Abstract
Evidence from a few genes in diverse species suggests that X-chromosome inactivation (XCI) in marsupials is characterized by exclusive, but leaky inactivation of the paternally derived X chromosome. To study the phenomenon of marsupial XCI more comprehensively, we profiled parent-of-origin allele-specific expression, DNA methylation, and histone modifications in fetal brain and extra-embryonic membranes in the gray, short-tailed opossum (Monodelphis domestica). The majority of X-linked genes (152 of 176 genes with trackable SNP variants) exhibited paternally imprinted expression, with nearly 100% of transcripts derived from the maternal allele; whereas 24 loci (14%) escaped inactivation, showing varying levels of biallelic expression. In addition to recently reported evidence of marsupial XCI regulation by the noncoding Rsx transcript, strong depletion of H3K27me3 at escaper gene loci in the present study suggests that histone state modifications also correlate strongly with opossum XCI. In contrast to mouse, the opossum did not show an association between X-linked gene expression and promoter DNA methylation, with one notable exception. Unlike all other X-linked genes examined, Rsx was differentially methylated on the maternal and paternal X chromosomes, and expression was exclusively from the inactive (paternal) X chromosome. Our study provides the first comprehensive catalog of parent-of-origin expression status for X-linked genes in a marsupial and sheds light on the regulation and evolution of imprinted XCI in mammals.
X-chromosome inactivation (XCI) is a chromosome-wide phenomenon, unique to therian mammals (mammals, exclusive of egg-laying prototherians), whereby most genes on one of the two X chromosomes of female embryos are rendered transcriptionally silent, and remain so in descendant somatic cells throughout life (Straub and Becker 2007; Payer and Lee 2008). XCI occurs in two distinct patterns in eutherian mammals: random XCI (rXCI) and paternally imprinted XCI (pXCI). In rXCI, the choice of X chromosome to be inactivated in any given cell is more or less random with regard to the parental source (Heard et al. 1997). In comparison, pXCI is decidedly nonrandom, in that the Xp (paternal X) is preferentially (or exclusively) inactivated, and the Xm (maternal X) remains active. These two forms of XCI can occur within a single species. In mouse, for example, rXCI occurs in epiblast cells, which develop from the inner cell mass of the embryo (Latham 2005; Okamoto and Heard 2006), whereas cells of the trophectoderm layer, which give rise to extra-embryonic structures including the placenta, display pXCI (Huynh and Lee 2001, 2005; Heard and Disteche 2006). pXCI has also been observed in placental tissues of rat (Wake et al. 1976) and cow (Xue et al. 2002; Dindot et al. 2004), but not in those of human, rabbit, horse, and mule, which exhibit rXCI in both embryonic and extra-embryonic cells (Moreira de Mello et al. 2010; Okamoto et al. 2011; Wang et al. 2012). Thus, in contrast with the highly uniform rXCI pattern in the eutherian soma, XCI patterns in trophectoderm-derived tissues are variable and lineage specific.
In contrast to the eutherian pattern, data from several species of metatherians (marsupials) indicate that females exhibit pXCI in embryonic, fetal, and adult somatic cells (for review, see Deakin et al. 2009). In addition, genes on the marsupial inactive Xp exhibit levels of “leaky” or “partial” expression (incomplete repression) that can vary across species, as well as across tissue types, developmental stages, and cultured cells within a species (for review, see VandeBerg et al. 1987; Cooper et al. 1990, 1993; see also Samollow et al. 1995; Hornecker et al. 2007). However, only four marsupial X-linked genes have been examined with regard to parent-of-origin allelic and leaky expression (Cooper et al. 1993), and simultaneous examination of expression from multiple X-linked loci has been reported for only one species (Samollow et al. 1987; Migeon et al. 1989). These limited data have not enabled many locus-by-locus comparisons within or among species, nor allowed extrapolation of the expression patterns of so few genes to the full X chromosome for any individual species. Thus, it remains unclear whether pXCI in marsupials is a concerted, chromosome-wide phenomenon or a piecemeal process that occurs on a region-by-region basis (Cooper et al. 1990; Riggs 1990; Al Nadaf et al. 2010).
Most genes on the inactive eutherian X chromosome (Xi) are strongly transcriptionally repressed, but ∼15% of human X-linked genes located outside the pseudo-autosomal region (PAR) are expressed at nontrivial (although not necessarily equal) levels from both alleles; i.e., they escape XCI (Disteche et al. 2002; Carrel and Willard 2005). In mouse, the proportion of such escaper genes is ∼3% in cultured cells derived from kidney (Yang et al. 2010), but in trophoblast stem cells, which exhibit pXCI, as many as 14% of X-linked genes are escapers, depending on the criteria used to classify relative allelic expression ratios (Calabrese et al. 2012). Judged from the limited information available, it appears that XCI escaper genes are common in marsupials, but the data are too sparse to enable estimation of the proportion of all Xp genes that escape inactivation or to discriminate species-specific differences in leaky expression from tissue- and developmental-stage-specific variation.
Information concerning molecular mechanisms of marsupial pXCI is also rudimentary. To date, single-gene bisulfite sequencing of CpG islands around selected X-linked genes has identified no differentially methylated regions (DMRs) (Kaslow and Migeon 1987; Loebel and Johnston 1996; Hornecker et al. 2007), suggesting that DNA methylation plays little or no role in marsupial XCI. Chromosome-level immunofluorescence staining studies have shown distinctive epigenetic profiles for the Xa (active X) and Xi in two distantly related marsupials, the gray, short-tailed opossum, Monodelphis domestica (Mahadevaiah et al. 2009; Chaumeil et al. 2011; Grant et al. 2012), and the brushtail possum, Trichosurus vulpecula (Rens et al. 2010); however, the relatively low resolution of these approaches makes it impossible to distinguish the locations of modified histones beyond the chromosome-band level, which precludes recognition of correlations among individual X-linked gene-expression levels and histone modification states. In an exciting development, an X-linked noncoding transcript gene that is expressed exclusively in female somatic cells was recently discovered in M. domestica. This gene, Rsx, appears to be the functional analog of the eutherian Xist gene, insofar that its transcript coats one of the X chromosomes (presumed Xp) and is suggested to act in cis to repress activity of its genes (Grant et al. 2012). Despite their apparent analogous functions, Rsx and Xist show no obvious sequence homology and might have arisen independently in marsupials and eutherians. Epigenetic characteristics of the active and inactive Rsx alleles have not been described.
To investigate some of the unknowns in marsupial XCI rigorously, we examined gene expression, DNA methylation, and histone modification data in fetal brain and extra-embryonic membranes (EEM) of offspring from two opossum laboratory stocks and their reciprocal crosses. The results of this study comprise the first comprehensive catalog of X-linked gene expression from the active Xm and inactive Xp in a marsupial species and the first chromosome-wide assessment of potential epigenetic mechanisms of pXCI in a marsupial at the individual gene level.
Results
Transcriptome-wide quantification of parent-of-origin allele-specific expression in opossum fetal brain and EEM
Relative expression levels of genes from the Xm and Xp of female opossums were determined by Illumina RNA-seq analyses of embryonic day 13 (E13, day 12–13 post-coitus) fetal brain and paired extra-embryonic membranes (EEM, counterpart of eutherian placenta) from reciprocal F1 hybrid and parental crosses of LL1 and LL2 stocks (Supplemental Figs. S1–S3; Supplemental Table S1). Opossum fetuses are born on E14 and approximate E11–E12 mouse fetuses in overall development (Supplemental Fig. S1; Supplemental Text S1). In total, we obtained 76.5 billion base pairs (bp) of sequences, >80% of which were uniquely mapped to the opossum reference genome (MonDom5) (Supplemental Table S2). Reads overlapped more than 10,000 expressed genes with FPKM (see Methods) ≥1 in both tissues, and 68,000 exonic SNPs were called from all samples combined. Parent-of-origin allelic expression ratios were quantified in each sample from the relative numbers of reads containing the reference and alternative alleles at high-quality SNP positions (Supplemental Methods).
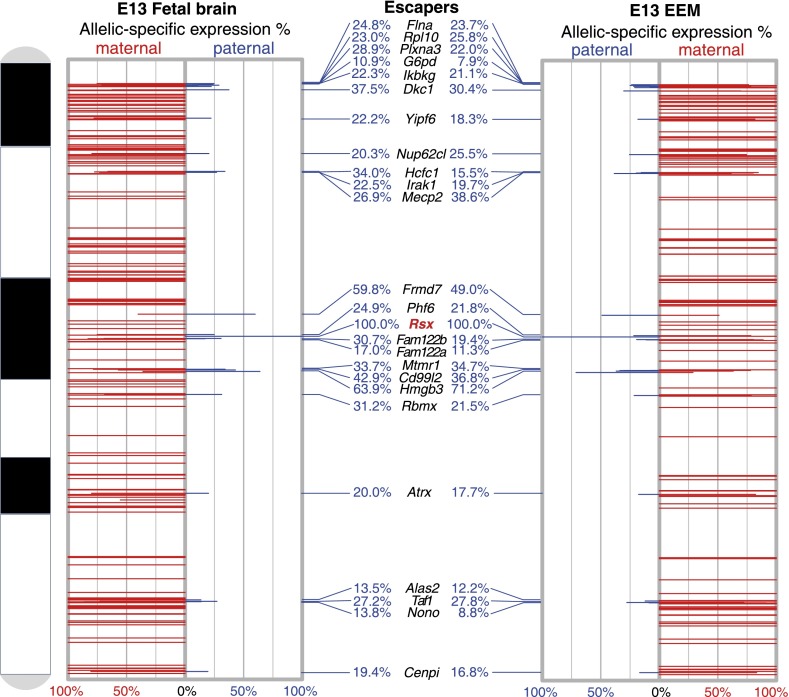
Three hundred and twelve (312) X-linked gene models were covered in female fetal brain samples with sufficient expression levels to call de novo SNPs (Supplemental Data S1). Nineteen (19) of these genes mapped to unanchored MonDom5 scaffolds (Supplemental Tables S3, S4) that exhibit extensive conserved synteny with the human X chromosome in the region corresponding to the conserved region of the eutherian X chromosome (XCR). Based on the strong conservation of gene content between eutherian and marsupial X chromosomes in this region (Deakin et al. 2008 and references therein), we presume these genes to be X-linked in the opossum as well. Of the 312 genes, 176 had a total of 512 informative heterozygous SNPs that could be used to quantify expression of the Xm and Xp alleles. Twenty-four (24) of these exhibited >10% paternal-allele expression and are considered escaper genes. These genes and the percentage of total transcripts derived from the Xp allele are shown in Figure 1.
Figure 1.
Chromosomal locations and allelic expression profiles of opossum X-linked genes in fetal brain and EEM. The x-axis shows allelic expression percentages: maternal expression on the left, and paternal expression on the right. The y-axis is the physical location of each gene along the opossum X chromosome (centromere at top). (Red bars) Maternal expression percentage; (blue bars) paternal expression percentage. The names of 24 genes that escape pXCI with >10% paternal expression (escaper genes) are indicated in the middle panel. The noncoding Rsx transcript gene (bold, red type) shows 100% expression from the paternal allele in both tissues.
The remaining 152 genes (non-escaper genes) were subject to virtually complete pXCI, with 100% or nearly 100% maternal-allele expression. Only three (2%) non-escaper genes showed detectable weak paternal expression (>3% paternal expression; >2 paternal allele counts in at least two informative individuals): Msn (5.2% paternal expression), Bcap31 (6.2%), and Pdzd11 (3.3%). In the EEM samples, 134 of 242 covered genes had informative SNPs, and the same 24 genes escaped pXCI with >10% paternal leakage (Fig. 1). Four (2%) of the 120 non-escaper genes in EEM samples displayed detectable weak paternal expression: Taz (5.0%), Fam3a (4.5%), Bcap31 (5.7%), and Timm8a (5.4%) (Supplemental Data S1).
To confirm XCI status independently, SNPs in escaper genes were also validated in multiple samples using Sanger sequencing and allele-specific pyrosequencing (Supplemental Table S5; Supplemental Data S2; Methods). This resulted in a 100% validation rate of the RNA-seq expression data (Fig. 2A–D; Supplemental Figs. S4–S28). Thus, it appears that most opossum X-linked genes show 100% monoallelic maternal expression in both tissues, and escaper genes are conserved between fetal brain and EEM.
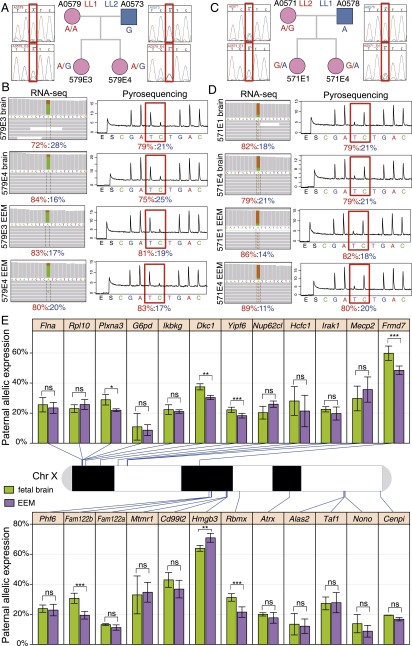
Figure 2.
Maternal versus paternal allelic expression ratios for 24 pXCI escaper genes in opossum female fetal brain and EEM. (A–D) SNP genotyping and pyrosequencing verification for example escaper gene Yipf6 in opossum fetal brain and EEM. Sanger sequencing genotypes confirmed that exonic SNP OMSNP0155110 was informative in all four F1 embryos: (A) A0579E3 and A0579E4 in LL1 (dam) × LL2 (sire) cross; (B) A0571E1 and A0571E4 in the reciprocal F1 cross LL2 (dam) × LL1 (sire). (C,D) Biallelic expression verified by RNA-seq (left) and allele-specific pyrosequencing strategies (right). The results for all 24 escaper genes, a non-escaper gene, and an autosomal control gene can be found in Supplemental Figures S4–S28. (E) Names, physical locations, and allelic expression percentages for the 24 escaper genes identified from RNA-seq on the opossum X chromosome (centromere to the left). For each gene, the mean and standard deviation of paternal-allele expression, as a percentage of total expression, is plotted for fetal brain (green) and EEM (purple). Statistical significance, indicated by asterisks, was assessed by a Mann–Whitney U-test; (ns) P-value > 0.05; (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001.
pXCI profile and comparison between brain and EEM
Approximately the same proportion of X-linked genes showed complete paternal allele repression in fetal brain (86%) and EEM (82%), and the same set of 24 escaper genes is shared between the two tissues (Fig. 1). The maternal/paternal allele-specific expression (M/P) ratio for escapers was considerably greater than 1.0 for most (22/24) of these genes, indicating that the maternal allele was the one preferentially expressed. However, M/P was <1.0 for two genes, Frmd7 and Hmgb3 (Fig. 2E). There was no significant difference in the level of paternal leakage between the two tissues for 17 of the escaper genes (Fig. 2E). However, six (Plxna3, Dkc1, Yipf6, Frmd7, Fam122b, and Rbmx) had significantly greater paternal expression in fetal brain than in EEM, and one (Hmgb3) showed significantly higher paternal expression in EEM than in brain (Mann-Whitney U-test, P-value < 0.05).
Relationship between escape from pXCI and total gene expression in females and males
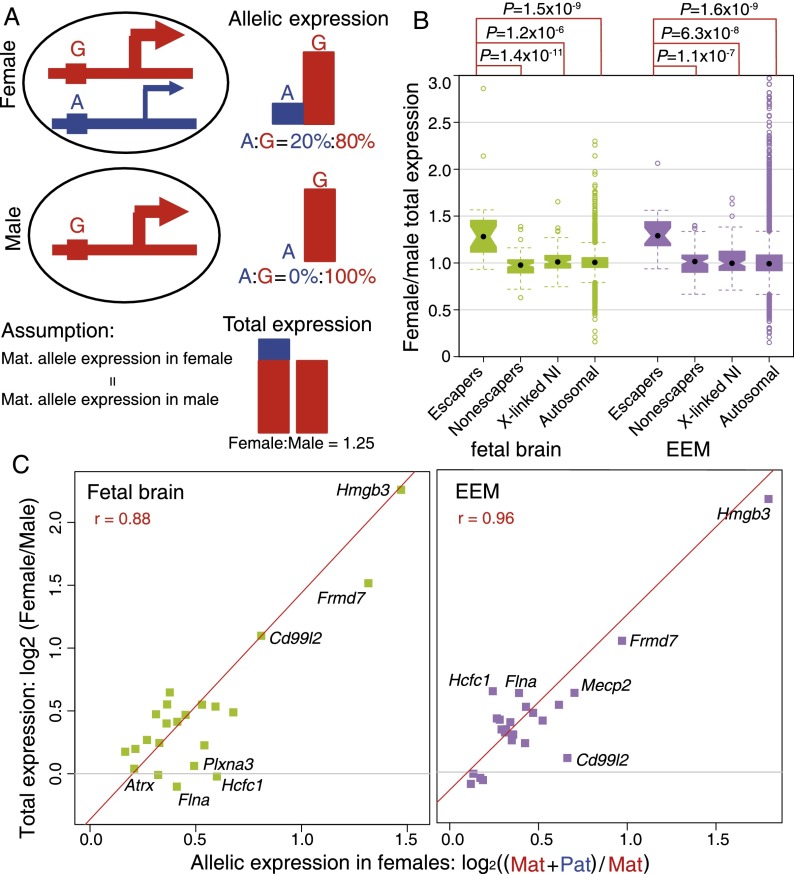
A primary effect of XCI in therian mammals is to “compensate” for the disparity in X-linked gene dosage between XX females and XY males. For opossum genes subject to complete pXCI (non-escaper genes), total expression levels are expected to be similar in females and males because only the maternal copy is expressed in each sex. Contrastingly, if expression from the maternal allele is the same in both sexes, without sexual dimorphism or feedback compensation mechanisms, escaper genes are expected to show higher total expression in females than in males, and the female/male (F/M) expression ratio should correlate with the degree of paternal leakage (Fig. 3A). To determine if this pattern holds for opossum escaper genes, we compared F/M expression ratios for escaper genes, non-escaper genes, noninformative X-linked genes (of which a small minority could be unidentified escaper genes that would slightly inflate the F/M expression ratio), and autosomal genes in fetal brain and EEM samples (Fig. 3B). As expected, the median F/M ratio is much higher (1.3) for escaper genes than for the other three groups of genes in both tissues (∼1.0), and the distribution for escaper genes is significantly different from those of the other three groups (P-value < 0.001, Kolmogorov-Smirnov test). In addition, a linear relationship was observed between increased female expression and the degree of paternal leakage in both fetal brain (Pearson's r = 0.88, P-value = 4.4 × 10−8) (Fig. 3C) and EEM (Pearson's r = 0.96, P-value = 1.5 × 10−13) (Fig. 3C). From these results, we conclude that opossum XCI escaper genes are expressed at significantly higher levels in females than in males and that this is the result of leaky expression of alleles from the “inactive” Xp.
Figure 3.
Correlation of paternal escaping percentages in females and female/male (F/M) total expression ratios. (A) Model for a hypothetical escaper gene showing individual allelic and total expression in female and male samples. (Red) The maternally derived (G) allele; (blue) the paternally derived (A) allele. In females (top panel), escaper genes have a fully active maternal allele, and the paternal allele accounts for 20% of the total expression. In males (middle panel), the single (maternal) allele accounts for 100% of expression. Assuming that maternally derived allele expression is the same in females and males, the estimated female/male total expression ratio would be (80% + 20%)/80% = 1.25 (bottom panel). For non-escaper genes, the paternal allele is repressed in females, so the F/M total expression ratio is 1. (B) Boxplot of female/male (F/M) total expression ratios for imprinted X-inactivation escaper genes, non-escaper genes, X-linked genes with no informative SNPs (X-linked NI), and autosomal genes in fetal brain (green) and EEM (purple). The median F/M total expression ratio is 1.3 for escaper genes and 1.0 for the other three groups of genes in both tissues. (C) Scatterplots of allele-specific expression percentages and female/male total expression ratios for escaper genes in fetal brain and EEM with normalized FPKM > 5. The y-axis is the female/male total expression ratio (log2 scale); the x-axis is (maternal + paternal)/maternal allele expression in females (log2 scale). A significant linear relationship is observed for escaper genes in both fetal brain (green squares) and EEM (purple squares) (see text).
Histone modification profiling by ChIP-seq
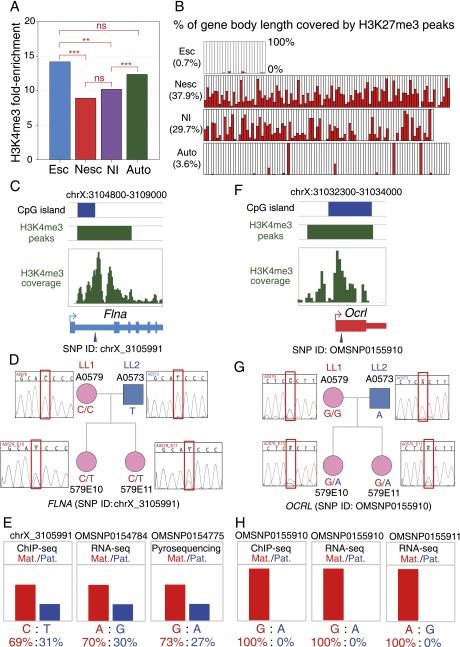
We performed native ChIP-seq experiments on fetal brain chromatin for two critical epigenetic marks that correlate with X-linked gene expression in eutherian mammals: the “on-mark” H3K4me3 (tri-methylation of lysine 4 of histone H3) and the “off-mark” H3K27me3 (tri-methylation of lysine 27 of histone H3) (Supplemental Methods). H3K4me3 is found at promoter regions and CpG islands and is strongly associated with active transcription in eukaryotes. H3K27me3 is most often associated with repressed transcription and is distributed throughout the gene body. We reasoned that if histone modifications are correlated with transcriptional states and potentially play an important role in the regulation of X-linked non-escaper genes in opossum, the promoters of the active maternal alleles should be enriched for H3K4me3, while repressed paternal alleles at the same loci should be enriched for H3K27me3 across the promoter and gene body (Supplemental Fig. S29A). For X-linked escaper genes (and biallelically expressed autosomal genes as well), the on-mark should be present and the off-mark absent, resulting in depletion of H3K27me3 marks compared with non-escaper genes (Supplemental Fig. S29B).
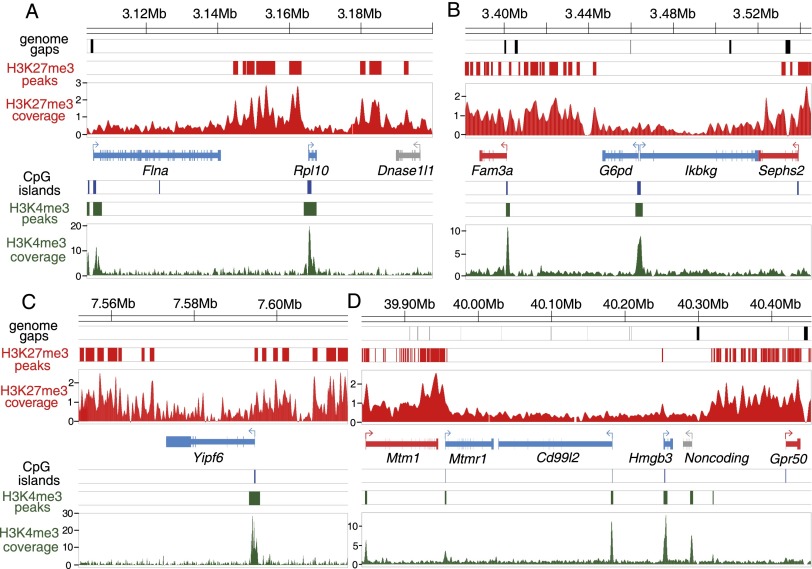
Female fetal brain chromatin showed significant H3K4me3 peaks overlapping promoter CpG islands of 23 of the 24 escaper genes. As expected, the 23 escaper genes with an on-mark peak were depleted of H3K27me3 peaks, beginning at the promoter and spanning the gene body, whereas both H3K4me3 and H3K27me3 marks were present at flanking non-escaper genes (Fig. 4A–D; Supplemental Figs. S30–S45). In control male fibroblast cell lines, the H3K4me3 mark was present and H3K27me3 marks were absent for all expressed X-linked genes (Supplemental Figs. S30–S45; Supplemental Data S3), which is consistent with the presence of a single, necessarily active, X chromosome in male cells. The remarkable association between escape of pXCI and depletion of H3K27me3 suggests that the H3K27me3 modification could be critical for the repression of genes on the opossum paternal X chromosome.
Figure 4.
Depletion of H3K27me3 marks at opossum XCI escaper genes. (A–D) H3K4me3 and H3K27me3 histone modification profiles at four X-chromosomal regions in female fetal brain samples from ChIP-seq experiments. The top three diagrams in each panel indicate the locations of genome assembly gaps (black bars), significant H3K27me3 peaks (red bars), and H3K27me3 coverage (red scans), respectively. Gene models are shown in the middle panel, color-coded according to pXCI status: (blue) escaper; (red) non-escaper; (gray) unknown due to lack of informative SNPs. The bottom three diagrams of each panel show CpG island locations (blue bars), significant H3K4me3 peaks (green bars), and H3K4me3 coverage profile (green scans). Numbers on y-axes indicate per base read depth using the spline smoothing method. For the escaper genes (Flna, Rpl10, G6pd, Ikbkg, Yipf6, Mtmr1, Cd99l2, and Hmgb3), the H3K4me3 mark is present at the promoter CpG island, while H3K27me3 marks are depleted across the gene body, consistent with the biallelic expression from both parental alleles. This pattern was seen for all 23 escaper genes with an H3K4me3 peak at the promoter region (Supplemental Figs. S30–S45).
To evaluate enrichment of on-marks, we quantified H3K4me3 peak intensity as “fold-enrichment” relative to background level (Supplemental Data S3). Enrichment was significantly higher for escaper genes than for non-escaper genes (P-value = 4.6 × 10−5, Kolmogorov-Smirnov test) and noninformative X-linked genes (P-value = 0.004) (Fig. 5A), but there was no significant difference between escaper genes and autosomal genes (P-value > 0.05). This finding is consistent with the hypothesis that biallelically expressed genes have stronger on-mark peaks due to the presence of the on-mark on both parental chromosomes (Supplemental Fig. S29A). For the off-mark, H3K27me3, average percentages of gene body length covered by significant peaks were calculated for escaper genes, non-escaper genes, noninformative X-linked genes, and 100 randomly selected autosomal genes. H3K27me3 peaks were completely absent on escaper genes, and most autosomal genes lacked off-mark peaks, although a small subset of non-expressed autosomal genes had 100% off-mark coverage (Fig. 5B). In contrast, most non-escaper and noninformative X-linked genes were extensively covered by off-mark peaks (Fig. 5B).
Figure 5.
Histone modifications are correlated with maternal- versus paternal-allele expression of X-linked escaper and non-escaper genes. (A) H3K4me3 peak fold-enrichment for pXCI escaper genes (Esc), non-escaper genes (Nesc), X-linked genes with no informative SNPs (X-linked NI), and autosomal genes (Auto) in female fetal brain samples. Statistical significance was assessed by a Kolmogorov-Smirnov test: (ns) P-value > 0.05; (**) P-value < 0.01; (***) P-value < 0.001. (B) Barplot of percent H3K27me3 peak coverage in the gene region for four classes of X-linked genes in female fetal brain samples. Gene class abbreviations are as in panel A. Numbers in parentheses are average percent significant H3K27me3 peak coverage of all genes in the corresponding class. Among the X-linked NI genes, 11 had histone modification profiles similar to escaper genes and are designated as potential escapers of pXCI in opossum (Supplemental Table S7; Supplemental Text S3). (C–E) Allele-specific H3K4me3 modification for escaper gene Flna in female fetal brain ChIP-seq data from LL1 × LL2 cross. (C, top to bottom) CpG island location (blue bar), H3K4me3 peak/coverage profile (green bar and scan), 5′-end gene model. (Blue arrowhead) The position of a SNP (chrX_3105991) under the H3K4me3 peak with sufficient coverage to infer parent-of-origin allele-specific histone modification. (D) Sanger sequencing genotyping results for fetuses A0579E10 and A0579E11 (used for ChIP-seq experiments) and their parents confirmed that the SNP was informative in both F1 offspring. (E) Skewed H3K4me3 ChIP-seq and RNA-seq reads from the maternal allele and paternal allele at chrX_3105991 suggest activity of both alleles. The histone profile is consistent with the allele-specific expression profile at SNP OMSNP0154784 in the RNA-seq data and SNP OMSNP0154775 from the allele-specific pyrosequencing results. (F–H) Allele-specific H3K4me3 modification for non-escaper gene Ocrl in female fetal brain ChIP-seq data from LL1 × LL2 cross. (F) Features and color codes as for panel E. (Blue arrowhead) Position of SNP (OMSNP0155910) under the H3K4me3 peak with sufficient coverage to infer individual allele histone modifications. (G) Sanger sequencing genotyping results in the two embryos (A0579E10 and A0579E11) and their parents confirmed that the SNP was informative in both fetuses. (H) Unimodal H3K4me3 reads from the maternal allele at OMSNP0155910 suggesting that the on-mark is present only at the maternal allele. This is consistent with the maternal-specific expression at OMSNP0155910 and OMSNP0155911 in the RNA-seq data.
To determine whether parent-of-origin allele-specific histone modification profiles were consistent with expression patterns, we examined three escaper genes (Flna, Yipf6, and Fam122b) and five non-escaper genes (Ocrl, Pnck, Gpc4, Itm2a, and Pdzd11) that had informative SNPs overlapping the H3K4me3 peaks. ChIP-seq reads for all three escaper genes contained both SNP alleles, indicating that the on-mark was present on both parental X chromosomes (Fig. 5C–E; Supplemental Figs. S46, S47). For the non-escaper genes, only maternal-allele-containing reads were found at the SNP positions, consistent with a close association between allele-specific epigenetic modification and monoallelic maternal expression (Fig. 5F–H; Supplemental Figs. S48–S51). From the overall and allele-specific histone modification profiles of escaper and non-escaper genes in fetal brain, we conclude that histone modifications are an important epigenetic feature of pXCI in opossum that could be involved directly in pXCI regulation.
Absence of promoter CpG island DNA methylation at opossum X-linked genes
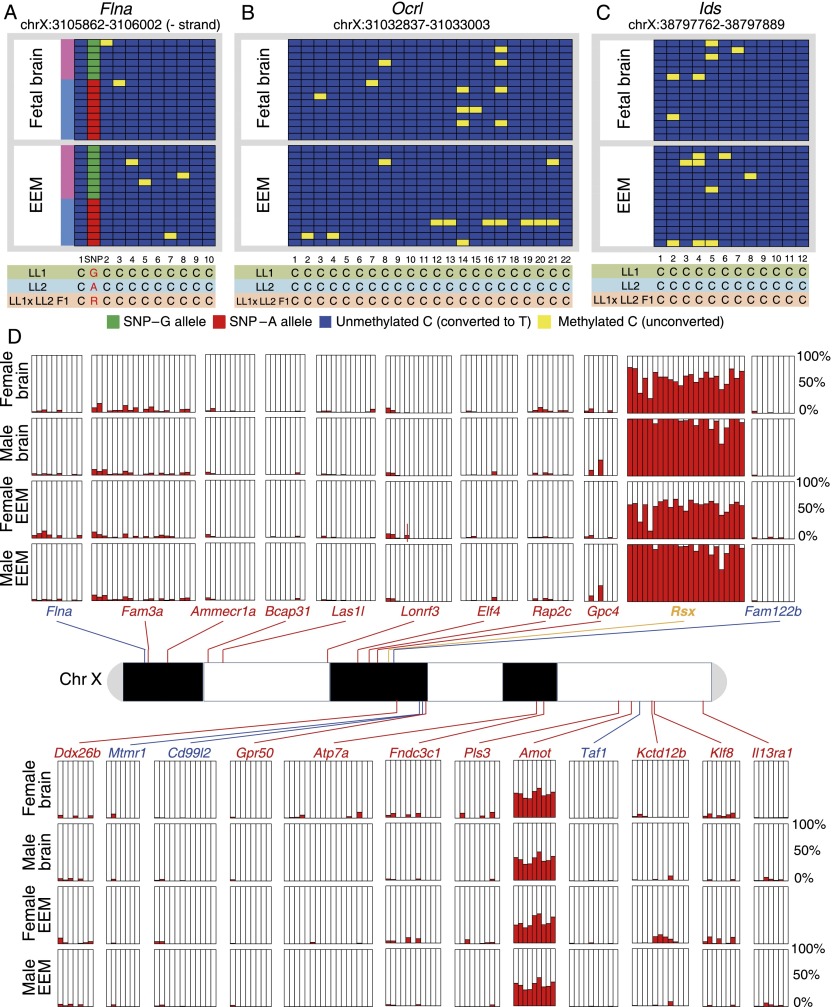
Differential DNA methylation is believed to be an important element in the maintenance of the inactive state of genes on the inactive X chromosome in eutherian females, wherein promoter CpG islands are methylated exclusively on the inactive allele and absent on the active one (Lock et al. 1986; Escamilla-Del-Arenal et al. 2011). If DNA methylation plays the same role in opossum XCI, differential methylation should be present at non-escaper genes, while little or no methylation would be expected for escaper genes, consistent with their biallelic expression. Lack of methylation would also be expected at promoters of all expressed X-linked genes in males, which have only one X chromosome. We examined this hypothesis by quantifying DNA methylation percentages at promoter CpG islands for 24 X-linked genes using bisulfite sequencing and PyroMark assays. Promoters of all five escaper genes assayed (Flna, Fam122b, Mtmr1, Cd99l2, and Taf1) showed <1% average methylation across all CpG sites in the promoter regions in female samples, which is consistent with biallelic expression (Fig. 6A; Supplemental Table S6; Supplemental Data S4). Surprisingly, 18 of the 19 non-escaper genes assayed at promoter CpG islands were also essentially unmethylated in both fetal brain and EEM tissues of both sexes, showing an overall average methylation percentage of 1.6% (Fig. 6B–D; Supplemental Table S6; Supplemental Data S4). The remaining gene (Amot) exhibited ∼40% methylation across promoter CpG sites, but there was no difference between the sexes (Fig. 6D). Thus, promoter CpG islands are unmethylated for both escaper and non-escaper genes, and there is no correlation between promoter CpG DNA methylation and X-inactivation status in opossum.
Figure 6.
Promoter CpG methylation profiles at X-linked escaper and non-escaper genes. (A–C) Bisulfite sequencing of the promoter CpG islands for an escaper gene Flna (A) and two non-escaper genes, Ocrl (B) and Ids (C), in opossum female fetal brain and EEM showing the absence of promoter methylation in these tissues. Each panel depicts replicate alignments of multiple CpG sites at the promoter for the corresponding gene/tissue. (Yellow boxes) Methylated CpGs; (blue boxes) unmethylated CpGs. A SNP in the Flna promoter (red and green boxes) enabled transmission direction and methylation on each allele to be inferred; the maternal allele is G (green boxes), and the paternal allele is A (red boxes). (D) Profile of promoter CpG island methylation status in five escaper genes (labeled in blue), 17 non-escaper genes (in red), and Rsx (in orange) using the PyroMark assay in female and male fetal brain and EEM samples. (Red bars) The methylation percentage (0%–100%) at one promoter CpG site in the corresponding CpG island.
Allele-specific expression and epigenetic profile for the noncoding Rsx gene
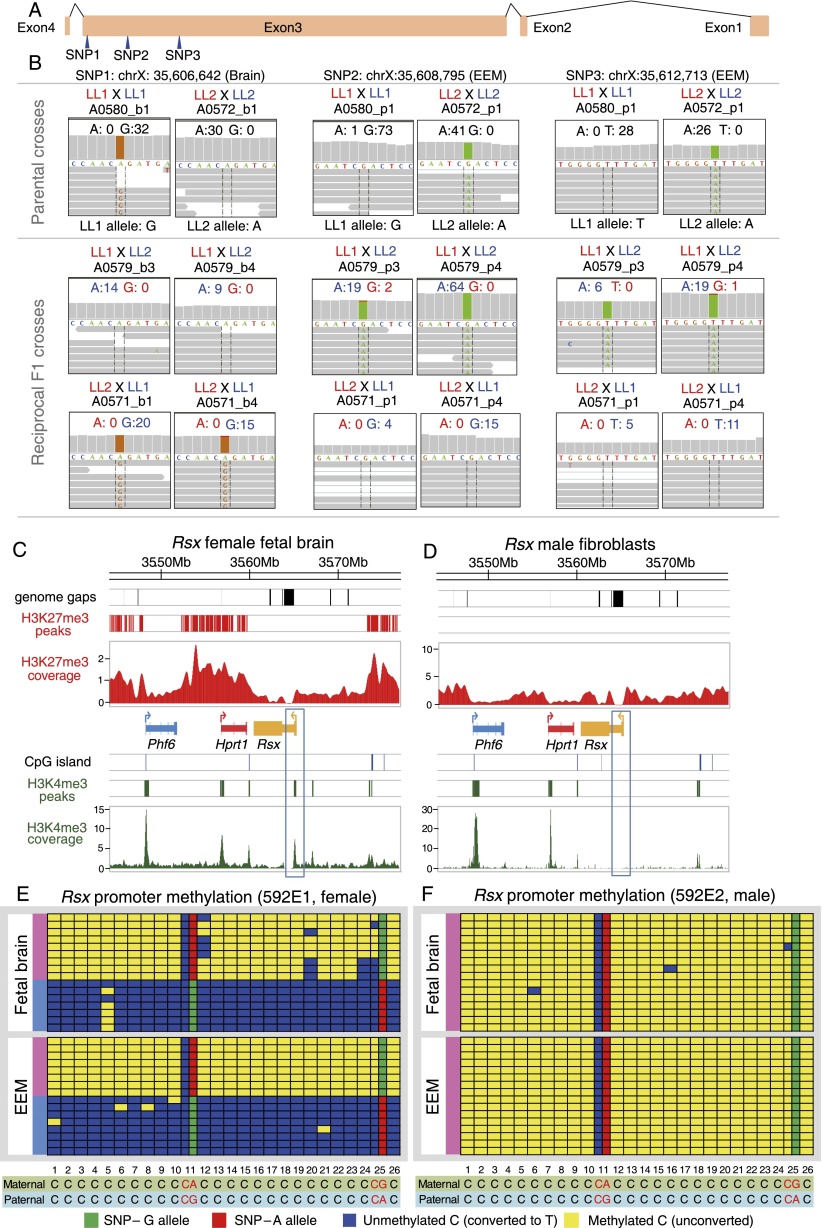
In eutherians, the X-linked noncoding Xist locus is transcribed exclusively from the Xi in females and coats it in cis to repress the expression of genes from this chromosome. The recently discovered marsupial noncoding transcript gene Rsx has functional properties reminiscent of Xist, including female-specific expression, coating of the inactive Xp by its noncoding transcripts, and the ability to silence genes in cis in transgenic mouse cells (Grant et al. 2012). We independently confirmed, in different tissues and developmental stages, that Rsx is indeed expressed exclusively in females, and we further demonstrated that it is expressed from the inactive Xp only. Using three informative SNPs in the nonrepetitive region of exon 3, we were able to directly quantify the parent-of-origin allelic expression level for Rsx in female E13 fetal brain and EEM (Fig. 7A). All reads detected in both tissues were exclusively from the paternal allele (Fig. 7B). The histone modification profile for Rsx shows the presence of the on-mark (H3K4me3) in females but not in males, which is consistent with female-specific expression (Fig. 7C,D). However, we did not observe off-mark (H3K27me3) peaks across the Rsx promoter or gene body in either sex (Fig. 7C,D). This finding contrasts with differential enrichment of this mark observed at the cytologic level (Grant et al. 2012), suggesting that accumulation of H3K27me3 occurs across the body of Rsx but not near transcription start sites.
Figure 7.
Allele-specific expression, allele-specific methylation, and histone modification profile for the noncoding RNA gene Rsx. (A) Exon model for the Rsx gene, with three SNPs between LL1 and LL2 opossum stocks indicated (blue arrowheads). (B) Allele-specific expression analysis of RNA-seq data for SNP1 in fetal brain and SNPs 2 and 3 in EEM in female offspring. Monoallelic paternal-allele expression was found at all three SNP positions in both tissues. (C,D) H3K4me3 and H3K27me3 histone modification profiles at the Rsx-containing region in female fetal brain (C) and control male fibroblast samples (D) from ChIP-seq experiments. In each panel, from top to bottom, are the genome gap locations (black bars), significant H3K27me3 peaks (red bars), H3K27me3 coverage (red scans), gene models, CpG island locations (blue bars), significant H3K4me3 peaks (green bars), and H3K4me3 coverage (green scans). (Blue) The escaper gene (Phf6) in this region; (red) the non-escaper gene (Hprt1); (orange) Rsx. (E,F) Bisulfite sequencing data for the Rsx promoter region in female (E) and male (F) fetal brain and EEM samples. The panel composition and colors are as described in Figure 6. Two SNPs found in this region (position 11 and 25) were genotyped in both parents by Sanger sequencing to infer the transmission direction and quantify methylation of the two alleles.
To assess the Rsx DNA methylation profile, we identified a previously unannotated CpG island at the proposed Rsx promoter and quantified DNA methylation percentages at all 24 CpG sites within this CpG island in fetal brain and EEM of both sexes. The CpG island was completely unmethylated in males, but unlike other X-linked genes that are subject to pXCI, it was strongly differentially methylated in females in both tissues. Maternal alleles showed nearly complete methylation, and paternal alleles showed virtually none (Fig. 7E,F; Supplemental Table S6; Supplemental Data S4), which parallels the situation at the Xist promoter in eutherian females (Beard et al. 1995). The similarity in differential methylation patterns of Rsx and Xist leads us to propose that Rsx parent-of-origin allelic expression is influenced, at least in part, by differential promoter methylation, but does not involve the accumulation of H3K27me3 that is coincident with X-linked non-escaper gene repression.
Discussion
XCI completely silences most, but not all, paternal X-linked alleles in opossum fetal brain and EEM
Paternally imprinted X-chromosome inactivation (pXCI) is characteristic of all somatic tissues of the adult, embryo proper, and EEM in marsupials. Pioneering studies of protein polymorphisms in several marsupial species (for review, see Cooper et al. 1990, 1993) indicated that inactivation of paternal X-linked genes was often incomplete, with locus-, species-, and tissue-specific levels of paternal allele leakage. However, it has remained unclear whether incomplete repression is the general pattern for marsupial X-linked genes or is a biased reflection of the small number of loci examined.
We used RNA-seq analyses of SNP-bearing transcripts to assess chromosome-wide relative expression of paternally and maternally derived genes in E13 fetal brain and EEM samples of two opossum laboratory stocks and their reciprocal F1 offspring. We found that 14% (24/176) of expressed opossum X-linked genes exhibited strong maternal allele expression accompanied by leaky expression from the paternal allele, while 86% (152/176) showed ∼100% monoallelic maternal expression. For most escapers (22/24), paternal-allele expression was considerably lower than that from the maternal allele. In addition, the X-inactivation status was remarkably similar between opossum E13 fetal brain and EEM, with exactly the same set of escaper genes in both tissues and similar paternal-allele expression for each gene (conserved escape). These results demonstrate for the first time that the large majority of genes on a marsupial X chromosome are subject to complete pXCI, with only a small minority of partial escapers in fetal brain and EEM. The implications of effective hemizygosity of X-linked genes in female opossums resulting from pXCI, and the lack of overlap of genes that escape XCI in opossums versus eutherian species, are discussed in Supplemental Text S2.
DNA methylation and histone modification profiles reveal potential regulation mechanisms of pXCI in opossum
Previous epigenetic studies of pXCI at the cytogenetic level in marsupials have found several histone modifications to be differentially associated with the active and inactive X chromosomes (Koina et al. 2009; Mahadevaiah et al. 2009; Rens et al. 2010; Chaumeil et al. 2011), suggesting possible involvement of histone modifications in marsupial XCI; however, there are fewer and less consistent results concerning the potential role of DNA methylation in this phenomenon (Kaslow and Migeon 1987; Loebel and Johnston 1993; Loebel and Johnston 1996; Rens et al. 2010).
Genome-wide ChIP-seq analyses enabled us to obtain base-pair resolution of histone modification states in opossum chromatin and showed both H3K4me3 (on-marks) and H3K27me3 (off-marks) to be strongly correlated with the activity status of X-linked genes. Depletion of H3K27me3 was observed only for escaper genes, consistent with expression from both alleles of these genes and with previous cytological studies.
Bisulfite sequencing analysis revealed that for all but one X-linked gene examined (Rsx), promoter DNA methylation was absent and, thus, not correlated with parent-of-origin differential allelic expression. This is consistent with previous single-gene analyses in marsupials (Kaslow and Migeon 1987; Loebel and Johnston 1996; Hornecker et al. 2007), but contrary to the differential methylation detected by two different methodologies at the whole-chromosome level (Loebel and Johnston 1993; Rens et al. 2010). This discrepancy suggests that while differential DNA methylation of marsupial X chromosomes can occur, it is not characteristic of the promoter regions of the majority of X-linked genes (at least in opossum), and therefore must occur in other regions of the chromosome.
The salient exception is the opossum Rsx gene. As reported for adult tissues (Grant et al. 2012), we found Rsx expression to be extremely female-biased in fetal brain and EEM and were able to demonstrate unequivocal monoallelic expression of the paternally derived Rsx allele in both tissues. Similar to X-linked escaper genes, H3K27me3 peaks were absent from the Rsx gene body, but in contrast to escaper genes, the CpG island at the Rsx promoter was differentially methylated in females, with ∼100% methylation of the maternal allele and virtually no methylation of the paternal allele. These findings suggest functional similarity in the epigenetic regulatory mechanisms of Rsx and the eutherian Xist gene despite the absence of sequence homology between these two noncoding transcripts.
The highly consistent epigenetic characteristics of XCI escaper genes in the present study suggest a strategy for identifying candidate escaper genes in the absence of SNP data, which can then be examined in animals or cells with different genetic backgrounds (Supplemental Figs. S32, S36, S42–S45; Supplemental Table S7; Supplemental Text S3).
A model for pXCI regulation in opossum
Based on our data and considerations of characteristics of imprinted XCI in mouse (Tada et al. 2000; Okamoto et al. 2005; Escamilla-Del-Arenal et al. 2011), we propose the following model for the epigenetic regulation of pXCI in opossum. At some stage of germline development or gametogenesis, the Rsx promoter is methylated in female, but not male, opossums, resulting in an imprinted Rsx on maternally derived X chromosomes. After fertilization, this imprint prevents transcription from the maternal Rsx allele, resulting in paternal-specific monoallelic expression of Rsx in the developing embryo. The Rsx transcript creates a transcriptionally unsupportive silencing domain, perhaps similar to that created by eutherian Xist (Calabrese et al. 2012; Engreitz et al. 2013) wherein most genes are transcriptionally repressed, and recruits the Polycomb group (PcG) proteins and other factors to target H3K27 for methylation on the paternally derived X chromosome. For reasons yet to be determined, escaper genes are avoided during this chromatin-remodeling process and remain active on both parental chromosomes, as is the case with eutherian escaper genes as well. In eutherian mammals, the promoter CpG islands of X-linked genes that are subject to imprinted expression in placenta are methylated on the paternal allele only, presumably to strengthen/stabilize their repression (Lock et al. 1987). In opossum, we did not observe promoter DNA methylation of X-linked genes (except Rsx) in EEM or the fetus proper, suggesting a decoupling of histone modification and promoter DNA methylation. In summary, we propose that pXCI in opossum arises from a Xist-like regulating mechanism via the noncoding Rsx gene and H3K27me3 histone modifications, but promoter DNA methylation does not play a direct role.
Presence of Y-linked homologs
No extensive sequence data from the opossum Y chromosome have been published, but Atrx and Rsp4 each have X- and Y-linked homologs in M. domestica (Jegalian and Page 1998; Carvalho-Silva et al. 2004), and 10 Y-linked homologs of X-linked genes have been detected in the tammar wallaby, Macropus eugenii (Murtagh et al. 2012). In our study, six of these marsupial X/Y homologous gene pairs (Atrx, Phf6, Hcfc1 Rbmx, Mecp2, and Rpl10) were found to have informative SNPs, and all six escaped pXCI in both fetal brain and EEM. Interestingly, four of them (Atrx, Phf6, Hcfc1, and Rbmx), had very similar levels of expression in males and females (F/M expression ratio < 1.2) in one or both tissues, unlike the other escaper genes, which had much higher expression in females. If these genes have retained their active Y-linked homologs in opossum, their expression from the Y chromosome could equalize dosage between the sexes and partially explain the lack of inactivation at these loci in females. As marsupial Y-chromosome data become more extensive, it will be important to see if any other escapers also have Y-linked homologs or, conversely, what proportion of genes with Y-linked homologs escape inactivation.
Evolution of pXCI in therian mammals
Because XCI occurs exclusively and almost universally (a few species eliminate the paternal X altogether) in therian mammals, the idea that the eutherian and metatherian patterns of XCI are descended from an ancient dosage-compensation mechanism in the common therian ancestor has long been attractive (Cooper 1971; Riggs 1990; Deakin et al. 2009; Rens et al. 2010; Chaumeil et al. 2011; Shevchenko et al. 2013). The occurrence of imprinted pXCI in marsupials and extra-embryonic tissues of rodents and cattle, as well as several epigenetic and functional similarities between them, have also fueled speculation that marsupial pXCI might represent the primitive state from which rXCI later arose in the inner cell mass, the uniquely eutherian developmental structure that gives rise to the embryo proper (Cooper 1971; McCarrey 2001; Huynh and Lee 2003, 2005). Moreover, due to meiotic sex-chromosome inactivation (MSCI), the Xp of all therian zygotes arrives in an inactive state, and lack of reactivation in the early embryo was often entertained as a potentially shared mechanism for pXCI in eutherians and marsupials (McCarrey 2001; Huynh and Lee 2003, 2005; Rens et al. 2010). However, it is now known that both opossums and mice reactivate the Xp early in embryogenesis, then subject it to imprinted or random XCI shortly thereafter (Mahadevaiah et al. 2009); and mounting evidence indicates that eutherian pXCI might be a derived state reflecting convergent exaptation of biologically widespread epigenetic processes that achieve the same functional end point in rodents and marsupials (Reik and Lewis 2005; Deakin et al. 2009; Mahadevaiah et al. 2009; Al Nadaf et al. 2010; Escamilla-Del-Arenal et al. 2011; Okamoto et al. 2011).
Nevertheless, pXCI in rodents is imprinted and dependent on imprinted Xist expression, which may be a consequence of the very early activation of the mouse embryonic genome, including Xist, relative to that of eutherian species that lack imprinted XCI (Escamilla-Del-Arenal et al. 2011). Thus, some form of memory of the parental source of the zygotic X chromosomes is retained in both mammalian groups. This memory could result from a mark on the maternal X chromosomes that renders it resistant to inactivation (Tada et al. 2000), or to an as-yet-undetected epigenetic marking of Xist itself during spermatogenesis (Okamoto et al. 2005, 2011). In any case, memory of parental source of X chromosomes in early embryonic cells of both therian groups sustains the possibility that marsupial and rodent imprinted XCI are fundamentally related.
The recent discovery of Rsx suggests that key epigenetic mechanisms operating during the inactivation of X chromosomes in somatic cells of marsupials and eutherians have converged to a remarkable degree. As is true for Xist, Rsx is a long, noncoding RNA expressed only in female somatic cells, but not germ cells, has a highly complex repeat structure, coats the inactive X in cis, and is capable of silencing genes (Grant et al. 2012). We have extended this list of similarities by demonstrating that Rsx is expressed exclusively from the inactive Xp and that, like Xist, is differentially methylated at its promoter, with methylation on the Xm only. Despite this demonstration of functionally analogous lncRNAs from independent origins, the restriction of XCI to therians, together with many epigenetic and mechanistic similarities of the XCI process in marsupials and eutherians (Reik and Lewis 2005; Chaumeil et al. 2011; Escamilla-Del-Arenal et al. 2011; Shevchenko et al. 2013), is pleasingly concordant with the hypothesis that they arose sequentially from an ancestral regulatory process shared by both groups. Unfortunately, the molecular similarities per se do not resolve the issue of the ancestral versus derived relationship between the two XCI patterns. Until sufficient data regarding patterns of XCI in EEM of a broader sampling of mammalian species become available to establish the evolutionary polarity of their functional and epigenetic characteristics, the proposal that rXCI arose from an ancestral pXCI mechanism remains highly attractive, but speculative.
Methods
Tissue dissection and RNA-seq analysis
Total RNA was extracted from E13 fetal brain and EEM dissected from reciprocal F1 offspring (Supplemental Figs. S1–S3; Supplemental Table S1) of two random-bred stocks of M. domestica, LL1 and LL2 (VandeBerg and Williams-Blangero 2010). Fetal sex was determined as described in the Supplemental Methods. RNA-seq libraries were made using the Illumina TruSeq RNA Sample Prep Kit and sequenced on a HiSeq 2000 platform (Illumina). Image analysis and base calling were performed using Illumina software. RNA-seq reads were aligned to the reference opossum genome assembly (MonDom5) using TopHat v1.4.1 (Trapnell et al. 2009) with three mismatches allowed. The metric for total expression level, FPKM (Fragments Per Kilobase-pair of exon Model), was calculated using Cufflinks v1.3.0 (Trapnell et al. 2010) based on all mapped reads.
Quantification of parent-of-origin allelic expression from RNA-seq data
Sixty-eight thousand SNPs were called in combined RNA-seq data from uniquely mapped reads using SAMtools software (Li et al. 2009). Allele-specific expression was quantified as the ratio of the number of reference allele–containing reads divided by the total coverage at each high-coverage SNP position (≥8×) in brain and EEM tissues (Wang et al. 2008; Wang et al. 2011, 2013). Allelic transmission direction was inferred from the parental crosses and supplemented using additional information from other LL1 individuals for which RNA-seq data were available. For details, see the Supplemental Methods.
Validation of X-linked escaper and non-escaper genes by allele-specific pyrosequencing
To verify escaping status, allele-specific pyrosequencing was performed on all 24 escaper genes (Supplemental Figs. S6–S28), one non-escaper gene (Hprt1) (Supplemental Fig. S4), and one autosomal control gene (Gpm6b) (Supplemental Fig. S5) at informative SNP positions confirmed by Sanger sequencing. For non-escaper genes, 20 SNPs (one per gene) were randomly selected and genotyped by Sanger sequencing. All 20 were verified as heterozygous in at least two of the four female samples (Supplemental Data S2). For details, see the Supplemental Methods.
ChIP-seq and data analysis
Native-ChIP was conducted on a primary fibroblast cell line, fetal brain, and EEM using a method modified from Dindot et al. (2009). Illumina libraries were constructed at Global Biologics, LLC (Columbia, MO). After sequencing, raw reads were quality filtered, trimmed, and aligned using Bowtie in the Galaxy suite (Giardine et al. 2005; Blankenberg et al. 2010; Goecks et al. 2010). Aligned reads were visualized on the UCSC Genome Browser and IGV (Robinson et al. 2011; Thorvaldsdottir et al. 2013), and significant peaks were called using the MACS algorithm (Zhang et al. 2008). For details, see the Supplemental Methods.
Bisulfite-sequencing and PyroMark assays on promoter DNA
Two micrograms of genomic DNA were treated with the EpiTect Bisulfite Kit from Qiagen and amplified using primers designed by Methyl Primer Express v 1.0 (Applied Biosystems). PCR products were cloned using the TOPO TA Cloning Kit (Life Technologies) and sequenced by Sanger sequencing. Methylation percentages at promoter CpG sites were also quantified using the PyroMark assay on a PSQ 96MA Pyrosequencer (Qiagen). For details, see the Supplemental Methods.
Data access
RNA-seq and ChIP-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) under accession nos. GSE45211 and GSE45186, respectively.
Acknowledgments
This work was supported in part by a Meinig Family Investigator Award to A.G.C. and grant RR014214 from the National Center for Research Resources of the National Institutes of Health (USA) to P.B.S. Development of the M. domestica stocks was supported by a grant to J.L.V. from the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation. We thank Jenny Xiang, Ying (Diana) Shao, Wei Zhang, Amanda Manfredo, and Li (Grace) Chi for assistance with Illumina sequencing and pyrosequencing experiments. We also thank Madhu Jasti and Abigail Wolff for assistance with ChIP and genotyping experiments. Opossum Sry primer sequences were provided by the Opossum Y-Chromosome Mapping Project (Daniel W. Bellott, Jennifer F. Hughes, Helen Skaletsky, Laura G. Brown, Tatyana Pyntikova, Gabriel Cho, Natalia Koutseva, Sara Zaghlul, and David C. Page at the Whitehead Institute for Biomedical Research, Cambridge, Massachusetts; and Madhu Jasti, Kory C. Douglas, William J. Murphy, and Paul B. Samollow at Texas A&M University, College Station, Texas), in collaboration with the Washington University Genome Sequencing Center, St. Louis, Missouri.
Author contributions: X.W., K.C.D., A.G.C., and P.B.S. designed the experiment; J.L.V. developed the LL1 and LL2 animal stocks; X.W. and K.C.D. performed the experiments; X.W., K.C.D., A.G.C., and P.B.S. analyzed the data; X.W., K.C.D., A.G.C., and P.B.S. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.161919.113.
References
- Al Nadaf S, Waters PD, Koina E, Deakin JE, Jordan KS, Graves JA 2010. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol 11: R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard C, Li E, Jaenisch R 1995. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev 9: 2325–2334 [DOI] [PubMed] [Google Scholar]
- Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J 2010. Galaxy: A web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 89: 19.10.1–19.10.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese JM, Sun W, Song L, Mugford JW, Williams L, Yee D, Starmer J, Mieczkowski P, Crawford GE, Magnuson T 2012. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell 151: 951–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF 2005. X-Inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404 [DOI] [PubMed] [Google Scholar]
- Carvalho-Silva DR, O'Neill RJ, Brown JD, Huynh K, Waters PD, Pask AJ, Delbridge ML, Graves JA 2004. Molecular characterization and evolution of X and Y-borne ATRX homologues in American marsupials. Chromosome Res 12: 795–804 [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Waters PD, Koina E, Gilbert C, Robinson TJ, Graves JA 2011. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: Epigenetic modifications in distantly related mammals. PLoS ONE 6: e19040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DW 1971. Directed genetic change model for X chromosome inactivation in eutherian mammals. Nature 230: 292–294 [DOI] [PubMed] [Google Scholar]
- Cooper DW, Johnston PG, Vandeberg JL, Robinson ES 1990. X-Chromosome inactivation in marsupials. Aust J Zool 37: 411–417 [Google Scholar]
- Cooper DW, Johnston PG, Watson JM, Graves JAM 1993. X-Inactivation in marsupials and monotremes. Semin Dev Biol 4: 117–128 [Google Scholar]
- Deakin JE, Koina E, Waters PD, Doherty R, Patel VS, Delbridge ML, Dobson B, Fong J, Hu Y, van den Hurk C, et al. 2008. Physical map of two tammar wallaby chromosomes: A strategy for mapping in non-model mammals. Chromosome Res 16: 1159–1175 [DOI] [PubMed] [Google Scholar]
- Deakin JE, Chaumeil J, Hore TA, Marshall Graves JA 2009. Unravelling the evolutionary origins of X chromosome inactivation in mammals: Insights from marsupials and monotremes. Chromosome Res 17: 671–685 [DOI] [PubMed] [Google Scholar]
- Dindot SV, Kent KC, Evers B, Loskutoff N, Womack J, Piedrahita JA 2004. Conservation of genomic imprinting at the XIST, IGF2, and GTL2 loci in the bovine. Mamm Genome 15: 966–974 [DOI] [PubMed] [Google Scholar]
- Dindot SV, Person R, Strivens M, Garcia R, Beaudet AL 2009. Epigenetic profiling at mouse imprinted gene clusters reveals novel epigenetic and genetic features at differentially methylated regions. Genome Res 19: 1374–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disteche CM, Filippova GN, Tsuchiya KD 2002. Escape from X inactivation. Cytogenet Genome Res 99: 36–43 [DOI] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Del-Arenal M, da Rocha ST, Heard E 2011. Evolutionary diversity and developmental regulation of X-chromosome inactivation. Hum Genet 130: 307–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. 2005. Galaxy: A platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J 2010. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11: R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, et al. 2012. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487: 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM 2006. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Heard E, Clerc P, Avner P 1997. X-Chromosome inactivation in mammals. Annu Rev Genet 31: 571–610 [DOI] [PubMed] [Google Scholar]
- Hornecker JL, Samollow PB, Robinson ES, Vandeberg JL, McCarrey JR 2007. Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis 45: 696–708 [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT 2001. Imprinted X inactivation in eutherians: A model of gametic execution and zygotic relaxation. Curr Opin Cell Biol 13: 690–697 [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT 2003. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426: 857–862 [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT 2005. X-chromosome inactivation: A hypothesis linking ontogeny and phylogeny. Nat Rev Genet 6: 410–418 [DOI] [PubMed] [Google Scholar]
- Jegalian K, Page DC 1998. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature 394: 776–780 [DOI] [PubMed] [Google Scholar]
- Kaslow DC, Migeon BR 1987. DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupials: Evidence for multistep maintenance of mammalian X dosage compensation. Proc Natl Acad Sci 84: 6210–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koina E, Chaumeil J, Greaves IK, Tremethick DJ, Graves JA 2009. Specific patterns of histone marks accompany X chromosome inactivation in a marsupial. Chromosome Res 17: 115–126 [DOI] [PubMed] [Google Scholar]
- Latham KE 2005. X chromosome imprinting and inactivation in preimplantation mammalian embryos. Trends Genet 21: 120–127 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock LF, Melton DW, Caskey CT, Martin GR 1986. Methylation of the mouse hprt gene differs on the active and inactive X chromosomes. Mol Cell Biol 6: 914–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock LF, Takagi N, Martin GR 1987. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell 48: 39–46 [DOI] [PubMed] [Google Scholar]
- Loebel DA, Johnston PG 1993. Analysis of DNase 1 sensitivity and methylation of active and inactive X chromosomes of kangaroos (Macropus robustus) by in situ nick translation. Chromosoma 102: 81–87 [DOI] [PubMed] [Google Scholar]
- Loebel DA, Johnston PG 1996. Methylation analysis of a marsupial X-linked CpG island by bisulfite genomic sequencing. Genome Res 6: 114–123 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM 2009. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol 19: 1478–1484 [DOI] [PubMed] [Google Scholar]
- McCarrey JR. 2001. X-chromosome inactivation during spermatogenesis: The original dosage compensation mechanism in mammals? In Gene families: Studies of DNA, RNA, enzymes, and proteins (ed. Xue G, et al.), pp. 59–72. World Scientific Publishing, Hackensack, NJ. [Google Scholar]
- Migeon BR, Jan de Beur S, Axelman J 1989. Frequent derepression of G6PD and HPRT on the marsupial inactive X chromosome associated with cell proliferation in vitro. Exp Cell Res 182: 597–609 [DOI] [PubMed] [Google Scholar]
- Moreira de Mello JC, de Araujo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV 2010. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS ONE 5: e10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtagh VJ, O'Meally D, Sankovic N, Delbridge ML, Kuroki Y, Boore JL, Toyoda A, Jordan KS, Pask AJ, Renfree MB, et al. 2012. Evolutionary history of novel genes on the tammar wallaby Y chromosome: Implications for sex chromosome evolution. Genome Res 22: 498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Heard E 2006. The dynamics of imprinted X inactivation during preimplantation development in mice. Cytogenet Genome Res 113: 318–324 [DOI] [PubMed] [Google Scholar]
- Okamoto I, Arnaud D, Le Baccon P, Otte AP, Disteche CM, Avner P, Heard E 2005. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438: 369–373 [DOI] [PubMed] [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, et al. 2011. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472: 370–374 [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Reik W, Lewis A 2005. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet 6: 403–410 [DOI] [PubMed] [Google Scholar]
- Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC 2010. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci 107: 17657–17662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD 1990. Marsupials and mechanisms of X-chromosome inactivation. Aust J Zool 37: 419–441 [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samollow PB, Ford AL, VandeBerg JL 1987. X-linked gene expression in the Virginia opossum: Differences between the paternally derived Gpd and Pgk-A loci. Genetics 115: 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samollow PB, Robinson ES, Ford AL, Vandeberg JL 1995. Developmental progression of Gpd expression from the inactive X chromosome of the Virginia opossum. Dev Genet 16: 367–378 [DOI] [PubMed] [Google Scholar]
- Shevchenko AI, Zakharova IS, Zakian SM 2013. The evolutionary pathway of X chromosome inactivation in mammals. Acta naturae 5: 40–53 [PMC free article] [PubMed] [Google Scholar]
- Straub T, Becker PB 2007. Dosage compensation: The beginning and end of generalization. Nat Rev Genet 8: 47–57 [DOI] [PubMed] [Google Scholar]
- Tada T, Obata Y, Tada M, Goto Y, Nakatsuji N, Tan S, Kono T, Takagi N 2000. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development 127: 3101–3105 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP 2013. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L 2010. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeBerg JL, Robinson ES, Samollow PB, Johnston PG 1987. X-linked gene expression and X-chromosome inactivation: Marsupials, mouse, and man compared. Isozymes Curr Top Biol Med Res 15: 225–253 [PubMed] [Google Scholar]
- VandeBerg JL, Williams-Blangero S 2010. The laboratory opossum (Monodelphis domestica). In UFAW handbook on the care and management of laboratory and other research animals, 8th ed. (ed. Hubrecht R, Kirkwood J), Chapter 19, pp. 246–261. Wiley, Chichester, UK [Google Scholar]
- Wake N, Takagi N, Sasaki M 1976. Non-random inactivation of X chromosome in the rat yolk sac. Nature 262: 580–581 [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE 3: e3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Soloway PD, Clark AG 2011. A survey for novel imprinted genes in the mouse placenta by mRNA-seq. Genetics 189: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Miller DC, Clark AG, Antczak DF 2012. Random X inactivation in the mule and horse placenta. Genome Res 22: 1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Miller DC, Harman R, Antczak DF, Clark AG 2013. Paternally expressed genes predominate in the placenta. Proc Natl Acad Sci 110: 10705–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue F, Tian XC, Du F, Kubota C, Taneja M, Dinnyes A, Dai Y, Levine H, Pereira LV, Yang X 2002. Aberrant patterns of X chromosome inactivation in bovine clones. Nat Genet 31: 216–220 [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20: 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]