2001) Botrytis cinerea endopolygalacturonase genes are differentially expressed in various plant tissues. Fungal Genet Biol 33: 97–105 [DOI] [PubMed] [Google Scholar]
- ten Have A, Mulder W, Visser J, van Kan JAL. (1998) The endopolygalacturonase gene Bcpg1 is required for full virulence of Botrytis cinerea. Mol Plant Microbe Interact 11: 1009–1016 [DOI] [PubMed] [Google Scholar]
- Van Damme M, Andel A, Huibers RP, Panstruga R, Weisbeek PJ, Van den Ackerveken G. (2005) Identification of Arabidopsis loci required for susceptibility to the downy mildew pathogen Hyaloperonospora parasitica. Mol Plant Microbe Interact 18: 583–592 [DOI] [PubMed] [Google Scholar]
- van den Brink J, de Vries RP. (2011) Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol 91: 1477–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen JW, Boer MP, Jansen RC, Maliepaard C (2002) MapQTL version 4.0: software for the calculation of QTL positions on genetic maps. Plant Research International, Wageningen, The Netherlands [Google Scholar]
- van Ooijen JW, Voorrips RE (2001) Joinmap 3.0: software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu CM, Woods-Tör A, Zipfel C, et al. (2008) A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol 147: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng Z-B (2006) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, http://statgen.ncsu.edu/qtlcart/WQTLCart.htm (September 25, 2006)
- Wubben JP, Mulder W, ten Have A, van Kan JAL, Visser J. (1999) Cloning and partial characterization of endopolygalacturonase genes from Botrytis cinerea. Appl Environ Microbiol 65: 1596–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YA, Zhang YX, Ding PT, Johnson K, Li X, Zhang YL. (2012) The ankyrin-repeat transmembrane protein BDA1 functions downstream of the receptor-like protein SNC2 to regulate plant immunity. Plant Physiol 159: 1857–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, van Kan JAL. (2013a) Botrytis cinerea mutants deficient in D-galacturonic acid catabolism have a perturbed virulence on Nicotiana benthamiana and Arabidopsis, but not on tomato. Mol Plant Pathol 14: 19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, van Kan JAL (2013b) Pectin as a barrier and nutrient source for fungal plant pathogens. In F Kempken, ed, The Mycota XI, Ed 2: Agricultural Applications. Springer-Verlag, Berlin, pp 361–375 [Google Scholar]
- Zhang W, Fraiture M, Kolb D, Löffelhardt B, Desaki Y, Boutrot FF, Tör M, Zipfel C, Gust AA, Brunner F. (2013) Arabidopsis RECEPTOR-LIKE PROTEIN30 and receptor-like kinase SUPPRESSOR OF BIR1-1/EVERSHED mediate innate immunity to necrotrophic fungi. Plant Cell 25: 4227–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Yang YA, Fang B, Gannon P, Ding PT, Li X, Zhang YL. (2010) Arabidopsis snc2-1D activates receptor-like protein-mediated immunity transduced through WRKY70. Plant Cell 22: 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Glazebrook J. (1999) Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11: 2419–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]