STATIS analysis of metabolic profiles discerns conserved metabolic processes during development and ripening across fruits.
Abstract
Computational analyses of molecular phenotypes traditionally aim at identifying biochemical components that exhibit differential expression under various scenarios (e.g. environmental and internal perturbations) in a single species. High-throughput metabolomics technologies allow the quantification of (relative) metabolite levels across developmental stages in different tissues, organs, and species. Novel methods for analyzing the resulting multiple data tables could reveal preserved dynamics of metabolic processes across species. The problem we address in this study is 2-fold. (1) We derive a single data table, referred to as a compromise, which captures information common to the investigated set of multiple tables containing data on different fruit development and ripening stages in three climacteric (i.e. peach [Prunus persica] and two tomato [Solanum lycopersicum] cultivars, Ailsa Craig and M82) and two nonclimacteric (i.e. strawberry [Fragaria × ananassa] and pepper [Capsicum chilense]) fruits; in addition, we demonstrate the power of the method to discern similarities and differences between multiple tables by analyzing publicly available metabolomics data from three tomato ripening mutants together with two tomato cultivars. (2) We identify the conserved dynamics of metabolic processes, reflected in the data profiles of the corresponding metabolites that contribute most to the determined compromise. Our analysis is based on an extension to principal component analysis, called STATIS, in combination with pathway overenrichment analysis. Based on publicly available metabolic profiles for the investigated species, we demonstrate that STATIS can be used to identify the metabolic processes whose behavior is similarly affected during fruit development and ripening. These findings ultimately provide insights into the pathways that are essential during fruit development and ripening across species.
While the field of transcriptomics has experienced revolution due to the advent of next-generation sequencing technologies, facilitating the identification of differentially expressed genes, alleles, and spliced transcripts (Alba et al., 2005; Vriezen et al., 2008; Bombarely et al., 2010; Matas et al., 2011; Rohrmann et al., 2011; Gordo et al., 2012; Kong et al., 2013), metabolomics allows the quantification of metabolites that are generally conserved across the kingdoms of life. Existing metabolomics technologies are routinely applied to obtain a (nontargeted) snapshot of biological systems operating under given environmental conditions (Schauer et al., 2006; Hanhineva et al., 2008, 2009; Do et al., 2010; Meléndez-Martínez et al., 2010; Moing et al., 2011; Osorio et al., 2011a; Lenucci et al., 2012; Toubiana et al., 2013; Wahyuni et al., 2013) and result in metabolic phenotypes (i.e. multivariate data sets) gathered from the same set of metabolites under various conditions and/or time domains. These metabolic phenotypes can readily be used in comparative analyses of metabolic processes across species to obtain conserved aspects of the cellular physiological status and, as such, may highlight common temporal aspects of biochemical regulation. However, the comparative analysis requires the development of methods for simultaneous analysis of multiple data sets (or data tables). Such methods, in turn, can be employed to determine preserved changes in the dynamics of metabolic processes across species. Thus, in this multispecies setting, the metabolomics-driven analysis goes a step further than the classical computational studies aimed at identifying biochemical components that exhibit differential expression under various scenarios (e.g. environmental and internal perturbations) within a single species.
The goal of our study is to introduce the STATIS approach into the analysis and interpretation of metabolomics data over the same set of metabolites in (not necessarily identical) fruit development and ripening stages of different species. The idea of STATIS is based on the integration of a given set of data tables into an optimum weighted average, called a compromise, which captures what is common to all or a subset of analyzed tables. The compromise is obtained based on principal component analysis (PCA) of a specially constructed matrix. In addition, enrichment analysis based on metabolite compound class and pathway participation can be carried out to facilitate further investigations of the resulting compromise. This approach has already been successfully used in the analysis of transcriptomics time-series data (Klie et al., 2012).
Fruit are generally classified into two physiological groups, climacteric and nonclimacteric, according to the presence or absence of ethylene biosynthesis peaks and respiratory bursts during ripening. Ethylene synthesis in climacteric fruits, such as tomato (Solanum lycopersicum), peach (Prunus persica), apple (Malus domestica), and banana (Musa spp.), is essential for normal fruit ripening; moreover, blocking either the synthesis or perception of this phytohormone prevents ripening (Giovannoni, 2001). Fruits such as strawberry (Fragaria × ananassa), pepper (Capsicum chilense), and grape (Vitis vinifera) have been classified as nonclimacteric, based on the low endogenous production of ethylene compared with standard climacteric fruits and due to their inability to accelerate fruit ripening by the external application of ethylene or ethylene-releasing compounds (Perkins-Veazie, 1995). Metabolomics data from different stages in the development and ripening of both climacteric and nonclimacteric fruits offer an excellent case for investigating which metabolic processes/functions show preserved dynamics and are regulated similarly in the two physiological groups. The main changes associated with ripening include color (i.e. loss of green color and increase in nonphotosynthetic pigments that vary depending on species and cultivar), firmness (i.e. softening by cell wall-degrading activities), taste (e.g. increase in sugar and decline in organic acids), and flavor (i.e. production of volatile compounds providing the characteristic aroma; Carrari and Fernie, 2006; Howard and Wildman, 2007; Fait et al., 2008; Zhang et al., 2011; Wahyuni et al., 2013). These transformations are the result of dynamic processes that involve a complex series of molecular and biochemical changes under genetic regulation and/or in response to environmental perturbations (Giovannoni, 2004; Do et al., 2010; Page et al., 2010; Klee and Giovannoni, 2011; Osorio et al., 2011a, 2011b, 2013a; Pan et al., 2012; Tieman et al., 2012).
To better understand fruit development and ripening mechanisms, numerous studies have focused on measuring transcript and metabolite levels in climacteric fruits, such as tomato (Alba et al., 2005; Carrari et al., 2006; Vriezen et al., 2008; Enfissi et al., 2010; Karlova et al., 2011; Osorio et al., 2011a) and peach (Borsani et al., 2009; Zhang et al., 2010; Lombardo et al., 2011; Li et al., 2012), and in nonclimacteric fruits, such as strawberry (Aharoni and O’Connell, 2002; Aharoni et al., 2002; Fait et al., 2008; Bombarely et al., 2010; Osorio et al., 2011b; Zhang et al., 2011), pepper (Kim et al., 2008; Lee et al., 2010; Osorio et al., 2012; Liu et al., 2013; Wahyuni et al., 2013), and grape (Deluc et al., 2007; Grimplet et al., 2007). These studies have been complemented by the investigation of transcriptomics, proteomics, and metabolomics data in the three dominant ripening mutants of tomato, ripening-inhibitor (rin), nonripening (nor), and never-ripe (Nr), along the developmental and ripening periods. While integration of genomics, transcriptomics, and metabolomics data during fruit development and ripening can give important insight into gene regulatory and metabolic events associated with these processes in a single species (Carrari et al., 2006; Grimplet et al., 2007; Enfissi et al., 2010; Zamboni et al., 2010; Osorio et al., 2011a, 2011b, 2012; Rohrmann et al., 2011), identifying the metabolic functions that are similarly regulated across different species has not yet been attempted.
RESULTS
Contribution of Species in STATIS Reveals the Influence of Physiologically Distinct Fruit Groups and Developmental Stages on the Compromise Space
Similar to the classical PCA, the analysis based on STATIS allows a description of the contributions of tables, observations, and variables. However, the advantage of STATIS is that it allows simultaneous investigation of multiple tables, which is not readily achievable by PCA. Here, the K tables contain the metabolic profiles from K = 6 organs (four from the following: two tomato cultivars, peach, and pepper; as well as strawberry fruit, which was separated into two organs: achenes, representing the true fruit, and receptacles), from which metabolomics data during fruit development and ripening were available, as detailed in “Materials and Methods.” The variables denote the 16 metabolites measured across species, while fruit development and ripening stages correspond to observations in the data tables.
Analysis of the contributions of the individual data tables to the compromise, S, then renders it possible to determine the species that has the strongest influence on the determined principal components and that, furthermore, reflects good overall similarity to the data tables from the remaining species. We note that the employed data tables (i.e. sampling time points of a given species; Fig. 1) differ in the coverage of the fruit development and ripening stages as well as in the number of tables from the considered species. Therefore, we first investigated not only the extent to which each data table contributes to the compromise but also the stability of the derived compromise upon the exclusion of a data table (i.e. sampling time point).
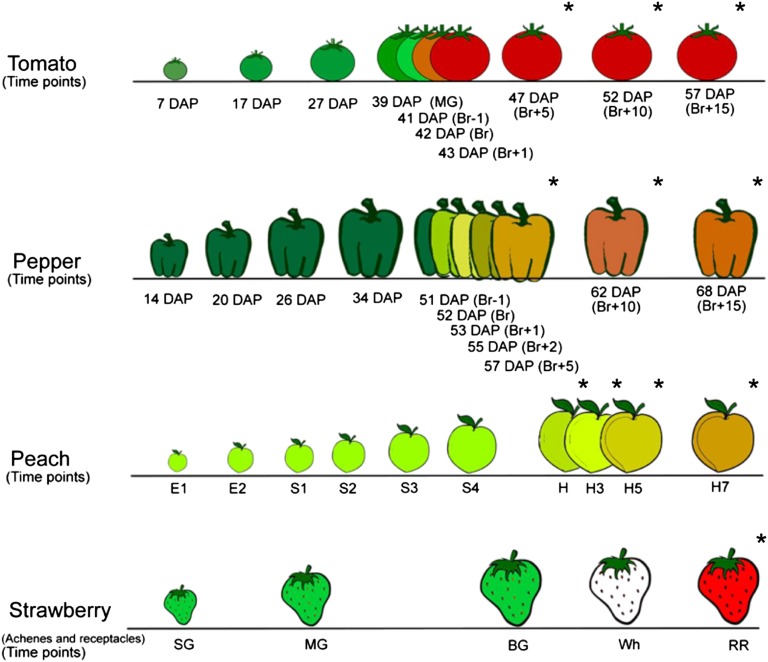
Figure 1.
Overview of the developmental stages used for the four species in the STATIS analysis. The STATIS analysis considers the following: tomato (two cultivars, M82 and Ailsa Craig), pepper, peach, and strawberry. The illustration details the stages at which the publicly available metabolomics profiles were obtained. Time points marked by stars are referred to as later ripening stages. Time points without stars denote the remaining stages. For tomato and pepper, Br = breaker; for strawberry, SG = small green, MG = medium green, BG = big green, Wh = white, and RR = red; and for peach, E1 (3, 5, 9 DAB), E2 = 17 DAB, S1 = 23, 29, and 37 DAB, S2 = 44, 51, 59 DAB, S3 = 74, 80, 83 and 87 DAB, S4 = 94 and 102 DAB; and Fruits were collected at 102 DAB (H) and kept in a chamber at 20°C and 90% relative humidity for 3 (H3), 5 (H5), and 7 (H7) days.
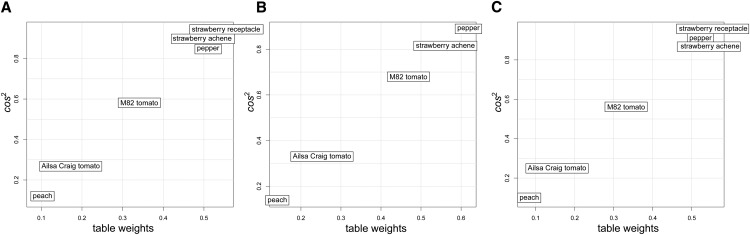
The contribution of a given table to the compromise is quantified by the obtained table weights αi, 1 ≤ i ≤ 6, while the similarity of each data table to the compromise is quantified by the corresponding (squared) Rv coefficients. By plotting each data table in the two-dimensional space determined by the table weights (x axis) and Rv2 coefficients (y axis), as illustrated in Figure 2A, it becomes apparent that for the nonclimacteric fruits, the table weights (and the Rv2 coefficients) are concentrated in the top right corner, while for climacteric fruits, these quantities are spread out. Moreover, we observe that the contribution of a data table to the compromise does not correspond to the number of stages (time points) included in each table, since the data table from pepper has 10 time points and the two data tables from strawberry have only five time points each, although all data are grouped together in Figure 2A.
Figure 2.
Analysis of the influence of data tables considered in STATIS. A, Typological values obtained for all six data tables used in STATIS. The x axis denotes the table weights, while the y axis illustrates the Rv2 coefficient. B, Leaving out the data set of the strawberry receptacle does not have a large influence on the typological values for the remaining data tables. This is supported by the order of the tables based on their weights (shown on the x axis). C, Likewise, after the removal of later ripening stages, all data sets have similar contributions to the compromise.
Since, in this analysis, we consider two data sets from strawberry, from the achene and the receptacle, we next ask if the obtained compromise is biased with respect to strawberry. To this end, we first determined the contribution of each table to the compromise obtained by leaving out the data table from strawberry receptacle and then compared it with the compromise obtained from all tables. As shown in Figure 2B, the new compromise remained largely unaffected. This analysis prompted us to investigate the stability of the compromise from all leave-one-out scenarios, whereby we determined the compromise with each of the tables omitted and quantified its similarity to the compromise with all tables included by using the Mantel correlation. Interestingly, the removal of each data table results in very similar compromise spaces, supported by the statistically significant Mantel correlation of high magnitude (Table I). This finding provides further evidence for the hypothesis of the preservation of metabolic patterns in fruit development and ripening irrespective of the subgroups of species used in the analysis per se.
Table I. Stability of the compromise from six data tables.
Values were obtained based on the Mantel correlation between the compromises projected on PC1 and PC2, obtained by removing either one or none (first column) of the six data sets. The bottom, left triangular matrix displays the P values, and the top, right triangular matrix displays the values of Mantel correlations.
| All Species | Tomato cv Ailsa Craig Removed | Tomato cv M82 Removed | Peach Removed | Pepper Removed | Strawberry Achene Removed | Strawberry Receptacle Removed | |
|---|---|---|---|---|---|---|---|
| All species | 0.999 | 0.932 | 1 | 0.954 | 0.936 | 0.956 | |
| Tomato cv Ailsa Craig removed | <0.001 | 0.93 | 1 | 0.951 | 0.935 | 0.952 | |
| Tomato cv M82 removed | <0.001 | <0.001 | 0.932 | 0.925 | 0.876 | 0.891 | |
| Peach removed | <0.001 | <0.001 | <0.001 | 0.952 | 0.936 | 0.954 | |
| Pepper removed | <0.001 | <0.001 | <0.001 | <0.001 | 0.911 | 0.936 | |
| Strawberry achene removed | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.979 | |
| Strawberry receptacle removed | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
This conclusion may appear counterintuitive given the observed difference between the contributions of the data tables from climacteric and nonclimacteric fruits to the compromise. Since the considered data tables partly differ in the included types of stages (i.e. development and ripening), we next investigated the effect of the removal of later ripening stages on the resulting compromise. To this end, we removed the time points corresponding to the later ripening stages (marked with stars in Fig. 1) in all six data sets. Strikingly, all four species still have similar contributions to the determined compromise, as seen in Figure 2C. This unexpected finding illustrates that metabolic profiles differ between the three climacteric fruits and two nonclimacteric fruits across the entirety of the developmental time course and not only during the ripening stages (with an increase of ethylene).
To further confirm the robustness of this result, we next considered applying STATIS on data tables from which earlier time points (i.e. those corresponding to the developmental stages not marked with stars in Fig. 1) were omitted. The compromise was largely unaffected, as supported by the Mantel correlation of 0.984, P = 0.0009, to the compromise of the unaltered data tables. Moreover, this was in line with the very small observed changes in the typological values for the modified data tables (compare Fig. 2 with Supplemental Fig. S1).
Altogether, this analysis confirms the stability of the derived compromise and points to some inherent differences between climacteric and nonclimacteric fruits based on their corresponding metabolic profiles. Therefore, in the following, the analysis will focus on the first scenario, illustrated in Figure 2A, including all species and all available time points. In this case, the data tables of highest weight correspond to nonclimacteric fruits (i.e. strawberry and pepper), followed by the climacteric fruits, including peach and the two tomato genotypes. The compromise space includes the effects of both climacteric and nonclimacteric fruits and thus could reveal coordinated key metabolic adjustments in fruit development and ripening across all investigated species.
Interstructure of the Data Tables Separates Climacteric from Nonclimacteric Fruits
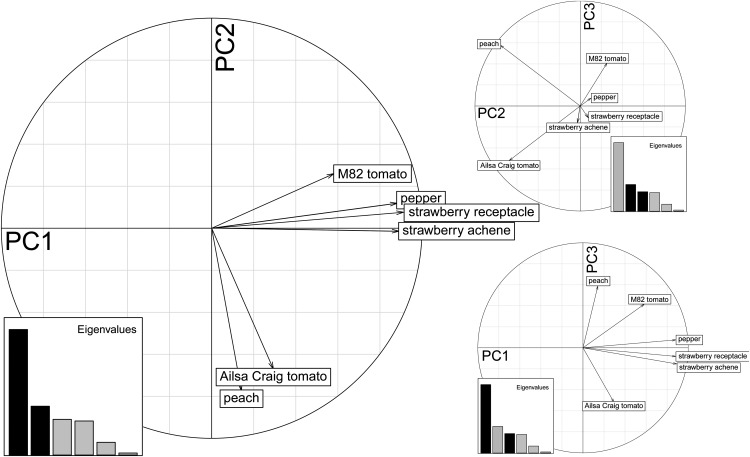
The effect of different organs/species on the compromise is further supported by investigating the contribution of the six data tables to the first, second, and third principal components (PC1–PC3; Fig. 3), also known as the table interstructure. The two data tables of strawberry and pepper have the largest contribution to PC1 while exhibiting a very minor contribution to PC2. In contrast, the data table of tomato cv Ailsa Craig together with the peach data set displays a strong contribution to PC2 with a weak contribution to PC1. The M82 tomato variety exhibits a contribution to PC2 of a different sign opposite to those from peach and tomato cv Ailsa Craig. Finally, PC3 separates peach and tomato cv Ailsa Craig.
Figure 3.
Interstructure of data tables. The visualization of the interstructure obtained via STATIS by analyzing the contribution of all six data tables onto PC1 and PC2 (left), PC2 and PC3 (right, top), and PC1 and PC2 (right, bottom) is shown. The contribution of each principal component to the variance explained is illustrated by the magnitude of the bars in the insets. The principal components used in the analysis of the contribution of all six tables are highlighted in black. The projection of the vector associated with each data table on the respective principal components indicates the (sign of) the contribution to the corresponding principal component.
The observed differences in cv M82 and cv Ailsa Craig tomato fruits in the contribution to PC1 and PC2 are likely due to the fact that they are quite distinctive cultivars. Both prominent genotypic and phenotypic differences have already been reported between these cultivars (Shirasawa et al., 2010; Kusano et al., 2011; Matas et al., 2011), which is perhaps not surprising since cv M82 is a processing or ketchup cultivar while cv Ailsa Craig has been bred as a salad variety. Further differences between these tomato cultivars are highlighted by investigating their more extensive data tables (with 23 metabolites) and relations to three tomato ripening mutants (see below).
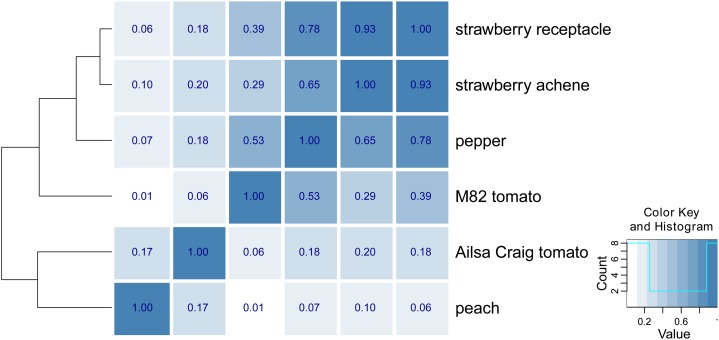
The STATIS analysis depends on determining the cross-product matrix of RV coefficients capturing the similarity between the data tables. As illustrated in Figure 4, we observed a clear metabolic distinction between climacteric and nonclimacteric fruits. Both data tables of strawberry exhibit greatest similarity with that of pepper, as all of them belong to the nonclimacteric fruit group. Within the climacteric fruits, similarities between peach and cv Ailsa Craig tomato fruits are the strongest. Perhaps surprisingly, although as seen previously by PCA, cv M82 tomato fruits exhibit stronger similarity on the metabolic level to the nonclimacteric species than to the cv Ailsa Craig tomato fruits (see below).
Figure 4.
Heat map illustrating the similarity of the metabolic patterns of the six data tables analyzed. The heat map illustrates the Rv coefficient for each pair of data tables (shown in the right-most column). A value of 1 for the Rv coefficient corresponds to equivalence (dark blue), while a value of 0 indicates complete dissimilarity (white; see key at right for the distribution of values). The separation between the climacteric (i.e. tomato and peach) and nonclimacteric (i.e. strawberry and pepper) fruits is already captured in this matrix of Rv coefficients, although, surprisingly, cv M82 indicates higher similarity to nonclimacteric fruits. [See online article for color version of this figure.]
Investigation of Metabolites by Inferential STATIS Reveals Housekeeping Metabolic Functions during Fruit Development and Ripening
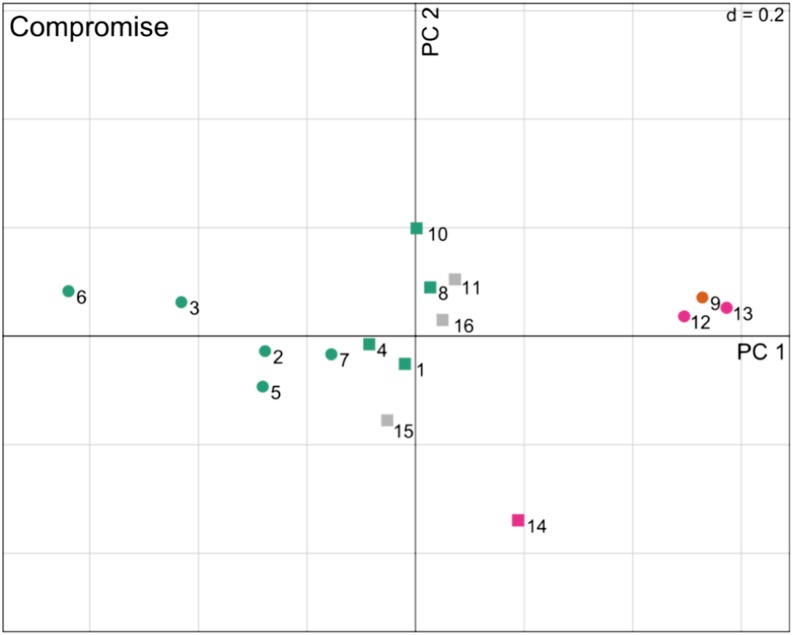
The characterization of variables is a pivotal step of any descriptive statistical method. We note that in our setting, the variables correspond to the 16 metabolites quantified across all developmental stages in all six of the investigated organs. To this end, STATIS allows not only the investigation of the contribution of tables (i.e. species) but also an analysis of the contributions of considered variables to the principal components. In particular, for the purpose of visualization, the contributions of individual variables, columns in matrix X, are obtained by projection onto the principal components of the compromise, as depicted in Figure 5.
Figure 5.
Visualization of the contribution of variables (i.e. metabolites) according to STATIS. Shown are the projections of the 16 analyzed metabolites on PC1 and PC2. Metabolites contributing significantly to either of the two components are denoted by circles, while the remaining metabolites are indicated as squares. The color code corresponds to the compound class of each metabolite as follows: sugars (magenta), amino acids (blue), organic acids (orange), and others (gray). The compounds are identified by the corresponding numbers given in Table II. [See online article for color version of this figure.]
To determine which metabolic functions are associated with the adjustments of metabolic content, a compound class/pathway enrichment analysis of ontology terms is typically performed (see “Materials and Methods”). However, an important intermediate step is to identify the variables (i.e. metabolites) that contribute significantly to either principal component. Here, bootstrap ratios (see “Materials and Methods”) were used to identify significant contributions at a significance level of α = 0.05. The metabolites found to significantly contribute to PC1, PC2, or PC3, illustrated as circles in Figure 5 and detailed in Table II, were next subjected to an enrichment analysis. Table III provides an overview of the pathways found to be overrepresented at significance levels of 0.01 and 0.05. We note that the very small “universe” (i.e. 16 metabolites) warrants caution when interpreting these values. On inspection of Table III, it becomes apparent that the metabolites whose contribution to PC1 is significant are enriched for the pathway of volatile organic compound biosynthesis as well as that of branched chain amino acid biosynthesis. More precisely, we find that (1) amino acids, such as Ile, Phe, Thr, Tyr, and Val, (2) sugars, such as Fru and Glc, as well as (3) citric acid are strongly contributing to PC1. On the other hand, PC2 is characterized by Glu, malate, and Suc. Finally, we observe that Asp strongly, albeit not significantly, contributes to PC3 (Supplemental Fig. S2), hinting at a difference of the two tomato varieties with respect to this metabolite.
Table II. Bootstrap ratios of metabolites (absolute values) for the six organs/species.
The bootstrap ratios of metabolites were obtained from 3,000 bootstrap samples for the first three principal components. Values denoted in boldface are significant at α = 0.01, while underlined values are significant at α = 0.05.
| No. | Metabolite | PC1 | PC2 | PC3 |
|---|---|---|---|---|
| 1 | Ala | 0.15 | 1.30 | 0.97 |
| 2 | Ile | 3.40 | 0.17 | 0.67 |
| 3 | Phe | 3.00 | 0.42 | 0.29 |
| 4 | Ser | 2.30 | 0.36 | 0.90 |
| 5 | Thr | 3.00 | 0.45 | 0.02 |
| 6 | Tyr | 3.10 | 0.28 | 0.79 |
| 7 | Val | 4.00 | 0.21 | 0.77 |
| 8 | Asp | 0.40 | 0.84 | 0.85 |
| 9 | Citric acid | 2.40 | 0.36 | 1.00 |
| 10 | Glu | 0.17 | 1.20 | 0.60 |
| 11 | Malic acid | 1.20 | 0.66 | 0.78 |
| 12 | Fru | 2.70 | 0.25 | 1.00 |
| 13 | Glc | 2.90 | 0.22 | 0.62 |
| 14 | Suc | 0.89 | 2.10 | 0.82 |
| 15 | Myoinositol | 0.40 | 1.10 | 0.92 |
| 16 | Phosphoric acid | 0.73 | 0.21 | 0.02 |
Table III. Pathway enrichment analysis for PC1.
Enriched pathways were determined for the significantly contributing metabolites to the respective principal components (compare with Fig. 3 and Table II). Note that PC2 and PC3 did not result in the enrichment of pathways. Values in boldface are significant at α = 0.01, while underlined values are significant at α = 0.05.
| Pathway Enrichment | P |
|---|---|
| Volatile organic compound biosynthesis | 9.62E-03 |
| Val, Leu, and Ile biosynthesis | 3.13E-02 |
| Phe metabolism | 8.75E-02 |
| Phe, Tyr, and Trp biosynthesis | 8.75E-02 |
| Phenylpropanoid biosynthesis | 8.75E-02 |
| Val, Leu, and Ile degradation | 8.75E-02 |
Climacteric ethylene biosynthesis includes the conversion of Asp to Met, the conversion of Met to ethylene, and the Met recycling pathway (Yang and Hoffman, 1984). Met and Phe as well as other amino acids, such as Ala, Leu, Ile, and Val, also have the potential to participate in volatile organic compound biosynthesis (a group of plant secondary metabolites; Maloney et al., 2010; Osorio et al., 2010; Dal Cin et al., 2011; El Hadi et al., 2013).
Based on our results, in nonclimacteric fruits, it is apparent that these precursor metabolites have an important role in the regulation of the shift in the flux to volatile organic compound biosynthesis concomitant with an increase of ethylene during climacteric fruit ripening. Malate and Asp have high bootstrap values for the contributions to PC2 and PC3 that separate the climacteric fruit group (cv M82, cv Ailsa Craig, and peach). Interestingly, malate, the precursor of Asp, has been described as an important metabolite for ethylene feedback regulation during tomato ripening (Osorio et al., 2011a) and is also an important contributor to cellular redox balance, which in turn defines highly important aspects of plastidial metabolism (Centeno et al., 2011; Osorio et al., 2013b).
Other metabolites that also have high bootstrap values for the contributions to PC2 and PC3 that separate the climacteric from nonclimacteric fruits include Suc, one of the major sugars in fruits, and myoinositol, which both relate to malate. Malate plays an important role in starch accumulation and in the total soluble sugar levels (Glc, Fru, and Suc) in developing fruits (Centeno et al., 2011; Osorio et al., 2013b). Phosphoenolpyruvate, a closely linked metabolite to malate, is transformed via the gluconeogenic pathway to sugar phosphates, which can subsequently be converted to starch (Lara et al., 2004). Evidence from radiolabel feedings (Halinska and Frenkel, 1991; Beriashvili and Beriashvili, 1996; Osorio et al., 2013b) suggests that gluconeogenesis does occur in grape and tomato fruits, particularly during ripening stages, when sugars are accumulating rapidly. Therefore, the combined data reveal that malate metabolism may have an effect on fruit ripening and strongly influences programs of fruit development. Interestingly, the oxidation of myoinositol provides an alternative starting point to a pathway furnishing uronosyl and pentosyl residues for cell wall biogenesis (Loewus, 2006), which may indicate that the flux through this pathway in climacteric and nonclimacteric fruits is differentially regulated during development and ripening. The results of this meta-analysis indicate that, while many metabolites display common dynamics across fruit development among the species tested, several divergent mechanisms likely underlie the overall dynamics of metabolism during development.
Analysis of Stages Suggests the Presence of Diverse Temporal Patterns in Fruit Development and Ripening
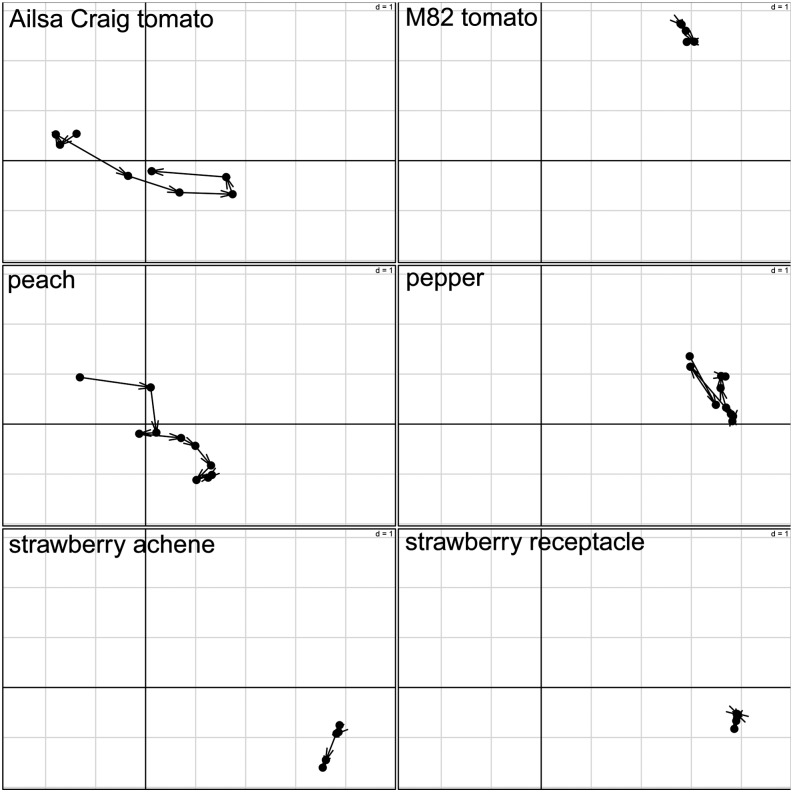
The analysis of the contributions of observations reflects the influence of the particular stage with respect to the overall metabolic adjustments of the investigated species during fruit development and ripening. Figure 6 shows the contributions of each stage per species on PC1 (x axis) and PC2 (y axis). Arrows between two successive stages illustrate the sequential progression of the contributions over time with the compromise space defined by PC1 and PC2. While all the trajectories show substantial contributions to either PC1 or PC2, clearly illustrating the ongoing transformations in the content of the levels of the investigated metabolites, of particular interest are the observations regarding the differences between and within climacteric and nonclimacteric fruits.
Figure 6.
Visualization of the contribution of observations (i.e. developmental and ripening stages) according to STATIS. The corresponding time stages are projected onto PC1 and PC2 for all six data tables. The position of a time point reflects the influence of the particular stage with respect to the overall metabolic adjustments of the investigated species during fruit development and ripening. Arrows between two successive stages illustrate the sequential progression of the contribution over time with the compromise space defined by PC1 and PC2. Although tomato cv M82 and peach are climacteric fruits, they exhibit different behavior with respect to the compromise, while peach and pepper exhibit similar behavior even though they belong to different categories of fruit.
Although tomato cv M82 and peach are climacteric fruits, they exhibit different behavior with respect to the compromise. The metabolic phenotypes of these species not only show opposite contributions to PC2, but also the progression of contributions to PC1 follows different directions, especially in the later stages, as illustrated in Figure 6. Moreover, peach exhibits a similar contribution of its stages of development and ripening to that of pepper with respect to PC2, although its contributions to PC1 are of a different sign in comparison with those of pepper (Fig. 6). In contrast, strawberry (achene and receptacle), as a nonclimacteric fruit, demonstrates an almost completely stable contribution of the different stages to the compromise, a pattern distinct in comparison with the other investigated species.
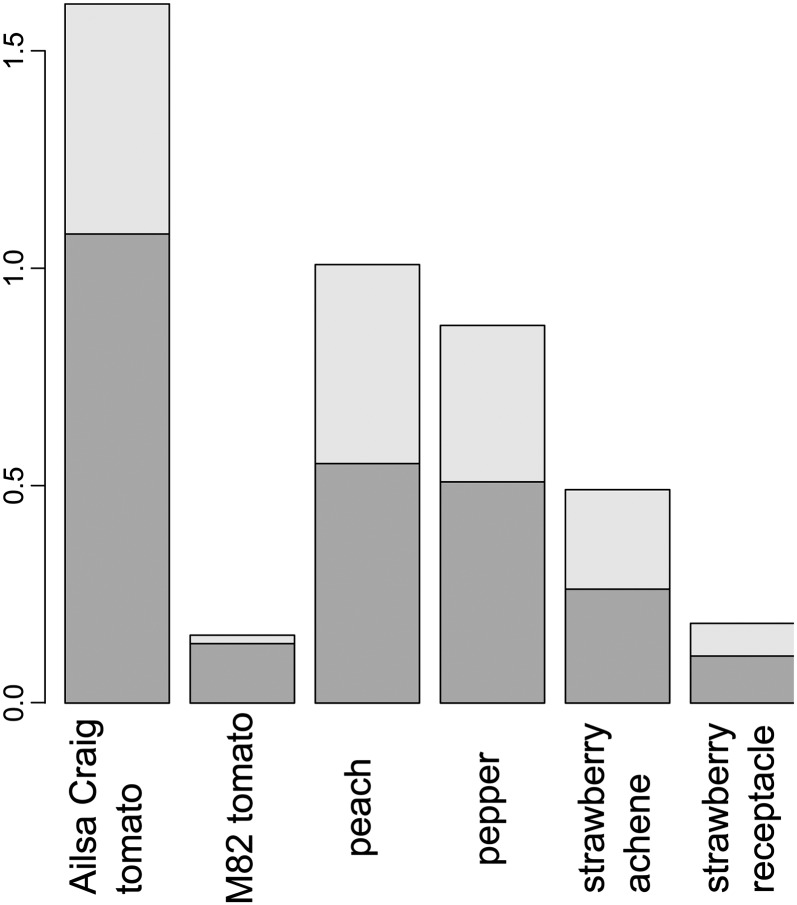
Finally, we investigated the distances between the consecutive time points on the resulting trajectories. Irrespective of the number of time points used, we observed that nonclimacteric fruit undergo less dramatic changes across development in comparison with climacteric fruits (Fig. 7). Moreover, all six species exhibited more severe changes during fruit development (Fig. 7, dark gray bars; corresponding to time points without stars in Fig. 1) in comparison with the changes during ripening (Fig. 7, light gray bars).
Figure 7.
Temporal progression of fruit developmental stages. A stacked bar plot illustrates the sum of distances of all consecutive time points in the compromise projection for all six data tables normalized for the number of time points available for each data set (compare with Fig. 6). Later ripening stages (indicated by stars in Fig. 1) are colored in lighter gray. Nonclimacteric fruit undergo less dramatic changes across development in comparison with climacteric fruits, with cv M82 again showing pronounced divergence from this pattern.
STATIS with Data Tables from Tomato Cultivars and Ripening Mutants
The previous findings from the six data tables were based on 16 metabolites measured across the different organs/species, which may be seen as a drawback in relating the computational findings to similarities of the underlying biochemical pathways. To demonstrate the power and robustness of the method, and to address the questions arising from the detected differences between the two tomato cultivars, we next focused on the analysis of data tables from the two tomato cultivars and three tomato ripening mutants, namely nor, Nr, and rin. Altogether, 23 metabolites were measured across these cultivars/mutants (Table IV) over eight (for cv Ailsa Craig, nor, and rin), five (cv M82), and four (Nr) time points (Supplemental Fig. S3).
Table IV. Bootstrap ratios of metabolites (absolute values) from the two tomato cultivars and the three ripening mutants.
The bootstrap ratios of metabolites were obtained from 3,000 bootstrap samples for the first three principal components. Values denoted in boldface are significant at α = 0.01, while underlined values are significant at α = 0.05.
| No. | Metabolite | PC1 | PC2 | PC3 |
|---|---|---|---|---|
| 1 | Ala | 1.66 | 1.00 | 0.66 |
| 2 | β-Ala | 0.95 | −1.28 | −0.58 |
| 3 | γ-Aminobutyrate | 0.25 | −0.54 | −1.95 |
| 4 | Gly | 0.40 | −2.33 | −0.50 |
| 5 | Ile | −1.54 | −2.23 | −0.24 |
| 6 | Phe | −1.35 | 0.18 | −1.56 |
| 7 | Ser | −1.81 | −0.81 | −0.80 |
| 8 | Thr | −1.46 | −0.75 | 0.15 |
| 9 | Val | −0.57 | −2.01 | −0.39 |
| 10 | Asp | −1.90 | 1.26 | 0.11 |
| 11 | Citric acid | 1.57 | 0.12 | −0.35 |
| 12 | Dehydroascorbic acid | 1.69 | 0.08 | 0.65 |
| 13 | Glu | −0.84 | −0.20 | 0.60 |
| 14 | Malic acid | 2.13 | 1.34 | 0.50 |
| 15 | Threonic acid | 1.31 | −1.01 | 1.33 |
| 16 | Fru | 1.62 | 0.22 | −0.53 |
| 17 | Glc | 1.00 | 1.42 | −1.28 |
| 18 | Rha | 1.38 | 0.40 | −0.31 |
| 19 | Suc | −0.32 | −2.12 | −0.57 |
| 20 | Xyl | 2.51 | 0.05 | 1.01 |
| 21 | Myoinositol | 1.12 | 0.07 | −0.90 |
| 22 | Hexadecanoic acid | 0.85 | 3.23 | −0.46 |
| 23 | Octadecanoic acid | 1.89 | −0.11 | 0.49 |
STATIS analysis of the five data tables revealed that the typological values for the nor and rin mutants were the highest and most similar to the obtained compromise (Supplemental Fig. S4A). This might be attributed to the fact that they are both in the cv Ailsa Craig background and that both are impaired in ripening. Interestingly, the typological value of cv Ailsa Craig is of intermediate magnitude; nevertheless, it is closer to those of the nor and rin mutants (with this variety as a background) and farthest from that of cv M82. Despite the common genomic background, the dissimilarity to nor and rin mutants could partly be explained by the impaired ripening in these mutants in comparison with cv Ailsa Craig. The Nr mutant is the most similar to cv M82. Nevertheless, we note that the data tables for Nr and cv M82, in comparison with the other data sets analyzed in this section, contained the smallest number of time points, excluding the early fruit development (i.e. starting with 42 and 39 d after pollination [DAP] for cv M82 and Nr, respectively).
With respect to the interstructure, shown in Supplemental Figure S4, B to D, PC1 and PC2 allow separation of the nor and rin mutants as well as cv Ailsa Craig (with intermediate contribution to PC1) from cv M82 and the Nr mutant (small contribution to PC1 and strong contribution to PC2), closely resembling the previously discussed table weights (i.e. typological values). Moreover, PC3 separates the Nr mutant from cv M82. Hence, the three principal components can distinguish between the data tables of the cultivars from those of the ripening mutants.
Furthermore, considering the bootstrap values of the contribution of metabolites to the compromise (PC1 and PC2; Supplemental Fig. S4), we identified that malate, previously discussed in the context of fruit ripening, while not exhibiting a very pronounced contribution to either PC1 or PC2, exhibited the second highest bootstrapped value for PC1 (Table IV). Moreover, Xyl, a cell wall-related compound, was the metabolite with the highest bootstrap support to PC1. This could be related to a recent finding that this abundant sugar, which is important in the formation of plant cell wall polymers, might be affected by down-regulation of cell wall-related gene expression in the ripening mutants (Gilbert et al., 2009; Osorio et al., 2011a). γ-Aminobutyrate was found to have the most pronounced bootstrapped value for PC3, which might be attributed to the pathways separating the Nr mutant from tomato cv M82. Finally, three amino acids of the Asp family, namely Ser, Thr, and Asp, exhibited strong negative contributions to PC1. This pathway has been implicated in the distribution of the carbon from Asp into the branches for the synthesis of Lys, Thr, Met, and Ile. It has already been reported that Asp metabolism in Arabidopsis (Arabidopsis thaliana) is tightly regulated; furthermore, Thr, the most sensitive variable in the system, has previously been suggested to have a regulatory role within the network of amino acid metabolism (Curien et al., 2009).
DISCUSSION AND CONCLUSION
The increased availability of high-throughput technologies (e.g. transcriptomics, proteomics, and metabolomics) facilitates comparative analyses across species with a focus on determining divergent behavior as well as conserved patterns in molecular entities at different levels of functional organization (Moore et al., 2005; Mutwil et al., 2011; Hampp et al., 2012; Lee et al., 2012; Osorio et al., 2012). Unlike transcripts, metabolites are not species specific. Therefore, data profiles from metabolomics technologies are particularly suitable for identifying biochemical pathways whose effects on an investigated biological function/process are conserved across species. Determining similarities in the dynamic fluctuations of biochemical pathways on the basis of a relatively small number of annotated metabolites (in comparison with full transcriptome coverage with next-generation sequencing technologies) represents a promising read out that may well reveal further similarities in the flow of biological information “upstream” of metabolism.
Here, we present an attempt to determine similarities in publicly available metabolic phenotypes, characterized by the collection of metabolic profiles, of six organs (five fruits and one receptacle) from four species, namely tomato (cv M82 and Ailsa Craig as well as three ripening mutants), peach, pepper, and strawberry (achene and receptacle). Our analysis is based on STATIS, an extension of classical PCA, which allows combining and simultaneously investigating multiple data tables. The crux of the approach is the concept of the compromise space, which not only captures the congruence between the investigated tables but also, as in PCA, facilitates quantification of the contribution of metabolites as well as developmental and ripening stages.
As we illustrate in this study, the findings from STATIS can readily be coupled with enrichment analysis to posit and test hypotheses based on the established biochemical knowledge gathered in pathway databases. Interestingly, although only 16 metabolites were unequivocally measured across the investigated organs from climacteric and nonclimacteric species, STATIS highlights the difference between climacteric and nonclimacteric fruits in the contributions of their temporally changing metabolic phenotypes to the compromise. Moreover, the stability of the compromise upon removal of any of the employed data tables suggests that there is a robust pattern of similarity, which merits further investigation.
The availability of well-characterized mutants in fruit development and ripening allowed us to investigate the metabolic differences during ripening. Here, we included the analysis of metabolic profiles from two mutants in the ripening-associated transcription factors, rin, which encoded a SEPALLATA MADS box gene (Vrebalov et al., 2002), and nor (GenBank accession no. AY573802; Tigchelaar et al., 1973), and a two-component His kinase ethylene receptor, ETR3 (Nr; Wilkinson et al., 1995; Yen et al., 1995). Growing evidence has indicated that nor and rin are necessary for ethylene biosynthesis and that both act upstream of Nr (Giovannoni et al., 1995; Osorio et al., 2011a). Moreover, the nor and rin mutants are phenotypically similar in that both fruits fail to produce climacteric ethylene and to complete normal ripening. In this context, STATIS revealed that some of the metabolites important for discriminating climacteric from nonclimacteric fruits, such as malate and three amino acids of the Asp family (i.e. Ser, Thr, and Asp), were also important for distinguishing the two tomato mutants (nor and rin) from the third ripening mutant, Nr, and cv M82. Therefore, these findings reinforce the importance of these precursor metabolites in the regulation of the shift in the flux to volatile organic compound biosynthesis concomitant with an increase during climacteric fruit ripening.
Degradation of plant cell wall polymers is another process directly associated with fruit ripening; therefore, the content of some cell wall-related metabolites, like Xyl, increases during this process (Rose and Bennett, 1999; Osorio et al., 2011a). Interestingly, we found that Xyl has a high bootstrap value for the contribution to PC1 that separates the nor and rin mutants from the third studied ripening mutant (Nr) and cv M82. This result could be explained by the evidence that the cell wall degradation-related genes are up-regulated during normal tomato ripening but not in equivalent-stage nor and rin fruits (Osorio et al., 2011a).
Similar to the conclusions drawn from PCA, one may conclude that the metabolites (i.e. variables) farthest from the origin in the compromise space are of highest importance. In such a setting, importance implies strong variation. More specifically, this would imply strong variation in the covariance patterns between data tables, since STATIS is based on (normalized) covariances of variables. Nevertheless, it is often the case that these variables do not have the highest bootstrap values: a metabolite with a strong (co)variance pattern that is conserved across the data tables may obtain a lower bootstrap value than a metabolite whose (co)variance pattern may be strongly affected by the removal of a particular data table, although its pattern may not be preserved. Therefore, the distance from the origin in the compromise should be understood only as a heuristic for judging the importance of a variable, suggesting that both forms of evidence should be taken into account when performing the data interpretation.
Our findings regarding the contributions of individual metabolites to the principal components of the compromise space in combination with the principal component-based separation of the species indicate that the climacteric and nonclimacteric fruits most significantly differ with respect to the metabolism of some sugars and amino acids. Moreover, our analysis indicates that climacteric and nonclimacteric fruits exhibit distinct patterns not only in ripening but also in the developmental stages (although more pronounced changes are observed in the ripening stages).
Most of these data sets come from different experimental setups followed by measurement of different metabolic profiles over varying fruit developmental and ripening stages obtained, nevertheless, with the same measuring technology. At present, obtaining larger coverage of metabolism for cross-species studies is severely hampered by the aforementioned heterogeneities in the publicly available data sets and would require experiments undertaken with the idea of simultaneous analysis across multiple fruits. Like other covariance-driven analyses, we note that our findings are contingent on the metabolites considered in the analysis.
Future efforts aimed at identifying preserved metabolic processes across fruits from different species would have to include careful experimental design with possibly aligned developmental and ripening states to further test the conclusions from our study. Recent technological advances mean that in excess of 1,000 metabolites can now be readily detected in biological samples (Giavalisco et al., 2011; Rogachev and Aharoni, 2012). Therefore, such an undertaking could additionally be anticipated to cover a larger portion of plant metabolism, including both central and specialized metabolisms (Higashi and Saito, 2013). Finally, analytes, which can be measured but are not yet annotated, can readily be included in the STATIS analysis. In combination with enrichment analysis, such comparative analyses may hint at the putative roles of nonannotated metabolites in preserved metabolic functions and may help in prioritizing which nonannotated metabolites would be most valuable to place into highly labor-intensive structural identification pipelines (Nakabayashi et al., 2009).
MATERIALS AND METHODS
Metabolic Data
Our study employs recently obtained metabolic phenotypes from two tomato (Solanum lycopersicum) cultivars, M82 and Ailsa Craig, as well as peach (Prunus persica ‘Dixiland’), as climacteric fruits, and pepper (Capsicum chilense ‘Habanero’) and strawberry (Fragaria × ananassa ‘Herut’), as nonclimacteric fruits. All metabolite data sets have been published previously. For ease of reference, the sampling strategies used for each species are given in brief here (Fig. 1). For tomato cv Ailsa Craig, the analyzed time points were 7, 17, 27, 39 (mature green), 41 (breaker−1), 42 (breaker), 43 (breaker+1), 47 (breaker+5), 52 (breaker+10), and 57 (breaker+15) DAP; while for tomato cv M82, the time points were 39 (mature green), 42 (breaker), 43 (breaker+1), 47 (breaker+5), and 52 (breaker+10) DAP. To illustrate the power and robustness of the employed method, we also used metabolic phenotypes from three tomato ripening mutants (for illustration of the sampling strategy, see Supplemental Fig. S3). For the nor and rin mutants (both in the cv Ailsa Craig background), the analyzed time points were 27, 39, 41, 42, 43, 47, 52, and 57 DAP, while for the Nr mutant, they were 42, 47, 52, and 57 DAP. Tomato data used are those published by Osorio et al. (2011a). The peach samples included early fruit developmental stages at 3, 5, 9 (for E1 samples) and 17 (for E2 samples) DAB (days after bloom); four developmental stages at 23, 29, and 37 DAB (for S1), 44, 51, 59, and 66 DAB (for S2), 74, 80, 83, and 87 DAB (for S3), 94 and 102 DAB (for S4); and four ripening stages (H, H3, H5, and H7). For these stages, fruits were collected at 102 DAB (H) and kept in a chamber at 20°C and 90% relative humidity for 3 (H3), 5 (H5), and 7 (H7) days. Peach data used are those published by Lombardo et al. (2011). For pepper, fruits were tagged at 14 DAP and harvested at one of the following 11 time points: 14, 20, 26, 34, 51 (breaker−1), 52 (breaker), 53 (breaker+1), 55 (breaker+2), 57 (breaker+5), 62 (breaker+10), and 68 (breaker+16) DAP. Pepper data used are those published by Osorio et al. (2012). Strawberry fruits were harvested at five different stages: small green, medium green, big green, white, and red. Then, for achenes and receptacle analysis, achenes were carefully removed from the corresponding receptacles with a scalpel tip from frozen fruit. Strawberry data used are those published by Fait et al. (2008).
Since the subsequent analysis requires simultaneous consideration of the same set of metabolites (i.e. variables), we focused only on the 16 metabolites, which were quantified across all considered species, given in the rows of Table I. The three tomato ripening mutants were not considered in this analysis in order to have a balanced representation of only the wild types from the two types of fruit. From these 16 metabolites, 13 were categorized to belong in one of the following three groups: sugars, amino acids, and organic acids; the remaining three metabolites could not be assigned a compound class based on BRITE hierarchies. In addition, the set of metabolites that were quantified across the tomato cultivars and mutants comprised 23 metabolites, as given in the rows of Table IV. The same compound class categorization was also applied to this richer data set, which was subsequently used to analyze similarity and differences only for the tomato varieties and mutants.
Overview of STATIS
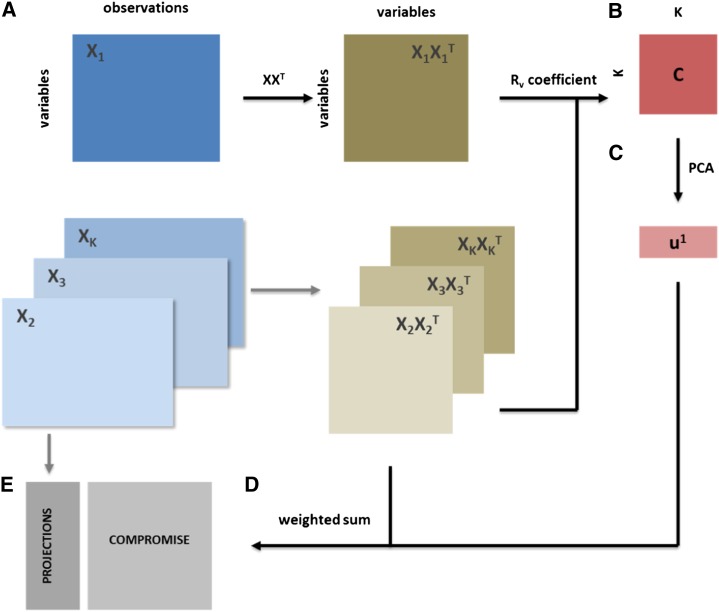
STATIS can be regarded as a generalization of PCA that allows simultaneous investigations of similarities and differences between multiple data tables over the same set of variables (e.g. metabolites), even when data about these variables have been gathered in a different number of experiments. The essence of the approach lies in combining the data tables into a common structure, called a compromise, which is then analyzed based on PCA. Hence, like PCA, STATIS allows for examining projections of the original data sets on the compromise and, thus, for quantifying the concordance between the data sets and the extent to which variables and observations contribute to it. The principal steps of STATIS are schematically represented in Figure 8 and consist of the following. First, a similarity transformation (i.e. computation of a scalar product matrix) is performed for every data table, where rows correspond to variables (Fig. 8A). Therefore, the resulting square matrix has as many rows as there are variables in the original data matrix. The cosine similarity (i.e. a value between 0 and 1) for each pair of the resulting matrices is then calculated based on the Rv coefficient. The obtained Rv coefficients yield a new matrix C (Fig. 8B). This captures the similarity between the data tables, and its eigenvalue decomposition can be employed for comparing the contribution of different data tables and for visual inspection (Fig. 8C). The entries of the scaled first eigenvector of C quantify the overall similarity of one data table to all others and are used as weights to combine the scalar product matrices into a single one as a weighted sum. This matrix is termed the compromise matrix (Fig. 8D) and is denoted by S in what follows. Finally, the resulting compromise matrix can also be investigated via PCA to obtain factor scores and loadings for the variables (Fig. 8E). The overview of the steps in STATIS indicates that the determined compromise matrix (and, thus, its investigation with PCA) is contingent on the variables and data tables used. For more technical details, see Supplemental Methods S1.
Figure 8.
Overview of STATIS. Six data tables are considered for this analysis: two tomato varieties, peach, pepper, and two strawberry organs; here, a data table is composed of 16 metabolites (variables) and developmental stages (observations). The scalar product captures the covariances of each metabolite pair (A). Based on data table similarity, a new matrix of data table similarity can be derived (B). Successively, a PCA (C) of this compromise yields weights for the combination of all six data tables (D) that allow the data tables to be combined in a common representation, the compromise (E). [See online article for color version of this figure.]
Enrichment Analysis Using Kyoto Encyclopedia of Genes and Genomes Pathway Information and BRITE Hierarchy Compound Classes
To infer which metabolic functions or compound classes are associated with the adjustments of metabolic content in the studied organs/species, we rely on enrichment analysis of ontology terms in combination with STATIS. Similar to a typical gene set enrichment analysis, for a given a set of metabolites, we aim at assessing whether certain associated pathway memberships or compound classes are overrepresented (i.e. overenriched; Subramanian et al., 2005; Rivals et al., 2007) in the investigated projections. Classically, hypergeometric tests are employed to assess which pathways or compound classes are statistically significantly overrepresented (Rivals et al., 2007). Enrichment results presented in this work were obtained using the previously mentioned hypergeometric test with corrected P values from Benjamini and Hochberg (1995) implemented using R. The significance level is set to α = 0.05. In this study, we rely on the Kyoto Encyclopedia of Genes and Genomes database (release 64.0, October 1, 2012; http://www.genome.jp/kegg/) to derive the pathway membership of the studied metabolites (Kanehisa and Goto, 2000; Kanehisa et al., 2012). To further reflect only plant-specific pathway information, we consider only pathways present in the model organism Arabidopsis (Arabidopsis thaliana; http://www.genome.jp/dbget-bin/get_linkdb?-t+pathway+genome:T00041). Furthermore, the Kyoto Encyclopedia of Genes and Genomes BRITE hierarchy (Tanabe and Kanehisa, 2002; Kanehisa et al., 2012) of compound classes is used to automatically assess the enrichment of particular compound classes (e.g. sugars or amino acids) for the given set of metabolites. The pathway membership as well as the corresponding compound classes for the 16 metabolites used in the analysis are given in Supplemental Table S1. We point out that, given that the metabolic pathways of the considered fruits substantially differ from the used reference species, manual curation was required. This curation affected only four pathways: “taurine and hypotaurine metabolism,” “tropane, piperidine, and pyridine alkaloid biosynthesis,” “indole alkaloid biosynthesis,” and “glucosinolate biosynthesis.” Three of them, taurine and hypotaurine metabolism, tropane, piperidine, and pyridine alkaloid biosynthesis, and indole alkaloid biosynthesis, were removed, whereas glucosinolate metabolism was renamed “volatile organic compound biosynthesis,” since it shares common precursors with the specialized genus Brassica pathway. As a further control, we inspected whether the 12 metabolites not assigned to the glucosinolate pathway belong to the volatile organic compound biosynthesis pathway and concluded that none of them belong to this pathway.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Typological values for the six data tables (from organs/species) after the removal of early fruit developmental stages.
Supplemental Figure S2. Projection of variables onto PC2 and PC3 of the compromise for the six data tables (organs/species).
Supplemental Figure S3. Sampling scheme for the tomato ripening mutants and the two tomato cultivars.
Supplemental Figure S4. STATIS on data tables from two tomato cultivars and three tomato ripening mutants: typological values and table interstructure.
Supplemental Table S1. Overview of metabolites, pathway membership, and BRITE hierarchies.
Supplemental Methods S1. Technical details about STATIS.
Glossary
- PCA
principal component analysis
- DAP
days after pollination
- DAB
days after bloom
References
- Aharoni A, O’Connell AP. (2002) Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J Exp Bot 53: 2073–2087 [DOI] [PubMed] [Google Scholar]
- Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goodenowe DB. (2002) Nontargeted metabolome analysis by use of Fourier transform ion cyclotron mass spectrometry. OMICS 6: 217–234 [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Beriashvili TV, Beriashvili LT. (1996) Metabolism of malic and tartaric acids in grape berries. Biochemistry (Moscow) 61: 1316–1321 [Google Scholar]
- Bombarely A, Merchante C, Csukasi F, Cruz-Rus E, Caballero JL, Medina-Escobar N, Blanco-Portales R, Botella MA, Muñoz-Blanco J, Sánchez-Sevilla JF, et al. (2010) Generation and analysis of ESTs from strawberry (Fragaria × ananassa) fruits and evaluation of their utility in genetic and molecular studies. BMC Genomics 11: 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani J, Budde CO, Porrini L, Lauxmann MA, Lombardo VA, Murray R, Andreo CS, Drincovich MF, Lara MV. (2009) Carbon metabolism of peach fruit after harvest: changes in enzymes involved in organic acid and sugar level modifications. J Exp Bot 60: 1823–1837 [DOI] [PubMed] [Google Scholar]
- Carrari F, Baxter C, Usadel B, Urbanczyk-Wochniak E, Zanor MI, Nunes-Nesi A, Nikiforova V, Centero D, Ratzka A, Pauly M, et al. (2006) Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol 142: 1380–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F, Fernie AR. (2006) Metabolic regulation underlying tomato fruit development. J Exp Bot 57: 1883–1897 [DOI] [PubMed] [Google Scholar]
- Centeno DC, Osorio S, Nunes-Nesi A, Bertolo AL, Carneiro RT, Araújo WL, Steinhauser MC, Michalska J, Rohrmann J, Geigenberger P, et al. (2011) Malate plays a crucial role in starch metabolism, ripening, and soluble solid content of tomato fruit and affects postharvest softening. Plant Cell 23: 162–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curien G, Bastien O, Robert-Genthon M, Cornish-Bowden A, Cárdenas ML, Dumas R. (2009) Understanding the regulation of aspartate metabolism using a model based on measured kinetic parameters. Mol Syst Biol 5: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RCH, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, et al. (2011) Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 23: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc LG, Grimplet J, Wheatley MD, Tillett RL, Quilici DR, Osborne C, Schooley DA, Schlauch KA, Cushman JC, Cramer GR. (2007) Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8: 429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do PT, Prudent M, Sulpice R, Causse M, Fernie AR. (2010) The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol 154: 1128–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hadi MAM, Zhang FJ, Wu FF, Zhou CH, Tao J. (2013) Advances in fruit aroma volatile research. Molecules 18: 8200–8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enfissi EM, Barneche F, Ahmed I, Lichtlé C, Gerrish C, McQuinn RP, Giovannoni JJ, Lopez-Juez E, Bowler C, Bramley PM, et al. (2010) Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22: 1190–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Hanhineva K, Beleggia R, Dai N, Rogachev I, Nikiforova VJ, Fernie AR, Aharoni A. (2008) Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol 148: 730–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L. (2011) Elemental formula annotation of polar and lipophilic metabolites using (13) C, (15) N and (34) S isotope labelling, in combination with high-resolution mass spectrometry. Plant J 68: 364–376 [DOI] [PubMed] [Google Scholar]
- Gilbert L, Alhagdow M, Nunes-Nesi A, Quemener B, Guillon F, Bouchet B, Faurobert M, Gouble B, Page D, Garcia V, et al. (2009) GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J 60: 499–508 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. (2001) Molecular biology of fruit maturation and ripening. Annu Rev Plant Physiol Plant Mol Biol 52: 725–749 [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. (2004) Genetic regulation of fruit development and ripening. Plant Cell (Suppl) 16: S170–S180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni JJ, Noensie EN, Ruezinsky DM, Lu X, Tracy SL, Ganal MW, Martin GB, Pillen K, Alpert K, Tanksley SD. (1995) Molecular genetic analysis of the ripening-inhibitor and non-ripening loci of tomato: a first step in genetic map-based cloning of fruit ripening genes. Mol Gen Genet 248: 195–206 [DOI] [PubMed] [Google Scholar]
- Gordo SM, Pinheiro DG, Moreira EC, Rodrigues SM, Poltronieri MC, de Lemos OF, da Silva IT, Ramos RT, Silva A, Schneider H, et al. (2012) High-throughput sequencing of black pepper root transcriptome. BMC Plant Biol 12: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimplet J, Deluc LG, Tillett RL, Wheatley MD, Schlauch KA, Cramer GR, Cushman JC. (2007) Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halinska A, Frenkel C. (1991) Acetaldehyde stimulation of net gluconeogenic carbon movement from applied malic acid in tomato fruit pericarp tissue. Plant Physiol 95: 954–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampp C, Richter A, Osorio S, Zellnig G, Sinha AK, Jammer A, Fernie AR, Grimm B, Roitsch T. (2012) Establishment of a photoautotrophic cell suspension culture of Arabidopsis thaliana for photosynthetic, metabolic, and signaling studies. Mol Plant 5: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanhineva K, Rogachev I, Kokko H, Mintz-Oron S, Venger I, Kärenlampi S, Aharoni A. (2008) Non-targeted analysis of spatial metabolite composition in strawberry (Fragaria × ananassa) flowers. Phytochemistry 69: 2463–2481 [DOI] [PubMed] [Google Scholar]
- Hanhineva K, Soininen P, Anttonen MJ, Kokko H, Rogachev I, Aharoni A, Laatikainen R, Kärenlampi S. (2009) NMR and UPLC-qTOF-MS/MS characterisation of novel phenylethanol derivatives of phenylpropanoid glucosides from the leaves of strawberry (Fragaria × ananassa cv. Jonsok). Phytochem Anal 20: 353–364 [DOI] [PubMed] [Google Scholar]
- Higashi Y, Saito K. (2013) Network analysis for gene discovery in plant-specialized metabolism. Plant Cell Environ 36: 1597–1606 [DOI] [PubMed] [Google Scholar]
- Howard LR, Wildman REC (2007) Antioxidant Vitamin and Phytochemical Content of Fresh and Processed Pepper Fruit (Capsicum annuum), Ed 2. CRC Press, Boca Raton, FL [Google Scholar]
- Kanehisa M, Goto S. (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: D109–D114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Baek KH, Lee SW, Kim J, Lee BW, Cho HS, Kim WT, Choi D, Hur CG. (2008) Pepper EST database: comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. BMC Plant Biol 8: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu Rev Genet 45: 41–59 [DOI] [PubMed] [Google Scholar]
- Klie S, Caldana C, Nikoloski Z. (2012) Compromise of multiple time-resolved transcriptomics experiments identifies tightly regulated functions. Front Plant Sci 3: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Lv W, Zhang D, Jiang S, Zhang S, Li D. (2013) Genome-wide identification and analysis of expression profiles of maize mitogen-activated protein kinase kinase kinase. PLoS ONE 8: e57714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Redestig H, Hirai T, Oikawa A, Matsuda F, Fukushima A, Arita M, Watanabe S, Yano M, Hiwasa-Tanase K, et al. (2011) Covering chemical diversity of genetically-modified tomatoes using metabolomics for objective substantial equivalence assessment. PLoS ONE 6: e16989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara MV, Drincovich MF, Andreo CS. (2004) Induction of a Crassulacean acid-like metabolism in the C(4) succulent plant, Portulaca oleracea L: study of enzymes involved in carbon fixation and carbohydrate metabolism. Plant Cell Physiol 45: 618–626 [DOI] [PubMed] [Google Scholar]
- Lee JJ, Park KW, Kwak YS, Ahn JY, Jung YH, Lee BH, Jeong JC, Lee HS, Kwak SS. (2012) Comparative proteomic study between tuberous roots of light orange- and purple-fleshed sweetpotato cultivars. Plant Sci 193-194: 120–129 [DOI] [PubMed] [Google Scholar]
- Lee S, Chung EJ, Joung YH, Choi D. (2010) Non-climacteric fruit ripening in pepper: increased transcription of EIL-like genes normally regulated by ethylene. Funct Integr Genomics 10: 135–146 [DOI] [PubMed] [Google Scholar]
- Lenucci MS, Serrone L, De Caroli M, Fraser PD, Bramley PM, Piro G, Dalessandro G. (2012) Isoprenoid, lipid, and protein contents in intact plastids isolated from mesocarp cells of traditional and high-pigment tomato cultivars at different ripening stages. J Agric Food Chem 60: 1764–1775 [DOI] [PubMed] [Google Scholar]
- Li X, Korir NK, Liu L, Shangguan L, Wang Y, Han J, Chen M, Fang J. (2012) Microarray analysis of differentially expressed genes engaged in fruit development between Prunus mume and Prunus armeniaca. J Plant Physiol 169: 1776–1788 [DOI] [PubMed] [Google Scholar]
- Liu S, Li W, Wu Y, Chen C, Lei J. (2013) De novo transcriptome assembly in chili pepper (Capsicum frutescens) to identify genes involved in the biosynthesis of capsaicinoids. PLoS ONE 8: e48156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA (2006) Inositol and plant cell wall polysaccharide biogenesis. In A Lahiri Majunder, BB Biswas, eds, Biology of Inositols and Phosphoinositides. Springer, Dordrecht, The Netherlands, pp 21–45 [DOI] [PubMed] [Google Scholar]
- Lombardo VA, Osorio S, Borsani J, Lauxmann MA, Bustamante CA, Budde CO, Andreo CS, Lara MV, Fernie AR, Drincovich MF. (2011) Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiol 157: 1696–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney GS, Kochevenko A, Tieman DM, Tohge T, Krieger U, Zamir D, Taylor MG, Fernie AR, Klee HJ. (2010) Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant Physiol 153: 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meléndez-Martínez AJ, Fraser PD, Bramley PM. (2010) Accumulation of health promoting phytochemicals in wild relatives of tomato and their contribution to in vitro antioxidant activity. Phytochemistry 71: 1104–1114 [DOI] [PubMed] [Google Scholar]
- Moing A, Aharoni A, Biais B, Rogachev I, Meir S, Brodsky L, Allwood JW, Erban A, Dunn WB, Kay L, et al. (2011) Extensive metabolic cross-talk in melon fruit revealed by spatial and developmental combinatorial metabolomics. New Phytol 190: 683–696 [DOI] [PubMed] [Google Scholar]
- Moore S, Payton P, Wright M, Tanksley S, Giovannoni J. (2005) Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J Exp Bot 56: 2885–2895 [DOI] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, Fernie AR, Usadel B, Nikoloski Z, Persson S. (2011) PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 23: 895–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R, Kusano M, Kobayashi M, Tohge T, Yonekura-Sakakibara K, Kogure N, Yamazaki M, Kitajima M, Saito K, Takayama H. (2009) Metabolomics-oriented isolation and structure elucidation of 37 compounds including two anthocyanins from Arabidopsis thaliana. Phytochemistry 70: 1017–1029 [DOI] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CM, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JK, Fei Z, et al. (2011a) Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol 157: 405–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Nikoloski Z, Kochevenko A, Fernie AR, Giovannoni JJ. (2012) Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol 159: 1713–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Bombarely A, Giavalisco P, Usadel B, Stephens C, Aragüez I, Medina-Escobar N, Botella MA, Fernie AR, Valpuesta V. (2011b) Demethylation of oligogalacturonides by FaPE1 in the fruits of the wild strawberry Fragaria vesca triggers metabolic and transcriptional changes associated with defence and development of the fruit. J Exp Bot 62: 2855–2873 [DOI] [PubMed] [Google Scholar]
- Osorio S, Muñoz C, Valpuesta V (2010) Physiology and biochemistry of fruit flavours. In YH Hui, ed, Handbook of Fruit and Vegetable Flavors. John Wiley & Sons, New York, pp 25–43 [Google Scholar]
- Osorio S, Scossa F, Fernie AR. (2013a) Molecular regulation of fruit ripening. Front Plant Sci 4: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Vallarino JG, Szecowka M, Ufaz S, Tzin V, Angelovici R, Galili G, Fernie AR. (2013b) Alteration of the interconversion of pyruvate and malate in the plastid or cytosol of ripening tomato fruit invokes diverse consequences on sugar but similar effects on cellular organic acid, metabolism, and transitory starch accumulation. Plant Physiol 161: 628–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page D, Gouble B, Valot B, Bouchet JP, Callot C, Kretzschmar A, Causse M, Renard CM, Faurobert M. (2010) Protective proteins are differentially expressed in tomato genotypes differing for their tolerance to low-temperature storage. Planta 232: 483–500 [DOI] [PubMed] [Google Scholar]
- Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G. (2012) An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep 31: 349–360 [DOI] [PubMed] [Google Scholar]
- Perkins-Veazie P (1995) Growth and ripening of strawberry fruit. Hortic Rev 17: 267–297 [Google Scholar]
- Rivals I, Personnaz L, Taing L, Potier MC. (2007) Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23: 401–407 [DOI] [PubMed] [Google Scholar]
- Rogachev I, Aharoni A. (2012) UPLC-MS-based metabolite analysis in tomato. Methods Mol Biol 860: 129–144 [DOI] [PubMed] [Google Scholar]
- Rohrmann J, Tohge T, Alba R, Osorio S, Caldana C, McQuinn R, Arvidsson S, van der Merwe MJ, Riaño-Pachón DM, Mueller-Roeber B, et al. (2011) Combined transcription factor profiling, microarray analysis and metabolite profiling reveals the transcriptional control of metabolic shifts occurring during tomato fruit development. Plant J 68: 999–1013 [DOI] [PubMed] [Google Scholar]
- Rose JKC, Bennett AB. (1999) Cooperative disassembly of the cellulose-xyloglucan network of plant cell walls: parallels between cell expansion and fruit ripening. Trends Plant Sci 4: 176–183 [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Shirasawa K, Isobe S, Hirakawa H, Asamizu E, Fukuoka H, Just D, Rothan C, Sasamoto S, Fujishiro T, Kishida Y, et al. (2010) SNP discovery and linkage map construction in cultivated tomato. DNA Res 17: 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Kanehisa M (2002) Using the KEGG database resource. In Current Protocols in Bioinformatics. John Wiley & Sons, New York, pp 1.12.1–1.12.54 [DOI] [PubMed] [Google Scholar]
- Tieman D, Bliss P, McIntyre LM, Blandon-Ubeda A, Bies D, Odabasi AZ, Rodríguez GR, van der Knaap E, Taylor MG, Goulet C, et al. (2012) The chemical interactions underlying tomato flavor preferences. Curr Biol 22: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Tigchelaar EC, Tomes M, Kerr EA, Barman RJ. (1973) A new fruit ripening mutant, non-ripening (nor). Rep Tomato Genet Coop 23: 33–34 [Google Scholar]
- Toubiana D, Fernie AR, Nikoloski Z, Fait A. (2013) Network analysis: tackling complex data to study plant metabolism. Trends Biotechnol 31: 29–36 [DOI] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. (2002) A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296: 343–346 [DOI] [PubMed] [Google Scholar]
- Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. (2008) Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol 177: 60–76 [DOI] [PubMed] [Google Scholar]
- Wahyuni Y, Ballester AR, Tikunov Y, de Vos RC, Pelgrom KT, Maharijaya A, Sudarmonowati E, Bino RJ, Bovy AG. (2013) Metabolomics and molecular marker analysis to explore pepper (Capsicum sp.) biodiversity. Metabolomics 9: 130–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen HC, Giovannoni JJ, Klee HJ. (1995) An ethylene-inducible component of signal transduction encoded by never-ripe. Science 270: 1807–1809 [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35: 155–189 [Google Scholar]
- Yen HC, Lee S, Tanksley SD, Lanahan MB, Klee HJ, Giovannoni JJ. (1995) The tomato Never-ripe locus regulates ethylene-inducible gene expression and is linked to a homolog of the Arabidopsis ETR1 gene. Plant Physiol 107: 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni A, Di Carli M, Guzzo F, Stocchero M, Zenoni S, Ferrarini A, Tononi P, Toffali K, Desiderio A, Lilley KS, et al. (2010) Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol 154: 1439–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Shen JY, Wei WW, Xi WP, Xu CJ, Ferguson I, Chen K. (2010) Expression of genes associated with aroma formation derived from the fatty acid pathway during peach fruit ripening. J Agric Food Chem 58: 6157–6165 [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang X, Yu O, Tang J, Gu X, Wan X, Fang C. (2011) Metabolic profiling of strawberry (Fragaria × ananassa Duch.) during fruit development and maturation. J Exp Bot 62: 1103–1118 [DOI] [PubMed] [Google Scholar]