Molecular and metabolic analysis in tobacco plants after virus-induced gene silencing uncovers different pathways of amino acid biosynthesis within the plastids.
Abstract
Chloroplasts and plastids of nonphotosynthetic plant cells contain two aspartate (Asp) aminotransferases: a eukaryotic type (Asp5) and a prokaryotic-type bifunctional enzyme displaying Asp and prephenate aminotransferase activities (PAT). We have identified the entire Asp aminotransferase gene family in Nicotiana benthamiana and isolated and cloned the genes encoding the isoenzymes with plastidic localization: NbAsp5 and NbPAT. Using a virus-induced gene silencing approach, we obtained N. benthamiana plants silenced for NbAsp5 and/or NbPAT. Phenotypic and metabolic analyses were conducted in silenced plants to investigate the specific roles of these enzymes in the biosynthesis of essential amino acids within the plastid. The NbAsp5 silenced plants had no changes in phenotype, exhibiting similar levels of free Asp and glutamate as control plants, but contained diminished levels of asparagine and much higher levels of lysine. In contrast, the suppression of NbPAT led to a severe reduction in growth and strong chlorosis symptoms. NbPAT silenced plants exhibited extremely reduced levels of asparagine and were greatly affected in their phenylalanine metabolism and lignin deposition. Furthermore, NbPAT suppression triggered a transcriptional reprogramming in plastid nitrogen metabolism. Taken together, our results indicate that NbPAT has an overlapping role with NbAsp5 in the biosynthesis of Asp and a key role in the production of phenylalanine for the biosynthesis of phenylpropanoids. The analysis of NbAsp5/NbPAT cosilenced plants highlights the central role of both plastidic aminotransferases in nitrogen metabolism; however, only NbPAT is essential for plant growth and development.
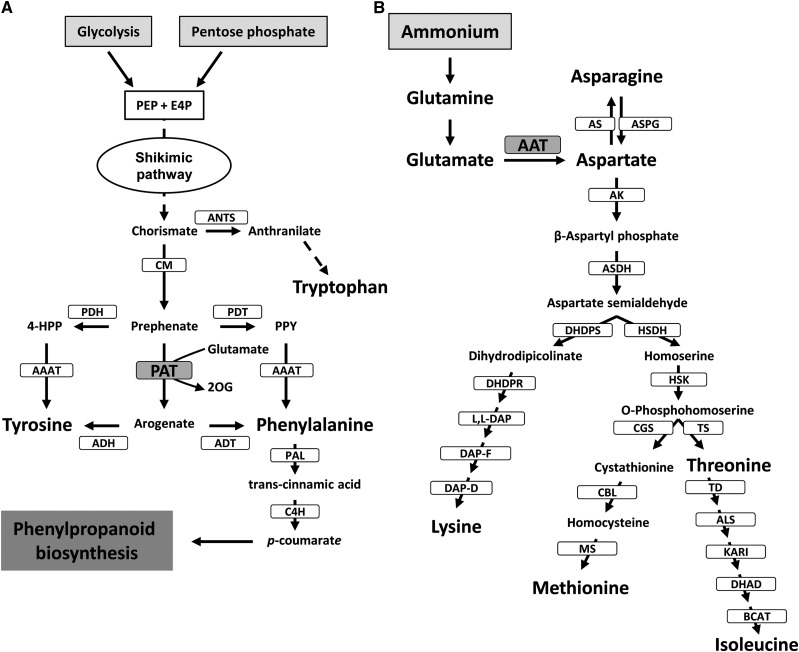
Inorganic nitrogen is primarily assimilated into the amino acids Gln, Glu, Asn, and Asp; these four amino acids serve as nitrogen metabolic precursors and nitrogen-transport compounds in most crops and higher plants (Buchanan et al., 2000). Glu is synthesized in the plastids and occupies a central position in plant amino acid metabolism, regulation, and signaling (Forde and Lea, 2007). Glu is the nitrogen donor for the biosynthesis of essential amino acids through the Asp and aromatic amino acid pathways, which are entirely located in plastids (Lea and Azevedo, 2003; Rippert et al., 2009; Fig. 1).
Figure 1.
Asp and aromatic amino acid biosynthetic pathways in plants. A, Postchorismate pathway leading to the biosynthesis of aromatic amino acids. CM, Chorismate mutase; E4P, d-erythrose 4-phosphate; 4-HPP, 4-hydroxyphenylpyruvate; PEP, phosphoenolpyruvate; PPY, phenylpyruvate. B, Asp-derived amino acid biosynthesis. ALS, Acetolactate synthase; AS, Asn synthetase; ASN, asparaginase; BCAT, branched-chain aminotransferase; CBL, cystathionine β-lyase; CGS, cystathionine γ-synthase; DAP-D, diaminopimelate decarboxylase; DAP-F, diaminopimelate epimerase; DHAD, dihydroxy acid dehydratase; DHDPR, dihydrodipicolinate reductase; DHDPS, dihydrodipicolinate synthase; HSDH, homoserine dehydrogenase; HSK, homoserine kinase; KARI, ketol-acid reductoisomerase; MS, Met synthase; TD, Thr deaminase; TS, Thr synthase.
The enzyme aspartate aminotransferase (AAT; EC 2.6.1.1) catalyzes a reversible transamination between Glu and oxaloacetate to yield Asp and 2-oxoglutarate and plays a crucial role channeling nitrogen from Glu to Asp in all living organisms. In plants, AAT is present as several isoenzymes located in different subcellular compartments: the cytosol, mitochondria, peroxisomes, and plastids (Ireland and Joy, 1985). In Arabidopsis (Arabidopsis thaliana), a small gene family encodes AAT isoenzymes: Asp2 and Asp4 (cytosolic), Asp1 (mitochondrial), Asp3 (peroxisomal), and Asp5 (plastidial; Schultz and Coruzzi, 1995; Wilkie and Warren, 1998). The function of cytosolic and plastidic AAT has been investigated using Arabidopsis mutants (Schultz et al., 1998; Miesak and Coruzzi, 2002). A specific nonredundant role has been proposed for Asp2 related to the biosynthesis of a specific Asp pool during the light phase that is used to produce Asn during the dark phase (Miesak and Coruzzi, 2002). The Asp2 gene has also been proposed to interact with plant defense responses in Arabidopsis plants infected with the necrotrophic pathogen Botrytis cinerea (Brauc et al., 2011). Asp5 mutants affected in the plastidic isoenzyme had no visible phenotype except that plants contained increased Gln levels either under light or dark growth conditions (Schultz et al., 1998; Miesak and Coruzzi, 2002). According to this, Asp5 might have a role in shuttling reducing equivalents, as proposed for the peroxisomal and mitochondrial isoenzymes (Liepman and Olsen, 2004).
In addition, de la Torre et al. (2006) reported the existence of a novel plastid-located AAT, a prokaryotic-type AAT unrelated to other eukaryotic AATs from plants and animals but closely related to cyanobacterial enzymes. The kinetic parameters of this enzyme were determined, highlighting its high affinity for Glu and Asp compared with other plant AATs (de la Torre et al., 2006). Recently, Graindorge et al. (2010) and Maeda et al. (2011) independently discovered that plant prokaryotic-type AATs displayed not only AAT activity but also prephenate aminotransferase (PAT) activity. PAT activity catalyzes the reversible transamination between Glu/Asp and prephenate to yield 2-oxoglutarate/oxaloacetate and arogenate. This metabolic reaction represents a key step in the biosynthesis of aromatic amino acids in the plastids. The product of PAT, arogenate, is the immediate precursor for the biosynthesis of Phe and Tyr using arogenate dehydratase (ADT) and arogenate dehydrogenase (ADH), respectively (Tzin and Galili, 2010; Maeda and Dudareva, 2012; Fig. 1). Recent genetic evidence has indicated that the arogenate pathway is the predominant route for Phe biosynthesis in plants (Maeda et al., 2010; Maeda and Dudareva, 2012). In maritime pine (Pinus pinaster), a Myb transcription factor (Myb8) regulates genes involved in the Phe pathway (Craven-Bartle et al., 2013).
It is important to remark that in the presence of Glu as nitrogen donor, PAT exhibits similar values of the specificity constant for oxaloacetate and prephenate, indicating that the enzyme can operate both as a PAT and as a classical AAT (Graindorge et al., 2010). Consequently, PAT could be involved not only in the biosynthesis of aromatic amino acids but also in the biosynthesis of Asp-derived amino acids. Asp is a precursor in two main pathways: (1) the biosynthesis of Asn in the cytosol (Lea and Azevedo, 2003; Lea et al., 2007); and (2) the biosynthesis of Lys, Met, Thr, and Ile through the so-called Asp metabolic pathway in the plastids (Azevedo et al., 2006; Jander and Joshi, 2009; Galili, 2011; Fig. 1).
In regard to PAT function, silencing of the gene encoding the enzyme was achieved in petals of petunia (Petunia hybrida) following an RNA interference (RNAi) strategy using a petal-specific promoter (Maeda et al., 2011). The targeted tissue was found to be impaired for Phe biosynthesis. However, the phenotypic and metabolic effects of PAT gene suppression in a whole plant were still unknown.
In this work, we report a new strategy to study the biochemical function of AAT and PAT activities in plastids, virus-induced gene silencing (VIGS), as a viable alternative experimental approach to obtain gene silencing in whole Nicotiana benthamiana plants. This strategy provided us with an efficient tool to study the in vivo role of the two plastidial aminotransferases encoded by NbAsp5 and NbPAT, respectively. Molecular and metabolic analysis of silenced plants revealed their contributions to different pathways of amino acid biosynthesis within the plastids.
RESULTS
Identification of the AAT Gene Family in N. benthamiana
A search in the EST databases of N. benthamiana (www.sgn.cornell.edu) allowed us to identify the complete AAT gene family. The N. benthamiana AAT gene family is similar to the well-characterized AAT gene family in Arabidopsis and includes five genes encoding eukaryotic-type AATs (NbAsp1–NbAsp5), predicted to be targeted to different subcellular compartments (mitochondria, cytosol, plastids, and peroxisomes) and an extra gene encoding a prokaryotic-type bifunctional PAT/AAT enzyme targeted to plastids (NbPAT). Therefore, N. benthamiana contains two enzymes located within the plastids housing AAT activity, NbAsp5 and NbPAT. These AAT proteins showed a high degree of identity with the corresponding counterparts from Arabidopsis (Supplemental Table S1).
VIGS of NbAsp5 and NbPAT
We developed a VIGS approach in order to study the phenotypic and metabolic impact associated with the loss of function of NbAsp5 and/or NbPAT proteins. The starting hypothesis was that these two enzymes are responsible for Asp and prephenate biosynthesis within the plastid. Using reverse transcription-PCR with primers based on NbAsp5 and NbPAT, we obtained and sequenced two complementary DNA (cDNA) fragments 469 and 440 bp in length, respectively, corresponding to specific regions in the 5′ open reading frame of both genes (Supplemental Fig. S1). We named these fragments Nb5′Asp5 and Nb5′PAT, respectively. These sequences had been previously selected as unique sequences, since in BLAST searches no other sequences were detected in the latest genome assembly of N. benthamiana that contained stretches longer than 21 to 24 nucleotides displaying 100% identity. This indicates the unlikelihood of cosilencing genes other than Asp5 and PAT using Nb5′Asp5 or Nb5′PAT in the VIGS assay. Fragments were inserted into pTRVGW, a Gateway-compatible tobacco rattle virus vector (Liu et al., 2002), courtesy of Savithramma P. Dinesh-Kumar and Dr. Olga del Pozo (pTRV-Asp5 and pTRV-PAT). In order to substantiate the specificity of their silencing phenotypes, we generated additional nonoverlapping silencing constructs for NbAsp5 (pTRV-Nb3′Asp5) and NbPAT (pTRV-NbMPAT and pTRV-Nb3′PAT; Supplemental Fig. S1). Furthermore, Nb5′Asp5 and Nb5′PAT fragments were PCR combined in a single DNA fragment, NbAsp5/NbPAT, and Gateway inserted into pTRVGW in a DNA construct for VIGS of both genes at the same time, pTRV-Asp5/PAT (Supplemental Fig. S1). Thus, N. benthamiana plantlets were silenced for NbAsp5, NbPAT, or both using pTRV-Asp5, pTRV-PAT, or pTRV-Asp5/PAT, respectively (Fig. 2). One month after initiation of VIGS, the morphology of the plants was observed and compared with the wild type and plants transfected with the empty vector, pTRV-EV. Silencing of the endogenous phytoene desaturase gene, NbPDS, was used as a control for the effectiveness of VIGS (Supplemental Fig. S2). When the NbAsp5 gene was targeted, no evident phenotype was detected in plants silenced using either pTRV-Asp5 or pTRV-Nb3′Asp5. In contrast, plants silenced for NbPAT using pTRV-PAT, or cosilenced for both NbPAT and NbAsp5 using pTRV-Asp5/PAT, exhibited severe reduction in growth, strong symptoms of chlorosis, and altered fresh weight-to-dry weight ratio in comparison with pTRV-EV or wild-type plants (Fig. 2; Supplemental Figs. S3 and S4). Similar effects were observed in plants silenced using alternative constructs for VIGS of NbPAT: pTRV-NbMPAT and pTRV-Nb3′PAT (data not shown).
Figure 2.
Phenotypes of N. benthamiana plants subjected to VIGS of NbAsp5 and/or NbPAT genes. Photographs of wild-type (WT) and control (pTRVGW-EV) plants are shown in the top row. Phenotypes of 6-week-old plants silenced for NbAsp5, NbPAT, or NbAsp-5/NbPAT are shown in the bottom row. Similar phenotypes were observed in six different experiments, each with five biological replicates. All photographs were taken under the same magnification.
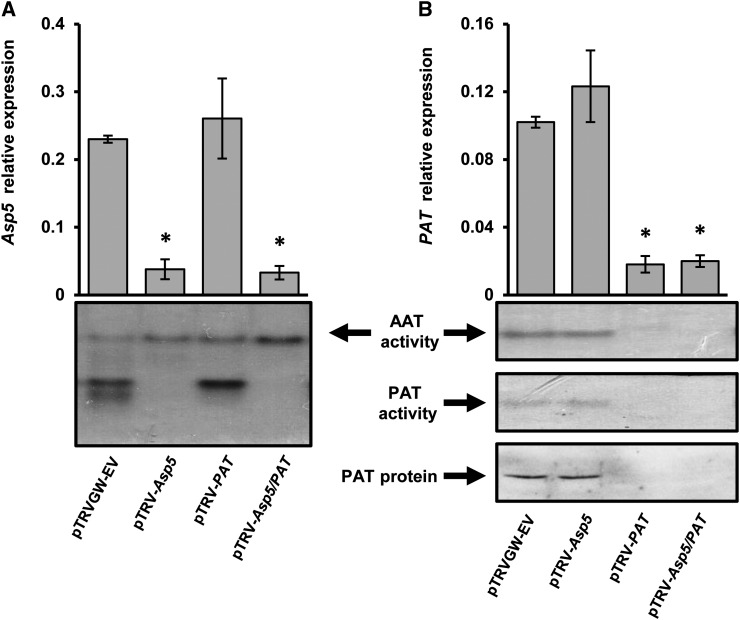
The degree of silencing observed was initially assessed by quantitative PCR (qPCR) determination of transcript abundance (Fig. 3). The transcript levels of both targeted genes, NbAsp5 (Fig. 3A) and NbPAT (Fig. 3B), were reduced approximately 90% both in single- and double-silenced N. benthamiana plants when compared with wild-type or pTRV-EV plants. Figure 3 also shows that the silencing of a single plastidic aminotransferase, either NbAsp5 or NbPAT, did not affect the transcript abundance of the other enzyme in the same plant. We also examined whether silencing of plastidic enzymes could alter the expression of other members of the gene family. As shown in Supplemental Figure S5, NbAsp5 and/or NbPAT silencing did not affect the transcript abundance of Asp1, Asp2, Asp3, and Asp4. The specific silencing of NbAsp5 and NbPAT was further analyzed by AAT activity determination on native gels. Our results clearly confirm effective suppression of the corresponding AAT activity bands in plants silenced for either NbAsp5 or NbPAT (Fig. 3). Moreover, when we conducted a similar experiment to test PAT activity on native gels, we observed complete suppression of the PAT activity band in plants silenced for NbPAT using both pTRV-PAT or the chimera containing both AAT sequences, pTRV-Asp5/PAT (Fig. 3B). The absence of the NbPAT polypeptide in these samples was further confirmed by western-blot analysis (Fig. 3B) using specific antibodies raised against the maritime pine protein (de la Torre et al., 2006).
Figure 3.
Molecular characterization of VIGS of NbPAT and/or NbAsp5 genes in N. benthamiana leaves. A, Relative NbAsp5 gene expression in silenced plants compared with controls. At bottom, AAT activity is shown on native gels with protein extracts prepared from control and silenced plants. B, Relative NbPAT gene expression in control and silenced plants. At bottom, AAT activity and PAT activity are shown in protein extracts prepared from control and silenced plants resolved by native gel electrophoresis; PAT protein levels were analyzed by western blotting using anti-PAT specific antibodies (de la Torre et al., 2006). The expression levels for all genes were normalized to that of NbActin2. Error bars represent se. Asterisks indicate significant differences compared with control plants. AAT activity corresponding to NbASP5 and NbPAT on the gels was determined using extracts from N. benthamiana leaves (Fig. 2) according to a previously described protocol (de la Torre et al., 2007).
Silencing of NbPAT Affects Lignin Deposition in Vascular Bundles
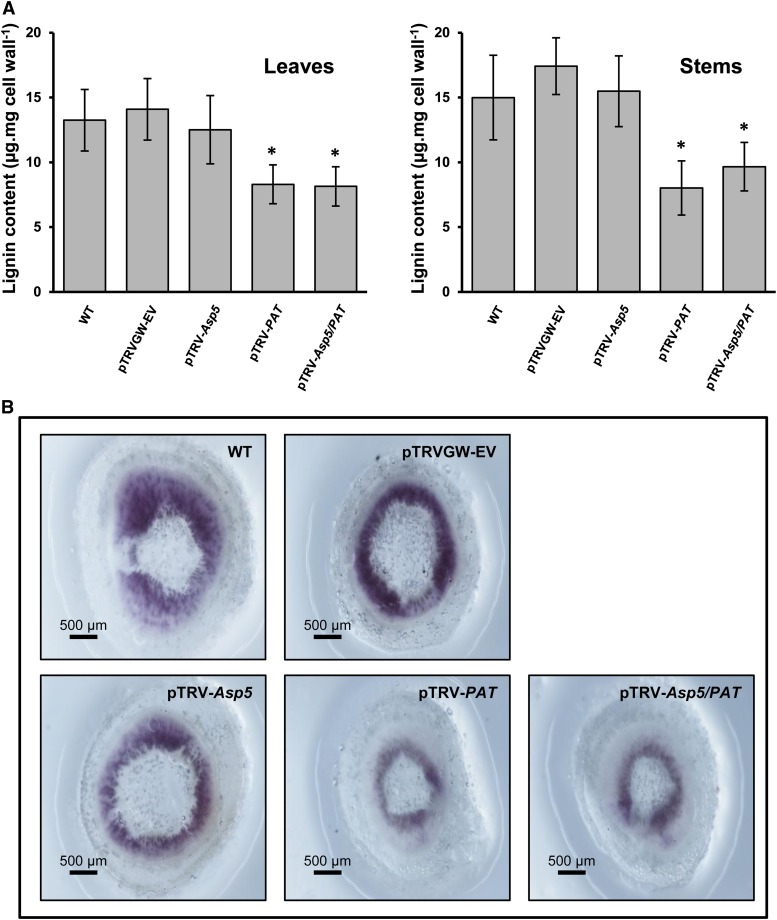
Recently, the arogenate pathway has been proposed to be the predominant route for Phe biosynthesis in plants (Maeda et al., 2011). Considering that the phenylpropane skeleton required for lignin biosynthesis is provided by the deamination of Phe, and the phenotype observed, we conducted an experiment in order to evaluate variations in lignin content in the NbPAT silenced plants (Fig. 4). The determination of lignin content showed a significant reduction compared with controls in leaves and stems of 6-week-old plants silenced for NbPAT (Fig. 4A). A similar reduction was observed in plants silenced using pTRV-Asp5/PAT (Fig. 4A). These results were further confirmed by in situ lignin detection in stem sections of plants silenced for NbAsp5, NbPAT, or both NbPAT and NbAsp5. The plants silenced for NbPAT or NbPAT and NbAsp5 showed greatly reduced phloroglucinol staining in comparison with control plants or plants silenced only for NbAsp5 (Fig. 4B).
Figure 4.
Lignin content and histological lignin staining in N. benthamiana plants. A, Quantification of lignin in leaves (left) and stems (right) from N. benthamiana plants. WT, Wild type; pTRVGW-EV, control vector; pTRV-Asp5, silenced NbAsp5; pTRV-PAT, silenced NbPAT; pTRV-Asp5/PAT, cosilenced NbAsp5/NbPAT. Six independent biological replicates were measured. Average values and sd were calculated. Error bars represent se, and asterisks indicate significant differences compared with control plants. B, Histological detection of lignin in fresh-cut stem sections of N. benthamiana corresponding to control and silenced plants. Red-violet color shows the reaction of phloroglucinol-HCl with cinnamaldehyde end groups of lignin. [See online article for color version of this figure.]
NbPAT Silencing Also Affects the Chlorophyll Content
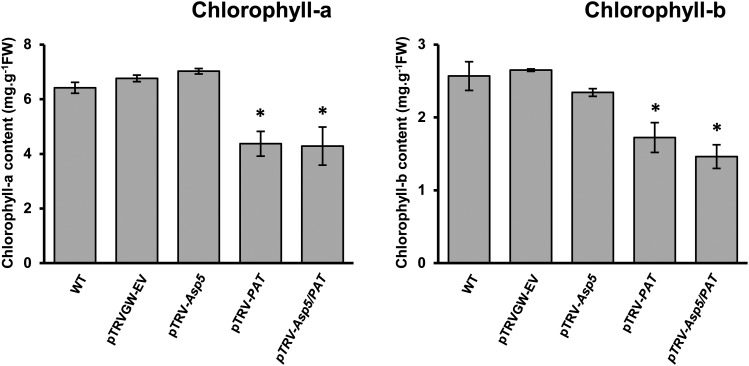
Concomitantly, and based on the observed chlorosis phenotype of plants silenced for NbPAT (Fig. 2; Supplemental Fig. S3), chlorophyll contents were also determined in the leaves of control and silenced plants. A significant reduction (30%–40%) in both chlorophyll a and b contents was observed in leaves of plants silenced for NbPAT and Asp5/PAT compared with control plants (Fig. 5). No comparable alteration in chlorophyll a or b could be detected in plants silenced for NbAsp5 (Fig. 5).
Figure 5.
Chlorophyll content in leaves of N. benthamiana plants. Chlorophyll a and b contents in N. benthamiana leaves were determined as described in “Materials and Methods.” For each measurement, the average value and sd were calculated using five independent biological replicates. Error bars represent se. Asterisks indicate significant differences compared with control plants. FW, Fresh weight; WT, wild type.
NbAsp5 and NbPAT Silencing Alter Carbohydrate Levels
Since the silencing of PAT activity affected chlorophyll levels, we next examined the relative abundance of Glc, Fru, Suc, and the polysaccharides cellulose and starch. Major changes in free sugar levels and starch were observed in cosilenced plants; meanwhile, no significant changes were observed in cellulose content (Supplemental Fig. S6).
Suppression of AAT Activity in Plastids of N. benthamiana Results in Altered Amino Acid Profiles
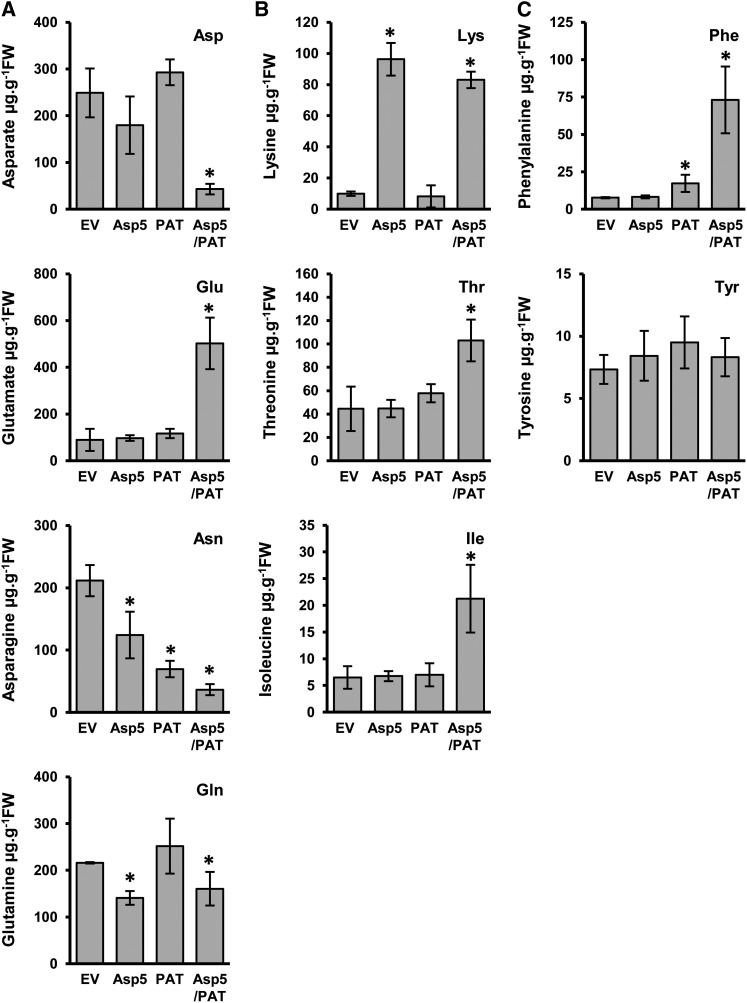
To get further insights into the metabolic role of plastidic AAT enzymes, we next examined the profiles of a set of amino acids. We first analyzed whether the silencing of NbPAT and/or NbAsp5 affected levels of key amino acids, Asp, Asn, Glu, and Gln (Fig. 6A). No significant changes were observed in the levels of Asp or Glu in plants silenced for either NbAsp5 or NbPAT. In contrast, plants silenced for both genes showed a dramatic reduction in the content of Asp and a concomitant increase in free Glu levels (approximately 5-fold; Fig. 6A). In addition, plants silenced alternately for NbAsp5 or NbPAT showed significant reductions in the levels of free Asn, close to 55% and 65%, respectively (Fig. 6A). As observed with Asp, the silencing of both genes simultaneously resulted in the additive reduction of the level of the amino acid of around 83% (Fig. 6). When we measured free Gln levels, only a small reduction was detected in plants silenced for NbAsp5 or NbAsp5/NbPAT. Absence of a major disturbance in Gln levels in plants silenced for NbAsp5 and/or NbPAT is consistent with the observed corresponding levels of Glu that guarantee its biosynthesis through the action of Gln synthetase.
Figure 6.
Amino acid content in plants silenced for NbPAT and/or NbAsp5. Amino acid profiles are shown in leaves from N. benthamiana silenced for NbPAT, NbAsp5, or both NbPAT and NbAsp5 compared with empty-vector controls (EV). Amino acid content was determined using an HPLC method with a UV detector developed for the determination of individual amino acids. A, Asp, Glu, Asn, and Gln. B, Lys, Thr, and Ile. C, Phe and Tyr. Data represent means of five plants. Significant differences between the control and silenced plants based on Student’s t test (P ≤ 0.05) are indicated with asterisks. FW, Fresh weight.
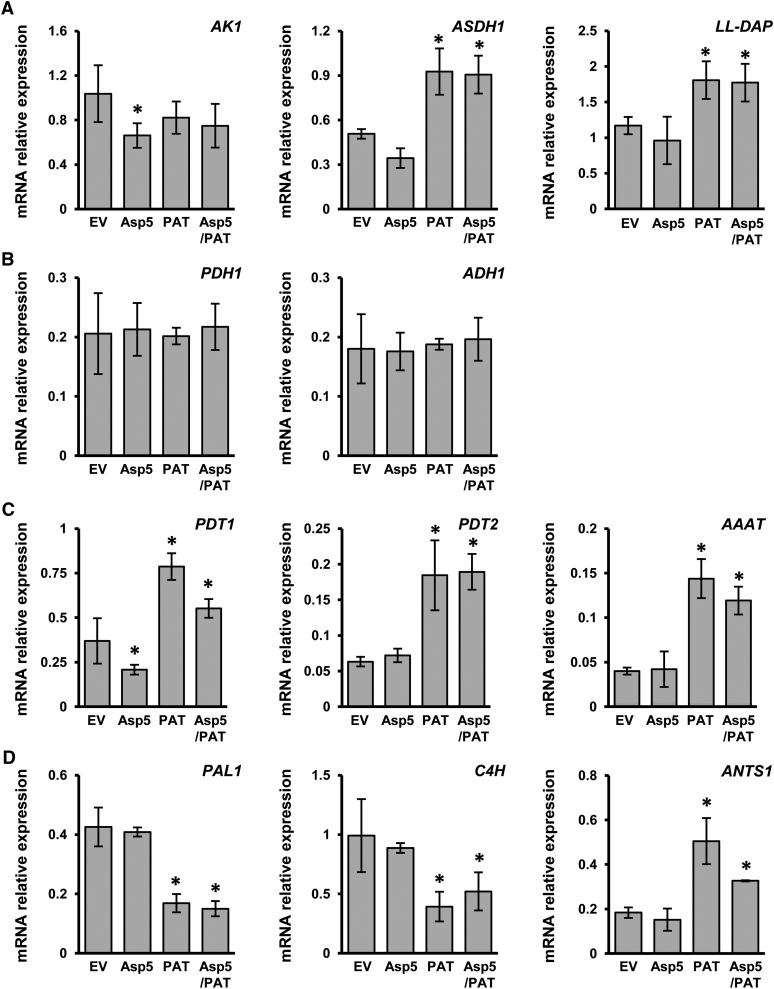
In order to investigate a putative role of NbPAT and/or NbAsp5 in providing Asp for the biosynthetic pathways of Asp-derived amino acids in plastids, we also determined the levels of Lys, Thr, and Ile in the leaves of plants silenced for both aminotransferases (Fig. 6B). Lys levels were dramatically increased in plants silenced for NbAsp5 or NbAsp5/NbPAT when compared with control plants. However, Lys levels in plants silenced for NbPAT were close to those observed in control plants (Fig. 6B). Figure 6B also shows that the levels of Thr and Ile in plants silenced alternately for NbPAT or NbAsp5 were similar to those observed in control plants. Nevertheless, the levels of both amino acids clearly increased (approximately 2-fold for Thr and 3-fold for Ile) in plants cosilenced for both aminotransferases when compared with control plants. To further study the impact of plastidic AAT silencing, the expression levels of genes encoding three key enzymes in the Asp metabolic pathway were determined: Asp kinase (AK1), Asp semialdehyde dehydrogenase (ASDH1), and ll-diaminopimelate aminotransferase (ll-DAP; Azevedo et al., 2006; Hudson et al., 2006). The relative abundance of AK1 transcripts was similar in all samples examined; however, the expression of ASDH1 and ll-DAP significantly increased in NbPAT and NbAsp5/NbPAT silenced plants (Fig. 7A).
Figure 7.
qPCR expression analysis in plants silenced for NbPAT and/or NbAsp5. Total RNA was extracted from N. benthamiana leaves as described. Transcript levels were determined by qPCR. A, NbAK1, NbASDH1, and NbLL-DAP. B, NbPDH1 and NbADH1. C, NbPDT1, NbPDT2, and NbAAAT. D, NbPAL1, NbC4H, and NbANTS1. The expression level for all genes was normalized to that of NbActin2. Values represent means of two assays of real-time qPCR analysis, with four biological replicates in each. Error bars represent se. Asterisks indicate significant differences compared with control plants.
Suppression of PAT Activity in N. benthamiana Leaves Affects Phe Metabolism
A database search analysis demonstrated the existence of a single locus coding for PAT in most plant genomes characterized (www.phytozome.net). Our VIGS-based approach revealed no detectable PAT activity in N. benthamiana leaves silenced for NbPAT, confirming that this enzyme is uniquely responsible for this activity in this tissue. In order to evaluate how the silencing of Asp5 and PAT genes affects the profiles of aromatic amino acids, Phe and Tyr levels were compared in control and silenced plants (Fig. 6C). No significant changes were observed in Phe levels in NbAsp5 silenced plants (Fig. 6C). In contrast, plants silenced for NbPAT, using either TRV-PAT or pTRV-Asp5/PAT, showed significant increases in the levels of Phe of 2- or 8-fold, respectively, compared with control plants. However, no significant alteration in Tyr levels was observed in plants silenced for NbPAT and/or NbAsp5 (Fig. 6C). This finding confirms the previous work of Maeda et al. (2011) reporting that the suppression of PAT in petals of petunia had no effect on the levels of free Tyr.
A putative alternate route for the biosynthesis of Phe could be the dehydration and decarboxylation of prephenate by arogenate/prephenate dehydratase (PDT) followed by transamination of phenylpyruvate to Phe using an aromatic-type aminotransferase. We have identified two full-length cDNAs encoding arogenate/PDTs expressed in N. benthamiana leaves, NbPDT1 and NbPDT2, as well as a full-length sequence for NbPPAT, the homolog of the Arabidopsis gene At5g36160, which encodes the single aminotransferase described so far that is able to catalyze the transamination of phenylpyruvate to Phe (Prabhu and Hudson, 2010). We conducted a set of qPCR measurements for transcripts encoded by NbPDT1, NbPDT2, and NbPPAT in plants silenced for NbPAT and/or NbAsp5. Our results showed a clear increase in the expression levels corresponding to these three genes in plants silenced for NbPAT using either pTRV-PAT or pTRV-Asp5/PAT (Fig. 7B). No significant alteration in expression levels was detected for the same genes in plants silenced only for NbAsp5 (Fig. 7B). Subsequently, we also analyzed the expression levels of two genes encoding the first two enzymes downstream of PAT in channeling Phe toward secondary metabolites: Phe ammonia lyase (NbPAL) and cinnamate 4-hydroxylase (NbC4H). The results indicated that the expression of these genes was clearly reduced in plants silenced for NbPAT (Fig. 7C), consistent with the observed reduction of lignin deposition in leaves and stems observed in plants silenced for NbPAT (Fig. 4). We also analyzed the expression levels of NbANTS1, coding for ANTHRANILATE SYNTHASE1, the first enzyme in the biosynthesis of Trp from chorismate, which showed significant overexpression in plants silenced for NbPAT (Fig. 7C).
Since Tyr levels were not altered significantly by silencing of NbPAT, we also analyzed the expression levels of NbPDH1, the gene encoding for PREPHENATE DEHYDROGENASE1, an enzyme that would be necessary in an alternate Tyr biosynthetic pathway using 4-hydroxyphenylpyruvate as intermediate. The qPCR analysis showed no significant alteration in NbPDH1 expression levels (Fig. 7D). Similarly, no alteration was detected in NbADH1, the gene encoding for AROGENATE DEHYDROGENASE1, the enzyme involved in the generation of Tyr from arogenate.
DISCUSSION
Plastids are subcellular organelles present in nearly all living plant cells and the exclusive site of many important biological processes, such as the major metabolic reactions involved in the biosynthesis of amino acids, including the Asp and aromatic amino acid pathways. In this paper, we analyze, to our knowledge for the first time, the effects on nitrogen metabolism following the suppression of plastidic AAT and PAT activities at the whole-plant level in N. benthamiana using VIGS of NbPAT and NbAsp5 enzymes as an experimental approach. Our results clearly demonstrated that VIGS is an effective method to study the metabolic impact of Asp5 and PAT suppression in planta given the effective gene silencing observed, close to 90% (Fig. 3).
Suppression of AAT Activities in Plastids of N. benthamiana
The analysis of the transcriptome of N. benthamiana allowed us to identify the complete AAT gene family, consisting of five genes encoding eukaryotic-type AAT (NbAsp1–NbAsp5) and a single gene encoding prokaryotic-type AAT (NbPAT). The existence of two AATs within the plastids of N. benthamiana matches the previously described models in Arabidopsis (Schultz et al., 1998) and Pinus pinaster (de la Torre et al., 2007). Despite efforts made by several groups during the past two decades, the role of each member of the AAT gene family has not been sufficiently established. The presence of various isoenzymes located in different subcellular compartments is an argument against a complete functional redundancy and suggests the existence of functional specificities. In Arabidopsis, a role for AtAsp2 was proposed in generating the bulk of Asp in the cytosol under illuminated conditions to be utilized later for the synthesis of Asn in the dark (Miesak and Coruzzi, 2002). Moreover, in the same article, the characterization of transgenic lines in Arabidopsis deficient for the plastidic isoenzyme AtAsp5 was described, without identifying any phenotypic change. Similarly, we found no evident phenotypic changes when NbAsp5 was suppressed in N. benthamiana, with no changes in growth or development, lignin deposition, or pigment contents (Figs. 2, 4, and 5). In this work, we focus on the analysis of plastidial AAT activity in N. benthamiana, considering the contributions of both the NbAsp5 and NbPAT enzymes. Silencing of NbPAT resulted in strong growth reduction and leaf chlorosis (Fig. 2; Supplemental Fig. S3), consistent with a role of the enzyme in the biosynthesis of amino acids needed not only for protein synthesis but also to serve as precursors for a wide range of secondary metabolites. The phenotype observed in plants silenced for NbPAT was similar to that observed in plants silenced for both the NbAsp5 and NbPAT genes (Fig. 2), indicating a minor contribution, if any, of NbAsp5 to the phenotype. In this respect, we observed that the levels of free Glu were not altered in plants individually silenced for NbAsp5 or NbPAT but were greatly increased in plants silenced for both enzymes at the same time (Fig. 6). Concomitantly, a decrease in free-Asp levels was observed in plants showing cosilencing of NbAsp5 and NbPAT, indicating that the bulk of Asp within the leaf is only reduced when none of these plastidic AAT enzymes is active. These data strongly suggest the participation of NbPAT in Asp biosynthesis and further support that cytosolic AAT (NbAsp2) has a limited contribution to the observed levels of Asp. Interestingly, the individual silencing of either NbAsp5 or NbPAT significantly reduced Asn levels even when Asp and Glu levels were not affected. It can be argued that NbAsp2 could be responsible for maintaining Asp levels in NbAsp5 silenced plants with a subsequent reduction in Asn biosynthesis. However, the strong reduction in Asn levels in NbPAT silenced plants is not consistent with this possibility (Fig. 6). Alternately, the above data may reflect a retention of Asp in the plastid to compensate a deficiency in its biosynthesis. Thus, our data suggest that when both AAT enzymes were blocked, Asn biosynthesis in the cytosol from Asp was greatly reduced. Gln levels did not show strong alterations in plants silenced for NbAsp5 and/or NbPAT, which is consistent with a stable flux from available Glu to assimilate ammonium through Gln synthetase activity.
These findings strongly suggest that both enzymes play a partially redundant role in the plastids. Furthermore, these results clearly point out the physiological relevance of the AAT activity housed by NbPAT and could explain why the single suppression of the eukaryotic-type plastidial AAT enzyme in N. benthamiana (this work) or in Arabidopsis (Miesak and Coruzzi, 2002) produced no phenotypic effects or alterations in free Asp levels.
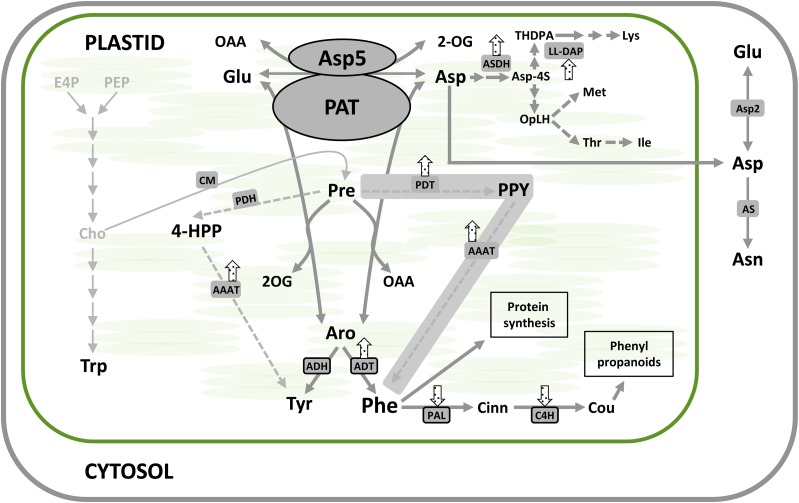
Lys levels clearly increased in plants silenced for NbAsp5 using pTRV-Asp5 or pTRV-Asp5/PAT but remained close to the control in plants silenced only for NbPAT (Fig. 6). This profile was not observed for other amino acids of the Asp metabolic pathway, such as Thr and Ile. These results suggest an essential, and likely exclusive, role of the eukaryotic-type AAT (NbAsp5) in the homeostasis of Lys. Furthermore, our studies regarding the plastidial AAT contribution to the Asp metabolic pathway suggest that the observed phenotype in plants silenced for NbPAT could be explained at least in part by the suppression of its AAT activity and not only by the suppression of its PAT activity. The expression levels observed for key genes in the Asp metabolic pathway are consistent with this assumption (Fig. 7). These results are compatible with a model previously proposed (de la Torre et al., 2009) according to which PAT enzymes could also be involved in the flux toward the Asp-derived pathway (Fig. 8). We cannot rule out that the metabolic effects generated by the suppression of PAT activity could be masking other effects corresponding to the suppression of the AAT activity housed by the same enzyme.
Figure 8.
Schematic representation of major metabolic effects of plastidic AAT and PAT silencing. VIGS resulted in altered amino acid profiles and altered expression of genes encoding enzymes involved in amino acid biosynthesis. Arrows indicate transcriptional up- or down-regulation of the corresponding genes under NbPAT silencing in N. benthamiana leaves. The alternate pathway leading to the biosynthesis of Phe is shadowed in gray. AAAT, Aromatic amino acid aminotransferase; Aro, arogenate; AS, Asn synthetase; ASDH, Asp semialdehyde dehydrogenase; Asp-4S, l-Asp 4-semialdehyde; Cho, chorismate; Cinn, trans-cinnamate; CM, chorismate mutase; Cou, p-coumarate; E4P, d-erythrose 4-phosphate; 4-HPP, 4-hydroxyphenylpyruvate; OAA, oxaloacetate; 2-OG, 2-oxoglutarate; OpLH, o-phospho-l-homoserine; PEP, phosphoenolpyruvate; PPY, phenylpyruvate; Pre, prephenate; THDPA, l-2,3,4,5-tetrahydrodipicolinate. [See online article for color version of this figure.].
The observed changes in nitrogen metabolism and carbohydrate levels in NbAsp5 and NbPAT cosilenced plants strongly suggest a deregulation of carbon/nitrogen balance triggered by suppression of the two plastidic enzymes.
Suppression of PAT Activity in N. benthamiana
PATs are bifunctional enzymes competent to function not only as PATs but also as AATs in Arabidopsis (Graindorge et al., 2010; Maeda et al., 2011), petunia (Maeda et al., 2011), tomato (Solanum lycopersicum; Dal Cin et al., 2011), and P. pinaster (de la Torre et al., 2006). The analysis of recombinant PAT protein indicates that the enzyme exhibits a high affinity for substrates involved in both PAT and AAT activities. The functional analysis performed in N. benthamiana silenced plants demonstrates that the enzyme is also able to display both activities in planta (Fig. 3). However, despite this knowledge of the biochemical characteristics and properties of the enzyme, its full metabolic role has not been fully elucidated. To a large extent, this is due to the lack of a system for efficient gene silencing in whole plants.
The difficulty in obtaining plants suppressed for the PAT enzyme was first supported by the work of Pagnussat et al. (2005), describing the identification of 130 transposon mutants of Arabidopsis with defects in female gametogenesis and embryo development. One of these lines, a mutant defective in the At2g22250 locus encoding PAT, was affected in early embryo development. This phenotype was consistent with the suppression of an essential enzymatic activity required for the biosynthesis of amino acids (de la Torre et al., 2009). Several attempts were carried out in our laboratory to obtain transgenic Arabidopsis plants with suppressed PAT gene expression using a targeted RNAi approach under the control of the constitutive 35S promoter, but all of them proved to be unsuccessful. PAT was locally silenced in petunia petals using an RNAi strategy, and the enzyme was strongly implicated in the biosynthesis of Phe (Maeda et al., 2011). However, analysis of PAT gene suppression in a whole plant is necessary to confirm the role of the enzyme without any masking effect provoked by the transfer of metabolites from other parts of the plant to offset the effect in a local tissue. In this work, the impact of silencing NbPAT was analyzed in a plantlet developmental stage to overcome its essential role during the embryonic phase.
Surprisingly, amino acid quantification registered a moderate increase in Phe levels in plants silenced for NbPAT. The increase in Phe was much more evident in plants silenced simultaneously for NbPAT and NbAsp5. Although silencing of NbPAT was not complete in our working model (close to 90%), these results appear inconsistent with a unique route for the synthesis of Phe in planta. Maeda et al. (2011), in petals of petunia silenced for PAT, observed similar levels of Phe to those measured in the same tissue in control plants. Both lines of evidence suggest the existence of an alternate source of Phe. An alternative pathway for the biosynthesis of Phe using phenylpyruvate as an intermediate has been proposed, a pathway known to exist in most microorganisms (Fig. 1). In this paper, we present a set of results that together indicate the existence of transcriptional reprogramming in plants silenced for NbPAT, resulting, on the one hand, in the activation of genes encoding enzymes whose activities are required in an alternate pathway based on phenylpyruvate as an intermediate (NbADT1, NbADT2, and NbAAAT), and on the other hand, in reduced expression of genes encoding enzymes involved in the subsequent use of Phe toward compounds such as lignins or phenylpropanoids, NbPAL1 and NbC4H (Fig. 8). NbANTS1, a gene encoding the first enzyme in the biosynthesis of Trp (Dubouzet et al., 2007), was also up-regulated in PAT suppressed plants, supporting a putative recycling of chorismate toward Trp in a physiological context with reduced demand for prephenate. These findings further confirm that Phe biosynthesis competes with Trp biosynthesis from their common precursor chorismate (Tzin et al., 2009). Overall, the reduction of PAT activity in these plants seems to be, at least partially, compensated by an alternate biosynthetic pathway and a reduced consumption toward secondary metabolism, probably ensuring a free-Phe reserve for the maintenance of basic functions such as protein synthesis. According to this model, reduced accumulation of lignin in the stems of plants silenced for NbPAT (Fig. 4) would be a direct consequence of a prioritized use of the Phe pool toward protein biosynthesis (Fig. 8). According to our model, the prephenate/arogenate pathway would be predominant in the biosynthesis of Phe, although the plants can use a functional alternate pathway using phenylpyruvate as an intermediate.
In summary, the roles of NbAsp5 and NbPAT were investigated in N. benthamiana using a VIGS approach. Phenotypic and metabolic analyses were conducted in silenced plants to investigate the specific roles of these enzymes in the biosynthesis of essential amino acids within the plastids. Our results indicate that NbPAT has an overlapping role with NbAsp5 in the biosynthesis of Asp and a key role in the production of Phe for the biosynthesis of phenylpropanoids.
MATERIALS AND METHODS
cDNA Cloning
Full-length NbPAT and NbAsp5 cDNAs were isolated from total RNA extracted from Nicotiana benthamiana leaves by reverse transcription-PCR using iScript Reverse Transcription Supermix (Bio-Rad) with NbPAT-1/NbPAT-2 and NbAsp5-1/NbAsp5-2, respectively (Supplemental Table S1). cDNA fragments were subcloned into the pJET1.2 vector and completely sequenced.
VIGS
cDNA fragments Nb5′PAT, NbMPAT, Nb3′PAT, NbAsp5, and Nb3′Asp-5 (Supplemental Fig. S1) were amplified using specific Gateway attb adapter primers (Nb5′PAT-F/Nb5′PAT-R, NbMPAT-F/NbMPAT-R, Nb3′PAT-F/Nb3′PAT-R, NbAsp5-F/NbAsp5-R, and Nb3′Asp-5-F/Nb3′Asp-5-R; Supplemental Table S1), recombined into pDONR207 (Invitrogen) and then cloned into the VIGS Gateway-adapted destination vector pTRVGW. Constructs were confirmed by sequencing using a CEQ 8000 (Beckman Coulter España). Subsequently, Agrobacterium tumefaciens strain C58C1 was transformed by electroporation with the new constructs. For silencing in N. benthamiana, seedlings with two emerging leaflets (10 d old) were syringe infiltrated with cultures containing pTRVGW-derived constructs mixed with cultures containing pTRV1, both with an optical density at 600 nm of 0.1, according to procedures described previously (Liu et al., 2002). Silencing of the corresponding genes was assessed by qPCR using specific primers (Supplemental Table S2). Plants were harvested for analyses 4 weeks after VIGS induction. Leaves and stems from silenced plants were immediately frozen in liquid nitrogen, ground in a mortar with pestle, and the resulting powder was stored at −80°C until use.
Expression Analysis
Total RNA was isolated following the protocol described by Liao et al. (2004), with minor modifications. The RNA samples were treated with RQ1 RNase-free DNase (Promega) to eliminate traces of genomic DNA, and cDNA synthesis was performed as described previously (Canales et al., 2012). qPCR was performed on a CFX-384 Real Time System (Bio-Rad) with SsoFast EvaGreen Supermix (Bio-Rad) under the following conditions: 95°C for 3 min (one cycle), and then 95°C for 1 s and 60°C for 5 s (45 cycles). After the final cycle, a melting curve analysis was performed over a temperature range of 65°C to 95°C in 0.5°C increments to verify the reaction specificity. cDNAs corresponding to 20 ng of reverse transcribed RNA were used as a template for each reaction (Supplemental Table S2).
The raw fluorescence data from each reaction were fitted to the Mass Action Kinetic2 model, which requires no assumptions about the amplification efficiency of a qPCR assay (Boggy and Woolf, 2010). The initial target concentrations (D0 parameter) for each gene were deduced from the Mass Action Kinetic2 model using the qpcR package for the R environment (Ritz and Spiess, 2008) and normalized to NbActin2.
In Gel AAT/PAT Activity
N. benthamiana tissues were frozen in liquid nitrogen, ground in a mortar and pestle, and the resulting powder was transferred into a tube containing extraction buffer consisting of 50 mm Tris-Cl, pH 7.5, 10% (v/v) glycerol, and 0.1% (v/v) Triton X-100. The samples were then subjected to native PAGE in a discontinuous system with a 4% (w/v) stacking buffer (29:1 acrylamide:bisacrylamide and 125 mm Tris-Cl, pH 6.8) and a 7% (w/v) separating buffer (29:1 acrylamide:bisacrylamide and 375 mm Tris-Cl, pH 8.8) in a Mini-Protean Tetra Cell module (Bio-Rad). Staining for AAT activity was carried out using Fast Blue BB salt (Sigma), which turns blue in the presence of oxaloacetate, as described previously (Wendel and Weeden, 1989; de la Torre et al., 2009). Using similar procedures, oxaloacetate generated through PAT activity was also detected on native gels.
Determination of Photosynthetic Pigments
Chlorophylls a and b were extracted with 100% methanol from leaf discs of plants grown for 6 weeks under greenhouse conditions, and the content was determined according to Lichtenthaler and Wellburn (1983).
Lignin Staining and Quantification
Cell wall preparation and lignin quantification were performed following the method described by Lange et al. (1995). Briefly, tubes containing about 200 mg of frozen samples were treated with 1 mL of methanol, shaken for 1 h, and then centrifuged at 7,500g for 5 min. The supernatant was removed and the pellet treated successively with 1.5 mL of the following solvents: methanol, 1 m NaCl, 1% (w/v) SDS, deionized water, and chloroform:methanol (1:1). Finally, the pellet was resuspended in 1.5 mL of tert-butyl methyl ether and lyophilized. Ten milligrams of the resulting powder was resuspended in 1 mL of 2 m HCl and 0.2 mL of thioglycolic acid and incubated at 95°C for 4 h. The solution was centrifuged at 18,000g for 10 min, and the resulting pellet was washed three times with deionized water, resuspended in 1.5 mL of 0.5 m NaOH, and shaken for 12 h. The samples were then centrifuged at 18,000g for 20 min, and the supernatant was mixed with 0.3 mL of 12 m HCl and incubated for 4 h at 4°C. After centrifugation for 40 min, the resulting pellets were dried using a speed vacuum concentrator, resuspended in 1 mL of 0.5 m NaOH, and the A280 was determined and compared with a previously determined lignin standard made with pure lignin from Sigma-Aldrich. Histochemical analysis of the accumulated lignin was performed using phloroglucinol staining of hand-cut sections of N. benthamiana stems. Sections were incubated for 10 min in a 1% phloroglucinol solution in ethanol:HCl (2:1) and visualized by light microscopy using a Nikon Eclipse E 800 microscope.
Free Amino Acid Analysis
Frozen plant material (100 mg) was ground in a mortar and extracted in 50 mm Tris-HCl, pH 8, 2 mm EDTA, and 10 mm 2-mercaptoethanol. The extract was centrifuged at 20,000g at 4°C, and the supernatant was recovered and centrifuged again as above. An aliquot of 400 μL was transferred to a fresh tube, to which 1 mL of methanol was added and mixed for 10 min at 4°C. After centrifugation as above, the supernatant was saved in a fresh tube and the pellet was resuspended in 200 μL of methanol:water (4:1, v/v), stirred for 10 min at 4°C, and centrifuged, and the supernatant was reserved. The last step was repeated twice, and all the reserved supernatants were combined. The volume was reduced to 200 μL by evaporation at 90°C in an oven. Finally, the extract was filtered through a 0.2-μm pore filter.
To determine the free amino acid pool content of the samples, amino acids were separated with no derivatization with a System Gold HPLC BioEssential high-performance liquid chromatograph (Beckman-Coulter) using a lithium citrate buffer system, followed by a postcolumn ninhydrin reaction detection system. For the identification and quantification of amino acids, the corresponding standards were used.
Carbohydrate Analysis
Carbohydrates were extracted from frozen powder corresponding to N. benthamiana leaves with 80% ethanol at 80°C for 30 min, followed by further washing with 50% ethanol at 80°C for 20 min and two additional washes with water. Combined supernatants were centrifuged to remove debris and lyophilized. The resulting powder was resuspended in water, and Suc, Glc, and Fru were measured enzymatically following the reduction of NADP at 340 nm after successive additions of the coupling enzymes Glc-6-P dehydrogenase (4 units mL−1), hexokinase (10 units mL−1), phosphoglucose isomerase (5 units mL−1), and invertase (Sekin, 1978). Starch was measured as Glc from the extracted pellet, following incubation at 37°C for 4 h with α-amylase (4 units mL−1) and amyloglucosidase (8 units mL−1). All enzymes were obtained from Roche Diagnostics. Cellulose content was determined by the anthrone method (Updegraff, 1969). Briefly, 100 mg of tissue powder was boiled in 1 mL acetic-nitric reagent (acetic acid:nitric acid:water, 8:1:2) for 30 min to remove lignin and hemicellulosic carbohydrates. After centrifugation at 4,500g for 20 min, the supernatant was removed and the remaining material was washed twice with 1 mL of distilled water. The cellulose samples were then hydrolyzed in 67% (v/v) sulfuric acid for 1 h at 25°C, and the Glc content of the samples was determined as follows: 5 μL of the sulfuric acid-hydrolyzed samples was mixed with 495 μL of water and 1 mL of 0.2% (w/v) anthrone in concentrated sulfuric acid on ice. The samples were boiled for 10 min, and then the absorbance was measured as optical density at 630 nm. Cellulose content was determined based on a previously determined standard using commercial cellulose from Sigma-Aldrich.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Open reading frame cloning of NbPAT, NbAsp5, and derivative VIGS constructs.
Supplemental Figure S2. Phenotypes of N. benthamiana leaves subjected to VIGS of NbPDS.
Supplemental Figure S3. Phenotypes of N. benthamiana leaves subjected to VIGS of NbPAT and/or NbAsp5 genes.
Supplemental Figure S4. Fresh weight-to-dry weight ratio in N. benthamiana plants silenced for NbPAT and/or NbAsp5.
Supplemental Figure S5. Expression analysis of the AAT gene family in plants silenced for NbPAT and/or NbAsp5.
Supplemental Figure S6. Analysis of carbohydrate content in N. benthamiana plants silenced for NbPAT and/or NbAsp5.
Supplemental Table S1. Amino acid sequence identity between Arabidopsis and N. benthamiana AAT.
Supplemental Table S2. Oligonucleotides used in this work.
Acknowledgments
We thank Savithramma P. Dinesh-Kumar (University of California [Davis]) and Dr. Olga del Pozo (Instituto Bioquímica Vegetal y Fotosíntesis, Consejo Superior de Investigaciones Científicas) for the pTRVGW silencing vector. We also thank Javier Canales (Universidad de Málaga) for technical advice with qPCR analysis.
Glossary
- AAT
aspartate aminotransferase
- RNAi
RNA interference
- VIGS
virus-induced gene silencing
- cDNA
complementary DNA
- qPCR
quantitative PCR
References
- Azevedo RA, Lancien M, Lea PJ. (2006) The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids 30: 143–162 [DOI] [PubMed] [Google Scholar]
- Boggy GJ, Woolf PJ. (2010) A mechanistic model of PCR for accurate quantification of quantitative PCR data. PLoS ONE 5: e12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauc S, De Vooght E, Claeys M, Höfte M, Angenon G. (2011) Influence of over-expression of cytosolic aspartate aminotransferase on amino acid metabolism and defence responses against Botrytis cinerea infection in Arabidopsis thaliana. J Plant Physiol 168: 1813–1819 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL (2000) Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD [Google Scholar]
- Canales J, Rueda-López M, Craven-Bartle B, Ávila C, Cánovas FM. (2012) Novel insights into regulation of asparagine synthetase in conifers. Front Plant Sci 3: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven-Bartle B, Pascual MB, Cánovas FM, Avila C. (2013) A Myb transcription factor regulates genes of the phenylalanine pathway in maritime pine. Plant J 74: 755–766 [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RC, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, et al. (2011) Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell 23: 2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre F, De Santis L, Suárez MF, Crespillo R, Cánovas FM. (2006) Identification and functional analysis of a prokaryotic-type aspartate aminotransferase: implications for plant amino acid metabolism. Plant J 46: 414–425 [DOI] [PubMed] [Google Scholar]
- de la Torre F, Moya-García AA, Suárez MF, Rodríguez-Caso C, Cañas RA, Sánchez-Jiménez F, Cánovas FM. (2009) Molecular modeling and site-directed mutagenesis reveal essential residues for catalysis in a prokaryote-type aspartate aminotransferase. Plant Physiol 149: 1648–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Torre F, Suárez MF, De Santis L, Cánovas FM. (2007) The aspartate aminotransferase family in conifers: biochemical analysis of a prokaryotic-type enzyme from maritime pine. Tree Physiol 27: 1283–1291 [DOI] [PubMed] [Google Scholar]
- Dubouzet JG, Ishihara A, Matsuda F, Miyagawa H, Iwata H, Wakasa K. (2007) Integrated metabolomic and transcriptomic analyses of high-tryptophan rice expressing a mutant anthranilate synthase alpha subunit. J Exp Bot 58: 3309–3321 [DOI] [PubMed] [Google Scholar]
- Forde BG, Lea PJ. (2007) Glutamate in plants: metabolism, regulation, and signalling. J Exp Bot 58: 2339–2358 [DOI] [PubMed] [Google Scholar]
- Galili G. (2011) The aspartate-family pathway of plants: linking production of essential amino acids with energy and stress regulation. Plant Signal Behav 6: 192–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graindorge M, Giustini C, Jacomin AC, Kraut A, Curien G, Matringe M. (2010) Identification of a plant gene encoding glutamate/aspartate-prephenate aminotransferase: the last homeless enzyme of aromatic amino acids biosynthesis. FEBS Lett 584: 4357–4360 [DOI] [PubMed] [Google Scholar]
- Hudson AO, Singh BK, Leustek T, Gilvarg C. (2006) An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol 140: 292–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland RJ, Joy KW (1985) Plant transaminases. In P Christen, DE Metzler, eds, Transaminases, Vol 2. Wiley, New York, pp 376–384 [Google Scholar]
- Jander G, Joshi V. (2009) Aspartate-derived amino acid biosynthesis in Arabidopsis thaliana. The Arabidopsis Book 7: e0121, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange BM, Lapierre C, Sandermann H., Jr (1995) Elicitor-induced spruce stress lignin: structural similarity to early developmental lignins. Plant Physiol 108: 1277–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea PJ, Azevedo RA (2003) Primary products: plant amino acids. In B Thomas, SJ Murphy, BG Murray, eds, Encyclopaedia of Applied Plant Sciences, Vol 3. Elsevier, Amsterdam, pp 871–883 [Google Scholar]
- Lea PJ, Sodek L, Parry MAJ, Shewry PR, Halford NG. (2007) Asparagine in plants. Ann Appl Biol 150: 1–26 [Google Scholar]
- Liao Z, Chen M, Guo L, Gong Y, Tang F, Sun X, Tang K. (2004) Rapid isolation of high-quality total RNA from taxus and ginkgo. Prep Biochem Biotechnol 34: 209–214 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR. (1983) Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 603: 591–592 [Google Scholar]
- Liepman AH, Olsen LJ. (2004) Genomic analysis of aminotransferases in Arabidopsis thaliana. Crit Rev Plant Sci 23: 73–89 [Google Scholar]
- Liu Y, Schiff M, Dinesh-Kumar SP. (2002) Virus-induced gene silencing in tomato. Plant J 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Maeda H, Dudareva N. (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Maeda H, Shasany AK, Schnepp J, Orlova I, Taguchi G, Cooper BR, Rhodes D, Pichersky E, Dudareva N. (2010) RNAi suppression of Arogenate Dehydratase1 reveals that phenylalanine is synthesized predominantly via the arogenate pathway in petunia petals. Plant Cell 22: 832–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Yoo H, Dudareva N. (2011) Prephenate aminotransferase directs plant phenylalanine biosynthesis via arogenate. Nat Chem Biol 7: 19–21 [DOI] [PubMed] [Google Scholar]
- Miesak BH, Coruzzi GM. (2002) Molecular and physiological analysis of Arabidopsis mutants defective in cytosolic or chloroplastic aspartate aminotransferase. Plant Physiol 129: 650–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V. (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Prabhu PR, Hudson AO. (2010) Identification and partial characterization of an L-tyrosine aminotransferase (TAT) from Arabidopsis thaliana. Biochem Res Int 2010: 549572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippert P, Puyaubert J, Grisollet D, Derrier L, Matringe M. (2009) Tyrosine and phenylalanine are synthesized within the plastids in Arabidopsis. Plant Physiol 149: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz C, Spiess AN. (2008) qpcR: an R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis. Bioinformatics 24: 1549–1551 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Coruzzi GM. (1995) The aspartate aminotransferase gene family of Arabidopsis encodes isoenzymes localized to three distinct subcellular compartments. Plant J 7: 61–75 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Hsu M, Miesak B, Coruzzi GM. (1998) Arabidopsis mutants define an in vivo role for isoenzymes of aspartate aminotransferase in plant nitrogen assimilation. Genetics 149: 491–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekin S. (1978) Enzymatic determination of glucose, fructose and sucrose in tobacco. Tobacco Science 23: 75–77 [Google Scholar]
- Tzin V, Galili G. (2010) New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol Plant 3: 956–972 [DOI] [PubMed] [Google Scholar]
- Tzin V, Malitsky S, Aharoni A, Galili G. (2009) Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in Arabidopsis. Plant J 60: 156–167 [DOI] [PubMed] [Google Scholar]
- Updegraff DM. (1969) Semimicro determination of cellulose in biological materials. Anal Biochem 32: 420–424 [DOI] [PubMed] [Google Scholar]
- Wendel JF, Weeden NF (1989) Visualization and interpretation of plant isoenzymes. In DE Soltis, PS Soltis, eds, Isoenzymes in Plant Biology. Dioscorides Press, Portland, OR, pp 5–45 [Google Scholar]
- Wilkie SE, Warren MJ. (1998) Recombinant expression, purification, and characterization of three isoenzymes of aspartate aminotransferase from Arabidopsis thaliana. Protein Expr Purif 12: 381–389 [DOI] [PubMed] [Google Scholar]