A genome-wide physical map of barley was constructed and anchored genetically by a novel method involving whole-genome resequencing of a mapping population.
Abstract
Barley (Hordeum vulgare) is an important cereal crop and a model species for Triticeae genomics. To lay the foundation for hierarchical map-based sequencing, a genome-wide physical map of its large and complex 5.1 billion-bp genome was constructed by high-information content fingerprinting of almost 600,000 bacterial artificial chromosomes representing 14-fold haploid genome coverage. The resultant physical map comprises 9,265 contigs with a cumulative size of 4.9 Gb representing 96% of the physical length of the barley genome. The reliability of the map was verified through extensive genetic marker information and the analysis of topological networks of clone overlaps. A minimum tiling path of 66,772 minimally overlapping clones was defined that will serve as a template for hierarchical clone-by-clone map-based shotgun sequencing. We integrated whole-genome shotgun sequence data from the individuals of two mapping populations with published bacterial artificial chromosome survey sequence information to genetically anchor the physical map. This novel approach in combination with the comprehensive whole-genome shotgun sequence data sets allowed us to independently validate and improve a previously reported physical and genetic framework. The resources developed in this study will underpin fine-mapping and cloning of agronomically important genes and the assembly of a draft genome sequence.
Barley (Hordeum vulgare) is an important source of human and animal nutrition and underpins the malting and brewing industries. It is among the earliest domesticated crop plants and is adapted to a wide range of environmental conditions. A wealth of genomic resources such as dense genetic maps, ESTs, complementary DNA libraries, and an atlas of gene expression have been developed in the past two decades (for review, see Schulte et al., 2009). Moreover, comprehensive germplasm collections from cultivars and wild accessions (van Hintum and Menting, 2003) as well as extensive mutant collections (Druka et al., 2011) provide a sound foundation for genetic studies into developmental and morphological processes. However, the full exploitation of these resources for basic research and crop improvement has been hampered by the lack of a reference genome sequence. The extent to which genomic research in a crop species may be spurred by the availability of a reference genome is aptly illustrated by the large number of agronomically important rice (Oryza sativa) genes that have been successfully cloned since the release of the rice genome sequence (for review, see Huang et al., 2013).
Before the advent of next-generation sequencing (NGS) technology, the scale of a barley genome project seemed daunting, owing to the large size (5.1 Gb) and the high repeat content. After a combination of sequencing of a small number of bacterial artificial chromosomes (BACs; Wicker et al., 2006; Steuernagel et al., 2009; Taudien et al., 2011) and shallow whole-genome sequencing (Wicker et al., 2008) demonstrated the utility of NGS for assembling a large and complex genome, the prospects of obtaining a high-utility sequence of the barley genome were considerably enhanced. In addition, the development of innovative sequence assembly algorithms made it possible to obtain robust whole-genome shotgun assemblies of mammalian genomes using only paired-end NGS sequencing reads of different insert sizes (Gnerre et al., 2011). Using these analytical approaches and libraries with various insert sizes, whole-genome shotgun assemblies of the bread wheat (Triticum aestivum) progenitors Triticum urartu (Ling et al., 2013) and Aegilops tauschii (Jia et al., 2013) have been published recently.
A major challenge in applying whole-genome shotgun sequencing to large and complex plant genomes is their highly repetitive structure. In barley, stretches of genomic sequence as large as several hundred kilobases may be entirely composed of nested transposable elements (Wicker et al., 2005). Further complications arise from highly similar families of paralogous genes, the remnants of recent or ancient polyploidization events (Levy and Feldman, 2002) or pseudogene formation (Wicker et al., 2011). As a result of these difficulties, so-called gene-space assemblies from whole-genome sequencing data have been widely adopted as an enabling alternative for many research applications, particularly when the majority of the gene complement has been successfully embedded into a broader genomic context. Such a milestone was recently completed for barley with the release of a gene-space assembly embedded in a sequence-enriched physical and genetic framework (The International Barley Genome Sequencing Consortium, 2012). Half of the physical contigs were genetically positioned through integration of a genome-wide physical map, with sequence data derived from BAC end sequencing, sequencing of complete BAC clones, whole-genome shotgun contigs, and shotgun sequencing of sorted chromosome arms. The integration of transcriptome sequence into this framework defined and provided context and definition for 26,159 high-confidence gene models.
Despite this achievement, the International Barley Genome Sequencing Consortium has continued to promote a map-based sequencing strategy encompassing restriction-based fingerprinting of deep-coverage BAC libraries, an approach that has had successful precedents in Arabidopsis (Arabidopsis thaliana), rice, and maize (Zea mays) and several animal genomes (Arabidopsis Genome Initiative, 2000; Lander et al., 2001; Waterston et al., 2002; International Rice Genome Sequencing Project, 2005; Schnable et al., 2009). Map-based or hierarchical sequencing approaches reduce the complexity of the sequence assembly process by partitioning the genome into smaller pieces. Although this involves the time-consuming and laborious steps of BAC library construction, restriction endonuclease fingerprinting, BAC contig assembly, and subsequent clone-by-clone sequencing, it currently remains indispensable in plant genome projects that have the goal of constructing contiguous pseudomolecules (Feuillet et al., 2011, 2012).
We recently developed a method termed POPSEQ (for population sequencing) for linearly ordering whole-genome shotgun sequence (WGS) assemblies in the absence of highly developed genomic resources. As a proof of concept, shallow sequencing two small biparental mapping populations was shown to greatly improve the genetic anchoring of the International Barley Genome Sequencing Consortium whole-genome shotgun assembly of the barley cv Morex (Mascher et al., 2013). While we demonstrated that POPSEQ anchoring of a whole-genome shotgun assembly is independent of physical map construction, we also argued that the data obtained should expedite the ongoing physical mapping project. In particular, the high marker density afforded by whole-genome sequencing should enable direct anchoring of the majority of fully sequenced BAC clones. Here, we extend and improve the previously established physical and genetic framework of barley (The International Barley Genome Sequencing Consortium, 2012). We describe the process of physical map construction and report the sequence-ready minimum tiling path (MTP) of overlapping clones that will be instrumental in producing a draft genome sequence. We illustrate the usefulness of our resource for map-based cloning and introduce POPSEQ as a new approach for genetic anchoring of physical maps.
RESULTS
Assembly of the Physical Map
Six large-insert BAC libraries of barley cv Morex had been constructed and characterized previously (Yu et al., 2000; Schulte et al., 2011). Additionally, gene-containing BACs had been identified in the library HVVMRXALLhA and rearrayed for fingerprinting (The International Barley Genome Sequencing Consortium, 2012). We analyzed 690,912 barley BAC clones from these six libraries by high-information content fingerprinting (HICF; Luo et al., 2003; Table I). Fingerprint profiles were checked for plate-wide, neighboring, or chloroplast contamination as well as for clones containing more than 250 or less than 30 fragments. A total of 571,007 (82.6%) clones representing approximately 14-fold haploid genome coverage passed our quality filters and were imported into the FingerPrint Contig (FPC) assembly program (Soderlund et al., 2000). For the initial assembly, we used very stringent parameters to avoid misassemblies due to highly similar repetitive regions present at high number in the barley genome and to construct a robust framework that was to be refined in further steps. This preliminary assembly organized 481,158 clones into 18,570 fingerprinted contigs (FP contigs), leaving 89,849 clones as singletons.
Table I. Fingerprinting statistics.
| BAC Librarya | Isolated Clones | Clones Exported into FPC | Clones in Contigs | Singleton Clones |
|---|---|---|---|---|
| HVVMRXALLhA/hB/hCb | 123,072 | 103,508 | 87,940 | 15,568 |
| HVVMRXALL83KhAc | 77,568 | 69,905 | 65,078 | 4,827 |
| HVVMRXALLeA | 147,456 | 122,819 | 117,529 | 5,289 |
| HVVMRXALLmA | 202,752 | 169,361 | 161,905 | 7,455 |
| HVVMRXALLrA | 140,064 | 105,414 | 84,750 | 20,664 |
| All libraries | 690,912 | 571,007 | 517,202 | 53,805 |
Naming is according to Schulte et al. (2011). The second to last letter encodes the mode of library construction (h, e, m, digest with HindIII, EcoRI, or MboI, respectively; r, random mechanic shearing). bCombined counts for all HindIII libraries are given. cHVVMRXALL83KhA is a subset of HVVMRXALLhA. Gene-bearing clones had been identified and rearrayed for fingerprinting (The International Barley Genome Sequencing Consortium, 2012).
Subsequently, further automatic assembly iterations were performed with a step-wise reduction of stringency (Supplemental Fig. S1), reducing the number of contigs to 9,436 and incorporating 37,050 additional clones. Manual editing of this contig set identified 171 misassembled contigs, each harboring two groups of markers from different genetic positions. These contigs were split up and rebuilt. Less stringent overlap criteria (Sulston score 1e-25) supported by genetic marker data allowed us to manually merge 130 contigs. Finally, 313 contigs composed only of two equivalent clones from BAC library HVVMRXALLhA and its rearrayed correspondent HVVMRX83KhA were considered as singletons. Manual editing resulted in a final build of 9,265 contigs with an average size of 538 kb (Table II). The assembly consists of approximately 4.0 million unique consensus bands and is estimated to cover 4.9 Gb (96%) of the 5.1-Gb genome of barley. In relation to genome size, the number of contigs in the final barley map is proportional to metrics of the rice physical map (1,019 contigs, approximately 450-Mb genome; Chen et al., 2002) and to the physical map of the 1-Gb bread wheat chromosome 3B (1,036 contigs; Paux et al., 2008).
Table II. Features of the barley physical map.
| Contigs | 9,265 |
| Clones in contigs | 517,202 |
| Singletons | 53,805 |
| Contigs containing | |
| >100 clones | 3,151 |
| 50–100 clones | 1,538 |
| 25–49 clones | 1,478 |
| 3–24 clones | 3,285 |
| 2 clones | 1,035 |
| Map length | 4.9 Gb |
| Average contig size | 538 kb |
| N50 | 904 kb |
Validation of the Physical Map
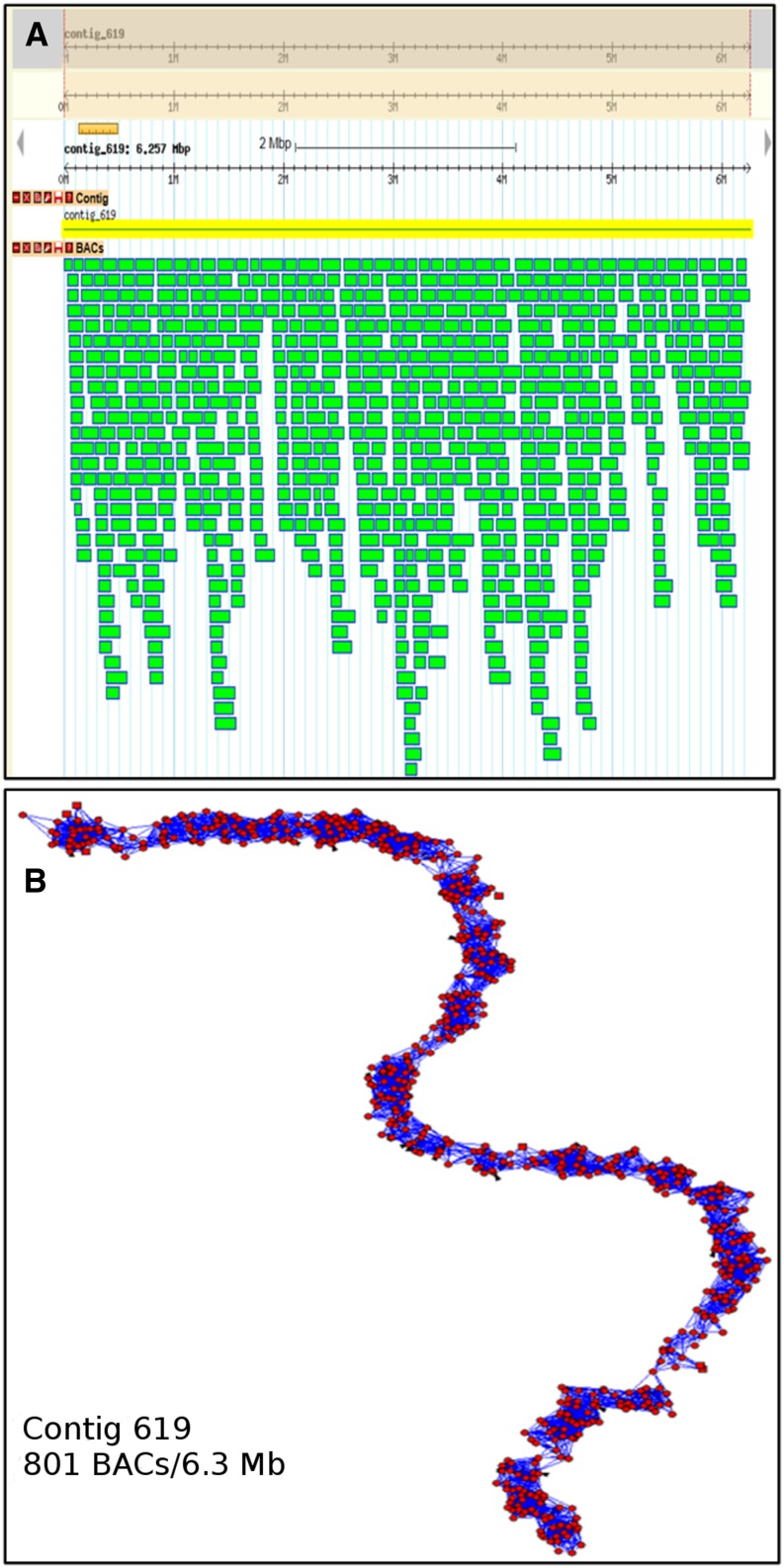
In addition to cross-checking contigs with genetic marker information during manual editing, the reliability of the map was corroborated by using an alternative program for the analysis of HICF data. The linear topology contigs (LTC) tool (Frenkel et al., 2010) had been specifically designed for the construction and verification of physical maps of complex genomes. In particular, LTC is able to detect misassembled contigs, where the net of clone overlaps has branched or cyclical topological structures contradicting the linearity of the chromosomes are observed. LTC analysis confirmed that about 90% of contigs constructed by FPC consisted only of reliably overlapping clones in a linear order (Fig. 1). The majority of the remaining 10% of putatively branched contigs contained only one pair of clones with unreliable overlap. Such contigs were considered as less problematic (hence retained) because single unreliable clone overlap can result from low-quality fingerprinting.
Figure 1.
BAC contig example and visualization. A, Visualization of FPcontig_619 and associated clones in Gbrowse (http://seacow.helmholtz-muenchen.de/cgi-bin/gb2/gbrowse/Barley_PhysMap). B, The topology of FPcontig_619 shown as a graph structure with the LTC tool.
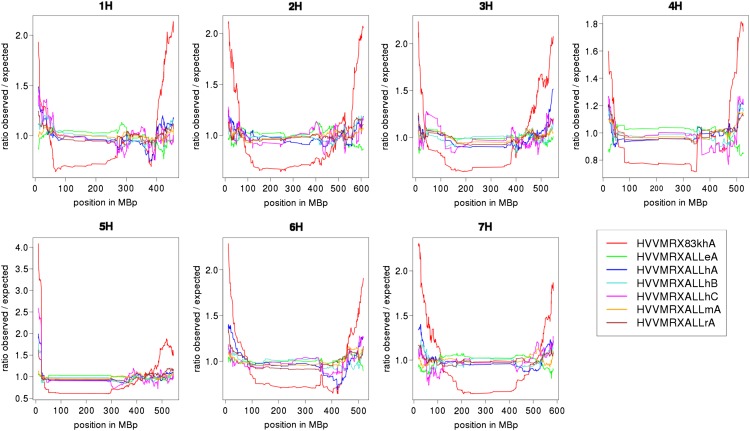
We checked the distribution of clones from different libraries across the genome (Fig. 2). Clones from the gene-enriched library HVVMRX83KhA were preferentially located in the distal part of the chromosomes, which have a higher gene density compared with pericentromeric regions (The International Barley Genome Sequencing Consortium, 2012). The other libraries showed a fairly equal distribution along the chromosomes.
Figure 2.
Normalized distribution of clones from different libraries along chromosomes. The distribution of clones from different libraries was plotted along the seven barley chromosomes. The counts of clones from different libraries were normalized by calculating the ratio between observed and expected clone number in a sliding window. Contigs toward telomeres were enriched for clones of library HVVMRX83khA, containing BAC clones identified for the presence of genes (Madishetty et al., 2007).
Establishment of an MTP
A MTP is a series of clones that covers the genome with a predefined minimal overlap between pairs of adjacent clones. MTPs are constructed to minimize the number of BAC clones that have to be sequenced for complete coverage of the genome. Estimating the exact order of contigs and the overlap between pairs of adjacent contigs only using fingerprinting profiles is computationally challenging. We applied the LTC tool to pick an MTP for each of the 9,265 BAC contigs. After recalculating the global order of clones in each contig, LTC selected an MTP consisting of 68,047 clones. Using the genetic anchoring of contigs, the MTP BACs were rearrayed into 178 microtiter plates, grouping together clones from the same chromosome and separating all clones of unanchored contigs. DNA from the clones for each chromosome was combined into plate, row, and column pools to facilitate future screening. Individual MTP clones as well as BAC pools can be obtained from the French Plant Genomic Resource Centre (CNRGV; http://cnrgv.toulouse.inra.fr/en/library/barley). A list of all MTP clones is available as Supplemental Table S1.
Anchoring the Physical Map by POPSEQ
Previously, we had anchored 4,556 physical contigs (3.9 Gb) to genetic positions through approximately 3,000 single-nucleotide polymorphism (SNP) markers and approximately 500,000 genotyping-by-sequencing markers (The International Barley Genome Sequencing Consortium, 2012). In addition, 1,881 contigs could be assigned to chromosome arms by using sequence data from flow-sorted chromosome arms (The International Barley Genome Sequencing Consortium, 2012). Most contigs without genetic or chromosomal positions were either short or lacked sequence or marker information. We recently introduced the POPSEQ approach to genetically anchor highly fragmented sequence assemblies by whole-genome sequencing of individuals of a segregating population (Mascher et al., 2013). The same methodology may be used to place BAC contigs or single BAC clones into a genetic framework. For this purpose, we first projected the genetically anchored WGS contigs onto the physical map. The WGS assembly is a necessary intermediary, as the sequence information attached to the physical map is incomplete and consequently cannot serve as an appropriate reference for short-read alignment and SNP-calling algorithms. We had anchored WGS contigs of barley cv Morex using two different mapping populations (Morex × Barke and the Oregon Wolfe Barleys [OWB]). The iSelect framework (Comadran et al., 2012) with 1,690 genetic bins and a genotyping-by-sequencing map with 983 bins were used for Morex × Barke and OWB, respectively. The iSelect framework was also used in the previous effort to anchor the physical map of barley (The International Barley Genome Sequencing Consortium, 2012).
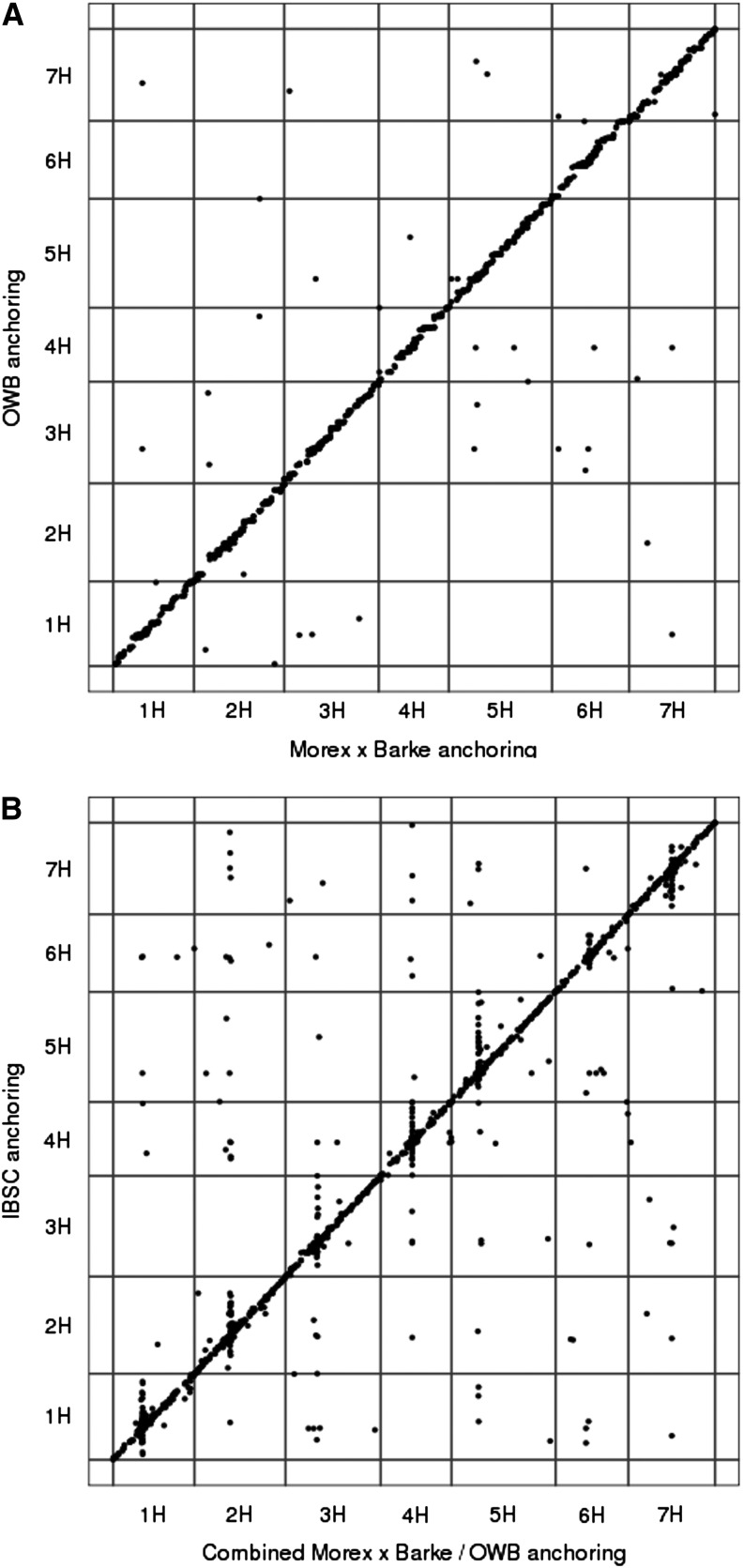
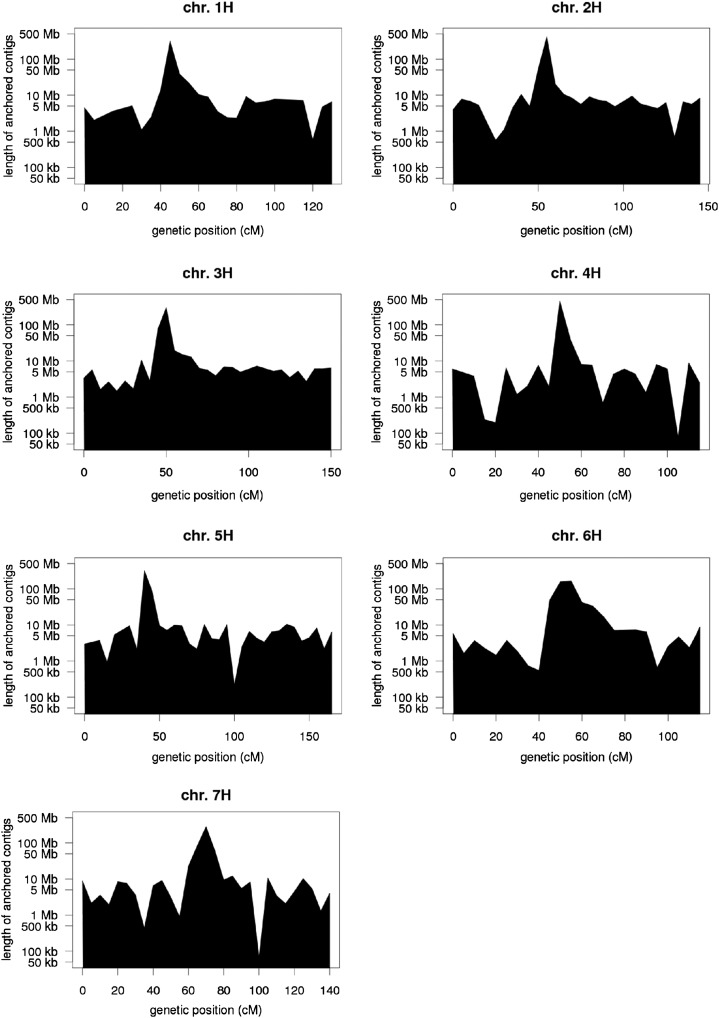
By stringent homology searches against fully sequenced BACs and BAC end sequences requiring at least 99.5% sequence identity and a minimum alignment length of 500 bp, we assigned 82,381 WGS contigs to 5,872 BAC contigs (72% of BAC contigs with associated sequence information). The genetic position of a physical contig was then set to the median genetic position of all POPSEQ-anchored WGS contigs assigned to it. A total of 4,920 and 5,002 BAC contigs could be anchored to the Morex × Barke and OWB maps, respectively. In both cases, three-quarters of contig positions were supported by at least two WGS contigs. Out of 4,411 BAC contigs anchored to both maps, 92.8% were positioned no farther than 5 centimorgan (cM) apart on the respective maps (Fig. 3A). The proportion of contigs anchored within 1 or 2 cM was 56.4% or 74.3%, respectively. A similar degree of agreement between different maps has already been reported for the anchoring of WGS contigs (The International Barley Genome Sequencing Consortium, 2012). This outcome is the result of the different resolutions of the underlying genetic maps as well as the procedures used for integration. Merging the anchoring results from both maps, we obtained a set of 5,193 anchored contigs (Table III). The number of anchored contigs varied considerably between distal and pericentromeric regions (Fig. 4). In distal regions, the ratio of physical to genetic distance was 1 to 10 Mb per cM, while it was 100 to 500 Mb in pericentromeric regions. Of all contigs anchored to the OWB or Morex × Barke framework, 3,830 (73.8%) with a cumulative length of 3.5 Gb are also anchored to the published physical and genetic framework (The International Barley Genome Sequencing Consortium, 2012). Chromosomal assignments between both maps agree in 97.6% of the cases, and cM coordinates disagreed in only 8.6% of cases (Fig. 3B). Similar to the anchoring of WGS contigs (Mascher et al., 2013), discordant contig placements mostly occurred in the genetic centromere. Although the POPSEQ anchoring contains 14% more contigs than the published physical and genetic framework (The International Barley Genome Sequencing Consortium, 2012), the cumulative length of all anchored contigs increases by only 1.3%. The high number of markers enabled us both to include shorter contigs (mean contig size of 761 versus 856 kb) and to exclude some longer contigs with inconsistent marker information. Furthermore, we applied more stringent alignment (500-bp minimum alignment length and 99.5% or greater identity) criteria compared with our previous effort (200-bp alignment length, 99% identity; The International Barley Genome Sequencing Consortium, 2012) to avoid confounding paralogous sequences when assigning WGS contigs to BAC sequences. The current POPSEQ anchoring of the barley physical map is available for download (http://dx.doi.org/10.5447/IPK/2013/1). BAC and marker sequences used by The International Barley Genome Sequencing Consortium (2012) can be retrieved from ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/. SNPs detected by whole-genome shotgun sequencing of the Morex × Barke and OWB populations and the resulting POPSEQ anchoring of WGS contigs can be downloaded from ftp://ftp.ipk-gatersleben.de/barley-popseq/.
Figure 3.
Agreement between different anchoring methods. Dot-plot comparisons of different genetic anchoring methods are shown. We anchored BAC contigs by POPSEQ to the Morex × Barke and OWB maps. A, Collinearity between the two genetic maps. B, Combined POPSEQ anchoring and the previously reported anchoring to various genetic maps (The International Barley Genome Sequencing Consortium, 2012).
Table III. POPSEQ anchoring statistics.
| POPSEQ Data |
BAC Contigs |
Sequenced Clones |
||
|---|---|---|---|---|
| Morex × Barke | OWB | Morex × Barke + OWB | Morex × Barke + OWB | |
| No. of all FP contigs/clones | 9,265 | 9,265 | 9,265 | 6,278 |
| No. of FP contigs/clones harboring WGS contigs | 5,872 | 5,872 | 5,872 | 6,243 |
| No. of FP contigs/clones harboring anchored WGS contigs | 5,295 | 5,417 | 5,720 | 6,189 |
| No. of anchored WGS contigs assigned to FP contigs/clones | 53,212 | 57,942 | 71,112 | 34,932 |
| No. of SNPs on anchored WGS contigs assigned to FP contigs/clones | 1,040,419 | 1,013,161 | 2,053,580 | 1,231,514 |
| No. of anchored contigs/clones | 4,920 | 5,002 | 5,193 | 5,591 |
| Length of anchored contigs/clones | 3.85 Gb | 3.90 Gb | 3.95 Gb | 703 Mb |
Figure 4.
Anchored sequence in 5-cM bins. The length of anchored physical contigs was calculated in 5-cM bins and plotted along the genetic map of each chromosome. Note that the y axis has a logarithmic scale.
Genetic Anchoring of Single BAC Clones
Led by the observation that POPSEQ is able to anchor shorter contigs, we attempted to anchor single, fully-sequenced BAC clones. Instead of aggregating anchoring information per physical contig, we averaged genetic positions at a per-BAC level. A total of 6,243 (99.4%) of all sequenced BACs harbored WGS contigs, and 5,591 (89.1%) could be anchored to the Morex × Barke or OWB framework (Table III). The genetic positions of BACs and their corresponding FP contigs agreed in 97.6% of cases. As the number of discordant chromosome assignments was three times higher than the number of discordant cM positions, disagreement between both anchoring methods arises most likely from single wrongly placed clones that are located on different chromosomes from their assigned physical contig. We found pairs of BACs on 71 FP contigs that were anchored to different chromosomes. For this analysis, BACs were required to harbor at least two WGS contigs that were consistently anchored in both the Morex × Barke and OWB frameworks. Among the anchored BACs, there were also 278 singleton clones that could now be assigned to chromosomal locations to guide their assignment to contigs based on sequence similarity.
DISCUSSION
A sequence-ready genome-wide physical map of the barley genome has been constructed. More than half a million clones representing 14-fold genome coverage have been fingerprinted and assembled into physical contigs. The MTP we have established will provide the framework for clone-by-clone sequencing of the barley genome. Restricting attention to single BAC clones reduces the algorithmic complexity of sequence assembly, thus enabling the use of short NGS reads and established assembly programs that would result in highly fragmented assemblies when applied on a whole-genome scale. The International Barley Genome Sequencing Consortium (2012) has already sequenced more than 6,000 clones by 454 or Illumina technology and will continue along this path to assemble a draft sequence.
Genetic anchoring of the barley physical map had been reported earlier (The International Barley Genome Sequencing Consortium, 2012). However, chromosomal locations could be assigned to only about half of all physical contigs through the integration of marker sequences from various genetic maps with BAC contigs. In this study, we applied a new approach for integrating the physical map with genetic maps by extending the POPSEQ method originally developed (Mascher et al., 2013) for fragmented whole-genome assemblies to physical contig assemblies. This novel straightforward approach was able to anchor more contigs than the complex, multilayered strategy presented earlier. The high number of informative markers obtained by high-throughput sequencing enabled genetic anchoring at a clone-by-clone scale and thereby provided an independent method of validating the physical map. This method could also complement other genome projects following a hierarchical shotgun strategy (Paux et al., 2008; Lin et al., 2010; Dohm et al., 2012). At present, POPSEQ anchoring of the barley physical map is limited by the paucity of high-quality sequence information from each individual BAC contig. Although approximately 300,000 BACs have been end sequenced, these sequences are shorter (less than 1,000 bp) than assembled sequence contigs and mostly originate from repetitive regions, because they are distributed randomly across the genome. As the physical length of all contigs anchored by POPSEQ amounts to 90% of the physical length of all contigs with associated WGS contigs, we anticipate that a substantial increase of anchoring efficiency can only be achieved when more BAC sequence information becomes available. The full power of POPSEQ to genetically anchor the physical map of barley will only be apparent when a completely sequenced MTP is available. Our preliminary analysis of approximately 6,200 fully sequenced clones showed that we can reasonably expect the vast majority of clones to harbor an anchored WGS contig. Alternatively, the sequenced MTP clones may serve as a reference for read mapping after removal of the sequence redundancy introduced by overlapping clones. SNP calling and genotyping could then be performed on the sequence scaffolds of the physical contigs, which would then be directly anchored to a genetic map without the intermediate step of a WGS assembly. The genetic anchoring of individual clones will enable us to further validate contig integrity, identify erroneously placed clones, and position singleton clones.
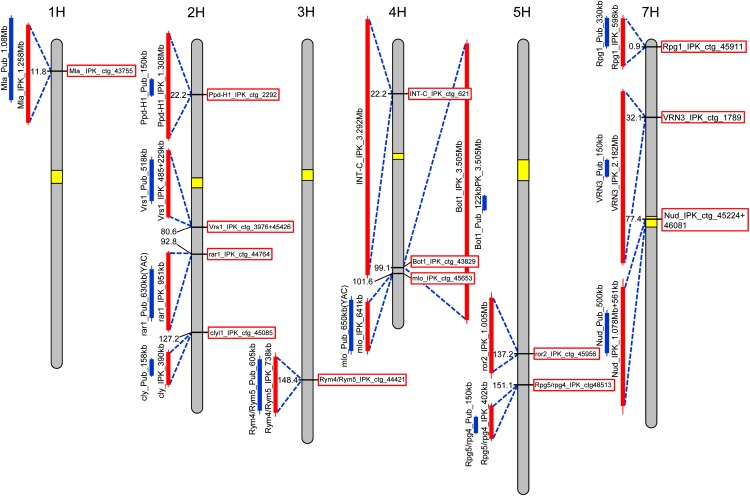
Apart from its value in assembling a draft genome sequence, the barley physical map presented here will assist genomic research by accelerating the isolation of genes underlying phenotypic traits. As an illustration, we explored the utility of the resource by determining the physical positions of 14 barley genes that had been isolated through positional cloning or genome-wide association studies (Fig. 5). All genes could be associated with contigs on the physical map, which were in most cases longer than the contigs of local restriction maps originally reported. For example, three BACs in the vicinity of Photoperiod-H1, a major regulator of photoperiod response, had been identified through BAC library screening (Turner et al., 2005). Photoperiod-H1 is annotated as a high-confidence gene on a whole-genome shotgun contig (morex_contig_94710). This contig has high sequence similarity (more than 99.9% over 5,000 bp) to the sequenced BAC HVVMRXALLhA0598A09, which is part of the physical map contig FP_contig_2992. The physical map also provides extensive local information for genes that were not found through positional cloning. For example, Required for mlo-specified resistance2, a gene involved in resistance to powdery mildew, was identified through a synteny-based approach, and Intermedium-c, a gene that modifies lateral spikelet fertility, was identified through association mapping and analysis of conserved gene order. Both genes were positioned on genetically anchored physical contigs that provide extended information about genomic context and local neighborhood. Therefore, the physical map of barley and its associated sequence and clone resources will serve as a hub for marker development and candidate gene identification in ongoing map-based cloning efforts.
Figure 5.
Positions of cloned barley genes in the physical map. The positions of 14 previously isolated barley genes on BAC contigs in the physical and genetic framework are visualized along chromosome ideograms. Contigs of the genome-wide physical map (pink bars) are compared with the contigs of the local physical maps from the original publications (blue bars). Centromere positions (marked yellow) are taken from Comadran et al. (2012).
The majority of barley genes cloned to date are located in the distal regions of the chromosomes, where the ratio of physical and genetic distances is tractable. In telomeric regions of barley, the ratio of genetic to physical distance is, on average, larger than 0.5 cM per Mb (Fig. 4). As half of the physical map is contained in contigs larger than 904 kb (Table II), finding markers flanking a target locus on both sides and located on a single or neighboring physical contig seems within reach for large mapping populations. In this situation, the genome-wide physical map of barley would obviate the need for chromosome walking. Instead, BAC contigs harboring flanking or cosegregating markers can be identified either by library screening or searching the sequence resources integrated with the physical map. Subsequently, the MTP of physical contigs can be sequenced and the sequencing data mined for candidate genes or additional markers.
One shortcoming of the current sequence-enriched framework is the lack of resolution in pericentromeric regions. Both positional cloning and genetic anchoring of the physical map are hampered by the severely reduced recombination frequency in the genetic centromere. It is not uncommon, even in large mapping populations, that closely flanking markers of a target gene residing in the genetic centromere are located on opposite chromosome arms (Shahinnia et al., 2012; Okagaki et al., 2013). Several hundred megabases (encompassing several dozen BAC contigs) may correspond to a genetic interval of less than 1 cM, and the ordering of physical contigs with respect to each other is lacking for these regions.
The limited resolution of our map in centromeric regions may be improved through populations that provide higher mapping resolution (e.g. a large number [more than 1,000] of recombinant inbred lines). Genome-wide high-density genotyping of several hundred or even thousands of individuals has been made possible by cost-effective genotyping by sequencing (Elshire at al., 2011; Poland et al., 2012). Apart from recombination-based mapping, radiation hybrid panels similar to those implemented in wheat (Kalavacharla et al., 2006) or obtained for barley by the activity of gametocidal chromosomes (Masoudi-Nejad et al., 2005) may be of value. Similarly, fluorescent in situ hybridization mapping (Cheng et al., 2001) and optical mapping (Zhou et al., 2009) hold some promise and may be used to order markers in recombinogenically inert regions. More importantly, once the MTP has been sequenced, overlapping adjacent physical contigs can be merged using sequence information and result in an improved linear order in the same genetic bin.
In summary, the genome-wide physical map of barley constitutes the backbone for map-based sequencing of the genome and facilitates high-resolution trait mapping and gene isolation. We expanded the POPSEQ method to anchoring of physical contigs and BAC clones. This can be adopted by current and future physical mapping projects of other large and complex genomes.
MATERIALS AND METHODS
HICF and Automatic Contig Assembly
Six BAC libraries were used in this study (Yu et al., 2000; Schulte et al., 2011). These had been either constructed by partial enzymatic fragmentation of high-Mr DNA with the restriction endonucleases HindIII, EcoRI, or MboI or after mechanical fragmentation and blunt-end ligation. Plasmid DNA was isolated from a total of 690,912 BAC clones as described previously (Schulte et al., 2011) and was subjected to HICF according to published procedures (Luo et al., 2003). Peak areas, peak heights, and fragment sizes of each BAC fingerprint profile were collected by the Applied Biosystems 3730xl data collection program. The sizing quality of raw fingerprinting data was assessed with GeneMapper version 4.0 software (Applied Biosystems), and fingerprint profiles were further edited with the tool FPminer as described previously (Schulte et al., 2011). Clones were tagged as neighbor or plate-wide contaminated and excluded from further analysis if the overall fragment identity of two clones at neighboring positions within one plate or at an identical position in subsequent plates of the library was higher than 50%. Chloroplast DNA contamination was determined by comparing the fingerprint profiles with a BAC clone containing the chloroplast genome (Saski et al., 2007) of barley (Hordeum vulgare) cv Morex. Clones sharing more than 50% of their fragments with this BAC were excluded from further analysis. Furthermore, all clones with less than 30 or more than 250 fragments were discarded in order to improve the contig assembly (Schulte et al., 2011). Fingerprinting profiles were assembled into contigs using the fingerprint assembly software tool FPC version 9.2 (Soderlund et al., 2000). The initial contig build was performed with a Sulston score threshold of 1e-90, a tolerance of 5, and the questionable clones (Q clones) parameter (Q clones are clones where the Contig Built algorithm of FPC cannot order at least 50% of the bands in the Contig Built map) was set to 10% (i.e. 10% of clones in a contig were tolerated to be questionable).
To incorporate further singleton clones into contigs, “single-to-end” and “end-to-end” merging (Match: 2; FromEnd: 55) was performed at nine successively reduced cutoffs (Sulston score threshold of 1e-85 to 1e-45) as described previously (Paux et al., 2008). Contigs with more than 10% Q clones (Q clones where FPC cannot order more than half of the bands) were reanalyzed with the DQer function of FPC as described previously (Soderlund et al., 2000; Paux et al., 2008).
Manual Editing
Physical contigs were genetically positioned or assigned to a chromosome arm using experimental as well as bioinformatics procedures as described previously (The International Barley Genome Sequencing Consortium, 2012). All contigs with genetic marker information or chromosome arm assignment were examined for marker distribution along the entire length of the contig and consistent position assignments among the diverse anchoring information. Contigs that contained at least one contradicting marker were verified manually by rebuilding the contig with increased stringency (Sulston score threshold of 1e-70). Putative chimeric contigs with clusters of markers from different chromosomes were manually disrupted at the appropriate site for marker consistency, and contig integrity was established by rebuilding the contigs (Soderlund et al., 2000). Final contig merging was performed with a Sulston score cutoff of 1e-25. At this stage, two contigs were merged only if a minimum of two BAC clones at the end of each contig matched in a reciprocal and unique manner and shared marker information with a maximum genetic distance of 5 cM. Finally, all contigs composed of only two BAC clones were examined to identify those consisting of equivalent clones from the library HVVMRXALLhA (Yu et al., 2000) and its rearrayed subset of gene-containing clones (Madishetty et al., 2007; named HVVMRX83KhA after rearraying). By comparing the average fragment number per clone in the final map with the average insert size determined by pulsed field gel electrophoresis or BAC sequencing (The International Barley Genome Sequencing Consortium, 2012), we determined the average size of an FPC consensus band to be 1.24 kb. Using this conversion factor, the cumulative length of all physical contigs was calculated.
Contig Validation and MTP Establishment
BAC contigs generated by FPC were further analyzed with the alternative physical mapping tool LTC (Frenkel et al., 2010) to trace misassembled contigs caused by chimerical clones and false clone overlaps. Misassemblies were detected by analysis of the topological structure of the net of clone overlaps within contigs (using a Sulston score cutoff of 1e-20): a contig was considered as putatively problematic if this net had a nonlinear structure. To check the linearity of the net, LTC automatically selected one of the possible diametric paths (the longest nonreducible path going through edges corresponding to significantly overlapping clones, with vertices corresponding to clones) and scored the ranks of all vertices relative to the vertices from this diametric path (vertices from the diametric path were ranked as 0, vertices connected to vertices from the diametric path by a single edge were ranked as 1, etc.). The presence of vertices having a rank of 2 and more points indicated the nonlinearity of the net (Frenkel et al., 2010). In the case of nonlinearity of the net structure, LTC automatically tries to repeat the analysis using a more stringent cutoff (with increasing cutoff stringency by 1 order of magnitude at each step). In many cases, this increasing stringency resulted in the exclusion of false overlaps, and the net of remaining clone overlaps was found to have a linear structure. To identify the presence of putative chimerical clones (composed by artificial fusion of DNA from distant genomic regions), the parallel clone overlaps were automatically searched for each clone of the contig (Frenkel et al., 2010). LTC was also used for the recalculation of clone end coordinates within FPC-based contigs (algorithms implemented in LTC enable reducing errors in the estimation of clone position and contig length caused by local optimization in FPC). The MTP was automatically selected by LTC. Following the application of the LTC algorithms, each contig in the selected MTP was based on clones corresponding to vertices from the diametric path used for contig testing (see above). In this manner, the MTP selection resulted in the identification of significant overlaps between neighboring MTP clones. For the selected cutoff stringency, each pair of neighbor MTP clones has overlap of about 35% of the clone length. The MTP clones were rearrayed into microtiter plates and grouped by chromosomes. Three-dimensional BAC DNA pools were established and validated for all chromosomes. The MTP clones and the three-dimensional BAC pools can be ordered from CNRGV (http://cnrgv.toulouse.inra.fr/en/library/barley).
Distribution of BAC Libraries
The expected number of clones from a library L per sliding window B on a chromosome C were calculated using the formula e = n(L,C) × (l(B)/l(L,C)), where l(L,C) is the combined length of all clones of library L on chromosome C, l(B) is the combined length of clones anchored to B, and n(L,C) is the number of all clones of library L that are contained in contigs anchored to chromosome C. The length of clones was derived from the number of FPC consensus bands. The normalized count of clones from a fixed library per genetic bin was calculated by dividing the observed number of clones by the expected number of clones. The size of a sliding window was chosen to include 30 adjacent positions in the genetic map.
BAC Contigs Harboring Previously Cloned Barley Genes
Sequences of cloned barley genes (Büschges et al., 1997; Shirasu et al., 1999; Collins et al., 2003; Shen et al., 2003; Stein et al., 2005; Turner et al., 2005; Brueggeman et al., 2006, 2008; Yan et al., 2006; Komatsuda et al., 2007; Sutton et al., 2007; Taketa et al., 2008; Nair et al., 2010; Ramsay et al., 2011) were searched for the presence of flanking or cosegregating markers and gene sequences with BLASTN (Altschul et al., 1990), or BAC clone identifiers from the original publications were identified in the FPC database of fingerprinted contigs.
POPSEQ Anchoring of Physical Contigs and BAC Clones
We had previously anchored the whole-genome shotgun assembly of barley cv Morex (Mascher et al., 2013). Sequences of anchored WGS contigs were compared with fully sequenced BACs and BAC end sequences of barley (The International Barley Genome Sequencing Consortium, 2012) by megablast (Zhang et al., 2000) and thus assigned to physical contigs. Similar to aggregating anchoring information of single SNPs on WGS contigs (Mascher et al., 2013), the genetic position of a physical contig or of a single clone was defined as the median genetic position of all WGS contigs assigned to it. We required that at least 80% of all WGS contigs assigned to an FP contig or BAC clone lie on the same chromosome and the median absolute deviation of their cM positions be less than 5 cM. Genetic positions in the OWB map were interpolated to the Morex × Barke iSelect map as described previously (Mascher et al., 2013). When combining coordinates, we required consistency between both maps (i.e. both genetic positions had to be no farther apart from each other than 5 cM). If contigs or clones were anchored to both framework maps, the Morex × Barke position was preferred.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Incremental assembly statistics.
Supplemental Table S1. MTP clones.
Acknowledgments
We acknowledge the excellent technical assistance of Ulrike Beyer, Anne Kusserow, Katrin Trnka, Jenny Knibbiche, and Jelena Perovic, Karin Lipfert (Leibniz Institute of Plant Genetics and Crop Plant Research [IPK Gatersleben]) for help with the figures, Dr. Tatjana Sretenovic (De Ruiter Seeds) for her help in the initial steps of the project, Dr. Hélène Berges, Arnaud Bellec, and Sonia Vautrin (CNRGV) for BAC library management and rearraying of MTP clones, Dr. William Nelson (University of Arizona) for adapting the FPC program to the large data set, and Timothy Close (University of California [Riverside]) for providing information to rearray the HVVMRX83khA library.
Glossary
- NGS
next-generation sequencing
- BAC
bacterial artificial chromosome
- POPSEQ
population sequencing
- WGS
whole-genome shotgun sequence
- HICF
high-information content fingerprinting
- FP contig
fingerprinted contig
- LTC
linear topology contigs
- MTP
minimum tiling path
- SNP
single-nucleotide polymorphism
- OWB
Oregon Wolfe Barleys
- cM
centimorgan
- Q clones
questionable clones
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Brueggeman R, Drader T, Kleinhofs A. (2006) The barley serine/threonine kinase gene Rpg1 providing resistance to stem rust belongs to a gene family with five other members encoding kinase domains. Theor Appl Genet 113: 1147–1158 [DOI] [PubMed] [Google Scholar]
- Brueggeman R, Druka A, Nirmala J, Cavileer T, Drader T, Rostoks N, Mirlohi A, Bennypaul H, Gill U, Kudrna D, et al. (2008) The stem rust resistance gene Rpg5 encodes a protein with nucleotide-binding-site, leucine-rich, and protein kinase domains. Proc Natl Acad Sci USA 105: 14970–14975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, van Daelen R, van der Lee T, Diergaarde P, Groenendijk J, et al. (1997) The barley Mlo gene: a novel control element of plant pathogen resistance. Cell 88: 695–705 [DOI] [PubMed] [Google Scholar]
- Chen M, Presting G, Barbazuk WB, Goicoechea JL, Blackmon B, Fang G, Kim H, Frisch D, Yu Y, Sun S, et al. (2002) An integrated physical and genetic map of the rice genome. Plant Cell 14: 537–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Presting GG, Buell CR, Wing RA, Jiang J. (2001) High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157: 1749–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Comadran J, Kilian B, Russell J, Ramsay L, Stein N, Ganal M, Shaw P, Bayer M, Thomas W, Marshall D, et al. (2012) Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat Genet 44: 1388–1392 [DOI] [PubMed] [Google Scholar]
- Dohm JC, Lange C, Holtgräwe D, Sörensen TR, Borchardt D, Schulz B, Lehrach H, Weisshaar B, Himmelbauer H. (2012) Palaeohexaploid ancestry for Caryophyllales inferred from extensive gene-based physical and genetic mapping of the sugar beet genome (Beta vulgaris). Plant J 70: 528–540 [DOI] [PubMed] [Google Scholar]
- Druka A, Franckowiak J, Lundqvist U, Bonar N, Alexander J, Houston K, Radovic S, Shahinnia F, Vendramin V, Morgante M, et al. (2011) Genetic dissection of barley morphology and development. Plant Physiol 155: 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6: e19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Leach JE, Rogers J, Schnable PS, Eversole K. (2011) Crop genome sequencing: lessons and rationales. Trends Plant Sci 16: 77–88 [DOI] [PubMed] [Google Scholar]
- Feuillet C, Stein N, Rossini L, Praud S, Mayer K, Schulman A, Eversole K, Appels R. (2012) Integrating cereal genomics to support innovation in the Triticeae. Funct Integr Genomics 12: 573–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel Z, Paux E, Mester D, Feuillet C, Korol A. (2010) LTC: a novel algorithm to improve the efficiency of contig assembly for physical mapping in complex genomes. BMC Bioinformatics 11: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, Sharpe T, Hall G, Shea TP, Sykes S, et al. (2011) High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA 108: 1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Lu T, Han B. (2013) Resequencing rice genomes: an emerging new era of rice genomics. Trends Genet 29: 225–232 [DOI] [PubMed] [Google Scholar]
- The International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436: 793–800 [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Kalavacharla V, Hossain K, Gu Y, Riera-Lizarazu O, Vales MI, Bhamidimarri S, Gonzalez-Hernandez JL, Maan SS, Kianian SF. (2006) High-resolution radiation hybrid map of wheat chromosome 1D. Genetics 173: 1089–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, et al. (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104: 1424–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Levy AA, Feldman M. (2002) The impact of polyploidy on grass genome evolution. Plant Physiol 130: 1587–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Pierce GJ, Bowers JE, Estill JC, Compton RO, Rainville LK, Kim C, Lemke C, Rong J, Tang H, et al. (2010) A draft physical map of a D-genome cotton species (Gossypium raimondii). BMC Genomics 11: 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, et al. (2013) Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496: 87–90 [DOI] [PubMed] [Google Scholar]
- Luo MC, Thomas C, You FM, Hsiao J, Ouyang S, Buell CR, Malandro M, McGuire PE, Anderson OD, Dvorak J. (2003) High-throughput fingerprinting of bacterial artificial chromosomes using the snapshot labeling kit and sizing of restriction fragments by capillary electrophoresis. Genomics 82: 378–389 [DOI] [PubMed] [Google Scholar]
- Madishetty K, Condamine P, Svensson JT, Rodriguez E, Close TJ. (2007) An improved method to identify BAC clones using pooled overgos. Nucleic Acids Res 35: e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Muehlbauer GJ, Rokhsar DS, Chapman J, Schmutz J, Barry K, Muñoz-Amatriaín M, Close TJ, Wise RP, Schulman AH, et al. (2013) Anchoring and ordering NGS contig assemblies by population sequencing (POPSEQ). Plant J 76: 718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoudi-Nejad A, Nasuda S, Bihoreau MT, Waugh R, Endo TR. (2005) An alternative to radiation hybrid mapping for large-scale genome analysis in barley. Mol Genet Genomics 274: 589–594 [DOI] [PubMed] [Google Scholar]
- Nair SK, Wang N, Turuspekov Y, Pourkheirandish M, Sinsuwongwat S, Chen G, Sameri M, Tagiri A, Honda I, Watanabe Y, et al. (2010) Cleistogamous flowering in barley arises from the suppression of microRNA-guided HvAP2 mRNA cleavage. Proc Natl Acad Sci USA 107: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okagaki RJ, Cho S, Kruger WM, Xu WW, Heinen S, Muehlbauer GJ. (2013) The barley UNICULM2 gene resides in a centromeric region and may be associated with signaling and stress responses. Funct Integr Genomics 13: 33–41 [DOI] [PubMed] [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, Korol A, Michalak M, Kianian S, Spielmeyer W, et al. (2008) A physical map of the 1-gigabase bread wheat chromosome 3B. Science 322: 101–104 [DOI] [PubMed] [Google Scholar]
- Poland JA, Brown PJ, Sorrells ME, Jannink JL. (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 7: e32253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Comadran J, Druka A, Marshall DF, Thomas WT, Macaulay M, MacKenzie K, Simpson C, Fuller J, Bonar N, et al. (2011) INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet 43: 169–172 [DOI] [PubMed] [Google Scholar]
- Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, Clarke JL. (2007) Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet 115: 571–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schulte D, Ariyadasa R, Shi B, Fleury D, Saski C, Atkins M, deJong P, Wu CC, Graner A, Langridge P, et al. (2011) BAC library resources for map-based cloning and physical map construction in barley (Hordeum vulgare L.). BMC Genomics 12: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP, et al. (2009) The International Barley Sequencing Consortium: at the threshold of efficient access to the barley genome. Plant Physiol 149: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinnia F, Druka A, Franckowiak J, Morgante M, Waugh R, Stein N. (2012) High resolution mapping of Dense spike-ar (dsp.ar) to the genetic centromere of barley chromosome 7H. Theor Appl Genet 124: 373–384 [DOI] [PubMed] [Google Scholar]
- Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. (2003) Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K, Lahaye T, Tan MW, Zhou F, Azevedo C, Schulze-Lefert P. (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99: 355–366 [DOI] [PubMed] [Google Scholar]
- Soderlund C, Humphray S, Dunham A, French L. (2000) Contigs built with fingerprints, markers, and FPC V4.7. Genome Res 10: 1772–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A. (2005) The eukaryotic translation initiation factor 4E confers multiallelic recessive bymovirus resistance in Hordeum vulgare (L.). Plant J 42: 912–922 [DOI] [PubMed] [Google Scholar]
- Steuernagel B, Taudien S, Gundlach H, Seidel M, Ariyadasa R, Schulte D, Petzold A, Felder M, Graner A, Scholz U, et al. (2009) De novo 454 sequencing of barcoded BAC pools for comprehensive gene survey and genome analysis in the complex genome of barley. BMC Genomics 10: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton T, Baumann U, Hayes J, Collins NC, Shi BJ, Schnurbusch T, Hay A, Mayo G, Pallotta M, Tester M, et al. (2007) Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science 318: 1446–1449 [DOI] [PubMed] [Google Scholar]
- Taketa S, Amano S, Tsujino Y, Sato T, Saisho D, Kakeda K, Nomura M, Suzuki T, Matsumoto T, Sato K, et al. (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105: 4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudien S, Steuernagel B, Ariyadasa R, Schulte D, Schmutzer T, Groth M, Felder M, Petzold A, Scholz U, Mayer KF, et al. (2011) Sequencing of BAC pools by different next generation sequencing platforms and strategies. BMC Res Notes 4: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034 [DOI] [PubMed] [Google Scholar]
- van Hintum T, Menting F 2003. Diversity in ex situ genebank collections of barley. In R von Bother, T van Hintum, H Knupffer, K Sato, eds, Diversity in Barley (Hordeum vulgare). Elsevier Science, Amsterdam, pp 247–257 [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Wicker T, Mayer KF, Gundlach H, Martis M, Steuernagel B, Scholz U, Simková H, Kubaláková M, Choulet F, Taudien S, et al. (2011) Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23: 1706–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Narechania A, Sabot F, Stein J, Vu GT, Graner A, Ware D, Stein N. (2008) Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics 9: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. (2006) 454 sequencing put to the test using the complex genome of barley. BMC Genomics 7: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Zimmermann W, Perovic D, Paterson AH, Ganal M, Graner A, Stein N. (2005) A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv-eIF4E locus: recombination, rearrangements and repeats. Plant J 41: 184–194 [DOI] [PubMed] [Google Scholar]
- Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103: 19581–19586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA. (2000) A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet 101: 1093–1099 [Google Scholar]
- Zhang Z, Schwartz S, Wagner L, Miller W. (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7: 203–214 [DOI] [PubMed] [Google Scholar]
- Zhou S, Wei F, Nguyen J, Bechner M, Potamousis K, Goldstein S, Pape L, Mehan MR, Churas C, Pasternak S, et al. (2009) A single molecule scaffold for the maize genome. PLoS Genet 5: e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]