Abstract
Many central vascular catheters (CVCs) are removed unnecessarily because current diagnostic methods for CVC-associated infection are unreliable. A quantitative PCR assay using primers and probe targeted to bacterial 16S ribosomal DNA was used to measure the levels of bacterial DNA in blood samples drawn through the CVC in a population of patients receiving intravenous nutrition. Bacterial DNA concentrations were raised in 16 of 16 blood samples taken during episodes of probable bacterial CVC-associated infection. Bacterial DNA concentrations were raised in 4 of 29 episodes in which bacterial CVC-associated infection was unlikely. The use of this technique has the potential to substantially reduce the unnecessary removal of CVCs.
In many patient populations, central vascular catheters (CVCs) are routinely used for the administration of drugs, fluids, or intravenous feeding solutions. CVCs are a major risk factor for bloodstream infections. The majority of hospital-acquired bloodstream infections are associated with the use of a CVC (3). These infections are associated with considerable costs for patients and health providers.
CVC infection is caused predominantly by bacteria, particularly staphylococci, with a small proportion of infections being attributable to fungi. Staphylococcus epidermidis is the most common cause of CVC-associated bacteremia. CVC infections caused by S. epidermidis can often be effectively treated with antibiotics infused through or locked in to the CVC (4, 15). Many of the other bacterial and fungal causes of infection respond poorly to antibiotic therapy alone, and patient outcome is improved by removal of the CVC.
Traditional methods of diagnosis of CVC-associated infection rely on clinical features and quantitative microbiology of blood samples (15) or on methods that can only be applied following CVC removal. The clinical features of CVC infection may be nonspecific, particularly in patients undergoing intensive care or in patients who are immunocompromised. A number of novel methods for the diagnosis of CVC infection have been proposed (1, 2, 4, 7-10). These methods have not been widely adopted, either because of poor performance in some groups of patients, such as those on antibiotics, or because of a requirement for invasive sampling of the CVC. Blot et al. (2) described a method that compared the differential times to positivity, as determined by a continuous automated blood culture monitoring system which compares the growth rates of organisms in blood collected from the catheter and a peripheral vein. Another method involves culture from the terminal 4-cm segment of the cannula, which is rolled over the entire surface of an agar plate five times, and counting of the resultant colonies (9, 12). However, interpreting the significance of the resultant growth may be problematic (9). In addition, quantitative microbiology is unreliable for patients exposed to antibiotics, so even with appropriate use of diagnostic methods there often remains considerable diagnostic uncertainty. As a result of this uncertainty, infection involving the CVC is confirmed for fewer than 25% of CVCs removed because of suspected infection (4).
We report an evaluation of a novel approach based on the quantitative detection of bacterial DNA in blood samples drawn through the CVC.
MATERIALS AND METHODS
Patients and samples.
A population of patients undergoing intravenous feeding were selected for study. These patients are not routinely treated with prophylactic antibiotics (which might compromise culture-based diagnostic methods) and have relatively few risk factors for disseminated bacterial infection other than the CVC. Samples collected from 46 patient episodes (13 patients) were analyzed. These 46 patient episodes included 16 episodes of probable bacterial CVC infection and one fungal CVC-associated infection. There were 52 CVC samples in total because in six episodes samples were collected from a second lumen of a double-lumen CVC. There were 28 paired blood samples (where blood was taken through the CVC and also from a peripheral vessel). Blood cultures were not collected from four episodes. For all but three patients, samples were taken when patients presented at the out-patient clinic for review (with or without signs or symptoms compatible with CVC-associated infection). Patients with signs or symptoms compatible with CVC-associated infection were reviewed within 24 h whenever possible. Ethical Approval for the study was obtained from the East London and City Health Authority Research Ethics Committee.
Four-milliliter blood samples were collected into EDTA anticoagulation tubes (Becton-Dickinson, Oxford, United Kingdom) directly from the CVC and also, when acceptable to the patient, from a peripheral vessel by aseptic technique. These samples were sent through the routine sample transport system (at ambient temperature) to the laboratory and stored in aliquots at −70°C. Sample analysis was performed by laboratory staff with no knowledge of the patient condition. Samples excluded from this analysis were those that were >72 h old on arrival in the laboratory, those collected within 14 days before or after the day of onset of an episode of probable CVC infection (those collected on the day of onset were included), and those collected while the patient was on intravenous antibiotic therapy.
Criteria for classifying episodes as probable CVC-associated infection.
In this evaluation, episodes were classified as fulfilling the criteria of probable CVC-associated infection or not (14). Probable CVC-associated infections were defined as those episodes with (i) two or more blood culture sets collected within a 72-h period positive for coagulase-negative staphylococci (category A); (ii) a positive blood culture in a patient with signs or symptoms of infection, a CVC in place, and without another potential source of infection and whose clinical condition improves in response to appropriate treatment for CVC infection (intravenous antibiotic treatment and/or CVC removal) (category B); (iii) a positive blood culture in a patient with signs or symptoms of infection and with an isolate indistinguishable from the CVC tip culture (category C); (iv) fever, chills, and/or hypotension associated with CVC manipulations and responding to treatment appropriate for CVC-associated infection (intravenous antibiotic treatment and/or CVC removal) (category D); or (v) inflammation extending beyond the subcutaneous cuff of the CVC in a patient with systemic signs or symptoms of infection (category E).
Development and evaluation of quantitative 16S rDNA detection methods. (i) Primer and probe design.
A total of 119 16S ribosomal DNA (rDNA) sequences comprising 56 species (Table 1) were retrieved from GenBank and aligned by using BioEdit (5), and highly conserved regions were identified. Possible primers and probes were selected and then validated by using the Primer Express Software provided by Applied Biosystems. We were unable to identify oligonucleotides with appropriate melting temperatures (Tms) for the TaqMan assay by using standard 6-carboxytetramethylrhodamine (TAMRA)-quenched probes; this was resolved by the use of a probe which contained a minor groove binding nonfluorescent quencher acting as a Tm enhancer, which permitted the use of a 17-bp probe targeted to a highly conserved region of the bacterial 16S gene family. The chosen primers and probes did not show any likely cross hybridization with human genome sequences. The performance of these primers and probes was compared to those of two other recently published sets of primers and probes (Table 2).
TABLE 1.
Organisms used in the design of 16S rRNA gene sequences and their GenBank accession numbers
| Organism | Accession no. |
|---|---|
| Staphylococcus aureus | X68417, AF146368, AF076030, D83353, D83355, D83357 |
| Staphylococcus capitis | AY030321 |
| Staphylococcus epidermidis | AY030342, D83363, D83362, X75944 |
| Staphylococcus auricularis | D83358 |
| Staphylococcus cohnii | AB009936, D83361 |
| Staphylococcus hominis | AY030318, X66101, L37601 |
| Staphylococcus simulans | D83373 |
| Staphylococcus saprophyticus | D83371, Z26902, L37596, L20250 |
| Staphylococcus arlettae | AB009933 |
| Staphylococcus condimenti | Y15750 |
| Staphylococcus delphini | AB009938 |
| Staphylococcus felis | D83364 |
| Staphylococcus gallinarum | D83366 |
| Staphylococcus intermedius | AF193882 |
| Staphylococcus kloosii | AB009940 |
| Staphylococcus lentus | D83370 |
| Staphylococcus linens | AF527483 |
| Streptococcus pneumoniae | X58312, AF003930 |
| Streptococcus agalactiae | AB023574 |
| Streptococcus sp. group G | AB002517 |
| Streptococcus milleri | U02917 |
| Streptococcus mitis | AY005045 |
| Streptococcus mutans | AF139603 |
| Streptococcus pyogenes | AB023575 |
| Streptococcus salivarius | U02923 |
| Streptococcus sanguinis | AF003928 |
| Bacillus subtilis | AB065370 |
| Lactobacillus gasseri | AF243165, AF243144 |
| Lactobacillus jensenii | AF243176, AF243153, AF243143 |
| Enterococcus faecium | AB018210, AF039901, AF070223, AJ276355, AJ291731, AJ291732, AJ301830, AJ420800, AY057055 |
| Nocardia otitidiscaviarum | X80611 |
| Kitasatospora setae | AB022868 |
| Pseudomonas aeruginosa | AF094720 |
| Stenotrophomonas maltophilia | AJ293474, AJ293469 |
| Acinetobacter baumannii | X81660 |
| Acinetobacter lwoffii | U10875 |
| Helicobacter pylori | AF302106, AF348617, AF363064, AF512997, AF535196, AF535197, AF535198, AY022898, AY062899, U00679, U01328, U01329, U01330, U01331, U01332 |
| Helicobacter rodentium | U96296, U96297 |
| Salmonella enterica serovar Typhi | U88545, Z47544 |
| Salmonella enterica serovar Paratyphi | X80682 |
| Salmonella enterica serovar Enteritidis | U90318 |
| Klebsiella pneumoniae | AB004753, AF130981, AF130982, AF130983, AF228918, AF228919, AJ23342, Y17654, Y17656, Y17657, X87276, X93214, |
| Klebsiella oxytoca | AB004754, AF129440, U78183, U78184, Y17655, Y17667, |
| Enterobacter cloacae | AJ251469 |
| Enterobacter aerogenes | AJ251468 |
| Escherichia coli | AB035920 |
| Yersinia pestis | Z75317 |
| Yersinia entercolitica | AF366378 |
| Yersinia pseudotuberculosis | AF366375 |
| Shigella dysenteriae | X96966, X80680 |
| Shigella sonnei | X96964 |
| Shigella boydii | X94965 |
| Shigella flexneri | X96963 |
| Haemophilus influenzae | AF224305, AF224306, AF224308, AF224309, |
| Haemophilus haemolyticus | M75045 |
| Haemophilus aegypticus | M75044 |
TABLE 2.
Primers and probes used in this study
| Reference | Primer or probe (name) | Sequence (5′→3′) | Tm(s)a (°C) | Position (bp)b | Fluorophore and quencherc | Amplicon length (bp) |
|---|---|---|---|---|---|---|
| This study | Forward primer (CVCf) | GGGCTACACACGTRCTACAATG | 59.2, 53.7 | 1220-1241 | 167 | |
| Reverse primer (CVCr) | CCGGGAACGTATTCACCG | 58 | 1386-1369 | |||
| Probe (CVC-P) | CGATTACTAGCGATTCC | 68.2 | 1353-1337 | 5′ VIC, 3′ MGBnfq | ||
| 17 | Forward primer (P891F) | TGGAGCATGTGGTTTAATTCGA | 59.1 | 943-964 | 159 | |
| Reverse primer (P1033R) | TGCGGGACTTAACCCAACA | 58.6 | 1101-1083 | |||
| Probe (UniProbe) | CACGAGCTGACGACARCCATGCA | 69.5, 67.3 | 1074-1052 | 5′ TET (original study used VIC), 3′ TAMRA | ||
| 13 | Forward primer (Nf) | TCCTACGGGAGGCAGCAGT | 59.4 | 340-358 | 467 | |
| Reverse primer (Nr) | GGACTACCAGGGTATCTAATCCTGTT | 58.1 | 806-781 | |||
| Probe (N-P) | CGTATTACCGCGGCTGCTGGCAC | 69.9 | 537-515 | 5′ 6-FAM, 3′ TAMRA |
Tms were calculated with Primer Express software (Applied Biosystems).
Numbered according to the E. coli O157:H7 rrsA gene (GenBank accession number AB035920).
MGBnfq, minor groove binder nonfluorescent quencher (ABI); TET, tetrachloro-6-carboxyfluorescein; 6-FAM, 6-carboxyfluorescein.
(ii) Assessment of 16S rDNA probes and primers with bacterial DNA standards.
The ability of the primers and probes to detect 10 pg of purified DNA from the following species was determined: Escherichia coli (National Collection of Type Cultures [NCTC] 11560), Pseudomonas aeruginosa (NCTC 10662), Stenotrophomonas maltophilia (clinical isolate), Enterococcus faecalis (ATCC 29212), Streptococcus agalactiae group B streptococcus control (clinical isolate), Haemophilus influenzae (NCTC 11315), Moraxella (Branhamella) catarrhalis (NCTC 11020) Streptococcus pneumoniae (ATCC 49619), Streptococcus oralis (NCTC 11427), Streptococcus salivarius (NCTC 8618), Streptococcus mitis (NCTC 12261), Streptococcus sanguis (NCTC 7863), Staphylococcus aureus (NCTC 6571), Staphylococcus haemolyticus (clinical isolate), Staphylococcus warneri (clinical isolate), Staphylococcus hominis (clinical isolate), Staphylococcus epidermidis (clinical isolate), Staphylococcus capitis (clinical isolate), Microbacterium oxydans (clinical isolate), Acinetobacter baumanii (clinical isolate), two Klebsiella spp. (clinical isolates), Klebsiella pneumoniae (clinical isolate), Enterobacter aerogenes (clinical isolate), Enterobacter cloacae (clinical isolate), Corynebacterium diphtheriae (NCTC 10356), Corynebacterium xerosis (NCTC 11861), Serratia marcescens (clinical isolate), Proteus mirabilis (clinical isolate), and Lactobacillus rhamnosus (clinical isolate). All organisms except C. diphtheriae, C. xerosis, and M. oxydans were detected.
(iii) Results of initial primer and probe development tests.
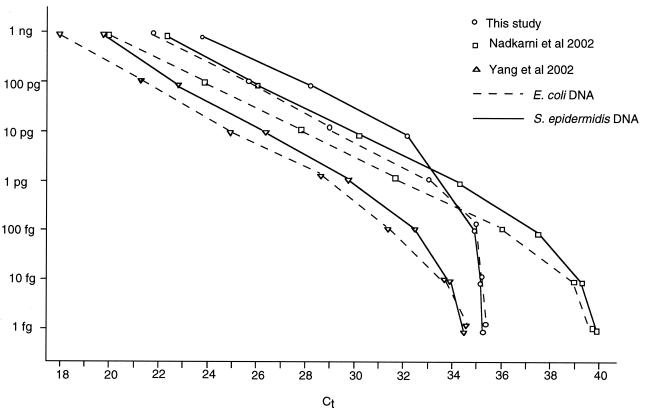
A comparison of the results obtained for the three quantitative DNA detection methods is shown in Fig. 1. For reasons of clarity, typical results for two organisms only, S. epidermidis and E. coli, are shown. With these bacteria, all three methods gave an approximately linear response with amounts of bacterial DNA down to about 1 pg. Below this level, the primers and probes designed by Nadkarni et al. (13) continued to give a linear response, which was maintained to below 100 fg. In preliminary experiments, the ability to detect DNA at amounts down to 1.0 pg was found to be critical in distinguishing probable CVC-associated infection from unlikely CVC-associated infection (M. R. Millar, S. Warwick, M. Wilks, J. Leeming, and McCarthy, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1883, 2003). The method of Nadkarni et al. was therefore used in this study.
FIG. 1.
Relationship between the apparent amounts of S. epidermidis and E. coli DNAs and the median Ct with three different sets of 16S rDNA primers and probes. The lowest Ct values indicate the highest concentrations of bacterial DNA.
(iv) DNA extraction from clinical and control samples.
DNA was extracted from 200-μl aliquots of EDTA-anticoagulated whole blood. The sample was mixed with 1,200 μl of freshly prepared 0.17 M ammonium chloride and incubated at room temperature for 30 min. Following centrifugation at 11,600 × g for 10 min, the pellet was washed twice with 500 μl of sterile saline (0.9%, wt/vol). By using the QIAamp DNA minikit (Qiagen), the pellet was resuspended in 180 μl of Qiagen ATL buffer (containing EDTA and sodium dodecyl sulfate) and exposed to six freeze-thaw cycles (cycling between −70°C and 50°C), with vortexing between, before being heated in a boiling water bath for 10 min. The remainder of the extraction procedure was performed according to the manufacturer's protocol. DNA was eluted in 50 μl of buffer following a 5-min incubation. Extracts were stored at −20°C before analysis.
A number of controls were routinely run with each batch of tests. These included blood samples from a healthy individual with and without spiking with bacteria. An extraction control of blood spiked with 10 CFU of S. epidermidis/μl was found to yield DNA levels close to the lower limit of detection. Bacterial DNA controls of known amounts (100 pg to 100 fg) and a negative control (with template DNA omitted to detect reagent contamination) were also included in each run.
(v) PCR conditions (TaqMan assay).
Real-time PCRs were carried out by using the ABI PRISM 7900HT sequence detection System (Applied Biosystems, Cheshire, United Kingdom) in optical 384-well plates. Ultrafiltration of reagents, as suggested by Yang et al. (17), was not performed. Reactions mixtures contained (1×) TaqMan universal PCR master mix (Applied Biosystems), a 300 nM concentration each of the forward and reverse primers, a 100 nM concentration of the fluorescent probe, 2 μl of template DNA, and water to give a final volume of 20 μl. The cycling conditions were 50°C for 2 min, 95°C for 10 min, and then 40 cycles of 95°C for 15 s and 60°C for 1 min.
The threshold cycle (Ct) value, which is inversely proportional to the log of the amount of target DNA initially present, was calculated by using SDS software version 2.0 (Applied Biosystems). All samples were run in triplicate.
Statistical methods.
The median Ct value from each triplicate was used in the analysis. The data were modeled in order to estimate detection rates, false-positive rates, and potential reductions in unnecessary removals of CVCs. The area under the curve and its 95% confidence interval were calculated by using Roctab in Stata (16).
RESULTS
Staphylococci were the predominant bacteria associated with episodes of probable bacterial CVC infection. The bacteria associated with CVC infection were coagulase-negative staphylococci, S. aureus, K. pneumoniae, S. maltophilia, and P. aeruginosa. The 16 episodes of probable bacterial CVC infection included two episodes of extensive tunnel infection with systemic signs and symptoms of infection (category E) from the exit site, of which S. aureus and P. aeruginosa were isolated. All of the other 14 episodes were associated with positive blood cultures and fell in to category A or B. The organisms isolated were coagulase-negative staphylococci (10 cases, including 1 mixed with S. maltophilia), K. pneumoniae (2 cases), E. cloacae (1 case), and S. aureus (1 case).
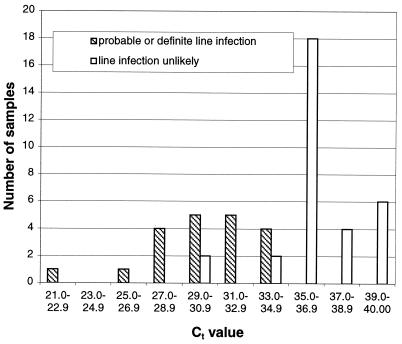
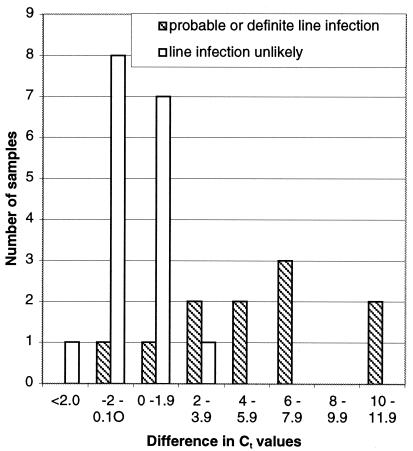
The results from the quantitative DNA detection methods of Nadkarni et al. (13) are presented here. The distributions of Ct values for blood samples collected through the CVC from patients with probable CVC-associated infection and those unlikely to have CVC infection are shown in Fig. 2. All of the episodes of probable CVC-associated infection gave Ct values of less than 35. Four of the episodes unlikely to be CVC-associated infection gave Ct values of less than 35. These included three episodes in two patients with exacerbations of severe inflammatory bowel disease and one episode in an otherwise clinically stable patient without fever or other stigmata of infection. The one episode of CVC-associated yeast infection gave a Ct value in the same range as those for the samples from patients thought to be unlikely to have CVC-associated infection. The distribution of the differences between Ct values for CVC and peripheral blood in patients with probable CVC-associated infection and those unlikely to have CVC infection is shown in Fig. 3. There were nine episodes of CVC-associated infection where peripheral and CVC blood samples were collected (11 CVC and 9 peripheral samples). In only two of the nine samples was the level of bacterial DNA raised in the peripheral blood sample.
FIG. 2.
Ct values in patients with probable CVC-associated bacterial infection and patients in whom CVC-associated infection was unlikely. Ct results are medians of three results.
FIG. 3.
Differences in median Ct values between CVC and peripheral blood sample. (Ct value for CVC blood − Ct value for peripheral blood).
In four episodes of probable CVC-associated infection there were double-lumen CVCs in place. In two of these three episodes there were large differences in the Ct values between the different lumens of the same CVC, although all were below 35 cycles.
Statistical analysis and use of a receiver-operating curve.
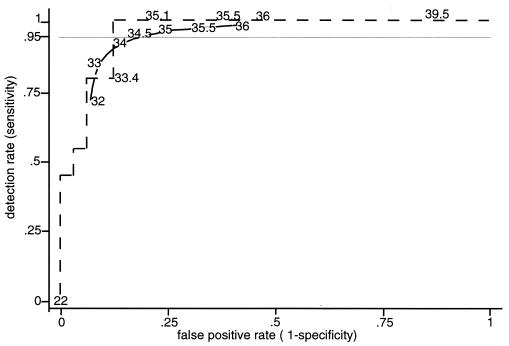
The data were examined to see whether it would be appropriate to model them by means of a normal distribution or a normal distribution above and below a cut point. Data for the group with probable CVC-associated infection and with low Ct values and data for the uninfected group with high Ct values will not affect the choice of cut point, so it was thought more useful to have the data at the other ends of these distributions modeled accurately. In the model for those episodes with probable CVC-associated infection, 5% would have CVC blood Ct values below 23 replications, while 95% would be distributed normally with a mean of 30.7 and a standard deviation of 2.48 cycles. For those unlikely to have CVC-associated infection, 6.25% would have Ct values below 32 and the rest would be distributed normally with a mean of 36.6 and standard deviation of 1.85 cycles. Figure 4 shows the receiver-operating curves for the actual data and the modeled data for Ct values from CVC blood samples. For both lines, the point nearest the top left corner is based on a cut point of around 33 to 33.5 cycles. These values minimize misclassification, but because it is less important to misclassify someone without a line infection than someone truly with one, the more useful cut points will be higher than this, and results for cut points of 34.5 and 35 cycles are described. The modeled data do not appear to overestimate the effects, as at the cut points of interest, the actual data give higher detection rates than the modeled data. The area under the curve for the actual data is 0.957, and the 95% confidence interval for the area under the curve is 0.91 to 1.0.
FIG. 4.
Receiver-operating curve plotting true-positive rate (sensitivity) against false-positive rate (1 − specificity), showing the actual data (dashed line) and the modeled receiver-operating curve (solid line).
DISCUSSION
The diagnosis of CVC-associated infection is often difficult because of dependence on quantitative microbial culture, which is compromised by use of antibiotics, or invasive sampling, or removal of the CVC. Microbial culture methods may take several days to give a result, and invasive sampling methods may result in embolization or bacteremia (1). We have evaluated a novel approach based on quantitative detection of bacterial DNA drawn through the CVC. The advantages of a DNA-based approach are that it can work even when patients have been treated with antibiotics (6) and can be automated. The principle of this approach is that the concentration of bacteria (and therefore bacterial DNA) is high in blood drawn through a colonized CVC. The method used in this evaluation would be unlikely to reliably detect the small numbers of bacteria in the blood of patients with bacteremia. The test is rapid, and the cost per test is low once the appropriate equipment is available. There are a number of published methods for the quantitative detection of 16S rDNA, and each will give a different Ct value that differentiates samples of blood from colonized and uncolonized CVCs. This discriminatory Ct value varies with the patient population, the choice of primers and probe, and the operating characteristics of the quantitative PCR analyses used (Fig. 1).
It is highly unlikely that one set of primers and probes will identify all bacteria that might cause CVC infections or bacteremia. The forward primer devised for this study had three mismatches with M. oxydans DNA, two mismatches with C. diphtheriae and C. xerosis DNA, and also one mismatch between the probe and the DNA from these organisms, which could not be detected in the assay. The forward primer devised by Yang et al. (17) also has mismatches with M. oxydans, C. diphtheriae, and C. xerosis DNA and also with S. maltophilia DNA. The UniProbe and reverse primer used by Yang et al. contain mismatches with P. aeruginosa and Helicobacter pylori, respectively. Similarly, the reverse primer for the method of Nadkarni et al. (13) shows a mismatch with Salmonella enterica serovar Enteritidis, and the probe has a mismatch with A. baumanii and Acinetobacter lwoffi.
Using the method of Nadkarni et al. (13) for a cut point of 34.5 or 35, the detection rates in our modeled data would be 94 and 96%, respectively, while the false-positive rate would be 18 and 24%, respectively. It is generally accepted that only one of every six CVCs that are removed because of a potential line infection actually show evidence of infection (3). If this test was applied to the hospital population, the percentage of CVCs removed unnecessarily could be reduced by 80% (from five to one for each real line infection) if the cut point was 34.5 and by 76% if the cut point was set at 35 cycles. This percent reduction in unnecessarily removed CVCs is not altered by more than 1% for a range of prevalences (10 to 50%) of probable CVC-associated infection. The difference in Ct values between CVC and peripheral samples was determined largely by the level of CVC blood positivity, and the difference between the CVC and peripheral blood samples did not improve the predictive value of this testing approach.
The results suggest that in this population of patients, a single sample from the CVC is as valuable in excluding the diagnosis of CVC bacterial infection as is comparison of the levels of bacterial DNA in the central and peripheral blood. In this study there were only a small number of samples from the multiple lumens of a colonized CVC, but we found variation in the amount of bacterial DNA in samples taken from the different lumens, suggesting that this test approach might be useful in differentiating which CVC is colonized in patients with multiple CVCs. There were four samples with positive test results from patient episodes not fulfilling criteria for CVC infection. We do not know if these were a result of sample contamination, in which case repeat samples might have produced negative results. Alternatively, these results might represent bacterial colonization of the infusion system, hub, or CVC without invasion of the vascular compartment; reflected bacteremia; or translocation of bacteria or bacterial DNA.
Although we deliberately excluded samples from patients who had received antibiotic therapy within the preceding 2 weeks. DNA from bacteria that have been killed by antibiotics can still be detected by PCR in the blood (6), suggesting that the technique may be useful in patient populations in which antibiotic treatment is more common. This approach needs to be evaluated for other patient groups, such as those in intensive care and those under treatment for cancer. In previous studies and further preliminary work using the method reported, here we have found high levels of bacterial DNA in the peripheral blood of some immunocompromised patients with bacteremias, even during antibiotic therapy (10). It may be that this testing approach would best be applied in an immunocompromised population by comparing bacterial DNA concentrations in blood drawn through all lumens and peripheral blood. Repeated sampling will also show whether bacterial DNA is transient or persistent. The target for the quantitative PCR is a region of the 16S rRNA gene which is highly conserved in most bacteria. This ensures that the test has the potential to detect virtually all of the bacterial causes of CVC infection. This approach may be further developed by combining it with methods utilizing genus- or species-specific targets (11, 17).
Acknowledgments
S.W. was supported by the Joint Research Board of St. Bartholomew's Hospital, London.
We thank the staff of the Genome Centre, Barts and The London School of Medicine and Dentistry, for their help with the TaqMan assays.
REFERENCES
- 1.Blot, F., A. Laplanche, B. Raynard, N. Germann, S. Antoun, and G. Nitenberg. 2000. Accuracy of totally implanted ports, tunnelled, single- and multiple-lumen central venous catheters for measurement of central venous pressure. Intensive Care Med. 26:1837-1842. [DOI] [PubMed] [Google Scholar]
- 2.Blot, F., G. Nitenberg, E. Chachaty, B. Raynard, N. Germann, S. Antoun, A. Laplanche, C. Brun-Buisson, and C. Tancrede. 1999. Diagnosis of catheter-related bacteraemia: a prospective comparison of the time to positivity of hub-blood versus peripheral-blood cultures. Lancet 354:1071-1077. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 2002. Guidelines for the prevention of intravascular catheter-related infections. Morb. Mortal. Wkly. Rep. 51:1-36. [Google Scholar]
- 4.Farr, B. M. 1999. Accuracy and cost-effectiveness of new tests for diagnosis of catheter-related bloodstream infections. Lancet 354:1487-1488. [DOI] [PubMed] [Google Scholar]
- 5.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 6.Heininger, A., M. Ulrich, G. Priebe, K. Unertl, A. Muller-Schauenburg, K. Botzenhart, and G. Doring. 2001. The effect of human serum DNAases on the ability to detect antibiotic-killed Escherichia coli in blood by PCR. J. Med. Microbiol. 50:243-248. [DOI] [PubMed] [Google Scholar]
- 7.Kite, P., B. M. Dobbins, M. H. Wilcox, W. N. Fawley, A. J. Kindon, D. Thomas, M. J. Tighe, and M. J. McMahon. 1997. Evaluation of a novel endoluminal brush method for in situ diagnosis of catheter related sepsis. J. Clin. Pathol. 50:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kite, P., B. M. Dobbins, M. H. Wilcox, and M. J. McMahon. 1999. Rapid diagnosis of central-venous-catheter-related bloodstream infection without catheter removal. Lancet 354:1504-1507. [DOI] [PubMed] [Google Scholar]
- 9.Kristinsson, K. G., I. A. Burnett, and R. C. Spencer. 1989. Evaluation of three methods for culturing long intravascular catheters. J. Hosp Infect. 14:183-191. [DOI] [PubMed] [Google Scholar]
- 10.Ley, B. E., C. J. Linton, D. M. Bennett, H. Jalal, A. B. Foot, and M. R. Millar. 1998. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 17:247-253. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig, W., and K.-H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556-562. [DOI] [PubMed] [Google Scholar]
- 12.Maki, D. G., C. E. Weise, and H. W. Sarafin. 1977. A semiquantitative culture method for identifying intravenous-catheter-related infection. N. Engl. J. Med. 296:1305-1309. [DOI] [PubMed] [Google Scholar]
- 13.Nadkarni, M. A., F. E. Martin, N. A. Jacques, and N. Hunter. 2002. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148:257-266. [DOI] [PubMed] [Google Scholar]
- 14.Raad, I. I., and G. P. Bodey. 1992. Infectious complications of indwelling vascular catheters. Clin. Infect. Dis. 15:197-208. [DOI] [PubMed] [Google Scholar]
- 15.Siegman-Igra, Y., A. M. Anglim, D. E. Shapiro, K. A. Adal, B. A. Strain, and B. M. Farr. 1997. Diagnosis of vascular catheter-related bloodstream infection: a meta-analysis. J. Clin. Microbiol. 35:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stata Corporation. 2001. Stata statistical software, release 7.0. Stata Corporation, College Station, Tex.
- 17.Yang, S., S. Lin, G. D. Kelen, T. C. Quinn, J. D. Dick, C. A. Gaydos, and R. E. Rothman. 2002. Quantitative multiprobe PCR assay for simultaneous detection and identification to species level of bacterial pathogens. J. Clin. Microbiol. 40:3449-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]