Virus-based microRNA silencing can be used for functional analysis of endogenous microRNAs in plants.
Abstract
MicroRNAs (miRNAs) play pivotal roles in various biological processes across kingdoms. Many plant miRNAs have been experimentally identified or predicted by bioinformatics mining of small RNA databases. However, the functions of these miRNAs remain largely unknown due to the lack of effective genetic tools. Here, we report a virus-based microRNA silencing (VbMS) system that can be used for functional analysis of plant miRNAs. VbMS is performed through tobacco rattle virus-based expression of miRNA target mimics to silence endogenous miRNAs. VbMS of either miR172 or miR165/166 caused developmental defects in Nicotiana benthamiana. VbMS of miR319 reduced the complexity of tomato (Solanum lycopersicum) compound leaves. These results demonstrate that tobacco rattle virus-based VbMS is a powerful tool to silence endogenous miRNAs and to dissect their functions in different plant species.
MicroRNAs (miRNAs) are genome-encoded 20- to 24-nucleotide small RNAs that act as posttranscriptional regulators in eukaryotes (Bartel, 2004; Vaucheret et al., 2004). In plants, mature miRNA is excised from the primary miRNA transcript by DICER-LIKE1 (RNase III-like endoribonucleases) in a stepwise manner. The mature miRNAs are selectively loaded into an RNA-induced silencing complex, which can cause either target mRNA degradation or translational repression directed by the miRNA with complete or partial complementarity to the target transcript (Sunkar et al., 2007; Ha et al., 2008).
Plant miRNAs play essential roles in various biological processes, such as development, signal transduction, protein degradation, response to abiotic and biotic stress, as well as the regulation of their own biogenesis (Zhang et al., 2006; Phillips et al., 2007; Sunkar et al., 2007; Jin, 2008; Lu et al., 2008; Shukla et al., 2008). To date, more than 6,800 miRNAs in approximately 62 plant species have been identified (miRBase, release 20.0, June 2013; Ambros et al., 2003; Griffiths-Jones et al., 2008). However, only a very limited number of miRNAs have been functionally characterized.
There are two reciprocal reverse genetic strategies to investigate the function of a particular miRNA (Jones-Rhoades et al., 2006). One is to enhance miRNA activity through transgenic overexpression of the miRNA in plants. The other is to block miRNA function, which can be accomplished by either identifying an individual mutant of a miRNA gene (Allen et al., 2007) or expressing a miRNA-resistant target, with silent mutations being introduced to avoid changing the encoded amino acids (Zhao et al., 2007). Recently, several alternative approaches have been developed for functional validation of miRNAs in plants, including miRNA target mimicry (Franco-Zorrilla et al., 2007), short tandem target mimic (STTM; Yan et al., 2012), transcriptional gene silencing of miRNA gene promoters (Vaistij et al., 2010), and artificial miRNA-directed silencing of miRNA precursors (Eamens et al., 2011). However, all the aforementioned approaches rely on time-consuming and costly processes to generate the stable transgenic plants, which limits their utility for high-throughput analysis.
Among various miRNA inhibition approaches, miRNA target mimicry and STTM have received more attention. The miRNA target mimicry was first reported to establish the mechanism of inhibition of miR399 activity by the non-protein-coding RNA INDUCED BY PHOSPHATE STARVATION1 (IPS1) in response to low inorganic phosphate (Franco-Zorrilla et al., 2007). The IPS1 mRNA contains a 23-nucleotide sequence partially complementary to miR399 with a three-nucleotide mismatch at the expected miRNA cleavage site of miR399. Because of the mismatched bulge region, IPS1 functions as a noncleavable target mimic of miR399 that sequesters miR399 and arrests its cleavage activity to the target PHOSPHATE2 mRNA. By replacing the miR399 bulged target of the IPS1 transcript with other miRNA target-mimic sequences, the noncleavable miRNA target mimics can be exploited to inhibit miRNAs other than miR399. Based on this strategy, two other miRNAs, miR156 and miR319, were sequestered by their target mimics (Franco-Zorrilla et al., 2007). Furthermore, target mimics of 15 out of the 75 miRNA families (20%) caused reproducible developmental defects in aerial tissues when expressed in transgenic Arabidopsis (Arabidopsis thaliana) plants (Todesco et al., 2010). On the other hand, the newly developed STTM technology is an important approach to block miRNA function (Yan et al., 2012). STTM consists of two mimicking small RNA target sequences separated by a 48- to 88-nucleotide artificially designed linker, whose expression leads to the degradation of targeted small RNAs by small RNA-degrading nucleases (Yan et al., 2012).
Plant viral vectors have been widely used for transient gene expression and for gene silencing in plants (Lu et al., 2003; Senthil-Kumar and Mysore, 2011; Hefferon, 2012). Viral vector-based techniques do not require the time-consuming procedure of generating stable transgenic plants, while they allow the characterization of phenotypes that might be lethal in stable transgenic lines. Therefore, these technologies have the potential to become an attractive and quick approach to uncover miRNA functions in plants, especially in those difficult for genetic transformation. Indeed, we have developed a cabbage leaf curl virus-based vector for the overexpression of miRNAs in plants (Tang et al., 2010). However, virus-based miRNA inactivation has not been reported.
Tobacco rattle virus (TRV) is a bipartite positive sense RNA virus that can infect a broad range of plants (MacFarlane et al., 1999). TRV-based vectors (Liu et al., 2002b) have been widely applied as virus-induced gene silencing (VIGS) to knock down gene expression in various plant species (Bachan and Dinesh-Kumar, 2012), and they have also been successfully modified for the expression of foreign genes in plants (MacFarlane and Popovich, 2000). TRV, like all successful viruses, can escape host RNA interference defense and infect host plants systemically because it encodes two weak gene-silencing suppressors (Martín-Hernández and Baulcombe, 2008; Deng et al., 2013). However, TRV induces only very mild symptoms in many host plants (Ratcliff et al., 2001), and it does not cause global deregulation of the miRNA regulatory pathway (Martínez-Priego et al., 2008). In this study, we modified the TRV vector (Liu et al., 2002b) into a TRV-based T-DNA expression vector. Furthermore, we developed a virus-based microRNA silencing (VbMS) system in which TRV-based expression of miRNA target mimics can effectively suppress endogenous miRNA activity in plants within a short period of time.
RESULTS
Development of a TRV-Based Expression Vector
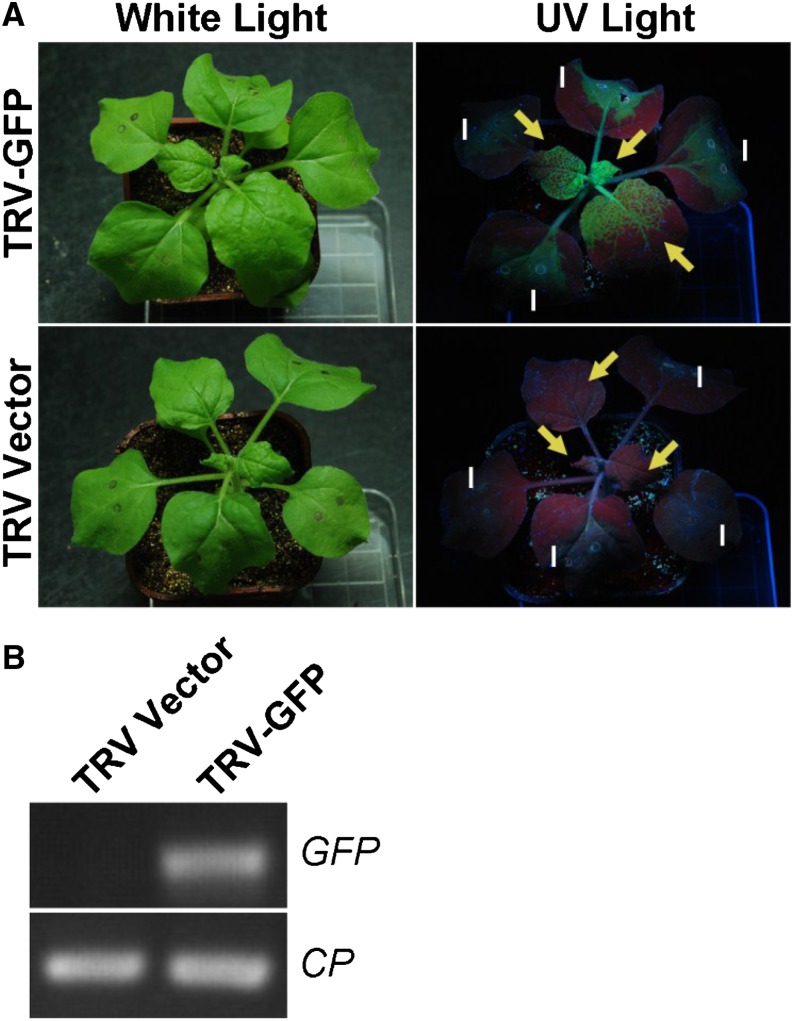
TRV vectors have been used for foreign gene expression by adding a fragment carrying the coat protein (CP) gene subgenomic promoter isolated from the pea early brown virus (PEBV) RNA genome (MacFarlane and Popovich, 2000). Here, we modified the TRV RNA2-derived vector pYL170 (Dong et al., 2007) by inserting the PEBV CP subgenomic promoter (Wang et al., 1997) and the ccdB (for control of cell death B) gene with ligation-independent cloning (LIC) adaptor sequences immediately downstream of the TRV CP gene to generate the TRV expression vector pTRV2e (Fig. 1). This vector can be used to express RNAs such as IPS1-based miRNA target mimics (MIM), STTM, and foreign protein-encoding genes as outlined in Figure 1. To assess whether pTRV2e can be used for gene expression in plants, we cloned a GFP coding sequence into pTRV2e to generate pTRV-GFP. When plants were infiltrated with Agrobacterium tumefaciens carrying pTRV-GFP and pTRV1 (Fig. 1), GFP fluorescence was visible in the upper noninoculated leaves at 4 d post inoculation (dpi), suggesting that the modified TRV vector can be used to express foreign genes in plants (Fig. 2A). Reverse transcription (RT)-PCR analysis further confirmed GFP expression (Fig. 2B). In addition, TRV-GFP RNA was detected in the infected plants undergoing vegetative to reproductive growth (data not shown). Moreover, the infected plants were symptomless or only showed extremely mild symptoms in Nicotiana benthamiana and tomato (Solanum lycopersicum) plants.
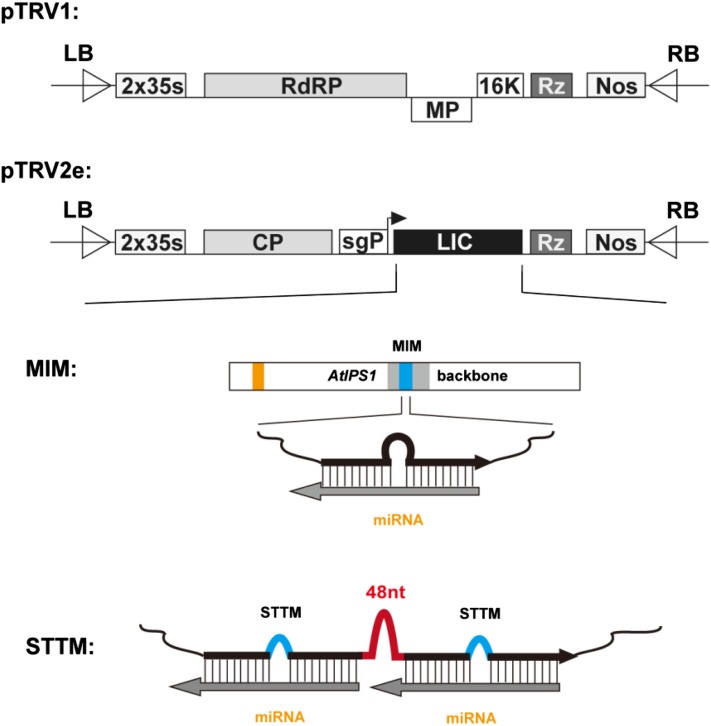
Figure 1.
Schemes of TRV-based VbMS vectors. pTRV1 is the TRV RNA1 T-DNA vector. pTRV2e is a PEBV CP subgenomic promoter (sgP)-containing TRV RNA2 T-DNA vector; the complementary DNA of TRV RNA2 is cloned between a cauliflower mosaic virus 35S promoter with the duplicated enhancers (2x35S) and NOS terminator (Nos). pTRV2e contains the LIC cassette for the insertion of the target gene sequences. LB, Left border of T-DNA; RdRP, RNA-dependent RNA polymerase; MP, movement protein; 16K, 16-kD Cys-rich protein; Rz, self-cleaving ribozyme; RB, right border of T-DNA. IPS1-based MIM and STTM sequences can be cloned into pTRV2e by the LIC reaction. MIM contains an AtIPS1 backbone, but the target mimic motif of miR399 is changed to that of corresponding miRNAs. STTM contains two tandem target mimics separated by a 48-nucleotide imperfect stem-loop linker (48nt). [See online article for color version of this figure.]
Figure 2.
Visualization of GFP expressed by the modified TRV vector in N. benthamiana. A, The TRV-GFP-infiltrated plants were photographed at 4 d post inoculation under white light (left) or UV illumination (right). Arrows indicate the upper uninfiltrated leaves, and “I” indicates the infiltrated leaves. Green color under UV light indicates the GFP signal. B, RT-PCR detection of TRV RNA in upper uninfiltrated leaves. RNA samples were extracted from TRV-GFP and TRV control plants, and RT-PCR was performed with GFP- and TRV CP- specific primers. [See online article for color version of this figure.]
VbMS of miR172 Caused Flower Developmental Defects in N. benthamiana
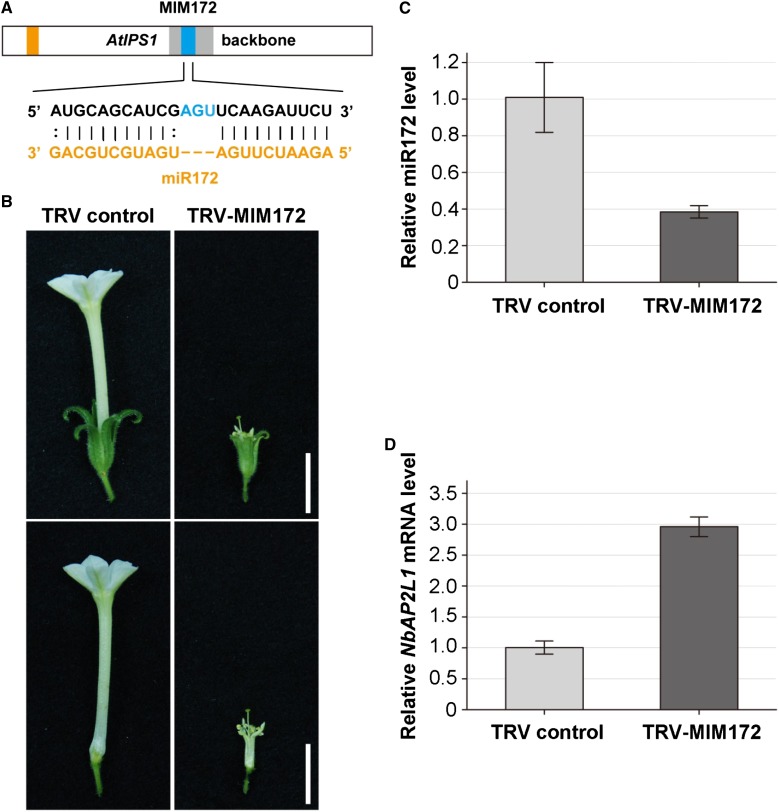
In Arabidopsis and N. benthamiana, miR172 regulates the expression of the floral homeotic gene APETALA2 (AP2). The AP2/miR172 regulatory circuit is conserved in higher plant species (Chen, 2004; Mlotshwa et al., 2006; Chuck et al., 2008; Zhu et al., 2009). Overexpression of miR172-resistant AP2 resulted in severe defects in floral patterning due to overaccumulated AP2 mRNA and protein (Chen, 2004; Mlotshwa et al., 2006). To test whether TRV-based VbMS via a miRNA target mimic can suppress miRNA activity, we used the modified TRV vector to express an IPS1-based target mimic against miR172 (MIM172) in N. benthamiana (Fig. 3A). In each plant expressing MIM172, more than one-third of the flowers had dramatically reduced petal size compared with flowers in controls (Fig. 3B, right column). The shortened petals could not cover the androecium and anthers extending out of the fringe of petals. In contrast, all flowers in control TRV plants showed normal developmental patterns (Fig. 3B, left column). These phenotypes were exactly the same as those observed in transgenic N. benthamiana lines overexpressing miR172-resistant AP2 (Mlotshwa et al., 2006). Furthermore, RT-PCR assays indicated that MIM172 was expressed in TRV-MIM172 plants with flower developmental defects (Supplemental Fig. S1A). Stem-loop RT-PCR assays indicated that the miR172 level was reduced in TRV-MIM172 plants (Fig. 3C). It is known that NbAP2-LIKE1 (NbAP2L1) is the target of miR172 in N. benthamiana (Mlotshwa et al., 2006). Thus, we used real-time RT-PCR to analyze the mRNA level of NbAP2L1. Indeed, the level of the NbAP2L1 mRNA was significantly higher in MIM172-expressing plants than in controls (Fig. 3D).
Figure 3.
VbMS of miR172 using ISP1-based miRNA target mimicry caused flower defects in N. benthamiana. A, Diagrammatic representation of MIM172. B, The flowers of plants infected with TRV control (left) and with TRV-MIM172 (right) were photographed at 12 d post flowering. Shown are typical flowers with sepal (top row) and sepal removed (bottom row). Bars = 1 cm. C, Stem-loop RT-PCR detection of miR172 level in plants infected with TRV control and with TRV-MIM172. D, Real-time RT-PCR analysis of mRNA levels of miR172 target NbAP2L1 in TRV control and plants expressing MIM172. Error bars show sd. [See online article for color version of this figure.]
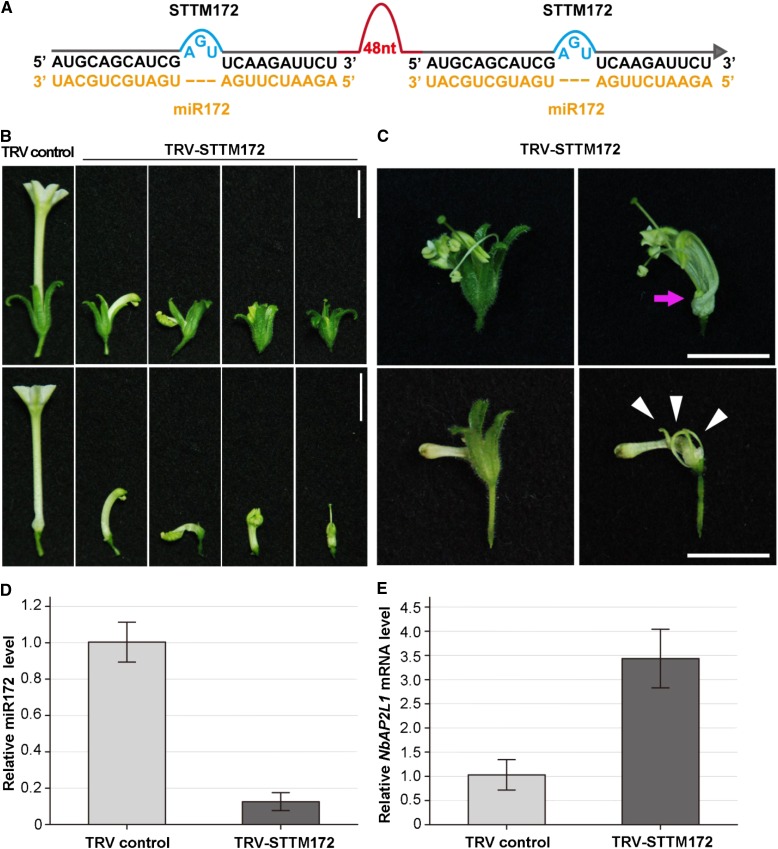
Furthermore, we tested whether VbMS can inhibit miRNA activity by TRV-based STTM expression. For this purpose, we generated STTM against miR172 (STTM172) and cloned it into pTRV2e to generate TRV-STTM172 (Fig. 4A). Similar to TRV-MIM172 plants, about one-third of flowers of TRV-STTM172-inoculated plants had short petals with varied morphologic patterns (Fig. 4B). In certain extremes, flowers in plants infected with TRV-STTM172 developed extra petals or unclosed petals that could not enclose the interior stamens and carpels (Fig. 4C), suggesting that TRV-based VbMS of miR172 using the STTM approach was very effective to suppress normal miR172 function and caused many abnormal flower phenotypes. Accompanied with viral expression of STTM172 (Supplemental Fig. S1B), miR172 level was lower (Fig. 4D) and the mRNA level of NbAP2L1 was higher (Fig. 4E) in STTM172-expressing plants than in controls. Furthermore, defective flowers were observed throughout the flowering periods.
Figure 4.
VbMS of miR172 using the STTM approach caused varied floral defects in N. benthamiana. A, Diagram of STTM172. 48nt, 48-nucleotid imperfect stem-loop linker. B, The varied morphology of flowers caused by TRV-based expression of STTM172. Bars = 1 cm. Each flower was photographed with sepal (top row) and sepal removed (bottom row). C, In some TRV-STTM172-infected plants, there are abnormal petals that could not enclose interior flower organs (top row, arrow) and ectopic genesis of petal-like tissues (bottom row, arrowheads). Bars = 1 cm. D, Stem-loop RT-PCR detection of miR172 level in plants infected with TRV control and with TRV-STTM172. E, Real-time RT-PCR analysis of miR172 target NbAP2L1 in VbMS plants. Error bars show sd. [See online article for color version of this figure.]
Taken together, these results suggest that TRV-based VbMS using either MIM or STTM can effectively suppress miRNA function in N. benthamiana.
VbMS of miR319 Caused Smaller Leaves and a Simpler Leaf Pattern in Tomato Plants
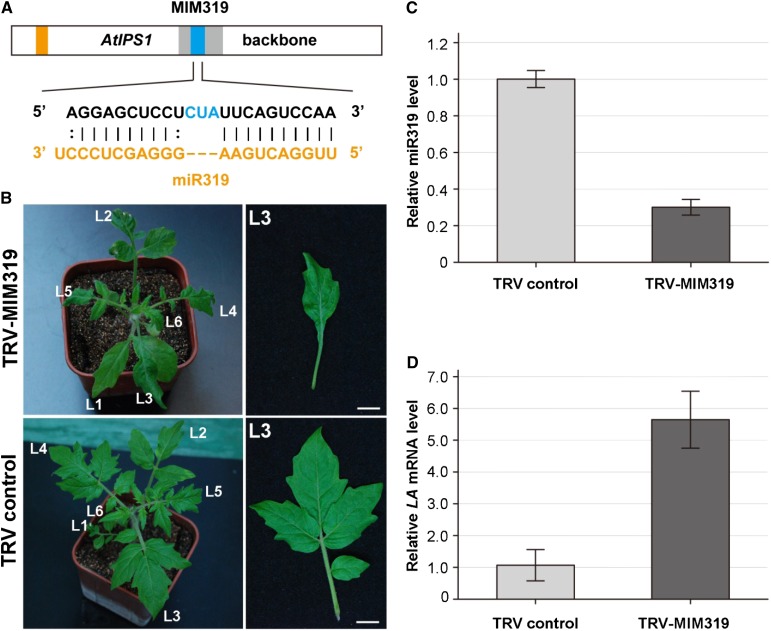
In tomato, miR319/LANCEOLATE (LA) is a well-defined miRNA/target pair, and misregulation of LA by miR319 led to a distinguishable phenotype in aerial organs (Ori et al., 2007). LA encodes a TEOSINTE BRANCHED/CYCLOIDEA/PROLIFERATING CELL FACTORS family transcription factor whose mRNA sequence contains a miR319-binding site. The dominant La mutant with mutation in the miR319-binding sequence conferred partial resistance against miR319-directed inhibition and led to elevated accumulation of LA protein, converting large compound leaves into small simple ones in tomato plants (Ori et al., 2007). To investigate whether VbMS works in tomato, we used the modified TRV vector to express an IPS1-based target mimic against miR319 (MIM319; Fig. 5A). Approximately 20% of TRV-MIM319 plants showed a similar phenotype to La mutant lines (Ori et al., 2007), with large compound leaves converted into small simple ones and reduced size of whole plants (Fig. 5B). RT-PCR confirmed that MIM319 was expressed in TRV-MIM319 plants (Supplemental Fig. S1C). Furthermore, the miR319 level was lower (Fig. 5C) and the LA mRNA level was obviously higher (Fig. 5D) in plants expressing MIM319 than in controls. We monitored the VbMS for more than 3 months, and the TRV-MIM319 plants continuously developed simplified leaves (Supplemental Fig. S2), indicating that VbMS of miR319 had an enduring impact on tomato leaf development.
Figure 5.
VbMS of miR319 using an ISP1-based target mimicry approach converted large compound leaves into small simple ones in tomato. A, Diagram of MIM319. B, The plants (left column) and the third leaf excised from the left-sided plants (right column) of TRV control (bottom row) and plants expressing MIM319 (top row) were photographed at 20 dpi. Leaf orders are indicated with numbers. Bars = 1 cm. C, Stem-loop RT-PCR detection of miR319 level in plants infected with TRV control and with TRV-MIM319. D, Real-time RT-PCR analysis of miRNA levels of the miR319 target LA in TRV and plants expressing MIM319. Error bars show sd. [See online article for color version of this figure.]
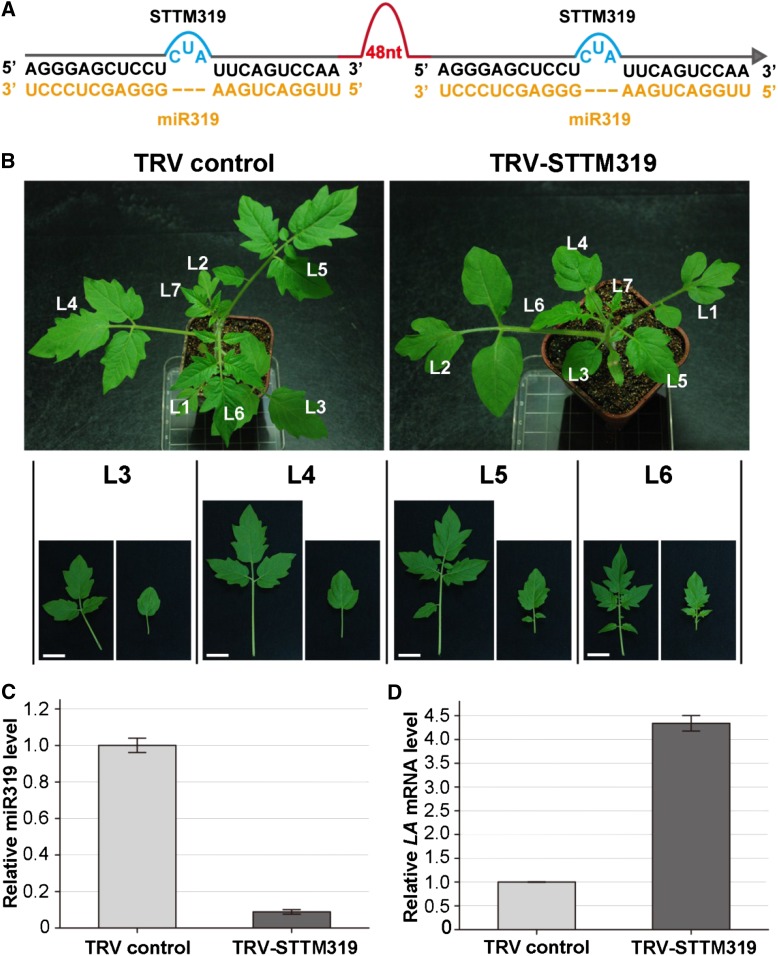
We also tested whether VbMS can inhibit miR319 activity by TRV-based expression of STTM targeting miR319 (STTM319; Fig. 6A). More than 30% of tomato plants infected with TRV-STTM319 showed a range of leaf simplification with reduced or no leaflets (Fig. 6B). RT-PCR confirmed that STTM319 was expressed in TRV-STTM319 plants (Supplemental Fig. S1D). Furthermore, the miR319 level was lower (Fig. 6C) and the LA mRNA level was evidently higher (Fig. 6D) in plants expressing STTM319 than in controls.
Figure 6.
VbMS of miR319 using the STTM approach caused smaller and simpler leaves in tomato plants. A, Diagram of STTM319. 48nt, 48-nucleotid imperfect stem-loop linker. B, The whole stature and the third to sixth leaves of plants infected with TRV (left column) and with TRV-STTM319 (right column) were photographed at 10 dpi. L1 to L7 indicate leaves 1 to 7. Bars = 1 cm. C, Stem-loop RT-PCR detection of miR319 level in plants infected with TRV control and with TRV-STTM319. D, Real-time RT-PCR analysis of miR319 target LA. Error bars show sd. [See online article for color version of this figure.]
Taken together, our results indicate that VbMS using either MIM or STTM can inhibit miRNA function in tomato.
VbMS of miR165/166 Reduced Apical Dominance in N. benthamiana
We have shown that VbMS can inhibit miRNA function quickly in N. benthamiana and tomato. To determine whether VbMS can be used to investigate the function of yet uncharacterized miRNAs, we performed VbMS of miR165/166 in N. benthamiana plants. The class III homeodomain-leucine zipper (HD-ZIP III) transcription factors have been clearly defined as miR165/166 target sets in Arabidopsis. miR165/166 target and repress the expression of the HD-ZIP III members, determining the behavior of the shoot apical meristem and the polarity of leaves (McConnell and Barton, 1998; Mallory et al., 2004; Kim et al., 2005; Jung and Park, 2007; Sakaguchi and Watanabe, 2012). In Nicotiana sylvestris, the ortholog of the HD-ZIP III transcription factor PHAVOLUTA was shown to be regulated by miR165/166-directed cleavage of its mRNA (McHale and Koning, 2004). miR165/166 also have been predicted and detected in N. benthamiana (Li et al., 2012). However, their exact function has not been characterized.
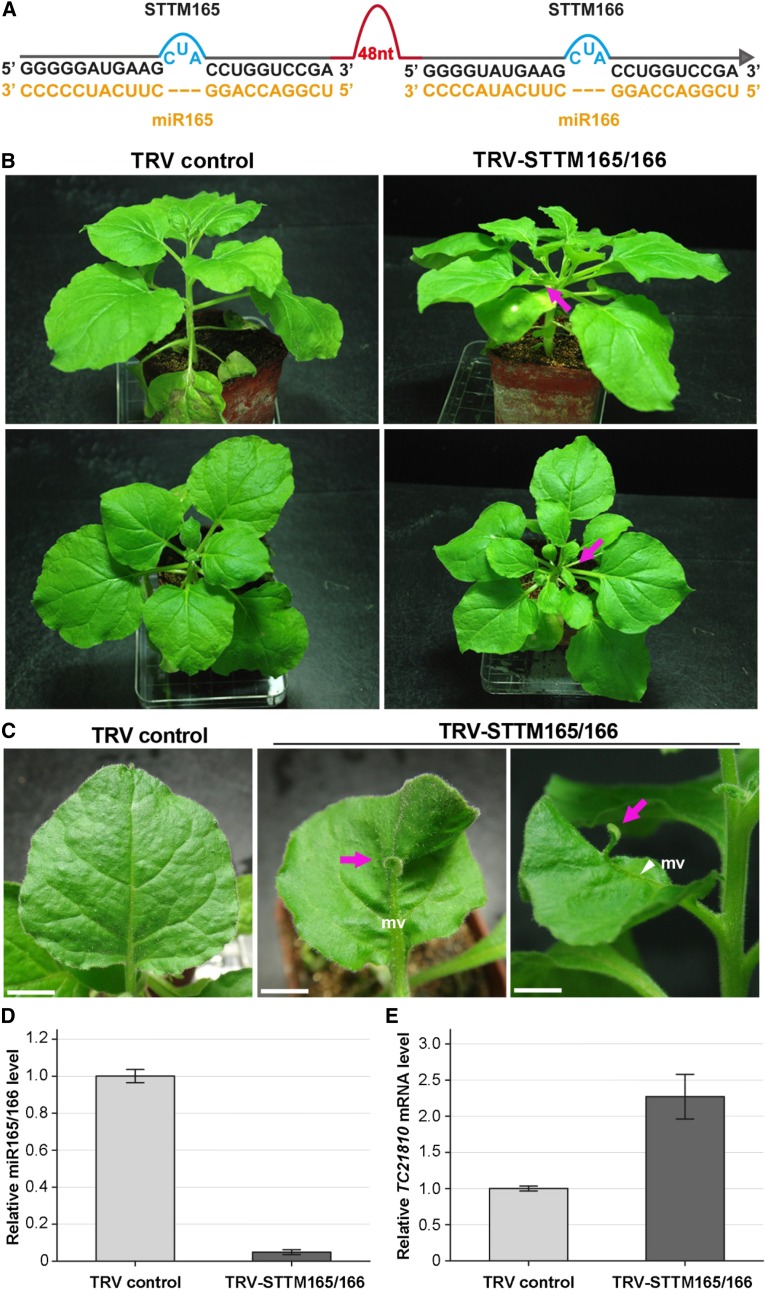
To determine the function of miR165/166, we performed TRV-based expression of STTM targeting miR165/166 (STTM165/166; Fig. 7A) in N. benthamiana plants. TRV control plants did not show any developmental defects. However, more than 20% of TRV-STTM165/166 plants showed reduced apical dominance and lacked the distinguishable main shoot (Fig. 7B). In certain extremes, ectopic leaf outgrowths on the leaf middle vein were observed (Fig. 7C, arrows). These observations reflected the disruption of apical meristermatic and leaf primordial functions caused by the inhibition of miR165/166 (Zhong and Ye, 2004; Yan et al., 2012). However, we did not observe obvious changes in leaf abaxial-adaxial polarity, as observed in Arabidopsis overexpressing STTM165/166 (Yan et al., 2012). RT-PCR confirmed that STTM165/166 was expressed in TRV-STTM165/166 plants (Supplemental Fig. S1E). Furthermore, the miR165/166 level was reduced in TRV-STTM165/166 plants (Fig. 7D). One of the HD-ZIP III transcription factors was predicted to be the miR165/166 cleavable target in N. benthamiana (TC21810; http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; N. benthamiana EST NbGI 4.0). Indeed, in TRV-STTM165/166 plants, the mRNA level of TC21810 was much higher compared with the controls (Fig. 7E). These results demonstrated that the TRV-based VbMS can be applied to study the function of uncharacterized miRNAs in plants, and there may be a conservation of the miR165/166-HD-ZIP III partner in plant species.
Figure 7.
VbMS of miR165/166 using the STTM approach caused the loss of apical dominance in N. benthamiana. A, Diagram of STTM165/166. 48nt, 48-nucleotid imperfect stem-loop linker. B, Whole plants infected with TRV-STTM165/166 (right column) or TRV (left column) were photographed at 28 dpi. Photographs were captured from top view (bottom row) and side view (top row). Arrows indicate the branched shoot apex. C, Outgrowth of an ectopic leaf. The ectopic leaf was photographed in front and side views. Arrows indicate ectopic leaf tissues, and “mv” and arrowhead indicate the middle vein. Bars = 1 cm. D, Stem-loop RT-PCR detection of miR165/166 level in plants infected with TRV control and with TRV-STTM165/166. E, Real-time RT-PCR analysis of relative mRNA levels of the putative miR165/166 target gene TC21810 in TRV control plants and TRV-STTM165/166 plants. Error bars show sd. [See online article for color version of this figure.]
DISCUSSION
In this study, we demonstrate that VbMS by TRV-based expression of miRNA target mimics can be used to block the function of miRNAs in plants. Using this system, we have successfully silenced miR172 in N. benthamiana and miR319 in tomato. VbMS of miR172 in N. benthamiana led to typical defects in flower organs, confirming that miR172 is a functionally conserved miRNA between tobacco and Arabidopsis. VbMS of miR319 in tomato reduced the complexity of tomato compound leaves. We also found that miR165/166 could regulate the development of the shoot apical meristem in N. benthamiana, which is consistent with their function in Arabidopsis (Eckardt, 2012; Yan et al., 2012). These results indicated the TRV-based VbMS by overexpressing miRNA target mimics was efficient to silence plant endogenous miRNAs. To the best of our knowledge, this is the first report of using a viral vector to investigate miRNA function by blocking miRNA activity in plants.
We found that only about 20% to 30% of plants expressing target mimics against miR319 and miR165/166 showed the expected phenotype. In addition, only about one-third of the flowers were defective in each plant expressing target mimics against miR172, although all plants had defective flowers. This could be caused by nonuniform and incomplete miRNA silencing, because TRV cannot infect 100% meristematic cells that finally divide and differentiate to form the tissues and organs of the plant. This is not surprising, because VIGS does not result in 100% uniform silencing (Liu et al., 2002a). There may be other unknown factors that contribute to this variability, because even different progeny from the same stable transgenic lines expressing target mimics gave various phenotypes (Todesco et al., 2010), and only about 60% of flowers exhibited defects in the transgenic plant lines expressing miR172-resistant AP2 (Mlotshwa et al., 2006).
Although transgenic plants expressing miR172-resistant AP2 show severe flower defects in both N. benthamiana and Arabidopsis (Chen, 2004; Mlotshwa et al., 2006), the flowers of transgenic plants expressing MIM172 are normal (Todesco et al., 2010). However, we found that TRV-based expression of either MIM172 or STTM172 caused abnormal flowers in N. benthamiana, suggesting that VbMS could be more effective to block miR172 function than stable transgene-expressing IPS1-based miRNA target mimics.
We found that VbMS of miR165/166 using STTM technology caused the loss of apical dominance. However, VbMS of miR165/166 using IPS1-based miRNA target mimics did not cause any visible phenotype in N. benthamiana (data not shown). These observations suggest that STTM is superior to IPS1-based miRNA target mimics at suppressing miRNA function through the viral approach. Indeed, disruption of miR165/166 function using the STTM approach also results in a more severe phenotype than that using an IPS1-based miRNA target mimic in transgenic Arabidopsis (Eckardt, 2012; Yan et al., 2012).
It should be noted that transgenic Arabidopsis plants expressing STTM165/166 exhibited severe loss of apical dominance and loss of leaf symmetry phenotypes (Yan et al., 2012). However, VbMS of miR165/166 using STTM technology caused the loss of apical dominance but no obvious changes in leaf symmetry in N. benthamiana. The phenotypic discrepancy caused by miR165/166 silencing between Arabidopsis and N. benthamiana is probably due to different miRNA silencing efficiencies between different approaches. It is possible that the STTM transgenic approach gives a stronger phenotype than virus-mediated STTM. Transgenic STTM blocks miRNAs in each cell of the transgenic plants from the very beginning without any difference between fast-dividing cells and developed cells, while the virus-mediated STTM has less of an effect in fast-dividing cells such as shoot apical meristems due to much diluted virus concentrations in fast-dividing cells. Thus, there may be a limitation of VbMS as a method to inhibit miRNA function due to its relatively weak phenotype. Nevertheless, VbMS is a great complementary method to the transgenic target mimics or STTM for a fast screening for functions of miRNAs.
Besides the aerial parts such as leaf and flower, TRV-based vectors are able to trigger VIGS in fruits (Hanania et al., 2007; Jia et al., 2011) and underground tissues (Valentine et al., 2004; Kaloshian, 2007) in various plant species. On the other hand, TRV-based vectors have been used to express foreign genes in systemically infected leaves (MacFarlane and Popovich, 2000) and in hairy roots (Larsen and Curtis, 2012). Furthermore, TRV-based expression has been successfully used in the functional characterization of scent-related genes, protein compartmentalization studies, and nontransgenic genome modification (Spitzer-Rimon et al., 2010, 2012, 2013). Therefore, TRV-based VbMS should also be able to inhibit miRNA in organs other than leaves and flowers.
Moreover, the currently described TRV-based VbMS possesses several advantages over other functional assays for plant miRNAs. First, VbMS is efficient and quick, and it usually can result in miRNA silencing-mediated phenotypes within 2 to 4 weeks. Second, VbMS does not require the tedious and time-consuming plant transformation procedure and only needs the simple agroinfitration technique for miRNA silencing. This is particularly useful for functional characterization of miRNAs whose knockout or knockdown might cause embryonic lethality in transgenic lines and for plant species that are not amenable to stable genetic transformation. Third, TRV has a wide host range, and TRV vectors have been applied in a wide range of plant species (Bachan and Dinesh-Kumar, 2012). Thus, TRV-based VbMS should be applicable for miRNA functional analysis in these plants.
Our finding also implies that many other available viral vectors, such as Apple latent spherical virus (Igarashi et al., 2009; Yamagishi et al., 2011), Brome mosaic virus (Ding et al., 2006), and Barley stripe mosaic virus (Holzberg et al., 2002), could be used in a similar strategy to the TRV VbMS vector for functional analysis of miRNAs in a diverse range of eudicot and monocot crops. In addition, besides IPS1-based miRNA target mimicry and STTM methods, several other techniques, such as transcriptional gene silencing of miRNA gene promoters (Vaistij et al., 2010), artificial miRNA-directed silencing of miRNA precursors (Eamens et al., 2011), and miRNA decoy (Ivashuta et al., 2011), have successfully been used to investigate miRNA/target interactions in transgenic plants. Given the performance of VbMS by TRV-based expression of miRNA target mimics, these miRNA-silencing approaches can be widely adapted to VbMS to elucidate small RNA functions in plants.
MATERIALS AND METHODS
Plasmid Construction
pTRV1 (pYL192) was described previously (Liu et al., 2002b). The pTRV2-derived expression vector pTRV2e was constructed as follows. The PEBV CP subgenomic promoter was amplified from a PEBV RNA2 vector (kindly provided by Daowen Wang). The ccdB gene with LIC adaptor sequences was amplified using pTRV2-LIC (Dong et al., 2007) as template. pTRV2e was obtained by cloning the digested DNA fragments of the PEBV CP subgenomic promoter and the ccdB gene containing LIC adaptor sequences into pYL170, a TRV RNA2 VIGS vector (Dong et al., 2007). pTRV1 can be obtained from the Arabidopsis Biological Resource Center (stock name is YL192), and pTRV2e will be available in the Arabidopsis Biological Resource Center (stock name is pTRV2e).
The GFP coding sequence was amplified from TMV-GFP (Liu et al., 2002b). Artificial miRNA target mimicry sequences were engineered into the IPS1 backbone by overlapping PCR as described (Franco-Zorrilla et al., 2007). A 48-nucleotide oligonucleotide was synthesized as a template for PCR amplification of STTM (Yan et al., 2012). Primers with LIC adaptor, corresponding target mimic of miRNA, and STTM optimal spacer were used to PCR amplify STTM molecules. The GFP, MIM, and STTM fragments were cloned into pTRV2e using the LIC protocol as described (Dong et al., 2007). All constructs were confirmed by DNA sequencing. Primers used in this study are listed in Supplemental Table S1.
Plant Growth, Agroinfiltration, and GFP Imaging
Nicotiana benthamiana and tomato (Solanum lycopersicum) plants (cv Moneymaker) were grown in pots at 25°C in growth chambers under a 16-h-light/8-h-dark cycle with 60% humidity. For TRV-based expression or VbMS, pTRV1 and pTRV2e or its derivatives were introduced into Agrobacterium tumefaciens strain GV3101 (An et al., 1988). A 5-mL culture was grown overnight at 28°C in the appropriate antibiotic selection medium, amplified in 50 mL of Luria-Bertani medium containing antibiotics, and grown overnight in a 28°C shaker. A. tumefaciens cells were harvested and resuspended in infiltration medium (10 mm MgCl2, 10 mm MES, and 200 μm acetosyringone), adjusted to an optical density at 600 nm of 1.0, and left at room temperature for 3 h before infiltration for N. benthamiana or adjusted to an optical density at 600 nm of 2.0 and incubated at room temperature for 6 h before infiltration for tomato. A. tumefaciens was infiltrated using a 1-mL syringe without needle into leaves of N. benthamiana before flowering or into cotyledons of tomato before true leaves emerged. The infiltrated plants were grown and observed until phenotypes appeared. For each VbMS construct, at least five plants were used for agroinfiltration and replicated at least six times. GFP imaging was illuminated under long-wavelength UV light, and photographs were taken using a digital camera.
RNA Isolation and RT-PCR Analysis
Total RNA was extracted from developing flowers (for VbMS of miR172), shoot apical tissues (for VbMS of miR165/166), and developing leaves (for VbMS of miR319) using the Trizol reagent (Invitrogen) and treated with RNase-free DNase I (Sigma-Aldrich). First-strand complementary DNA was synthesized using 2 to 5 μg of total RNA with oligo(dT) primer or TRV-specific primer and Moloney murine leukemia virus reverse transcriptase (Promega). Primers were designed with Primer Premier 9.0 and are listed in Supplemental Table S1.
Real-Time RT-PCR Analysis
Real-time RT-PCR was performed using Power SYBR Green PCR master mix (ABgene), and eIF4a and Tubulin were used as internal controls for N. benthamiana and tomato, respectively, for normalization. Stem-loop RT-PCR was performed as described with the SYBR Green assay (Varkonyi-Gasic and Hellens, 2011), and the miRNA complementary regions in reverse transcript primers were elongated to cross the cleavage site to exclude STTM contamination. Primers were designed with Primer Expression 3.0 (Applied Biosystems) and are listed in Supplemental Table S1. The values were calculated using the comparative normalized cycle threshold method, and all the experiments were repeated at least three times. Data were analyzed and plotted with Origin 8.1. Data shown are from at least three repeated experiments.
Sequence data from this article can be found in the GenBank data libraries under the following accession numbers: PEBV subgenomic promoter (NC_001368); TRV CP (AF406991); GFP (SCU87973); AtIPS1 (NM_180219); NbAP2L1 (CK287095); LA (EF091571); and tomato Tubulin (XM_004244485). The eIF4a sequence can be found in the Dana-Farber Cancer Institute N. benthamiana Gene Index under TC Annotator TC19454. miRNA sequence can be found in miRBase (http://www.mirbase.org).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR confirmation of TRV infection and expression of target mimics in VbMS plants.
Supplemental Figure S2. VbMS of miR319 caused developmental defects of tomato plants in later growth stages.
Supplemental Table S1. Primers used in vector construction and PCR analysis.
Acknowledgments
We thank Daowen Wang at the Institute of Genetics and Developmental Biology, Chinese Academy of Science, for providing PEBV vectors.
Glossary
- miRNA
microRNA
- STTM
short tandem target mimic
- TRV
Tobacco rattle virus
- VIGS
virus-induced gene silencing
- VbMS
virus-based microRNA silencing
- CP
coat protein
- PEBV
the pea early brown virus
- LIC
ligation-independent cloning
- dpi
days post inoculation
- RT
reverse transcription
- HD-ZIP III
class III homeodomain-leucine zipper
References
- Allen RS, Li J, Stahle MI, Dubroué A, Gubler F, Millar AA. (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104: 16371–16376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. (2003) A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G, Ebert PR, Mitra A, Ha SB (1988) Binary vectors. In SB Gelvin, RA Schilperoort, DP Verma, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp A3:1–A3:19 [Google Scholar]
- Bachan S, Dinesh-Kumar SP. (2012) Tobacco rattle virus (TRV)-based virus-induced gene silencing. Methods Mol Biol 894: 83–92 [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Chen X. (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S. (2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019 [DOI] [PubMed] [Google Scholar]
- Deng X, Kelloniemi J, Haikonen T, Vuorinen AL, Elomaa P, Teeri TH, Valkonen JP. (2013) Modification of Tobacco rattle virus RNA1 to serve as a VIGS vector reveals that the 29K movement protein is an RNA silencing suppressor of the virus. Mol Plant Microbe Interact 26: 503–514 [DOI] [PubMed] [Google Scholar]
- Ding XS, Schneider WL, Chaluvadi SR, Mian MA, Nelson RS. (2006) Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol Plant Microbe Interact 19: 1229–1239 [DOI] [PubMed] [Google Scholar]
- Dong Y, Burch-Smith TM, Liu Y, Mamillapalli P, Dinesh-Kumar SP. (2007) A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol 145: 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eamens AL, Agius C, Smith NA, Waterhouse PM, Wang MB. (2011) Efficient silencing of endogenous microRNAs using artificial microRNAs in Arabidopsis thaliana. Mol Plant 4: 157–170 [DOI] [PubMed] [Google Scholar]
- Eckardt NA. (2012) A new tool for investigating small RNA function. Plant Cell 24: 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Pang M, Agarwal V, Chen ZJ. (2008) Interspecies regulation of microRNAs and their targets. Biochim Biophys Acta 1779: 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanania U, Velcheva M, Or E, Flaishman M, Sahar N, Perl A. (2007) Silencing of chaperonin 21, that was differentially expressed in inflorescence of seedless and seeded grapes, promoted seed abortion in tobacco and tomato fruits. Transgenic Res 16: 515–525 [DOI] [PubMed] [Google Scholar]
- Hefferon KL. (2012) Plant virus expression vectors set the stage as production platforms for biopharmaceutical proteins. Virology 433: 1–6 [DOI] [PubMed] [Google Scholar]
- Holzberg S, Brosio P, Gross C, Pogue GP. (2002) Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J 30: 315–327 [DOI] [PubMed] [Google Scholar]
- Igarashi A, Yamagata K, Sugai T, Takahashi Y, Sugawara E, Tamura A, Yaegashi H, Yamagishi N, Takahashi T, Isogai M, et al. (2009) Apple latent spherical virus vectors for reliable and effective virus-induced gene silencing among a broad range of plants including tobacco, tomato, Arabidopsis thaliana, cucurbits, and legumes. Virology 386: 407–416 [DOI] [PubMed] [Google Scholar]
- Ivashuta S, Banks IR, Wiggins BE, Zhang Y, Ziegler TE, Roberts JK, Heck GR. (2011) Regulation of gene expression in plants through miRNA inactivation. PLoS ONE 6: e21330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H. (2008) Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett 582: 2679–2684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57: 19–53 [DOI] [PubMed] [Google Scholar]
- Jung JH, Park CM. (2007) MIR166/165 genes exhibit dynamic expression patterns in regulating shoot apical meristem and floral development in Arabidopsis. Planta 225: 1327–1338 [DOI] [PubMed] [Google Scholar]
- Kaloshian I. (2007) Virus-induced gene silencing in plant roots. Methods Mol Biol 354: 173–181 [DOI] [PubMed] [Google Scholar]
- Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Narry Kim V, et al. (2005) MicroRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 42: 84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JS, Curtis WR. (2012) RNA viral vectors for improved Agrobacterium-mediated transient expression of heterologous proteins in Nicotiana benthamiana cell suspensions and hairy roots. BMC Biotechnol 12: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. (2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002a) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Marathe R, Dinesh-Kumar SP. (2002b) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30: 415–429 [DOI] [PubMed] [Google Scholar]
- Lu R, Martin-Hernandez AM, Peart JR, Malcuit I, Baulcombe DC. (2003) Virus-induced gene silencing in plants. Methods 30: 296–303 [DOI] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. (2008) Stress-responsive microRNAs in Populus. Plant J 55: 131–151 [DOI] [PubMed] [Google Scholar]
- MacFarlane SA, Popovich AH. (2000) Efficient expression of foreign proteins in roots from tobravirus vectors. Virology 267: 29–35 [DOI] [PubMed] [Google Scholar]
- MacFarlane SA, Vassilakos N, Brown DJ. (1999) Similarities in the genome organization of tobacco rattle virus and pea early-browning virus isolates that are transmitted by the same vector nematode. J Gen Virol 80: 273–276 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP. (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23: 3356–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Priego L, Donaire L, Barajas D, Llave C. (2008) Silencing suppressor activity of the Tobacco rattle virus-encoded 16-kDa protein and interference with endogenous small RNA-guided regulatory pathways. Virology 376: 346–356 [DOI] [PubMed] [Google Scholar]
- Martín-Hernández AM, Baulcombe DC. (2008) Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J Virol 82: 4064–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. (1998) Leaf polarity and meristem formation in Arabidopsis. Development 125: 2935–2942 [DOI] [PubMed] [Google Scholar]
- McHale NA, Koning RE. (2004) MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 16: 1730–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa S, Yang Z, Kim Y, Chen X. (2006) Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol Biol 61: 781–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, et al. (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39: 787–791 [DOI] [PubMed] [Google Scholar]
- Phillips JR, Dalmay T, Bartels D. (2007) The role of small RNAs in abiotic stress. FEBS Lett 581: 3592–3597 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC. (2001) Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Sakaguchi J, Watanabe Y. (2012) miR165⁄166 and the development of land plants. Dev Growth Differ 54: 93–99 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS. (2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16: 656–665 [DOI] [PubMed] [Google Scholar]
- Shukla LI, Chinnusamy V, Sunkar R. (2008) The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta 1779: 743–748 [DOI] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Cna’ani A, Vainstein A. (2013) Virus-aided gene expression and silencing using TRV for functional analysis of floral scent-related genes. Methods Mol Biol 975: 139–148 [DOI] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Farhi M, Albo B, Cna’ani A, Ben Zvi MM, Masci T, Edelbaum O, Yu Y, Shklarman E, Ovadis M, et al. (2012) The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 24: 5089–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B, Marhevka E, Barkai O, Marton I, Edelbaum O, Masci T, Prathapani NK, Shklarman E, Ovadis M, Vainstein A. (2010) EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. Plant Cell 22: 1961–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. (2007) Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci 12: 301–309 [DOI] [PubMed] [Google Scholar]
- Tang Y, Wang F, Zhao J, Xie K, Hong Y, Liu Y. (2010) Virus-based microRNA expression for gene functional analysis in plants. Plant Physiol 153: 632–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. (2010) A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet 6: e1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Elias L, George GL, Jones L. (2010) Suppression of microRNA accumulation via RNA interference in Arabidopsis thaliana. Plant Mol Biol 73: 391–397 [DOI] [PubMed] [Google Scholar]
- Valentine T, Shaw J, Blok VC, Phillips MS, Oparka KJ, Lacomme C. (2004) Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol 136: 3999–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Hellens RP. (2011) Quantitative stem-loop RT-PCR for detection of microRNAs. Methods Mol Biol 744: 145–157 [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Vazquez F, Crété P, Bartel DP. (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, MacFarlane SA, Maule AJ. (1997) Viral determinants of pea early browning virus seed transmission in pea. Virology 234: 112–117 [DOI] [PubMed] [Google Scholar]
- Yamagishi N, Sasaki S, Yamagata K, Komori S, Nagase M, Wada M, Yamamoto T, Yoshikawa N. (2011) Promotion of flowering and reduction of a generation time in apple seedlings by ectopical expression of the Arabidopsis thaliana FT gene using the apple latent spherical virus vector. Plant Mol Biol 75: 193–204 [DOI] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G. (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289: 3–16 [DOI] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X. (2007) miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J 51: 840–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. (2004) amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol 45: 369–385 [DOI] [PubMed] [Google Scholar]
- Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA. (2009) Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol 9: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]