Abstract
Transcription by all three eukaryotic RNA polymerases involves the assembly of a large preinitiation complex (PIC) at gene promoters. The PIC comprises several general transcription factors (GTFs), including TBP, and the respective RNA polymerase. It has been suggested that some GTFs remain stably bound at active promoters to facilitate multiple transcription events. Here we used two complementary approaches to show that, in G1-arrested yeast cells, TBP exchanges very rapidly even at the most highly active RNA Pol II promoters. A similar situation is observed at RNA Pol III promoters. In contrast, TBP remains stably bound at RNA Pol I promoters. We also provide evidence that, unexpectedly, PIC dynamics are neither the cause nor the consequence of nucleosome exchange at most of the RNA Pol II promoters we analyzed. These results point to a stable reinitiation complex at RNA Pol I promoters and suggest independent PIC and nucleosome turnover at many RNA Pol II promoters.
Transcription in all eukaryotes is carried out by three distinct RNA polymerases (Vannini and Cramer 2012). RNA polymerase I (Pol I) transcribes rRNA genes, RNA Pol II transcribes mRNA genes, while RNA Pol III synthesizes tRNA and other small RNA species. Transcription by the three RNA polymerases involves the assembly of a large preinitiation complex (PIC) at gene promoters. The PIC consists of a set of general transcription factors (GTFs) that are specific for each RNA polymerase, and the TATA-binding protein (TBP) that is required by all three RNA polymerases (for review, see Vannini and Cramer 2012). PIC assembly is regulated by activators and allows the recruitment of the appropriate RNA polymerase to the promoter and transcription to initiate.
Although it is well established that during the course of gene activation, that is, prior to the initial transcription event, a complete PIC must assemble at the promoter, at least in yeast (Pokholok et al. 2002), much less is known about the dynamics of the PIC during continuous transcription. Does the complex disassemble completely after each round of transcription, or do some GTFs remain at the promoter to facilitate multiple rounds of transcription? Does the nature of the activator and/or core promoter type influence the process? Most of our current knowledge on PIC dynamics stems from fluorescence recovery after photobleaching (FRAP) experiments. In these studies, a transcription factor of interest is fused to a fluorescent protein, and the dynamics of its interaction with DNA is assessed either by photobleaching an arbitrary region of the nucleus or by using an artificial construct containing tandemly repeated reporter elements (Karpova et al. 2008). Such experiments originally suggested that several PIC components of the RNA Pol I machinery are continuously assembling and disassembling at the promoter of actively transcribed rRNA genes in mammalian cells (Dundr et al. 2002). The same was reported for RNA Pol II both in yeast (Sprouse et al. 2008) and in mammalian cells (Chen et al. 2002). However, these approaches have some limitations (Stasevich and McNally 2011). In particular, it is unclear if photobleaching randomly selected nuclear regions in cells overexpressing the fluorescently tagged GTF actually measures binding dynamics at authentic transcription start sites rather than nonspecific association with chromosomal DNA (Karpova et al. 2008; Hager et al. 2009). Indeed, more recent FRAP studies in cultured cells (de Graaf et al. 2010) and in living tissues (Giglia-Mari et al. 2009) revealed that a significant fraction of TBP and other GTFs remain immobile when the tagged factor is expressed at physiological levels. This is more consistent with some PIC components remaining promoter-associated during ongoing transcription, as suggested by other in vivo observations (Christova and Oelgeschlager 2002; Xing et al. 2008) and in agreement with earlier in vitro studies (Yudkovsky et al. 2000).
TBP functions as a GTF for all three RNA polymerases, and its interaction with promoter sites is a prerequisite for PIC assembly at all promoters. Recent chromatin immunoprecipitation (ChIP) studies in dividing yeast cells suggest a dynamic interaction of TBP at RNA Pol II genes that depends on the promoter type (van Werven et al. 2009). We revisited this notion in G1-arrested yeast cells using two distinct and complementary approaches. We show that, under these conditions, TBP remains stably bound at RNA Pol I promoters but exchanges rapidly at all RNA Pol II genes tested regardless of promoter type and transcription rates. In addition, we investigated the role of chromatin in PIC formation and dynamics. We provide evidence that, unexpectedly, the PIC and nucleosomes turn over independently of each other at most RNA Pol II promoters we investigated.
Results
Competition approach
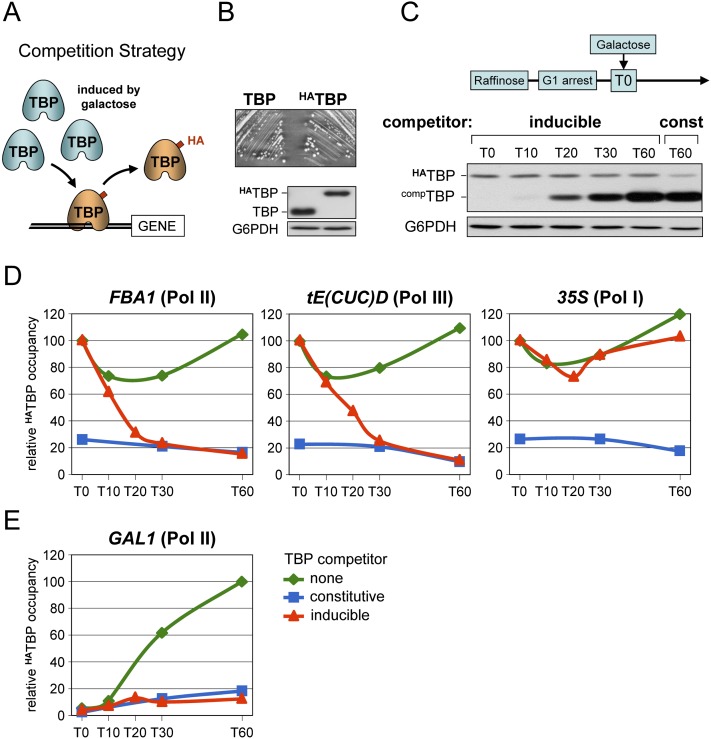
To explore the dynamics of TBP-promoter interaction in living yeast cells, we first used a competition approach (Fig. 1A). We employed a yeast strain containing a chromosomally integrated hemagglutinin (HA) epitope-tagged version of TBP (encoded by SPT15) placed under the control of its own promoter as the sole source of TBP. The strain behaves like wild-type, and HATBP is expressed at close to physiological levels (Fig. 1B). We then induced expression of a native TBP in this strain using a galactose-inducible GAL1 promoter (Fig. 1C). The amount of HATBP detected at active promoters either before or at various time points after induction of the TBP competitor was assessed by quantitative ChIP analysis using antibodies against the HA-tag (Fig. 1A). The expectation was that TBP produced in excess will replace HATBP at promoters where the protein is constantly exchanging, and that the rate at which this occurs will give an indication of the residence time of HATBP at the promoter. As a control, we used two strains constitutively expressing either TBP or no TBP competitor. To exclude any replication-associated events, cells were arrested at the G1 phase of the cell cycle by treatment with alpha factor (Jamai et al. 2007).
Figure 1.
A ChIP-based competition assay reveals high TBP turnover at RNA Pol II and Pol III promoters and stable binding at RNA Pol I promoters. See also Supplemental Figure S1 for additional promoters and Supplemental Figure S9 for biological replicates. (A) Schematic diagram of the competition approach. See text for details. (B) The starting yeast strain expressing an HA-tagged TBP (HATBP) from its native locus and its isogenic wild-type counterpart (TBP) were examined for growth on YPD plates. TBP protein levels were monitored by Western blot analysis using anti-TBP antibodies and anti-G6PDH antibodies as a loading control. (C) Cells expressing HATBP and a plasmid-encoded native TBP competitor (compTBP) from the galactose-inducible GAL1 promoter were grown overnight in raffinose and arrested in G1 by treatment with alpha factor. Expression of native TBP was then induced by directly adding galactose to the medium. Culture aliquots were removed just prior to (T0) and at the indicated time points (in minutes) after galactose addition, and directly processed for Western blot and ChIP analyses (D,E). HATBP and TBP protein levels in whole cell extracts prepared before cross-linking were evaluated using anti-TBP antibodies. G6PDH served as a loading control. The last sample on the right is from cells constitutively expressing TBP (const) from the DED1 promoter. Note that HATBP expression remains constant throughout the experiment. (D) The levels of HATBP occupancy at the indicated promoters and time points (in minutes) were measured by quantitative ChIP analysis using anti-HA antibodies. The strains were treated in parallel as in C and express the TBP competitor from the galactose-inducible GAL1 promoter (inducible), the constitutively active DED1 promoter (constitutive), or no TBP competitor (none). The ChIP signals are relative to those measured at T0 in each strain. These were set to a value of 100, except for the constitutively expressing TBP strain for which the T0 value is relative to the signal detected in the strain expressing no TBP competitor. (E) Same as in D but showing HATBP occupancy at the galactose-induced GAL1 promoter. The ChIP signals are expressed relative to those measured at T60 (=100) in the strain with no TBP competitor (none). Note that the slight increase in HATBP occupancy at T20 following galactose induction of TBP was not observed in an independent experiment (Supplemental Fig. S9).
We focused on highly transcribed RNA Pol II genes showing high TBP occupancy (Kim and Iyer 2004), as TBP is more likely to remain stably bound at the promoter of these genes. Figure 1D shows that HATBP occupancy decreased rapidly at the FBA1 promoter, reaching minimal levels at 30 min after induction of the TBP competitor. The same was observed at other highly expressed RNA Pol II genes, regardless of their dependency on the TBP-containing TFIID or SAGA complex (Supplemental Fig. S1A; Bhaumik 2011). The decrease in HATBP ChIP signals was not due to the amounts of HATBP varying during the experiment. Expression of HATBP remained constant (Fig. 1C), and comparable amounts of the protein were immunoprecipitated for ChIP analysis at all time points (Supplemental Fig. S1C). We also tested the GAL1 gene, which is induced by galactose. Evidently, the only TBP protein present at the onset of galactose induction is HATBP, which accumulated rapidly at the GAL1 promoter in the absence of a TBP competitor (Fig. 1E). Yet there was no increase, even transient, in the HATBP ChIP signals above those seen in cells constitutively expressing TBP when the TBP competitor was induced by addition of galactose (Fig. 1E). This is consistent with the HATBP originally bound at the GAL1 promoter at the onset of induction being rapidly replaced by untagged TBP. HATBP occupancy also decreased abruptly at tRNA gene promoters, although a generally slower kinetics was observed at RNA Pol III genes (Fig. 1D; Supplemental Fig. S1B), in agreement with a previous study (van Werven et al. 2009). Strikingly, no reduction in HATBP occupancy was detected at RNA Pol I promoters (Fig. 1D), unless cells were allowed to divide (Supplemental Fig. S2). Thus, TBP remains stably bound at RNA Pol I promoters in G1-arrested cells while exchanging rapidly at all other promoters tested.
Nuclear depletion approach
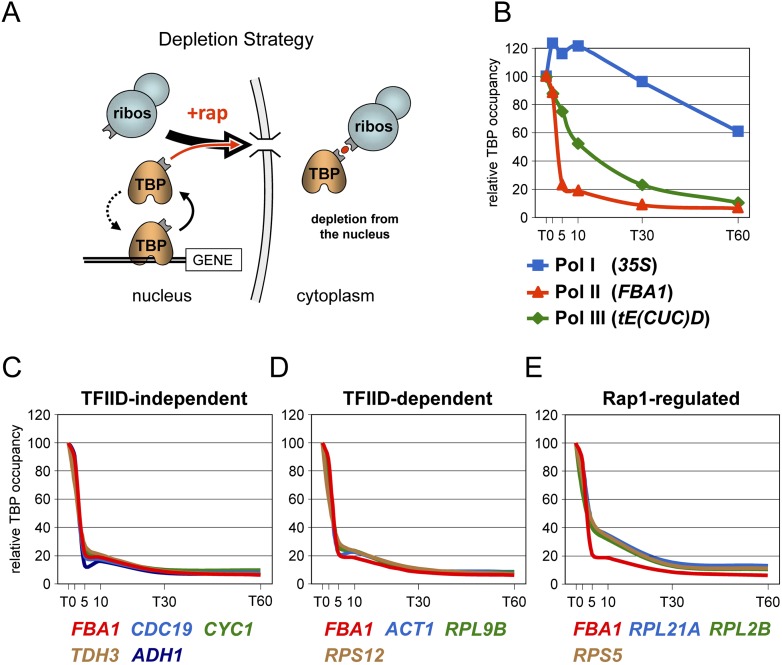
To confirm and extend these studies, we employed the anchor-away method that permits a conditional and rapid depletion of TBP from the nucleus (Haruki et al. 2008). The approach exploits the massive export of ribosomal subunits out of the nucleus to translocate TBP fused to a rapamycin binding domain (FRB) into the cytoplasm through its rapamycin-dependent interaction with a ribosomal protein (Fig. 2A; Supplemental Fig. S3; Haruki et al. 2008). The expectation was that depletion of the free pool of TBP would lead to a decrease in TBP occupancy at promoters at which TBP normally exchanges, and that the rate at which this occurs would provide an indication of the binding duration of TBP at the promoter (Fig. 2A). Addition of rapamycin to cells expressing FRB-tagged TBP caused a marked and rapid drop in TBP occupancy at the FBA1 gene (Fig. 2B). The same was observed at other TFIID-independent (or SAGA-dependent) genes (Fig. 2C), both in G1-arrested and in dividing cells (Supplemental Fig. S4A), and at three TFIID-dependent genes (Fig. 2D). This effect required the rapamycin-binding domain FRB as no major decrease in occupancy occurred in a strain expressing an untagged TBP protein (Supplemental Fig. S4B). Furthermore, loss of TBP from promoters was paralleled by a similar decrease in RNA Pol II density over transcribed regions, consistent with TBP being continuously required during ongoing transcription (Supplemental Fig. S4C). We also considered a few genes for which indirect evidence suggests that TBP may remain stably bound at the promoter. These include the CYC1 gene (Fig. 2C), whose promoter is occupied by TBP even in the absence of transcription (Martens et al. 2001), and the PMA1 gene (Supplemental Fig. S4D), which forms a loop between the promoter and termination regions to allow for RNA Pol II recycling during ongoing transcription, a scenario consistent with a stable PIC at the promoter (for review, see Shandilya and Roberts 2012). Furthermore, we tested two genes showing comparable TFIIB promoter occupancy and either high or low RNA Pol II occupancy (Supplemental Fig. S4E). The latter situation may be indicative of stable PIC formation in the absence of RNA Pol II. However, the same rapid disappearance of TBP was observed at all these promoters. Only three RNA Pol II genes reproducibly showed a slightly reduced rate of TBP disappearance (Fig. 2E; Supplemental Fig. S5). These genes are under the control of Rap1, an activator known to recruit TBP at the promoter (Mencia et al. 2002). Nevertheless, TBP occupancy still decreased rapidly at these genes, indicating that TBP is very dynamic even when tethered to the promoter by the activator. We also examined a few RNA Pol III genes. As in the competition approach (Fig. 1D; Supplemental Fig. S1B), TBP showed a dynamic behavior at tRNA genes but the trend was generally slower at these promoters (Fig. 2B; Supplemental Fig. S5). Also in agreement with the competition approach, TBP occupancy at RNA Pol I promoters remained largely unaffected, decreasing slightly only at the late time points (Fig. 2B). This, however, is likely a consequence of secondary effects due to transcriptional shutdown because the same occurred upon nuclear depletion of RNA Pol II (Supplemental Fig. S4F). Overall, these results point to a highly dynamic interaction of TBP at RNA Pol II promoters and stable binding at RNA Pol I promoters throughout the G1 phase of the cell cycle.
Figure 2.
TBP residence time at RNA Pol I, Pol II, and Pol III promoters as measured by the anchor-away assay. See also Supplemental Figures S4 and S5. (A) Schematic diagram of the TBP anchor-away approach (Haruki et al. 2008). Upon addition of rapamycin (+rap, red dot), TBP fused to the rapamycin-binding domain FRB is exported out of the nucleus by the flow of ribosomal subunits (ribos) through its interaction with a ribosomal protein bearing the complementary FKBP rapamycin-binding domain (for details, see Haruki et al. 2008). (B) FRB-tagged TBP occupancy at the indicated promoters was measured just before and at the indicated time points after rapamycin addition by quantitative ChIP using anti-TBP antibodies. Note that the experiment includes a 2-min time point. Because TBP occupancy varies among promoters, results are expressed relative to the T0 value, which was taken as 100, to allow for direct comparison. (C) Same as in B at other TFIID-independent RNA Pol II promoters. The results for FBA1 are from B and serve as a comparison. The individual data points are as in B and are not shown in this and the next two panels to facilitate visual comparison. (D) Same as in C at TFIID-dependent promoters. (E) Same at three genes regulated by the Rap1 activator, which is known to recruit the TBP-containing TFIID factor to the promoter (Mencia et al. 2002). FBA1 is from B.
Competition between PICs and nucleosomes
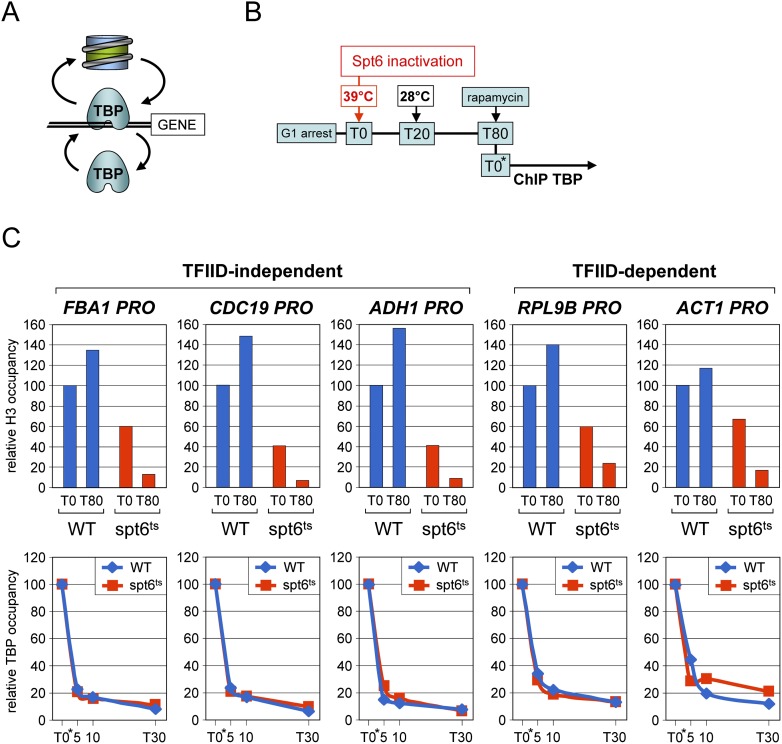
The highly dynamic behavior of TBP at all RNA Pol II genes tested is intriguing in light of the long promoter residence time of TBP and its further stabilization by activators in vitro (Yudkovsky et al. 2000). However, the DNA template in vivo is embedded within nucleosomes, which exchange at a high rate at the promoters of transcriptionally active genes (Dion et al. 2007; Jamai et al. 2007; Rufiange et al. 2007). It is therefore possible that the PIC and nucleosomes are in competition, with the PIC being constantly evicted from the promoter by new nucleosomes (Fig. 3A). If so, depleting nucleosomes from promoters should increase the average binding duration of TBP. To explore this possibility, we generated a TBP-anchor-away strain carrying a temperature-sensitive mutation in the histone chaperone Spt6 (spt6-14) (Bortvin and Winston 1996), which has been implicated in nucleosome assembly at promoters (Adkins and Tyler 2006). Wild-type and mutant cells were arrested in the G1 phase as before, shifted for 20 min to the nonpermissive (39°C) temperature to inactivate Spt6, and then returned to the permissive temperature (Fig. 3B). Nucleosome occupancy is partially reduced in the mutant strain even at the permissive temperature and rapidly drops further upon Spt6 inactivation (Fig. 3C, upper panels; data not shown). Unexpectedly, nucleosome occupancy remained low even after a 60-min recovery period at permissive temperature (Fig. 3C; data not shown). This allowed us to perform the experiment long after Spt6 heat inactivation to exclude any effects associated with the temperature shift. Rapamycin was added to deplete TBP from the nucleus, and the amount of TBP detected at promoter sites was measured by quantitative ChIP. Strikingly, the same rapid decrease in TBP levels was observed in the wild-type strain harboring normal nucleosome occupancy and in the spt6 mutant strain after nucleosome loss at all five promoters tested (Fig. 3C, lower panels). Consistently, nucleosome loss upon Spt6 nuclear depletion also had no effect on native TBP steady-state occupancy (Supplemental Fig. S6). This argues against chromatin being responsible for the TBP promoter interaction dynamics.
Figure 3.
TBP turnover at active promoters in the absence of nucleosomes. See also Supplemental Figure S6. (A) Schematic diagram illustrating the possibility that TBP and nucleosomes compete dynamically for promoter binding. (B) Cells from TBP-anchor-away strains carrying either wild-type or a temperature-sensitive spt6 allele (spt6-14) (Bortvin and Winston 1996) were arrested in G1, and rapidly shifted to 39°C for 20 min in order to inactivate Spt6. At T20, cells were brought back to the permissive temperature. After a 60-min recovery period (T80), rapamycin was added to the culture to deplete TBP from the nucleus. (C) (Upper panel) Nucleosome occupancy just before (T0) and at 80-min (T80) after transient heat inactivation of Spt6 was measured at the indicated TFIID-independent and TFIID-dependent promoters by quantitative ChIP using antibodies against core histone H3. The ChIP signals for each gene are expressed relative to those measured at T0 (=100) in the wild-type Spt6 strain (WT). (Lower panel) TBP occupancy at the same promoters was determined prior to (T0*) and at the indicated time points after addition of rapamycin, as in Figure 2. Note that T0* corresponds to the T80 time point in the upper panel. This experiment was performed in a strain bearing a wild-type TOR1 allele (see Supplemental Methods).
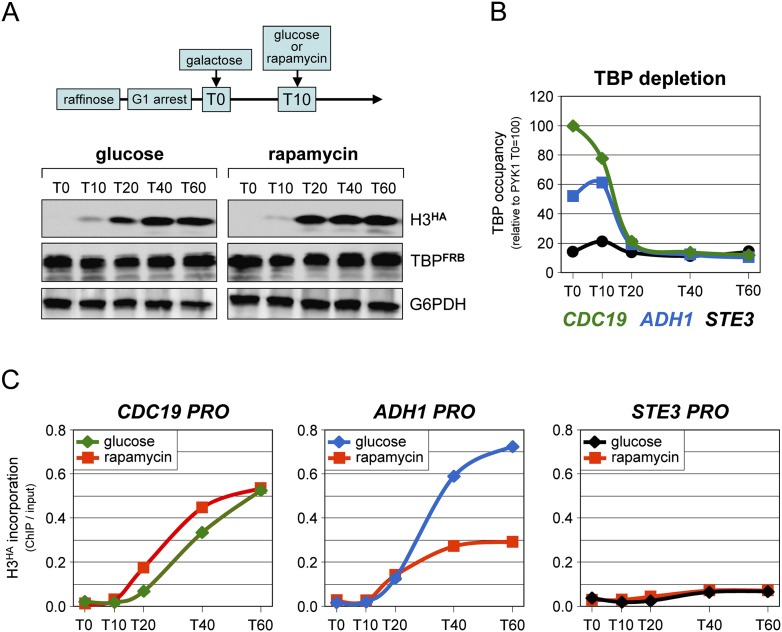
We then addressed the opposite possibility, namely, that PIC dynamics triggers nucleosome exchange. We determined whether loss of TBP from promoters would affect the incorporation of newly synthesized histones, which is a measure of nucleosome exchange (Dion et al. 2007; Jamai et al. 2007; Rufiange et al. 2007). Yeast cells were arrested at G1 with alpha factor as before. Expression of an HA-tagged histone H3 (H3HA) was then induced by the addition of galactose (Fig. 4A; Jamai et al. 2007). Shortly after, rapamycin was added to deplete TBP, or glucose was added as a control to block further expression of the tagged histone. An abrupt decrease in TBP ChIP signals was observed at the active CDC19 and ADH1 promoters, rapidly reaching the background levels detected at the silent STE3 promoter (Fig. 4B). Unexpectedly, incorporation of newly synthesized H3HA, which under normal conditions occurs at active promoters but not at inactive genes such as STE3 (Fig. 4C; Jamai et al. 2007), persisted or even increased after complete depletion of TBP at the CDC19 (Fig. 4C) and other promoters (Supplemental Fig. S7C). This was not linked to an increase in nucleosome steady-state occupancy, which remained largely unaffected following TBP dissociation (Supplemental Fig. S7A,B), consistent with previous studies (Fan et al. 2010). The only exception was ADH1, where a clear reduction in H3HA incorporation was observed in the absence of TBP (Fig. 4C). This suggests that nucleosome exchange at the ADH1 promoter is due, at least in part (see Supplemental Fig. S7D), to nucleosomes dynamically competing with the PIC for promoter binding, whereas exchange at all other promoters tested is caused by an event independent of PIC formation.
Figure 4.
Nucleosome turnover in the absence of TBP. See also Supplemental Figure S7. (A) Cells from a TBP-anchor-away strain expressing a galactose-inducible HA-tagged version of histone H3 (H3HA) from a single copy plasmid were grown overnight in raffinose before being arrested in G1 with alpha factor. Galactose was then added directly to the medium at T0 to induce expression of H3HA. Glucose was added 10 min later (T10) to suppress further expression of H3HA, or rapamycin was added to deplete TBP from the nucleus. Culture aliquots were removed at these and subsequent time points and directly processed for Western blot and ChIP analyses. H3HA and TBPFRB protein levels in whole-cell extracts prepared before cross-linking were assessed using anti-HA and anti-TBP antibodies. G6PDH served as a loading control. Note that the left and right panels are from the same gel exposure for each protein and are directly comparable. (B) TBP promoter occupancy at the highly transcribed CDC19 and ADH1 genes and at the silent STE3 gene was measured at the indicated time points after galactose addition, as in Figure 2. The results are expressed relative to the T0 value for CDC19, which was set to 100. (C) H3HA incorporation at the same promoters and time points after addition of glucose or rapamycin (red curves) to deplete TBP. The values are expressed as percentage of input DNA recovered.
Discussion
The competition and anchor-away strategies are complementary
We made use of two independent strategies—a competition approach (Fig. 1A) and the recently described anchor-away method (Fig. 2A; Haruki et al. 2008)—to explore the dynamic interaction of TBP at specific promoters in living yeast cells. The two strategies are complementary and yielded fully consistent results. They both revealed that in G1-arrested cells, TBP exchanges rapidly at all RNA Pol II promoters tested while remaining stably associated at RNA Pol I promoters.
A main advantage of the competition approach over the anchor-away technique or FRAP analysis is that the HA-tag is small and therefore less likely to interfere with transcription complex formation and dynamics (Stasevich and McNally 2011). However, the relatively slow induction kinetics of the TBP competitor by galactose limits the temporal resolution of the approach. Furthermore, reliable measurement is limited to promoters showing high levels of TBP occupancy and thus a significant difference in HATBP ChIP signals in the absence or presence of the TBP competitor. Another potential limitation is that overexpression of the TBP competitor may titrate out other factors and thereby indirectly impact on the PIC dynamics.
In contrast, the anchor-away strategy does not rely on protein overexpression, results in a surprisingly rapid block in TBP binding to promoters, and makes use of a yeast strain that is mutated in the TOR pathway (Haruki et al. 2008), thus minimizing indirect effects of rapamycin on global gene expression. However, inactivation of other transcription factors may not be as rapid as observed here with TBP. For example, experiments performed with the large subunit of RNA Pol II resulted in a significantly slower disappearance rate of RNA Pol II from active genes (data not shown). This, however, may reflect recycling of the same RNA Pol II molecule to favor multiple rounds of initiation events (for review, see Shandilya and Roberts 2012)
Rapid exchange of TBP at RNA Pol II promoters during steady-state transcription
We found that TBP is very dynamic at all active RNA Pol II promoters examined. These include some of the most highly expressed genes, which show elevated TBP occupancy (Kim and Iyer 2004) and are therefore the best candidates for having a TBP stably bound at their promoters during ongoing transcription. The same is likely true at more weakly expressed genes, since these genes show lower steady-state TBP occupancy (Kim and Iyer 2004), implying that TBP is not always present at the promoter. The anchor-away approach, which allows for a more accurate estimation of the residence time of TBP at promoters, revealed that within the temporal resolution of the experiment and with the possible exception of Rap1-regulated genes (see below), all RNA Pol II promoters tested showed indistinguishable rapid rates of TBP turnover. We failed to observe a difference noticed in a broader competition ChIP study (van Werven et al. 2009) between TFIID-independent and TFIID-dependent promoters at the few genes we investigated. We also considered other genes for which indirect evidence suggests that TBP may remain stably bound at the promoter. However, the same rapid TBP turnover was also observed. Noticeably, the loss of TBP was accompanied by a parallel decrease in RNA Pol II occupancy within the promoter region, consistent with an absence of promoter proximal RNA Pol II pausing in yeast (data not shown; Adelman and Lis 2012). Taken together, these findings suggest that TBP turns over rapidly at RNA Pol II genes regardless of promoter type.
The highly dynamic behavior of TBP contrasts with the reported long-lived promoter interaction of some transcriptional activators when engaged in activating transcription. For example, the activator Gal4 interacts transiently with its cognate regulatory element under noninducing conditions but becomes stably bound after transcriptional activation, with a half-life on the order of 1 h (Nalley et al. 2006). Stable binding during active transcription has also been reported for Cup2 (also known as Ace1) (Karpova et al. 2008) and for the heat shock transcription factor (HSF) in Drosophila (Yao et al. 2006). Another recent example is Rap1, which has an estimated residence time of >1 h at highly expressed genes (Lickwar et al. 2012). This is far longer than what we observed for TBP at Gal4- and Rap1-regulated genes, even if TBP may turn over at slightly reduced rates at Rap1-regulated genes (Figs. 1E, 2E). The case of Rap1 is particularly intriguing, since Rap1 is known to activate transcription by interacting directly with the TBP-containing TFIID complex (Mencia et al. 2002). One would therefore expect the two factors to bind promoter DNA cooperatively. This suggests either that the residence time of Rap1 has been overestimated, as recently suggested for Gal4 (Collins et al. 2009), or that stabilization of the activator-promoter interaction involves another mechanism.
Evidence against an RNA Pol II reinitiation scaffold in vivo
In yeast, transcription initiation from constitutively active promoters occurs as single, temporally distinct events (Zenklusen et al. 2008). As most active yeast genes are transcribed at rates below 30 mRNAs per hour (Pelechano et al. 2010), this implies that individual initiation events are separated in time by ≥2 min on average at most promoters. The five- to 10-fold decrease in TBP occupancy observed within the first 5 min in the anchor-away assay points to an average promoter residence time for TBP of ∼2 min. This estimate is conservative, because any delay in rapamycin-mediated depletion of TBP would lead to an overestimation. It is therefore reasonable to speculate that, at most yeast promoters, TBP does not remain long enough to support more than one round of transcription; and thus that the entire PIC falls apart and reassembles for each transcription event.
Stable association of TBP at RNA Pol I promoters
In contrast to RNA Pol II and Pol III promoters, RNA Pol I promoters show stable binding of TBP in G1 cells. Similar observations were made in dividing cells (van Werven et al. 2009). The reason for this difference remains elusive. It may relate to the unusual chromatin structure of ribosomal RNA genes (Goetze et al. 2010), reflect some unique feature of the TBP-containing SL1 complex that is required for promoter recognition by RNA Pol I, or result from the extremely high transcription rate of these genes (French et al. 2003). Interestingly, FRAP experiments in HeLa cells revealed a dynamic behavior of RNA Pol I even under conditions of maximal transcription, with a residence time at the promoter in the range of only a few minutes (Gorski et al. 2008). This contrasts with the stable binding of TBP that we observed in yeast. Although this may reflect species differences, an interesting possibility is that a stable TBP-containing reinitiation scaffold forms at RNA Pol I promoters to allow for rapid reloading of the RNA polymerase (for review, see Russell and Zomerdijk 2005).
Independent TBP and nucleosome turnover
TBP can interact stably with TATA sequences in vitro, and together with other GTFs it assembles into a long-lived reinitiation scaffold at the promoter to facilitate multiple rounds of transcription (Yudkovsky et al. 2000). What then triggers TBP turnover in vivo? An attractive possibility is that the PIC competes with and is constantly evicted by nucleosomes, which continuously assemble and disassemble at the promoters of actively transcribed genes (Dion et al. 2007; Jamai et al. 2007; Rufiange et al. 2007). Indeed, in vivo histone depletion results in increased gene expression (Han and Grunstein 1988; Gossett and Lieb 2012), and transcriptional activation coincides with a decrease in nucleosome occupancy at the promoter (Zanton and Pugh 2006). Furthermore, this event generally occurs proportionally to the rate at which the gene is transcribed (Bernstein et al. 2004; Lee et al. 2004; Pokholok et al. 2005) and is abolished when PIC formation is blocked (Kristjuhan and Svejstrup 2004). Moreover, artificial recruitment of the RNA Pol II holoenzyme is sufficient to trigger a loss of promoter nucleosomes (Gaudreau et al. 1997; Imoberdorf et al. 2006). These observations all point to the PIC and nucleosomes competing for promoter binding. Yet unexpectedly, within the limits of the temporal resolution of our measurements, TBP showed the same rapid turnover when nucleosomes were depleted from promoters and, conversely, TBP depletion, and thus RNA Pol II disappearance, did not affect histone exchange at most promoters tested. Thus, the PIC and nucleosomes turn over in a highly dynamic fashion, but they apparently largely do so independently of each other. This is consistent with previous findings suggesting that nucleosome eviction is not a prerequisite for PIC assembly at most promoters (Zanton and Pugh 2006). It will be interesting to decipher the mechanisms involved in RNA Pol II PIC instability, which may play a role in the regulation of transcription (Sprouse et al. 2009), and the role of activators and/or promoter sequences in nucleosome exchange at promoters in vivo.
Methods
Yeast strains and growth conditions
The parental yeast strain used in the competition assay is derived from FY104 (relevant genotype MATa, Ura3-52, Trp1Δ63) (kindly provided by Fred Winston, Harvard Medical School). The chromosomal TBP (SPT15) gene was replaced by an allele carrying three tandem copies of the HA epitope tag at the N terminus by two-step homologous recombination. To facilitate G1 arrest by alpha factor, the BAR1 gene encoding a protease that cleaves and inactivates alpha factor was disrupted by one-step replacement with the loxP–KlURA3–loxP cassette amplified by PCR from plasmid pUG72 (Euroscarf No. P30117). Disruption was confirmed by PCR and “shmoo” phenotype analyses. The resulting strain was then transformed with the parental vector pRS314 carrying a TRP1 selectable marker, a derivative expressing native TBP under control of the constitutively active DED1 promoter, or with a TRP1 marked multicopy plasmid YEplac112 expressing native TBP from the GAL1 promoter.
Cells were grown overnight at 30°C to a density at OD600 nm of about 0.4–0.5 in Casamino acid medium lacking tryptophan and uracil supplemented with 2% raffinose and 0.1% glucose. Cells were arrested in G1 by treatment with 400 ng/mL alpha factor (Primm srl, Italy) for 3 h, at which time >95% of cells displayed the elongated “shmoo” phenotype associated with G1 arrest. Expression of the native TBP was then induced by adding 2% galactose. Culture aliquots were removed just prior to (T0) and at the indicated time points after galactose addition and immediately processed for ChIP and Western blot analyses.
The anchor-away strains (kindly provided by Hirohito Haruki and Ulrich Laemmli, University of Geneva) expressing native TBP (HHY221), FRB-tagged TBP (HHY154), or FRB-tagged Rpb1 (HHY170) have been described (Haruki et al. 2008). To allow G1 arrest by alpha factor, strains were switched to MATa by transient expression of the HO endonuclease from a GAL1-HO plasmid, and the BAR1 gene was disrupted as described above. The mating type switch was confirmed by conventional crossing to tester strains. The strains used in Figure 3 and Supplemental Figure S6 that contain, respectively, a temperature-sensitive spt6 allele (spt6-14) (Bortvin and Winston 1996) or FRB-tagged Spt6, were constructed as described in the Supplemental Methods. All strains were confirmed by PCR, sequencing, and phenotypic analyses. In Figure 4, the MATa TBP-anchor-away strain was transformed with a C-terminal triple-HA-tagged histone H3 construct (Jamai et al. 2007) cloned into a derivative of the single-copy plasmid pRS316 in which URA3 has been replaced with ADE2, between a GAL1–CYC1 hybrid promoter and the destabilizing MFA2 3′-untranslated region. Details of the plasmid constructions and strains are available in the Supplemental Methods.
Cells were grown as above, except that the initial overnight cultures were in medium containing 2% glucose instead of raffinose in Figure 2 and associated Supplemental Figures, in YPD medium in Figure 3 and Supplemental Figures S3 and S6, and in raffinose medium lacking adenine in Figure 4. In all experiments, cells were arrested in G1 with 800 ng/mL of alpha factor. In Figure 3, cells expressing wild-type or the temperature sensitive spt6-14 mutant were grown overnight at 28°C and then shifted to 39°C for 20 min to inactivate Spt6. In Figure 4, H3HA expression was transiently induced in the control strain by adding galactose and then glucose 10 min later, both at a final concentration of 2%. The FRB-tagged proteins were depleted from the nucleus by addition of 4 μg/mL (10 μg/mL in Fig. 3) rapamycin (LC Laboratories). No deleterious effect on growth of the parental anchor-away strain HHY221 was observed with rapamycin concentrations of up to 20 μg/mL (Supplemental Fig. S8). Culture aliquots were removed just prior to time 0 and at the indicated time points after heat shock, galactose, and/or rapamycin addition and immediately processed for Western blot and ChIP analyses.
Western blotting
Whole-cell extracts were obtained by the classical glass beads breakage method using equal volumes of glass beads and RIPA Buffer (50 mM HEPES [pH 7.5], 2 mM EDTA, 0.25 M NaCl, 0.1% SDS, 0.1% DOC, 1% Triton X-100) supplemented with 0.5 mM phenylmethylsulfonyl fluoride. The membranes were probed with 1:10,000 anti-HA monoclonal antibody (clone 16B12, Covance), 1:10,000 rabbit polyclonal anti-TBP antibodies (a kind gift from Laurie Stargell, Colorado State University), or 1:10,000 rabbit polyclonal anti-G6PDH antibodies (Sigma, A9521) as an internal standard. Horseradish peroxidase–conjugated goat anti-mouse (Bio-Rad, 1:10,000) or anti-rabbit (Biorad, 1:10,000) IgG were used as secondary antibodies. Detection was carried out with Immobilon Western Chemiluminescent HRP substrate (Millipore). In Supplemental Figure S1C, aliquots of the immunoprecipitated formaldehyde cross-linked chromatin samples were directly mixed 2:1 with fourfold concentrated SDS sample buffer, boiled for 10 min, and diluted for SDS-PAGE electrophoresis.
Chromatin immunoprecipitation (ChIP) and quantitative PCR
ChIP and quantitative PCR analysis were performed as described in the Supplemental Methods. Immunoprecipitation was with anti-HA antibodies (clone 16B12, Covance), anti-TBP antibodies (Laurie Stargell), anti-Rpb1 (RNA Pol II) antibodies (clone 8WG16, Covance), or antibodies to core histone H3 (ab1791, Abcam). All data are representative of at least two completely independent experiments. Independent data sets for all main figures are provided in Supplemental Figure S9. See also the Supplemental Table for technical replicates. Note that the efficiency and kinetics of galactose activation and rapamycin-mediated nuclear depletion vary between experiments, leading to significant variations in the absolute ChIP values. However, the relative effects within each experiment are very reproducible (for example, see Supplemental Fig. S5). This is why representative experiments are shown in the figures.
Acknowledgments
We are most grateful to Fred Winston for providing us with the yeast strain FY957 (spt6-14), to Ulrich Laemmli for the anchor-away yeast strains, to Robbie Loewith for the YcpLac33::Tor1 plasmid and very helpful suggestions, to Laurie Stargell for the anti-TBP antibodies, and to Joseph Curran for critical reading of the manuscript. This work was supported by a grant from the Swiss National Science Foundation (31003A-127384) to M.S. and by the Canton of Geneva.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.157792.113.
Freely available online through the Genome Research Open Access option.
References
- Adelman K, Lis JT 2012. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet 13: 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkins MW, Tyler JK 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell 21: 405–416 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL 2004. Global nucleosome occupancy in yeast. Genome Biol 5: R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR 2011. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim Biophys Acta 1809: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476 [DOI] [PubMed] [Google Scholar]
- Chen D, Hinkley CS, Henry RW, Huang S 2002. TBP dynamics in living human cells: Constitutive association of TBP with mitotic chromosomes. Mol Biol Cell 13: 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R, Oelgeschlager T 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat Cell Biol 4: 79–82 [DOI] [PubMed] [Google Scholar]
- Collins GA, Lipford JR, Deshaies RJ, Tansey WP 2009. Gal4 turnover and transcription activation. Nature 461: E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf GP, Mousson F, Geverts B, Scheer E, Tora L, Houtsmuller AB, Timmers HT 2010. Chromatin interaction of TATA-binding protein is dynamically regulated in human cells. J Cell Sci 123: 2663–2671 [DOI] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408 [DOI] [PubMed] [Google Scholar]
- Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T 2002. A kinetic framework for a mammalian RNA polymerase in vivo. Science 298: 1623–1626 [DOI] [PubMed] [Google Scholar]
- Fan X, Moqtaderi Z, Jin Y, Zhang Y, Liu XS, Struhl K 2010. Nucleosome depletion at yeast terminators is not intrinsic and can occur by a transcriptional mechanism linked to 3′-end formation. Proc Natl Acad Sci 107: 17945–17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Cioci F, Nomura M, Beyer AL 2003. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol 23: 1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreau L, Schmid A, Blaschke D, Ptashne M, Hörz W 1997. RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell 89: 55–62 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Theil AF, Mari PO, Mourgues S, Nonnekens J, Andrieux LO, De WJ, Miquel C, Wijgers N, Maas A, et al. 2009. Differentiation driven changes in the dynamic organization of Basal transcription initiation. PLoS Biol 7: e1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze H, Wittner M, Hamperl S, Hondele M, Merz K, Stoeckl U, Griesenbeck J 2010. Alternative chromatin structures of the 35S rRNA genes in Saccharomyces cerevisiae provide a molecular basis for the selective recruitment of RNA polymerases I and II. Mol Cell Biol 30: 2028–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski SA, Snyder SK, John S, Grummt I, Misteli T 2008. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. Mol Cell 30: 486–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossett AJ, Lieb JD 2012. In vivo effects of histone H3 depletion on nucleosome occupancy and position in Saccharomyces cerevisiae. PLoS Genet 8: e1002771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T 2009. Transcription dynamics. Mol Cell 35: 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Grunstein M 1988. Nucleosome loss activates yeast downstream promoters in vivo. Cell 55: 1137–1145 [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK 2008. The anchor-away technique: Rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932 [DOI] [PubMed] [Google Scholar]
- Imoberdorf RM, Topalidou I, Strubin M 2006. A role for gcn5-mediated global histone acetylation in transcriptional regulation. Mol Cell Biol 26: 1610–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Imoberdorf RM, Strubin M 2007. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell 25: 345–355 [DOI] [PubMed] [Google Scholar]
- Karpova TS, Kim MJ, Spriet C, Nalley K, Stasevich TJ, Kherrouche Z, Heliot L, McNally JG 2008. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science 319: 466–469 [DOI] [PubMed] [Google Scholar]
- Kim J, Iyer VR 2004. Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol Cell Biol 24: 8104–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristjuhan A, Svejstrup JQ 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J 23: 4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905 [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD 2012. Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Krett B, Laybourn PJ 2001. RNA polymerase II and TBP occupy the repressed CYC1 promoter. Mol Microbiol 40: 1009–1019 [DOI] [PubMed] [Google Scholar]
- Mencia M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol Cell 9: 823–833 [DOI] [PubMed] [Google Scholar]
- Nalley K, Johnston SA, Kodadek T 2006. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442: 1054–1057 [DOI] [PubMed] [Google Scholar]
- Pelechano V, Chavez S, Perez-Ortin JE 2010. A complete set of nascent transcription rates for yeast genes. PLoS ONE 5: e15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A 2007. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell 27: 393–405 [DOI] [PubMed] [Google Scholar]
- Russell J, Zomerdijk JC 2005. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem Sci 30: 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya J, Roberts SG 2012. The transcription cycle in eukaryotes: From productive initiation to RNA polymerase II recycling. Biochim Biophys Acta 1819: 391–400 [DOI] [PubMed] [Google Scholar]
- Sprouse RO, Karpova TS, Mueller F, Dasgupta A, McNally JG, Auble DT 2008. Regulation of TATA-binding protein dynamics in living yeast cells. Proc Natl Acad Sci 105: 13304–13308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprouse RO, Wells MN, Auble DT 2009. TATA-binding protein variants that bypass the requirement for Mot1 in vivo. J Biol Chem 284: 4525–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich TJ, McNally JG 2011. Assembly of the transcription machinery: Ordered and stable, random and dynamic, or both? Chromosoma 120: 533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Werven FJ, van Teeffelen HA, Holstege FC, Timmers HT 2009. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat Struct Mol Biol 16: 1043–1048 [DOI] [PubMed] [Google Scholar]
- Vannini A, Cramer P 2012. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 45: 439–446 [DOI] [PubMed] [Google Scholar]
- Xing H, Vanderford NL, Sarge KD 2008. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol 10: 1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT 2006. Dynamics of heat shock factor association with native gene loci in living cells. Nature 442: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408: 225–229 [DOI] [PubMed] [Google Scholar]
- Zanton SJ, Pugh BF 2006. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev 20: 2250–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Larson DR, Singer RH 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]