Abstract
RNA-guided endonucleases (RGENs), derived from the prokaryotic Type II CRISPR-Cas system, enable targeted genome modification in cells and organisms. Here we describe the establishment of gene-knockout mice and zebrafish by the injection of RGENs as Cas9 protein:guide RNA complexes or Cas9 mRNA plus guide RNA into one-cell-stage embryos of both species. RGENs efficiently generated germline transmittable mutations in up to 93% of newborn mice with minimal toxicity. RGEN-induced mutations in the mouse Prkdc gene that encodes an enzyme critical for DNA double-strand break repair resulted in immunodeficiency both in F0 and F1 mice. We propose that RGEN-mediated mutagenesis in animals will greatly expedite the creation of genetically engineered model organisms, accelerating functional genomic research.
The clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated (Cas) system induces an adaptive immune response against invading phages and plasmids in bacteria and archaea (Wiedenheft et al. 2012). Cas9, a protein derived from Streptococcus pyogenes, is complexed with CRISPR RNA (crRNA), whose sequence is originated from phages and plasmids, and trans-activating crRNA (tracrRNA) to form an active DNA-cleaving endonuclease whose specificity is determined by Watson–Crick base pairing between crRNA and the target DNA of 20 bp in length and by the NGG-trinucleotide protospacer adjacent motif (PAM) recognized by Cas9. A single-guide RNA (sgRNA) generated by fusing crRNA and tracrRNA is also functional and can reprogram Cas9 protein to cleave a specific DNA sequence (Fig. 1A; Jinek et al. 2012). Thus, Cas9 is an RNA-guided endonuclease (RGEN) that protects microbial cells from invading phages and plasmids by digesting their DNA site-specifically, reminiscent of restriction endonuclease-mediated protection of host cells from foreign genetic elements. Unlike restriction endonucleases, however, the Cas9 specificity is readily reprogrammed by replacing crRNA, making these nucleases ideal tools for genome engineering in higher eukaryotic cells and organisms.
Figure 1.
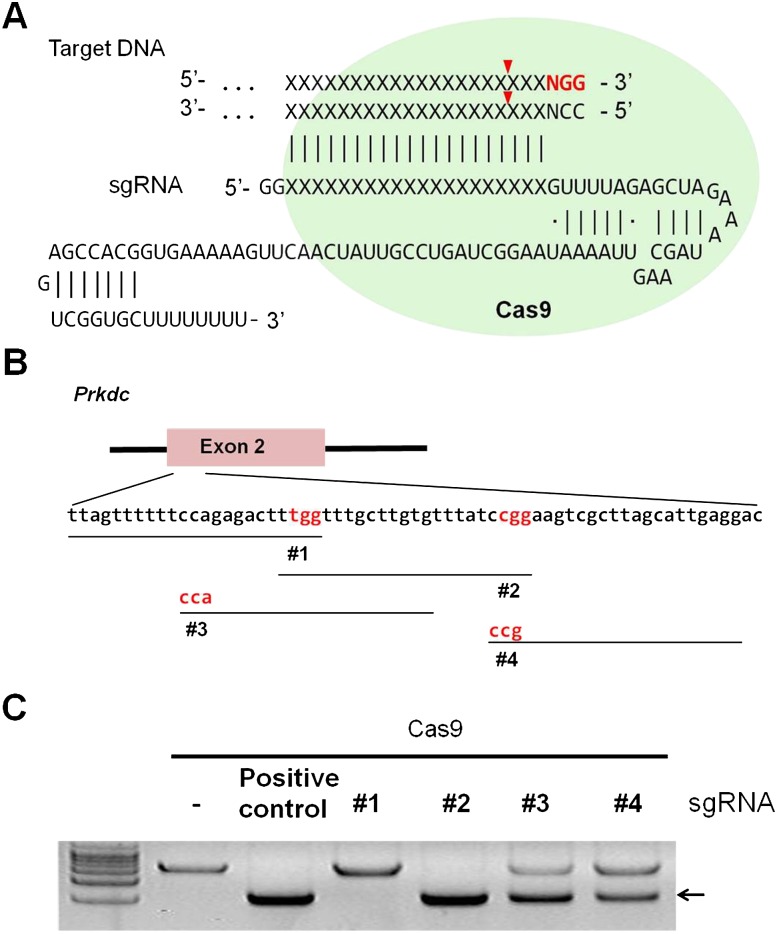
Generation of a RGEN specific for the Prkdc gene. (A) A schematic diagram depicting a target-specific single guide RNA (sgRNA). (B) RGEN target sites in exon 2 of the mouse Prkdc gene. PAMs are shown in red and targets are denoted by black lines. (C) In vitro cleavage assays evaluating Prkdc-RGEN activity. The arrow indicates bands cleaved by RGENs.
Recent studies have shown that Cas9-derived RGEN is, indeed, a new member in the growing family of genome editing nucleases (Bassett et al. 2013; Chang et al. 2013; Cho et al. 2013a; Cong et al. 2013; DiCarlo et al. 2013; Friedland et al. 2013; Hwang et al. 2013b; Jiang et al. 2013; Jinek et al. 2013; Mali et al. 2013; Shen et al. 2013; Wang et al. 2013; Xiao et al. 2013), which include zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs). These programmable nucleases induce site-specific DNA double-strand breaks (DSBs) in cells, whose repair via high-fidelity homologous recombination or error-prone nonhomologous end-joining (NHEJ) gives rise to targeted mutations (Bibikova et al. 2003; Kim et al. 2009; Kim et al. 2010; Miller et al. 2010) or chromosomal rearrangements (Brunet et al. 2009; Lee et al. 2009, 2011). For example, Shen et al. (2013) demonstrated that RGENs induced mutations in a reporter gene in transgenic mouse lines. Wang et al. (2013) used RGENs to induce multiple mutations in a single step in mice. Although these initial pioneering results are encouraging, it is unknown whether and how efficiently the RGEN-induced mutations detected in embryos or pups can be transmitted to the next generation, a process required for the establishment of gene-knockout (KO) animals. Here, we show that the injection of Cas9 protein complexed with guide RNA or Cas9 mRNA plus guide RNA into one-cell embryos gives rise to germline-transmittable mutations in vertebrate animals.
Results
RGEN design and production
To examine the gene knockout potential of RGENs in pronuclear (PN)–stage mouse embryos, we selected the forkhead box N1 (Foxn1) gene, which is important for thymus development and keratinocyte differentiation (Nehls et al. 1996), and the protein kinase, DNA activated, catalytic polypeptide (Prkdc) gene, which encodes an enzyme critical for DNA DSB repair and recombination (Taccioli et al. 1998). We designed several sgRNAs specific to exon 2 in the Foxn1 gene (Supplemental Fig. 1A) and exon 2 in the Prkdc gene (Fig. 1B). First, we tested the in vitro DNA-cleavage activities of RGENs composed of synthetic sgRNA and recombinant Cas9 protein expressed in and purified from Escherichia coli (Fig. 1C; Supplemental Fig. 1B). The Foxn1- and Prkdc-specific sgRNAs that manifested the highest activities were chosen for the subsequent in vivo experiments (#3 and #2, respectively) (Fig. 1C; Supplemental Fig. 1B).
Generation of founder mice with RGEN-induced mutations
To evaluate the genome-editing activity of the Foxn1-RGEN, we injected Cas9 mRNA (10 ng/μL solution) with various doses of the sgRNA into the cytoplasm of PN-stage mouse embryos and conducted T7 endonuclease I (T7E1) assays (Kim et al. 2009) using genomic DNAs obtained from in vitro–cultivated embryos (Supplemental Fig. 2A). Mutant fractions (the number of mutant embryos/the number of total embryos) were dose dependent, ranging from 31% (1 ng/μL sgRNA) to 87% (100 ng/μL) (Supplemental Fig. 2B). Sequence analysis confirmed mutations in the Foxn1 gene; most mutations were small deletions (Supplemental Fig. 2C), reminiscent of those induced by ZFNs and TALENs (Kim et al. 2013b). Notably, ∼90% of the RGEN-injected embryos developed to blastocysts and then hatched from the zona pellucida, suggesting that RGENs were not cytotoxic under our experimental conditions (Supplemental Fig. 2B).
Encouraged by the high mutant frequencies and low cytotoxicity induced by RGENs, we produced live animals by transferring the mouse embryos into the oviducts of pseudo-pregnant foster mothers (Supplemental Fig. 3A–C; Table 1). Notably, the birth rates were very high, ranging from 58% to 73%, and were not affected by the increasing doses of Foxn1-sgRNA (Table 1). Out of 147 newborns, we obtained 99 mutant founder mice (Table 1). Consistent with the results observed in cultivated embryos (Supplemental Fig. 2B), mutant fractions were proportional to the doses of Foxn1-sgRNA, and reached up to 93% (100 ng/μL Foxn1-sgRNA) (Table 1). To generate Prkdc-targeted mice and to test whether higher doses of RGENs are tolerated in embryos, we applied a fivefold higher concentration of Cas9 mRNA (50 ng/μL) with increasing doses of Prkdc-sgRNA (50, 100, and 250 ng/μL). Again, the birth rates were very high, ranging from 37% to 60%, enough to produce a sufficient number of newborns for the analysis (Fig. 2; Table 1). The mutant fraction was 57% (21 mutant founders among 37 newborns) at the maximum dose of Prkdc-sgRNA (Table 1). These birth rates obtained with RGENs were approximately twofold to 10-fold higher than those obtained with TALENs targeting two unrelated genes, which were reported in our previous study (Sung et al. 2013), on par with those of transgenic mouse production (Nagy et al. 2003). These results demonstrate that RGENs are potent gene-disrupting reagents with minimal toxicity.
Table 1.
RGEN-mediated gene knockout in FVB/NTac mice
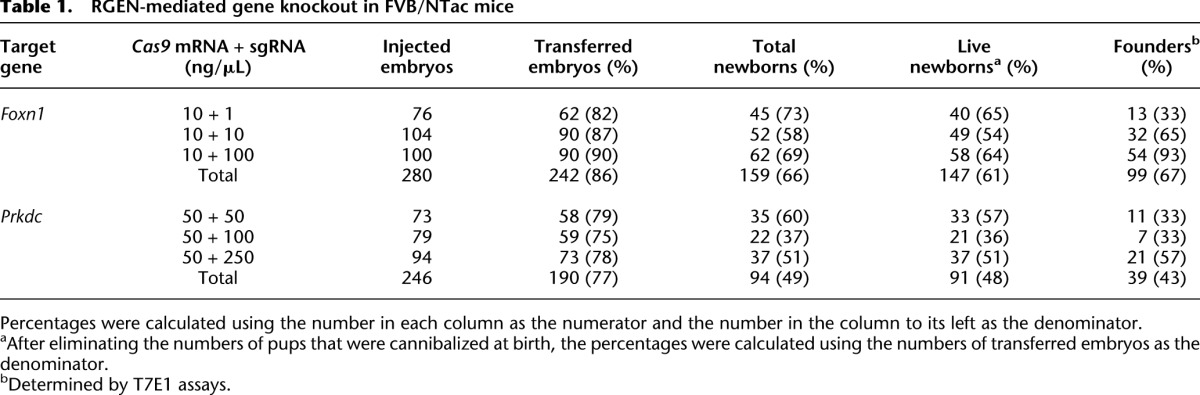
Figure 2.
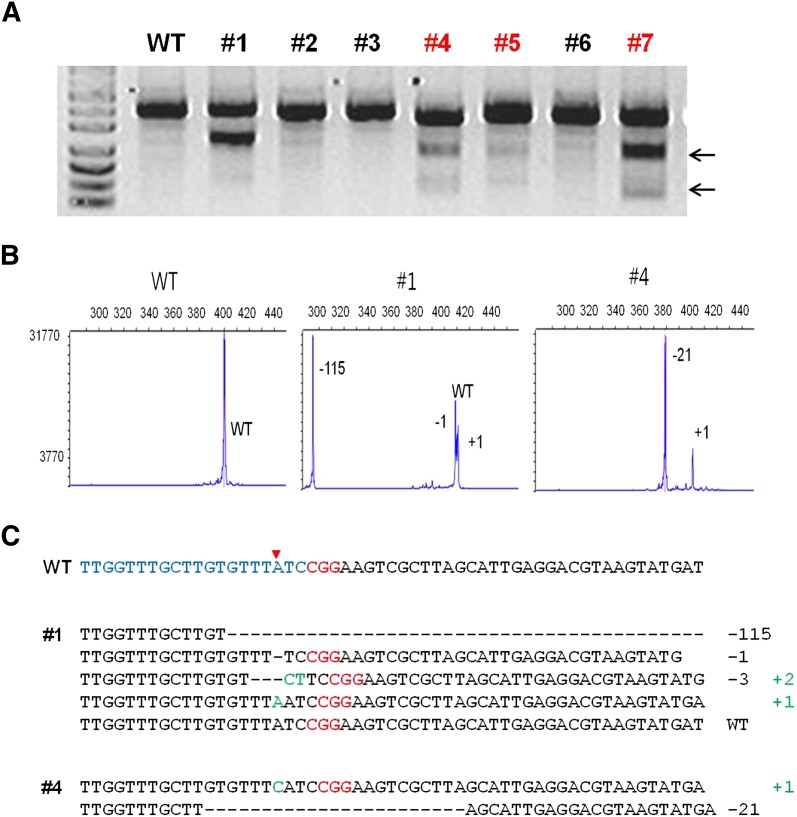
RGEN-induced Prkdc gene disruption in mice. (A) Representative T7E1 assays demonstrating mutagenesis efficiencies of Cas9 mRNA plus Prkdc-specific sgRNA that were delivered via intracytoplasmic injection into one-cell-stage mouse embryos. Numbers indicate independent founder mice generated from the highest dose. Arrows indicate bands cleaved by T7E1. fPCR results (B) and DNA sequences of mutant alleles (C) observed in three Prkdc mutant founders denoted in red in A. The red arrowhead in C indicates the cleavage site by the RGEN.
Fluorescent PCR (fPCR) (Fig. 2B; Supplemental Fig. 3B; Supplemental Tables 1, 2; Schuelke 2000) and Sanger sequencing analyses (Fig. 2C; Supplemental Fig. 3C; Supplemental Tables 3, 4) revealed distinct characteristics of RGEN-induced mutations in founder mice. Most Foxn1 mutants were mosaic (22 out of 33 founders, 67%), and exhibited up to five alleles (founder #66), indicating that the RGEN activity persisted later than the first cleavage of one-cell embryos (Supplemental Table 1). Notably, nine mice (27%) harbored bi-allelic mutations with no wild-type allele. Similar mosaic patterns were also observed in Prkdc mutant mice (20/39 founders, 51%) (Supplemental Table 2). In addition, identical mutant alleles were often observed for both Foxn1 and Prkdc genes (Supplemental Tables 3, 4). For example, we identified three founder mice with homozygous mutations in the Foxn1 gene (Supplemental Table 1).
Gene disruption by Cas9 protein–sgRNA complex in mice and zebrafish
Alternatively, we directly injected the RGEN in the form of recombinant Cas9 protein harboring a nuclear localization signal (NLS) (0.3–30 ng/μL) complexed with Foxn1-specific sgRNA (0.14–14 ng/μL) into the cytoplasm or pronucleus of one-cell mouse embryos and analyzed mutations in the Foxn1 gene using in vitro cultivated embryos (Supplemental Fig. 4). These injection doses and methods minimally affected the survival and development of mouse embryos in vitro: >70% of RGEN-injected embryos normally hatched from the zona pellucida in both experiments. Again, mutant fractions obtained with Cas9 protein injection were dose dependent, and reached up to 88% at the highest dose via pronucleus injection and to 71% via intracytoplasmic injection (Supplemental Fig. 4A,B). Similar to the mutation patterns induced by Cas9 mRNA plus sgRNA (Supplemental Fig. 3C), those induced by the Cas9 protein–sgRNA complex were mostly small deletions (Supplemental Fig. 4C). These results clearly demonstrate that the proteinaceous RGEN has high gene-disrupting activity in mouse embryos.
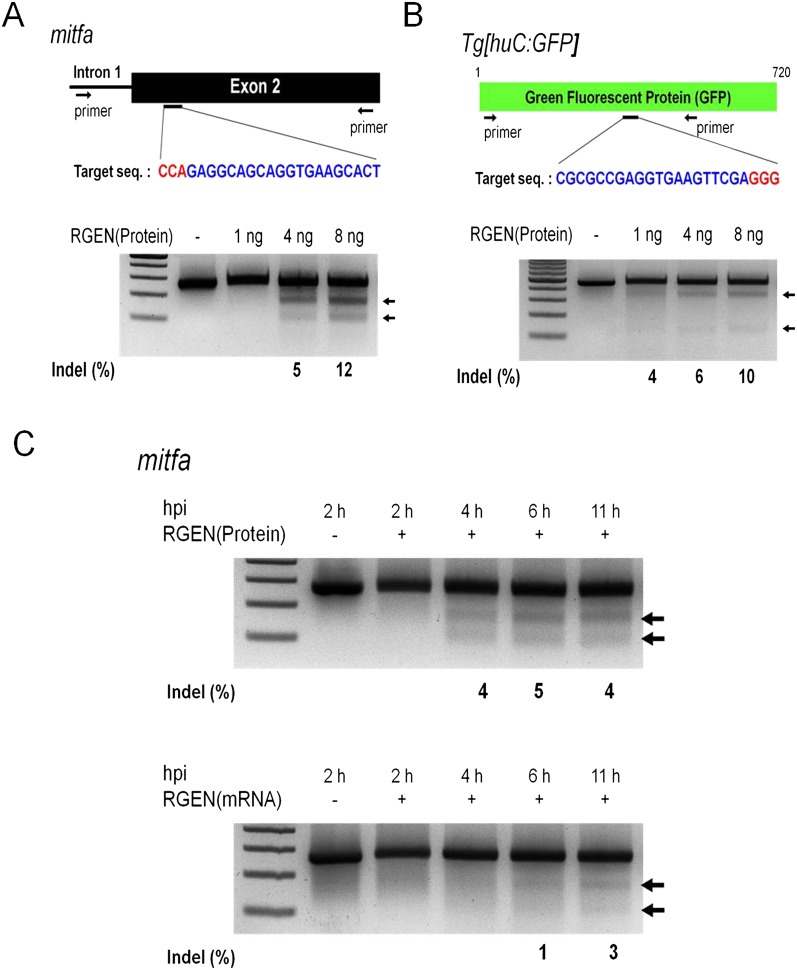
We also tested whether proteinaceous RGENs could induce mutations in zebrafish embryos. We designed RGENs that target the exogenous GFP reporter gene or an endogenous gene, microphthalmia-associated transcription factor a (mitfa), which encodes a master regulator of melanocyte development in vertebrates (Fig. 3A,B; Lister et al. 1999; Park et al. 2000). Injection of up to 8 ng of Cas9 protein complexed with mitfa-specific sgRNA did not perturb normal development in >80% of the embryos (Supplemental Fig. 5). When embryos were injected with Cas9 protein plus mitfa- or GFP-specific sgRNA, mutations were detected at target sites in a dose-dependent manner (Fig. 3A,B). A time-course experiment revealed that the Cas9 protein:sgRNA complex triggered mutations as early as at 4 h post-injection stage, 2 h faster than did Cas9 mRNA plus sgRNA (Fig. 3C). A subset of zebrafish embryos coinjected with Cas9 protein:mitfa-specific sgRNA manifested defects in melanocyte development (Supplemental Fig. 7), demonstrating that some of their somatic tissues were nullizygous for the mitfa gene. These results indicate that RGENs injected as Cas9 protein:sgRNA complexes can exert sufficient gene-disrupting activities to induce null mutations at both endogenous and exogenous genomic loci at an early developmental stage in zebrafish embryos.
Figure 3.
RNA-guided mutagenesis in zebrafish embryos. (A,B) Target sequences (upper) and RGEN-induced indels detected by T7E1 assays (lower). The Cas9 protein–sgRNA complex was microinjected into one-cell-stage embryos with concentrations of 1 ng, 4 ng, or 8 ng per embryo. (C) Time courses of Cas9 protein:sgRNA and Cas9 mRNA:sgRNA injection into embryos during early development. Arrows indicate bands cleaved by T7E1. (hpi) Hours post-injection.
Analysis of off-target effects of RGENs
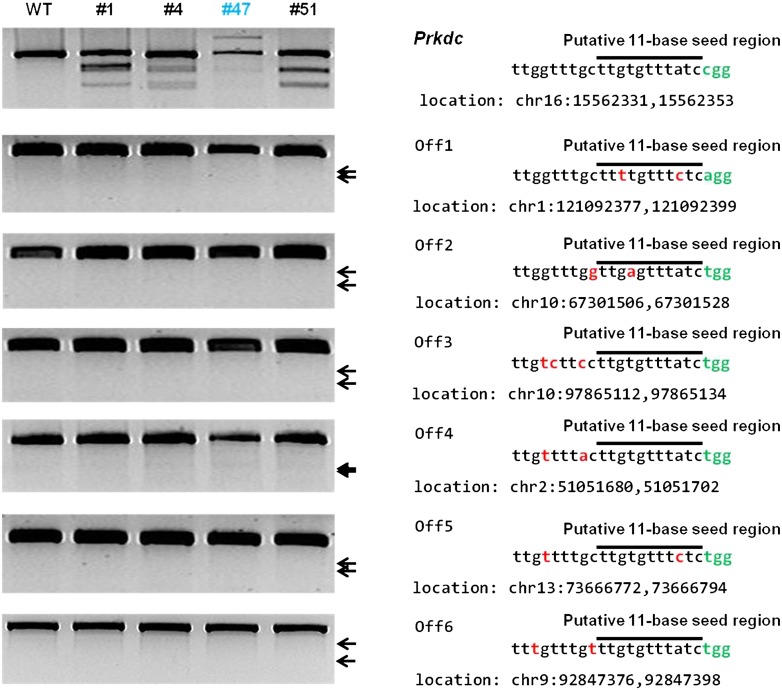
We examined whether the RGENs generated off-target mutations in the mouse genome. We searched for possible off-target sites that included 1- to 3-bp mismatches with the sgRNA target sequence. A total of four and 63 such sites were found for the Foxn1 and Prkdc target sequences, respectively, in the entire mouse genome. T7E1 assays were conducted using genomic DNA samples from the founder mice obtained with the highest doses of Foxn1- and Prkdc-sgRNAs. These founder mice were bi-allelic or mosaic mutants (Supplemental Tables 1, 2). Even though these mutant founders were produced using high levels of RGENs in one-cell embryos, no off-target activities were detected in any of the four genomic loci that were highly homologous with the target sequence of Foxn1-RGEN or any of the six such sites of the Prkdc-RGEN (Fig. 4; Supplemental Fig. 8). Of note is the absence of T7E1-detectable mutations at sites with a single-base mismatch in the putative 11-base seed region near the GG-dinucleotide PAM (e.g., Prkdc Off5 and Foxn1 Off3 in Fig. 4 and Supplemental Fig. 8) or with a perfect match in the seed region and 2- or 3-base mismatches upstream of the seed region (Prkdc Off3, Off4, and Off6 in Fig. 4).
Figure 4.
Analysis of off-target activity of RGENs in Prkdc-mutant founders. T7E1 assays were conducted using genomic DNA samples from the indicated Prkdc mutant founders. The Prkdc gene and putative off-target sites bearing 1 to 3-bp mismatches that were selected with Bowtie 0.12.9 (Langmead et al. 2009) were examined. Bi-allelic mutants containing large insertions are shown in blue (Supplemental Table 2), mismatched bases in red, and PAM in green; black lines indicate putative 11-bp seed regions.
Phenotype analysis and germline transmission of Prkdc-mutant mice
Targeted deficiency of the Prkdc gene in mice results in severe combined immunodeficiency (Taccioli et al. 1998). For the functional validation of RGEN-induced mutations of the Prkdc gene in mice, we evaluated the immunological phenotypes of F0 mutant founders using peripheral blood mononuclear cells (PBMCs). The mosaic founder #1 carried a wild-type allele (Supplemental Table 2) and showed comparable B- and T-cell populations to those of the wild-type BALB/c mouse (Supplemental Fig. 9). On the other hand, the mutant founder #47 possessed bi-allelic null mutations in the Prkdc gene, and these immune cell populations were significantly lowered in the PBMC (Supplemental Fig. 9), indicating the functional loss of the Prkdc gene.
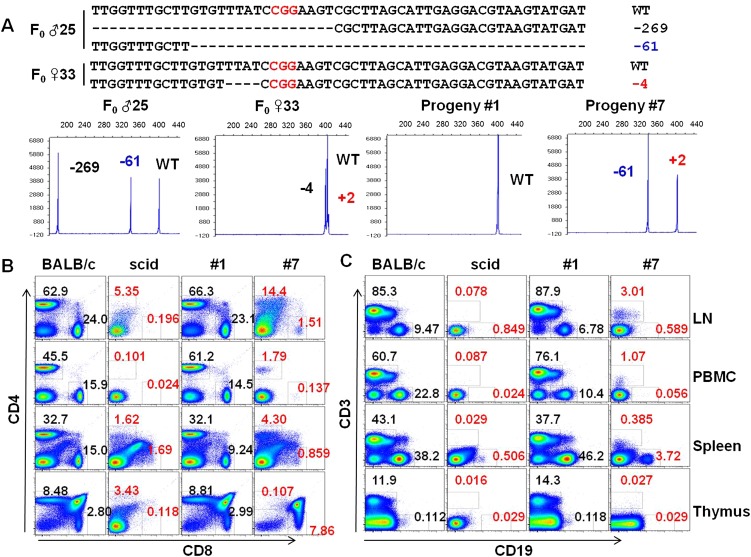
To test the germline transmission of the mutant alleles of the Prkdc gene, we crossed the male founder #25 (Δ269/Δ61/WT) with female founder #33 (Δ4/+2/WT) (Supplemental Table 2) and monitored the genotypes of the F1 offspring (Fig. 5A). As expected, all the progenies were wild type or inherited one or two mutant alleles from their parents (Fig. 5A). We also confirmed the germline transmission of Foxn1 mutant alleles from the Foxn1 founder #108 and observed the segregation of the mutant alleles in the progenies (Supplemental Fig. 10).
Figure 5.
Germline transmission and mutant phenotypes of the Prkdc mutant mouse. (A) Detailed genotypes of F0 mutants (♂ 25 and ♀ 33, upper panel) and determination of genotypes of their F1 progenies (#1 and #7) by fPCR (lower panel). Note that a mutant allele (+2) of female founder #33 was only detected by fPCR (Supplemental Table 2). Using lymphocytes isolated from peripheral blood (PBMC), lymph node (LN), spleen, and thymus, frequencies of CD4+ and CD8+ T cells (B) and those of CD3+ T cells and CD19+ B cells (C) were determined. The abnormal frequencies of immune cells are denoted in red. BALB/c and scid mice, positive and negative controls for B and T lymphocytes, respectively.
As expected, the bi-allelic Prkdc mutant progeny showed a null phenotype (Fig. 5A). Compared with the wild-type BALB/c mouse and the progeny #1 carrying a wild-type allele, the bi-allelic mutant progeny #7 exhibited defects in B- and T- cell populations (Fig. 5B,C). Thus, lymphocytes isolated from PBMC and from the primary and secondary lymphoid organs including thymus, spleen, and lymph node were defective in B- and T-cell populations (Fig. 5B). Compared with severe combined immunodeficiency (scid) mice, the mutant mouse exhibited phenotypic leakiness. For example, the F1 progeny had 1.79% CD4+ and 1.07% CD3+ T cells, whereas the T-cell populations examined were <1% in the scid mouse (Fig. 5B). These small differences between scid and the bi-allelic mutant mice might reflect the differences in their genetic backgrounds, ages, or genotypes. Overall, these results clearly proved the functional loss of the Prkdc gene by RGEN-mediated gene knockout in mice.
Discussion
Our study provides strong evidence that RGENs hold great promise for generations of knockout mice and zebrafish. Although there were recent reports describing the (EA) generations of mutant mice using RGENs (Shen et al. 2013; Wang et al. 2013), germline transmission of the mutations induced by RGENs has not been confirmed. We showed that RGEN-induced mutant alleles were stably transmitted to F1 progenies in vertebrate animals and confirmed the mutant phenotypes using both founder mice and F1 mutant progenies. In addition, we also showed that recombinant Cas9 protein complexed with sgRNA induced mutations in both mice and zebrafish, which potentially broadens the range of the RGEN application for in vivo mutagenesis. Nuclease proteins might have different kinetics and half-lives in cells, when delivered directly, as compared with being produced by gene- or mRNA-mediated expression, potentially alleviating off-target effects (Izmiryan et al. 2011; Gaj et al. 2012). Unlike recombinant ZFNs and TALENs whose purification is cumbersome, time-consuming, and laborious, new proteinaceous RGENs are prepared simply by mixing Cas9 protein with different sgRNAs or dualRNAs. These features strongly support the notion that RGENs are convenient and efficient tools for gene knockout in model organisms.
The specificity of RGENs is of concern. Because RGENs are derived from prokaryotes, whose genomes are much smaller than those of higher eukaryotes, RGENs may have off-target effects in animals, plants, and cultured human cells such as stem cells. Furthermore, unlike ZFNs and TALENs, which function as dimers that enhance specificity, RGENs act as monomers. Recent in vitro studies conducted using cell lines have provided the evidence of off-target mutations induced by RGENs (Cong et al. 2013; Fu et al. 2013; Hsu et al. 2013; Pattanayak et al. 2013). Fortunately, however, our analyses of off-target effects suggest that RGENs might not be associated with highly frequent off-target mutations in vivo or in clones. Unlike in vitro experiments using cell populations, our in vivo experiments analyzed individual offspring or clones. Off-target mutations present in cell populations can be diluted out in individual clones. In addition, some of the rare off-target mutations in the founders can be removed during mating. Although it is possible that the RGENs induced off-target mutations at sites that are not analyzed in this study, our results suggest that RGENs are specific enough to allow targeted mutagenesis in the mouse. A careful choice of target sites that lack highly homologous sequences elsewhere in the genome should be useful for avoiding or minimizing unwanted off-target mutations as much as possible (Hsu et al. 2013; Kim et al. 2013a).
On the basis of these results, we propose that RGEN-mediated in vivo mutagenesis will greatly expedite the creation of genetically engineered model organisms and thereby accelerate functional genomic research. Recently, three groups (Fujii et al. 2013; Hwang et al. 2013a; Li et al. 2013) have independently reported germline transmission of RGEN-induced mutations in zebrafish, mice, and rats, demonstrating broad utility of RNA-guided genome editing in vertebrate model organisms. We also note that we have used proteinaceous RGENs to disrupt genes in Caenorhabditis elegans (Cho et al. 2013b).
Methods
RGEN components
Cas9 mRNA and sgRNAs were synthesized in vitro from linear DNA templates using the mMESSAGE mMACHINE T7 Ultra kit (Ambion) and MEGAshortscript T7 kit (Ambion), respectively, according to the manufacturer's instructions, and were diluted with appropriate amounts of diethyl pyrocarbonate (DEPC, Sigma-Aldrich)–treated injection buffer (0.25 mM EDTA, 10 mM Tris at pH 7.4). Templates for sgRNA synthesis were generated using the oligonucleotides listed in Supplemental Table 5 as described previously (Cho et al. 2013a). Recombinant Cas9 protein (Cho et al. 2013a) was obtained from ToolGen, Inc.
In vitro cleavage reactions
A restriction enzyme–treated linearized plasmid (5 nM) was incubated for 60 min at 37°C with Cas9 protein (20 nM), and sgRNA (40 nM) in 1× NEB 3 buffer. Reactions were stopped with 6× stop solution containing 30% glycerol, 1.2% SDS, and 100 mM EDTA. Products were resolved with 1% agarose gel electrophoresis and were visualized with ethidium bromide (EtBr) staining.
Microinjection of RGENs into mouse embryos
All animal experiments were performed in accordance with the Korean Food and Drug Administration (KFDA) guidelines. Protocols were reviewed and approved by the Institutional Animal Care and Use Committees (IACUC) of the Laboratory Animal Research Center at Yonsei University (Permit Number: 2013-0099). All mice were maintained in the specific pathogen–free (SPF) facility of the Yonsei Laboratory Animal Research Center.
FVB/NTac (Taconic) and ICR (DBL, Korea) mouse strains were used as embryo donors and foster mothers, respectively. Female FVB/NTac mice (7–8 wk old) were superovulated by intraperitoneal injections of 5 IU of pregnant mare serum gonadotropin (PMSG, Sigma-Aldrich) and 5 IU of human chorionic gonadotropin (hCG, Sigma-Aldrich) at a 48-h interval. The superovulated female mice were mated to FVB/NTac stud males, and fertilized embryos were collected from oviducts.
Cas9 mRNA and sgRNAs in M2 medium (Sigma-Aldrich) were injected into the cytoplasm of fertilized eggs with well-recognized pronuclei using a Piezo-driven micromanipulator (Prime Tech). The recombinant Cas9 protein:Foxn1-sgRNA complex was diluted with DEPC-treated injection buffer (0.25 mM EDTA, 10 mM Tris at pH 7.4) and injected into male pronuclei using a TransferMan NK2 micromanipulator and a FemtoJet microinjector (Eppendorf). The manipulated embryos were transferred into the oviducts of pseudopregnant foster mothers to produce live animals or were cultivated in vitro for further analyses.
Fluorescent PCR
One hundred nanograms of genomic DNA was amplified by PCR using 5′-6-carboxyfluorescein (6-FAM)-labeled primers (F2) (Supplemental Table 6). PCR products were resolved using an ABI 3730xl DNA analyzer, and fPCR results were analyzed using a Peak scanner v1.0.
Genotyping, sequence analyses, and phenotyping of mutant mice
To screen F0 mice and in vitro–cultivated mouse embryos with RGEN-induced mutations, T7E1 assays were performed as previously described using genomic DNA samples from tail biopsies and lysates of whole embryos (Cho et al. 2013a). Briefly, the genomic region encompassing the RGEN target site was PCR-amplified, melted, and reannealed to form heteroduplex DNA, which was treated with T7 endonuclease 1 (New England Biolabs) and then analyzed by agarose gel electrophoresis. For the newborn mice that were negative in the T7E1 assays, additional T7E1 assays were conducted by mixing equal amounts of wild-type PCR products. Potential off-target sites were identified by searching with Bowtie 0.12.9 (Langmead et al. 2009) and were also similarly monitored by T7E1 assays. The primer pairs used in the T7E1 assays were represented in Supplemental Tables 6 and 7.
Mutant founders identified by the T7E1 assay were further analyzed by fPCR. For routine PCR genotyping of F1 progenies, the following primer pairs were used for both wild-type and mutant alleles: 5′-CTACTCCCTCCGCAGTCTGA-3′ and 5′-CCAGGCCTAGGTTCCAGGTA-3′ for the Foxn1 gene, 5′-CCCCAGCATTGCAGATTTCC-3′ and 5′-AGGGCTTCTTCTCTACAATCACG-3′ for the Prkdc gene.
Characterization of immune cells by flow cytometry
BALB/c mice were purchased from DBL (Korea), and C.B.-17 scid mice (C.B-Igh-1b/IcrTac-Prkdcscid) were imported from Taconic. Lymphocytes were prepared and processed as previously described (Park et al. 2000). PerCP-, APC-, V605-, V421-, PE-Cy7-, and PerCP-Cy5.5-conjugated antibodies specifically detecting CD3e (145-2C11), CD19 (1D3), CD4 (RM4-5), and CD8 (53-6.7), respectively, were purchased from BD Biosciences or Biolegend. Flow cytometry was performed with FACS Calibur and FACS CantoII (BD Biosciences), and the data were analyzed using the FlowJo software (Tree Star).
Procedures of RGEN-mediated mutagenesis in zebrafish embryos
Wild-type and Tg[huC:GFP] transgenic zebrafish were maintained at 28.5°C on a 14-h light/10-h dark cycle. Embryonic stages were determined by the post-fertilization hour and microscopic observation. Animal work was approved by the internal animal ethics committee at Chungnam National University (CNU-00191).
One-cell-stage zebrafish embryos were injected with the indicated doses of Cas9 protein solution (7 μg/μL) containing crRNA (1 μg/μL), tracrRNA (2 μg/μL), or sgRNA (2.5 μg/μL). The injected embryos were incubated at 28.5°C for the indicated points, and their phenotypes and genotypes were examined. Genomic DNA was extracted from developing zebrafish embryos using a genomic DNA purification system (Promega) following the manufacturer's instructions. Targeted genomic loci were amplified from five pooled embryos using primers designed to flank the Cas9 complex target site and cloned into the pGEM T-easy vector (Promega). The cloned segments were then sequenced (Genotech). T7E1 assays were performed as described above.
Acknowledgments
H.-W.L. was supported by the Ministry of Education, Science, and Technology (20120006489, 20120000174, 2012009607) and by the Ministry of Health & Welfare (A085136), Republic of Korea. J.-S.K. was supported by the National Research Foundation of Korea (2013000718).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.163394.113.
References
- Bassett AR, Tibbit C, Ponting CP, Liu JL 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4: 220–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D 2003. Enhancing gene targeting with designed zinc finger nucleases. Science 300: 764. [DOI] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M 2009. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci 106: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ 2013. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res 23: 465–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS 2013a. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 31: 230–232 [DOI] [PubMed] [Google Scholar]
- Cho SW, Lee J, Carroll D, Kim JS 2013b. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9-sgRNA ribonucleoproteins. Genetics doi: 10.1534/genetics.113.155853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41: 4336–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10: 741–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD 2013. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31: 822–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii W, Kawasaki K, Sugiura K, Naito K 2013. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res doi: 10.1093/nar/gkt772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Guo J, Kato Y, Sirk SJ, Barbas CF III 2012. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods 9: 805–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. 2013. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR 2013a. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS ONE 8: e68708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK 2013b. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol 31: 227–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izmiryan A, Basmaciogullari S, Henry A, Paques F, Danos O 2011. Efficient gene targeting mediated by a lentiviral vector-associated meganuclease. Nucleic Acids Res 39: 7610–7619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31: 233–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J 2013. RNA-programmed genome editing in human cells. Elife 2: e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee MJ, Kim H, Kang M, Kim JS 2010. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat Methods 8: 7. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim A, Chon JK, Yoo JY, Kim HJ, Kim S, Lee C, Jeong E, Chung E, et al. 2013a. A library of TAL effector nucleases spanning the human genome. Nat Biotechnol 31: 251–258 [DOI] [PubMed] [Google Scholar]
- Kim Y, Kweon J, Kim JS 2013b. TALENs and ZFNs are associated with different mutation signatures. Nat Methods 10: 185. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kim E, Kim JS 2009. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res 20: 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Kweon J, Kim E, Kim S, Kim JS 2011. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res 22: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, et al. 2013. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 31: 681–683 [DOI] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW 1999. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126: 3757–3767 [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. 2010. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29: 143–148 [DOI] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R 2003. Manipulating the mouse embryo: A laboratory manual, p. 255. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Nehls M, Kyewski B, Messerle M, Waldschutz R, Schuddekopf K, Smith AJ, Boehm T 1996. Two genetically separable steps in the differentiation of thymic epithelium. Science 272: 886–889 [DOI] [PubMed] [Google Scholar]
- Park HC, Kim CH, Bae YK, Yeo SY, Kim SH, Hong SK, Shin J, Yoo KW, Hibi M, Hirano T, et al. 2000. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol 227: 279–293 [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR 2013. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31: 839–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuelke M 2000. An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18: 233–234 [DOI] [PubMed] [Google Scholar]
- Shen B, Zhang J, Wu H, Wang J, Ma K, Li Z, Zhang X, Zhang P, Huang X 2013. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res 23: 720–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW 2013. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol 31: 23–24 [DOI] [PubMed] [Google Scholar]
- Taccioli GE, Amatucci AG, Beamish HJ, Gell D, Xiang XH, Torres Arzayus MI, Priestley A, Jackson SP, Marshak Rothstein A, Jeggo PA, et al. 1998. Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 9: 355–366 [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA 2012. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482: 331–338 [DOI] [PubMed] [Google Scholar]
- Xiao A, Wang Z, Hu Y, Wu Y, Luo Z, Yang Z, Zu Y, Li W, Huang P, Tong X, et al. 2013. Chromosomal deletions and inversions mediated by TALENs and CRISPR/Cas in zebrafish. Nucleic Acids Res 41: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]