Abstract
Small RNA control of gene expression is critical for developmental processes in vertebrate embryos. To determine the dynamics of small RNA expression and to uncover novel small RNAs in the early vertebrate embryo, we performed high-throughput sequencing of all small RNAs in Xenopus tropicalis embryos at three developmental time points and in dissected halves of gastrula embryos. This analysis allowed us to identify novel microRNAs and we show that microRNA expression is highly dynamic and spatially localized in early embryos. In addition, we have developed a microRNA prediction pipeline and demonstrate that it has the power to predict new miRNAs that are experimentally detectable in frogs, mice, and humans. By combining the small RNA sequencing with mRNA profiling at the different developmental stages, we identify a new class of small noncoding RNAs that we name siteRNAs, which align in clusters to introns of protein-coding genes. We show that siteRNAs are derived from remnants of transposable elements present in the introns. We find that genes containing clusters of siteRNAs are transcriptionally repressed as compared with all genes. Furthermore, we show that this is true for individual genes containing siteRNA clusters, and that these genes are enriched in specific repressive histone modifications. Our data thus suggest a new mechanism of siteRNA-mediated gene silencing in vertebrates, and provide an example of how mobile elements can affect gene regulation.
Small noncoding RNAs have emerged as critical regulators of gene expression, both at the post-transcriptional and epigenetic level. Based on biogenesis criteria, there are three main classes of small RNAs: small interfering RNAs (siRNAs), microRNAs (miRNAs), and Piwi-interacting RNAs (piRNAs), although small RNAs with novel functions continue to be discovered.
siRNAs are ∼21- to 24-nucleotide (nt) RNAs that have antisense complementarity to target transcripts and are thought to act predominantly by direct endonucleolytic cleavage (“slicing”) of the target. siRNAs were first identified in response to viral infection in plants and have since been sequenced in Drosophila, Caenorhabditis elegans, mouse oocytes, and embryonic stem cells, and most recently in Xenopus tropicalis (Hamilton and Baulcombe 1999; Ambros et al. 2003b; Babiarz et al. 2008; Czech et al. 2008; Watanabe et al. 2008; Armisen et al. 2009). In a number of organisms they have also been shown to regulate chromatin structure (Olovnikov et al. 2012). In plants and C. elegans, siRNAs are thought to silence mobile elements, but the role of endogenous siRNAs in vertebrates is not fully understood. Recent work in mouse oocytes showed that disruption of siRNA biogenesis led to an increase in transposon and protein-coding transcripts, suggesting that mammalian endogenous siRNAs protect against transposons and negatively regulate gene expression (Watanabe et al. 2008).
miRNAs are ∼22-nt small RNAs found in species ranging in complexity from unicellular algae to humans. miRNAs negatively regulate gene expression post-transcriptionally via base-pairing to the 3′ UTR of target mRNAs (Bartel 2009), affecting mRNA stability and/or translation (Baek et al. 2008; Selbach et al. 2008). There are 18,226 miRNAs listed in the miRNA registry (miRbase 18), of which 1527 are human and 210 are Xenopus miRNAs (Kozomara and Griffiths-Jones 2011). A relatively small number of miRNAs have been shown to regulate a variety of processes in early vertebrate development, including the clearance of maternal mRNAs at the maternal-zygotic transition (Giraldez et al. 2006; Lund et al. 2009), modulation of embryonic signaling pathways such as Nodal signaling (Choi et al. 2007; Martello et al. 2007), and promotion of cell differentiation and control of cell fate, in particular, germ layer formation (Rosa et al. 2009), neural differentiation (Yoo et al. 2011), and regulation of epithelial-to-mesenchymal transition (Bracken et al. 2008).
Vertebrate piRNAs are 25- to 30-nt small RNAs that function via interaction with Piwi proteins of the Argonaute family and were initially discovered in Drosophila (Aravin et al. 2003). piRNAs are complementary to intergenic repeats and mediate silencing of transposons both by inducing cleavage of transcripts and through regulation of the chromatin structure by acting as guides to target enzymes required for methylation of genomic sequences of active transposons and neighboring regions (Aravin et al. 2001; Houwing et al. 2007; Siomi et al. 2011; Olovnikov et al. 2012; Sienski et al. 2012; Huang et al. 2013). piRNAs have been sequenced in the germ cells of many species including mouse, zebrafish, and X. tropicalis (Aravin et al. 2006; Houwing et al. 2007; Armisen et al. 2009; Lau et al. 2009; Faunes et al. 2012), where they are required to maintain genome integrity (Malone and Hannon 2009). In addition, they have recently been sequenced in somatic tissues of the mouse, rhesus macaque, and human (Yan et al. 2011; Li et al. 2012), suggesting additional functions outside of the germline. In Xenopus, piRNAs and transposable element (TE) transcripts have been identified in gastrula embryos and during neural tissue development, but piRNAs were not found in adult skin or liver (Armisen et al. 2009; Faunes et al. 2011, 2012).
In this study, we characterize the small RNA profile of early X. tropicalis development by deep sequencing small RNA libraries generated from blastula, gastrula, and neurula stage embryos. The libraries were designed to contain miRNAs, endogenous siRNAs, piRNAs, and any other unknown small RNAs. Our aim was to uncover the dynamics of small RNA expression and discover novel small RNAs in the early vertebrate embryo. Embryos were also dissected at the gastrula stage to provide information on the spatial localization of small RNAs during germ layer formation. We report the temporal and spatial expression profiles of 167 miRNAs and three entirely novel miRNAs. We show that miRNAs and other known small RNAs comprise <9% of the small RNAs present in the early embryo. Our analysis of the small RNA profile that did not correspond to known small RNA species (“unannotated” small RNAs) identified a new class of small RNAs that align to introns of protein-coding genes. We show that they are derived from remnants of TEs present in the introns. To reflect this and the location to which they align, we name them siteRNAs (small intronic transposable element RNAs). We find that genes containing clusters of siteRNAs are transcriptionally repressed both on a genome-wide scale and at the individual gene level. Furthermore, we demonstrate a correlation between the presence of siteRNA clusters and an enrichment of repressive histone modifications. Taken together, our data suggest a novel siteRNA-mediated mechanism of coordinated epigenetic and transcriptional repression.
Results
Overview of the early X. tropicalis embryo small RNAome
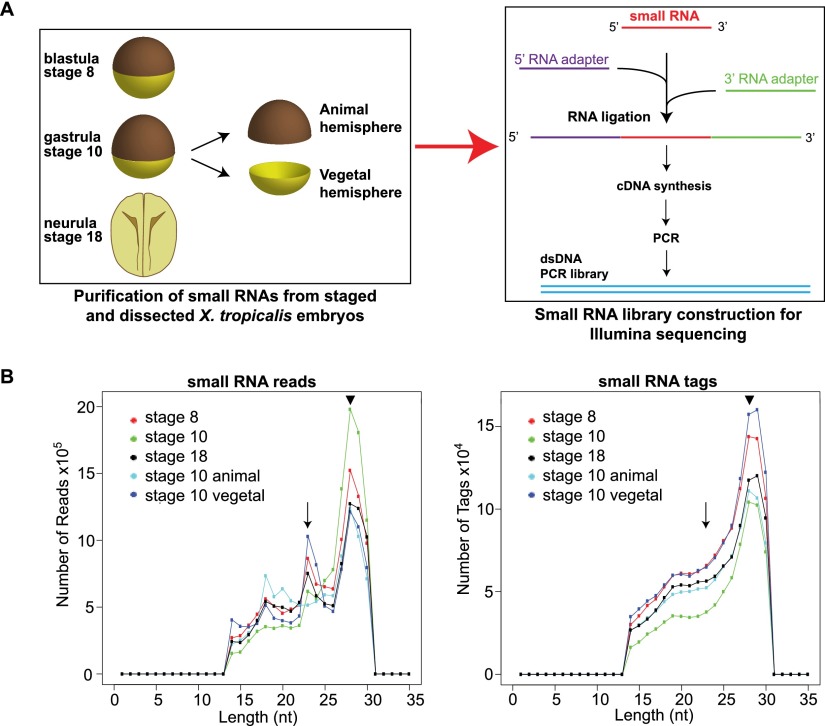
To sequence all small RNAs in the developing vertebrate embryo, we collected blastula (stage 8), gastrula (stage 10), and late neurula (stage 18) X. tropicalis embryos and also dissected gastrula embryos into animal and vegetal hemispheres. Small RNA libraries were prepared and subjected to high-throughput sequencing (Fig. 1A). Over 14 million raw reads were obtained per library, of which 45%–59% aligned perfectly to version 4.1 of the X. tropicalis genome (Hellsten et al. 2010). We sequenced between 358,909 and 834,281 unique small RNAs, which we refer to as tags (Armisen et al. 2009), depending on the library (Supplemental Table S1). The quality and reproducibility of the high-throughput sequencing was confirmed by resequencing the small RNA libraries in a technical repeat (Supplemental Tables S1, S2). An immediate picture of the quality, content, and heterogeneity of the libraries is given by the length distribution of small RNA species sequenced (Fig. 1B). Peaks in the length distribution suggest the presence of different classes of small RNAs in the libraries (Armisen et al. 2009). A major peak of 28 nt is present in all libraries and a lesser peak of 23 nt is seen in most libraries. These peaks are the correct size to contain piRNAs and miRNAs, respectively (Armisen et al. 2009). The 28-nt peak does not collapse when raw reads are converted into tags, indicating a complex population of small RNAs. In contrast, the collapsibility of the 23-nt peak suggests the presence of miRNAs, as highly abundant miRNAs usually compress into relatively few unique sequence tags (Armisen et al. 2009).
Figure 1.
Small RNA library preparation and quality control. (A) Schematic of library preparation. Small RNAs of 18–30 nt were purified from X. tropicalis embryos, and Illumina RNA adapters were ligated by an RNA ligation reaction. The adapter sequence is needed to attach the template DNA to the flow cell in the sequencing machine. The ligated RNAs were reverse-transcribed and the cDNA used as the template for a large-scale PCR. The PCR library was then subjected to single-end deep sequencing. (B) Length distributions of small RNA sequencing reads and unique sequence tags. Small RNA read and tag numbers are plotted against length for the five small RNA libraries. The sequencing reads are trimmed to remove sequencing adapters and mapped to the genome with zero mismatches. The arrow indicates the miRNA population that collapses when reads are converted to tags, and the arrowhead indicates the population of 28-nt RNAs.
Next we annotated the small RNAome by aligning the small RNA sequences to databases of known noncoding RNAs (ncRNAs) including miRbase (Griffiths-Jones et al. 2006), Rfam (Griffiths-Jones et al. 2003), RNAdb (Pang et al. 2005), and others (Supplemental Table S2). In total, miRNA reads comprised <1% of the small RNAome at blastula and gastrula stages, maximally accounting for ∼1.6% of sequencing reads at late neurula stage (Supplemental Table S2). Between 1.1% and 3.1% of the reads aligned to ncRNAs in the Rfam database. Of these, the most abundant were fragments of ribosomal RNAs (∼0.40%–0.75%) (Quast et al. 2013), tRNAs (∼0.03%–0.79%) (Lowe and Eddy 1997), and small nucleolar RNAs (∼0.04%–0.1%) (Griffiths-Jones et al. 2003). Similarly, small RNA reads aligning perfectly to a piRNA sequence in RNAdb (Pang et al. 2005) accounted for <1.1% of small RNAs in the early embryo. Interestingly, we identified 51 small RNAs that were classified as piRNAs in human, mouse, and rat, but which also matched specific fragments of tRNAs, suggesting that they could be the recently described tRFs (Lee et al. 2009). Significantly, depending on the library, between 91% and 98% of the small RNA sequence reads remained unannotated following alignment to known noncoding RNA databases (Supplemental Table S2).
Dynamics and localization of miRNA expression
After applying stringent filters (see Methods), we identified a total of 167 different miRNA sequences, of which 53 had been previously identified in Xenopus. It is important to note that the 167 miRNA sequences identified are not necessarily all distinct miRNAs. Indeed, 64 sequences (marked by an asterisk in Supplemental File 1) differ only by 1–2 nt at the 3′ or 5′ end compared with a previously identified homolog, which could be explained by trimming to remove sequencing adapters or by inherent variation in Dicer processing (Seitz et al. 2008) and thus might only correspond to 28 distinct miRNAs. In other cases, however, such as mmu-miR-182 and hsa-let-7d, the miRNA sequence cannot be formed by 5′ or 3′ trimming of another miRNA sequence, suggesting these may be new X. tropicalis miRNA family members. The five most frequently sequenced miRNAs in each library are presented in Supplemental Table S3. Small RNA sequencing is semi-quantitative; therefore, sequencing frequency correlates with abundance (Creighton et al. 2009). Sequencing reads were normalized to allow comparison of miRNA expression at three time points in early frog development and in the animal and vegetal hemispheres of the embryo at gastrula stage (Supplemental File 1). A selection of miRNA expression levels from the high-throughput sequencing were experimentally validated using small RNA qPCR, giving us confidence in the quantitative nature of the small RNA-seq data set (Supplemental Fig. S1).
Heatmap analysis revealed relatively low-level miRNA expression at the blastula stage, whereas a small number of miRNAs are highly expressed at gastrulation and late neurula stage (Fig. 2). For example, X. tropicalis miR-206 (xtr-miR-206) dominates the neurula stage miRNA profile (42% of all miRNA reads, respectively; see Supplemental Table S3). Other miRNAs are enriched in the animal hemisphere at gastrulation, for instance the Xenopus homologs of Bombyx mori and Drosophila melanogaster miR-184 (bmo-miR-184 and dme-miR-184). We also analyzed the global trends of miRNA expression dynamics and localization in more detail. On average, miRNAs increase in abundance 75-fold from blastula to late neurula stage, although individual miRNAs in the data set do decrease in abundance (Fig. 3A). Thirteen miRNAs increase >100-fold between blastula and neurula stage, including xtr-miR-206. This agrees with the previous conclusion that a small number of highly abundant miRNAs dominate the miRNA profile at stage 18. In addition, miRNAs are clearly enriched in the animal hemisphere of gastrula-stage embryos (Fig. 3B).
Figure 2.
Heatmap analysis of miRNA expression in early Xenopus development. miRNA tags of 22–23 nt identical in sequence to a miRbase miRNA were clustered by expression similarity. miRNA expression is displayed using a color key where blue corresponds to low and red to high numbers of miRNA normalized reads. The miRNAs labeled are those mentioned in the text and/or the most abundant in the five libraries (Supplemental Table S3).
Figure 3.
miRNA expression is highly dynamic and localized. (A) Normalized read numbers for individual miRNAs at stages 10 and 18 are shown relative to the stage 8 values. The red line indicates the mean. (B) Normalized miRNA reads in the stage 10 animal and stage 10 vegetal libraries are plotted on a logarithmic scale with y = x superimposed to show the animal–vegetal spatial distribution of miRNAs at gastrulation.
Novel miRNA prediction and validation
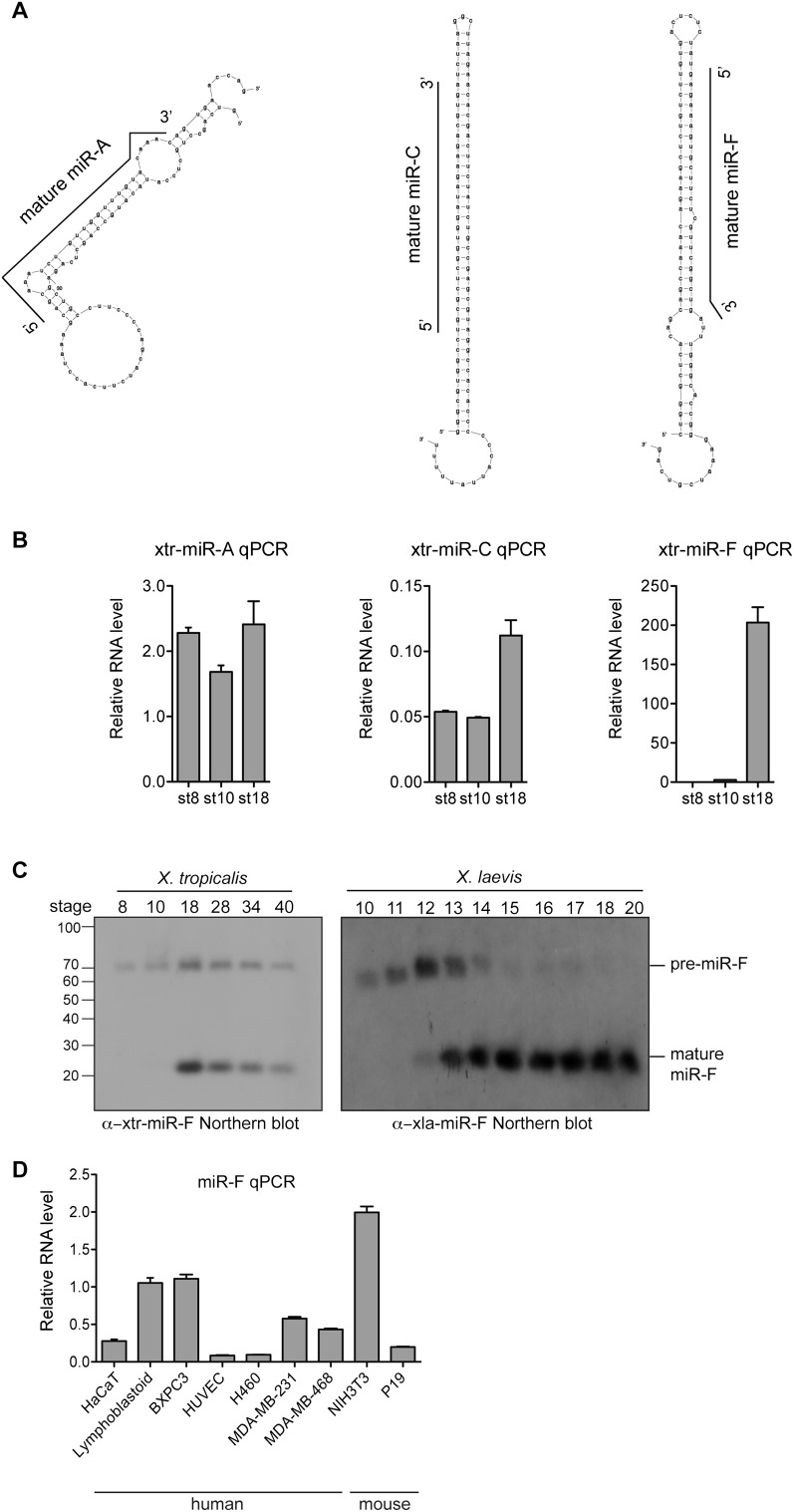
To reveal novel miRNAs within the unannotated small RNA high-throughput sequencing reads, we designed a de novo miRNA prediction pipeline. Briefly, unannotated small RNA tags were filtered (see Methods for full details) and the surrounding genomic sequence was extracted to form a candidate miRNA precursor sequence (pre-miRNA), whose secondary structure was predicted using Mfold (Zuker 2003). Secondary structures were screened for the following features: a hairpin secondary structure without large bulges, a mature miRNA sequence contained within one arm of the hairpin, and at least 16 bp of the mature miRNA sequence base-pairing with the candidate miRNA-star sequence (Ambros et al. 2003a). Three novel miRNAs (provisionally named miR-A, C, and F) passed these criteria and were detected by qPCR in X. tropicalis (Fig. 4A,B). Interestingly, miR-F shows regulated pre-miRNA processing, as mature miR-F expression begins at neurula stage and persists to tadpole stage (Fig. 4C, left panel), whereas pre-miR-F is present from blastula stages. This regulated processing was also seen in X. laevis embryos (Fig. 4C, right panel). miR-F is evolutionarily conserved, as it was also detected in mouse and human cell lines (Fig. 4D). In summary, three novel X. tropicalis miRNAs were confidently identified, including a highly conserved miRNA, miR-F. miR-A, C, and F were named xtr-miR-9406, xtr-miR-9407, and xtr-miR-428b, respectively, by miRBase when the manuscript was accepted for publication.
Figure 4.
Identification of novel miRNAs. (A) Predicted pre-miRNA structures of X. tropicalis miR-A, C, and F. The black line indicates the mature miRNA sequence within the hairpin precursor. (B) qPCR detection of candidate novel miRNAs in X. tropicalis. Relative RNA levels to odc were calculated and error bars are standard deviations from triplicate repeats. (C) Northern blot for miR-F on RNA from X. tropicalis embryos of stages 8–40 (left panel) and X. laevis embryos of stages 10–20 (right panel). Both the mature miRNA (23 nt) and pre-miRNA (70 nt) are detected. Comparison to decade markers (Ambion) revealed the size of the small RNAs. (D) qPCR detection of miR-F in human and mouse cell lines. miR-F RNA levels relative to gapdh were calculated. Error bars are standard deviations of triplicate repeats.
Identification of small intronic RNAs from unannotated reads
The vast majority (>91%) of the small RNAs that we sequenced in X. tropicalis embryos were not present in known small ncRNA databases (Supplemental Table S2). To gain insight into their possible function, we investigated where they align in the genome. Between 44% and 46% of them aligned (with no mismatches) to introns of protein-coding genes in each library, whereas between 2%–3% and 52%–55% aligned to exons and intergenic regions, respectively (Fig. 5A). The overrepresentation of clusters aligning in introns, which represent 16% of the genome, led us to focus our analysis on this specific class. The small intronic RNAs were conserved in all five libraries, shown for example for the tdp1 gene (Supplemental Fig. S2A). This conservation is also evident in the Venn diagram (Fig. 5B). The size range of the small intronic RNAs is 23–29 nt (data not shown). Further examples of small intronic RNAs, as well as small RNAs mapping within 5 kb upstream of a transcription start site (TSS) are shown in Supplemental Figure S2B,C.
Figure 5.
siteRNAs are conserved, cluster in introns, and align to TE remnants. (A) The percentage of unannotated small RNA tags that map to intergenic regions, exons, and introns (using RefSeq gene annotation) is shown for the stage 8, 10, 18, stage 10 animal, and stage 10 vegetal libraries. (B) Venn diagram showing the number and overlap of genes with small intronic noncoding RNAs in the stage 8, stage 10, and stage 18 libraries. (C) Small RNA alignments and mRNA-seq alignments (tracks I–III) to the sptlc1 gene at stage 10 shown in the Integrated Genome Viewer (IGV). Exons are indicated by dark blue boxes, introns by blue lines, and an arrow shows the direction of transcription. Small RNA tags are colored red for a sense alignment and blue for an antisense alignment to the genome, respectively. The gray histograms show peaks of read density with the scale indicated (e.g., 0–137 reads, track I). Track I shows small RNA-seq data before filtering by window-scanning analysis to reveal regions of high siteRNA density (track II). Track III shows mRNA reads, which align to exons. Reads that map over an exon–exon boundary are connected by a blue line. (D,E) Diagrams are shown for map7d2 and tdp1 to indicate the relationship between the siteRNA clusters and the TE remnants present in the introns. The representation of the genes is as in C. Small RNA reads are shown in the track labeled “small RNA-seq” in gray, or in green for those which passed the window scanning filter. The scale indicated is as in C. The green bars below indicate individual siteRNA clusters and the red arrow indicates the direction of transcription of the small RNA reads. In the track labeled “transposon,” the TE remnants are shown as colored boxes, with the names given below. The TE remnant from which the siteRNAs are derived is shown in red, and this entire TE is shown to scale above. The red box in the TE is the region of almost identical sequence to the intronic TE remnant. The purple line shows the protein-coding region and the arrow gives the direction of transcription of the entire transposon. Note that the small RNA reads are antisense relative to the direction of TE transcription.
To find regions of high small intronic RNA density in the genome which we reasoned were more likely to be functional, we applied a sliding window filter to the small RNA-seq data set (Fig. 5C, cf. tracks I and II; Akkers et al. 2009; Konig et al. 2011). A total of 1930 intronic regions passed the sliding window filter in the stage 8, stage 10, and stage 18 small RNA libraries (Supplemental File 2; Fig. 5C). We obtained small RNA clusters that were sense or antisense (52% and 48%, respectively) relative to the gene, such as the cluster in sptlc1 (Fig. 5C) and that in gnai2, respectively (Supplemental Fig. S2B). Using the sliding window filter, we also identified 465 small upstream ncRNA clusters (Supplemental File 2).
An analysis of the sequences of the intronic small RNA clusters revealed that almost all of them (92%) could be aligned in 41 different groups based on sequence similarity (Table 1; Supplemental Fig. S3). Further analysis revealed that the sequences represented by these groups were related to those of TEs of both major classes: retrotransposons (both LTR and non-LTR) and DNA transposons (Table 1; Faunes et al. 2011; Siomi et al. 2011). To reflect their alignment to introns and their relationship to TEs, we named these small RNAs siteRNAs. Interestingly, only specific regions of the TEs align with siteRNA clusters. The siteRNA clusters in the same group all display high sequence similarity, but as they all map uniquely in the genome, none of the sequences are identical. We next investigated, for representatives from each group, whether the alignment in the intron corresponded to the entire TE, or to a fragment. The siteRNA clusters aligned to remnants of TEs and 85% were antisense, relative to the TE (Fig. 5D,E). In addition, we noted that the alignment was not necessarily to the coding portion of the TE, and also that not all TE remnants in a given intron aligned to siteRNAs (Fig. 5D,E).
Table 1.
Grouping of the siteRNA clusters according to the regions of TEs from which they are derived

Genome-wide mRNA-seq reveals a correlation between siteRNAs and low transcriptional activity of the genes to which they map
To investigate whether siteRNA clusters might regulate the genes to which they map, we performed mRNA-seq transcriptome profiling at the same stages of X. tropicalis development. We compared the mRNA expression profile of all the transcriptome (all RefSeq genes Release 39; Jan 23 2010) to that of genes containing small upstream RNA clusters or genes containing siteRNA clusters. We found that the expression of genes containing siteRNA clusters was statistically significantly decreased compared with the whole transcriptome (P < 1 × 10−5) (Fig. 6A). In contrast, there was no statistically significant difference in the expression of small upstream RNA cluster genes and the transcriptome (P > 0.15) (Fig. 6B). Therefore, on a genome-wide level, siteRNA clusters correlate with low expression of the genes to which they map.
Figure 6.
siteRNA clusters are associated with low expression of the gene to which they map. (A) mRNA expression levels obtained by RPKM normalization of mRNA-seq reads for all RefSeq genes (“transcriptome”) and genes containing siteRNA clusters (Supplemental File 2) are compared for stages 8–18. The box and whisker plot displays the median, first, and third quartiles and the whiskers indicate 1.5× the interquartile range. Pairwise Wilcoxon tests were performed to assess changes in the RPKM values between “transcriptome” and siteRNA cluster genes at each stage, and Bonferroni-corrected P-values are indicated. (B) mRNA expression levels for all RefSeq genes (“transcriptome”) and genes containing upstream RNA clusters are compared for stages 8–18 as in A. (C) siteRNA clusters can be amplified using specific qRT-PCR primers at stages 8, 10, and 18 in X. tropicalis embryos. The bars labeled “common” correspond to qRT-PCR using primers that would recognize RNA from all of the group 7 TE remnants. RNA levels relative to odc were calculated and error bars are standard deviations from duplicates. The groups refer to the TE sequence clusters shown in Supplemental Figure S3 and Table 1. (D) mRNA expression levels detected by qPCR at stages 8–18. mRNA levels relative to odc were calculated. Error bars are standard deviations from triplicates. mRNA levels of the developmentally regulated gene xbra are also plotted as a reference.
To investigate these siteRNA clusters in more detail, we focused on siteRNA clusters representing three of the TE groups (Supplemental Fig. S3). Because the small RNA reads spanned the entire intronic TE remnant, we hypothesized that the siteRNAs might be processed from a longer transcript encompassing the TE remnant. To test this, we designed qRT-PCR primers that would allow specific amplification of a longer RNA derived from the individual TE remnants, and also a pair of primers that would amplify a common product for the group 7 siteRNA cluster. In all cases, qRT-PCR revealed weak expression of these longer RNA precursors at stages 8 and 10 and strong up-regulation at stage 18, which must be a result of zygotic transcription (Fig. 6C). As observed in the genome-wide analysis, the siteRNA clusters were associated with genes whose expression is repressed. Using levels of the mesoderm-specific xbra transcript at stage 8 as a reference for a repressed gene and levels of xbra at stages 10 and 18 as a reference for an active gene (Smith et al. 1991), we assessed the levels of gene expression for all the genes for which we analyzed the siteRNA clusters. All of these genes were expressed at low levels, or not at all (Fig. 6C,D). In addition, for the genes map7d2, galk2, and kiaa1468 that were slightly more strongly expressed at stage 8 and 10, their mRNA levels decreased at stage 18 coincident with the increase in siteRNA cluster expression (Fig. 6C,D). This indicates that the presence of siteRNAs correlated with low steady-state mRNA levels both at the genome-wide and individual transcript level. As the introns to which siteRNAs align are spliced out in mature mRNA, this repression is more likely to occur at the transcriptional, rather than the post-transcriptional level.
The presence of siteRNA clusters correlates with combinations of repressive histone modifications
Small RNA-directed transcriptional silencing via formation of heterochromatin has been observed in other organisms and, moreover, can occur at TEs (Buhler and Moazed 2007; Hawkins et al. 2009; Verdel et al. 2009; Kanhere et al. 2010; Olovnikov et al. 2012; Sienski et al. 2012). We therefore investigated whether the genes containing siteRNA clusters were enriched in repressive histone modifications. A genome-wide analysis in gastrula-stage X. tropicalis embryos for RNA polymerase II, H3K4me3 (Santos-Rosa et al. 2002), and another histone modification associated with transcriptional repression, H3K27me3 (Cao et al. 2002), has previously been published (Akkers et al. 2009). In a comparative analysis of this data set with our genome-wide siteRNA cluster data set we found no overlap between the siteRNA clusters and the H3K27me3 histone mark (data not shown). We therefore performed chromatin immunoprecipitation (ChIP) on stage 10 and stage 18 X. tropicalis embryos for other repressive histone marks, H3K9me3 and H4K20me3 (Stewart et al. 2005; Wongtawan et al. 2011). We also performed ChIP for H3K4me3 to include an epigenetic mark that is mainly found at promoters. It also serves as a positive control for comparison with the previous ChIP-seq analysis (Akkers et al. 2009). Strikingly, we observed H3K9me3 and H4K20me3 deposition at the siteRNA cluster region and at the TSS for all of the genes analyzed, and in many cases the enrichment was increased at stage 18 (Fig. 7). Thus these siteRNA cluster genes all share a similar repressive epigenetic profile, consistent with their low expression levels.
Figure 7.
siteRNA clusters are enriched for repressive H3K9me3 and H4K20me3 epigenetic marks. ChIP-qPCR profiles for tri-methylated H3K4 (H3K4me3), H3K9me3, and H4K20me3 histone modifications at the TSS and siteRNA cluster region. Chromatin was prepared from whole X. tropicalis embryos at stages 10 and 18. The quantity relative to the input after qPCR was calculated for each IP. The bars marked “beads” correspond to a control IP without antibody. Means and standard deviations of duplicates from a representative experiment are shown.
Discussion
In this study we characterize the small RNA profile of early X. tropicalis development and analyze the spatial localization of small RNAs in the vertebrate embryo by high throughput sequencing. We identify a total of 167 miRNAs in X. tropicalis and show that miRNAs increase in abundance and diversity from late blastula to neurula stage. This result is consistent with the role of miRNAs as repressors of pluripotency in the embryo, as the specification of tissues progresses between gastrulation and neurulation. We found that miRNAs were enriched in the animal hemisphere compared with the vegetal hemisphere at gastrulation, which may contribute to the differentiation of neural tissue from prospective ectoderm, as neural-specific expression of miRNAs has been widely reported (Wienholds et al. 2005; Walker and Harland 2008). miRNA localization to the animal hemisphere could also be required to attenuate the expression of mesodermal and endodermal inducers in the animal hemisphere to maintain ectodermal cell fates.
We found that >91% of the small RNAs present in the early X. tropicalis embryo could not be identified by comparison with known small RNAs. These are unlikely to be fragments of mRNA, as we observed very few sequences corresponding to very abundant transcripts. This suggested that there are many small RNAs and possible regulatory mechanisms in the early embryo that we do not yet understand. To start addressing this, we designed and validated a miRNA prediction pipeline that processes unannotated small RNA reads into experimentally detectable, novel miRNAs. Interestingly, while we were writing this paper the sequence of miR-F appeared in a list of putative miRNAs from X. laevis, further validating our prediction pipeline (Ambady et al. 2012).
Furthermore, by analyzing where the unannotated small RNAs map in the genome, we uncovered a novel class of small RNAs that cluster in introns of protein-coding genes that we term “siteRNA clusters.” We detected hundreds of high-density siteRNA clusters. Since they can be sense or antisense with respect to the gene to which they map, we concluded that they cannot be by-products of mRNA processing, mirtrons (Okamura et al. 2007; Ruby et al. 2007), or the recently identified sisRNAs (Gardner et al. 2012). Analysis of the sequences of the siteRNA clusters revealed that the vast majority aligned to regions of TEs, both DNA transposons and retrotransposons. Contrary to a previous report showing that X. tropicalis small RNAs mapped predominantly to L1-55–XT, HAT-10–XT, and Piggybac-1–XT (Faunes et al. 2012), we observed that the siteRNAs are derived from a distinct TE set (Table 1). To understand in more detail the origin of the siteRNAs and their function, we focused on siteRNA clusters that aligned uniquely in the genome. It is clear that 85% of the clusters align antisense to TE remnants in the introns. Using primers specific for the unique siteRNA cluster we could show that a longer precursor, encompassing the TE remnant, is strongly up-regulated at stage 18. This result contrasts with the approximately equal levels of individual siteRNAs derived from these elements in the stage 8, 10, and 18 libraries. We thus speculate that siteRNAs are laid down prior to stage 8, possibly by being processed from longer precursors, and their levels are maintained as a result of zygotic transcription between stage 10 and stage 18. As yet, we do not understand the mechanism by which siteRNAs are synthesized or processed.
Genome-wide transcriptome profiling showed that genes with siteRNA clusters are transcriptionally repressed compared with all genes in the transcriptome. This was also true at the level of individual genes. Importantly, we found a correlation between the presence of the siteRNA clusters in a gene and deposition of repressive H3K9me3 and H4K20me3 histone modifications, but not the repressive H3K27me3 mark. Small RNA interaction with chromatin to direct transcriptional silencing has been observed in plants (Zilberman et al. 2003), yeast (Verdel et al. 2009), Drosophila (Pal-Bhadra et al. 2004; Sienski et al. 2012; Huang et al. 2013), and in certain situations, at promoters of mammalian genes (Gonzalez et al. 2006; Janowski et al. 2006; Weinberg et al. 2006; Gonzalez et al. 2008; Kim et al. 2008; Schmitz et al. 2010). In these cases, small RNAs target enzymes involved in DNA methylation or histone methylation either through direct recruitment to DNA via triplex formation (Schmitz et al. 2010), or via complementarity to nascent transcripts (Olovnikov et al. 2012). Our work shows that siteRNA clusters coincide with the deposition of repressive epigenetic marks to effect transcriptional repression in the early vertebrate embryo of groups of genes characterized by specific TE remnants in their introns. We suggest that the siteRNAs act predominantly in a cis mechanism, being both produced as a result of transcription of the TE remnants, and acting as guides to modify chromatin structure. It will be important in the future to analyze small RNA sequencing data sets in other species to determine if this mechanism is deployed in other vertebrates to regulate gene expression during embryonic development.
Methods
Animal procedures
X. tropicalis and X. laevis embryos were obtained by in vitro fertilization, de-jellied in 2.2% cysteine, and cultured in 1/20 × MMR with gentamycin until the indicated stage as described (Khokha et al. 2002). Dissections were carried out in 1 × NAM supplemented with 0.1% (w/v) BSA.
Small RNA and mRNA sequencing, alignment, and normalization
Details of small RNA and mRNA library preparation and sequencing are given in the Supplemental Methods. Small RNA reads were trimmed to remove sequencing adapters and aligned to the X. tropicalis genome, Joint Genome Institute, assembly version 4.1 (Hellsten et al. 2010) allowing for zero mismatches using the BWA alignment program (Li and Durbin 2009). mRNA reads were aligned using bowtie (Langmead et al. 2009) and those over splice junctions were aligned using TopHat (Trapnell et al. 2009). Up to two sequence mismatches were permitted in mRNA-seq read alignments.
Small RNA reads were normalized using a linear scaling whereby the raw reads were multiplied by the greatest number of genomic alignments in any library divided by the number of genomic alignments in the library of interest. mRNA reads were quantified using the reads per kilobase per million of exon (RPKM) method for each gene annotated in RefSeq (Release 39; Jan 23 2010) to normalize for transcript length and total read number (Mortazavi et al. 2008).
miRNA heatmaps and a novel miRNA prediction pipeline
Trimmed reads of length 22–23 nt or 25–30 nt with a perfect match to a miRbase miRNA sequence and present at the minimum level of 10 reads in at least one library were retained. Heatmaps were generated using a modified version of the heatmap2 function from R (http://www.R-project.org) based on a dendrogram of small RNA expression similarity. Color keys represent log10 (normalized read numbers +1). For the novel miRNA prediction pipeline, see Supplemental Methods.
Detection of high-density small RNA clusters
A modified version of a sliding window approach was used (Akkers et al. 2009; Konig et al. 2011) with a window of 100 nt sliding in 50-nt increments across the unannotated small RNA-seq reads aligning to an “upstream” genomic sequence (defined as within 5 kb 5′ of a TSS of a gene) or an intronic genomic sequence using RefSeq (Oct. 4, 2010). The mean nucleotide coverage per window was calculated and a cutoff of 30 was used. Overlapping windows in the same upstream or intronic region were merged and coverage was recalculated. The underlying genomic sequence passing the cutoff was extracted and reported as a small RNA cluster.
Alignment of siteRNA sequences to TEs and clustering
siteRNA clusters were blasted against the reference collection of repeat sequences from Repbase (www.girinst.org/repbase/) using CENSOR (Kohany et al. 2006). The siteRNA cluster groups were aligned by Clustal Omega (Sievers et al. 2011).
Quantitative RT-PCR
Small RNA qRT-PCR was performed essentially as described (Shi and Chiang 2005) using the Affinityscript kit (Stratagene) and EXPRESS SYBR Green reagents (Invitrogen). qRT-PCR was performed using cDNA generated from random hexamer priming of total RNA. See Supplemental Methods for oligonucleotide sequences used for all qPCR and for details of cell culture.
Small RNA Northern blotting
Total RNA (15 μg) prepared by TRIzol extraction and DNase I treatment from X. tropicalis embryos or X. laevis embryos was fractionated by denaturing gel electrophoresis and transferred to a nylon membrane, which was chemically cross-linked (Pall and Hamilton 2008). The small RNA probe (32P radiolabeled complementary DNA oligonucleotide; sequences listed in the Supplemental Information) was then hybridized to the previously equilibrated membrane overnight under rotation at 39°C, before washing and detection of signal.
ChIP assays and antibodies
ChIPs were performed as previously described (Blythe et al. 2009; Heliot and Cereghini 2012) with the following modifications. Chromatin was prepared in radioimmunoprecipitation assay buffer, and 4 min of medium intensity sonication using a Biodisruptor sonicator was sufficient to yield 200- to 500-bp DNA fragments. Magnetic protein A or G beads were used as appropriate (Invitrogen). The following antibodies were used: anti-H3K4me3 (Abcam ab8580), anti-H3K9me3 (Abcam ab8898), and anti-H4K20me3 (Abcam ab9053). The primers used for ChIP qPCR are listed in the Supplemental Information.
Data access
The data have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (Edgar et al. 2002) under accession number GSE38605. The novel miRNAs miR-A, C, and F have been submitted to miRBase and named xtr-miR-9406, xtr-miR-9407, and xtr-miR-428b, respectively.
Acknowledgments
We thank the Cambridge Research Institute for small RNA sequencing and Nik Matthews and the LRI Advanced Sequencing Facility for the small RNA-seq technical repeat and mRNA-seq. We thank the LRI Equipment Park for technical support, and Hiro Mahbubani, Darren Martin, and Chris Sergeant for maintaining the Xenopus colony. We are grateful to George Gentsch for the xbra primers sequences. We thank members of the Developmental Signalling laboratory, Aengus Stewart, and Mike Gilchrist for useful discussions and/or comments on the manuscript. This work was supported by Cancer Research UK and the European Commission Network of Excellence EpiGeneSys (HEALTH-F4-2010-257082). L.B.Z. was funded by MRC U117560482 and NIH 1R01 DC011901-01.
Author contributions: J.L.H., C.H., and C.S.H. conceived the study. J.L.H., S.H., and C.H. performed the experiments and analyzed the data with help from J.A., L.B.Z., N.M.L., and E.A.M. The paper was written by J.L.H., C.H., and C.S.H. with input from the other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.144469.112.
Freely available online through the Genome Research Open Access option.
References
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ 2009. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev Cell 17: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambady S, Wu Z, Dominko T 2012. Identification of novel microRNAs in Xenopus laevis metaphase II arrested eggs. Genesis 50: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, et al. 2003a. A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D 2003b. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T 2003. The small RNA profile during Drosophila melanogaster development. Dev Cell 5: 337–350 [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Armisen J, Gilchrist MJ, Wilczynska A, Standart N, Miska EA 2009. Abundant and dynamically expressed miRNAs, piRNAs, and other small RNAs in the vertebrate Xenopus tropicalis. Genome Res 19: 1766–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS 2009. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev Dyn 238: 1422–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68: 7846–7854 [DOI] [PubMed] [Google Scholar]
- Buhler M, Moazed D 2007. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol 14: 1041–1048 [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Choi W-Y, Giraldez AJ, Schier AF 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318: 271–274 [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Reid JG, Gunaratne PH 2009. Expression profiling of microRNAs by deep sequencing. Brief Bioinform 10: 490–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faunes F, Sanchez N, Moreno M, Olivares GH, Lee-Liu D, Almonacid L, Slater AW, Norambuena T, Taft RJ, Mattick JS, et al. 2011. Expression of transposable elements in neural tissues during Xenopus development. PLoS ONE 6: e22569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faunes F, Almonacid LI, Melo F, Larrain J 2012. Characterization of small RNAs in Xenopus tropicalis gastrulae. Genesis 50: 260–270 [DOI] [PubMed] [Google Scholar]
- Gardner EJ, Nizami ZF, Talbot CC Jr, Gall JG 2012. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev 26: 2550–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Klatt P, Delgado S, Conde E, Lopez-Rios F, Sanchez-Cespedes M, Mendez J, Antequera F, Serrano M 2006. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature 440: 702–706 [DOI] [PubMed] [Google Scholar]
- Gonzalez S, Pisano DG, Serrano M 2008. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle 7: 2601–2608 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR 2003. Rfam: An RNA family database. Nucleic Acids Res 31: 439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hawkins PG, Santoso S, Adams C, Anest V, Morris KV 2009. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res 37: 2984–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heliot C, Cereghini S 2012. Analysis of in vivo transcription factor recruitment by chromatin immunoprecipitation of mouse embryonic kidney. Methods Mol Biol 886: 275–291 [DOI] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, Ovcharenko I, Putnam NH, Shu S, Taher L, et al. 2010. The genome of the Western clawed frog Xenopus tropicalis. Science 328: 633–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H 2013. A major epigenetic programming mechanism guided by piRNAs. Dev Cell 24: 502–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Huffman KE, Schwartz JC, Ram R, Nordsell R, Shames DS, Minna JD, Corey DR 2006. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol 13: 787–792 [DOI] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. 2002. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn 225: 499–510 [DOI] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snove O Jr, Rossi JJ 2008. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci 105: 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J 2006. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Luscombe NM, Ule J 2011. Protein-RNA interactions: New genomic technologies and perspectives. Nat Rev Genet 13: 77–83 [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S 2011. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–D157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD 2009. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J 28: 2945–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Shibata Y, Malhotra A, Dutta A 2009. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23: 2639–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang HY, Wan FC, Liu FJ, Liu J, Zhang N, Jin SH, Li JY 2012. Deep sequencing analysis of small non-coding RNAs reveals the diversity of microRNAs and piRNAs in the human epididymis. Gene 497: 330–335 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE 2009. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15: 2351–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ 2009. Small RNAs as guardians of the genome. Cell 136: 656–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, et al. 2007. MicroRNA control of Nodal signalling. Nature 449: 183–188 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC 2007. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov I, Aravin AA, Fejes Toth K 2012. Small RNA in the nucleus: The RNA-chromatin ping-pong. Curr Opin Genet Dev 22: 164–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672 [DOI] [PubMed] [Google Scholar]
- Pall GS, Hamilton AJ 2008. Improved northern blot method for enhanced detection of small RNA. Nat Protoc 3: 1077–1084 [DOI] [PubMed] [Google Scholar]
- Pang KC, Stephen S, Engstrom PG, Tajul-Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y, Mattick JS 2005. RNAdb—a comprehensive mammalian noncoding RNA database. Nucleic Acids Res 33: D125–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO 2013. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH 2009. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16: 517–527 [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP 2007. Intronic microRNA precursors that bypass Drosha processing. Nature 448: 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, Grummt I 2010. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24: 2264–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD 2008. Argonaute loading improves the 5′ precision of both microRNAs and their miRNA* strands in flies. Curr Biol 18: 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N 2008. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Shi R, Chiang VL 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39: 519–525 [DOI] [PubMed] [Google Scholar]
- Sienski G, Donertas D, Brennecke J 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA 2011. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Smith JC, Price BM, Green JB, Weigel D, Herrmann BG 1991. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell 67: 79–87 [DOI] [PubMed] [Google Scholar]
- Stewart MD, Li J, Wong J 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 25: 2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Vavasseur A, Le Gorrec M, Touat-Todeschini L 2009. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol 53: 245–257 [DOI] [PubMed] [Google Scholar]
- Walker JC, Harland RM 2008. Expression of microRNAs during embryonic development of Xenopus tropicalis. Gene Expr Patterns 8: 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV 2006. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA 12: 256–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH 2005. MicroRNA expression in zebrafish embryonic development. Science 309: 310–311 [DOI] [PubMed] [Google Scholar]
- Wongtawan T, Taylor JE, Lawson KA, Wilmut I, Pennings S 2011. Histone H4K20me3 and HP1α are late heterochromatin markers in development, but present in undifferentiated embryonic stem cells. J Cell Sci 124: 1878–1890 [DOI] [PubMed] [Google Scholar]
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, et al. 2011. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res 39: 6596–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR 2011. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE 2003. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]