A basic leucine zipper family transcription factor is an important transcriptional regulator of abscisic acid-dependent grape berry ripening.
Abstract
In grape (Vitis vinifera), abscisic acid (ABA) accumulates during fruit ripening and is thought to play a pivotal role in this process, but the molecular basis of this control is poorly understood. This work characterizes ABSCISIC ACID RESPONSE ELEMENT-BINDING FACTOR2 (VvABF2), a grape basic leucine zipper transcription factor belonging to a phylogenetic subgroup previously shown to be involved in ABA and abiotic stress signaling in other plant species. VvABF2 transcripts mainly accumulated in the berry, from the onset of ripening to the harvesting stage, and were up-regulated by ABA. Microarray analysis of transgenic grape cells overexpressing VvABF2 showed that this transcription factor up-regulates and/or modifies existing networks related to ABA responses. In addition, grape cells overexpressing VvABF2 exhibited enhanced responses to ABA treatment compared with control cells. Among the VvABF2-mediated responses highlighted in this study, the synthesis of phenolic compounds and cell wall softening were the most strongly affected. VvABF2 overexpression strongly increased the accumulation of stilbenes that play a role in plant defense and human health (resveratrol and piceid). In addition, the firmness of fruits from tomato (Solanum lycopersicum) plants overexpressing VvABF2 was strongly reduced. These data indicate that VvABF2 is an important transcriptional regulator of ABA-dependent grape berry ripening.
Grape (Vitis vinifera) is a nonclimacteric fruit that is important worldwide for wine production and fresh consumption. The ripening of grape berry is a complex process involving the catabolism of organic acids, the accumulation of soluble sugars, flavonoids, and aromatic compounds, and an increase in berry softness (Seymour et al., 1993; Ribéreau-Gayon et al., 2000). Berry composition and quality largely depend on the processes that coordinate these biochemical, physiological, and anatomical changes during ripening. While major progress has been made in the understanding of the key mechanisms supporting ethylene-mediated ripening of climacteric fruits (e.g. tomato [Solanum lycopersicum]; Adams-Phillips et al., 2004; Giovannoni, 2004, 2007), the events controlling the ripening of nonclimacteric fruits are less investigated and known.
Grape berry ripening involves the integration of multiple hormone signals. Classical plant hormones such as abscisic acid (ABA), auxin (indole-3-acetic acid [IAA]), brassinosteroids, and, to a lesser extent, ethylene have previously been implicated in this process (Davies et al., 1997; Chervin et al., 2004, 2008; Symons et al., 2006). ABA content of grape berries gradually and strongly increases just before the onset of ripening, called véraison (Coombe and Hale, 1973; Scienza et al., 1978; Davies et al., 1997; Antolín et al., 2003; Deluc et al., 2009). In relation with its accumulation profile, numerous reports suggested that ABA may play a major role in controlling several ripening-associated processes of grape berry, including coloration, sugar accumulation, and softening (Coombe, 1992; Davies et al., 1997; Giovannoni, 2001; Rodrigo et al., 2003; Yu et al., 2006; Wheeler et al., 2009; Gambetta et al., 2010; Giribaldi et al., 2010; Gagné et al., 2011).
In recent years, much progress has been made in the understanding of ABA signal transduction pathways in Arabidopsis (Arabidopsis thaliana; Umezawa et al., 2009; Cutler et al., 2010; Weiner et al., 2010). The ABA signal is perceived by multiple receptors (Shen et al., 2006; Liu et al., 2007; Fujii et al., 2009; Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Pandey et al., 2009; Santiago et al., 2009; Shang et al., 2010; Chai et al., 2011; Jia et al., 2011; Sun et al., 2011), which trigger downstream signaling cascades resulting in physiological effects. Among these ABA receptors, only the PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) proteins have been well characterized. ABA promotes the interaction of its receptor (PYR1) to PROTEIN PHOSPHATASE2C (PP2C), which results in the inactivation of PP2C and the activation of SUCROSE NONFERMENTING-RELATED KINASE2 (SnRK2). Activated SnRK2 turns on ABA signaling through the phosphorylation of downstream targets such as AREB/ABF (for ABA-response element-binding factor) transcription factors, which in turn activate several sets of genes (Fujii et al., 2009).
AREB/ABFs are ABA-responsive transcription factors containing a basic leucine zipper family (bZIP)-type DNA-binding domain that binds the ABA-responsive element (T/CACGTGGC) and have a pivotal role in ABA-dependent gene activation (Choi et al., 2000; Uno et al., 2000; Hattori et al., 2002; Kang et al., 2002; Gómez-Porras et al., 2007). Among the AREB/ABFs, AREB1/ABF2, AREB2/ABF4, and ABF3 were shown to be master transcription factors that cooperatively regulate ABA response element (ABRE)-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation (Yoshida et al., 2010).
Although the ABA regulation of fleshy fruit development has been studied extensively, the molecular mechanisms underlying ABA perception and signal transduction in these fruits remain unclear. In this study, we characterized and functionally studied VvABF2, an AREB/ABF-like transcription factor from grape, previously described as GRAPE RIPENING-INDUCED PROTEIN55 (GRIP55; Davies and Robinson, 2000), which accumulates in cv Cabernet Sauvignon berries from véraison until the end of the ripening phase. VvABF2 expression is induced by ABA. A transcriptomic analysis made on transgenic grape cells overexpressing VvABF2 treated or not with ABA led to the identification of putative target genes for VvABF2 mediated by ABA-dependent or -independent pathways. VvABF2 overexpression in grape cells also strongly increased the accumulation of stilbenes. Its overexpression in tomato accelerated fruit ripening. Altogether, this work shows that VvABF2 is involved in the ABA signaling pathway and may affect grape berry ripening by activating several processes, including the synthesis of some phenolic compounds and fruit softening.
RESULTS
Identification of VvABF2, a bZIP Transcription Factor from Grape
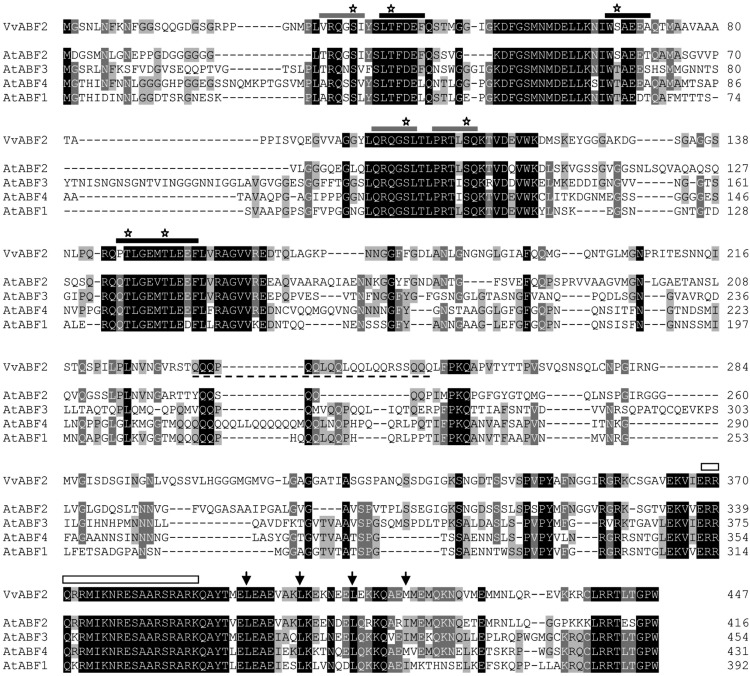
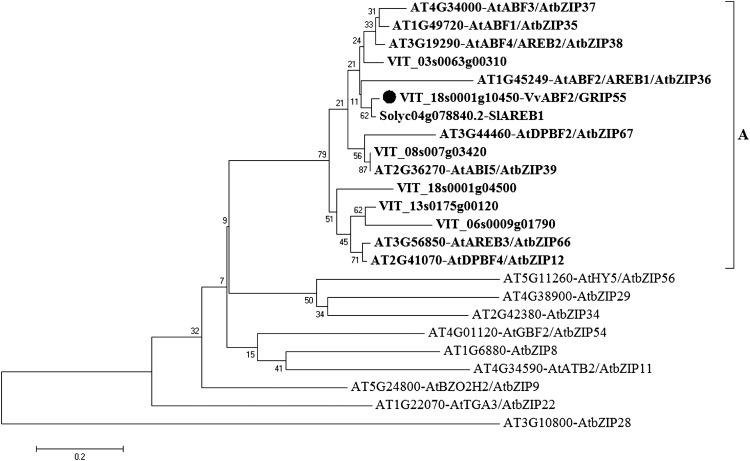
Data from the literature describing changes in mRNA profiles during grape berry ripening led to the identification of a number of GRIP complementary DNAs (cDNAs) whose transcripts accumulate during berry development (Davies and Robinson, 2000). Among these GRIP genes, GRIP55 (VIT_18s0001g10450, Q9M4H1), a transcription factor of the bZIP family, was selected for further analysis. This choice was driven by the fact that members from this family are responsive to ABA (Choi et al., 2000; Uno et al., 2000; Amir Hossain et al., 2010), a hormone playing a crucial role in grape berry development and ripening (Giribaldi et al., 2010; Koyama et al., 2010). The GRIP55 full-length cDNA was amplified by PCR using mRNAs extracted from cv Cabernet Sauvignon grape berries at véraison. The corresponding transcript is 1,341 bp long and encodes a protein of 447 amino acids. Amino acid sequence analysis further confirmed that this protein belongs to the transcription factors of the bZIP family characterized by a typical DNA basic-binding region, a Leu zipper dimerization motif located at the C-terminal region (amino acids 367–418; Jakoby et al., 2002), and conserved domains predicted as phosphorylation sites involved in stress or ABA signaling (Furihata et al., 2006; Fig. 1). A phylogenetic analysis revealed that this protein belongs to group A of bZIP transcription factors, previously shown to be involved in ABA and abiotic stress signaling (Choi et al., 2000; Uno et al., 2000; Amir Hossain et al., 2010; Fig. 2). Compared with Arabidopsis, bZIP group A from grape contains six members, among which only two belong to the possible groups of orthologs A5 (Corrêa et al., 2008; Fig. 1). The close homology of GRIP55 with AtAREB1/AtABF2 from Arabidopsis led us to rename this protein as VvABF2 (Figs. 1 and 2).
Figure 1.
Sequence analysis of VvABF2. Full-length sequence comparison of VvABF2 and its closest orthologs from Arabidopsis, AtABF2 (AF093445), AtABF3 (AF093546), AtABF4 (AF093547), and AtABF1 (AF093544), using the Clustal Omega program (Sievers et al., 2011). Conserved residues are shaded in black, conserved residues in four out of five of the sequences are indicated in dark gray shading, and conserved residues in three out of five of the sequences are shown by a light gray shading. The basic regions and the Leu repeats are indicated by white rectangles and arrows, respectively. The Gln-rich region, commonly found in transcriptional activation domains, is underlined (dashed line) for VvABF2. The recognition sites for calmodulin-dependent protein kinase II (XRXXSX) and casein kinase II [X(S/T)XX(D/E)X] are indicated, respectively, by gray and black lines on the top of the alignment. Putative phosphorylated amino acids in the VvABF2 sequence are marked by stars.
Figure 2.
Phylogenetic analysis of VvABF2. The phylogenetic tree represents VvABF2 (black circle) and its orthologs (boldface) from the A subgroup of bZIP transcription factors in Arabidopsis (AT) and grape (VIT). The closest ortholog of VvABF2 from tomato (SlAREB1; Bastías et al., 2011) and representative Arabidopsis bZIP transcription factors from other bZIP subgroups (subgroup G, AtGBF2; H, AtHY5; I, AtbZIP29; E, AtbZIP34; D, AtTGA3; S, AtATB2; B, bZIP28; and C, AtBZO2H2) are also reported according to Jakoby et al. (2002) and Corrêa et al. (2008). The gene identifiers as well as synonyms (published names) are given. Multiple sequence alignments were generated using Clustal Omega (Sievers et al., 2011) on full-length proteins as implemented by MEGA software, version 5.0 (Tamura et al., 2011). The phylogenetic tree was constructed by neighbor joining with complete deletions as implemented by MEGA. Reliability values at each branch represent bootstrap samples (2,000 replicates).
Expression Analysis of VvABF2 in Grapevine and in Response to ABA
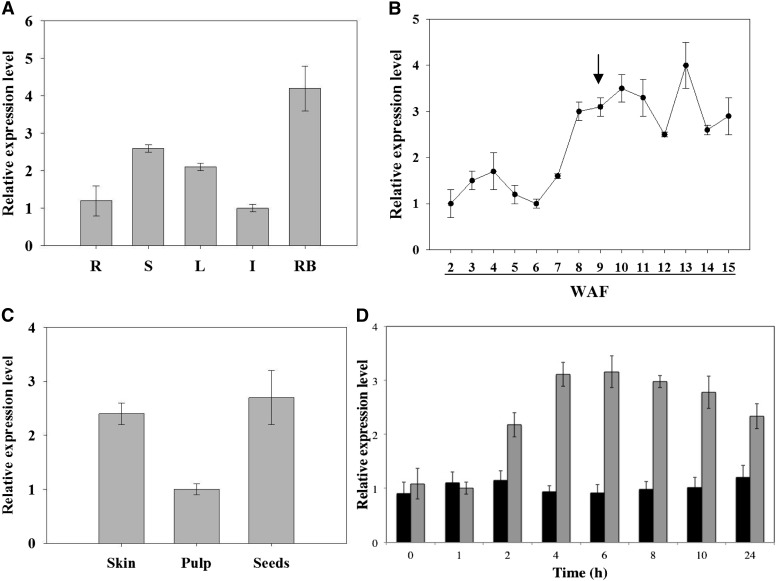
The expression profile of VvABF2 was determined in different grapevine organs by real-time reverse transcription (RT)-PCR with RNA extracted from cv Cabernet Sauvignon roots, stems, leaves, flowers, and mature berries (11 weeks after flowering [WAF]). VvABF2 was ubiquitously expressed in the different grape organs, but its relative expression depended on the organ (Fig. 3A). In decreasing order, VvABF2 transcript accumulation was highest in ripening berries, stems, leaves, roots, and inflorescences.
Figure 3.
Quantitative real-time RT-PCR analysis of VvABF2 expression patterns in grapevine cv Cabernet Sauvignon plants and ABA-treated cells. A, VvABF2 expression in grapevine organs: roots (R), stems (S), leaves (L), inflorescences (I), and ripening berries at 11 WAF (RB). Error bars were calculated as sd for three independent experiments. Gene expression was normalized with VvEF1γ (V. vinifera ELONGATION FACTOR1; Q9SPF8). B, VvABF2 expression at different stages of berry development, from 2 WAF to mature berries at 15 WAF. The arrow indicates the véraison stage. Error bars were calculated as the sd for four replicates from two independent experiments (summer 2006 and 2009). Gene expression was normalized with VvEF1γ. C, VvABF2 expression in different tissues from ripening berries at 11 WAF. Error bars were calculated as the sd for three independent experiments. Gene expression was normalized with VvEF1γ. D, VvABF2 transcript accumulation in cv Cabernet Sauvignon suspension cells treated with 20 μm ABA (gray bars) or with the same amount of ethanol (control; black bars). Error bars were calculated as sd for three independent experiments. Gene expression was normalized with VvEF1γ.
VvABF2 transcript accumulation was also assessed during berry development (Fig. 3B). Interestingly, VvABF2 expression increased just before the onset of grape berry ripening (8 WAF). Additionally, VvABF2 transcripts accumulated more than 2-fold during the ripening stage (9–15 WAF) when compared with the herbaceous phase (2–7 WAF; Fig. 3B).
VvABF2 gene expression was also analyzed in the different berry compartments (seeds, pulp, and skin) after véraison. VvABF2 transcripts were more abundant in both seeds and skin than in pulp (Fig. 3C).
The phylogenetic analysis revealed that this protein belongs to group A of bZIP transcription factors, previously shown to be involved in ABA signaling (Amir Hossain et al., 2010). Therefore, we investigated the effect of ABA on VvABF2 expression by treating Cabernet Sauvignon berry (CSB) cell suspensions with 20 µm ABA. In these treated cells, VvABF2 transcripts accumulated within the first 2 h of treatment and reached a maximum at 6 h, before slightly declining until 24 h after ABA supply (Fig. 3D).
VvABF2 Encodes a Functional Transcription Factor
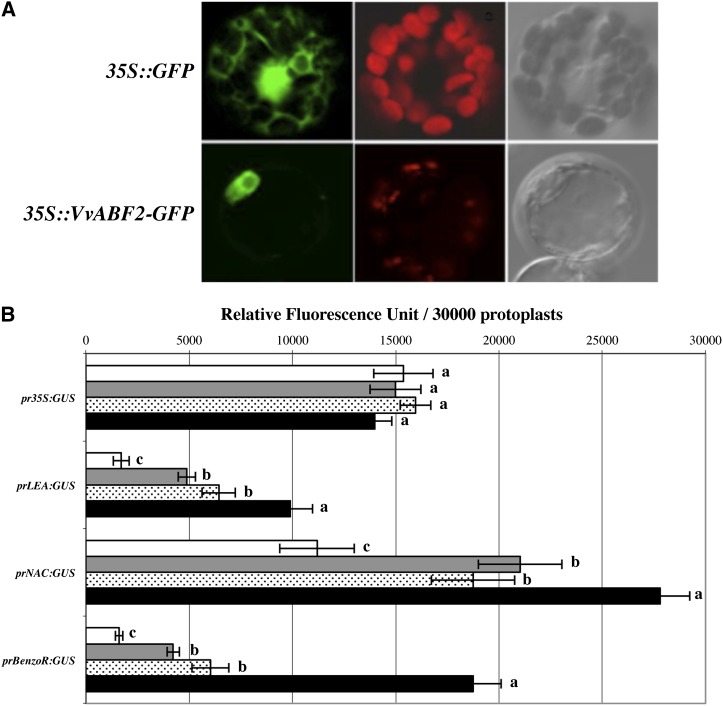
The presence of a bipartite nuclear localization signal between amino acids 357 and 373 suggests that VvABF2 is probably targeted to the nucleus (Fig. 1). To confirm this prediction, a GFP was fused in frame to the C terminus of VvABF2, and the resulting protein was expressed in tobacco (Nicotiana tabacum) mesophyll protoplasts under the control of the cauliflower mosaic virus 35S promoter. Unlike the GFP control, in which green fluorescence was seen throughout the cell, green fluorescence from VvABF2-GFP was only detected in the nucleus (Fig. 4A), consistent with a putative role of this protein in the control of transcription. In order to examine the transactivation ability of VvABF2, GUS reporter gene assays were performed by transient expression in tobacco protoplasts. The promoters of three known ABA up-regulated genes were selected: VvLEA (V. vinifera LATE EMBRYOGENESIS ABUNDANT; VIT_08s0007g04240), VvNAC (V. vinifera NO APICAL MERISTEM [NAM], ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR [ATAF], CUP-SHAPED COTYLEDON [CUC]; VIT_19s0014g03290), and VvBenzoR (Vitis vinifera Benzodiazepine Receptor; VIT_07s0005g00140; Wang et al., 2011) and fused to the GUS reporter gene. Quantitative GUS expression analysis was assessed by using these constructs and VvABF2 as an effector protein in protoplasts treated or not with 20 μm ABA (Fig. 4B). All promoters tested displayed enhanced activities upon ABA treatment. Interestingly, coexpression of VvABF2 significantly increased all promoter activities in the presence or absence of ABA (Fig. 4B). As multiple ABRE motifs were found in the promoter regions of these three VvABF2 putative target genes (Supplemental Table S1), the direct binding of VvABF2 was tested by electrophoretic mobility shift assay. Our results showed that VvABF2 binds specifically to these promoter regions, but only when ABRE motifs are present (Supplemental Fig. S1, A and C). Indeed, in the absence of these promoter elements, VvABF2 failed to bind to the promoter sequence (Supplemental Fig. S1B). Taken together, these results indicate that VvABF2 can recognize the ABRE sequences in the target promoters and activate these in response to ABA, under which activation levels were strongly enhanced. Therefore, VvABF2 is a positive regulator modulating downstream ABA signaling pathways.
Figure 4.
Subcellular localization and transactivation ability of VvABF2. A, Nuclear localization of the GFP-VvABF2 fusion protein in tobacco leaf protoplasts. These confocal microscopy images indicate, from left to right, protoplast sections analyzed for GFP fluorescence, the same sections analyzed for chloroplast autofluorescence, and the transmission light image from the same protoplast sections. B, Promoter activation by VvABF2 of selected ABA-regulated genes (Wang et al., 2011), VvLEA (VIT_08s0007g04240), VvNAC (VIT_19s0014g03290), and VvBenzoR (VIT_07s0005g00140), in tobacco protoplasts. pr35S::GUS was used as a positive control. White bars indicate GUS activity without additional construct or treatment, gray bars indicate GUS activity after transformation with the 35S::VvABF2 construct, dotted bars indicate GUS activity after 20 μm ABA treatment, and black bars indicate GUS activity after transformation with the 35S::VvABF2 construct coupled with 20 μm ABA treatment. Data from three independent experiments were pooled and analyzed. Error bars indicate sd. Statistical significance was assessed by one-way ANOVA followed by Tukey’s honestly significant difference post-hoc test (P ≤ 0.05).
Production of Transgenic Cell Lines Overexpressing VvABF2
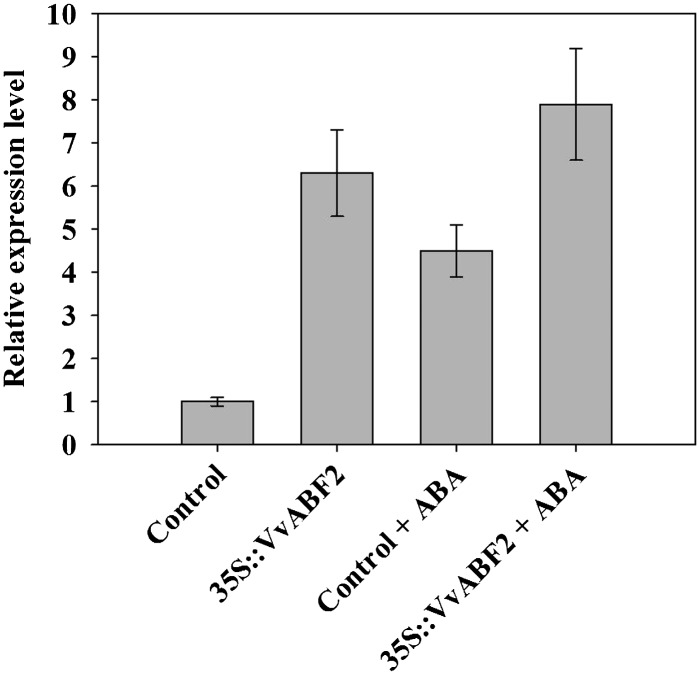
Transgenic grape cells overexpressing VvABF2 were produced using a 35S-driven VvABF2 construct. After stabilization of the cell suspension by subculture in glycerol-maltose-naphthoxyacetic acid culture medium supplemented with paromomycin and cefotaxime, the expression of VvABF2 was tested by real-time RT-PCR using VvABF2-specific primers. In cells expressing the 35S::VvABF2 construct, VvABF2 transcript accumulated six times more than in cells expressing the empty vector (Fig. 5). A 1-h ABA treatment (20 µm) of control cells dramatically stimulated VvABF2 expression. ABA also further stimulated VvABF2 expression in transgenic cells, but to a lesser extent (Fig. 5).
Figure 5.
Relative expression level of VvABF2 in transgenic grape 41B cells. VvABF2 transcript level was assessed by quantitative real-time PCR in control (pFB8 empty vector) and VvABF2-overexpressing (35S::VvABF2) 41B cell lines, treated or not with 20 μm ABA for 1 h. Gene expression was normalized with VvEF1γ. Data are means of three independent experiments, and error bars are sd.
Transcriptomic Analysis of VvABF2-Overexpressing Cell Lines upon ABA Supply
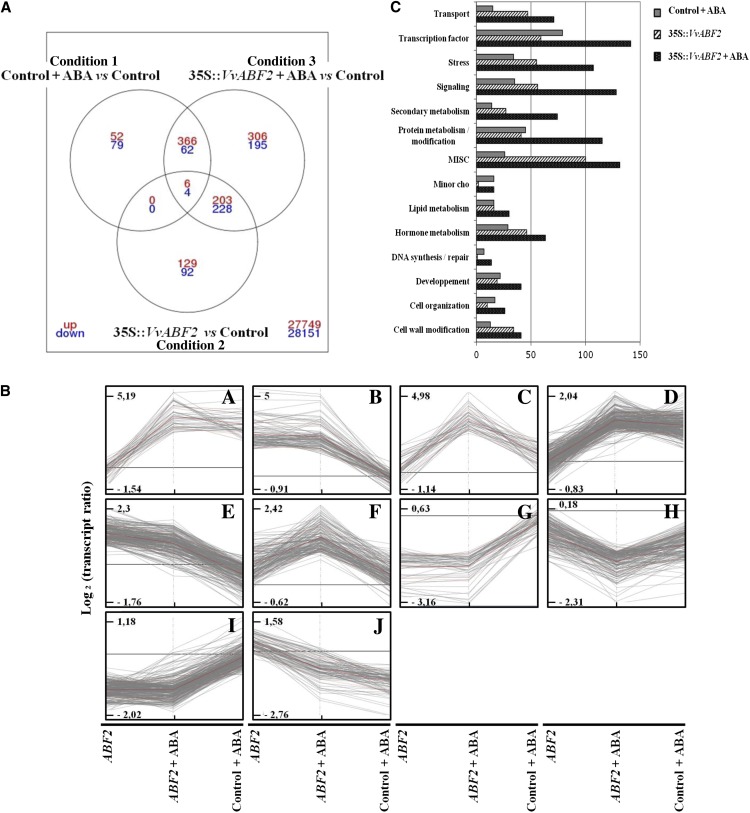
Total RNAs extracted from 41B cells treated or not with 20 µm ABA for 1 h were hybridized with 60-mer oligoarrays bearing a set of probes for 29,582 unigenes (NimbleGen Gene Expression 12x135K Arrays). Analysis of differentially expressed genes was performed using a 2-fold expression change and an adjusted P < 0.05 (with false discovery rate correction) as a cutoff from three independent experiments. Differential gene expression analysis was assessed through microarray data comparison of three different experimental conditions: ABA-treated versus untreated control cell lines (condition 1), untreated 35S::VvABF2 versus untreated control cell lines (condition 2), and ABA-treated 35S::VvABF2 versus untreated control cell lines (condition 3). The overlap in genes differentially expressed in these three conditions was depicted with a three-way Venn diagram (Fig. 6A). The results showed that 1,722 genes were differentially expressed in at least one of the three conditions. More specifically, ABA treatment of control cell lines (condition 1) regulated 569 (1.9%) of the 29,582 unigenes represented on the microarray slide. Among these, 424 (74.5%) were up-regulated and 145 (25.5%) were down-regulated. In the absence of ABA treatment, overexpression of VvABF2 led to the differential expression of 662 (2.2%) genes when compared with the control (condition 2). Three hundred thirty-eight (51%) genes were up-regulated and 324 (49%) were down-regulated (Fig. 6A). Treatment of 35S::VvABF2 transgenic cells with ABA resulted in the differential expression of 1,370 (4.6%) genes when compared with the untreated control cells (condition 3). Eight hundred eighty-one (64.4%) genes were up-regulated and 489 (35.6%) were down-regulated (Fig. 6A). The comparison of all three conditions showed that only 10 genes were commonly affected. Four hundred forty-one genes were common between conditions 2 and 3, and 438 genes were common between conditions 1 and 3. Moreover, 501 genes were specific for 35S::VvABF2 cells treated with ABA (condition 3; Fig. 6A).
Figure 6.
Overlap, expression profile clustering, and functional categorization of the 1,722 differentially expressed genes in the three experimental conditions. A, Three-way Venn diagram showing the overlap of differentially expressed genes in the three experimental conditions: condition 1 (control + ABA versus control), condition 2 (35S::VvABF2 versus control), and condition 3 (35S::VvABF2 + ABA versus control). B, Ten clusters have been created using MapMan (Thimm et al., 2004) on transcript ratios for the 1,722 differentially expressed genes under condition 1, condition 2, and condition 3. C, Classification of the 1,722 differentially expressed genes within selected MapMan ontology classes. The x axis indicates the number of genes within the different functional categories (y axis) for each condition: condition 1, condition 2, and condition 3. MISC, Miscellaneous.
The differentially expressed genes identified in our experiments were clustered based on expression ratios from conditions 1, 2, and 3 into 10 clusters of specific expression profiles (Fig. 6B). The clusters A, D, and J contained genes regulated by ABA in both control and VvABF2-overexpressing cells. Genes were up-regulated by ABA in clusters A and D and down-regulated in cluster J. Clusters B, E, G, and I included genes affected by VvABF2 overexpression (up-regulated [B and E] and down-regulated [G and I]) independently of the presence of ABA. Clusters C, F, and H contained genes regulated by ABA and/or ABF2. Clusters C and F encompassed genes significantly stimulated in 35S::VvABF2 cells treated with ABA, whereas cluster H corresponded to genes that were repressed in the same condition. Thus, clusters C, F, and H contained genes regulated by ABA in a VvABF2-dependent manner.
To functionally classify the 1,722 genes differentially expressed in all three types of conditions, we also performed an analysis using MapMan functional categories (Thimm et al., 2004; Fig. 6C). This classification suggested that most transcripts were linked to “secondary metabolism,” “hormone metabolism,” “protein metabolism,” “transport,” “signaling,” “stress,” and “transcription” when cells were supplemented with ABA (Fig. 6C). Among these ABA-stimulated groups, overexpression of VvABF2 led to the further enrichment of most categories and, more particularly, of the groups linked to secondary metabolism (enrichment factor 5.5×), transport (4.5×), signaling (3×), stress (3×), and cell wall modification (3×).
The more altered biological functions with statistical significance among the 1,722 genes differentially expressed were identified using a MapMan Wilcoxon test (Table I). In condition 1, genes involved in calcium signaling (P = 4.4 × 10−16), minor CHO (carbohydrate) metabolism (P = 3.19 × 10−10), AP2/ERF (for APETALA2/Ethylene Responsive Element Binding Factor) transcription factor family proteins (P = 4.44 × 10−10), and phenylpropanoid metabolism (P = 5.1 × 10−9) were among the highest differentially expressed genes (Tables I and II). These genes followed the expression profile of cluster A or D (Fig. 6B). In addition, other modulated genes previously reported to exhibit ABA-regulated expression in grape berry (Koyama et al., 2010) and other plant species (Seki et al., 2002; Rabbani et al., 2003; Buchanan et al., 2005; Matsui et al., 2008), such as LEA and biotic/abiotic stress-related proteins, were also identified (Table II). In condition 2, genes involved in secondary metabolism (P = 1.1 × 10−13), protein degradation and modification (P = 4.7 × 10−4 and 2.2 × 10−7, respectively), hormone metabolism and signaling (auxin and ethylene; P = 7.63 × 10−6 and 4.1 × 10−7, respectively), and cell wall degradation (P = 9.3 × 10−5) were significantly affected by overexpression of VvABF2 (Table I). These genes, which characterize the effect of VvABF2 on grape cells, belong to cluster B, E, G, or I (Fig. 6B). Some of these genes are shown in Table III. Among these (Table III), numerous genes encode cell wall hydrolytic enzymes (e.g. pectinesterases and polygalacturonases), secondary metabolism enzymes (e.g. flavonoid-O-glycosyltransferase, isoflavone reductases, dihydroflavonol 4-reductase, and isoflavone 2′-hydroxylase), hormone metabolism- and signaling-related proteins (e.g. IAA-amido synthetase and Ser/Thr protein kinase BRASSINOSTEROID INSENSITIVE1 [BRI1]-like), protein degradation-related proteins (e.g. F-box protein), sugar transport- and metabolism-related proteins (e.g. hexose transporter and invertase), transport-related proteins (e.g. multidrug and toxic compound extrusion [MATE], amino acids, and potassium transporter), transcription factors (e.g. ERF, MYELOBLASTOSIS [MYB], basic HELIX-LOOP-HELIX [bHLH], and HOMEODOMAIN LEUCINE ZIPPER), and stress-related proteins (e.g. pathogenesis-related [PR] protein, endochitinases, and peroxidases). Finally, in condition 3, the clustering into functional categories showed that genes involved in the synthesis of secondary metabolites were dramatically affected (P < 1 × 10−20). The enrichment was also observed for genes (Table I) corresponding to calcium signaling (P < 1 × 10−20), stress (P < 1 × 10−20), hormone metabolism and signaling (auxin and ethylene; P = 2.5 × 10−8 and 1.77 × 10−8, respectively), and WRKY transcription factor (P = 1.25 × 10−7). Clusters C, F, and H contained genes affected by ABA in VvABF2-overexpressing cells (Fig. 6B; Table IV). The transcript abundance of key enzymes involved in secondary metabolism and, more particularly, in the first steps of the phenylpropanoid pathway was considerably increased in condition 3. These enzymes correspond to phenylalanine ammonia lyase (PAL) and cinnamate 4-hydroxylase. Interestingly, several stilbene synthase genes followed a similar expression pattern (Table IV). Numerous genes encoding proteins related to hormone metabolism and signaling (e.g. 1-aminocyclopropane-1-carboxylate oxidase, GA receptor, and GA 2-oxidase), protein degradation (e.g. F-box protein, ubiquitin-protein ligase, and RING-H2 finger protein), and transport (e.g. ATP-binding cassette [ABC] transporter G family member and Glu receptor) were also strongly increased in the same condition. Finally, the transcript abundance of transcription factors (e.g. dehydration-responsive element-binding protein [DREB], WRKY, and MYB) and stress-related proteins (e.g. PR proteins, endochitinases, peroxidases, and receptor-like kinase) was also increased in VvABF2-overexpressing cells supplied with ABA (Table IV).
Table I. P values of selected categories identified by the MapMan Wilcoxon test to be significantly different from all other differentially expressed genes.
| Category Name | P Value, Condition 1 (Control + ABA Versus Control) | P Value, Condition 2 (35S::VvABF2 Versus Control) | P Value, Condition 3 (35S::VvABF2 + ABA Versus Control) |
|---|---|---|---|
| Calcium signaling | 4.4 × 10−16 | 0.4 × 10−2 | <1–10−20 |
| Secondary metabolism | |||
| Phenylpropanoids/lignins | 5.1 × 10−9 | 6.8 × 10−8 | 6.8 × 10−9 |
| Flavonoids/stilbenes | 1.1 × 10−4 | 1.1 × 10−13 | <1–10−20 |
| Minor CHO metabolism | 3.2 × 10−10 | 1.9 × 10−4 | 1.3 × 10−8 |
| AP2-ERF transcription factor | 4.4 × 10−10 | 5.5 × 10−3 | 6.6 × 10−11 |
| WRKY transcription factor | 9.3 × 10−5 | 8.7 × 10−2 | 1.2 × 10−7 |
| Cell wall degradation | 1.3 × 10−1 | 9.3 × 10−5 | 4 × 10−5 |
| Protein degradation/proteasome 26S | 4.2 × 10−2 | 4.7 × 10−4 | 1.1 × 10−3 |
| Auxin metabolism and signaling | 8.4 × 10−2 | 7.63 × 10−6 | 2.5 × 10−8 |
| Ethylene metabolism and signaling | 2.2 × 10−5 | 4.1 × 10−7 | 1.77 × 10−8 |
| Protein modification/kinase | 1.3 × 10−3 | 2.2 × 10−7 | 7.1 × 10−4 |
| Stress | 1.6 × 10−5 | 1.3 × 10−4 | <1–10−20 |
Table II. Selected ABA-induced genes from clusters A and D associated with minor CHO metabolism, signaling, flavonoid metabolism, stress, development, and transcription functional categories.
Ratio values are presented with P < 0.05.
| Gene Name | Locus Name | Ratio, Condition 1 (Control + ABA Versus Control) | Ratio, Condition 2 (35S::VvABF2 Versus Control) | Ratio, Condition 3 (35S::VvABF2 + ABA Versus Control) |
|---|---|---|---|---|
| Minor CHO metabolism | ||||
| Raffinose synthase | VIT_17s0000g08960 | 5 | 1.2 | 4 |
| Trehalose-6-phosphate synthase | VIT_11s0037g00720 | 2.2 | 1 | 2.1 |
| Signaling | ||||
| PP2C | VIT_00s0179g00140 | 4 | 1 | 4 |
| VIT_11s0016g03180 | 4 | 1 | 3 | |
| SNF1-related protein kinase | VIT_03s0038g04580 | 3 | 1.1 | 4 |
| Calmodulin-stimulated protein kinase | VIT_18s0001g06180 | 5 | 1.1 | 6 |
| Calmodulin | VIT_17s0000g02480 | 2.3 | 0.9 | 2.3 |
| VIT_01s0010g02950 | 3 | 0.8 | 3.5 | |
| VIT_16s0100g00620 | 2 | 0.9 | 2 | |
| Secondary metabolism | ||||
| Anthocyanidin 3-O-glucosyltransferase | VIT_19s0085g00750 | 4 | 1 | 3 |
| Flavanone 3-hydroxylase | VIT_18s0001g03510 | 4 | 1 | 4 |
| Stress | ||||
| Heat stress transcription factor | VIT_00s0179g00150 | 10 | 0.9 | 10 |
| Pathogenesis-related protein | VIT_06s0004g04010 | 3.5 | 0.8 | 3.5 |
| 17.2-kD class II heat shock protein | VIT_09s0002g00640 | 2.5 | 1 | 2.5 |
| Aquaporin TIP1-1 | VIT_08s0007g04780 | 3.2 | 1.2 | 3.3 |
| Desiccation-related protein | VIT_05s0077g00610 | 2.1 | 1 | 2.1 |
| Senescence-associated protein | VIT_00s2814g00010 | 3 | 1.1 | 3 |
| Development | ||||
| Late embryogenesis abundant protein | VIT_13s0067g01250 | 5 | 1.2 | 5 |
| VIT_08s0007g06420 | 3 | 1.1 | 3 | |
| VIT_16s0115g00170 | 4.5 | 1.1 | 5 | |
| Transcription | ||||
| AP2/ERF transcription factor | VIT_07s0031g00720 | 34 | 0.7 | 34 |
| VIT_04s0008g02230 | 5 | 0.9 | 6 | |
| VIT_15s0046g00310 | 2.5 | 1.1 | 3 | |
| DREB protein | VIT_18s0001g13320 | 7 | 1 | 7 |
| VIT_13s0067g01960 | 2.3 | 1 | 2.3 | |
| NAC transcription factor | VIT_19s0014g03290 | 10 | 0.9 | 10 |
| VIT_02s0236g00100 | 8 | 0.9 | 8 | |
| ABI5 | VIT_03s0063g00310 | 2.3 | 1 | 2.3 |
Table III. Selected VvABF2-regulated genes from clusters B, E, G, and I associated with cell wall metabolism, secondary metabolism, hormonal metabolism and signaling, protein degradation, sugar metabolism and transport, transcriptional regulation, transport, and stress response functional categories.
Ratio values are presented with P < 0.05.
| Gene Name | Locus Name | Ratio, Condition 1 (Control + ABA Versus Control) | Ratio, Condition 2 (35S::VvABF2 Versus Control) | Ratio, Condition 3 (35S::VvABF2 + ABA Versus Control) |
|---|---|---|---|---|
| Cell wall metabolism | ||||
| Pectinesterase | VIT_18s0001g12670 | 1 | 11 | 11 |
| VIT_16s0022g00710 | 1 | 8 | 8 | |
| VIT_03s0038g04740 | 1 | 5 | 5 | |
| Polygalacturonase | VIT_14s0066g01060 | 1 | 5.9 | 3.5 |
| Rhamnogalacturonate lyase | VIT_00s0346g00030 | 1 | 3.7 | 4 |
| Expansin | VIT_07s0005g02310 | 0.7 | 3 | 2 |
| Endoglucanase | VIT_02s0087g00930 | 0.8 | 0.25 | 0.25 |
| VIT_14s0036g01040 | 1 | 0.25 | 0.25 | |
| Secondary metabolism | ||||
| Flavonoid 5,3-O-glucosyltransferase | VIT_18s0041g00900 | 0.8 | 7 | 5 |
| VIT_18s0041g00800 | 1 | 5 | 5 | |
| VIT_18s0041g00970 | 0.8 | 3.5 | 3 | |
| Isoflavone reductase | VIT_02s0033g00260 | 1 | 3.1 | 3.5 |
| Dihydroflavonol 4-reductase | VIT_02s0025g01260 | 1 | 3 | 2.9 |
| VIT_01s0011g03480 | 1 | 0.3 | 0.3 | |
| Isoflavone 2′-hydroxylase | VIT_09s0002g06450 | 1 | 3.2 | 2 |
| Laccase | VIT_18s0164g00170 | 1.1 | 3.3 | 2 |
| Hormonal metabolism and signaling | ||||
| IAA-amido synthetase | VIT_12s0134g00230 | 0.9 | 12.5 | 8.8 |
| VIT_01s0150g00300 | 0.7 | 3.7 | 3 | |
| AUXIN/IAA family protein | VIT_05s0020g04690 | 0.9 | 6.5 | 4.6 |
| SAUR family protein | VIT_03s0038g01220 | 0.9 | 5.8 | 4.3 |
| PIN family protein | VIT_11s0052g00440 | 0.4 | 4.9 | 2.3 |
| 9-cis-Epoxycarotenoid dioxygenase | VIT_02s0087g0093 | 0.9 | 0.25 | 0.3 |
| Ser/Thr protein kinase BRI1-like | VIT_16s0013g01500 | 1.1 | 3 | 3.7 |
| VIT_00s0316g00010 | 0.9 | 3.2 | 3.2 | |
| Protein degradation | ||||
| F-box protein | VIT_01s0011g0122 | 1 | 11 | 5 |
| Ubiquitin family protein | VIT_14s0219g00210 | 1 | 4.6 | 4.9 |
| Sugar transport and metabolism | ||||
| Vacuolar invertase | VIT_16s0022g00670 | 1 | 3.1 | 3 |
| Hexose carrier protein (HT4) | VIT_16s0013g01950 | 1 | 3 | 3 |
| Polyol transporter | VIT_03s0063g02250 | 1 | 3.5 | 3.2 |
| Transport | ||||
| Amino acid permease | VIT_18s0001g01850 | 1 | 3.6 | 3 |
| Vacuolar amino acid transporter | VIT_19s0027g01890 | 0.6 | 3 | 4 |
| Peptide transporter PTR2 | VIT_18s0001g11280 | 1 | 4 | 3.5 |
| MATE efflux family protein | VIT_13s0064g00940 | 1 | 5.5 | 5.1 |
| VIT_16s0100g00460 | 0.7 | 10 | 4 | |
| Potassium transporter | VIT_01s0011g03020 | 1 | 7.2 | 6 |
| Potassium channel KAT3 | VIT_04s0008g04510 | 1.2 | 3 | 3 |
| Aquaporin PIP1.1 | VIT_13s0067g00220 | 1 | 3.5 | 5 |
| ABC transporter family protein | VIT_19s0085g00060 | 1 | 0.01 | 0.01 |
| MATE efflux family protein | VIT_08s0056g00870 | 1 | 0.3 | 0.3 |
| GDP-Man transporter | VIT_16s0022g00370 | 0.6 | 0.01 | 0.01 |
| Transcription | ||||
| Ethylene-responsive transcription factor | VIT_10s0003g00590 | 0.8 | 6 | 5 |
| Homeobox-Leu zipper protein HB40 | VIT_04s0023g01330 | 1.1 | 8 | 10 |
| MYB transcription factor MYB36 | VIT_11s0016g02780 | 0.8 | 4 | 5 |
| Transcription factor bHLH68 | VIT_11s0016g03560 | 1 | 6 | 4.5 |
| Dof zinc finger protein | VIT_07s0255g00020 | 1 | 4.5 | 4 |
| VIT_10s0003g00040 | 0.9 | 4.5 | 3 | |
| bHLH transcription factor | VIT_00s0274g00070 | 0.8 | 0.3 | 0.3 |
| VIT_07s0205g00190 | 1 | 0.3 | 0.4 | |
| MYB transcription factor | VIT_09s0070g00410 | 0.9 | 0.2 | 0.3 |
| Homeobox-Leu zipper protein KNAT1 | VIT_18s0001g08380 | 0.8 | 0.2 | 0.2 |
| Stress response | ||||
| PR protein | VIT_03s0088g00700 | 0.8 | 32 | 26 |
| VIT_08s0040g02170 | 1.1 | 4.5 | 4.5 | |
| VIT_13s0147g00150 | 1.4 | 4 | 4.5 | |
| VIT_18s0041g02190 | 1.1 | 0.3 | 0.3 | |
| Endochitinase | VIT_05s0094g00280 | 1.3 | 21 | 20 |
| VIT_16s0050g02210 | 0.9 | 4 | 3.6 | |
| Peroxidase | VIT_07s0104g01100 | 1.1 | 5 | 3.6 |
| VIT_10s0116g00340 | 1 | 5 | 4.2 | |
| VIT_11s0016g05280 | 0.6 | 0.3 | 0.2 | |
| Accelerated cell death | VIT_14s0081g00350 | 1 | 3 | 3.3 |
| Protein kinase resistance-like | VIT_12s0028g01850 | 0.8 | 0.2 | 0.2 |
Table IV. Selected genes transcriptionally regulated in VvABF2-overexpressing cells treated by ABA, belonging to clusters C, F, and H, and associated with secondary metabolism, hormonal metabolism and signaling, protein degradation, transport, transcriptional regulation, and stress response functional categories.
Ratio values are presented with P < 0.05.
| Gene Name | Locus Name | Ratio, Condition 1 (Control + ABA Versus Control) | Ratio, Condition 2 (35S::VvABF2 Versus Control) | Ratio, Condition 3 (35S::VvABF2 + ABA Versus Control) |
|---|---|---|---|---|
| Secondary metabolism | ||||
| Stilbene synthase | VIT_10s0042g00840 | 1.2 | 2.8 | 5.5 |
| VIT_16s0100g00780 | 1.5 | 2.2 | 6.5 | |
| VIT_16s0100g01020 | 1.7 | 1.3 | 13 | |
| VIT_16s0100g00960 | 1.6 | 1.3 | 8 | |
| Cinnamate 4-hydroxylase | VIT_11s0078g00290 | 1.8 | 1.3 | 7 |
| VIT_11s0065g00350 | 2 | 1.3 | 13 | |
| CAD | VIT_18s0122g00450 | 1.4 | 3.2 | 5 |
| PAL | VIT_16s0039g01170 | 2 | 0.9 | 6 |
| VIT_16s0039g01320 | 2.1 | 0.9 | 5 | |
| VIT_00s2508g00010 | 2.1 | 0.8 | 4.5 | |
| VIT_08s0040g01710 | 1.6 | 0.9 | 3 | |
| Hormonal metabolism and signaling | ||||
| AUXIN/IAA family protein | VIT_11s0016g04490 | 1.8 | 1.5 | 3 |
| 1-Aminocyclopropane-1-carboxylate oxidase | VIT_18s0001g01840 | 1.1 | 12 | 19 |
| GA receptor GID1 | VIT_01s0011g03270 | 1.2 | 3.6 | 10 |
| GA 2-oxidase | VIT_19s0140g00120 | 3.5 | 2.3 | 8 |
| ABA 8′-hydroxylase | VIT_06s0004g05050 | 1.9 | 1.1 | 3 |
| Cytokinin-O-glucosyltransferase | VIT_13s0019g03120 | 1.2 | 1.6 | 3.5 |
| Ser/Thr protein kinase BRI1-like | VIT_09s0018g00780 | 1.3 | 1.3 | 4.5 |
| Protein degradation | ||||
| F-box protein | VIT_00s1386g00020 | 1.1 | 1 | 22 |
| VIT_10s0116g01290 | 1.2 | 1 | 19 | |
| Ubiquitin-protein ligase | VIT_08s0105g00190 | 2.7 | 1 | 11 |
| VIT_18s0166g00190 | 1.2 | 0.8 | 3 | |
| Cupin superfamily protein | VIT_07s0005g04620 | 1 | 1.2 | 3 |
| RING-H2 finger protein | VIT_05s0077g01970 | 2 | 0.8 | 3.7 |
| Transport | ||||
| ABC transporter G family member | VIT_06s0061g01490 | 1.5 | 1.1 | 4 |
| Glu receptor | VIT_05s0051g00780 | 1.5 | 1 | 3 |
| Amino acid transporter | VIT_03s0038g03530 | 1.2 | 1.1 | 3 |
VvABF2 Overexpression Stimulates ABA Induction of Stilbene Production
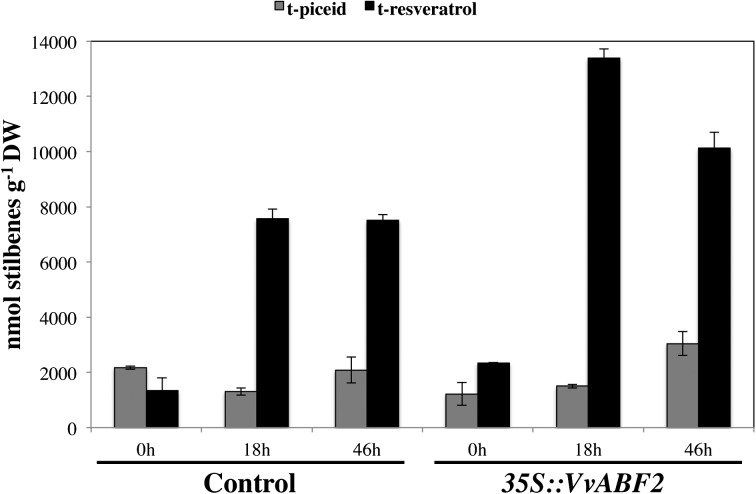
Stilbenes, resveratrol, and its derivatives such as trans-piceid (a glycosylated form of resveratrol) represent some of the major forms of phytoalexins in grapevine (Pezet et al., 2004). They are important for plant defense, and resveratrol also has beneficial effects on human health (Bradamante et al., 2004; Hofseth et al., 2010; Szkudelska and Szkudelski, 2010). Stilbene production in grapevine transgenic cell suspensions was monitored after the addition of 20 µm ABA. Before treatment, control and 35S::VvABF2 transgenic cells secreted different amounts of each stilbene in the culture medium. Indeed, in control cells, trans-resveratrol was present at concentrations representing 57.6% of its amount in VvABF2 transgenic cells. By contrast, in nontreated VvABF2-overexpressing cells, trans-piceid only represented 56% of that present in nontreated control cells (Fig. 7). However, the total amount of stilbenes is similar for both conditions (Supplemental Fig. S2).
Figure 7.
Time course of stilbene production (nmol g−1 dry weight [DW]) in the culture medium of control and 35S::VvABF2 transgenic cells supplied with 20 μm ABA. Total trans-piceid (gray bars) and trans-resveratrol (black bars) were measured in the extracellular medium. Values represent means ± sd of triplicate assays of one representative experiment out of three.
After the addition of ABA to control cell suspensions, trans-resveratrol strongly accumulated in the culture medium (5.6-fold more), whereas the trans-piceid amount was maintained (Fig. 7). By contrast, in the culture medium of 35S::VvABF2 cell suspensions, ABA treatment increased both trans-resveratrol and trans-piceid contents (Fig. 7). Indeed, trans-resveratrol and trans-piceid contents were multiplied by maximal factors of 6.2 and of 2.5, respectively. Finally, ABA treatment increased the total stilbene amount by 2.7-fold in control cells and by 4.1-fold in the transgenic cells (Supplemental Fig. S2).
Phenotype of Tomato Fruits Overexpressing VvABF2
To investigate the involvement of VvABF2 in fruit development and ripening processes, transgenic tomatoes overexpressing VvABF2 were produced. The full-length coding region of VvABF2 was cloned downstream of the cauliflower mosaic virus 35S promoter and used for transformation. Five independent 35S::VvABF2 transgenic plants were selected for their successful integration of the transgene (Supplemental Fig. S3). VvABF2 overexpression did not result in apparent developmental defects. Flowers and fruits developed normally until the turning stage, with kinetics comparable to the control plants. By contrast, after the turning stage and at the later ripening stages, fruits from all 35S::VvABF2 lines exhibited dramatic phenotypes. Indeed, as soon as 20 d after turning, the firmness of the tomatoes from VvABF2-overexpressing lines was strongly reduced in comparison with the control fruits. This reduction in firmness is illustrated in Supplemental Figure S3, showing the phenotype observations and the Durofel indices that were obtained for the control and the VvABF2-overexpressing fruits 20 or 50 d after turning.
DISCUSSION
Ripening of nonclimacteric fruit is usually associated with ABA, a phytohormone that gradually accumulates from the onset of ripening (Coombe, 1992; Giribaldi et al., 2010; Koyama et al., 2010). Two different ABA receptors from strawberry (Fragaria spp.), FaCHLH/ABAR (for Fragaria ananassa H subunit of magnesium chelatase/ABA receptor; Jia et al., 2011) and FaPYR1 (Chai et al., 2011), and one receptor from grape, VvPYL1 (Li et al., 2012), were recently described as positive regulators of fruit ripening in response to ABA. In this context, AREB/ABF bZIP transcription factors that act as regulators of ABA and stress responses in plants are good candidates for the regulation of ABA-mediated fruit ripening. Moreover, a possible role of AREB/ABF transcription factors has been reported for the ripening climacteric fruits (Bastías et al., 2011).
This work gives further insight into the role of this class of transcription factor during the ABA-mediated ripening of grape berries. In this context, we isolated and characterized VvABF2, previously known as GRIP55 (VIT_18s0001g10450, Q9M4H1). VvABF2 belongs to group A of bZIP transcription factors, which are involved in ABA and abiotic stress signaling (Choi et al., 2000; Uno et al., 2000; Amir Hossain et al., 2010). VvABF2 was ubiquitously expressed in different grape organs, and its transcript accumulated just before the onset of grape berry ripening (Fig. 3), when ABA concentrations increase (Coombe, 1992). These data fit well with the observation that VvABF2 expression was regulated by ABA (Fig. 3D) and underline a putative role for this transcription factor in influencing ABA-regulated grape berry ripening processes. The nuclear localization of VvABF2 (Fig. 4A) and its ability to transactivate selected promoters (Fig. 4B) are consistent with its function as a transcriptional activator. Additionally, the transactivation experiments performed in protoplasts further support the hypothesis of the involvement of VvABF2 in ABA-mediated signaling pathways. Indeed, VvABF2 was able to bind and transactivate several known ABA-regulated genes (VvLEA, VvNAC, and VvBenzoR), and its transcriptional activating role was further increased by ABA supply (Fig. 4B).
The function of VvABF2 and its role in ABA signaling were further analyzed by a transcriptomic approach comparing the response to ABA of transgenic grape cells that overexpressed VvABF2 with that of control cells. Overexpression of VvABF2 (condition 2) modulates the expression of many grape genes in the absence of ABA. Only a few (10) genes were common with those observed in ABA-treated control cells (condition 1). This might be the consequence of an ABA treatment (1 h) that was too short to affect all ABA-regulated genes in control cells. This hypothesis was strengthened by the fact that many of the VvABF2-regulated genes (condition 2) belong to families previously shown to be involved in ABA responses. This also indicates that the ectopic expression of VvABF2 was sufficient to affect some of the ABA-dependent genes. In addition, ABA treatment of 35S::VvABF2 transgenic cells (condition 3) regulated many other genes, including 438 genes that were common with ABA-treated control cells (condition 1). This represents 77% of the total number of genes affected by the ABA treatment in the control cells (among which 65.4% were up-regulated and 11.6% were down-regulated). By comparison, ABA treatment of 35S::VvABF2-overexpressing cells resulted in the modification of the expression of 32% of all the genes affected by VvABF2 overexpression (27.2% up-regulated and 4.8% down-regulated). These results indicated that the expression of some genes that follow an ABA-dependent pathway were mainly mediated by VvABF2, whereas others were specifically dependent on VvABF2 and independent of ABA treatment. The identification and in silico analysis of 1,344 bp of the VvABF2 promoter region using the Genomatix suite of programs (Quandt et al., 1995) led to the identification of several putative cis-acting regulatory elements within the 5′ regulatory region of VvABF2 (Supplemental Table S2). These transcription factor-binding sites were associated with plant development, hormonal regulation, and biotic and abiotic stress responses. Their presence occurred at different frequencies and are described in Supplemental Table S2. This result suggests the existence of both ABA-dependent and -independent pathways in the control of VvABF2 expression.
ABA treatment of grape cells led to the regulation of many genes known to be ABA and stress related. For instance, genes specifically affected by the ABA treatment encode (1) proteins involved in osmotic stress, desiccation (raffinose synthase and LEA), and biotic stress (PR protein) responses, (2) proteins known to improve stress tolerance to abiotic stresses (AP2-DREB, bZIP, and NAC), and (3) members of ABA signaling (PP2C) and calcium signaling (calmodulin and calmodulin-stimulated protein kinase; Desikan et al., 2001; Yu et al., 2006; Nishimura et al., 2007; Nakashima et al., 2009; Park et al., 2009; Wang et al., 2011; Table II). Our results confirm data from the literature and validate these experiments.
As mentioned before, VvABF2 overexpression can affect some ABA-dependent genes in the absence of ABA treatment (Table III). For instance, several transcripts encoding PR proteins such as endochitinases were strongly up-regulated in VvABF2 transgenic cells. These genes are induced during berry development and ripening (Davies and Robinson, 2000; Pilati et al., 2007). Genes encoding potassium transporters and channels were also strongly up-regulated (Table III). These genes are known ABA targets that are also important for berry ripening (Davies et al., 2006). Indeed, together with sugar accumulation, K+ influx might help the turgor-driven berry expansion (Davies et al., 2006), and the K+ channel VvK1.1 may play a major role in K+ loading into berry tissues, especially upon drought stress (Cuéllar et al., 2010, 2013).
Effects on Ripening-Associated Processes
ABA promotes sugar metabolism and accumulation in fleshy fruits (Yamaki and Asakura, 1991; Kobashi et al., 1999; Richings et al., 2000; Pan et al., 2005). In tomato, overexpression of SlAREB1 stimulates hexose accumulation in correlation with increased expression of vacuolar invertases (Bastías et al., 2011). In VvABF2-overexpressing cells, the stimulated expression of vacuolar invertase and hexose transporter genes is consistent with this hypothesis (Table III).
The softening that accompanies the last stages of fleshy fruit ripening is typically attributed to changes in cell wall properties (Li et al., 2010). Several studies have underlined the role of ABA in enhancing softening in both tomato (Zhang et al., 2009b) and grapevine (Gambetta et al., 2010). Indeed, it was suggested by Koyama et al. (2010) that in grape berry, the modification of cell walls occurring under ABA treatment may be reflected by the differential transcript abundances of cell wall proteins and pectin-modifying enzymes. The high transcript levels of genes encoding cell wall hydrolytic enzymes (pectinesterase, polygalacturonase, and rhamnogalacturonase) in VvABF2-overexpressing cells suggest a role for VvABF2 in fruit softening (Table III). This hypothesis is further strengthened by the observation that in tomato fruits overexpressing VvABF2, the softening process is dramatically accelerated (Supplemental Fig. S3).
Responses to oxidative stress have been reported during fruit development in several species, including tomato (Jiménez et al., 2002), strawberry (Aharoni and O’Connell, 2002), and grape (Pilati et al., 2007). Reactive oxygen species (ROS) amounts are accurately controlled by both their production and the antioxidant systems. During grape berry development, Pilati et al. (2007) showed that oxidative processes and enzyme-mediated scavenging systems are activated during the ripening stage. In agreement with this work, our microarray data showed that both ROS production and scavenging systems were affected (Supplemental Table S4). Indeed, in at least one condition (in the presence of ABA and/or VvABF2), the expression of 30 genes involved in ROS production was significantly affected. These genes belong to the RBOH (respiratory burst oxidase homologue) and secretory peroxidase (class III) families that are responsible for the production of ROS and of some secondary metabolites leading to the synthesis of antifungal products or the reinforcement of cell walls. In the same conditions, the expression of more than 80 genes related to ROS scavenging systems was also altered. These include ascorbate and glutathione peroxidases, peroxiredoxins, thioredoxins, glutaredoxins, and glutathione S-transferases. The fine-tuned control of this gene network by ABA and/or VvABF2 may contribute to the regulation of the redox balance and to the production of some metabolites that are necessary for ripening.
The role of ABA in enhancing the synthesis of phenolic compounds during fruit ripening has been reported extensively (Ban et al., 2003; Jeong et al., 2004; Peppi et al., 2006; Wheeler et al., 2009; Gambetta et al., 2010). Our transcriptomic analysis revealed that ABA regulates some genes encoding secondary metabolism enzymes and that VvABF2 overexpression does it as well: for example, treatment of control cells with ABA up-regulated genes involved in flavonoid/anthocyanin biosynthesis (Table II). Among these, flavanone 3-hydroxylase was previously shown to be induced in berry skin after ABA treatment (Koyama et al., 2010). Likewise, VvABF2 overexpression also led to the stimulation of this pathway (Table III). Indeed, the expression of genes encoding proteins involved in the biosynthesis of anthocyanins (UDP-glucose:flavonoid 3-O-glucosyltransferase and dihydroflavonol reductase) and isoflavones (isoflavone hydroxylase and isoflavone reductase) is modulated by VvABF2. Finally, treatment of 35S::VvABF2 cells by ABA also up-regulated secondary metabolism and, more particularly, the first steps of the phenylpropanoid pathway (PAL and cinnamic acid 4hydroxylase), the lignin biosynthesis pathway (cinnamyl alcohol dehydrogenase [CAD]), and the phytoalexin pathway (stilbene synthase). The strong up-regulation of these genes in ABA-treated 35S::VvABF2 cells by comparison with ABA-treated control cells suggests that their expression depends on both ABA and VvABF2. Measurements of stilbene production in 35S::VvABF2 transgenic cells corroborate our microarray data. Indeed, ABA treatment of control and VvABF2-overexpressing cells stimulated this production and particularly that of trans-resveratrol (Fig. 7). Therefore, trans-resveratrol secretion is stimulated by ABA in both control and 35S::VvABF2 cells (5.6- and 6.2-fold increases, respectively, when compared with nontreated cells). By contrast, trans-piceid amounts did not significantly change in ABA-treated control cells, whereas its amount was more than doubled in 35S::VvABF2 transgenic cells treated with ABA.
Overexpression of VvABF2 also affected the accumulation of MATE transporter transcripts. Since this gene family is involved in the transport of flavonoids (Gomez et al., 2009; Zhao and Dixon, 2009), these data indicate that VvABF2 may also be involved in the regulation of flavonoid transport.
Effects on Genes Controlling Hormonal Balance
Hormone balance is important for fruit development and ripening. For example, the maintenance of precise amounts of cytokinins is needed to achieve fruit ripening (Mok, 1994). Low levels of free IAA (maintained by Gretchen Hagen3 [GH3]) are also needed to allow berry ripening (Böttcher et al., 2010, 2011), and ABA stimulates this conjugating enzyme (Seo et al., 2009). Ethylene plays an important role in climacteric fruit ripening (Giovannoni, 2007), and ABA is thought to control the ripening of climacteric fruit through the activation of ethylene biosynthesis (Zhang et al., 2009b). Although the role of ethylene in nonclimacteric fruits remains unclear, its involvement in grapevine berry ripening has been suggested (Chervin et al., 2004, 2008). Finally, brassinosteroids are positive hormonal regulators of berry ripening (Symons et al., 2006) that interact with other phytohormones (ABA and auxin) to achieve their biological function (Hardtke, 2007; Zhang et al., 2009). Our results suggest a fine-tuning of hormone amounts and signaling pathways by ABA and underline a putative role of VvABF2 in this control. Indeed, VvABF2-overexpressing cells accumulated transcripts of genes involved in hormone biosynthesis and/or signaling (Tables III and IV). Two sets of genes could be identified. The first one corresponded to genes stimulated by ABA in control cells, and their expression was further induced by VvABF2 overexpression. The second set included genes that were already affected in 35S::VvABF2 cells and whose accumulation was further affected by ABA supply, including many genes involved in the control of hormone amounts and, more particularly, their biosynthesis (9-CIS-EPOXYCAROTENOID DIOXYGENASE and 1-AMINOCYCLOPROPANE-1-CARBOXYLATE OXIDASE) and catabolism (GH3, GA 2-OXIDASE, ABA 8′-HYDOXYLASE, and CYTOKININ-O-GLUCOSYLTRANSFERASE; Tables III and IV). Other genes corresponding to proteins involved in hormone signaling were also affected, including genes involved in hormone sensing (Gibberellin Insensitive Dwarf1 [GID1] and BRI1) and transduction (AUXIN/IAA and small auxin up RNA [SAUR]).
The ubiquitin/26S proteasome pathway plays a key role in the perception and transmission of environmental and hormonal signals (Smalle and Vierstra, 2004; Liu and Stone, 2010; Antoni et al., 2011). In this work, the importance of this pathway in ABA signaling is highlighted by the response of several genes encoding F-box proteins and other components of the proteasome pathway in 35S::VvABF2 transgenic lines and after ABA supply (Table III).
Effects on Genes Involved in Stress Responses
Numerous transcription factors are regulated by ABA or ABA-regulated stresses, even though their specific roles in ABA signaling remain unknown (Nemhauser et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006). ABA-regulated members of the DREB, WRKY, and MYB transcription factor families were up-regulated when VvABF2 was overexpressed (Table III), reinforcing the role of this transcription factor in regulating ABA-mediated pathways. Thus, among the known homologs of the transcription factor genes identified, AtWRKY28 and AtWRKY40 are involved in oxidative stress, cold, high-salinity, and osmotic stress responses (Seki et al., 2002) and AtWRKY33 and AtWRKY53 are involved in hydrogen peroxide and ozone responses (Tosti et al., 2006; Vanderauwera et al., 2007). Additionally, it has been suggested that AtWRKY33 may regulate plant responses to both abiotic stress and ABA (Jiang and Deyholos, 2009). In addition, AtWRKY40 binds the promoters of several members of the AREB/ABF transcription factor subfamily, and its activity is inhibited by ABA perception by the magnesium-chelatase H subunit receptor (CHLH/ABAR; Shang et al., 2010).
Taken together, these data show that VvABF2 overexpression not only activates new gene networks but mainly functions by exacerbating and/or modifying existing networks related to ABA responses. Indeed, ABA treatment of VvABF2-overexpressing cells often led to an enhanced response compared with ABA treatment of control cells (Fig. 6B, clusters C, F, and H). This was particularly observed for genes involved in the phenylpropanoid pathway. Several genes encoding enzymes involved in the first step of this pathway (PAL and C4H) and several stilbene synthases were strongly induced. These data indicate that in the presence of ABA, VvABF2 is an important regulator of the phenylpropanoid pathway leading to stilbene biosynthesis. This result is consistent with data from the literature showing an up-regulation of several stilbene synthase genes paralleling an increase in resveratrol concentrations in ABA-treated berries (Koyama et al., 2010). In addition, VvABF2 seems to affect lignin biosynthesis by the stimulation of CAD and laccase genes (Tables III and IV). The induction of lignin biosynthetic genes by ABA was previously illustrated in Arabidopsis (Østergaard et al., 2001; Seki et al., 2002)
Finally, several reports indicate that fruit softening is hormonally regulated by ABA and ethylene (Jiang et al., 2000; Zhang et al., 2009a, 2009b). Inhibitors of ABA biosynthesis delay tomato ripening and softening (Zhang et al., 2009a, 2009b). Treatment of berries with ABA or ethephon (an ethylene analog) increases softening (Peppi et al., 2006). In this work, ABA treatment led to modification of the expression of some softening-related genes. Among these, we can cite various xyloglucan endotransglycosylase genes, cellulose synthase genes, and pectinesterase genes. Additionally, genes involved in galactinol (galactinol synthase, stachyose synthase, and seed imbibition protein) and trehalose (trehalose-6-P phosphatase and trehalose-6-P synthase) metabolism were up-regulated. Both microarray data and the results obtained with transgenic tomatoes strongly indicate the involvement of the ABA-regulated transcription factor VvABF2 in stimulating fruit maturation and softening.
In summary, this work characterizes VvABF2, a transcription factor of the AREB/ABF family sensitive to ABA. This transcription factor mediates at least in part several ABA-mediated pathways controlling both maturation processes and responses to environmental stresses. These include the regulation of secondary metabolism, cell wall metabolism, hormone metabolism and signaling, and stress responses. In addition to its putative role in fruit maturation and because of its general pattern of expression, VvABF2 could be a key component of ABA-mediated grape development and adaptation to environmental cues.
MATERIALS AND METHODS
VvABF2 cDNA Isolation and Production of Constructs for Plant Transformation
A full-length VvABF2 clone was produced from a cDNA library isolated from grape (Vitis vinifera ‘Cabernet Sauvignon’) berries at the véraison stage. PCR was performed using synthetic oligonucleotide primers designed to begin and end at the start and stop codons of the open reading frame of VvABF2 (forward primer, 5′-ATGGGGAGTAATTTGAACTTCAAAAACTTC-3′; reverse primer, 5′-CCAGGGGCCAGTCAGTGTGCGTCTCAAGCAA-3′). The complete open reading frame was amplified and cloned into the pGEM-T Easy vector (Promega) for DNA sequencing, prior to subcloning into a stable expression binary vector downstream of the 35S promoter of the Cauliflower mosaic virus. pFB8 and Pk7m34GW binary vectors (Gateway; Karimi et al., 2002) were used to generate VvABF2-overexpressing 41B cells and tomato (Solanum lycopersicum) plants, respectively.
Plant Transformation and Culture Conditions
Grapevine transformations were made in 41B rootstock (cv Chasselas × Vitis berlandieri) according to Lecourieux et al. (2010). An embryogenic cell suspension culture was initiated as described previously (Coutos-Thévenot et al., 1992a, 1992b). This cell suspension was subcultured weekly in 25 mL of glycerol-maltose culture medium (Coutos-Thévenot et al., 1992b) supplemented with synthetic auxin (naphthoxyacetic acid) at 1 mg L−1 in the dark. Embryogenic cells were transformed using an Agrobacterium tumefaciens cocultivation method (Mauro et al., 1995), and after selection, the transgenic cells were subcultured in the same condition in a medium supplemented with paromomycin at 2 mg mL−1 and cefotaxime at 200 mg mL−1 (Duchefa).
Transgenic tomato plants (cv Wva106) were generated by A. tumefaciens-mediated transformation of tomato cotyledons as described by Gonzalez et al. (2007). Tomato plants were grown in a culture chamber with a 14-h-day/10-h-night cycle. The temperature was 25°C during the day and 20°C during the night. Individual flowers were tagged on the day of anthesis (flower opening).
Quantification of Stilbenes
Stilbenes from the culture medium were obtained by a triple ethyl acetate extraction using 5 mL of culture medium. Stilbenoid samples were filtered through 0.45-μm polytetrafluoroethylene membrane filters (Fioroni). Analysis of stilbenes was performed by HPLC on a 250- × 4-mm Prontosil C18 (5 µm) reverse-phase C18 column (Bischoff Chromatography) protected by a guard column of the same material. The HPLC device was coupled to an Esquire 3000 Plus ion-trap mass spectrometer using an electrospray ionization source (Bruker-Daltonics). The chromatographic conditions were not modified, and the HPLC output was split 1:10 in the mass spectrometry detector. Data analysis was performed with Bruker Data Analysis 3.2 software. Separation was performed at a flow rate of 1 mL min−1 with a mobile phase composed of water:1% trifluoroacetic acid (97.5:2.5, v/v; A) and acetonitrile:A (80:20, v/v; B). The run was set as follows: 0 to 1 min, 20% B; 1 to 8 min, from 20% to 24% B; 8 to 10 min, from 24% to 25% B; 10 to 13 min, 25% B; 13 to 18 min, from 25% to 30% B; 18 to 35 min, from 30% to 50% B; 35 to 37 min, from 50% to 100% B; 37 to 41 min, 100% B; 41 to 42 min, from 100% to 20% B; and then 20% B for 4 min. UV detection was performed at 286 and 306 nm. Absolute trans-resveratrol and trans-piceid contents were estimated from calibration curves prepared with pure standards. Trans-resveratrol and trans-piceid were purchased from Sigma.
RNA and cDNA Production
Roots, shoots, leaves, and inflorescences were collected from cv Cabernet Sauvignon fruit cuttings grown in a greenhouse. Berries (cv Cabernet Sauvignon) were harvested in Domaine de la Grande Ferrade, Villenave d’Ornon, France. In order to compare berries at the same developmental stage, berries were sorted by weight before véraison and on a NaCl density gradient after véraison.
All collected samples were quickly frozen in liquid nitrogen, ground to a fine powder with a Dangoumau blender, and stored at –80°C prior to use. Total RNA from grape organs and berries was extracted according to Lecourieux et al. (2010). Total RNA from grape cells was extracted from 100 mg of starting tissue using the Spectrum Plant Total RNA Kit (Sigma) following the manufacturer’s protocol. RNA isolation was followed by DNaseI treatment. For each sample, RT was performed from 2 µg of purified RNA using Moloney murine leukemia virus reverse transcriptase (Promega) according to the manufacturer’s instructions. The cDNA obtained was diluted (1:20) in distilled water.
Gene Expression Analysis
Quantitative real-time RT-PCR expression analysis was carried out using the CFX96 Real-Time PCR Detection system (Bio-Rad). Ten-microliter reaction mixes were prepared, which included 5 µL of iQ SYBR Green Supermix (Bio-Rad), 0.2 µm of each primer, and 2 µL of diluted cDNA. Gene transcripts were quantified with normalization to VvEF1γ (grape experiments [Q9SPF8]; Nicolas et al., 2013) and SlEiF4α (tomato experiments [SGN-U578071]) as internal standards. All biological samples were tested in triplicate, and sd values were calculated using standard statistical methods. Specific oligonucleotide primer pairs were designed with Beacon Designer 7 software (Premier Biosoft International). Specific annealing of the oligonucleotides was controlled by dissociation kinetics performed at the end of each PCR run. The efficiency of each primer pair was measured on a PCR product serial dilution. Quantitative real-time RT-PCR primer sequences are listed in Supplemental Table S1.
Protoplast Isolation and Transient Expression Assays
The coding sequence of VvABF2 was cloned as a C-terminal fusion in frame with the GFP into the pRT101 vector (Kiegerl et al., 2000) and expressed under the control of the cauliflower mosaic virus promoter.
The promoter regions of VvLEA (VIT_08s0007g04240), VvNAC (VIT_19s0014g03290), and VvBenzoR (VIT_07s0005g00140) were inserted upstream of the GUS reporter gene into pAM35 (Guerineau et al., 2003). Primer sequences used for promoter cloning are listed in Supplemental Table S1.
Tobacco (Nicotiana tabacum ‘SR1’) in vitro plants used for protoplast preparation were cultivated in a growth chamber with a constant temperature of 25°C and a 14-h/10-h day/night photoperiod. Protoplasts were obtained from young leaves of 15-d-old subcultured plants according to Hosy et al. (2005). A 10-µg aliquot of each plasmid DNA was used for polyethylene glycol-mediated cotransformation. Transfected protoplasts were incubated for 16 h at 25°C.
GFP fluorescence was analyzed by confocal microscopy. A fluorometric GUS assay was performed following the protocol described by Jefferson et al. (1987). Protoplasts were centrifuged briefly for 30 s and vortexed with a GUS extraction buffer containing 50 mm sodium phosphate (pH 7.0), 10 mm β-mercaptoethanol, 10 mm Na2-EDTA (pH 8.0), and 0.1% Triton X-100. The extracts were centrifuged for 15 min in a microcentrifuge at 4°C, and the supernatants were incubated at 37°C for GUS assay using 1 mm 4-methylumbelliferyl-β-d-glucuronide as a substrate. The reaction was stopped with 0.2 m Na2CO3. The amount of methylumbelliferone production was determined using a fluorometer (Versafluor fluorometer; Bio-Rad). Total protein content of the samples was determined by the method of Bradford (1976).
Microarray Experiments and Analyses
Three biological replicates of each sample (untreated control, control + ABA, 35S::VvABF2, and 35S::VvABF2 + ABA) were hybridized on NimbleGen microarray 090818 Vitis exp HX12 (Roche, NimbleGen), bearing a set of probes for 29,582 unigenes based on the 12X grapevine V1 gene model prediction (http://genomes.cribi.unipd.it/). The chip probe design is available at http://ddlab.sci.univr.it/FunctionalGenomics/. Robust multiarray average-processed data are available at PLEXdb (Wise et al., 2007) with accession number VV30:VvABFOx. Data analyses were performed using R/Bioconductor (Gentleman et al., 2004). Expression intensities were background corrected, quantile normalized, and summarized using the robust multiarray average function of the oligo package (Carvalho and Irizarry, 2010). Differentially expressed genes were identified using the Limma package (Smyth, 2004) for the following contrasts: 35S::VvABF2 versus control, 35S::VvABF2 + ABA versus control, and control + ABA versus control. Genes with absolute log2-fold changes above 1 (ratio of 2) and false discovery rate (Benjamini and Hochberg, 1995)-corrected P values below 0.05 were considered significant. Expression profiles of these genes were clustered using the MapMan software (Thimm et al., 2004). Gene models were aligned against the UnirRef100-2011-07 database (Suzek et al., 2007) using the BLAST program (Altschul et al., 1997). Genes considered as differentially expressed were associated with the MapMan Ontology (Thimm et al., 2004) using the Mercator Online tool (http://mapman.gabipd.org/web/guest/app/mercator). The most extreme categories within these differentially expressed genes were identified using a Wilcoxon rank-sum test implemented in the MapMan tool (Usadel et al., 2005).
Sequence Analysis
Full-length amino acid sequences of bZIP transcription factors from several species were retrieved from public databases. Multiple amino acid sequence alignment was generated using the Clustal Omega alignment algorithm with default parameters (Sievers et al., 2011; http://www.ebi.ac.uk/Tools/msa/clustalo/). The phylogenetic tree was constructed from the Clustal Omega alignment using the MEGA5 package (Tamura et al., 2011). The evolutionary history was inferred using the neighbor-joining method. The optimal tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and were in units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). There were a total of 117 positions in the final data set.
In Silico VvABF2 Promoter Region Identification and Analysis
The identification and in silico analysis of the VvABF2 promoter region were conducted using the Genomatix suite of programs (http://www.genomatix.de [Genomatix Software]; Quandt et al., 1995). The Gene2promotor program from the Genomatix software package was used to define 1,344 bp upstream of the transcription start site of the VvABF2 promoter region. The corresponding sequence was then used as the target sequence for transcription factor recognition site identification using the MatInspector version 8.06 program (Cartharius et al., 2005). The parameters used were the Matrix Family Library version 8.4 (June 2011), the standard (0.75) core similarity, and the optimized matrix similarity.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. VvABF2 binds to promoter fragments containing ABRE motifs.
Supplemental Figure S2. Time course of total stilbene production (nmol g−1 dry weight) in the culture medium of control and 35S::VvABF2 transgenic cells after the addition of 20 μm ABA.
Supplemental Figure S3. Mature fruit phenotypes of VvABF2-overexpressing tomato plants.
Supplemental Table S1. Potential ABRE cis-acting elements identified in the 5′ regulatory region of VvLEA, VvBenzoR, and VvNAC.
Supplemental Table S2. Potential cis-acting elements identified in the 5′ regulatory region of VvABF2.
Supplemental Table S3. Genes involved in the response to oxidative stress and affected in the response to ABA and/or VvABF2 overexpression.
Supplemental Table S4. PCR primers used to amplify gene- and promoter-specific regions for cloning and expression analyses.
Acknowledgments
We thank Zhanwu Dai for the statistical analysis and the Bordeaux Imaging Center for the confocal facility.
Glossary
- ABA
abscisic acid
- IAA
indole-3-acetic acid
- bZIP
basic leucine zipper family
- ABRE
ABA response element
- cDNA
complementary DNA
- WAF
weeks after flowering
- RT
reverse transcription
- PAL
phenylalanine ammonia lyase
- ABC
ATP-binding cassette
- DREB
dehydration-responsive element-binding protein
- PR
pathogenesis-related
- ROS
reactive oxygen species
References
- Adams-Phillips L, Barry C, Giovannoni J. (2004) Signal transduction systems regulating fruit ripening. Trends Plant Sci 9: 331–338 [DOI] [PubMed] [Google Scholar]
- Aharoni A, O’Connell AP. (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir Hossain M, Lee Y, Cho JI, Ahn CH, Lee SK, Jeon JS, Kang H, Lee CH, An G, Park PB. (2010) The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol Biol 72: 557–566 [DOI] [PubMed] [Google Scholar]
- Antolín MC, Baigorri H, De Luis I, Aguirrezábal F, Geny L, Broquedis M, Sánchez-Díaz M. (2003) ABA during reproductive development in non-irrigated grapevines (Vitis vinifera L. cv. Tempranillo). Aust J Grape Wine Res 9: 169–176 [Google Scholar]
- Antoni R, Rodriguez L, Gonzalez-Guzman M, Pizzio GA, Rodriguez PL. (2011) News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr Opin Plant Biol 14: 547–553 [DOI] [PubMed] [Google Scholar]
- Ban T, Ishimaru M, Kobayashi S, Goto-Yamamoto SN, Horiuchi S. (2003) Abscisic acid and 2,4-dichlorophenoxyacetic acid affect the expression of anthocyanin biosynthetic pathway genes in ‘Kyoho’ grape berries. J Hortic Sci Biotechnol 78: 586–589 [Google Scholar]
- Bastías A, López-Climent M, Valcárcel M, Rosello S, Gómez-Cadenas A, Casaretto JA. (2011) Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol Plant 141: 215–226 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57: 289–300 [Google Scholar]
- Böttcher C, Boss PK, Davies C. (2011) Acyl substrate preferences of an IAA-amido synthetase account for variations in grape (Vitis vinifera L.) berry ripening caused by different auxinic compounds indicating the importance of auxin conjugation in plant development. J Exp Bot 62: 4267–4280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher C, Keyzers RA, Boss PK, Davies C. (2010) Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J Exp Bot 61: 3615–3625 [DOI] [PubMed] [Google Scholar]
- Bradamante S, Barenghi L, Villa A. (2004) Cardiovascular protective effects of resveratrol. Cardiovasc Drug Rev 22: 169–188 [DOI] [PubMed] [Google Scholar]
- Buchanan CD, Lim S, Salzman RA, Kagiampakis I, Morishige DT, Weers BD, Klein RR, Pratt LH, Cordonnier-Pratt MM, Klein PE, et al. (2005) Sorghum bicolor’s transcriptome response to dehydration, high salinity and ABA. Plant Mol Biol 58: 699–720 [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21: 2933–2942 [DOI] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA. (2010) A framework for oligonucleotide microarray preprocessing. Bioinformatics 26: 2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai YM, Jia HF, Li CL, Dong QH, Shen YY. (2011) FaPYR1 is involved in strawberry fruit ripening. J Exp Bot 62: 5079–5089 [DOI] [PubMed] [Google Scholar]
- Chervin C, El-Kereamy A, Roustan JP, Latche A, Lamon J, Bouzayen M. (2004) Ethylene seems required for the berry development and ripening in grape, a non-climacteric fruit. Plant Sci 167: 1301–1305 [Google Scholar]
- Chervin C, Tira-Umphon A, Terrier N, Zouine M, Severac D, Roustan JP. (2008) Stimulation of the grape berry expansion by ethylene and effects on related gene transcripts, over the ripening phase. Physiol Plant 134: 534–546 [DOI] [PubMed] [Google Scholar]
- Choi H, Hong J, Ha J, Kang J, Kim SY. (2000) ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275: 1723–1730 [DOI] [PubMed] [Google Scholar]
- Coombe BG. (1992) Research on development and ripening of the grape berry. Am J Enol Vitic 43: 101–110 [Google Scholar]
- Coombe BG, Hale CR. (1973) The hormone content of ripening grape berries and the effects of growth substance treatments. Plant Physiol 51: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa LG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M. (2008) The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS ONE 3: e2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutos-Thévenot P, Goebel-Tourand I, Mauro MC, Jouanneau JP, Boulay M, Deloire A, Guern J. (1992a) Somatic embryogenesis from grapevine cells. I. Improvement of embryo development by changes in culture conditions. Plant Cell Tissue Organ Cult 29: 125–133 [Google Scholar]
- Coutos-Thévenot P, Maës O, Jouenne T, Mauro MC, Boulay M, Deloire A, Guern J. (1992b) Extracellular protein patterns of grapevine cell suspensions in embryogenic and non-embryogenic situations. Plant Sci 86: 137–145 [Google Scholar]
- Cuéllar T, Azeem F, Andrianteranagna M, Pascaud F, Verdeil JL, Sentenac H, Zimmermann S, Gaillard I. (2013) Potassium transport in developing fleshy fruits: the grapevine inward K(+) channel VvK1.2 is activated by CIPK-CBL complexes and induced in ripening berry flesh cells. Plant J 73: 1006–1018 [DOI] [PubMed] [Google Scholar]
- Cuéllar T, Pascaud F, Verdeil JL, Torregrosa L, Adam-Blondon AF, Thibaud JB, Sentenac H, Gaillard I. (2010) A grapevine Shaker inward K(+) channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J 61: 58–69 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Davies C, Boss PK, Robinson SP. (1997) Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol 115: 1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Robinson SP. (2000) Differential screening indicates a dramatic change in mRNA profiles during grape berry ripening: cloning and characterization of cDNAs encoding putative cell wall and stress response proteins. Plant Physiol 122: 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies C, Shin R, Liu W, Thomas MR, Schachtman DP. (2006) Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. J Exp Bot 57: 3209–3216 [DOI] [PubMed] [Google Scholar]
- Deluc LG, Quilici DR, Decendit A, Grimplet J, Wheatley MD, Schlauch KA, Mérillon JM, Cushman JC, Cramer GR. (2009) Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genomics 10: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, A-H-Mackerness S, Hancock JT, Neill SJ. (2001) Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol 127: 159–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103: 1988–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagné S, Cluzet S, Merillon JM, Gény L. (2011) ABA initiates anthocyanin production in grape cell cultures. J Plant Growth Regul 30: 1–10 [Google Scholar]
- Gambetta GA, Matthews MA, Shaghasi TH, McElrone AJ, Castellarin SD. (2010) Sugar and abscisic acid signaling orthologs are activated at the onset of ripening in grape. Planta 232: 219–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerineau F, Benjdia M, Zhou DX. (2003) A jasmonate-responsive element within the A. thaliana vsp1 promoter. J Exp Bot 54: 1153–1162 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ. (2007) Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol 10: 283–289 [DOI] [PubMed] [Google Scholar]
- Giribaldi M, Gény L, Delrot S, Schubert A. (2010) Proteomic analysis of the effects of ABA treatments on ripening Vitis vinifera berries. J Exp Bot 61: 2447–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez C, Terrier N, Torregrosa L, Vialet S, Fournier-Level A, Verriès C, Souquet JM, Mazauric JP, Klein M, Cheynier V, et al. (2009) Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol 150: 402–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Porras JL, Riaño-Pachón DM, Dreyer I, Mayer JE, Mueller-Roeber B. (2007) Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8: 260–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A. (2007) The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J 51: 642–655 [DOI] [PubMed] [Google Scholar]
- Hardtke CS. (2007) Transcriptional auxin-brassinosteroid crosstalk: who’s talking? Bioessays 29: 1115–1123 [DOI] [PubMed] [Google Scholar]
- Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A. (2002) Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol 43: 136–140 [DOI] [PubMed] [Google Scholar]
- Hofseth LJ, Singh UP, Singh NP, Nagarkatti M, Nagarkatti PS. (2010) Taming the beast within: resveratrol suppresses colitis and prevents colon cancer. Aging (Albany NY) 2: 183–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosy E, Duby G, Véry AA, Costa A, Sentenac H, Thibaud JB. (2005) A procedure for localisation and electrophysiological characterisation of ion channels heterologously expressed in a plant context. Plant Methods 1: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka M. (2004) Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci 167: 247–252 [Google Scholar]
- Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. (2011) Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol 157: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69: 91–105 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Joyce DC, Macnish AJ. (2000) Effect of abscisic acid on banana fruit ripening in relation to the role of ethylene. J Plant Growth Regul 19: 106–111 [DOI] [PubMed] [Google Scholar]
- Jiménez A, Creissen G, Kular B, Firmin J, Robinson S, Verhoeyen M, Mullineaux P. (2002) Changes in oxidative processes and components of the antioxidant system during tomato fruit ripening. Planta 214: 751–758 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, Liwosz A, Eklöf S, Till S, Bögre L, Hirt H, et al. (2000) SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell 12: 2247–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobashi K, Gemma H, Iwahori S. (1999) Sugar accumulation in peach fruit as affected by abscisic acid treatment in relation to some sugar metabolizing enzymes. J Jpn Soc Hortic Sci 68: 465–470 [Google Scholar]
- Koyama K, Sadamatsu K, Goto-Yamamoto N. (2010) Abscisic acid stimulated ripening and gene expression in berry skins of the Cabernet Sauvignon grape. Funct Integr Genomics 10: 367–381 [DOI] [PubMed] [Google Scholar]
- Lecourieux F, Lecourieux D, Vignault C, Delrot S. (2010) A sugar-inducible protein kinase, VvSK1, regulates hexose transport and sugar accumulation in grapevine cells. Plant Physiol 152: 1096–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xin H, Zheng XF, Li S, Hu Z. (2012) Identification of the abscisic acid receptor VvPYL1 in Vitis vinifera. Plant Biol (Stuttg) 14: 244–248 [DOI] [PubMed] [Google Scholar]
- Li X, Xu CJ, Korban SS, Chen KS. (2010) Regulatory mechanisms of textural changes in ripening fruits. Crit Rev Plant Sci 29: 222–243 [Google Scholar]
- Liu H, Stone SL. (2010) Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22: 2630–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yue Y, Li B, Nie Y, Li W, Wu WH, Ma L. (2007) A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315: 1712–1716 [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Mauro MC, Toutain S, Walter B, Pinck L, Otten L, Coutos-Thévenot P, Deloire A, Barbier P. (1995) High efficiency regeneration of grapevine plants transformed with the GFLV coat protein gene. Plant Sci 112: 97–106 [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Miyakawa T, Sawano Y, Kubota K, Kang H-J, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. (2009) Structural basis of abscisic acid signalling. Nature 462: 609–614 [DOI] [PubMed] [Google Scholar]
- Mok MC (1994) Cytokinins and plant development: an overview. In DWS Mok, MC Mok, eds, Cytokinins: Chemistry, Activity and Function. CRC Press, Boca Raton, FL, pp 155–156 [Google Scholar]
- Nakashima K, Fujita Y, Kanamori N, Katagiri T, Umezawa T, Kidokoro S, Maruyama K, Yoshida T, Ishiyama K, Kobayashi M, et al. (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol 50: 1345–1363 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Nicolas P, Lecourieux D, Gomès E, Delrot S, Lecourieux F. (2013) The grape berry-specific basic helix-loop-helix transcription factor VvCEB1 affects cell size. J Exp Bot 64: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, Schroeder JI, Getzoff ED. (2009) Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326: 1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N, Yoshida T, Kitahata N, Asami T, Shinozaki K, Hirayama T. (2007) ABA-Hypersensitive Germination1 encodes a protein phosphatase 2C, an essential component of abscisic acid signaling in Arabidopsis seed. Plant J 50: 935–949 [DOI] [PubMed] [Google Scholar]
- Østergaard L, Lauvergeat V, Naested H, Mattsson O, Mundy J. (2001) Two differentially regulated Arabidopsis genes define a new branch of the DFR superfamily. Plant Sci 160: 463–472 [DOI] [PubMed] [Google Scholar]
- Pan QH, Li MJ, Peng CC, Zhang N, Zou X, Zou KQ, Wang XL, Yu XC, Wang XF, Zhang DP. (2005) Abscisic acid activates acid invertases in developing grape berry. Physiol Plant 125: 157–170 [Google Scholar]
- Pandey S, Nelson DC, Assmann SM. (2009) Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 136: 136–148 [DOI] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peppi MC, Fidelibus MW, Dokoozlian N. (2006) Abscisic acid application timing and concentration affect firmness, pigmentation, and color of ‘Flame Seedless’ grapes. Hortic Sci 41: 1440–1445 [Google Scholar]
- Pezet R, Gindro K, Viret O, Spring JL. (2004) Glycosylation and oxidative dimerization of resveratrol are respectively associated to sensitivity and resistance of grapevine cultivars to downy mildew. Physiol Mol Plant Pathol 65: 297–303 [Google Scholar]
- Pilati S, Perazzolli M, Malossini A, Cestaro A, Demattè L, Fontana P, Dal Ri A, Viola R, Velasco R, Moser C. (2007) Genome-wide transcriptional analysis of grapevine berry ripening reveals a set of genes similarly modulated during three seasons and the occurrence of an oxidative burst at vèraison. BMC Genomics 8: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribéreau-Gayon Y, Glories Y, Maujean A, Dubourdieu D. (2000) Handbook of Enology, Vol 2: The Chemistry of Wine and Stabilization and Treatments. John Wiley & Sons, Chichester, England [Google Scholar]
- Richings EW, Cripps RF, Cowan AK. (2000) Factors affecting ‘Hass’ avocado fruit size: carbohydrate, abscisic acid and soprenoid metabolism in normal and phenotypically small fruit. Physiol Plant 109: 81–89 [Google Scholar]
- Rodrigo MJ, Marcos JF, Alférez F, Mallent MD, Zacarías L. (2003) Characterization of Pinalate, a novel Citrus sinensis mutant with a fruit-specific alteration that results in yellow pigmentation and decreased ABA content. J Exp Bot 54: 727–738 [DOI] [PubMed] [Google Scholar]
- Santiago J, Dupeux F, Round A, Antoni R, Park SY, Jamin M, Cutler SR, Rodriguez PL, Márquez JA. (2009) The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462: 665–668 [DOI] [PubMed] [Google Scholar]
- Scienza A, Miravalle CV, Fregoni M. (1978) Relationships between seed number, gibberellin and abscisic acid levels and ripening in ‘Cabernet Sauvignon’ grape berries. Vitis 17: 361–368 [Google Scholar]
- Seki M, Ishida J, Narusaka M, Fujita M, Nanjo T, Umezawa T, Kamiya A, Nakajima M, Enju A, Sakurai T, et al. (2002) Monitoring the expression pattern of around 7,000 Arabidopsis genes under ABA treatments using a full-length cDNA microarray. Funct Integr Genomics 2: 282–291 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim S-G, Lee YH, Park WJ, Park CM. (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Taylor JE, Tucker GA. (1993) Biochemistry of Fruit Ripening. Chapman and Hall, London [Google Scholar]
- Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q, Wu FQ, Wang XF, Du SY, Jiang T, et al. (2010) The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22: 1909–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YY, Wang XF, Wu FQ, Du SY, Cao Z, Shang Y, Wang XL, Peng CC, Yu XC, Zhu SY, et al. (2006) The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443: 823–826 [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen DG, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55: 555–590 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: e3. [DOI] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. (2011) Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J Exp Bot 62: 5659–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. (2007) UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23: 1282–1288 [DOI] [PubMed] [Google Scholar]
- Symons GM, Davies C, Shavrukov Y, Dry IB, Reid JB, Thomas MR. (2006) Grapes on steroids: brassinosteroids are involved in grape berry ripening. Plant Physiol 140: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkudelska K, Szkudelski T. (2010) Resveratrol, obesity and diabetes. Eur J Pharmacol 635: 1–8 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tosti N, Pasqualini S, Borgogni A, Ederli L, Falistocco E, Crispi S, Paolocci F. (2006) Gene expression profiles of O3-treated Arabidopsis plants. Plant Cell Environ 29: 1686–1702 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D, et al. (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderauwera S, De Block M, Van de Steene N, van de Cotte B, Metzlaff M, Van Breusegem F. (2007) Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci USA 104: 15150–15155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RS, Pandey S, Li S, Gookin TE, Zhao Z, Albert R, Assmann SM. (2011) Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner JJ, Peterson FC, Volkman BF, Cutler SR. (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler S, Loveys B, Ford C, Davies C. (2009) The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust J Grape Wine Res 15: 195–204 [Google Scholar]
- Wise RP, Caldo RA, Hong L, Shen L, Cannon EK, Dickerson JA (2007) BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens. Methods Mol Biol 406: 347–363 [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yamaki S, Asakura T. (1991) Stimulation of the uptake of sorbitol into vacuoles from apple fruit flesh by abscisic acid and into protoplasts by indoleacetic acid. Plant Cell Physiol 32: 315–318 [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Yu XC, Li MJ, Gao GF, Feng HZ, Geng XQ, Peng CC, Zhu SY, Wang XJ, Shen YY, Zhang DP. (2006) Abscisic acid stimulates a calcium-dependent protein kinase in grape berry. Plant Physiol 140: 558–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Leng P, Zhang G, Li X. (2009a) Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J Plant Physiol 166: 1241–1252 [DOI] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P. (2009b) The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot 60: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Cai Z, Wang X. (2009) The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA 106: 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dixon RA. (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21: 2323–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]