Abstract
RNA polymerase III (Pol III) occurs in two versions, one containing the POLR3G subunit and the other the closely related POLR3GL subunit. It is not clear whether these two Pol III forms have the same function, in particular whether they recognize the same target genes. We show that the POLR3G and POLR3GL genes arose from a DNA-based gene duplication, probably in a common ancestor of vertebrates. POLR3G- as well as POLR3GL-containing Pol III are present in cultured cell lines and in normal mouse liver, although the relative amounts of the two forms vary, with the POLR3G-containing Pol III relatively more abundant in dividing cells. Genome-wide chromatin immunoprecipitations followed by high-throughput sequencing (ChIP-seq) reveal that both forms of Pol III occupy the same target genes, in very constant proportions within one cell line, suggesting that the two forms of Pol III have a similar function with regard to specificity for target genes. In contrast, the POLR3G promoter—not the POLR3GL promoter—binds the transcription factor MYC, as do all other promoters of genes encoding Pol III subunits. Thus, the POLR3G/POLR3GL duplication did not lead to neo-functionalization of the gene product (at least with regard to target gene specificity) but rather to neo-functionalization of the transcription units, which acquired different mechanisms of regulation, thus likely affording greater regulation potential to the cell.
The three main nuclear eukaryotic RNA polymerases (pols) are issued from a common ancestor and have remained highly similar to each other during eukaryotic evolution (Werner and Grohmann 2011). They consist of a 10-subunit core containing five common subunits and five subunits related among the three enzymes, as well as additional subcomplexes (for a compilation of the various subunit names in Homo sapiens, Mus musculus, and Saccharomyces cerevisiae, see Supplemental Table S1; for reviews, see Cramer et al. 2008; Vannini and Cramer 2012). The subcomplex forming the RNA polymerase stalk consists of two subunits that have little sequence conservation among polymerases but are clearly related in their three-dimensional structure. Another two-subunit subcomplex present in Pol I and Pol III has structural similarity to the two-subunit TFIIF Pol II general transcription factor (Kuhn et al. 2007; Carter and Drouin 2010; Geiger et al. 2010). The third subcomplex has partial structural similarity to TFIIE (Geiger et al. 2010; Lefevre et al. 2011). In Pol III, this subcomplex, which is detachable from the rest of the enzyme (Werner et al. 1992, 1993; Wang and Roeder 1997), contains the subunits POLR3C (RPC3/RPC62) and POLR3F (RPC6/RPC39), with structural similarities to GTF2E1 (TFIIE-A) and GTF2E2 (TFIIE-B), respectively, as well as the subunit POLR3G (RPC7/RPC32-alpha), the only polymerase subunit without an identified counterpart in the other two transcription machineries.
Pol III is recruited to its target promoters through the formation of transcription initiation complexes that invariably contain TFIIIB. In yeast, TFIIIB consists of three subunits, the TATA box binding protein Spt15 (Tbp), the SANT domain protein Bdp1, and the TFIIB-related factor Brf1. In mammalian cells, two forms of TFIIIB exist, BRF1-TFIIIB and BRF2-TFIIIB, in which BRF1 is replaced by another TFIIB-related factor, BRF2 (Geiduschek and Kassavetis 2001; Schramm and Hernandez 2002; Jawdekar and Henry 2008). The trimeric POLR3C (RPC3)-POLR3F (RPC6)-POLR3G (RPC7) complex plays a role in transcription initiation complex formation (Thuillier et al. 1995; Brun et al. 1997; Wang and Roeder 1997), at least in part through direct contacts with TFIIIB: The yeast homolog of human POLR3F, Rpc34, has been shown to associate with Brf1 (Werner et al. 1993), and human POLR3F with both BRF1 and TBP (Wang and Roeder 1997). Moreover, down-regulation of POLR3F in mammalian cells prevents Pol III association with its target genes (Kenneth et al. 2008). Consistent with the structural similarities of POLR3C and POLR3F with TFIIE subunits, the trimeric complex stabilizes the open preinitiation complex (Brun et al. 1997).
Recently, an isoform of POLR3G, RPC32-beta or POLR3GL (RPC7-Like, RPC7L), encoded by a separate gene, was identified by database searches (Haurie et al. 2010). Interestingly, the two isoforms were found to be differentially expressed, with POLR3G (RPC32-alpha) decreasing during differentiation and increasing during cellular transformation relative to POLR3GL (Haurie et al. 2010). Indeed, POLR3G is one of the most highly up-regulated genes in undifferentiated human stem cells relative to differentiated cells (Enver et al. 2005), and decreasing its levels results in loss of pluripotency (Wong et al. 2011). Suppression of each isoform by siRNA suggested that POLR3GL, but not POLR3G, is essential for cell survival. Moreover, ectopic expression of POLR3G, but not POLR3GL, leads to anchorage-independent growth in partially transformed human IMR90 fibroblasts (Haurie et al. 2010). Together, these results suggest that POLR3G and POLR3GL carry out different functions in the cell, but what these functions may be is unclear.
We identified POLR3GL during a mass spectrometry analysis of Pol III highly purified from HeLa cells, and determined that these cells contain two forms of Pol III, one containing POLR3G and the other POLR3GL, consistent with previous results (Haurie et al. 2010). We show that POLR3G and POLR3G arose from a DNA-based gene duplication, probably in a common ancestor of vertebrates, and we describe the genome-wide occupancy of these two forms of Pol III in IMR90 cells, a nontransformed and nonimmortalized human cell line, as well as in normal mouse liver and mouse hepatocarcinoma cells. The results allow us to refine the list of Pol III–occupied loci in human and mouse cells, and confirm that only a small number of SINEs or nonannotated (NA) loci are clearly occupied by Pol III in addition to known Pol III genes. They also show that the large majority of Pol III–occupied loci are more occupied in hepatocarcinoma cells compared with mouse liver cells, consistent with the idea that Pol III transcription is up-regulated in cancer cells. Most importantly, the results indicate that both forms of Pol III occupy the same target genes but that POLR3G and POLR3GL expression is differentially regulated, most likely at least in part by the transcription factor MYC. The POLR3G/POLR3GL gene duplication seems thus to have led to neo-functionalization of the transcription units, which have acquired different mechanisms of regulation, rather than to neo-functionalization of the gene products.
Results
Identification of POLR3GL (RCP7L) in highly purified Pol III
We used a HeLa cell line (9-8) expressing a Flag- and His-tagged POLR3D (RPC4) Pol III subunit (Hu et al. 2002) to purify Pol III extensively, as summarized in Supplemental Figure S1A. The resulting preparations, purified either through the Flag tag or through both the Flag and His tags (Supplemental Fig. S1B), were subjected to global mass spectrometry analysis. In addition to all the previously described Pol III subunits, a subunit sharing 49% amino acid identities with POLR3G (RPC7), POLR3GL (RPC7L), was detected in both singly and doubly affinity chromatography-purified material. As shown in Supplemental Figure S1C, the peptides detected were all specific to either POLR3G or POLR3GL, excluding any ambiguity as to the identity of the corresponding protein sequence. The ratios of POLR3GL over POLR3G, as determined by normalized spectral abundance factor (see Methods), were 0.45 and 0.53 in the singly and doubly purified material, respectively, indicating a lower amount of POLR3GL than POLR3G in these transformed cells. POLR3G and POLR3GL proteins correspond to the RPC32-alpha and RPC32-beta proteins described by Haurie et al. (2010) and are encoded by two different genes: POLR3G located on chromosome 5 and POLR3GL located on chromosome 1, respectively.
The discovery of a POLR3G-related gene prompted us to examine whether other, so far undetected, Pol III subunit homologs might exist. We examined the human genome for sequences potentially encoding proteins with homology with any of the known Pol III subunits. Apart from POLR3GL and the genes encoding known Pol I and Pol II paralogs of Pol III subunits, we detected ORFs encoding putative homologs of POLR3K (RPC10) and POLR2K (RPABC4) (Supplemental Fig. S1D). However, unlike the POLR3GL ORF, these ORFs were not interrupted by introns, suggesting that they arose by recent retroduplication, and neither sequence could be found in the EST database, indicating that, consistent with the lack of any corresponding peptides in our Pol III preparation, they are unlikely to be expressed.
Evolution of the POLR3G and POLR3GL genes
The POLR3G and POLR3GL genes code for proteins with 46% amino acid identities, strongly suggesting that they arose from duplication. To examine whether the duplication arose through an RNA- or DNA-based event, we compared the genomic structure of the human POLR3G and POLR3GL genes. Although the intron sequences are not conserved between the two genes, the division of the protein-coding sequence into seven exons is close to identical in the two genes, as shown in Figure 1, A and B. This implies that the duplication did not occur by retroduplication, giving rise to a processed gene that would have then acquired introns, but rather by a DNA-based event.
Figure 1.
Evolution of the POLR3G and POLR3GL genes. (A) The genomic organization of the POLR3G (top line) and POLR3GL (bottom line) genes is shown with coding parts of exons as thick boxes, noncoding parts of exons as thinner boxes, and introns as lines with the arrowheads indicating the sense of transcription. The corresponding POLR3G and POLR3GL protein sections are schematized in the middle of the panel. (B) Alignment of POLR3G and POLR3GL protein sequences showing the borders (arrowheads) of corresponding exons. (C) Number of POLR3G and BRF homologs in different species. Species were classified according to species phylogeny. The numbers of detected POLR3G and BRF-related genes are indicated on the right.
We then examine the number of POLR3G/POLR3GL genes in some of the available genomes, as well as the number of BRF1/BRF2 genes, which, like POLR3G/POLR3GL, code for a subunit of a complex required for Pol III transcription, in this case TFIIIB. As shown in Figure 1C, we could identify only one BRF gene in S. cerevisiae, as expected, as well as in Caenorhabditis elegans and Drosophila melanogaster. We also found only one gene in Ciona intestinalis, a representative of the vase tunicates, the closest parents of vertebrates, and in Petromyzon marinus, a representative of agnaths (jawless vertebrates), a very ancient vertebrate lineage. In contrast, all gnathostomes genomes examined contained two genes except for the fish Takifugu rubripes, which contained three, two of which are close to BRF1 and one to BRF2. In the case of the POLR3G/POLR3GL genes, we found one gene in S. cerevisiae, C. elegans, and D. melanogaster; one gene also in C. intestinalis and P. marinus; and one, two, or three genes in other vertebrates. These observations suggested that the POLR3G/POLR3GL duplication might have occurred in the common ancestor of vertebrates or of gnathostomes.
In an attempt to time the POLR3G/POLR3GL duplication, we performed protein alignments and phylogeny reconstruction using the PhyML (Phylogenetic estimation using Maximum Likelihood) software (Guindon et al. 2010) with 1000 bootstraps. As shown in Supplemental Figure S2, the tree revealed two major clusters, the upper one containing sequences resembling POLR3G and the lower one containing sequences resembling POLR3GL, with the D. melanogaster, C. elegans, and the C. intestinalis proteins falling outside of these two clusters. These observations are consistent with the duplication occurring in a common ancestor of vertebrates, followed by loss of one gene in P. marinus, the fishes Gasterosteus aculeatus, Oryzias latipes, and T. rubripes, and the birds Gallus gallus, Meleagris gallopavo, and Taeniopygia guttata (zebra finch), and separate events of duplication in Danio rerio and Spermophilus tridecemlineatus.
To examine whether the ancestral protein more closely resembled POLR3G or POLR3GL, we directly compared the C. intestinalis and P. marinus proteins to human POLR3G and POLR3GL (Supplemental Fig. S3). Although the C. intestinalis protein fell outside of the POLR3G- and POLR3GL-resembling clusters in the phylogeny reconstruction tree (Supplemental Fig. S2), it was closer to human POLR3GL than human POLR3G (51% vs. 37% amino acid identities) (see Supplemental Fig. S3A,B) when directly aligned with these two proteins, as was the P. marinus protein (63% vs. 45% amino acid identities) (see Supplemental Fig. S3C,D). This is consistent with the ancestral gene being closer to the POLR3GL than the POLR3G gene.
Two forms of Pol III
The detection of the POLR3GL polypeptide in purified Pol III preparations suggested that two variants of Pol III, containing either POLR3G or POLR3GL, might coexist in HeLa cells. Indeed, antibodies specific for either POLR3G or POLR3GL coimmunoprecipitated POLR3C but not the other POLR3G subunit, indicating that POLR3G and POLR3GL lie in separate complexes (Supplemental Fig. S4A). We then fractionated a HeLa whole-cell extract by gel filtration chromatography and analyzed the resulting fractions by Western blot with antibodies against POLR3A and POLR3C (Supplemental Fig. S4B,C). POLR3C eluted in two main peaks, the first also containing POLR3A and eluting with an apparent size corresponding to the full Pol III complex (Supplemental Fig. S4C, fractions 10–13), and the second lacking POLR3A and eluting with a smaller apparent size (fractions 18–21). We then used these fractions for immunoprecipitations with either a preimmune or an anti-POLR3C antibody. The anti-POLR3C antibody specifically coimmunoprecipitated POLR3A, POLR3G, and POLR3GL in fractions from the first peak, and POLR3G and POLR3GL, but not POLR3A, in fractions from the second peak (Supplemental Fig. S4D). This further suggests that the first peak corresponds to the full enzyme and that the second peak contains the trimeric POLR3C/POLR3F/POLR3G subcomplex described previously (Wang and Roeder 1997). Thus, POLR3GL, similar to POLR3G, can be incorporated into the full enzyme, probably as part of the detachable trimeric subcomplex. Indeed, in GST pull-down experiments with immobilized GST-POLR3G, GST-POLR3GL, or GST-GFP as a control, we observed that both POLR3G subunits directly interacted with POLR3C (Supplemental Fig. S4E). They did not detectably interact with POLR3F or with BRF1 and BRF2, suggesting that (1) within the trimeric complex, POLR3G/POLR3GL have strong interactions only with POLR3C, and (2) the trimeric complex interacts with BRF1 and BRF2 mostly through subunits other than POLR3G/POLR3GL, consistent with previous reports showing interactions between POLR3F and BRF1 (Werner et al. 1993; Wang and Roeder 1997). Most importantly, for all interactions tested, POLR3GL behaved similarly as POLR3G.
POLR3G- and POLR3GL-containing Pol III on human genes
The results described above as well as previous results (Haurie et al. 2010) indicate that there are two forms of Pol III. To determine whether POLR3G- and POLR3GL-containing Pol III target different Pol III genes, we performed ChIP-seq with IMR90 cells and antibodies directed against POLR3D, POLR3G, POLR3GL, and BDP1 to map these two forms onto the human genome. We aligned tags and calculated scores as described in the Methods. As noted previously (Canella et al. 2010), our anti-POLR3D antibody was the most sensitive and scored a total of 494 loci as significantly occupied, whereas the anti-BDP1 antibody scored 222 loci as significantly occupied. All loci showing significant BDP1 scores also had significant POLR3D scores. Supplemental Table S2 lists all annotated tRNA genes (whether occupied by Pol III or not) as well as all other loci found significantly occupied by POLR3D together with their scores. We compared the results with our previous results obtained in the slightly different IMR90Tert cell line (Canella et al. 2010). As indicated in column E of Supplemental Table S2, we found 29 additional loci occupied by Pol III compared with our previous list, including one RN5S-related sequence on chromosome 10 (number 482 in column A); the BCYRN1 gene (number 181), which codes for BC200 RNA and is mostly expressed in neurons (Martignetti and Brosius 1993); one tRNA-derived sequence (number 715); 19 SINEs; and seven other loci (see Supplemental Table S2). This may reflect the much greater sequencing depth of this study and/or the use of a different cell line as starting material. On the other hand, 57 loci annotated as SINEs, LINEs, or “other” we had flagged as potentially occupied in our previous work were below the cutoff in this study (listed on the last page of Supplemental Table S2); all of these loci had displayed very low Pol III scores and no detectable BRF1 or BDP1 in our previous study (see Supplemental Fig. S7; Canella et al. 2010) and are, therefore, unlikely to be much transcribed even in IMR90Tert cells. Thus, consistent with our previous results, we find that only a limited number of genomic loci are occupied by Pol III besides the known Pol III genes.
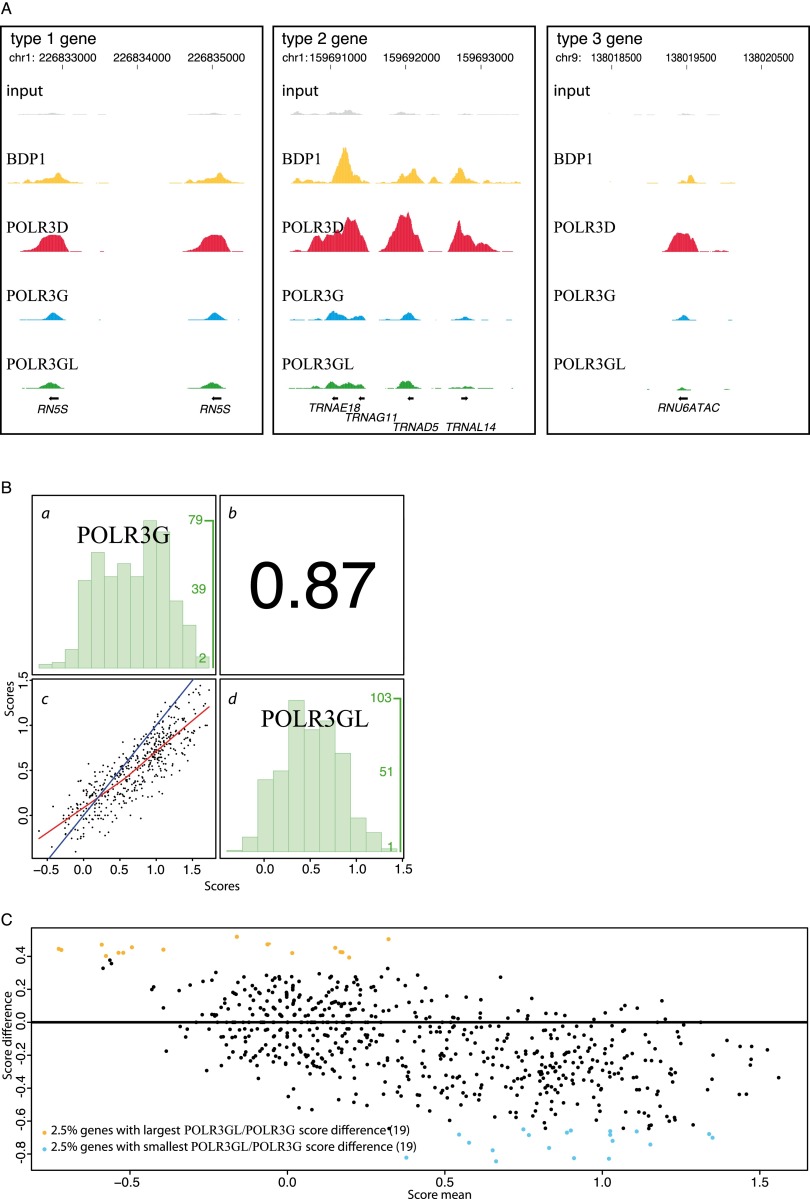
We then examined POLR3G and POLR3GL occupancy. We observed a total of 293 occupied loci occupied by POLR3GL and/or POLR3G (Supplemental Table S2). Since these loci correspond to those with the highest POLR3D scores, it is highly likely that, as for the anti-BDP1 antibody, our anti-POLR3G and anti-POLR3GL antibodies are less sensitive than the anti-POLR3D antibody, although it is possible that some loci are occupied by partial Pol III complexes. As shown in Figure 2A, we observed both POLR3G and POLR3GL on Pol III genes with type 1, 2, and 3 promoters. Moreover, for all Pol III–occupied loci, POLR3G and POLR3GL scores were highly correlated (Spearman correlation coefficient = 0.87) (Fig. 2B). Most genes displayed higher occupancy by POLR3G compared with POLR3GL except for some genes with very low scores, as visualized by the regression line (red) crossing the x = y line (blue) (Fig. 2B). This is also visualized in the MvA plot in Figure 2C, showing, first, most genes with a negative POLR3GL/POLR3G score difference and, second, the 2.5% genes displaying the largest (orange dots) and smallest (light blue dots) POLR3GL/ POLR3G score differences toward the lower and higher end of the score means, respectively. In fact, among the 19 loci with the largest POLR3GL/POLR3G ratio in Figure 2C, only one (indicated in orange in columns I–L of Supplemental Table S2, page 1) had either a POLR3G or a POLR3GL score above the cutoff, and it corresponds to a locus of unknown function. Among the 19 loci with the smallest POLR3GL/POLR3G ratio, 18 (indicated in turquoise in columns I–L of Supplemental Table S2) had either a POLR3G or a POLR3GL score above the cutoff; one is a RNU6 gene (U6-9), another the VTRNA1-2 gene (HVG 2), and all others are tRNA genes.
Figure 2.
POLR3G and POLR3GL occupy largely the same loci in human IMR90 cells. (A) POLR3G and POLR3GL occupy all three types of Pol III promoters. UCSC Browser view of type 1 (RN5S), type 2 (TRNA), and type 3 (RNU6ATAC) Pol III genes showing occupancy by BDP1, POLR3D, POLR3G, and POLR3GL, as well as the input. The x-axis shows the genomic location; the y-axis shows sequence tag accumulation. The scales on the y-axes are similar for all factors. (B) Spearman's rank correlation of the POLR3G versus POLR3GL scores. (c) x-axis, POLR3G scores; y-axis, POLR3GL scores; in blue the x = y line; in red, the regression line. (b) Correlation coefficient. (a) Distribution histogram representing, for each POLR3G score interval of 0.2 (see x-axis scale at the bottom of c), the number of genes in that interval (y-axis at the right of the panel: The numbers in green correspond to the lowest, middle, and highest number of genes). (d) As in a, but for each POLR3GL score interval of 0.2. (C) MvA plot with the score means [(POLR3GL score + POLR3G score)/2] on the x-axis and the score difference (POLR3GL score − POLR3G score) on the y-axis. All scores are in log2 (see Supplemental Table S2).
Thus, not only were both forms of the polymerase present on the large majority of Pol III–occupied loci, the proportion of each form was mostly constant from gene to gene, with perhaps a small bias toward more POLR3GL-containing RNA Pol III on the weakly occupied loci and more POLR3G-containing RNA Pol III on the highly occupied loci. These data do not offer strong support to the possibility that the two forms of Pol III might specifically target different genes. Nevertheless, it could still be possible that the two forms occupy different genes in different types of cells or tissues, in particular when one of the Pol III forms is much more present than the other.
POLR3G and POLR3GL are differentially expressed under different conditions and in different cell types
To identify conditions or cell types likely to have different amounts of POLR3G- and POLR3GL-containing Pol III, we measured POLR3G and POLR3GL mRNA levels by real-time PCR in different cell types and under different conditions. In human IMR90Tert cells, serum starvation (Supplemental Fig. S5A) resulted in a decrease in the ratio of POLR3G over POLR3GL mRNA, as did increasing confluency (Supplemental Fig. S5B). Moreover, the POLR3G-over-POLR3GL ratio was smaller in a primary culture of human foreskin tissue (4A cells) consisting of fully differentiated fibroblasts from young individuals than in a primary culture of human fetal dermal fibroblasts (Feo cells) that has large expansion capabilities (Supplemental Fig. S5C). To have access to a fully differentiated, normal tissue, we tested mouse liver and, as a comparison, mouse hepatocarcinoma cells (Hepa 1-6). Since POLR3G mRNA had not been detected in human liver (Haurie et al. 2010), for controls for the mouse liver experiment we used Ucp1 and Pdk4, two genes that are silent in mouse liver. As shown in Supplemental Figure S5, D and E, Polr3g mRNA was clearly present in mouse liver, but the Polr3g-over-Polr3gl ratio was much lower in normal liver compared with Hepa 1-6 cells. Thus, in all cases tested, POLR3GL mRNA was relatively more abundant than POLR3G mRNA in nondividing cells compared with highly dividing cells.
Pol III–occupied loci in mouse liver and mouse hepatocarcinoma cells
We chose to compare genome-wide POLR3G and POLR3GL occupancy in mouse liver and Hepa 1-6 cells, as (1) these two samples displayed very different Polr3g over Polr3gl mRNA ratios and (2) we were interested in determining POLR3G and POLR3GL occupancy in a normal tissue in addition to cultured cells. We performed ChIP-seq with antibodies directed against POLR3D, POLR3G, and POLR3GL in biological replicates (two pools of three mouse livers and two cultures of Hepa 1-6 cells). We identified significantly occupied regions and calculated scores as above for the human sample (see Methods).
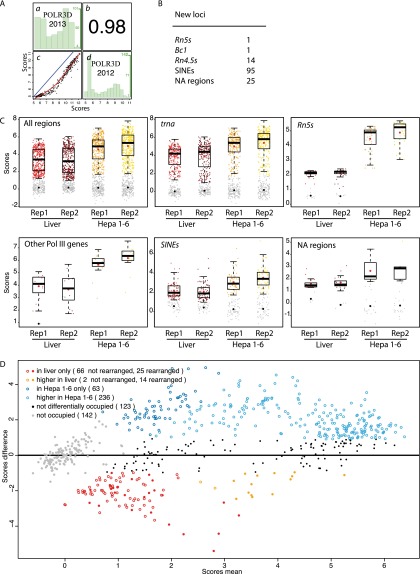
Supplemental Table S3 shows all the annotated tRNA genes (whether occupied or not) as well as all loci found to be significantly occupied by Pol III in mouse liver, mouse Hepa 1-6 cells, or both. We first compared the results obtained for POLR3D in mouse liver with our previously published POLR3D results in the same tissue (Canella et al. 2012). As shown in Figure 3A, the correlation of scores for the tRNA genes and SINEs identified by Canella et al. (2012) was extremely high (Spearman correlation coefficient 0.98), which is remarkable given that the two experiments were done at different times, by different people, and were sequenced on different machines (Illumina Genome Analyzer II in Canella et al. [2010, 2012] and Illumina HiSeq 2000 in this work). However, seven Rn5s (5S) and seven tRNA genes above the cutoff in Canella et al. (2012) were below the cutoff in this study (note, however, that all but n-Tg4 [number 93] and n-Te18 [number 355] are above the cutoff in Hepa 1-6 cells) (indicated in yellow in column D of Supplemental Table S3) and reciprocally for 26 tRNA genes (indicated in light green in column D of Supplemental Table S3).
Figure 3.
Pol III–occupied loci in mouse liver and Hepa 1-6 cells. (A) Spearman's rank correlation of scores obtained by Canella et al. (2012) and in this work. The loci considered include all tRNAs and SINEs. (c) x-axis, POLR3D scores in this work; y-axis, POLR3D scores by Canella et al. (2012); in blue, the x = y line; in red, the regression line. (b) Correlation coefficient. (a) Distribution histogram representing, for each POLR3D 2013 score interval of 1 (see x-axis at the bottom of c), the number of genes in that interval (y-axis at the right of the panel: The numbers in green correspond to the lowest, middle, and highest number of genes). (d) As in a but for each POLR3D 2012 score interval of 0.5. (B) List of additional, Pol III–occupied loci identified in this work compared with that of Canella et al. (2012). (C) Box plots showing scores in replicate 1 (Rep1) and replicate 2 (Rep2) samples from liver or Hepa 1-6 cells, as indicated on the x-axis. The y-axis shows scores in log2. Genes with scores below the cutoff (see Methods) are represented by gray dots. The median is indicated by the black horizontal bar, the mean of genes above the cutoff by the red dot, and the mean of genes below the cutoff by the black dot. The genes shown on the various panels correspond to the lists on the various pages of Supplemental Table S3. (D) MvA plot illustrating differential Pol III occupation in liver versus Hepa 1-6 cells. The x-axis shows score means [(POLR3D mean score in liver + POLR3D mean score in Hepa 1-6 cells)/2], and the y-axis score differences (POLR3D mean score in Hepa 1-6 cells − POLR3D mean score in liver). All scores are in log2 (see Supplemental Tables S3, S4).
When considering loci significantly occupied either in the liver or in Hepa 1-6 cells or in both, we uncovered another 136 loci (indicated in column D of Supplemental Table S3), including one Rn5s locus on chromosome 6, outside of the cluster of Rn5s genes on chromosome 8, and encoding a divergent 5S RNA; the Bc1_Mm_scRNA locus, encoding Bc1 RNA, a transcript previously described as neural-specific (Martignetti and Brosius 1995) and corresponding to human BCYRN1 (BC200) (Martignetti and Brosius 1993); 14 Rn4.5s loci; 95 SINEs, most of them from the B2 family; and 25 NA loci (Fig. 3B). The Rn4.5s loci (also referred to as 4.5S RNAH), whose function is unknown, are intriguing; they are located, like the ones we previously described (Canella et al. 2012), on chromosome 6 but are embedded in a tandemly repeated 4.3-kb sequence (which may exist at a much higher copy number than represented in the genome assembly) (Schoeniger and Jelinek 1986). Note that Rn4.5s loci are different from the 4.5S RNAI genes described by Gogolevskaya and Kramerov (2010), which correspond to B4A SINEs (see Supplemental Table S3, lines 243, 246, and 250; Canella et al. 2012), and from the 4.5S HybRNA genes described by Trinh-Rohlik and Maxwell (1988), which we could not find in the Mm9 genome assembly. Another 16 LINEs, five Rn (rRNA) repeats and nine NA loci (labeled in red in columns A and C of Supplemental Table S3) were considered unreliable (see Methods) and were excluded, therefore, from the analyses below. Supplemental Table S3 thus lists 701 loci, of which we consider 529 significantly and reliably occupied by Pol III either in mouse liver or hepatocarcinoma cells or in both, with the rest representing unoccupied tRNA genes and unreliable loci.
Differential genomic occupation by Pol III in mouse liver and Hepa 1-6 cells
We then examined whether loci were differentially occupied in mouse liver and Hepa 1-6 cells by considering the POLR3D scores as a measure of occupancy by both Pol III forms. When all loci were considered, there was an increase in the median and the mean of Pol III occupancy scores in Hepa 1-6 cells relative to liver cells, as shown in the upper left panel in Figure 3C. Similarly, there was an increase when tRNA genes, Rn5s genes, other Pol III genes, SINEs, or NA regions were considered separately (Fig. 3C).
To get an idea of the behavior of individual loci, we applied the limma linear model fitting on the genes scores (Smyth 2004, 2005) to determine adjusted P-values of the fold change in Hepa 1-6 cells versus liver (Supplemental Table S3, columns AD, AE, and AF), and we then plotted the score differences over the score means, as shown in Figure 3D. As indicated by the box plots in Fig. 3C, the large majority of loci were either only (dark blue circles; for list, see Supplemental Table S4) or more (light blue circles; for list, see Supplemental Table S4) occupied in Hepa 1-6 cells compared with liver. Among the more occupied were mostly tRNA (161) and Rn5s genes (42), the rest corresponding to “other Pol III genes” (13 out of 15, the only missing ones being the Rn7s genes, which remained unchanged), SINEs (19), and Rn4.5s loci (two). Thus, most tRNA, Rn5S, and other Pol III genes were more occupied in Hepa 1-6 cells than in liver, consistent with the idea that Pol III transcription is overactivated in cancer cells (White 2005; Johnson et al. 2008). Among the 63 loci only occupied in Hepa 1-6 cells were mostly SINEs and NA loci (39 loci), with only 16 tRNA and eight Rn5s gene loci.
Nevertheless, a number of genes appeared either exclusively (Fig. 3D, red circles and dots; for list, see Supplemental Table S4) or preferentially (Fig. 3D, orange circles and dots; for list, see Supplemental Table S4) occupied in liver cells. In examining these loci further, we noticed that several of them appeared deleted or rearranged in Hepa 1-6 cells, as suggested by interrupted peaks or total or near total absence of tags, even in the input sample. A few examples of apparent complete (three upper panels) or partial (three lower panels) deletions are shown in Supplemental Figure S6. Strikingly, nearly all of the 105 tRNA genes in the large cluster on chromosome 13, extending over 2.37 million base pairs (from position 21252654 to 23622288), appeared heavily altered/rearranged in Hepa 1-6 cells, and the same was true for several other tRNA genes. Because such rearranged regions give rise to tags that cannot be aligned to the reference genome, the resulting Hepa 1-6 scores for these regions are artificially low. When these loci (indicated as red and orange dots in Fig. 4B and listed in Supplemental Table S4) were removed from the picture, we were left with 66 loci only occupied, and two loci more occupied, by Pol III in liver: 45 of which were SINEs, 16 were NA loci, and seven were tRNA genes.
Figure 4.
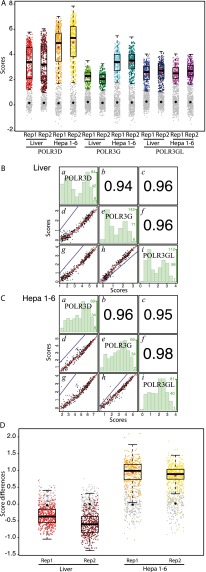
POLR3G and POLR3GL occupy largely the same loci. (A) Box plots showing scores in replicate 1 (Rep1) and replicate 2 (Rep2) samples from liver or Hepa 1-6 cells, as indicated on the x-axis. The y-axis shows scores in log2. Genes with scores below the cutoff (see Methods) are represented by gray dots. The median is indicated by the black horizontal bar, the mean of genes above the cutoff by the red dot, and the mean of genes below the cutoff by the black dot. The four box plots on the left are reproduced from Figure 3C, upper left panel. (B) Spearman's rank correlation of POLR3D and POLR3G (d), POLR3D and POLR3GL (g), or POLR3G and POLR3GL (h) scores in liver cells: In blue, the x = y line; in red, the regression line. (b,c,f) The correlation coefficients corresponding to panels d, g, and h, respectively. (a) Distribution histogram representing, for each POLR3D score interval of 0.5 (see x-axis at the bottom of g), the number of genes in that interval (y-axis at the right of the panel: The numbers in green correspond to the lowest, middle, and highest number of genes). (e) As in a but for each POLR3G score interval of 0.5 (see x-axis at the bottom of h). (i) As in a but for each POLR3GL score interval of 075. (C) As in B but in Hepa 1-6 cells. (D) Box plots showing POLR3G-POLR3GL score differences (in log2) in replicate 1 (Rep1) and replicate 2 (Rep2) samples from liver or Hepa 1-6 cells, as indicated on the x-axis. The y-axis shows score differences (in log2). Genes with scores below the cutoff (see Methods) are represented by gray dots. The median is indicated by the black horizontal bar, the mean of genes above the cutoff by the red dot, and the mean of genes below the cutoff by the black dot.
Thus, the large majority of Pol III–occupied loci, including most tRNA genes, are more occupied in Hepa 1-6 cells than in liver, and among those more occupied in liver, most are SINEs of unknown function. Moreover, of the 158 tRNA genes that were unoccupied in liver, all but 16 remained so in Hepa 1-6 cells, suggesting that for the vast majority of tRNA genes, transformation results in higher occupation of active genes rather than activation of genes that were silent. As an example, chromosome 6 contains a large cluster of 51 n-TC genes (tRNA cysteine genes), all of which are silent in liver except for the first one, n-Tc57, which is separated by >150,000 bp from the rest of the cluster. In Hepa 1-6 cells, all of these genes remain silent except for n-Tc57, which is more occupied. In contrast, a number of SINEs silent in liver were apparently activated de novo in Hepa 1-6 cells.
POLR3G and POLR3GL occupy the same loci in mouse liver and mouse hepatocarcinoma cells
Having identified Pol III–occupied loci in mouse liver and Hepa 1-6 cells, we then compared POLR3G and POLR3GL occupancy on these loci. As described above for human cells, we identified fewer loci occupied by POLR3G and/or POLR3GL compared with POLR3D, and these loci again corresponded to the loci with the highest POLR3D scores, consistent with the anti-POLR3G and anti-POLR3GL antibodies being less efficient than the anti-POLR3D antibodies. Figure 4A reproduces the POLR3D box plots shown in Figure 3C, upper left panel, and shows similar box plots for POLR3G and POLR3GL. Similar to what was observed for total Pol III occupancy as reflected by POLR3D scores, POLR3G scores were on average higher in mouse Hepa 1-6 cells than in mouse liver cells. In contrast, POLR3GL average scores were very similar in both types of cells. This suggests that the increase in Pol III occupancy in Hepa 1-6 cells compared with liver is provided by POLR3G-containing Pol III.
We then selected the loci considered as occupied by Pol III as determined by the presence of POLR3D, and examined whether the usage of one Pol III form compared with the other one was similar among genes and in the two different cell types. As shown in Figure 4, B and C, the occupancy by each of these subunits, whether in mouse liver or in Hepa 1-6 cells, was highly correlated with POLR3D occupancy scores, indicating that the more a gene is POLR3D-occupied and, by extension, probably transcribed in a given cell type, the more it is occupied by both POLR3G and POLR3GL. Indeed, in each cell type, the correlation between POLR3G and POLR3GL occupancy was similarly very high (liver: 0.96; Hepa 1-6: 0.98). Importantly, in mouse liver, the regression line (red line) for the POLR3G and POLR3GL score correlation was above the x = y line (blue line), indicating that in this tissue, scores were almost always higher for POLR3GL than for POLR3G, whereas in Hepa 1-6 cells, the opposite was observed, indicating that in this case, scores were almost always higher for POLR3G than for POLR3GL. Indeed, the log2 ratios of POLR3G over POLR3GL scores were almost always negative in mouse liver and almost always positive in hepatocarcinoma cells (Fig. 4D).
To determine whether any particular gene escaped the general trend described above, we used limma (Smyth 2004) to compare the POLR3G-over-POLR3GL score ratios in liver versus Hepa 1-6 cells (Supplemental Table S3, columns AG, AH, and AI). When considering the loci that were (1) significantly occupied by POLR3D in at least one cell type and (2) significantly occupied by either POLR3G or POLR3GL in at least one cell type, we found a majority (293) of loci with a higher POLR3G/POLR3GL ratio in Hepa 1-6 compared with liver, as expected. One locus (NA_21) had a slightly decreased ratio (LogFC = 0.882) in Hepa 1-6 compared with liver, and the remaining 46 loci showed no significant change (see Supplemental Table S3, column AI).
Together, these results show, first, that both subunits are generally present at all Pol III–occupied loci in the same proportion. We did not find any locus significantly occupied only by POLR3G or POLR3GL or displaying a very different ratio of occupancy by these two subunits compared with the bulk of Pol III–occupied loci. Thus, the pattern of genome-wide occupancy by POLR3G- and POLR3GL-containing Pol III argues against POLR3G or POLR3GL serving to target Pol III differentially to certain genes. Second, the results indicate that the general ratio of POLR3G and POLR3GL occupancy varies in different tissues and is higher in cancer liver cells compared with normal liver cells.
The promoters of the genes encoding POLR3G and POLR3GL are differentially occupied by the oncogene MYC
The observation that the POLR3G and POLR3GL genes are differentially expressed under various conditions and in different cell types indicates that they are controlled by different mechanisms. Our results (see Supplemental Fig. S5) and those of others (Haurie et al. 2010; Wong et al. 2011) suggest that cell proliferation is likely to be one of the factors affecting their expression. The oncogene MYC is a transcription factor broadly involved in the cellular mitogenic and growth responses and is very often up-regulated in cancerous cells (Eilers and Eisenman 2008; Meyer and Penn 2008; Dang 2012). MYC binds as a dimer with MAX to sequences referred to as E-boxes. The E-box consensus sequence is CANNTG, with the palindromic sequence CACGTG constituting a high affinity site. Although we did not find CACGTG sequences close to POLR3G and POLR3GL transcription start sites (TSSs), we observed multiple CANNTG sequences. We thus took advantage of recently published data (Lin et al. 2012) describing genome wide MYC occupancy in P493-6 cells, which can be induced to express ectopic MYC, as well as in other cell lines, to examine MYC presence close to the POLR3G and POLR3GL TSSs.
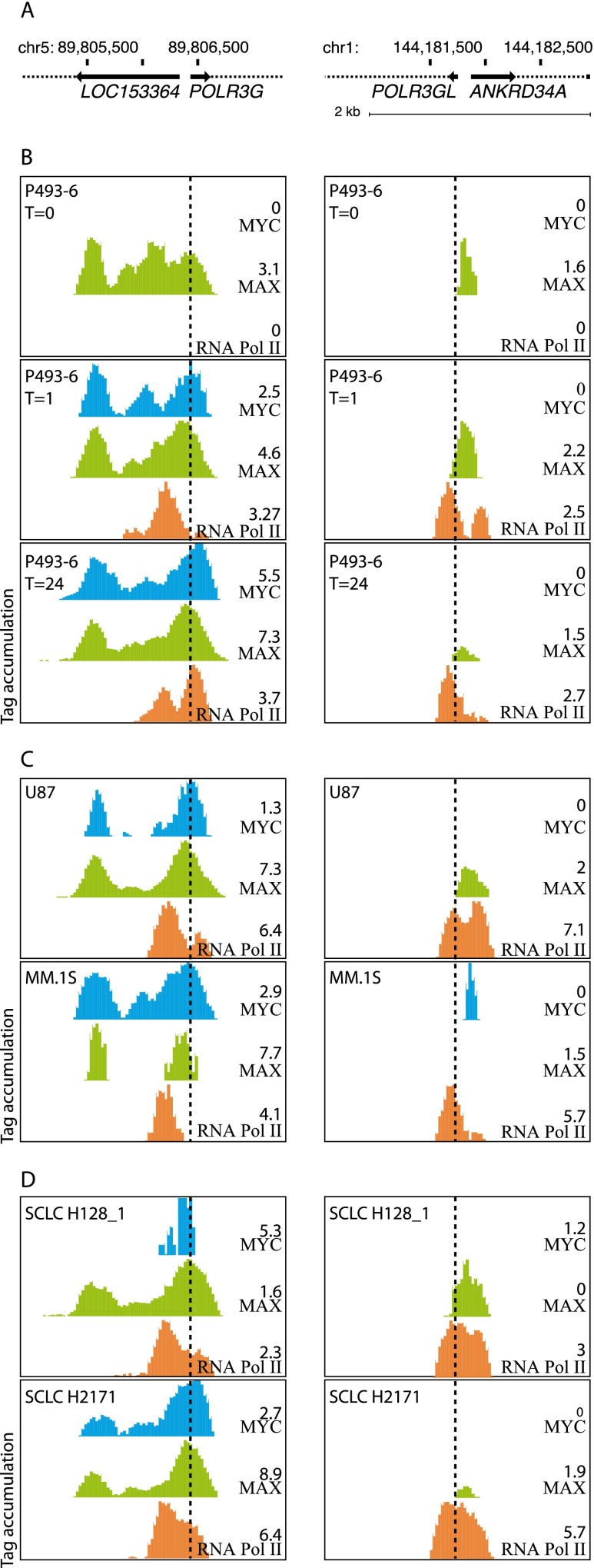
Both the POLR3G and POLR3GL genes are under the control of bidirectional promoters with closely located divergent TSSs (Fig. 5A). Nevertheless, it is clear that upon induction of ectopic MYC in P493-6 cells, there is accumulation of MYC and RNA Pol II at the POLR3G TSS. In contrast, there is no detectable MYC at the POLR3GL TSS, even after 1 h and 24 h of MYC induction (Fig. 5B, cf. middle and lower panels to upper panel), when the ectopic MYC protein accumulates to 76,500 and 362,000 molecules per cell, respectively (Lin et al. 2012). We also examined MYC occupancy in two pairs of cell lines studied by Lin et al. (2012): (1) primary glioblastoma U87 cells and MM.1S malignant B-lymphocytes, which are both transformed cell lines but differ in their MYC expression levels, with U87 cells containing 4.5 times fewer MYC molecules per cell than MM.1S cells (Lin et al. 2012), and (2) H128_1 and H2171 cells, two subtypes of small-cell lung carcinoma lines, again exhibiting lower and higher levels of MYC, respectively (Lin et al. 2012). In all these cases, MYC was detected exclusively at the POLR3G TSS, with the possible exception of MM.1S cells, where a very small peak was detected downstream from, albeit not at, the POLR3GL TSS (Fig. 5C,D).
Figure 5.
MYC binds to POLR3G but not to POLR3GL TSS. (A) Schematic of the POLR3G and POLR3GL genomic regions. (B) UCSC Browser views showing MYC, MAX, and Pol II (antibody directed against the N-terminus of POLR2A, Santa Cruz sc-899) tag accumulation, as indicated on the right, on the POLR3G and POLR3GL promoter regions, in P493-6 cells at time 0, 1 h, and 24 h after induction of MYC, as indicated on the left. The scales on the y-axes were adjusted to the maximum height of the peaks in each track, which is indicated on the right of each track, just above the track identity. (Based on the data of Lin et al. [2012]). (C) As in B but in U87 and MM.1S cells. (D) As in A but in SCLC H128_1 and SCLC H2171 cells.
The presence of MYC on the POLR3G but not the POLR3GL TSS, suggests that the POLR3G promoter is activated by MYC to allow for increased production of a POLR3G subunit when the cells require higher levels of Pol III. We checked whether we could detect MYC on the promoters of the genes encoding all the other Pol III subunits. Indeed, we observed MYC binding over the TSSs of all other Pol III subunit-encoding genes, namely, POLR3A, -B, -C, -D, -E, -F, -H, -K, and CRCP (RPC9) in cells expressing high levels of MYC (Supplemental Fig. S7). For the promoters of genes encoding subunits common to Pol III and either Pol I (POLR1C/AC1, POLR1D/AC2) or Pol I and II (POLR2E/ABC1, POLR2F/ABC2, POLR2H/ABC3, POLR2K/ABC4, POLR2L/ABC5), MYC occupancy was less prominent but could be observed in all cases in at least one of the cell lines (data not shown). Thus, the POLR3GL gene is exceptional among genes encoding Pol III subunits in that its TSS is apparently not bound by MYC, even in cells with high MYC levels.
Discussion
The availability of genome sequences from many organisms has revealed that gene duplication followed by retention of two functional copies is widespread (for reviews, see Prince and Pickett 2002; Long et al. 2003; see also Chen et al. 2010; Ross et al. 2013). According to classical models, duplicated genes can have two main fates. The one considered most likely is the degeneration or loss through genome remodeling of one of the copies, a process known as nonfunctionalization. The other, which is expected to be much less frequent, is neo-functionalization, i.e., the acquisition, through mutations in either the coding or the regulatory sequence, of a new function. However, the frequency of functional duplicated genes in genomes is much higher than would be expected from this model alone (Prince and Pickett 2002). An alternative model, known as the duplication-degeneration-complementation (DDC) model, provides an explanation for the prevalence of duplicated genes (Force et al. 1999; for review, see Prince and Pickett 2002). In the DDC model, each duplicated gene can acquire independent degenerative mutations affecting one of several subfunctions, which are still provided by the other copy (subfunctionalization). Given the combinatorial mechanism of transcription regulation, in which several short binding sites provide, alone or in combinations, different functions such as tissue- or stage-specific expression, regulatory sequences have been proposed to be a likely target for such mutations (Force et al. 1999).
Human cells contain two genes encoding two versions of the Pol III POLR3G subunit with 46% identical residues, each of which can be incorporated into RNA Pol III (Haurie et al. 2010; this work). The genomic structure of the two genes, which display close to identical exon organization with respect to the protein-coding sequence, indicates a DNA-based duplication event. A comparison of available sequences from a number of organisms suggests that the two genes arose from the duplication of a common ancestor gene in vertebrates. This may have been as a result of a small duplication or one of the two genome-wide duplications commonly thought to have occurred in vertebrate genomes (2R hypothesis) (Makalowski 2001; Kasahara 2007), before the divergence of the ancestral lamprey and Gnathostomata lineages (Smith et al. 2013).
The POLR3G/POLR3GL duplication was apparently followed by the loss of a gene in some organisms (although we cannot exclude the possibility that the apparent lack of a second gene reflects in some cases problems with genome assemblies) and a second duplication in others. Among vertebrates, the gene present in the agnath P. marinus and the fishes G. aculeatus, O. latipes, and T. rubripes codes for a protein with higher amino acid sequence identity to human POLR3GL than human POLR3G, whereas in the birds G. gallus, M. gallopavo, and T. guttata, the remaining copy codes for a protein closer to POLR3G. This may reflect a period during which the two genes, although structurally distinguishable, remained functionally redundant in some species, allowing loss of one or the other copy. In contrast, all eutherians, metatherians, and prototherians examined have at least two copies, consistent with both genes having acquired and fixed separate functions in the common ancestor of mammals.
A different function for the POLR3G and POLR3GL genes is experimentally supported by the work of Haurie et al. (2010), who observed that suppression of POLR3GL expression by siRNA resulted in cell death, whereas suppression of POLR3G had no deleterious effect under normal growth conditions but inhibited the formation of colonies in soft agar. Here, we have examined whether POLR3G-containing Pol III and POLR3GL-containing Pol III recognize different target genes, in the same way as BRF1 and BRF2 recognize specifically type 1 and 2 Pol III promoters, in the first case, and type 3 promoters, in the second (Canella et al. 2010; Carriere et al. 2012; James Faresse et al. 2012). For this purpose, we have performed genome-wide ChIP-seq experiments with anti-Pol III antibodies in human and mouse cultured cells as well as in mouse liver. From these experiments, we can refine our previous lists of Pol III–occupied loci in both human and mouse cells and confirm that apart from the known Pol III genes, relatively few loci are clearly occupied by Pol III; we find 26 SINEs clearly occupied by Pol III in human cells, 31 in both Hepa 1-6 and mouse liver cells, 36 only in Hepa 1-6, and 60 only in liver cells. These numbers are lower (Barski et al. 2010; Moqtaderi et al. 2010; Kutter et al. 2011; Carriere et al. 2012) or grossly similar (Oler et al. 2010; Raha et al. 2010) to those reported by others, which may reflect biological differences in the cell lines used, as well as the stringency of the filters applied. We suspect that our lists contain very few false positives but may well be missing loci occupied at very low levels.
When comparing Pol III occupancy in liver and Hepa 1-6 cells, we observed some SINEs more, or only, expressed in liver. However, consistent with previous findings reporting increased RNA Pol III activity in cancer cells (for a review, see White 2005; Johnson et al. 2008), the large majority of Pol III–occupied loci were more occupied in hepatocarcinoma cells than in liver cells. For example, 5S genes were collectively more occupied by Pol III in Hepa 1-6 cells compared with liver cells. Notably, eight 5S genes in the chromosome 8 cluster that scored below the cutoff in liver (and that scored among the nine lowest occupied 5S genes in Canella et al. 2012) were clearly occupied in Hepa 1-6 cells. Of note, most tRNA genes that were silent in liver cells (as opposed to occupied at low levels) remained so in Hepa 1-6 cells, suggesting that they are embedded in a deeply repressed chromatin environment. In contrast, some SINEs were de novo transcribed in Hepa 1-6 cells, perhaps reflecting genomic rearrangements in these cells rather than differential regulation in the same environment. Among the loci appearing more occupied in liver compared with Hepa 1-6 cells, many, in particular tRNA genes, were clearly deleted, rearranged, or otherwise changed in Hepa 1-6 cells, which is likely to lead to underestimated scores in these cells. Despite these score uncertainties, combining the scores for tRNA genes by isotype revealed increased Pol III occupancy for all amino acids except for selenocysteine, histidine, and asparagine: In these cases, the corresponding tRNA genes did not appear rearranged in Hepa 1-6 cells, and yet the combined scores were barely higher (SeC) or lower (His and Asn) in Hepa 1-6 cells compared with liver (data not shown). It will be interesting to determine whether this is also the case in other malignant cells or whether it reflects a particularity of Hepa 1-6 cells. In the case of selenocysteine, this may be quite general as there are only two genes, n-Ts1 and n-Ts2, one of which (n-Ts1) is silent in liver and remains so in Hepa 1-6 cells.
To determine whether the POLR3G- and POLR3GL-containing forms of Pol III might specialize to recognize different targets, we compared POLR3G and POLR3GL scores in human IMR90 cells. Most Pol III–occupied loci yielded higher scores for POLR3G compared with POLR3GL, but because the anti-POLR3G and anti-POLR3GL antibodies may have different affinities, the absolute numbers cannot be interpreted. Importantly, however, the proportion of POLR3G and POLR3GL was very similar on all but a few genes. This result strongly suggests that within this cell line, POLR3G- and POLR3GL-containing RNA polymerases are recruited to the very same promoters. Since ChIP-seq results, like any biochemical experiment, reflect the average situation in all cells, it is possible that some genes are transcribed exclusively by, for example, POLR3G-containing Pol III in some cells and POLR3GL-containing Pol III in others. It remains that unlike the BRF1 and BRF2 transcription factors, which recognize specifically different subsets of Pol III promoters, both forms of polymerases have the capacity to recognize the same promoters.
Since the two isoforms were found to be differentially expressed (Haurie et al. 2010), we examined POLR3G and POLR3GL expression in various cells and conditions. Our results show higher POLR3G expression relative to POLR3GL expression in dividing cells compared with resting cells, consistent with the observation that POLR3G (RPC32-alpha) increases relative to POLR3GL (RPC32-beta) during cellular transformation and decreases during differentiation (Haurie et al. 2010). In particular, we found much higher Polr3gl expression than Polr3g in normal mouse liver, where hepatocytes are in G0, a situation that was reversed in mouse hepatocarcinoma cells, which divide approximately every 24 h. We thus used these two types of cells to perform a genome-wide ChIP-seq experiment with antibodies directed against POLR3D, POLR3G, and POLR3GL.
In both types of mouse cells, we observed a high correlation between POLR3D and POLR3G, and POLR3D and POLR3GL, occupancy scores, with POLR3G scores generally lower than POLR3GL scores in liver cells and higher than POLR3GL scores in Hepa 1-6 cells. Although we cannot interpret these results in terms of absolute amounts, given that different antibodies with potentially different affinities were used, it is clear that the relative ratios of both subunits change in the two cell lines. Nevertheless, however, within one cell line, the ratios of POLR3G and POLR3GL scores were, like in human IMR90 cells, highly constant from one locus to another, as illustrated by the high correlation between POLR3G and POLR3GL occupancy in both types of cells. Together, the genome-wide localization results of POLR3G and POLR3GL do not support the idea that the incorporation of POLR3G or POLR3GL into Pol III confers a different specificity to the enzyme for its target genes.
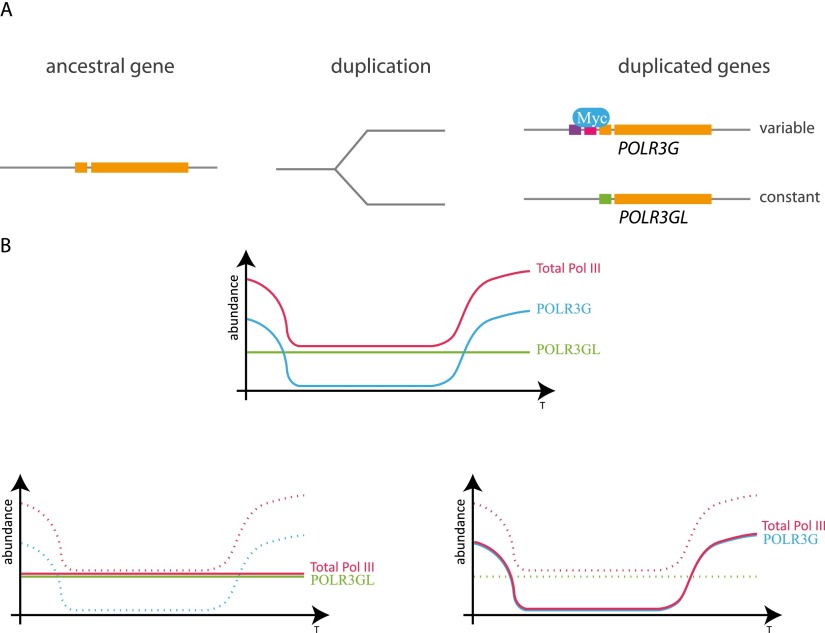
If the different functions of the two forms of Pol III are not related to transcription of different target genes, what are they? One possibility is that the two enzymes respond differently to regulators such as MAF1, which represses Pol III transcription by direct binding to the enzyme (Upadhya et al. 2002; Oficjalska-Pham et al. 2006; Reina et al. 2006; Vannini et al. 2010). Another is that POLR3GL or POLR3G has a function completely unrelated to its role as a subunit of Pol III, for example, as part of another complex. However, the DDC model argues that complementary mutations in duplicated genes will frequently affect regulatory elements rather than the function of the protein itself (Force et al. 1999). Indeed, we found MYC bound to the TSS of all genes encoding Pol III subunits, suggesting that they are responsive to activation by MYC, with the notable exception of the POLR3GL gene. Perhaps MYC activation of the POLR3GL promoter is not desirable because it might lead to activation of the closely spaced ANKRD34A promoter. It is the presence of the POLR3G gene then that allows the cell to produce increased levels of this subunit when needed. Thus, the POLR3G gene duplication did not lead to two forms of Pol III with different specificities for target genes but rather to two transcription units with different regulation potentials, as illustrated in Figure 6A. If this is the main function of the duplication, the cell death observed by Haurie et al. (2010) upon suppression of the POLR3GL gene most likely results from very low levels of POLR3G expression under certain conditions, as illustrated in Figure 6B. It seems likely, then, that the POLR3G/POLR3GL duplication was retained as a result of DDC, perhaps with neo-functionalization of the regulatory regions related to the proximity for each of the two genes of a divergent promoter.
Figure 6.
The POLR3G gene duplication leads to neo-functionalization of promoter sequences rather than gene product. (A) The POLR3G and POLR3GL promoters are differentially regulated, probably at least in part through exclusive binding of MYC to the POLR3G TSS. (B) Model of POLR3G and POLR3GL regulation. The model assumes a constant cellular level of POLR3GL and a variable level of POLR3G that allows adaptation of total Pol III levels to cell growth and proliferation conditions. If the cell loses POLR3G, the constant expression level of POLR3GL allows survival. If the cell loses POLR3GL, it can encounter conditions when POLR3G expression levels are too low for survival.
The transcriptional apparatus is highly conserved, with multi-subunit RNA polymerases displaying a strikingly similar catalytic core and some general transcription factors easily recognizable in organisms as remote from each other as bacteria, archae, and mammals (see Carter and Drouin 2010; Vannini and Cramer 2012, and references therein). Nevertheless, gene duplications have led to new protein functions, as for BRF1 and BRF2, which recognize different subsets of promoters (Schramm et al. 2000; Geiduschek and Kassavetis 2001; Schramm and Hernandez 2002; Jawdekar and Henry 2008; Carriere et al. 2012), or RNA polymerases IV and V, which appeared in plants through several gene duplications and are specifically involved in noncoding RNA-mediated gene silencing processes (Haag and Pikaard 2011), as well as to transcription units with new regulation potentials, of which the POLR3G/POLR3GL duplication is an example.
Methods
Cell lines
HeLa, IMR90, IMR90Tert (kindly provided by Greg Hannon, Cold Spring Harbor Laboratory), FEO and 4A (kindly provided by Lee Ann Laurent-Appelgate, University of Lausanne), and Hepa 1-6 (kindly provided by David Gatfield, University of Lausanne) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 4 mM glutamine, 100 units/mL of penicillin, and 100 μg/mL of streptomycin. For serum deprivation (Supplemental Fig. S5A), the medium was removed, and the cells were washed two times and then incubated with fresh medium lacking FBS for 4 and 8 h before cell harvesting. Control cells were handled similarly, but after medium removal, normal Dulbecco's modified Eagle's medium supplemented with 10% FBS was used. Similar results were obtained with serum deprivation protocols performed with medium containing 0.5% FBS rather than 0% FBS. For analyses of cell density effects (Supplemental Fig. S5B), growth curves were first established for each cell line in test experiments. Cells were then seeded at the appropriate concentration to obtain the expected density at the time of harvesting, 18 h later.
Animals
Two pools of three C57BL/6 12-wk-old male mice were killed, and livers were collected as described below for RNA preparation and ChIP experiments. All animal care and handling was performed according to Swiss law for animal experimentation.
Pol III purification
Pol III was purified from whole-cell extracts prepared from 48 liters of the clonal cell line HeLa 9-8 as previously described (Hu et al. 2002). Briefly, the extracts were first fractionated by ammonium sulfate precipitation. All buffers used after the ammonium sulfate precipitation except for buffer D100 were supplemented with 0.5 mM PMSF, 1 μM pepstatin A, 5 mM β-mercaptoethanol, and Sigma protease inhibitor cocktail (cat no P8849) diluted 1:10000. The proteins precipitated between 18% and 40% ammonium sulfate were dissolved in TBS120 buffer (50 mM Tris-HCl at pH 8.0, 120 mM NaCl, 5% glycerol) to a final salt concentration of 150 mM and were loaded onto anti-Flag immunoaffinity beads (Sigma). The anti-Flag beads were rotated overnight at 4°C and washed with 30 bead volumes of TBS300 and 20 bead volumes of TBS150. The bound proteins were eluted with five bead volumes of a solution containing 300 μg/mL of Flag peptide in TBS150 buffer. The fractions were pooled, adjusted to 300 mM NaCl and 10 mM imidazole, and incubated with Ni2-nitrilotriacetic acid (NTA) agarose beads (Qiagen) overnight at 4°C. The beads were washed with buffer B (50 mM NaH2PO4, 20 mM imidazole, pH 8.0). The bound proteins were eluted with five bead volumes of buffer B containing 300 mM imidazole. The fractions were dialyzed against buffer D100 (50 mM HEPES [pH 7.9], 0.2 mM EDTA, 20% glycerol, 0.1% Tween 20, 100 mM KCl, 3 mM DTT, 0.5 mM PMSF).
Mass spectrometry analysis
TCA-precipitated protein pellets were analyzed at the Proteomics Center of the Stowers Institute for Medical Research (Kansas City, MI, USA) for mass spectrometry analysis. The pellets were solubilized in Tris-HCl (pH 8.5) and 8 M urea and then reduced and alkylated with TCEP (tris-(2-carboxylethyl)-phosphine hydrochloride, Pierce) and iodoacetamide (Sigma), respectively. Proteins were digested with endoproteinase Lys-C (Roche) at 1:100 w/w, followed by Trypsin (Promega) at 1:100 w/w. Formic acid was added to 5% to stop the reactions. Peptides were loaded on triple-phase fused-silica microcapillary columns (McDonald et al. 2002) and placed in-line with a Deca-XP ion trap mass spectrometer (ThermoScientific), coupled with a quaternary Agilent 1100 series HPLC. A fully automated seven-step chromatography run was carried out for each sample, as described by Florens and Washburn (2006). The MS/MS data sets were searched using SEQUEST (Eng and McCormack 1994) against a database of 61,318 sequences, consisting of 34,521 H. sapiens nonredundant proteins (released by NCBI on 2012-08-27), 160 usual contaminants (such as human keratins, IgGs, and proteolytic enzymes), and, to estimate false-discovery rates (FDRs), randomized sequences derived from each nonredundant protein entry. Peptide/spectrum matches were sorted, selected, and compared using DTASelect/CONTRAST (Tabb et al. 2002). Combining all runs, proteins had to be detected by at least two peptides, leading to FDRs of 1% and 0.2% at the protein and spectral levels, respectively. To estimate relative protein levels, normalized spectral abundance factors (dNSAFs) were calculated for each detected protein, as described by Zhang et al. (2010).
Homology and phylogeny analysis
Homology search was performed in a stepwise procedure. A BLAST search was first performed in genome assemblies available in Ensembl with human POLR3G (or BRF1) as the query. Candidate homologs identified in other genomes were then filtered manually. Proteins sequences were aligned with MUSCLE (Edgar 2004) and cleaned using BMGE (Criscuolo and Gribaldo 2010). Phylogenetic trees were constructed using the PhyML (Guindon et al. 2010) and drawn with Phylodendron (see http://iubio.bio.indiana.edu/treeapp/). All software was used with the default parameters.
Antibodies
The antibodies used in this work were rabbit polyclonal antibodies, as follows: anti-human POLR3G SZ3070 antibody (Ab), raised against peptide DYKPVPLKTGEGEEYML; anti-human POLR3GL SZ3072 Ab, raised against peptide RPPKTTEDKEETIK; anti-mouse POLR3G ZCH10075-1430 Ab, raised against peptide VGFSRGEKLPDVVLK; anti-mouse POLR3GL ZCH10079-1434 Ab, raised against peptide RPPKSTDDKEETIQK; anti-POLR3D (mouse and human) CS681 Ab, raised against peptide CSPDFESLLDHKHR (Chong et al. 2001); anti-human POLR3C CS2125 Ab, raised against peptide DEDAAGEPKAKRPKY; and anti-human BDP1: CS914, raised against peptide CSDRYRIYKAQK, like CS913 (Schramm et al. 2000).
Gel filtration
One milliliter of HeLa whole-cell extract was fractionated on a Superose 6 10/300 GL column (GE Healthcare UK) by FPLC. The column was first equilibrated with two volumes of TBS300, the sample was loaded, and 500 μL fractions were collected. Molecular weights were determined from the elution profile of high molecular weight standards performed under similar conditions furnished by the manufacturer. Fifteen microliters of each fraction was loaded on a 12% SDS-polyacrylamide gel, and fractionated proteins were detected with various antibodies. Another 200 μL of the same fractions was used for immune-precipitation with anti-POLR3C serum or a preimmune serum as negative control.
GST-pull downs
Recombinant GST-POLR3G, GST-POLR3GL, and GST-GFP were produced in the BL-21 Escherichia coli strain. After overnight induction, cells were harvested and resuspended in 50 mL of suspension buffer (25 mM Hepes [pH 7.9], 100 mM KCl, 20% Glycerol, 0.5 mM PMSF, EDTA-free complete tablet [Roche], and 10 mM β-mercaptoethanol). Cells were incubated with lysozyme at a final concentration of 100 μg/mL for 20 min on ice. After the addition of NP-40 to a final concentration of 0.1%, the lysate was homogenized with a Dounce homogenizer. GST-tagged proteins were bound to glutathione-agarose beads, and the sample was rotated overnight at 4°C. Beads were then washed with 30 bead volumes of HEMGN buffer (25 mM Hepes [pH7.9], 150 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 10% glycerol, 0.1% NP-40 and 0.5 mM PMSF, EDTA-free Complete tablet [Roche], and 1 mM dithiothreitol) and 30 bead volumes of TBS150 (containing 0.5 mM PMSF, EDTA-free Complete tablet [Roche], and 1 mM dithiothreitol).
The human POLR3C, POLR3F, BRF1, and BRF2 proteins were produced with the TnT Quick Coupled Transcription/Translation System (Promega). Similar amounts of recombinant GST-POLR3G, GST-POLR3GL, or GST-GFP (negative control) bound to beads were then incubated overnight with 15 μL of the TnT-produced proteins. Columns were washed four times with 10 volumes of TBS150 (with 0.5 mM PMSF, EDTA-free complete tablet [Roche], and 1 mM dithiothreitol). Proteins bound to the column were then eluted in Laemmli buffer (60 mM Tris-Cl at pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue) and analyzed by autoradiography.
RNA isolation
Cell lines
About 5 million cells were scraped in 5 mL of TRIzol reagent (Invitrogen) and incubated for 5 min at room temperature. One milliliter of chloroform was then added; the tubes were inverted several times to mix the samples and then centrifuged for 15 min at 12,000g at 4°C. The RNA-containing aqueous phase was recovered and added to 500 μL of EtOH for precipitation. RNA was recovered after centrifugation for 30 min at 12,000g at 4°C. The pellets were washed with 70% of EtOH and resuspended in DEPC-treated water. RNA quality was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies) as well as by fractionation on a 1% agarose gel.
Mouse liver
About 100 mg of snap-frozen liver tissue was disrupted in 1 mL of TRIzol reagent (Invitrogen) with a TissueLyser (Qiagen). The homogenate solution was then processed as described above for the cell lines.
Quantitative RT-PCR
One microgram of total RNA was used for reverse transcription with oligo-dT primers and the Improm-II reverse transcription system (Promega). Two microliters of the resulting cDNA was amplified with 0.4 mM specific cDNA (exon–exon junction) primers. The reactions, which contained the Fast SYBR Green Master Mix (Roche), were analyzed by quantitative PCR on a Rotor-Gene-3000 (Corbett, Life Science). The thermal cycling conditions were optimized according to the manufacturer's protocol. The results were analyzed with the software provided with the instrument, using the comparative quantification function. The quantification was normalized to PCRs performed with POLR3C mRNA-specific primers (Supplemental Fig. S5A–C). PCR reactions were repeated three times with independent cDNA preparations.
ChIP
Approximately 10 million subconfluent IMR90 cells were used per ChIP. The protocol used was similar to the one described by O'Geen et al. (2006). Cells were directly crosslinked in the culture medium for 7 min with 1% formaldehyde. The chromatin was sonicated to an average size of 200–600 bp. Livers were perfused with 5 mL of PBS through the spleen, immediately homogenized in PBS containing 1% formaldehyde, and then processed as described by Ripperger and Schibler (2006). Aliquots of sonicated chromatin were mixed with different antibodies and incubated overnight at 4°C on a rotating wheel. Immunoprecipitated material was recovered by addition of 15 μL of protein A agarose beads (preblocked with 10 μg/mL of BSA and 10 μg/mL of salmon sperm DNA) and incubation for 1 h at room temperature on a rotating wheel. The beads were washed with dialysis and wash buffer (O'Geen et al. 2006). De-crosslinking, RNase A treatment, proteinase K treatment, and DNA purification were performed as described by O'Geen et al. (2006). The optimal amount of each antibody was determined in test ChIPs analyzed by q-PCR.
Ultra-high-throughput sequencing
Ten nanograms of immunopurified chromatin as well as input DNA was used to prepare sequencing libraries with the ChIP-seq Sample Preparation Kit (Illumina; catalog no. IP-102-1001) according to the protocol supplied by the manufacturer. Sequencing libraries were loaded onto one lane of a Genome Analyzer flow cell (human IMR90 cells) or onto one lane of a HiSeq 2000 flow cell (mouse liver and Hepa 1-6 cells).
Analysis of ChIP-seq data
Tag alignment
The sequence tags obtained after ultra-high-throughput sequencing were first mapped onto the UCSC Genome Browser versions mentioned in Supplemental Tables S2 and S3 via the eland_extended mode of ELAND v2e in the Illumina CASSAVA pipeline version 1.8.2. The tags with multiple matches in the genome were then mapped with the “fetchGWI” software (www.isrec.isb-sib.ch/tagger/) (Iseli et al. 2007). As in our previous work, we included tags matching up to 500 times in the genome but did not allow any mismatch for tag alignment (Canella et al. 2012). The numbers of tags sequenced with and without redundancy, the numbers of tags aligned onto the genome, as well as the percentage of tags falling in the list of loci in Supplemental Tables S2 and S3 are listed in Supplemental Table S5.
Identification of enriched genomic regions
To identify Pol III–enriched regions in each sample (human IMR90 cells, mouse liver, and mouse Hepa 1-6 cells), we divided the genome into 400 nucleotide (nt) bins and then compared the tag counts in the POLR3D immunoprecipitations with the tag counts in the input in each of those bins. After eliminating the bins with a similar enrichment level in both IP and Input, we calculated scores for the remaining 400-nt regions and extracted those regions with a score above the cutoff (see below). For regions corresponding to known Pol III genes, the scores were then calculated over the RNA coding region, as well as upstream and downstream sequences as described below. For the regions that did not correspond to known Pol III genes, the scores were recalculated over a window of minus to plus 200 nt around the peak maxima.
For the IMR90 cells, the method identified 917 enriched regions, of which 568 had a POLR3D score above the cutoff. Of these, we removed 47 loci in satellite regions, as well as 27 regions containing peak trails (i.e., corresponding to bins falling toward the end of a peak) or peaks with strange shapes. This left a total of 494 loci clearly occupied by POLR3D. For both the mouse liver and Hepa 1-6 cells, the method identified a total of 1084 enriched regions, of which we kept 589 with scores above the cutoff (see below) in at least one type of cell. Thirty of these regions displayed peaks with unusual (often rectangular) shapes and were thus not considered in the analysis (indicated in red in columns A and D of Supplemental Table S3), leaving 559 loci clearly occupied by Pol III, plus another two above the cutoff in the work by Canella et al. (2012) but below the cutoff in both liver and Hepa 1-6 cells in this study, for a total of 561 loci.
Score calculations
Tags with up to 500 matches in the genome were attributed a weight corresponding to the number of times they were sequenced divided by the number of matches in the genome. For tags sequenced multiple times, we established a cutoff at 50, i.e., tags sequenced more than 50 times were counted as 50. This allowed us to include >99% of the data. As shown in Supplemental Figure S8, the correlation between scores calculated with all redundant tags counted up to 50 times and all redundant tags counted only once was very high. Scores were calculated as log2[(immunoprecipitation tag counts + 30)/(input tag counts + 30)] over regions encompassing the RNA coding sequence as well as 150 bp upstream of and 150 bp downstream from the RNA coding region. When two Pol III–occupied loci were closer to each other than 300 bp, the region separating the coding regions was divided into two equal parts, and each half was attributed to the closest gene for score determination.
Cutoffs
The cutoffs were calculated on the data scaled to a total amount of 25 million tags for the IMR90 cell data and 150 million tags for the mouse liver and Hepa 1-6 cell data. For each experiment, the cutoff for genes considered as not occupied was calculated as follows: (1) The whole genome was split into 400-bp (the mean size of regions used to calculate Pol III scores) bins; (2) the scores of these regions were calculated as described above; (3) the mean and standard deviation of these scores were calculated, and each region was attributed a P-value by applying the mean and standard deviation to a normal distribution and comparing it with the real data distribution; (4) the P-value was adjusted with the Benjamini-Hochberg method to obtain the FDR; and (5) the cutoff chosen was the lowest score giving a FDR of 0.001 on the whole genome.
Data access
The ChIP-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE47849.
Acknowledgments
We thank Pascal Cousin for help with the gel filtration column. We thank Donatella Canella and Nicolas Bonhoure for help with chromatin preparations. We thank Henrik Kaessmann and Diego Cortez for help with the evolution and phylogeny analysis. We thank Keith Harshman, director of the Lausanne Genomic Technologies Facility, where all the high-throughput sequencing was performed. M.R. thanks Ioannis Xenarios for discussion and support. This work was funded by the University of Lausanne and by SNSF grant 31003A_132958 to N.H.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.161570.113.
Freely available online through the Genome Research Open Access option.
References
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K 2010. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol 17: 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun I, Sentenac A, Werner M 1997. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J 16: 5730–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canella D, Praz V, Reina JH, Cousin P, Hernandez N 2010. Defining the RNA polymerase III transcriptome: Genome-wide localization of the RNA polymerase III transcription machinery in human cells. Genome Res 20: 710–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canella D, Bernasconi D, Gilardi F, LeMartelot G, Migliavacca E, Praz V, Cousin P, Delorenzi M, Hernandez N 2012. A multiplicity of factors contributes to selective RNA polymerase III occupancy of a subset of RNA polymerase III genes in mouse liver. Genome Res 22: 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carriere L, Graziani S, Alibert O, Ghavi-Helm Y, Boussouar F, Humbertclaude H, Jounier S, Aude JC, Keime C, Murvai J, et al. 2012. Genomic binding of Pol III transcription machinery and relationship with TFIIS transcription factor distribution in mouse embryonic stem cells. Nucleic Acids Res 40: 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R, Drouin G 2010. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol Biol Evol 27: 1035–1043 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhang YE, Long M 2010. New genes in Drosophila quickly become essential. Science 330: 1682–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SS, Hu P, Hernandez N 2001. Reconstitution of transcription from the human U6 small nuclear RNA promoter with eight recombinant polypeptides and a partially purified RNA polymerase III complex. J Biol Chem 276: 20727–20734 [DOI] [PubMed] [Google Scholar]
- Cramer P, Armache KJ, Baumli S, Benkert S, Brueckner F, Buchen C, Damsma GE, Dengl S, Geiger SR, Jasiak AJ, et al. 2008. Structure of eukaryotic RNA polymerases. Annu Rev Biophys 37: 337–352 [DOI] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S 2010. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 10: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV 2012. MYC on the path to cancer. Cell 149: 22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers M, Eisenman RN 2008. Myc's broad reach. Genes Dev 22: 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J, McCormack AL 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Amer Mass Spectrom 5: 976–989 [DOI] [PubMed] [Google Scholar]
- Enver T, Soneji S, Joshi C, Brown J, Iborra F, Orntoft T, Thykjaer T, Maltby E, Smith K, Abu Dawud R, et al. 2005. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum Mol Genet 14: 3129–3140 [DOI] [PubMed] [Google Scholar]
- Florens L, Washburn MP 2006. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol 328: 159–175 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA 2001. The RNA polymerase III transcription apparatus. J Mol Biol 310: 1–26 [DOI] [PubMed] [Google Scholar]
- Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P 2010. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol Cell 39: 583–594 [DOI] [PubMed] [Google Scholar]
- Gogolevskaya IK, Kramerov DA 2010. 4.5SI RNA genes and the role of their 5′-flanking sequences in the gene transcription. Gene 451: 32–37 [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Haag JR, Pikaard CS 2011. Multisubunit RNA polymerases IV and V: Purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12: 483–492 [DOI] [PubMed] [Google Scholar]
- Haurie V, Durrieu-Gaillard S, Dumay-Odelot H, Da Silva D, Rey C, Prochazkova M, Roeder RG, Besser D, Teichmann M 2010. Two isoforms of human RNA polymerase III with specific functions in cell growth and transformation. Proc Natl Acad Sci 107: 4176–4181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Wu S, Sun Y, Yuan CC, Kobayashi R, Myers MP, Hernandez N 2002. Characterization of human RNA polymerase III identifies orthologues for Saccharomyces cerevisiae RNA polymerase III subunits. Mol Cell Biol 22: 8044–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iseli C, Ambrosini G, Bucher P, Jongeneel CV 2007. Indexing strategies for rapid searches of short words in genome sequences. PLoS ONE 2: e579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N 2012. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet 8: e1003028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawdekar GW, Henry RW 2008. Transcriptional regulation of human small nuclear RNA genes. Biochim Biophys Acta 1779: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Dubeau L, Johnson DL 2008. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem 283: 19184–19191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M 2007. The 2R hypothesis: An update. Curr Opin Immunol 19: 547–552 [DOI] [PubMed] [Google Scholar]
- Kenneth NS, Marshall L, White RJ 2008. Recruitment of RNA polymerase III in vivo. Nucleic Acids Res 36: 3757–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn CD, Geiger SR, Baumli S, Gartmann M, Gerber J, Jennebach S, Mielke T, Tschochner H, Beckmann R, Cramer P 2007. Functional architecture of RNA polymerase I. Cell 131: 1260–1272 [DOI] [PubMed] [Google Scholar]
- Kutter C, Brown GD, Goncalves A, Wilson MD, Watt S, Brazma A, White RJ, Odom DT 2011. Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat Genet 43: 948–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre S, Dumay-Odelot H, El-Ayoubi L, Budd A, Legrand P, Pinaud N, Teichmann M, Fribourg S 2011. Structure-function analysis of hRPC62 provides insights into RNA polymerase III transcription initiation. Nat Struct Mol Biol 18: 352–358 [DOI] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA 2012. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, Betran E, Thornton K, Wang W 2003. The origin of new genes: Glimpses from the young and old. Nat Rev Genet 4: 865–875 [DOI] [PubMed] [Google Scholar]
- Makalowski W 2001. Are we polyploids? A brief history of one hypothesis. Genome Res 11: 667–670 [DOI] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J 1993. BC200 RNA: A neural RNA polymerase III product encoded by a monomeric Alu element. Proc Natl Acad Sci 90: 11563–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti JA, Brosius J 1995. BC1 RNA: Transcriptional analysis of a neural cell-specific RNA polymerase III transcript. Mol Cell Biol 15: 1642–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WH, Ohi R, Miyamoto DT, Mitchison TJ, Yates JR 2002. Comparison of three directly coupled HPLC MS/MS strategies for identification of proteins from complex mixtures: Single-dimension LC-MS/MS, 2-phase MudPIT, and 3-phase MudPIT. Int J Mass Spectrom 219: 245–251 [Google Scholar]
- Meyer N, Penn LZ 2008. Reflecting on 25 years with MYC. Nat Rev Cancer 8: 976–990 [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K 2010. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol 17: 635–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oficjalska-Pham D, Harismendy O, Smagowicz WJ, Gonzalez de Peredo A, Boguta M, Sentenac A, Lefebvre O 2006. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol Cell 22: 623–632 [DOI] [PubMed] [Google Scholar]
- O'Geen H, Nicolet CM, Blahnik K, Green R, Farnham PJ 2006. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41: 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17: 620–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince VE, Pickett FB 2002. Splitting pairs: The diverging fates of duplicated genes. Nat Rev Genet 3: 827–837 [DOI] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina JH, Azzouz TN, Hernandez N 2006. Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS ONE 1: e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U 2006. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet 38: 369–374 [DOI] [PubMed] [Google Scholar]
- Ross BD, Rosin L, Thomae AW, Hiatt MA, Vermaak D, de la Cruz AF, Imhof A, Mellone BG, Malik HS 2013. Stepwise evolution of essential centromere function in a Drosophila neogene. Science 340: 1211–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeniger LO, Jelinek WR 1986. 4.5S RNA is encoded by hundreds of tandemly linked genes, has a short half-life, and is hydrogen bonded in vivo to poly(A)-terminated RNAs in the cytoplasm of cultured mouse cells. Mol Cell Biol 6: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev 16: 2593–2620 [DOI] [PubMed] [Google Scholar]
- Schramm L, Pendergrast PS, Sun Y, Hernandez N 2000. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev 14: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Kuraku S, Holt C, Sauka-Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genetics 45: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: Article3. [DOI] [PubMed] [Google Scholar]
- Smyth GK, ed. 2005. Limma: Linear models for microarray data. Springer, New York [Google Scholar]
- Tabb DL, McDonald WH, Yates JR 2002. DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res 1: 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillier V, Stettler S, Sentenac A, Thuriaux P, Werner M 1995. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J 14: 351–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh-Rohlik Q, Maxwell ES 1988. Homologous genes for mouse 4.5S hybRNA are found in all eukaryotes and their low molecular weight RNA transcripts intermolecularly hybridize with eukaryotic 18S ribosomal RNAs. Nucleic Acids Res 16: 6041–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM 2002. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell 10: 1489–1494 [DOI] [PubMed] [Google Scholar]
- Vannini A, Cramer P 2012. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell 45: 439–446 [DOI] [PubMed] [Google Scholar]
- Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P 2010. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143: 59–70 [DOI] [PubMed] [Google Scholar]
- Wang Z, Roeder RG 1997. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev 11: 1315–1326 [DOI] [PubMed] [Google Scholar]
- Werner F, Grohmann D 2011. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol 9: 85–98 [DOI] [PubMed] [Google Scholar]
- Werner M, Hermann-Le Denmat S, Treich I, Sentenac A, Thuriaux P 1992. Effect of mutations in a zinc-binding domain of yeast RNA polymerase C (III) on enzyme function and subunit association. Mol Cell Biol 12: 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M, Chaussivert N, Willis IM, Sentenac A 1993. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem 268: 20721–20724 [PubMed] [Google Scholar]
- White RJ 2005. RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol 6: 69–78 [DOI] [PubMed] [Google Scholar]
- Wong RC, Pollan S, Fong H, Ibrahim A, Smith EL, Ho M, Laslett AL, Donovan PJ 2011. A novel role for an RNA polymerase III subunit POLR3G in regulating pluripotency in human embryonic stem cells. Stem Cells 29: 1517–1527 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wen ZH, Washburn MP, Florens L 2010. Refinements to label free proteome quantitation: How to deal with peptides shared by multiple proteins. Anal Chem 82: 2272–2281 [DOI] [PubMed] [Google Scholar]