Meta-analysis of differentially expressed rice genes under different stress conditions accurately classified them using machine learning approaches and identified genes likely to confer broad resistance to multiple abiotic and biotic stresses.
Abstract
Abiotic and biotic stress responses are traditionally thought to be regulated by discrete signaling mechanisms. Recent experimental evidence revealed a more complex picture where these mechanisms are highly entangled and can have synergistic and antagonistic effects on each other. In this study, we identified shared stress-responsive genes between abiotic and biotic stresses in rice (Oryza sativa) by performing meta-analyses of microarray studies. About 70% of the 1,377 common differentially expressed genes showed conserved expression status, and the majority of the rest were down-regulated in abiotic stresses and up-regulated in biotic stresses. Using dimension reduction techniques, principal component analysis, and partial least squares discriminant analysis, we were able to segregate abiotic and biotic stresses into separate entities. The supervised machine learning model, recursive-support vector machine, could classify abiotic and biotic stresses with 100% accuracy using a subset of differentially expressed genes. Furthermore, using a random forests decision tree model, eight out of 10 stress conditions were classified with high accuracy. Comparison of genes contributing most to the accurate classification by partial least squares discriminant analysis, recursive-support vector machine, and random forests revealed 196 common genes with a dynamic range of expression levels in multiple stresses. Functional enrichment and coexpression network analysis revealed the different roles of transcription factors and genes responding to phytohormones or modulating hormone levels in the regulation of stress responses. We envisage the top-ranked genes identified in this study, which highly discriminate abiotic and biotic stresses, as key components to further our understanding of the inherently complex nature of multiple stress responses in plants.
The need to breed robust and high-productivity crops is more important than ever due to increasingly adverse environmental conditions and scarce natural resources. Food productivity has to be raised by as much as 70% to 100% to meet the nutritional needs of the growing population, which is expected to rise to 9 billion by 2050 (Godfray et al., 2010; Lutz and Samir, 2010). Rice (Oryza sativa) is both a major food crop and a model organism that shares extensive synteny and collinearity with other grasses. Thus, the development of rice that can sustain a wide variety of adverse conditions is vital to meet the imminent global energy demands.
A broad range of stress factors divided into two major categories, namely abiotic stresses encompassing a variety of unfavorable environmental conditions, such as drought, submergence, salinity, heavy metal contamination or nutrient deficiency, and biotic stresses caused by infectious living organisms, such as bacteria, viruses, fungi, or nematodes, negatively affect the productivity and survival of plants. Advancements in whole-genome transcriptome analysis techniques like microarrays and RNA sequencing have revolutionized the identification of changes in gene expression in plants under stress, making it possible now to chart out individual stress-specific biomolecular networks and signaling pathways. However, in field conditions, plants are often subjected to multiple stresses simultaneously, requiring efficient molecular mechanisms to perceive a multitude of signals and to elicit a tailored response (Sharma et al., 2013). Increasing evidence from experimental studies suggests that the cross talk between individual stress response signaling pathways via key regulatory molecules, resulting in the dynamic modulation of downstream effectors, is at the heart of multiple stress tolerance. A number of studies have identified many genes, especially transcription factors (TFs) and hormone response factors, that play a central role in multiple stresses and manifest a signature expression specific to the stress condition. For example, abscisic acid (ABA) response factors are up-regulated in the majority of abiotic stresses, activating an oxidative response to protect cells from reactive oxygen species damage, but were found to be down-regulated in a number of biotic conditions, possibly suppressed by immune response molecules (Cao et al., 2011).
The wide range of abiotic and biotic stress factors and their numerous combinations in natural conditions generate a customized stress response. This suggests that the identification and characterization of key genes and their coexpression partners, which show an expression profile that discriminates abiotic and biotic stress responses, would increase our understanding of plant stress response manyfold and provide targets for genetic manipulation to improve their stress tolerance. The availability of multiple genome-wide transcriptome data sets for the same stress condition provides an opportunity to identify, compare, and contrast the stress-specific gene expression profile of one stress condition with other stresses. Meta-analysis combining similar studies provides a robust statistical framework to reevaluate original findings, improve sensitivity with increased sample size, and test new hypotheses. Meta-analysis of microarray studies is widely used, especially in clinical research, to improve statistical robustness and detect weak signals (Liu et al., 2013; Rung and Brazma, 2013). For instance, thousands of samples belonging to hundreds of cancer types were combined, which provided new insights into the general and specific transcriptional patterns of tumors (Lukk et al., 2010). Microarray studies are burdened with a high dimensionality of feature space, also called the “curse of dimensionality” (i.e. the availability of very many variables [genes] for very few observations [samples]). Machine learning algorithms (supervised and unsupervised), such as principal component analysis (PCA), decision trees, and support vector machines (SVM), provide a way to efficiently classify two or more classes of data. Further feature selection procedures like recursive-support vector machines (R-SVM) provide means to identify the top features contributing most to the accuracy of classification.
In this study, we performed a meta-analysis of stress response studies in rice using publicly available microarray gene expression data conducted on a single platform (AffymetrixRiceArray). Meta-analysis of abiotic and biotic stresses was performed separately to identify differentially expressed genes (DEGs) involved in multiple stress conditions. The lists of abiotic and biotic DEGs were then compared to identify common genes with conserved and nonconserved gene expression (i.e. whether up-regulated, down-regulated, or oppositely regulated in both the categories), revealing the broad patterns of their involvement in the stress response. In order to test the efficiency of identified common DEGs in the classification of abiotic and biotic stresses as well as individual stresses within abiotic and biotic stresses, we systematically investigated various classification and machine learning techniques, including PCA, partial least squares discriminant analysis (PLS-DA), SVM, and random forest (RF). We characterized the shared DEGs through functional enrichment analysis of gene ontologies, metabolic pathways, TF families, and microRNAs (miRNAs) targeting them. We also analyzed the correlation of coexpression between the common DEGs to find sets of genes showing high coexpression and identify hub genes that show the greatest number of edges over a very high cutoff value.
RESULTS
DEGs Common to Abiotic and Biotic Stresses
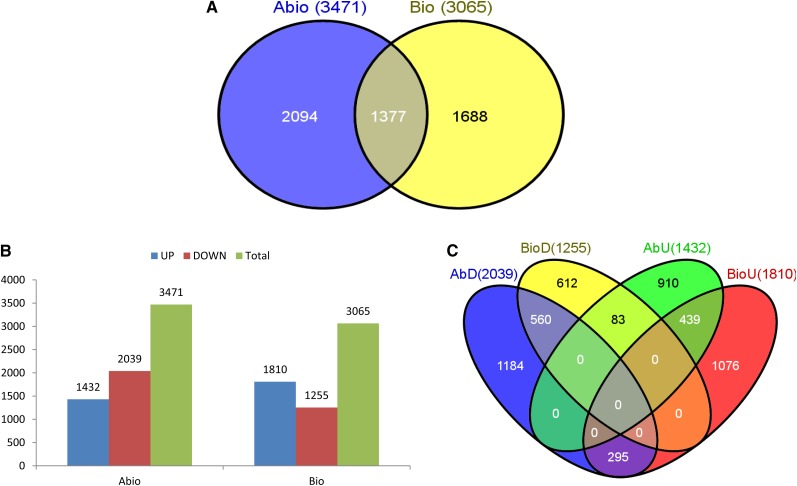
We analyzed 559 microarray samples (219 from abiotic and 340 from biotic stresses) from 13 stress conditions, of which seven were abiotic (cold, drought, heat shock, metal, nutrient, salt, and submergence) and six were biotic (bacterium, fungus, insect, nematode, virus, and weed; Supplemental Table S1A). Meta-analysis by combinatorial analysis of seven abiotic stresses from 15 different studies together identified 3,471 DEGs and six biotic stresses from 17 different studies revealed 3,065 DEGs, with a false discovery rate (FDR) ≤ 0.01 (Fig. 1A; Supplemental Table S2). About 60% of DEGs in abiotic stresses were down-regulated, while 60% of DEGs in biotic stresses were up-regulated (Fig. 1B). This broad pattern indicates that a wide variety of biological processes are down-regulated under abiotic stress, as it affects the whole system, thus driving the plant to a protective and energy-conserving mode. On the other hand, biotic stresses are often localized, especially at the early stages, and require an array of defense response molecules and metabolites to be synthesized and orchestrated, such as in systemic acquired resistance, to execute a resistance response against a specific infectious organism. Among the DEGs, more than 26% or 1,377 genes were common to abiotic and biotic stresses, indicating that these genes, which are just 3.5% of all non-transposable element genes in rice (Michigan State University [MSU] Rice Genome Annotation Project release 7), are affected by a diverse set of stress conditions and possibly play significant roles in multiple stress responses (Supplemental Table S3). Our major objective in this study is to analyze the stress-responsive genes involved in multiple stresses that regulate cross talk between abiotic and biotic stresses. Therefore, we focused on the 1,377 common DEGs for our study.
Figure 1.
Comparison of DEGs under abiotic and biotic stress responses. A, Two-way Venn diagram showing the common DEGs between abiotic and biotic stresses. B, Number of up-regulated and down-regulated DEGs in all identified abiotic and biotic stresses. C, Four-way Venn diagram showing the number of genes with conserved and nonconserved expression patterns. [See online article for color version of this figure.]
We found 72% or 999 out of 1,377 common DEGs with conserved expression between abiotic and biotic stresses, suggesting that most of these genes and their associated biological processes are regulated in a similar fashion in the vast majority of stress conditions. Among the 28% of DEGs showing nonconserved expression, 21% or 295 genes were down-regulated in abiotic and up-regulated in biotic stress (Fig. 1C). About 16% or 221 of these genes are annotated as “expressed protein” and approximately 7% or 96 have no GOSlim assignment, revealing that many of stress-responsive genes are still poorly understood. Studies elucidating the functional roles of these genes would be vital for a comprehensive understanding of the stress response in rice.
Machine Learning Approaches Based on Common DEGs Classified Abiotic and Biotic Stresses into Two Classes with High Accuracy
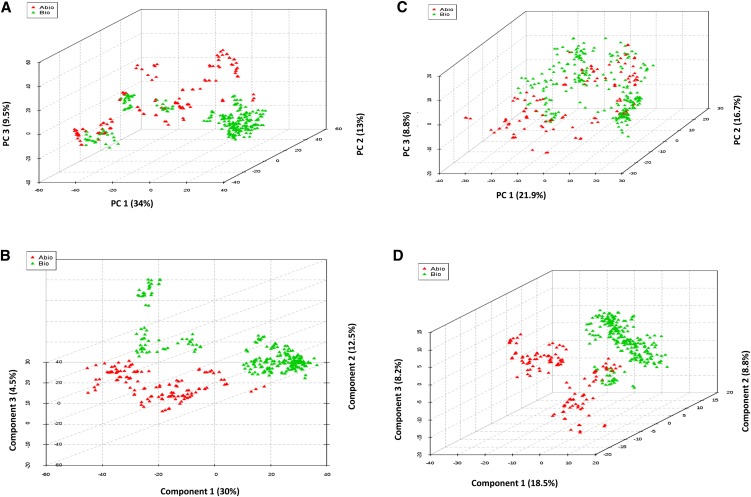
We investigated if the different stress conditions can be accurately classified using the common DEGs employing machine learning approaches. Initially, we investigated the performance of PCA in discriminating abiotic stresses from biotic stresses as two classes using all of the 1,377 common DEGs. The first three principal components (PCs) captured 56.4% of variance between the samples. The three-dimensional (3D) PCA plot of the top three PCs showed clear separation of abiotic and biotic classes for the majority of the samples, although both classes were widely dispersed across components (Fig. 2A). Nonetheless, there were some samples showing considerable overlap between the classes. We then analyzed the data set using PLS-DA, a technique that is specifically suited for the analysis of data sets with high feature dimensions and multicollinearity (Pérez-Enciso and Tenenhaus, 2003). Many of the published microarray studies have found PLS-DA to be a highly efficient method for multiclass classification (Student and Fujarewicz, 2012). PLS-DA resulted in five components that captured approximately 62% variance between the two classes and separated them with a very high accuracy of 0.99 (r2 = 0.95 [goodness of fit], Q2 = 0.93 [predictive value], P < 0.01) upon 10-fold cross validation. The 3D plot of PLS-DA showed clear separation of all the samples between abiotic and biotic stresses (Fig. 2B). The important genes contributing most to the PLS-DA separation can be identified using a variable importance in projection (VIP) score, which is a weighted sum of squares of partial least squares loadings (Pérez-Enciso and Tenenhaus, 2003). There were 177 genes with a VIP score (component 1) cutoff value of 1.5 or greater (Zhang et al., 2013) and 33 genes with values of 2 or greater (Supplemental Table S4).
Figure 2.
3D plots of two-class classification of abiotic and biotic stresses. A and B, 3D plots based on the top three components by PCA and PLS-DA, respectively using 1,377 common DEGs. C and D, 3D plots based on the top three components by PCA and PLS-DA, respectively, using the top 540 genes ranked by SVM return 100% accuracy of classification. The axes of B are rotated 90°, which shows the best possible separation of two groups. [See online article for color version of this figure.]
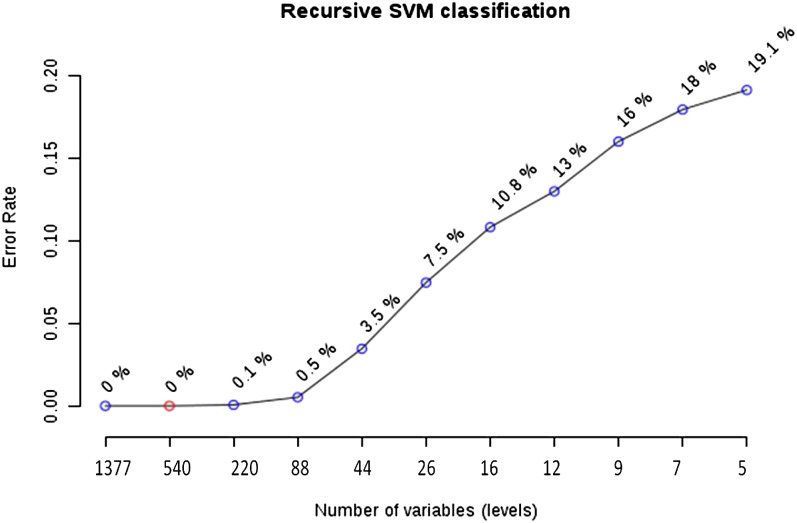
Next, we analyzed the same data set using another very popular supervised learning technique for microarray data classification, R-SVM, which identified 540 genes (39.2% out of 1,377) that can classify abiotic and biotic stresses with 100% accuracy and 88 (6%) genes with 95% accuracy after rigorous cross validation using leave-one-out cross validation (Fig. 3). These 540 genes included a number of hormone response and stress response signaling genes. All five of the MYB TFs, which are important regulators of development and defense responses in plants (Yanhui et al., 2006), found in the common DEGs were among these 540 genes. Furthermore, 103 (19%) of the 540 genes were part of a recently published database of stress-responsive TFs (Stress Responsive Transcription Factor Database version 2 [STIFDB2]; Naika et al., 2013), which provides a list of stress-responsive genes (1,118 genes of rice subspecies japonica) identified through biocuration and genomic data mining. Out of 540 genes, 178 (33%) were genes with nonconserved expression patterns between abiotic and biotic stresses, which is slightly higher compared with the 28% of genes showing nonconserved expression in all of the common DEGs. Although PCA based on these 540 genes resulted in poor separation of the classes, with 47.4% variance captured by the top three PCs, PLS-DA showed clear separation of the two classes (Fig. 2, C and D). The top five components of PLS-DA captured 53% of variance with classification accuracy of 0.97 (r2 = 0.91, Q2 = 0.87, P < 0.01), which is slightly less than the 0.99 accuracy obtained using all 1,377 common DEGs. There were 79 genes (14% of 540) with VIP ≥ 1.5 and 27 genes with VIP ≥ 2. There were two genes with VIP ≥ 3, which code for xylanase inhibitor and glycosyl hydrolase, both showing conserved up-regulation.
Figure 3.
Classification error rates of different subsets of common DEGs upon 10-fold cross validation using R-SVM. The error rate using all of 1,377 or 540 common DEGs was 0% (100% accuracy of classification) and 0.1% (99% accuracy) using 220 genes and 0.5% (95% accuracy) using 88 genes. [See online article for color version of this figure.]
Analysis of Shared DEGs Identified Top Genes with Discordant Behavior among Multiple Stresses
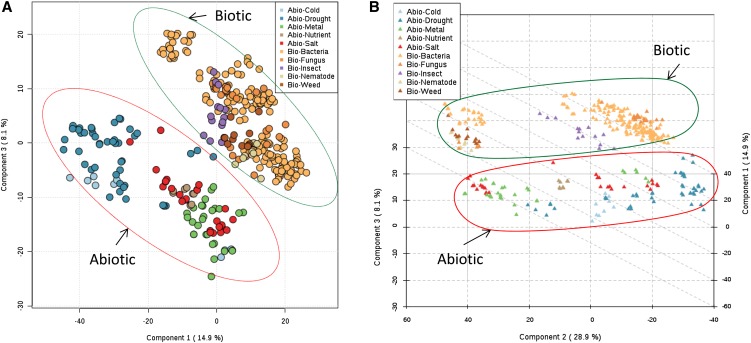
From the 13 stress conditions analyzed, we selected the top 10 stresses (five abiotic stresses [drought, metal, salt, cold, and nutrient] and five biotic stresses [bacterium, fungus, insect, weed, and nematode]) based on a higher number of microarray samples. We analyzed these data using the normalized and pareto-scaled intensities of 1,377 DEGs to assess the performance of these genes for the classification of different stress conditions. The top five components of PLS-DA captured 62.9% of variance between various stresses and showed classification accuracy of 0.77 (r2 = 0.92, Q2 = 0.88, P < 0.01). There were 196 and 53 genes with VIP scores (component 1) of 1.5 and 2 or greater. The relatively low classification accuracy reflects the inherently similar expression patterns between different stresses. Nonetheless, components 1 and 3 as shown in the two-dimensional score plot and the top three components as shown in the 3D score plot were able to clearly separate abiotic and biotic stresses as two major groups (Fig. 4). The two-dimensional and 3D plots also showed wide dispersion of drought stress and closeness with the majority of cold stress samples. Similarly, the 3D plot showed higher overlap between salt and metal stresses than other stresses, suggesting a higher similarity of the gene expression profile between them. The nutrient stress samples can be observed as a distinct group, although closer to other abiotic stresses. Bacterial stress samples show two major groups. One of the groups with the most bacterial samples showed overlap with fungal stress samples only. The other group was closer to weed, nematode, and fungal stress samples. Insect stress was observed as a distinct group closer to the group with bacterial and fungal samples.
Figure 4.
Multiclass classification of 10 stress conditions by PLS-DA. All five abiotic stresses are circled by red ovals and all five biotic stresses are circled by green ovals. A, Two-dimensional plot between PLS-DA components 1 (14.9%) and 3 (8.1%). B, 3D plot between PLS-DA components 1 (14.9%), 2 (28.9%), and 3 (8.1%). [See online article for color version of this figure.]
The same data set was analyzed using another classification technique, RF, which classified eight of the 10 stresses with 100% accuracy with an overall out-of-box (OOB) error rate of 0.0087, which is an unbiased estimate of classification error based on the one-third of samples left out (test samples) after bootstrap sample selection (Table I). Two of the stresses with less than 100% accuracy of classification were salt, with one wrongly classified sample (error rate of 0.037), and fungal stress, with two wrongly classified samples (error rate of 0.08). RF also provides a measure of variable importance by evaluating the increase in OOB error rate upon permutations called mean decrease in accuracy (Hsueh et al., 2013). The top 15 significant genes based on mean decrease in accuracy are shown in Supplemental Figure S1, including LOC_Os02g45170 (error rate of 0.0056), a bHLH TF, and LOC_Os05g31040, which codes for a cytokinin dehydrogenase precursor.
Table I. Classification of multiple stresses using the RF method.
The overall OOB error rate was 0.0087.
| Abiotic, Cold | Abiotic, Drought | Abiotic, Metal | Abiotic, Nutrient | Abiotic, Salt | Biotic, Bacterium | Biotic, Fungus | Biotic, Insect | Biotic, Nematode | Biotic, Weed | Class Error | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Abiotic, cold | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Abiotic, drought | 0 | 46 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Abiotic, metal | 0 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Abiotic, nutrient | 0 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Abiotic, salt | 0 | 1 | 0 | 0 | 26 | 0 | 0 | 0 | 0 | 0 | 0.037 |
| Biotic, bacterium | 0 | 0 | 0 | 0 | 0 | 166 | 0 | 0 | 0 | 0 | 0 |
| Biotic, fungus | 0 | 0 | 0 | 0 | 0 | 2 | 23 | 0 | 0 | 0 | 0.08 |
| Biotic, insect | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 |
| Biotic, nematode | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 |
| Biotic, weed | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 |
Functional Enrichment Analysis Revealed Molecular Mechanisms and Gene Families in Conserved and Nonconserved Gene Sets
Gene Ontology (GO) enrichment analysis of the 560 genes showing conserved down-regulation in abiotic and biotic stresses identified major biological and cellular processes: photosynthesis (FDR = 1.40E-07), electron carrier activity (FDR = 3.60E-06), small molecule biosynthetic process (FDR = 2.10E-05), and cellular nitrogen compound metabolic process, which is the parent term for a number of amino acids and nucleobase-containing compounds to be overrepresented. The terms transcription repressor activity (FDR = 0.0008) and response to oxidative stress (FDR = 0.034) were also found to be significant (Supplemental Fig. S2; Supplemental Table S4). On the other hand, 439 genes showing conserved up-regulation revealed a number of terms related to regulatory processes. The most significant innermost child terms are Ser-type endopeptidase inhibitor activity (FDR = 2.2E-06) and chitin catabolic process (FDR = 0.00013). Ser proteases perform diverse physiological roles in plants, important among which are induction after pathogen attack leading to the hypersensitivity response, regulation of Rubisco proteolysis, stomata development, perception of growth hormones, symbiosis, and senescence (Antão and Malcata, 2005; van der Hoorn, 2008). Significant enrichment of inhibitors of Ser-type endopeptidases in diverse stress conditions indicates the induction of several activities repressed by Ser proteases as part of the stress response. Furthermore, Ser protease inhibitors were also found to act as defense proteins by suppressing the activity of bowel proteinases in insects and plant pathogenic microorganisms (Mosolov and Valueva, 2011). Among the genes showing nonconserved expression, the set of genes down-regulated in abiotic stresses and up-regulated in biotic stresses were enriched with GO terms that include extracellular region (FDR = 5.30E-06) and catalytic activity (FDR = 5.3E-05).
Metabolic pathway enrichment analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) revealed a number of pathways specifically enriched in one of the sets. In the conserved down-regulated gene set, there were four annotation clusters with enrichment scores greater than 2.0 related to porphyrin and chlorophyll metabolism, transcription repressor activity via the Nmr-A-like domain, which is involved in posttranslational modification of the GATA TFs, which bind the DNA sequence GATA (Stammers et al., 2001), photosynthesis, and nicotianamine synthase activity. There were three annotation clusters with enrichment scores greater than 2.0 in the conserved up-regulated gene set related to heat shock protein Hsp20, Val, Leu, and Ile degradation, and the Bowman-Birk proteinase inhibitor family of Ser protease inhibitors. In rice, Bowman-Birk proteinase inhibitor genes were reported previously to be induced in multiple stresses like wounding, infection, and hormonal stress (Rakwal et al., 2001; Qu et al., 2003). The top annotation clusters in the nonconserved abiotic down-regulated and biotic up-regulated gene set were made up of a number of interpro domain terms, glycoprotein, metal-ion binding, plant peroxidases (POXs), and glycoside hydrolases.
There were 97 TF and regulator genes in the common DEGs (7%) belonging to 24 gene families that showed a distinct pattern (Supplemental Table S5). The major TF families NAC (for no apical meristem, Arabidopsis transcription activation factor, and cup-shaped cotyledon domain), HSF (for heat shock transcription factor), WRKY (for WRKY amino acid signature at the N-terminus), MYB (for MYB DNA binding domain), and MYB related were part of conserved down-regulated genes and nonconserved genes down-regulated under abiotic stresses and up-regulated under biotic stresses. Similarly, the TF families ERF (for ethylene response factor), bZIP (for basic leucine zipper), bHLH (for basic helix-loop-helix), and three others were among the conserved up-regulated genes and/or nonconserved genes up-regulated under abiotic stresses and down-regulated under biotic stresses. Twelve out of 13 ERFs were found in conserved up-regulated gene sets. These APETALA2 (AP2) domain-containing ERFs are well known for their role in both abiotic and biotic stress responses and were also shown to enhance multiple stress tolerance (Xu et al., 2011). Nine out of 12 WRKY TFs were among the nonconserved genes down-regulated under abiotic stresses and up-regulated under biotic stresses, which suggests that these TFs are the major regulatory factors that determine the direction of the molecular machinery and ultimately the cellular fate under simultaneous multiple stresses. MYB along with NAC TFs are reported to control antagonism between hormone-mediated abiotic stress and pathogen response pathways (Atkinson and Urwin, 2012). On the other hand, all five GOLDEN2 (G2)-like TF family members, which also contain a MYB-like DNA-binding domain, were part of the conserved up-regulated gene set. The G2-like TFs are required for chloroplast development and were shown to influence nuclear photosynthetic gene expression (Waters et al., 2009). We found a dearth of studies on the role of G2-like TFs under stress conditions. Down-regulation of photosynthetic mechanisms under stress is well established, as also observed in the enriched GO terms in our conserved down-regulated gene set. Genetic manipulation of G2-like TFs would shed further light on the regulation of photosynthesis under stress and reveal novel mechanisms to enhance stress tolerance. Out of the five lesion-simulating disease (LSD; Dietrich et al., 1997) family members reported in rice subspecies japonica by Plant Transcription Factor Database (PlnTFDB), two were part of the conserved down-regulated gene set. LSD TFs act as negative regulators of programmed cell death in a hypersensitive response (Epple et al., 2003). Transgenic suppression of LSD orthologs in rice resulted in a dwarf phenotype due to a deficiency of bioactive gibberellic acid, while overexpression of LSD enhanced resistance to rice bacterial blight (Xu and He, 2007). Based on our finding, studying LSD TFs under simultaneous abiotic and biotic stresses would provide vital clues on stress cross talk and the modulation of programmed cell death.
We analyzed the miRNAs predicted to target the 1,377 common DEGs using the database PMRD (Zhang et al., 2010). Out of the 456 experimentally verified miRNAs (miRBase; Griffiths-Jones et al., 2006) in rice, 142 (31%) miRNAs belonging to 50 miRNA families were found to target one or more common DEGs (Supplemental Table S6). Recently, 35 miRNAs from 31 miRNA families were found to be differentially expressed under abiotic stresses, drought, salt, and cold (Shen et al., 2010). Eighteen of these 31 stress-responsive miRNA families were among the 50 miRNA families targeting the common DEGs. The miRNA osa-miR1436 was found to target five of the conserved up-regulated genes, including LOC_Os09g23620, a MYB TF, while osa-miR446 was found to target five of the conserved down-regulated genes.
Coexpression Analysis Revealed Two Dense Clusters of Positively and Negatively Correlated Genes under Multiple Stresses
We conducted coexpression analysis using the normalized gene expression values of the common DEGs from stressed microarray samples and calculating Pearson correlation coefficient (r) between them. Out of the 947,376 possible edges (coexpression gene pairs) between the common DEGs, we found 8,924 edges with very high correlation (r ≥ 0.9 = 4,254 and r ≤ −0.7 = 4,670 edges, P = 0.01) in abiotic stress samples and 21,229 edges (r ≥ 0.9 = 7,673 and r ≤ −0.7 = 13,656 edges, P = 0.01) in biotic stress samples. A very high number of negative edges were observed in biotic stresses compared with abiotic stresses. For instance, there were 88 edges in biotic stresses with r ≤ −0.9 but only four edges in abiotic stresses with r ≤ −0.9. There were 3,701 shared edges between the two data sets with r ≥ 0.9 and r ≤ −0.7, out of which 2,684 (72%) were positive edges and 1,017 were negative edges. These 3,701 edges were between 381 genes, out of which 257 (67%) genes showed conserved down-regulation, 54 genes showed conserved up-regulation, and 49 genes showed down-regulation in abiotic stresses and up-regulation in biotic stresses. The 2,684 positive edges were between 208 genes, out of which 194 (93%) genes showed conserved down-regulation. Among the 381 genes, 15 had more than 75 high-correlation edges. The top three genes with the highest number of edges were LOC_Os02g22480 (glycosyltransferase; 142 edges), LOC_Os11g47840 (putative rhomboid homolog; 120 edges), and LOC_Os03g57200 (glutathione S-transferase; 93 edges). All three of these genes showed conserved up-regulation. Among the 14 TFs with significant edges, three TFs belonging to Nuclear Factor Y (a histone-like CCAAT-binding domain TF), G2-like, and bHLH TF families had the highest number of significant edges (79, 37, and 20, respectively). The majority of these edges were positive edges with other genes that showed conserved down-regulation.
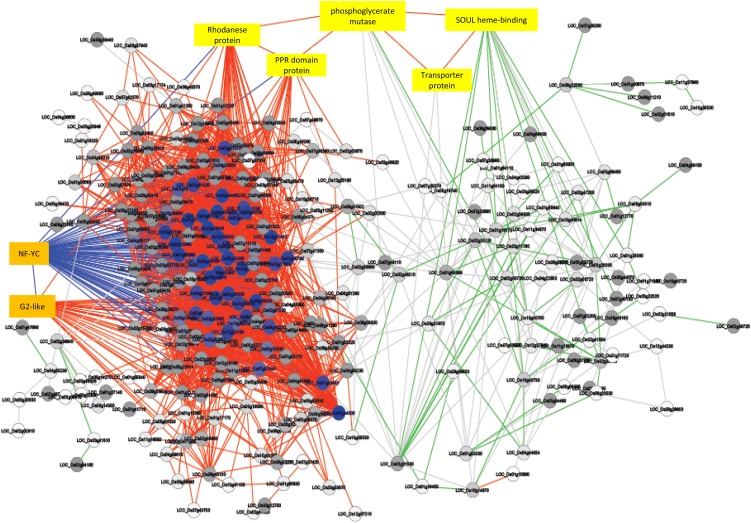
We analyzed the 3,701 significant edges using the plugin NetworkAnalyzer in the network analysis platform Cytocape 2.8.3 (Shannon et al., 2003), which revealed a dense cluster of positive edges (edges with r ≥ 0.95 are shown in red) and included most of the nodes with more than 75 edges (shown in blue) and a sparse cluster of negative edges (edges with r ≤ −0.9 are shown in green; Fig. 5). The two positive edge- and negative edge-rich clusters were found to be bridged by the gene LOC_Os01g13570, coding for phosphoglycerate mutase, with a positive edge to the SOUL heme-binding protein that was highly connected to the negative edge-rich cluster, and positive edges with rhodanese and pentatricopeptide domain-containing proteins, which were highly connected to the positive edge-rich cluster.
Figure 5.
Coexpression network of common DEGs. The edges with r ≥ 0.95 are shown in red and those with r ≤ −0.9 are shown in green. Nodes with more than 75 edges are shown in blue and those with more than 25 edges are shown in gray. The edges of the Nuclear Factor Y (NF-YC) TFs are shown in blue. [See online article for color version of this figure.]
High Overlap among Genes Identified by Different Classification Techniques, Coexpression, and Functional Enrichment Analysis
We compiled the significance of the common DEGs based on various criteria, including feature importance identified by different classification techniques, count of the number of coexpression edges, PlnTFDB gene, and STIFDB2 gene (Supplemental Table S3). We found that many of the PLS-DA two-class significant genes (177 genes with VIP ≥ 1.5) were also significant in PLS-DA multiclass (36% or 71 out 196) and RF’s top 100 genes (68%) but showed poor overlap with the 540 significant genes found by SVM (2% or nine out of 540), TF genes (9% or nine out of 97), and STIFDB2 genes (10% or 27 out of 259). However SVM’s 540 genes showed high overlap with PLS-DA multiclass (50% or 99 out of 196), TF genes (45% or 44 out of 97), and STIFDB2 genes (40% or 103 out of 259). Taken together, the 196 top genes of PLS-DA multiclass showed overlap with most of the other significant gene lists, of which 43 (22%) were also part of the STIFDB2 list. Out of 1,118 japonica rice genes reported as stress-responsive genes in STIFDB2 (Naika et al., 2013), 259 (23%) were part of common DEGs. Furthermore, out of 97 TF genes in the common DEGs, only 12 were part of STIFDB2 and none of the major WRKY and MYB TF genes, including those previously reported as stress-responsive genes (Atkinson and Urwin, 2012), were part of STIFDB2’s list. The top 10 of these 196 genes are given in Table II. The top-most gene encodes a CCCH zinc finger domain-containing TF known to control embryogenesis (Li and Thomas, 1998) and involved in multiple abiotic stresses (Sun et al., 2007; Kim et al., 2008). A homolog of this gene (LOC_Os05g10670), which was also part of the 1,432 up-regulated genes in our meta-analysis of abiotic stresses, was recently reported to confer delayed senescence and improved tolerance to high-salt and drought stresses by regulating reactive oxygen species homeostasis and metal homeostasis (Jan et al., 2013). One gene that was part of all feature selection lists was LOC_Os11g26780, a dehydrin gene that had one significant positive edge with another dehydrin gene (LOC_Os11g26790; r = 0.97 and 0.93 in abiotic and biotic stresses, respectively), both of which showed conserved up-regulation.
Table II. Top 10 genes with the highest VIP score in multiclass classification by PLS-DA.
| MSU Identifier | Annotation | PLS-DA Multiple Stress (VIP component 1) | PLS-DA Abiotic Versus Biotic (VIP component 1) | RF Top 100 (Mean Decrease in Accuracy) | PLS-DA Two-Class (SVM 540) | SVM 540 (Frequency) |
|---|---|---|---|---|---|---|
| LOC_Os01g09620 | Zinc finger/CCCH TF | 2.899 | 2.1512 | 0 | 0 | 0 |
| LOC_Os11g11970 | Expressed protein | 2.8141 | 2.1813 | 0.003567 | 0 | 0 |
| LOC_Os11g26780 | Dehydrin | 2.8021 | 1.6348 | 0.002054 | 2.5939 | 358 |
| LOC_Os07g48020 | POX | 2.7992 | 1.8946 | 0 | 0 | 0 |
| LOC_Os06g24990 | Xylanase inhibitor protein1 | 2.7427 | 2.2645 | 0 | 3.593 | 358 |
| LOC_Os11g32890 | Expressed protein | 2.718 | 1.6966 | 0 | 2.6919 | 358 |
| LOC_Os06g48300 | Protein phosphatase 2C | 2.6559 | 0 | 0 | 2.2334 | 358 |
| LOC_Os10g40040 | Expressed protein | 2.5979 | 0 | 0 | 2.3424 | 358 |
| LOC_Os09g07350 | Fasciclin-like arabinogalactan protein8 | 2.5059 | 0 | 0 | 1.9348 | 358 |
| LOC_Os05g06920 | RelA-SpoT like protein RSH4 | 2.5036 | 2.8245 | 0.004242 | 0 | 0 |
Comparison of the common DEGs with the list of 1,922 hormone-related genes of Arabidopsis (Arabidopsis thaliana) as reported in the Arabidopsis Hormone Database 2.0 (Jiang et al., 2011) using putative orthologous genes found by GreenPhylDB (Rouard et al., 2011) revealed 31 common DEGs that were orthologous to 51 Arabidopsis hormone genes (Supplemental Table S3). A summary table of the expression status of hormone-related genes in the common DEGs (78 genes) based on Arabidopsis hormone database orthologs and paralogs with the same annotation and expression status in both abiotic and biotic stresses (except TFs) or name of the hormone in the gene annotation provided by MSU Rice Genome Annotation Project release 7 is given in Table III. Overall, the expression status of various hormone-related genes was very similar to the one proposed in a recent review (Atkinson and Urwin, 2012). For instance, nine out 12 ABA-responsive genes showed conserved up-regulation, while six out of 10 ethylene (ET)-responsive genes showed nonconserved down-regulation under abiotic stress and up-regulation under biotic stress. Most of the conserved auxin down-regulated genes were related to auxin biosynthesis and response factors, while conserved up-regulated genes were related to auxin-repressed factors, which indicates extensive down-regulation of auxin-induced biological processes. A recent study analyzed the transcriptome of rice under bacterial stress by Xanthomonas oryzae pv oryzae and compared the DEGs with those found in seven other microarray studies conducted on AffymetrixRiceArray (Narsai et al., 2013). The authors reported 240 genes (212 loci) as differentially expressed in multiple stresses. Out of these loci, 110 (51.8%) were part of our common DEGs list, most of which belonged to the conserved up-regulation gene set (64%) and included many important genes, such as WRKY, AP2/EREBP (for ethylene-responsive element binding protein) family TFs, ATP-binding cassette transporter, multidrug resistance, and universal stress genes.
Table III. Expression status of various hormone-related genes in the common DEGs.
The number of orthologs of Arabidopsis plant hormone database genes are shown in parentheses
| Hormone | Total Genes | Conserved, Down | Conserved, Up | Nonconserved (Abiotic Up-Biotic Down) | Nonconserved (Abiotic Down-Biotic Up) |
|---|---|---|---|---|---|
| ABA | 12 (9) | – | 9 (3) | – | 3 (1) |
| Auxin | 21 (6) | 5 (1) | 11 (3) | 3 (1) | 2 (1) |
| Brassinosteroid | 11 (5) | – | 7 (4) | – | 4 (1) |
| Cytokinin | 4 (2) | 4 (2) | – | – | – |
| ET | 10 (5) | 3 (2) | 1 (1) | – | 6 (2) |
| Gibberellic acid | 8 (3) | 2 (1) | 5 (2) | 1 (0) | – |
| JA | 7 (3) | 4 (1) | – | 1 (1) | 2 (1) |
| SA | 5 (3) | – | 4 (2) | – | 1 (1) |
DISCUSSION
The multiple stress response in plants has been a hot topic of research, as many studies, including those involving genetic manipulation and chemical intervention, reported that increased resistance to one stress resulted in heightened susceptibility to other abiotic and biotic stress conditions (Atkinson and Urwin, 2012; Sharma et al., 2013). Furthermore, it was suggested that plant hormones are the key determinants of genetic switches and cellular adjustments in a multistress environment. Different plant hormones are broadly categorized to play central roles in different kinds of stress responses. For instance, within biotic stresses, (hemi)biotrophic pathogens commonly activate the salicylic acid (SA)-dependent defense response, while necrotrophic pathogens activate jasmonic acid (JA)- and ET-dependent signaling pathways (Sharma et al., 2013). SA and JA/ET often act antagonistically and propagate opposing influences (Pieterse et al., 2009). On the other hand, ABA is well established as the major player of the abiotic stress response. ABA is increasingly found to also play a critical role in biotic stresses by negatively regulating plant immunity. Many studies found that abiotic stresses enhance plant susceptibility to pathogen attacks due to weakening of defense systems. Thus, it was proposed that plants prioritize abiotic stress tolerance over the biotic stress response, with ABA as the molecular switch between the two responses to minimize the damage (Lee and Luan, 2012). Recently, however, contrary studies where biotic stress takes precedence have been reported (Kim et al., 2011; Mang et al., 2012; Sánchez-Vallet et al., 2012). Thus, in light of these recent developments, which revealed a rather complicated picture of multiple stress responses, we embarked on the identification of DEGs in abiotic and biotic stress environments separately and performed comparative analysis of the shared stress-responsive genes, which would provide vital clues on the causative factors behind the cross talk resulting in the observed synergistic and antagonistic regulation of known abiotic and biotic stress response pathways.
Our study identified 1,377 differentially expressed common genes under a wide spectrum of abiotic and biotic stress conditions, and their expression status can be considered as a representation of their overall involvement in the stress response to nonliving factors and living organisms. Thus, this list of genes forms an ideal gene set to objectively investigate the similarities and differences between abiotic and biotic stress responses. Although more than 70% of common DEGs showed conserved differential expression, we were able to classify different stresses, including abiotic and biotic stresses, with high accuracy, indicating that their subtle expression differences can be exploited to effectively discriminate between various stress conditions.
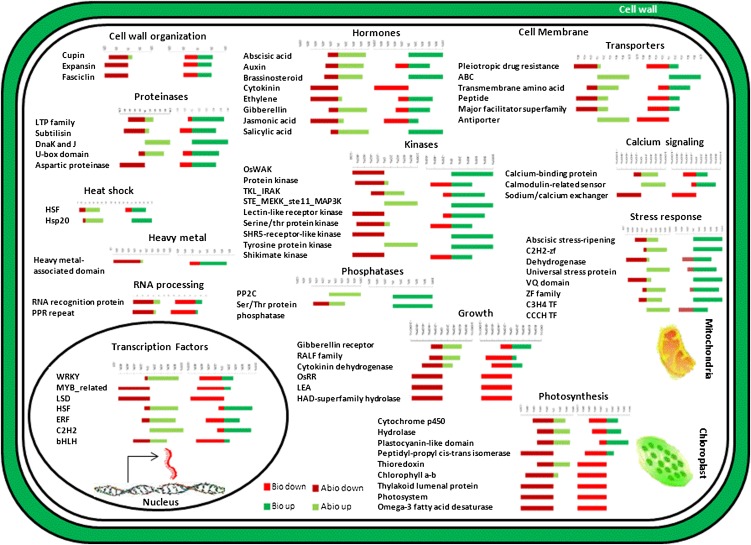
A closer look at chloroplast- and photosynthesis-related genes in the common DEGs revealed conserved down-regulation of 17 out of 18 PSII, chlorophyll a/b-binding, and thylakoid lumenal genes (Fig. 6). A diverse set of 40 chloroplast precursor enzymes that contain an N-terminal transit peptide for import into chloroplast (Jarvis, 2008) were also part of the common DEGs, 26 (65%) of which showed conserved down-regulation. Furthermore, a number of cytochrome P450 genes (29 genes) that encode parent compounds for a number of secondary metabolites involved in plant defense (Jirschitzka et al., 2013) were part of the common DEGs. Fourteen of these 29 (approximately 48%) genes showed conserved down-regulation, while seven showed conserved up-regulation, and the rest showed nonconserved differential expression. Thus, exploring the nonconserved DEGs would shed further light on the cross talk of stress responses via metabolic adjustments. The cell wall is the first line of plant defense in response to external stimuli. A number of important gene families involved in cell wall synthesis and modifications showed distinct patterns of expression under abiotic and biotic stresses. For instance, there were six OsWAK (for wall-associated kinase) genes in common DEGs, all of which showed nonconserved down-regulation under abiotic stresses and up-regulation under biotic stresses (Fig. 6). WAKs are part of the transmembrane receptor-like kinase superfamily, which perceive stimuli using extracellular domains with signal transmission through their cytoplasmic kinase domains (Li et al., 2009). There are currently 144 rice genes regarded as WAKs (MSU Rice Genome Annotation Project release 7) compared with 26 genes in Arabidopsis, which is most likely due to lineage-specific gene duplications (Zhang et al., 2005). However, very little is known about the function of these genes in rice except OsWAK1, whose overexpression increased resistance to the blast fungus Magnaporthe oryzae (Li et al., 2009; Kohorn and Kohorn, 2012). FAS1 (for fasciclin-like) domain-containing genes are another group of transmembrane genes involved in cell adhesion (Johnson et al., 2003; Ma and Zhao, 2010). All five fasciclin domain genes in the common DEGs showed conserved down-regulation. Similarly, most cupin, expansin, and aquaporin genes involved in cell wall synthesis and organization showed conserved down-regulation.
Figure 6.
Visual representation of different gene families and functional categories based on expression between abiotic and biotic stresses. For each annotation, the stacked bars represent up-regulated genes (total) scaled to 100% and down-regulated genes scaled to −100%. [See online article for color version of this figure.]
A number of transporter and kinase/phosphatase genes showed clear patterns of coordinated expression under abiotic and biotic stresses. All three of the genes coding for pleiotropic drug resistance-type ATP-binding cassette transporter proteins, which were found to be induced by ABA, SA, and JA in rice (Moons, 2008), showed conserved up-regulation. Reversible protein phosphorylation executed by kinases and phosphatases is a fundamental mechanism that facilitates the orchestration of some of the most sophisticated signaling pathways. A number of different kinds of kinases and phosphatases were found in the list of common DEGs, out of which Ser/Thr protein kinases and phosphatases showed high distinction between the two stresses, as also found by the GO analysis (Supplemental Table S4). All five of the protein phosphatase 2C genes, which are key players in ABA signaling pathways, showed conserved up-regulation (Fig. 6). Four of these protein phosphatase 2C genes were part of the significant genes found by both SVM and PLS-DA multiclass, indicating that these genes show distinct patterns of expression in different stress conditions and can be considered as some of the most important genes to study the multiple stress response.
A number of transporter and POX precursor genes showed clear patterns of difference in expression between abiotic and biotic stresses. For instance, two out of three major facilitator superfamily antiporter genes showed nonconserved up-regulation under abiotic stresses. As many as 23 POX precursor genes were part of common DEGs, out of which 13 (56%) showed nonconserved down-regulation under abiotic stresses. Furthermore, nine and 12 of these 23 POX genes were part of SVM and PLS-DA multiclass significant features, respectively. A study of rice infected with blast fungus showed that 10 POX genes redundantly respond to multiple stresses (Sasaki et al., 2004). Our findings suggest that the functionalities of many of the POX genes are specific to biotic stresses and are promising candidates to decipher the cross talk between stresses.
The domain family with the highest number of conserved up-regulated genes was the zinc finger family, including C2H2 (for zinc finger domain with two cysteines and two histidines), C3H (for CCCH zinc finger domain) TFs, C3HC4 (for cysteine-rich zinc finger), and ZIM (for zinc-finger protein expressed in inflorescence meristem) domain-containing members, with 14 and 15 members out of 17 showing overexpression in abiotic and biotic stresses, respectively. All of the pentatricopeptide domain genes (11), which play essential roles in RNA editing, organelle biogenesis (Yuan and Liu, 2012), and plant development by coordinating interaction between mitochondria and chloroplasts (Toda et al., 2012), showed conserved down-regulation except LOC_Os07g36450, which showed conserved up-regulation. Thus, this gene would be an important candidate to further explore and understand their specific role under stress conditions and determine what makes it different from other pentatricopeptide genes. Another interesting gene family showing high distinction between the two stress categories was the protease inhibitor/seed storage/lipid transfer protein family, with five out of nine members showing nonconserved down-regulation in abiotic stresses. VQ domain-containing proteins were recently found to interact with WRKY TFs (WRKY33) in Arabidopsis. Furthermore, knockout or overexpression of VQ genes substantially altered the defense response (Cheng et al., 2012). There are five VQ domain genes in common DEGs, out of which four showed nonconserved biotic up-regulation. Furthermore, WRKY24, which is the rice ortholog of WRKY33, also showed nonconserved biotic up-regulation. The striking contrast in these genes in their behavior between abiotic and biotic stresses suggests them as important candidates to explore the multiple stress response.
A list of studies that overexpressed or suppressed 10 of the common DEGs that significantly altered the stress response is provided in Table IV. Seven of these are TF genes (four genes code for the WRKY family of TFs) and are part of significant features found by SVM. Overexpression of a gene (LOC_Os01g55940) coding for an indole-3-acetic acid-amido synthetase conferred broad-spectrum resistance to Magnaporthe grisea, X. oryzae pv oryzae, and X. oryzae pv oryzicola (Fu et al., 2011). Overexpression of two genes coding for NAC TFs enhanced tolerance to multiple abiotic stresses such as drought and salinity (LOC_Os03g60080) as well as cold (LOC_Os11g03300; Hu et al., 2006; Jeong et al., 2010). The expression of these three genes and LOC_Os07g40290, an auxin-responsive gene coexpressed with 40 other common DEGs, was up-regulated in both biotic and abiotic stresses. Furthermore, we compared the common DEGs against a recently released database of Arabidopsis loss-of-function mutants (Lloyd and Meinke, 2012) using orthologous identifiers, which revealed 138 orthologous mutant genes out of which 33 showed increased resistance or sensitivity to a variety of stresses (Supplemental Table S7). Two genes (LOC_Os06g44010 and LOC_Os12g16720) are common between the DEGs in Table IV and their orthologs with loss-of-function mutants in Arabidopsis. The first gene encodes a WRKY TF that is up-regulated under biotic stress and down-regulated under abiotic stress. The second gene codes for a cytochrome P450 monooxygenase whose inactivation leads to Sekiguchi lesion mutant rice (Fujiwara et al., 2010). This gene is part of a conserved up-regulated gene set. Experimental analysis involving the overexpression or knockout of the stress-responsive genes described above has often focused on one or a few stresses. Genetically engineering rice plants with the top candidate genes identified in our study, singly or in combination, would identify their role in conferring broad-range resistance to multiple abiotic and biotic stresses.
Table IV. List of common DEGs that showed alteration in stress response upon overexpression/suppression.
| MSU Identifier | Annotation | Phenotype | Reference |
|---|---|---|---|
| LOC_Os01g55940 | OsGH3.2, probable indole-3-acetic acid-amido synthetase | Enhanced broad-spectrum disease resistance | Fu et al. (2011) |
| LOC_Os02g08440 | WRKY71 | Enhanced defense response | Liu et al. (2007) |
| LOC_Os03g60080 | NAC domain-containing protein67 | Increased drought and salt tolerance | Hu et al. (2006) |
| LOC_Os05g25770 | WRKY45 | Increased susceptibility to bacteria | Tao et al. (2009) |
| LOC_Os06g44010 | WRKY28 | Enhanced disease resistance | Peng et al. (2010) |
| LOC_Os07g40290 | OsGH3.8, probable indole-3-acetic acid-amido synthetase | Enhanced disease resistance | Ding et al. (2008) |
| LOC_Os08g06280 | LSD1 zinc finger domain-containing protein | Increased resistance to blast fungus | Wang et al. (2005) |
| LOC_Os09g25070 | WRKY62 | Increased bacterial susceptibility | Peng et al. (2008) |
| LOC_Os11g03300 | NAC domain TF | Increased drought tolerance and yield | Jeong et al. (2010) |
| LOC_Os12g16720 | Cytochrome P450 71A1 | Enhanced fungal resistancea | Fujiwara et al. (2010) |
Suppression of gene expression by knockout.
CONCLUSION
The availability of large volumes of genome-scale gene expression data and advanced computational techniques enabled us to dissect the complex nature of the stress response and examine in depth the overlap between abiotic and biotic stress responses. The plethora of novel insights reported in this work revealed the overarching roles of major stress regulatory molecules, including phytohormones such as ABA and JA/ET, parent compounds of small metabolites like shikmate, TFs like WRKY and MYB, and signaling genes like WAKs, which are central to the fine-tuning of stress response pathways. Furthermore, the expression patterns exhibited by these genes provided a molecular basis to classify different stress conditions with high accuracy. The top regulatory and signaling genes identified in this study are likely to be involved in cross talk between biotic and abiotic stress responses and to provide potential candidates crucial for the development of a rice variety with broad-range stress tolerance. Furthermore, mechanistic insights gained in rice on multiple stress responses would provide anchor points to explore specific stress signaling pathways and orthologous genes in other cereal crops.
MATERIALS AND METHODS
Selection of Stress Response Microarray Studies and Identification of DEGs
All of the microarray studies performed on the Affymetrix Rice Genome Array and deposited at the Gene Expression Omnibus under the platform GPL2025 were manually searched to identify and categorize 13 stress conditions (seven abiotic and six biotic stresses), as shown in Supplemental Table S1. Two meta-analysis studies were performed combining abiotic and biotic stresses separately. Briefly, the raw intensity CEL files of the selected samples were downloaded from the Gene Expression Omnibus, and intensity values were extracted from the CEL files using the bioconductor package Affy in R (Gautier et al., 2004), quality checked using the package ArrayQualityMetrics (Kauffmann et al., 2009), and the samples failing quality tests were removed.
The samples of each stress were normalized together using the robust multichip average method (Irizarry et al., 2003). The probes were then matched to their loci based on annotation provided at http://www.ricechip.org. Probes with no match or ambiguously matching multiple loci were discarded. The retained probes and their normalized intensity values were then loaded into a oneChannelGUI environment to perform nonspecific filtering of probes with relatively small signal distribution using an interquartile range filter at the most stringent setting (0.5) and probes with very low intensity values [probes below threshold log2(50) = 5.64 in 90% or more of arrays]. DEGs were identified using the rank product method (Breitling et al., 2004). We used the function RPadvance of the bioconductor package RankProd (Hong et al., 2006), which is specifically designed for meta-analysis by taking into consideration the different origins of samples. The number of permutation tests was set to 250. The function topGene with a percentage of false positives cutoff value of 0.01 or less was used to output DEGs. Among multiple probes matching the same locus, the probe identifier with the highest fold change was retained.
Classification Methods
We used a number of classification and machine learning techniques to assess the performance of identified common DEGs between abiotic and biotic stresses in the classification of different stresses. We extracted the robust multichip average-normalized intensity values of the identified common DEGs between abiotic and biotic stresses from stress-treated microarrays (126 abiotic and 232 biotic arrays) and scale adjusted using mean centering and dividing by the square root of the sd of each variable (pareto scaling; Supplemental Fig. S3). Pareto scaling was chosen as it keeps the data structure partially intact while reducing the relative importance of large values (van den Berg et al., 2006).
PCA is a nonsupervised (i.e. it does not make use of class labels) dimensionality reduction procedure that performs an orthogonal transformation of the original variables into a set of linearly uncorrelated variables such that the largest variance between the classes is captured in the transformed variables, also called PCs (Yeung and Ruzzo, 2001). The PCs are numbered in decreasing order, and the top one (PC1) captures the maximal variance between different classes. PLS-DA is a supervised (i.e. it makes use of class labels) projection method that separates groups by rotating the PCs such that a maximum separation among classes is obtained (Zhang et al., 2013).
SVM classifies binary training data by drawing a hyperplane (linear or nonlinear based on the type of kernel selected) that maximally separates the two categories (Furey et al., 2000). R-SVM performs this type of classification recursively using different feature subsets and selects the best-performing features based on cross-validation error rates. Although SVM based on microarray data is widely used to classify and predict disease status in humans (Hedenfalk et al., 2001) and identify important features (Zhang et al., 2006), only a few studies have used R-SVM to identify stress-responsive genes in plants (Liang et al., 2011). We performed R-SVM classification using a linear kernel with genes (features) in columns and samples in rows. We utilized a leave-one-out cross-validation procedure to determine the accuracy of the classification, in which features are randomly partitioned into training and test sets and the poorly performing features with higher cross-validation error rate are recursively eliminated. RF is a decision tree-based algorithm that grows the branches of an ensemble of classification trees by selecting random subsets of features from bootstrap samples and makes class predictions based on the majority vote of the ensemble. A number of characteristics of RF make it ideal for our data set, including its use for multiclass problems, it is less affected by noise, and it does not overfit the training data (Díaz-Uriarte and Alvarez de Andrés, 2006). The statistical packages and tools provided by R, WEKA (Frank et al., 2004), and Metaboanalyst (Xia et al., 2012) were utilized to implement different analytical procedures.
Functional Enrichment Analysis
GO analysis was carried out using the Singular Enrichment Analysis tool offered by agriGO (Du et al., 2010) at default settings of Fisher’s t test (P < 0.05), FDR correction by the Hochberg method, and five minimum mapping entries against species-specific precomputed background references. Metabolic pathway enrichment analysis was carried out using the tool DAVID version 6.7 (Huang et al., 2009). This functional annotation tool performs enrichment analysis of various annotation resources including gene ontologies, protein domains, and pathways using a modified Fisher’s exact test called EASE. Furthermore, it clusters significant annotation terms using κ statistics and fuzzy heuristic clustering based on the degree of common genes between two annotations and provides an enrichment score for each annotation cluster. Information on TF genes in rice (Oryza sativa) was obtained from the database PlnTFDB (Pérez-Rodríguez et al., 2010) and analyzed for the enrichment of TF families. The miRNAs predicted to target stress-responsive genes were obtained from PMRD, the plant miRNA database (Zhang et al., 2010).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Top 15 genes found by RF based on mean decreasing accuracy.
Supplemental Figure S2. Top GO terms of common DEGs.
Supplemental Figure S3. Box plot and density distribution of common DEGs before and after pareto normalization.
Supplemental Table S1. Number of microarray studies and samples analyzed for different stress conditions and description of the samples in different studies.
Supplemental Table S2. DEGs in abiotic and biotic stresses.
Supplemental Table S3. List of common DEGs.
Supplemental Table S4. Top functional annotation clusters found by the tool DAVID.
Supplemental Table S5. Distribution of TF families in conserved and nonconserved DEGs.
Supplemental Table S6. List of miRNAs targeting common DEGs.
Supplemental Table S7. List of common DEGs with loss-of-function mutant Arabidopsis orthologs and their mutant phenotypes.
Glossary
- ABA
abscisic acid
- PCA
principal component analysis
- SVM
support vector machines
- R-SVM
recursive-support vector machines
- DEGs
differentially expressed genes
- PLS-DA
partial least squares discriminant analysis
- RF
random forest
- FDR
false discovery rate
- PCs
principal components
- 3D
three-dimensional
- VIP
variable importance in projection
- TF
transcription factor
- OOB
out-of-box
- GO
Gene Ontology
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- miRNA
microRNA
- SA
salicylic acid
- JA
jasmonic acid
- ET
ethylene
- POX
peroxidase
References
- Antão CM, Malcata FX. (2005) Plant serine proteases: biochemical, physiological and molecular features. Plant Physiol Biochem 43: 637–650 [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Urwin PE. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63: 3523–3543 [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Cao FY, Yoshioka K, Desveaux D. (2011) The roles of ABA in plant-pathogen interactions. J Plant Res 124: 489–499 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zhou Y, Yang Y, Chi YJ, Zhou J, Chen JY, Wang F, Fan B, Shi K, Zhou YH, et al. (2012) Structural and functional analysis of VQ motif-containing proteins in Arabidopsis as interacting proteins of WRKY transcription factors. Plant Physiol 159: 810–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Uriarte R, Alvarez de Andrés S. (2006) Gene selection and classification of microarray data using random forest. BMC Bioinformatics 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL. (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. (2008) Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20: 228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64– W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Mack AA, Morris VR, Dangl JL. (2003) Antagonistic control of oxidative stress-induced cell death in Arabidopsis by two related, plant-specific zinc finger proteins. Proc Natl Acad Sci USA 100: 6831–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank E, Hall M, Trigg L, Holmes G, Witten IH. (2004) Data mining in bioinformatics using Weka. Bioinformatics 20: 2479–2481 [DOI] [PubMed] [Google Scholar]
- Fu J, Liu H, Li Y, Yu H, Li X, Xiao J, Wang S. (2011) Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol 155: 589–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Maisonneuve S, Isshiki M, Mizutani M, Chen L, Wong HL, Kawasaki T, Shimamoto K. (2010) Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem 285: 11308–11313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. (2000) Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 16: 906–914 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. (2010) Food security: the challenge of feeding 9 billion people. Science 327: 812–818 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, et al. (2001) Gene-expression profiles in hereditary breast cancer. N Engl J Med 344: 539–548 [DOI] [PubMed] [Google Scholar]
- Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. (2006) RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827 [DOI] [PubMed] [Google Scholar]
- Hsueh HM, Zhou DW, Tsai CA. (2013) Random forests-based differential analysis of gene sets for gene expression data. Gene 518: 179–186 [DOI] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: 12987–12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K. (2013) OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 161: 1202–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. (2010) Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol 153: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Liu X, Peng Z, Wan Y, Ji Y, He W, Wan W, Luo J, Guo H. (2011) AHD2.0: an update version of Arabidopsis Hormone Database for plant systematic studies. Nucleic Acids Res 39: D1123–D1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirschitzka J, Mattern DJ, Gershenzon J, D’Auria JC. (2013) Learning from nature: new approaches to the metabolic engineering of plant defense pathways. Curr Opin Biotechnol 24: 320–328 [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. (2003) The fasciclin-like arabinogalactan proteins of Arabidopsis: a multigene family of putative cell adhesion molecules. Plant Physiol 133: 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. (2009) ArrayQualityMetrics: a bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G. (2008) SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20: 1260–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Hauser F, Ha T, Xue S, Böhmer M, Nishimura N, Munemasa S, Hubbard K, Peine N, Lee BH, et al. (2011) Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol 21: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kohorn SL. (2012) The cell wall-associated kinases, WAKs, as pectin receptors. Front Plant Sci 3: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Luan S. (2012) ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ 35: 53–60 [DOI] [PubMed] [Google Scholar]
- Li H, Zhou SY, Zhao WS, Su SC, Peng YL. (2009) A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol Biol 69: 337–346 [DOI] [PubMed] [Google Scholar]
- Li Z, Thomas TL. (1998) PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell 10: 383–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Zhang F, Wang J, Joshi T, Wang Y, Xu D. (2011) Prediction of drought-resistant genes in Arabidopsis thaliana using SVM-RFE. PLoS ONE 6: e21750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bai X, Wang X, Chu C. (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164: 969–979 [DOI] [PubMed] [Google Scholar]
- Liu Z, Xie M, Yao Z, Niu Y, Bu Y, Gao C. (2013) Three meta-analyses define a set of commonly overexpressed genes from microarray datasets on astrocytomas. Mol Neurobiol 47: 325–336 [DOI] [PubMed] [Google Scholar]
- Lloyd J, Meinke D. (2012) A comprehensive dataset of genes with a loss-of-function mutant phenotype in Arabidopsis. Plant Physiol 158: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkilä J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. (2010) A global map of human gene expression. Nat Biotechnol 28: 322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz W, Samir KC. (2010) Dimensions of global population projections: what do we know about future population trends and structures? Philos Trans R Soc Lond B Biol Sci 365: 2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhao J. (2010) Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J Exp Bot 61: 2647–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang HG, Qian W, Zhu Y, Qian J, Kang HG, Klessig DF, Hua J. (2012) Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24: 1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moons A. (2008) Transcriptional profiling of the PDR gene family in rice roots in response to plant growth regulators, redox perturbations and weak organic acid stresses. Planta 229: 53–71 [DOI] [PubMed] [Google Scholar]
- Mosolov VV, Valueva TA. (2011) [Inhibitors of proteolytic enzymes under abiotic stresses in plants (review)]. Prikl Biokhim Mikrobiol 47: 501–507 [PubMed] [Google Scholar]
- Naika M, Shameer K, Mathew OK, Gowda R, Sowdhamini R. (2013) STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol 54: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R, Wang C, Chen J, Wu J, Shou H, Whelan J. (2013) Antagonistic, overlapping and distinct responses to biotic stress in rice (Oryza sativa) and interactions with abiotic stress. BMC Genomics 14: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Canlas P, Ronald PC. (2010) OsWRKY IIa transcription factors modulate rice innate immunity. Rice 3: 36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC. (2008) OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1: 446–458 [DOI] [PubMed] [Google Scholar]
- Pérez-Enciso M, Tenenhaus M. (2003) Prediction of clinical outcome with microarray data: a partial least squares discriminant analysis (PLS-DA) approach. Hum Genet 112: 581–592 [DOI] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B. (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38: D822–D827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Leon-Reyes A, Van der Ent S, Van Wees SC. (2009) Networking by small-molecule hormones in plant immunity. Nat Chem Biol 5: 308–316 [DOI] [PubMed] [Google Scholar]
- Qu LJ, Chen J, Liu M, Pan N, Okamoto H, Lin Z, Li C, Li D, Wang J, Zhu G, et al. (2003) Molecular cloning and functional analysis of a novel type of Bowman-Birk inhibitor gene family in rice. Plant Physiol 133: 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwal R, Kumar Agrawal G, Jwa NS. (2001) Characterization of a rice (Oryza sativa L.) Bowman-Birk proteinase inhibitor: tightly light regulated induction in response to cut, jasmonic acid, ethylene and protein phosphatase 2A inhibitors. Gene 263: 189–198 [DOI] [PubMed] [Google Scholar]
- Rouard M, Guignon V, Aluome C, Laporte MA, Droc G, Walde C, Zmasek CM, Périn C, Conte MG. (2011) GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res 39: D1095–D1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rung J, Brazma A. (2013) Reuse of public genome-wide gene expression data. Nat Rev Genet 14: 89–99 [DOI] [PubMed] [Google Scholar]
- Sánchez-Vallet A, López G, Ramos B, Delgado-Cerezo M, Riviere MP, Llorente F, Fernández PV, Miedes E, Estevez JM, Grant M, et al. (2012) Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol 160: 2109–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Iwai T, Hiraga S, Kuroda K, Seo S, Mitsuhara I, Miyasaka A, Iwano M, Ito H, Matsui H, et al. (2004) Ten rice peroxidases redundantly respond to multiple stresses including infection with rice blast fungus. Plant Cell Physiol 45: 1442–1452 [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC. (2013) Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant 6: 250–260 [DOI] [PubMed] [Google Scholar]
- Shen J, Xie K, Xiong L. (2010) Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol Genet Genomics 284: 477–488 [DOI] [PubMed] [Google Scholar]
- Stammers DK, Ren J, Leslie K, Nichols CE, Lamb HK, Cocklin S, Dodds A, Hawkins AR. (2001) The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J 20: 6619–6626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Student S, Fujarewicz K. (2012) Stable feature selection and classification algorithms for multiclass microarray data. Biol Direct 7: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C. (2007) The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol 48: 1148–1158 [DOI] [PubMed] [Google Scholar]
- Tao Z, Liu H, Qiu D, Zhou Y, Li X, Xu C, Wang S. (2009) A pair of allelic WRKY genes play opposite roles in rice-bacteria interactions. Plant Physiol 151: 936–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Fujii S, Noguchi K, Kazama T, Toriyama K. (2012) Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing of nad5 transcripts in mitochondria. Plant J 72: 450–460 [DOI] [PubMed] [Google Scholar]
- van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. (2006) Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn RA. (2008) Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol 59: 191–223 [DOI] [PubMed] [Google Scholar]
- Wang L, Pei Z, Tian Y, He C. (2005) OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant Microbe Interact 18: 375–384 [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. (2012) MetaboAnalyst 2.0: a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40: W127– W133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, He C. (2007) The rice OsLOL2 gene encodes a zinc finger protein involved in rice growth and disease resistance. Mol Genet Genomics 278: 85–94 [DOI] [PubMed] [Google Scholar]
- Xu ZS, Chen M, Li LC, Ma YZ. (2011) Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol 53: 570–585 [DOI] [PubMed] [Google Scholar]
- Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60: 107–124 [DOI] [PubMed] [Google Scholar]
- Yeung KY, Ruzzo WL. (2001) Principal component analysis for clustering gene expression data. Bioinformatics 17: 763–774 [DOI] [PubMed] [Google Scholar]
- Yuan H, Liu D. (2012) Functional disruption of the pentatricopeptide protein SLG1 affects mitochondrial RNA editing, plant development, and responses to abiotic stresses in Arabidopsis. Plant J 70: 432–444 [DOI] [PubMed] [Google Scholar]
- Zhang PJ, Broekgaarden C, Zheng SJ, Snoeren TA, van Loon JJ, Gols R, Dicke M. (2013) Jasmonate and ethylene signaling mediate whitefly-induced interference with indirect plant defense in Arabidopsis thaliana. New Phytol 197: 1291–1299 [DOI] [PubMed] [Google Scholar]
- Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux PG. (2005) Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol 139: 1107–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lu X, Shi Q, Xu XQ, Leung HC, Harris LN, Iglehart JD, Miron A, Liu JS, Wong WH. (2006) Recursive SVM feature selection and sample classification for mass-spectrometry and microarray data. BMC Bioinformatics 7: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Yu J, Li D, Zhang Z, Liu F, Zhou X, Wang T, Ling Y, Su Z. (2010) PMRD: plant microRNA database. Nucleic Acids Res 38: D806–D813 [DOI] [PMC free article] [PubMed] [Google Scholar]