Abstract
Genetic imprinting is a specific epigenetic phenomenon in which a subset of genes is expressed depending on their parent-of-origin. Two types of chromatin modifications, DNA methylation and histone modification, are generally believed to be involved in the regulation of imprinting. However, the genome-wide correlation between allele-specific chromatin modifications and imprinted gene expression in maize remains elusive. Here we report genome-wide high resolution allele-specific maps of DNA methylation and histone H3 lysine 27 trimethylation (H3K27me3) in maize endosperm. For DNA methylation, thousands of parent-of-origin dependent differentially methylated regions (pDMRs) were identified. All pDMRs were uniformly paternally hypermethylated and maternally hypomethylated. We also identified 1131 allele-specific H3K27me3 peaks that are preferentially present in the maternal alleles. Maternally expressed imprinted genes (MEGs) and paternally expressed imprinted genes (PEGs) had different patterns of allele-specific DNA methylation and H3K27me3. Allele-specific expression of MEGs was not directly related to allele-specific H3K27me3, and only a subset of MEGs was associated with maternal-specific DNA demethylation, which was primarily located in the upstream and 5′ portion of gene body regions. In contrast, allele-specific expression of a majority of PEGs was related to maternal-specific H3K27me3, with a subgroup of PEGs also associated with maternal-specific DNA demethylation. Both pDMRs and maternal H3K27me3 peaks associated with PEGs are enriched in gene body regions. Our results indicate highly complex patterns of regulation on genetic imprinting in maize endosperm.
Genetic imprinting is an epigenetic phenomenon, where genes are expressed in a parent-of-origin dependent manner in many plant species and mammals. Although first discovered in plants (Kermicle and Alleman 1990), research on genetic imprinting is much more advanced in mammals in terms of the number of imprinted genes identified and the understanding of their regulatory mechanisms (Koerner and Barlow 2010; Barlow 2011; Bartolomei and Ferguson-Smith 2011). Only a small number of imprinted genes were observed in plants for a long time since the phenomenon is highly specific to the triploid endosperm (Raissig et al. 2011). However, recent studies have indicated that genetic imprinting in plants is much more prevalent than previously thought, with hundreds of genes shown to be imprinted in several plant species (Gehring et al. 2011; Hsieh et al. 2011; Luo et al. 2011; Waters et al. 2011; Zhang et al. 2011). In contrast to mammals, where the regulation of genetic imprinting has been extensively studied (Koerner and Barlow 2010; Barlow 2011; Abramowitz and Bartolomei 2012), the understanding of regulation of parental imprinting in plants is highly limited.
DNA methylation is one of the primary modifications reported to be associated with genetic imprinting. In Arabidopsis, DNA methylation around several maternally expressed imprinted protein-coding genes (MEG) including FWA, FIS2, and MPC was shown to be important for their maternally preferred expression, as all these genes exhibited biallelic expression in endosperm fertilized with met1 pollen (Kinoshita et al. 2004; Jullien et al. 2006; Tiwari et al. 2008). Two studies using RNA-seq in Arabidopsis also showed that a number of MEGs exhibited biallelic expression in paternal met1 endosperm, and the maternal alleles of dozens of paternally expressed imprinted genes (PEGs) were reactivated in maternal dme endosperm (Hsieh et al. 2011; Wolff et al. 2011). In maize, five confirmed endosperm MEGs (Fie1, Fie2, Mez1, Meg1, and Mee1) contain differentially methylated regions (DMRs) (Gutierrez-Marcos et al. 2004; Gutierrez-Marcos et al. 2006; Haun et al. 2007), and activation of the Fie1 maternal allele in the endosperm requires DNA demethylation of the maternal allele (Hermon et al. 2007).
Another modification associated with genetic imprinting involves histone methylation. Polycomb repressive complex 2 (PRC2) is known to mediate the trimethylation of histone H3 lysine 27 (H3K27me3) (Schuettengruber and Cavalli 2009). Results on PHE1, the only well-studied PEG in plants, indicated that silencing of its maternal allele depends on a functional PRC2 complex in addition to DNA demethylation (Kohler et al. 2003, 2005; Makarevich et al. 2008). Recently, a number of MEGs and PEGs were shown to be biallelically expressed in maternal fie or fis2 endosperm (Hsieh et al. 2011; Wolff et al. 2011).
Although the studies above suggest that DNA methylation and the PRC2 complex could be responsible for monoallelic expression of imprinted genes, there is not yet any general rule for the function of DNA and histone methylation on the regulation of genetic imprinting in plants. A high resolution genome-wide map of allele-specific DNA methylation and allele-specific histone modification will be crucial to gain better understanding of the regulation of genetic imprinting. Recently, several genome-wide studies have provided evidence that allele-specific patterns of DNA methylation or parent-of-origin dependent differentially methylated regions (pDMRs) are associated with some imprinted genes in mice and plants (Zhang et al. 2011; Ibarra et al. 2012; Xie et al. 2012; Rodrigues et al. 2013). Several studies in mammals also suggest a mutually exclusive relationship between allele-specific DNA methylation and histone modification or among different histone modifications (Xin et al. 2001; Fournier et al. 2002; Carr et al. 2007; Lindroth et al. 2008; Singh et al. 2010; Hon et al. 2012).
Here we report a genome-wide analysis of allele-specific DMRs, H3K27me3, and imprinted gene expression in maize endosperm. Thousands of pDMRs and allele-specific H3K27me3 peaks were identified. Correlation of pDMRs, allele-specific H3K27me3 profile, and the expression of imprinted genes showed that MEGs and PEGs have different patterns of DNA methylation and H3K27me3. This study reveals complex patterns of genetic imprinting regulation in maize endosperm.
Results
Global analysis of maize endosperm DNA methylome
To investigate the allele-specific DNA methylation pattern of maize endosperm on a genome-wide scale, we performed MethylC-seq for shoot, embryo, and endosperm tissue 12 d after pollination (DAP) of inbred B73, and the endosperm tissue 12 DAP of reciprocal crosses B73 × Mo17 (BM) and Mo17 × B73 (MB) (Supplemental Table S1). The parental origins of the two alleles in the hybrids were distinguished using 4.17 million SNPs identified from whole genome resequencing between B73 (Schnable et al. 2009) and Mo17 (Lai et al. 2010; Jiao et al. 2012). Cytosine methylations were inferred by mapping of the MethylC-seq sequences to the reference B73 genome (Schnable et al. 2009). The overall methylation level in maize endosperm compared with shoot and embryo tissue was very similar, except for CHG methylation, which was lower in the endosperm (Supplemental Fig. S1). Direct comparison of methylation between two parental genomes showed that the levels of methylation of the maternal and paternal genomes are almost the same in both CG and CHG contexts, with the methylation level of the maternal genome slightly lower than that of the paternal genome in CG context (Supplemental Fig. S2).
Identification and analyses of parent-of-origin dependent DMRs
To understand the relationship of DNA methylation and genetic imprinting, we scanned the genome for pDMRs using a sliding-window strategy (see Methods). As a result, 6910 and 4456 pDMRs were identified in CG and CHG context, respectively. It is worth noting that pDMRs identified in our study were uniformly paternally hypermethylated and maternally hypomethylated. For validation, pDMR of Fie1 identified in this study is consistent with the previous allelic methylation study of this gene (Supplemental Fig. S3; Hermon et al. 2007). These pDMRs in the CG context (CG_pDMR) and pDMRs in the CHG context (CHG_pDMR) account for 1.5% and 1%, respectively, of the maize endosperm genome where allelic methylation can be analyzed.
Both CG_ and CHG_pDMRs exhibited lower CG content (Supplemental Fig. S4), a feature that was also reported for DME targets in Arabidopsis (Ibarra et al. 2012) and a greater enrichment in short transposable elements compared with the whole genome analyzed (P-value < 2.2 × 10−16). Further analysis showed that CG_pDMR preferentially occurred in genic regions (Supplemental Fig. S5).
To further understand the relationship between CG_pDMRs and CHG_pDMRs, we investigated the methylation pattern (both CHG and CG) of the corresponding regions of our identified CG_pDMRs and CHG_pDMRs. We found that the majority of CHG_pDMRs (73.5%) exhibited differential methylation in their CG context, while only a small portion of CG_pDMRs (39%) showed differential methylation between two parental alleles in their CHG context. In other words, CHG_pDMRs are usually accompanied by differential methylation of the CG context. A group of CG_pDMRs had a very low level of CHG methylation in both of their parental alleles and did not show differential methylation in the CHG context (Supplemental Fig. S6).
Next, we examined the methylation status of these pDMRs in the shoot and embryo. All the pDMR regions exhibited much higher methylation levels in the shoot and embryo tissues compared with that of the endosperm, and the levels of methylation of paternal alleles were similar to those of the shoot and embryo (Supplemental Fig. S7).
Identification of additional imprinted genes in maize endosperm
A recent study (Zhang et al. 2011) suggested that ∼1.7% of genes expressed in maize endosperm are imprinted and the currently identified imprinted genes (Waters et al. 2011; Zhang et al. 2011) are still only a small part of all imprinted genes in maize endosperm, due to the coverage of sequencing and developmental stages of samples used. Hence, we performed additional RNA-seq for samples from 12-DAP and 10-DAP endosperm of both reciprocal crosses between inbreds B73 and Mo17 (Supplemental Table S1). A total of 304 imprinted transcripts were identified (including the imprinted genes previously identified) using previously reported high stringent criteria (Zhang et al. 2011) where the expression level of actively expressed alleles must be at least five times that of the repressed alleles. Among these 304 transcripts, there were 125 PEGs, 143 MEGs, eight paternally expressed noncoding RNAs (PNCs), and 28 maternally expressed noncoding RNAs (MNCs).
Among our newly identified 28 MNCs, 18 of them were located partially or wholly in intronic regions of annotated genes (Supplemental Table S2). Interestingly, 13 of the 18 intronic MNCs are from potential PEGs, five of which were identified with reduced criteria (parental bias in both reciprocal crosses was only required to deviate from 2:1 using χ2 test at α = 0.05). However, we did not find any PNCs in the vicinity of MEGs (Supplemental Table S2). Our results suggested a potential functional relationship between imprinted noncoding RNA (particularly MNCs) and PEGs. By combining our data with that of earlier reports (Waters et al. 2011; Zhang et al. 2011) we were able to identify 107 imprinted genes (68 PEGs and 39 MEGs) that have imprinted expression consistently in three endosperm tissue stages (10 DAP, 12 DAP, and 14 DAP) (Supplemental Table S3). These 107 high quality imprinted genes will be used for our further analysis of the correlation of allele-specific DNA methylation and H3K27me3 with the expression of imprinted genes. The average length of PEGs (7718 bp) was longer than that of MEGs (4180 bp). The difference in length was largely due to differences in intron size (MEG: 2295 bp; PEG: 6117 bp), with PEGs and MEGs having similar lengths of coding sequences (MEG: 1441 bp; PEG: 2031 bp). In addition, compared with MEGs, PEGs tend to have lower CG content (Supplemental Fig. S8).
Relationship between differential DNA methylation and imprinted genes
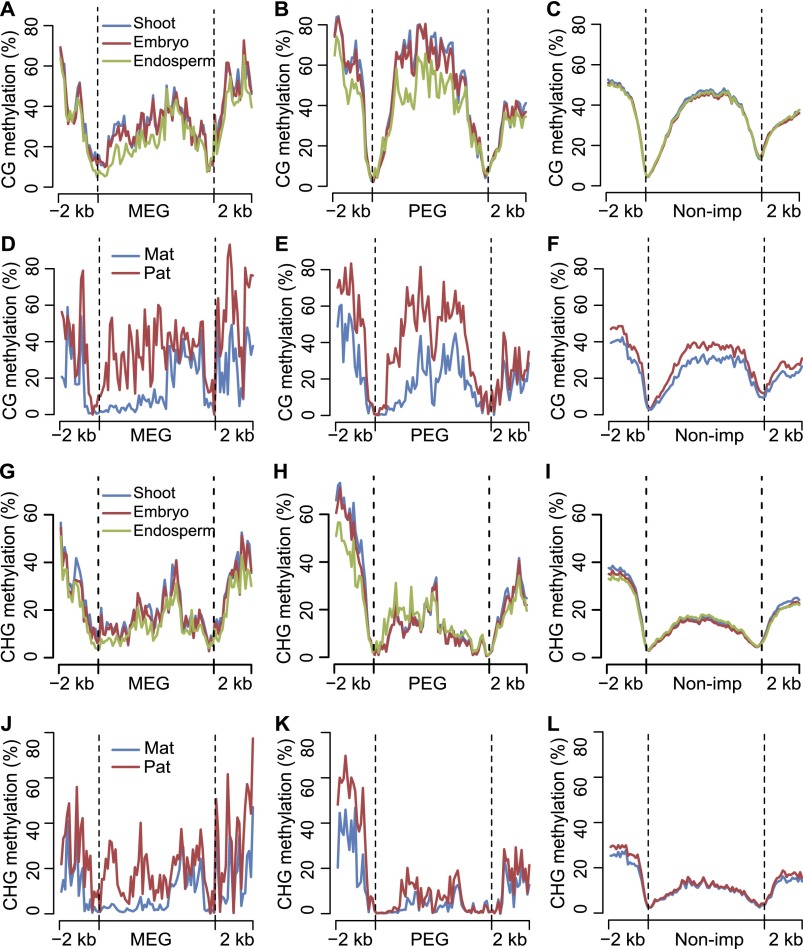
The availability of imprinted genes and whole genome allele-specific DNA methylome data in maize endosperm allowed us to investigate the correlation between DNA methylation and expression of the imprinted genes. We first looked at the DNA methylation pattern in CG context. In all three tissues examined (endosperm, shoot, and embryo), the overall CG methylation level for PEGs was higher than that of MEGs, while that of MEGs was about the same as nonimprinted genes (Fig. 1A–C). Additionally, the overall levels of DNA methylation of PEGs in the upstream and gene body regions were slightly lower in the endosperm than that in shoot and embryo tissues. There was not much difference in the DNA methylation level for either MEGs or nonimprinted genes among the three tissues (Fig. 1A–C). When the DNA methylation levels of the two parental alleles were examined in the reciprocal crosses, methylation levels of maternal and paternal alleles of MEGs were about the same, except for the 5′ portion of gene body regions where there was maternal demethylation compared with the paternal alleles or nonimprinted genes (Fig. 1D–F). The DNA methylation levels of the maternal alleles of PEGs were lower than their paternal alleles along their upstream and gene body regions (Fig. 1E). Examples of extensive maternal CG demethylation through nearly entire gene bodies were shown for two confirmed PEGs (Zhang et al. 2011): GRMZM2G028366 and GRMZM2G406553 (Supplemental Fig. S9). For CHG context, slight maternal DNA demethylation was also seen in the 5′ parts of gene bodies of MEGs (Fig. 1J). For PEGs, maternal DNA demethylation in endosperm was seen only in the upstream regions (Fig. 1K). Similar upstream DNA demethylation can also be seen when patterns of methylation of PEGs in the endosperm are compared with those in shoot and embryo tissues (Fig. 1G,H).
Figure 1.
Differential DNA methylation among tissues and between two parental genomes for maternally and paternally expressed imprinted genes. (A–C) Average DNA methylation levels of MEGs, PEGs, and nonimprinted genes (Non-imp) for shoot, embryo, and endosperm in CG context throughout the gene body and its 2-kb up- and downstream regions. (D–F) Comparison of average DNA methylation levels between two parental genomes of MEGs, PEGs, and nonimprinted genes (Non-imp) in CG context throughout the gene body and its 2-kb up- and downstream regions. (G–I) Average DNA methylation levels of MEGs, PEGs, and nonimprinted genes (Non-imp) for shoot, embryo, and endosperm in CHG context throughout the gene body and its 2-kb up- and downstream regions. (J–L) Comparison of average DNA methylation levels between two parental genomes of MEGs, PEGs, and nonimprinted genes (Non-imp) in CHG context throughout the gene body and its 2-kb up- and downstream regions. (A–L) Gene body regions were separated into 60 bins, and extended 2-kb up- and downstream regions were separated into 20 bins. The average methylation levels were calculated with the same method as in Supplemental Figure S7.
Next, we analyzed the relationship between pDMRs we identified and the high quality imprinted genes (68 PEGs and 39 MEGs). Our results show that, compared with nonimprinted genes, both PEGs and MEGs are highly correlated with the occurrence of CG_pDMRs, or regions with both CG_pDMR and CHG_pDMR. Among 68 PEGs (including 2-kb up- and downstream regions), 41 (60.3%) of them possessed pDMRs (CG or CHG), including one PEG with CHG_pDMR only (Supplemental Tables S4, S5). Similarly, among 39 MEGs, 21 (53.8%) MEGs were associated with pDMRs (CG or CHG), including one MEG with CHG_pDMR only (Supplemental Tables S4, S5). Overall, the proportions of MEGs and PEGs associated with pDMRs were very similar. It is worth noting that, due to sequencing coverage and the availability of SNPs, the proportion of imprinted genes containing pDMRs may be underestimated. CG_pDMRs in MEGs were enriched in the upstream and 5′ portion of gene body regions (Fig. 1D; Supplemental Fig. S10), while for PEGs, CG_pDMRs tended to be enriched in the upstream and gene body regions (Fig. 1E; Supplemental Fig. S10).
Identification of parent-of-origin dependent H3K27me3 peaks in maize endosperm
Histone modifications such as H3K27me3 have long been known to be involved in imprinting regulation in plants. To investigate the potential relationship between allele-specific H3K27me3 and the regulation of imprinted genes in maize, we performed ChIP-seq using an antibody of H3K27me3 for both 12-DAP BM and MB endosperm tissues (Supplemental Table S1).
Using MACS software (Feng et al. 2012), 17,652 and 22,962 H3K27me3 peaks were identified in BM and MB, respectively. Among them, 12,125 peaks identified in both BM and MB were kept for further analysis. Quantitative real-time PCR were performed for four randomly selected candidate peaks and all of them exhibited significant enrichment for the H3K27me3 antibody compared with the control (Supplemental Fig. S11). Comparing with previously reported H3K27me3 ChIP-seq data from B73 shoot (Wang et al. 2009), we found only 24.3% peaks overlapped between shoot and endosperm, indicating that the targets of PRC2 in different development stages (vegetative and reproductive) exhibit high tissue specificity, which is similar as in Arabidopsis (Weinhofer et al. 2010).
We developed a protocol to screen for allele-specific H3K27me3 peaks (see Methods). With our stringent criteria, 1131 parent-of-origin dependent H3K27me3 peaks were identified. Interestingly, all of the allele-specific H3K27me3 peaks identified were enriched in maternal alleles. Two resulting allele-specific peaks were selected for experimental validation. They both were confirmed to be maternally preferred (Supplemental Fig. S12).
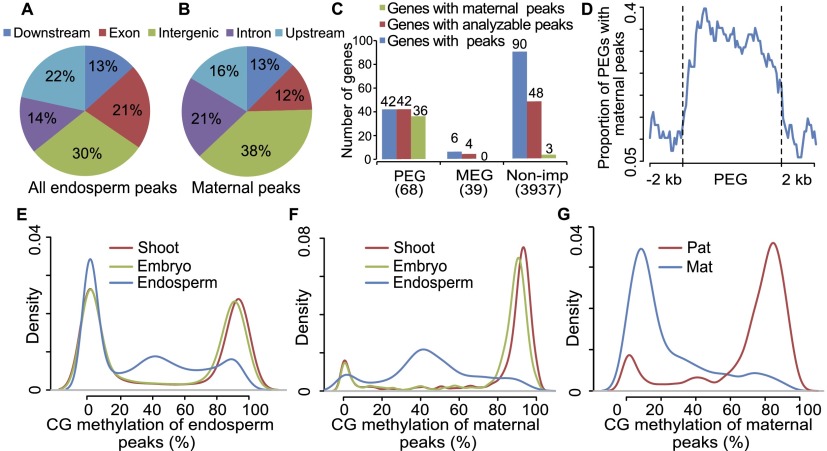
Further analysis showed that only 52 of 1131 (4.6%) maternally preferred H3K27me3 peaks overlapped with shoot H3K27me3 peaks, suggesting that most of these peaks are very likely endosperm specific. The genomic distribution (using the working gene set) for the identified peaks showed that maternally preferred peaks had some enrichment in introns and intergenic regions compared with all peaks identified in the endosperm (P-value < 2.2 × 10−16) (Fig. 2A,B). The 1131 maternally preferred peaks also exhibited lower CG content when compared with all peaks in endosperm, and more so when compared with the peaks identified from shoot tissue (Supplemental Fig. S13).
Figure 2.
Analysis of maize H3K27me3 peaks. (A,B) Genomic distribution of 12,125 H3K27me3 peaks (A) and 1131 maternal peaks (B) identified from hybrid endosperm of reciprocal crosses. (C) Overlap between maternal peaks and MEGs/PEGs/nonimprinted genes (Non-imp). “Genes with peaks” indicates the genes that are overlapped with H3K27me3 peaks identified in the endosperm of both BM and MB. “Genes with analyzable peaks” refers to genes that had SNPs within the H3K27me3 enriched region. “Genes with maternal peaks” refers to genes that contained maternal-specific H3K27me3 peaks identified in this study. The number in parentheses indicates the number of genes that were analyzed. (D) The distribution of maternal-specific H3K27me3 peaks located in PEGs and their 2-kb up- and downstream regions. (E,F) The distributions of DNA methylation levels for maize endosperm H3K27me3 peaks (E) and maternal-specific peaks (F) in shoot, embryo, and endosperm of B73. (G) The distribution of parental DNA methylation of maternal peaks.
Correlation between maternally preferred H3K27me3 peaks and imprinted genes
To understand the relationship of maternally preferred H3K27me3 peaks and genetic imprinting, we investigated the distribution of maternally preferred H3K27me3 peaks around MEGs, PEGs, and nonimprinted genes. We found that maternally preferred H3K27me3 peaks were strongly associated with PEGs, but not with MEGs or nonimprinted genes (Fig. 2C). Among 68 high quality PEGs, 42 (61.8%) of them overlapped with H3K27me3 peaks we identified, and all 42 PEGs contained peaks that can be tested for their allelic preferential enrichment. Thirty six (85.7%) out of the 42 PEGs have maternal-specific H3K27me3 peaks (Fig. 2C; Supplemental Table S6) according to our high stringency criteria of at least 85% of reads derived from one parent in both reciprocal hybrids. The other six PEGs also have at least 75% of H3K27me3 peaks in their maternal alleles in both BM and MB hybrid endosperm. In contrast, among 39 MEGs, only six (15.4%) MEGs overlapped with H3K27me3 peaks we identified. Although four of the six MEGs overlapped with H3K27me3 peaks that can be analyzed for allelic preference, none of them contained an H3K27me3 peak that showed apparent parental preference (Fig. 2C). Similarly, among 3937 nonimprinted genes, there are only 90 (2.3%) genes that have H3K27me3 peaks. Forty-eight of them contained peaks that can be analyzed for allelic preference, and only three (6.3%) genes had maternally preferred H3K27me3 peaks (Fig. 2C). In summary, H3K27me3 peaks in endosperm tend to be enriched around imprinted genes, especially around PEGs. More importantly, allele-specific H3K27me3 peaks we identified only occur around PEGs. We also found that maternally preferred peaks were located throughout the gene body regions of PEGs (Fig. 2D).
Due to the high stringency criteria used, the number of allele-specific peaks and the proportion of PEGs containing H3K27me3 peaks were underestimated. For example, if including peaks that existed only in BM or MB endosperm, 54 (79.4%) PEGs possessed peaks. Accumulation of additional data generated from both BM and MB endosperm may reveal that a majority of PEGs will be associated with peaks.
We then investigated the imprinting status of all the genes that overlapped with maternally preferred H3K27me3 peaks. For the 246 genes (including regions of 2 kb up- and downstream of genes) that overlapped with at least one maternally preferred H3K27me3 peak, there are 115 genes for which imprinting status can be called from our transcriptome data. Of the 115 genes, 75 (65.2%) are PEGs. This proportion is increased to 89.6% using lower criteria for identifying imprinted genes, where parental bias is only required to deviate from 2:1 by χ2 test (α = 0.05) in both reciprocal crosses. Our results suggest that the majority of genes overlapping with maternally preferred H3K27me3 peaks exhibit paternally preferred expression.
Regions with maternally preferred H3K27me3 peaks tend to exhibit allele-specific differential DNA methylation
Previous studies have revealed that DNA methylated regions usually exclude the PRC2 targeting (Weinhofer et al. 2010; Deleris et al. 2012). However, the relationship of parent-of-origin dependent allele-specific H3K27me3 peaks and allele-specific DNA methylation has never been tested at the genome scale for any plant. All the H3K27me3 peaks identified in endosperm exhibited extreme hypermethylation (>80%) or extreme hypomethylation (<20%) in both shoot and embryo. However, a portion of regions with peaks in the endosperm had medium levels of CG methylation in the endosperm (Fig. 2E). These regions with medium levels of DNA methylation correspond mostly to maternal-specific H3K27me3 peaks and these maternal peaks are hypermethylated in both shoot and embryo (Fig. 2F). When the two parental alleles in endosperm were examined separately, the paternal alleles of these maternally preferred peaks were hypermethylated, while their maternal alleles were hypomethylated (Fig. 2G). DNA methylation in CHG context for these maternal peaks exhibited a very similar pattern as that of CG context (Supplemental Fig. S14). On one hand, 62% (70%) of CG_pDMRs (CHG_pDMRs) that overlap with H3K27me3 peaks had H3K27me3 preferentially target to maternal alleles. On the other hand, 59% (65%) of regions of maternally preferred H3K27me3 peaks where CG (CHG) methylation can be analyzed contained CG_pDMRs (CHG_pDMRs). These numbers are much larger than comparisons using all of the analyzable H3K27me3 peaks (20% for CG_pDMRs, 15.5% for CHG_pDMRs). Our results suggest that a considerable portion of allelic differential DNA methylation and allelic H3K27me3 tend to occur together in the genome.
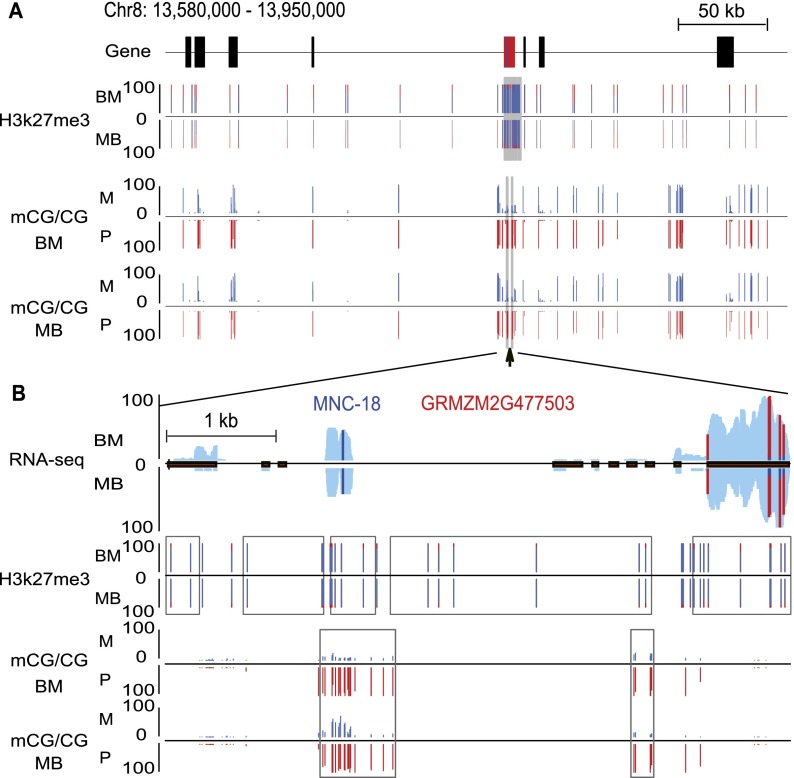
Figure 3 shows an integrated view of a selected genomic region of ∼400 kb of chromosome 8. This region includes an experimentally confirmed PEG (GRMZM2G477503), one maternally expressed intronic noncoding RNA MNC-18 (Zhang et al. 2011), and nine other genes (three nonimprinted genes and six genes with unknown imprinting status) (Fig. 3A). Globally, both DNA methylation and H3K27me3 in most SNPs examined (covered by at least five reads) in the region were biallelically distributed without significant parental bias. However, several maternally preferred H3K27me3 peaks were found to cover nearly the entire PEG (GRMZM2G477503). Additionally, two pDMRs were identified, with one of them located in the longest intron of GRMZM2G477503 (Fig. 3A,B). The intronic noncoding RNA MNC-18 resides within the CG_pDMR region of the PEG and overlaps with maternal-specific H3K27me3 peaks (Fig. 3B).
Figure 3.
Allelic views of H3K27me3 and DNA methylation in a genomic region. (A) Allelic levels of H3K27me3 and CG DNA methylation are shown for an ∼400-kb region in both BM and MB endosperm. The region contained nine genes with unknown or nonimprinting status (black block), one PEG (red block), and one MNC (blue block). (B) A zoomed-in allelic view of RNA-seq, H3K27me3, and CG DNA methylation for a confirmed PEG that overlapped with a MNC. The overall expression level of transcribed regions is shown in light blue for both BM and MB. The relative expression levels, the percentage of allelic reads of H3K27me3 ChIP-seq data, and the DNA methylation level for specific SNP sites are shown for both maternal and paternal alleles, with red lines for the paternal allele (P) and blue lines for the maternal allele (M). Black rectangle, exon; black line, intron. The gray rectangles highlight the maternal H3K27me3 peaks and pDMRs identified in this region. The lines (or dots) correspond to all analyzable sites.
Discussion
The endosperm has been subjected to extensive epigenetic studies, as it is the primary tissue where genetic imprinting occurs in higher plants. In this study, we present genome-wide high resolution allele-specific maps of both DNA methylation and H3K27me3 in maize endosperm. Integration of 6910 CG_pDMRs, 4456 CHG_pDMRs, 1131 maternally preferred H3K27me3 peaks and the expression of 107 imprinted genes provides an unprecedented opportunity to uncover global patterns of association between allele-specific chromatin modifications (DNA methylation and H3K27me3) and genetic imprinting.
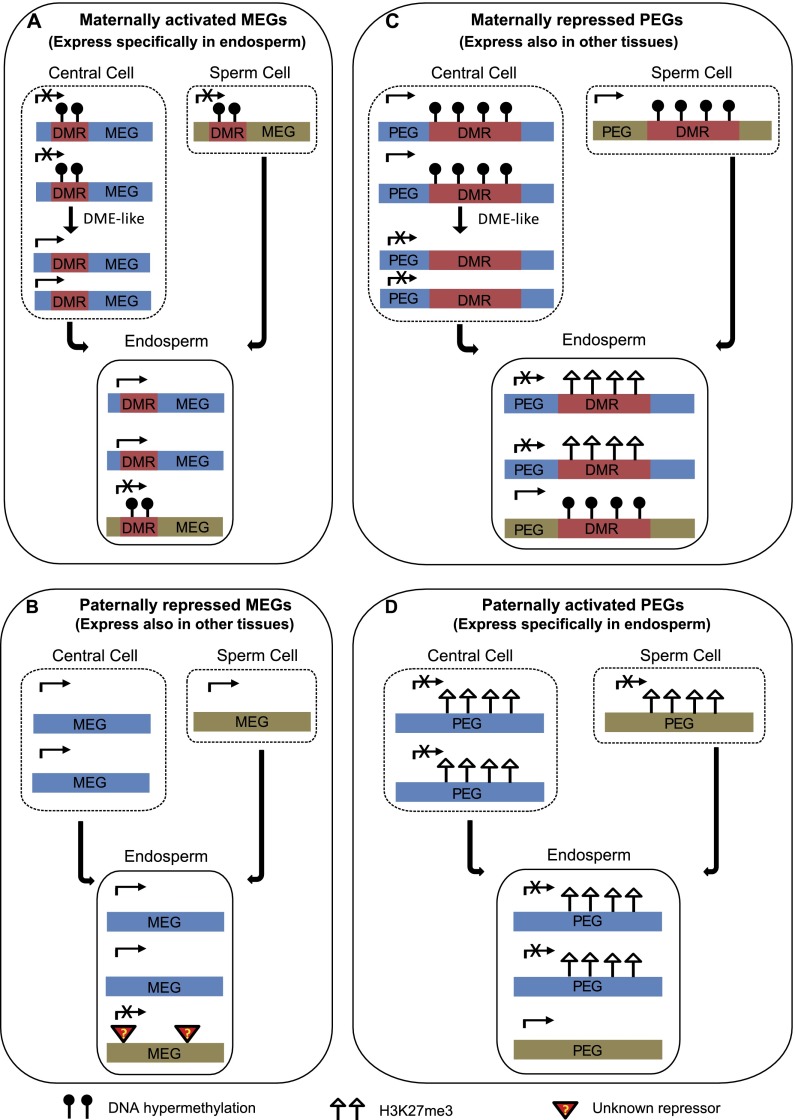
Several models for the regulation of genetic imprinting in the endosperm have been proposed in recent reviews (Raissig et al. 2011; Jiang and Kohler 2012; Kohler et al. 2012), which were primarily based on the regulatory mechanisms of a few imprinted genes or genome-wide transcriptome analysis in mutants such as dme, fis2 (Hsieh et al. 2011; Wolff et al. 2011). In these models, it has been generally discussed that DNA methylation and H3K27me3 modification are both involved in the regulation of PEGs and MEGs. Based on our data, we present an updated model for the regulation of genetic imprinting with emphasis on the involvement of DNA methylation and/or H3K27me3 in particular subgroups of MEGs and PEGs (Fig. 4).
Figure 4.
Models proposed for the regulation of imprinting in maize endosperm. (A) Model for maternally activated MEGs. MEGs expressing specifically in endosperm are probably maternally activated. Most of these MEGs are associated with pDMRs located around the upstream and 5′ portion of gene body regions. In central cells, the methylation of pDMR regions can be removed by DME-like demethylase, but not in sperm cells. After fertilization, pDMRs that were identified in the endosperm exhibit maternal DNA demethylation and paternal DNA hypermethylation. Maternal DNA demethylation of these MEGs results in their maternally preferred expression. (B) Model for paternally repressed MEGs. MEGs expressing in endosperm but also in other tissues are probably paternally repressed. The nonexpression of the paternal alleles of these MEGs may have resulted from paternally specific unknown repressor(s). (C) Model for maternally repressed PEGs. PEGs expressing in endosperm but also in other tissues are probably maternally repressed. Paternal preferred expressions of these PEGs require both DNA methylation and H3K27me3. In central cells, DNA methylation in pDMRs is removed, but not in sperm cells. In the endosperm, H3K27me3 only targeted to the maternal alleles with DNA demethylation of the pDMR regions (demonstrated by maternal-specific H3K27me3). As the paternal alleles remain hypermethylated, they cannot be targeted by H3K27me3. Repression of the maternal alleles by H3K27me3 led to the paternal preferred expression of these PEGs. (D) Model for paternally activated PEGs. PEGs expressing specifically in endosperm are probably paternally activated (de-repressed) PEGs. In other tissues, these PEGs possibly possess biallelic H3K27me3 peaks. As only maternal-specific H3K27me3 peaks were identified in endosperm, it is likely that for these PEGs, their paternal alleles have lost their H3K27me3, which resulted in paternally specific expression. The dashed boxes around the central cell and sperm cell indicate that the gene expression and chromatin modification in these tissues have not been supported by experimental data.
For any imprinted gene, its monoallelic expression can be the result of either activation or repression of one allele. For example, MEGs can be either due to maternal activation or paternal repression, while PEGs can be either due to maternal repression or paternal activation. It is reasonable to hypothesize that most MEGs expressed specifically in endosperm are maternally activated, while MEGs expressed also in other tissues are paternally repressed. Similarly, PEGs expressed only in the endosperm are likely to be paternally activated, while PEGs expressed also in other tissues can be maternally repressed. Using the RNA-seq data of 27 samples of various tissues from previous studies (Supplemental Table S7) (Jia et al. 2009; Wang et al. 2009; Li et al. 2010; Davidson et al. 2011; Zhang et al. 2011; Bolduc et al. 2012; Kakumanu et al. 2012) and data of endosperm and embryo from this study, MEGs and PEGs can be respectively divided into two subgroups: genes expressed primarily in endosperm and genes also expressed in other tissues (Supplemental Fig. S15). About 61.5% (24/39) of MEGs and 76.5% (52/68) of PEGs are expressed in other tissues. The subgroups of MEGs and PEGs have distinct DNA methylation profiles (Supplemental Fig. S16) and different extents of association with pDMRs (Table 1). Among 15 MEGs with endosperm-specific expression, 11 (73.3%) of them possessed pDMRs, compared with 10 (41.7%) of the 24 MEGs expressed in other tissues (Table 1). In contrast, four (25.0%) of the 16 PEGs with endosperm-specific expression and 37 (71.2%) of the 52 PEGs expressed in other tissues possessed pDMRs (Table 1). The two subgroups of PEGs are highly associated with allele-specific H3K27me3 (Table 1).
Table 1.
Subgroups of imprinted genes and their association with pDMRs and H3K27me3 peaks
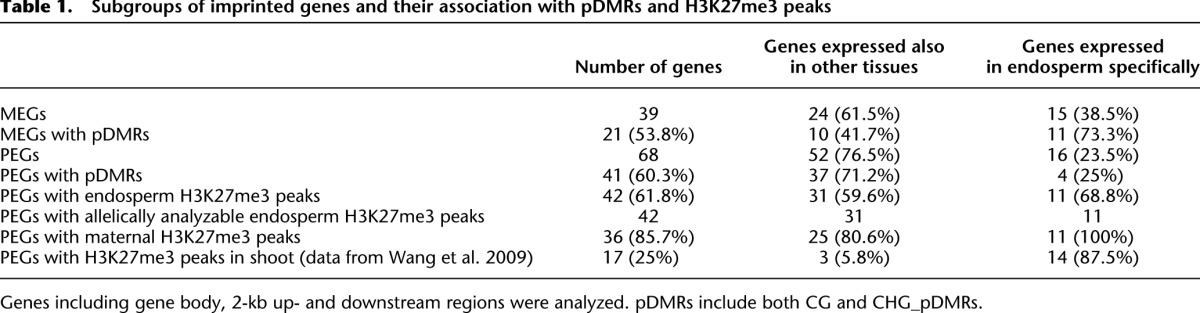
For maternally activated MEGs (estimated to be ∼38% of total MEGs), we propose that maternal DNA demethylation around the upstream and the 5′ portion of their gene body regions leads to maternal-specific expression (Fig. 4A), as a majority (>70%) of them are associated with pDMRs. Consistent with our data, results in Arabidopsis showed that the met1 and dme mutants affect a relatively small number of MEGs (Hsieh et al. 2011; Wolff et al. 2011). However, for the paternally repressed MEGs subgroup (∼62% of total MEGs based on our estimation) expressed also in other tissues, maternal-specific expression would require unknown repressor(s), as they are less likely associated with pDMRs and rarely with allele-specific H3K27me3. Still, ∼41.7% of this paternally repressed MEGs subgroup contained pDMRs, implying that the current expression data are not optimal for separation of the two subgroups. The maternal-specific expression of some MEGs has been proposed to be simply a result of mRNA transported from maternal tissues (Hsieh et al. 2011). Interestingly, the imprinting status of some MEGs was also changed in fie or fis2 mutants in Arabidopsis (Hsieh et al. 2011; Wolff et al. 2011). We argue that this could be due to an indirect effect. According to our criteria, no parental-specific H3K27me3 is shown to be associated with any MEG, although potential enrichment (much lower than our criteria) of paternal H3K27me3 for a few MEGs has been previously reported (Haun and Springer 2008).
For maternally repressed PEGs (∼77% of total PEGs), we propose that they require both maternal-specific DNA demethylation and maternal-specific H3K27me3 peaks. We showed that >70% of these PEGs are associated with pDMRs and >80% are associated with maternal-specific H3k27me3. Maternal DNA demethylation is likely to occur before maternal H3K27me3. The “constitutive” PEGs, which displayed gene body DNA hypermethylation in shoot and embryo (Supplemental Fig. S16), are generally not targets of H3K27me3 in vegetative tissues according to a recent study (Makarevitch et al. 2013). Previously reported ChIP-seq data (Wang et al. 2009) showed that only three out of 52 PEGs had H3K27me3 peaks in shoot. Our analysis suggests that there is another subgroup of PEGs, the paternally activated PEGs subgroup (estimated to be ∼23% of total PEGs), for which the imprinted expression can be due to paternal-specific removal of H3K27me3. These PEGs are expressed specifically in the endosperm and a majority of them are associated with maternal-specific H3K27me3. These PEGs were shown to have already been targeted (14 out of 16 PEGs) by H3K27me3 in shoot (Wang et al. 2009). Both subgroups of PEGs involve H3K27me3: the de novo addition of maternal H3K27me3 or paternal-specific removal of H3K27me3. Our model fits very well with the recently reported H3K27me3 distribution of PEGs in several maize vegetative and reproductive tissues (Makarevitch et al. 2013). Our model is also consistent with results in Arabidopsis where most PEGs identified in their study expressed biallelically in fie or fis2 mutants (Hsieh et al. 2011; Wolff et al. 2011).
It has been suggested that DNA demethylation is required but not sufficient for targeting of PRC2 complex (Mathieu et al. 2005; Stroud et al. 2013). Since several noncoding RNAs (particularly MNCs) were shown to be associated with PEGs, we speculate that maternal DNA demethylation around PEGs may potentially lead to the transcription of MNCs, which then facilitate the PRC2 complex, with later function in directing H3K27me3. Long noncoding RNAs have long been regarded as an important regulator of imprinting in mammals (Pauler et al. 2007; Peters and Robson 2008). Several long noncoding RNAs such as Kcnqlot1, Air, and Xist were shown to have regulatory function in an imprinting cluster (Mohammad et al. 2009). Among 18 intronic MNCs identified, 13 of them were transcribed from intronic regions of 12 PEGs. Among these 12 PEGs containing MNCs, 11 PEGs (including 12 MNCs) overlapped with pDMRs. However, due to the limited sequencing depth of ChIP-seq data, only two of these PEGs with MNCs overlapped with maternal-specific H3K27me3 peaks. A mediation function of long noncoding RNAs on the PRC2 complex has been reported in several studies of mammals (Tsai et al. 2010; Zhao et al. 2010; Kotake et al. 2011; Margueron and Reinberg 2011; Deleris et al. 2012) and plants (Heo and Sung 2011), though the exact regulatory role of imprinted noncoding transcripts on imprinting in maize remains to be tested.
Methods
Tissue collection
Shoot, 12-DAP embryo, and 12-DAP endosperm tissue from the inbred B73 and 10-DAP and 12-DAP endosperm from reciprocal crosses of B73 and Mo17 were harvested. The seeds of B73 were planted in the incubator for 14 d, and the aerial parts of seedlings from at least three plants were collected for shoot tissue. Embryo and endosperm tissues were collected from at least three plants in the field by manual dissection and the embryos were washed three times after separation from the endosperm. We did not perform biological replication for each cross in our analysis of imprinted genes, as DNA methylation and H3K27me3 patterns were consistent in the reciprocal crosses.
Library construction for RNA-seq, MethylC-seq, and ChIP-seq
RNA-seq was performed as described in our previous study (Zhang et al. 2011), using 12-DAP and 10-DAP endosperm of reciprocal crosses of B73 and Mo17.
The libraries for MethylC-seq using shoot, embryo, and endosperm of 12 DAP of inbred B73, and 12-DAP endosperm of reciprocal crosses of B73 and Mo17, were constructed as described in our previous study (Zhang et al. 2011).
ChIP was done following the protocol described in Liu et al. (2008). The libraries for ChIP-seq using 12-DAP endosperm of reciprocal crosses of B73 and Mo17 were constructed by the normal library construction protocol for ChIP-seq.
Pipeline for analysis of MethylC-seq
MethylC-seq reads were done using the recommended workflow (Krueger et al. 2012). Low quality bases were filtered using SolexaQA software (Cox et al. 2010). Next, processed reads were mapped to the B73 reference using Bismark (Krueger and Andrews 2011).
Informative SNPs were used for allele assignment of reads from MethylC-seq. For SNPs that may be changed due to bisulfite conversion, their bisulfite-converted forms were considered. When more than one SNP existed in a read or a read pair, the results of allele assignment of all the SNPs should be identical. For CG and CHG contexts, the reads from forward and reverse strand were merged together. Cytosine sites covered by at least five reads were used in subsequent analyses. Statistically significant alellic methylation bias was assessed using Fisher's exact test. Resulting P-values were corrected with the Q-value method (Storey and Tibshirani 2003) and a FDR of 1% was accepted.
Identification of pDMRs
Different criteria were formulated to identify CG_pDMR and CHG_pDMR. First, due to the limit of sequencing depth, we adopted a sliding-window approach with window size of 200 bp and slide step of 20 bp to find DMRs across the whole genome. Windows containing more than five CGs/CHGs supported with at least five reads were kept. Second, the significance of allele bias of each window was weighed by P-value using Fisher's exact test. Resulting P-values were converted to Q-values and a FDR of 1% was accepted. Finally, the remaining windows were filtered by the following criteria: the level of methylation between two alleles differed >30% (the actual level, not their relative percentage) and, for CG context, the hypermethylated alleles had methylation levels >40%. The candidate pDMRs were then further filtered using a smaller window size of 50 bp, and pDMRs within 200 bp were merged.
Identification of imprinted genes
Imprinted genes were identified using a similar pipeline as described in Zhang et al. (2011) except that mapping software TopHat (Trapnell et al. 2012) was used. SNPs between B73 and Mo17 genomes were also updated. Candidate imprinted protein-coding genes and noncoding RNAs were manually analyzed.
Analysis of H3K27me3 ChIP-seq data
ChIP-seq reads were mapped to B73 reference genome using Bowtie (Langmead et al. 2009). H3K27me3 peaks were identified using MACS (Feng et al. 2012). The following parameters in MACS were used to call the H3K27me3 peak: (1) P-value ≤ 1 × 10−5; (2) mfold (10–30); (3) genome size, g = 2.06 × 109.
Identification of parent-of-origin dependent H3K27me3 peaks
ChIP-seq reads from two reciprocal hybrids were mapped to B73 and simulated Mo17 genome, respectively. A simulated Mo17 genome was constructed by substituting Mo17 bases for B73 bases in the SNP sites. Peaks with at least two SNPs and with at least 10 reads on both B73 and Mo17 alleles were used for allele-specific peak analysis. As the endosperm has two copies of maternal alleles and only one paternal allele, two-tailed χ2 tests were performed for each peak to test parental bias greater or less than 2:1 (α = 0.05) in both reciprocal hybrids. Final parent-of-origin dependent H3K27me3 peaks were identified for those with at least 85% of reads derived from one parent in both reciprocal hybrids.
Classification of imprinted genes based on expression patterns
RNA-seq data of 27 samples from several previous studies (Supplemental Table S7; Jia et al. 2009; Wang et al. 2009; Li et al. 2010; Davidson et al. 2011; Zhang et al. 2011; Bolduc et al. 2012; Kakumanu et al. 2012) and our endosperm and embryo data were aligned to maize B73 reference (V2) using TopHat (Trapnell et al. 2012). Reads per kilobase of transcript per million mapped reads (RPKM) for each gene were calculated. Hierarchical clustering analysis using MeV (http://www.tm4.org/mev.html) was conducted on the relative expression value by setting parameters: average linkage and Euclidean distance. For a gene in a special sample, relative expression value was the RPKM normalized by the maximum RPKM value of the gene over all samples. Based on the results of cluster, we extracted MEGs (or PEGs) primarily expressed in endosperm as a subgroup and the rest of MEGs (or PEGs) assigned to the other subgroup.
Data access
Data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra) under accession number SRP011991.
Acknowledgments
Research is supported by the National Basic Research Program (973 program) (2009CB118400), 863 Project (2012AA10A305), and NSF (31225020).
Author contributions: M.Z., S.X., and J.L. designed the experiments. X.Z. and H.L conducted ChIP. M.Z., J.C., and G.W. prepared all the materials used. M.Z. performed the bisulfite treatment. S.X., X.D., B.Z., and W.Y. performed analysis of MethylC-seq, RNA-seq, and ChIP-seq data. M.Z., J.C., and G.W. conducted experimental validation for ChIP data. M.Z. and J.L. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.155879.113.
Freely available online through the Genome Research Open Access option.
References
- Abramowitz LK, Bartolomei MS 2012. Genomic imprinting: Recognition and marking of imprinted loci. Curr Opin Genet Dev 22: 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP 2011. Genomic imprinting: A mammalian epigenetic discovery model. Annu Rev Genet 45: 379–403 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O'Connor D, Grotewold E, Hake S 2012. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr MS, Yevtodiyenko A, Schmidt CL, Schmidt JV 2007. Allele-specific histone modifications regulate expression of the Dlk1-Gtl2 imprinted domain. Genomics 89: 280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ 2010. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, Childs KL, Lin H, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM, Jiang N, et al. 2011. Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- Deleris A, Stroud H, Bernatavichute Y, Johnson E, Klein G, Schubert D, Jacobsen SE 2012. Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet 8: e1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Qin B, Zhang Y, Liu XS 2012. Identifying ChIP-seq enrichment using MACS. Nat Protoc 7: 1728–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier C, Goto Y, Ballestar E, Delaval K, Hever AM, Esteller M, Feil R 2002. Allele-specific histone lysine methylation marks regulatory regions at imprinted mouse genes. EMBO J 21: 6560–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M, Missirian V, Henikoff S 2011. Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6: e23687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O'Sullivan DM, Wormald M, Perez P, Dickinson HG 2004. maternally expressed gene1 is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16: 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Marcos JF, Costa LM, Dal Pra M, Scholten S, Kranz E, Perez P, Dickinson HG 2006. Epigenetic asymmetry of imprinted genes in plant gametes. Nat Genet 38: 876–878 [DOI] [PubMed] [Google Scholar]
- Haun WJ, Springer NM 2008. Maternal and paternal alleles exhibit differential histone methylation and acetylation at maize imprinted genes. Plant J 56: 903–912 [DOI] [PubMed] [Google Scholar]
- Haun WJ, Laoueille-Duprat S, O'Connell MJ, Spillane C, Grossniklaus U, Phillips AR, Kaeppler SM, Springer NM 2007. Genomic imprinting, methylation and molecular evolution of maize Enhancer of zeste (Mez) homologs. Plant J 49: 325–337 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hermon P, Srilunchang KO, Zou J, Dresselhaus T, Danilevskaya ON 2007. Activation of the imprinted Polycomb Group Fie1 gene in maize endosperm requires demethylation of the maternal allele. Plant Mol Biol 64: 387–395 [DOI] [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. 2012. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res 22: 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Shin J, Uzawa R, Silva P, Cohen S, Bauer MJ, Hashimoto M, Kirkbride RC, Harada JJ, Zilberman D, et al. 2011. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci 108: 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA, Feng X, Schoft VK, Hsieh TF, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. 2012. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Lisch DR, Ohtsu K, Scanlon MJ, Nettleton D, Schnable PS 2009. Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet 5: e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kohler C 2012. Evolution, function, and regulation of genomic imprinting in plant seed development. J Exp Bot 63: 4713–4722 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Zhao H, Ren L, Song W, Zeng B, Guo J, Wang B, Liu Z, Chen J, Li W, et al. 2012. Genome-wide genetic changes during modern breeding of maize. Nat Genet 44: 812–815 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Kinoshita T, Ohad N, Berger F 2006. Maintenance of DNA methylation during the Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18: 1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakumanu A, Ambavaram MM, Klumas C, Krishnan A, Batlang U, Myers E, Grene R, Pereira A 2012. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 160: 846–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermicle JL, Alleman M 1990. Gametic imprinting in maize in relation to the angiosperm life cycle. Dev Suppl 1990: 9–14 [PubMed] [Google Scholar]
- Kinoshita T, Miura A, Choi Y, Kinoshita Y, Cao X, Jacobsen SE, Fischer RL, Kakutani T 2004. One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303: 521–523 [DOI] [PubMed] [Google Scholar]
- Koerner MV, Barlow DP 2010. Genomic imprinting—an epigenetic gene-regulatory model. Curr Opin Genet Dev 20: 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U 2003. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Page DR, Gagliardini V, Grossniklaus U 2005. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30 [DOI] [PubMed] [Google Scholar]
- Kohler C, Wolff P, Spillane C 2012. Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol 63: 331–352 [DOI] [PubMed] [Google Scholar]
- Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y 2011. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 30: 1956–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR 2011. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Kreck B, Franke A, Andrews SR 2012. DNA methylome analysis using short bisulfite sequencing data. Nat Methods 9: 145–151 [DOI] [PubMed] [Google Scholar]
- Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M, et al. 2010. Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42: 1027–1030 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. 2010. The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Park YJ, McLean CM, Dokshin GA, Persson JM, Herman H, Pasini D, Miro X, Donohoe ME, Lee JT, et al. 2008. Antagonism between DNA and H3K27 methylation at the imprinted Rasgrf1 locus. PLoS Genet 4: e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yue W, Li D, Wang RR, Kong X, Lu K, Wang G, Dong Y, Jin W, Zhang X 2008. Structure and dynamics of retrotransposons at wheat centromeres and pericentromeres. Chromosoma 117: 445–456 [DOI] [PubMed] [Google Scholar]
- Luo M, Taylor JM, Spriggs A, Zhang H, Wu X, Russell S, Singh M, Koltunow A 2011. A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet 7: e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G, Villar CB, Erilova A, Kohler C 2008. Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci 121: 906–912 [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Eichten SR, Briskine R, Waters AJ, Danilevskaya ON, Meeley RB, Myers CL, Vaughn MW, Springer NM 2013. Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu O, Probst AV, Paszkowski J 2005. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J 24: 2783–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Kanduri C 2009. Epigenetics of imprinted long noncoding RNAs. Epigenetics 4: 277–286 [PubMed] [Google Scholar]
- Pauler FM, Koerner MV, Barlow DP 2007. Silencing by imprinted noncoding RNAs: Is transcription the answer? Trends Genet 23: 284–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Robson JE 2008. Imprinted noncoding RNAs. Mamm Genome 19: 493–502 [DOI] [PubMed] [Google Scholar]
- Raissig MT, Baroux C, Grossniklaus U 2011. Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JA, Ruan R, Nishimura T, Sharma MK, Sharma R, Ronald PC, Fischer RL, Zilberman D 2013. Imprinted expression of genes and small RNA is associated with localized hypomethylation of the maternal genome in rice endosperm. Proc Natl Acad Sci 110: 7934–7939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. 2009. The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G 2009. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542 [DOI] [PubMed] [Google Scholar]
- Singh P, Cho J, Tsai SY, Rivas GE, Larson GP, Szabo PE 2010. Coordinated allele-specific histone acetylation at the differentially methylated regions of imprinted genes. Nucleic Acids Res 38: 7974–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE 2013. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152: 352–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Schulz R, Ikeda Y, Dytham L, Bravo J, Mathers L, Spielman M, Guzman P, Oakey RJ, Kinoshita T, et al. 2008. MATERNALLY EXPRESSED PAB C-TERMINAL, a novel imprinted gene in Arabidopsis, encodes the conserved C-terminal domain of polyadenylate binding proteins. Plant Cell 20: 2387–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW 2009. Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AJ, Makarevitch I, Eichten SR, Swanson-Wagner RA, Yeh CT, Xu W, Schnable PS, Vaughn MW, Gehring M, Springer NM 2011. Parent-of-origin effects on gene expression and DNA methylation in the maize endosperm. Plant Cell 23: 4221–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhofer I, Hehenberger E, Roszak P, Hennig L, Kohler C 2010. H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet 6: e1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff P, Weinhofer I, Seguin J, Roszak P, Beisel C, Donoghue MT, Spillane C, Nordborg M, Rehmsmeier M, Kohler C 2011. High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet 7: e1002126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B 2012. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 148: 816–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin Z, Allis CD, Wagstaff J 2001. Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader-Willi syndrome imprinting center. Am J Hum Genet 69: 1389–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zhao H, Xie S, Chen J, Xu Y, Wang K, Zhao H, Guan H, Hu X, Jiao Y, et al. 2011. Extensive, clustered parental imprinting of protein-coding and noncoding RNAs in developing maize endosperm. Proc Natl Acad Sci 108: 20042–20047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]