Abstract
Background
Oxidative damage and DNA repair dysfunction are associated with carcinogenesis. 8-OHdG is one of the major oxidative DNA adducts. Present work aims to investigate whether the expression of 8-OHdG and its key repair gene hOGG1 play distinctive role in two types of serous ovarian cancer.
Materials and methods
8-OHdG level in DNA from tumor and matched tumor-adjacent normal tissue in 48 high-grade papillary serous carcinomas (HG-SOC), 24 low-grade papillary serous carcinomas (LG-SOC), 20 serous cystadenomas, and 16 non-tumor control ovaries was tested. The Cox proportional hazards model and the log-rank test were used to assess the associations between the 8-OHdG level in two types of serous cancer and patients’ survival. Real-time polymerase chain reaction and protein immunoblot were employed to detect hOGG1 mRNA and protein levels in tumor and adjacent normal tissues. Immunohistochemistry was used to determine the expression of hOGG1 and p53.
Results
There was no difference of average 8-OHdG/106dG DNA level either between HG-SOC (27.8 ± 8.9), LG-SOC (25.2 ± 7.4) and benign serous cystadenoma (26.5 ± 7.7, p = 0.35); or between the tumor-adjacent normal tissue of HG-SOC (18.8 ± 5.2), LG-SOC (21.4 ± 6.5), benign serous cystadenoma (20.5 ± 9.1) and non-tumor ovary (21.6 ± 4.9, p = 0.62). The 8-OHdG/106dG level was significantly higher in tumor comparing to that in matched normal tissue adjacent to carcinoma in HG-SOC (1.52 ± 0.52, p = 0.02), but not in LG-SOC or benign serous cystadenoma. Increased level of 8-OHdG in tumor DNA was an independent factor of overall survival in serous ovarian carcinoma upon multivariate analysis (p < 0.01). Increased level of 8-OHdG in tumor DNA indicates poorer overall and progression-free survival durations than counterparts (47.3 vs 105.7 months and 13.5 vs 45.3 months, respectively). Protein levels of hOGG1 were remarkably decreased in HG-SOC (p < 0.01), but not in LG-SOC and serous cystadenoma compared with the tissue adjacent to carcinoma. A positive result on p53 immunostaining was associated with lower hOGG1 expression in HG-SOC (p = 0.04).
Conclusion
Increased 8-OHdG level and decreased expression of hOGG1 in tumor were found in HG-SOC but not LG-SOC. Increased 8-OHdG level in tumor DNA was significantly associated with poorer overall survival and progression-free survival in serous ovarian carcinoma.
Keywords: hOGG1 gene, 8-OHdG, Epithelial ovarian cancer, Serous carcinoma, p53
Background
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancy in USA, and the second rank in China while the incidence rate of EOC is evidently increasing during the past decades [1,2]. The fact that the more aggressive high-grade subgroup accounts for more than 50% of EOCs is regarded as the major reason for the poor survival rate of this disease [3-5]. A recent defined model revealed that high-grade serous ovarian carcinoma (HG-SOC) can progress through greater genetic instability that leads to rapid metastasis without an identifiable precursor lesion aside from a stepwise mutation process in low-grade serous ovarian carcinoma (LG-SOC) [6]. HG-SOC was thought to be driven predominantly by multiple amplification and deletion; and nearly all cases have inactivating mutations of p53 [7], whereas 40% exhibit mutations of BRCA1 or BRCA2 [8], while LG-SOC has few copy number abnormalities, but it frequently exhibits activating mutations of Ras, Raf, and PTEN.
Defects in response to DNA damage was one of the central pathogeneses of human malignancies [9]. BRCA1 and BRCA2, involved in DNA repair and cellular response to DNA damage, were reported to be responsible for approximately 5% to 10% of EOCs in patients with a family history [10]. However, whether other DNA repair-associated genes confer risk to the development of sporadic EOC remains uncertain. The genetic variation in base excision repair (BER) system-induced genetic instability linked to malignancy susceptibility is substantial at present [11,12]. The 8-hydroxy-2′-deoxyguanosine (8-OHdG) is one of the best-characterized oxidized bases. 8-OHdG in DNA could lead to mis-incorporation of adenines opposite the 8-OHdG lesion thus inducing G:C to T:A mutations in genomic DNA. 8-oxoguanine DNA glycosylase 1 (OGG1), a bifunctional glycosylase, is mainly involved in repairing 8-OHdG from oxidative damage. Polymorphisms and loss of heterozygozity of hOGG1 gene, as susceptibility factors for sporadic EOC, have been revealed in several case–control studies [13-16]. In our previous study, we found that variations in the 5′UTR of hOGG1 gene conferred risk to type II but not to type I EOC [17]. We also suggested that hOGG1 variations are more frequent in p53 positive breast and ovarian cancer [18,19].
Studies revealed that increasing 8-OHdG level is associated with higher stage and non-optimal surgical outcome in EOC [20,21], and may confer risk to high grade subtype. Comprehensive mapping of TP53 mutation rates in a homogeneous group of EOC patients revealed that mutant TP53 is a driver mutation in HG-SOC pathogenesis [22]. However, there was still no evidence exploring the relationship between 8-OHdG or hOGG1 alteration and the p53 mutation in serous ovarian cancer.
In the present study, we explored 8-OHdG level and compared hOGG1 mRNA and protein levels in the two types of SOC, benign serous cystadenomas, and normal ovaries (non-tumor control). We also analyzed the relationship between 8-OHdG expression and the survival in serous ovarian cancer. To eliminate differences in base repair ability among individuals, a match pair study of tumor and adjacent non-tumor tissue was conducted.
Materials and methods
Subjects and samples
All patients were clinically diagnosed sporadic ovarian tumor and underwent primary treatment in the Department of Gynecologic Oncology, Jiangsu Institute of Cancer Research (JICR), the University of Nanjing Medical University between January 2003 and December 2010. The exclusion criterions were summarized as below: histology confirmed to be other than ovarian tumor; a history of prior cancer, diabetes; an inability to give informed consent, or was either pregnant or breastfeeding. Patients were genetically unrelated ethnic Han Chinese women from Nanjing City and its surrounding regions. All subjects had complete clinical data or follow-up information as shown in Table 1.
Table 1.
Patient characteristics of the population in the present study
| Characteristic | n (%)/median (range) |
|---|---|
| Age (years) |
61.2 years (26.0 - 82.5) |
| BMI (index) |
26.3 (17.2 - 36.7) |
| Histology, n = 108 |
|
| HG-SOC |
48(44.4 %) |
| LG-SOC |
24(22.2 %) |
| Serous cystadenoma |
20(18.5 %) |
| Normal ovary |
16(14.8 %) |
| FIGO stage, n = 72 |
|
| I |
13(18.0 %) |
| II |
7(9.7 %) |
| III |
37(51.4 %) |
| IV | 15(20.8 %) |
HG-SOC, High-grade serous ovarian cancer.
LG-SOC, Low-grade serous ovarian cancer.
BMI, Body mass index.
FIGO, International federation of gynecology and obstetrics.
Tumor and matched non-tumors ovarian tissues were collected from 24 LG-SOCs, 48 HG-SOC, 20 serous cystadenomas, and 16 non-tumor control ovaries. The median age of patients was 55 years (38 to 77 years). Samples were preserved in RNAlater reagent (QIAGEN) and stored at −80°C until further analysis. Tumor adjacent non-tumor tissue is defined as at least 1 cm far away from cancer tissue boundary and pathologically confirmed non-tumor. Normal samples were pathologically excluded tumor in ovary. All resection specimens were reviewed by two pathologists from JICR (XY. Xu and N. Hou). Tumor tissues available for 8-OHdG level, mRNA and protein expression assays were all tissue slides confirmed for quality control. The study was approved by the Local Ethic Committee of the Jiangsu Cancer Institute, and each patient provided a written consent.
8-OHdG levels in genomic DNA of tissue
Based on the salting out method, DNA extraction from normal ovary, tumor, and adjacent non-tumor tissues (10 mL, with EDTA added to prevent coagulation) was performed within one hour after collection. Eppendorf BioPhotometer Plus (Eppendorf, North America) was used to check the purity of the DNA sample through OD260 nm/OD280 nm, and OD260 nm/OD230 nm. After dissolving into 135 μL water, DNA (200 μg) was mixed with sodium acetate (15 μL, 200 mM) and nuclease P1 (15 μL, 6 units, Sigma, USA) and incubated at 37°C for 30 min. After another 30 min at 37°C, Tris–HCl buffer (15 μL, 1 M, pH 7.4) and alkaline phosphatase (7 μL, 2 units, TAKARA, Shiga, Japan) were added. An ELISA kit (Highly Sensitive 8-OHdG Check, JaICA, Fukuroi, Shizuoka, Japan) was used to measure 8-OHdG according to protocol. Results were converted to 8-OHdG/106 dG according to a previously reported method [23].
Immunohistochemical staining for hOGG1 and p53 in EOC
Polyclonal anti-hOGG1 (1:1000, Novus Biologicals, NB 100–106) and monoclonal anti-p53 (1:100, Novocastra, DO-7) antibodies were obtained commercially. Mouse IgG prepared against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was obtained from Chemicon International, Inc. (Temecula, CA). The hOGG1 immunostaining results were divided semiquantitatively into four groups: 0 = no immunostaining present; 1 = weak immunostaining (< 10% positive staining cells); 2 = moderate immunostaining (10-50% positive staining cells); 3 = strong immunostaining (> 50% positive staining cells). We define “0”, “1” as “negative” and “2”, “3” as “positive” [24]. Immunohistochemical analysis of paraffin-embedded section (IHC-P) staining of p53 was assessed semi-quantitatively using a previously reported standard [17]. Ten percent of neoplastic cells were used as a cutoff from negative expression for p53 immunostaining. Staining evaluation was performed by two independent pathologists.
hOGG1 expression through protein immunoblot
In accordance with the instructions for the total protein extraction kit, total protein was extracted from 100 mg specimens. Protein concentrations were assayed via the Bradford method, and specimens were adjusted to the same protein concentration, packaged, and preserved at −70°C for later use. With a prestained marker serving as an index, required gels were selected after polyacrylamide gel electrophoresis was performed, and a nitrocellulose filter was used for the transfer print. The primary antibody concentration was 1:1000 and the secondary antibody was 1:2000. Using alkaline phosphatase coloration, the protein hybridization band was scanned with a GIS-2020 digital image analysis system and the absorbance value was assayed. The ratios of hOGG1 and GAPDH were calculated for the semi-quantitative analysis.
hOGG1 mRNA expression from EOC tissue
Total RNA from tissue samples was extracted using Trizol (Invitrogen, Carlsbad, CA) and ethanol precipitation, according to the instructions of the manufacturer. Primers and probes for hOGG1 mRNAs were 5′-TGACACCTCACCTCACCCAC-3′, 5′-CACTGTCTTCCGCAAGTTCAC −3′, and TaqMan 5′ (6-FAM)-ACCCTGGTCCGAGGTGTCCCTGAG-(TAMRA-Q) 3′. Primers for β-actin mRNA were 5′- CCAACCGCGAGAAGATGA −3′ and 5′- CCAGAGGCGTACAGGGATAG −3′. Analysis of hOGG1 expression was carried out via RT-PCR. Quantitative detection of hOGG1 mRNA was performed using the Quantitect Multiplex RT-PCR Kit (QIAGEN), following the instructions of the manufacturer. Reaction was performed in a 25 μL solution containing 1x Master mix, 0.25 μL RT mix, 0.4 μM of each primer pair, 0.2 μM probe, and 250 ng RNA. Reaction mixture was reversely transcribed at 50°C for 20 min, and heated at 95°C for 15 min before being amplified, using 40 cycles of thermo cycling, with each cycle at 94°C for 45 s and at 60°C for 1.15 min. Samples were normalized based on β-actin mRNA content. Amplification of β-actin was also performed using the kit form QIAGEN. PCR was performed in a 20 μL solution containing 1x SYBR RT Master Mix buffer, 0.2 μL Quantitect RT mix, 0.4 μM of each primer, and 200 ng RNA. PCR occurred following reverse transcription at 50°C for 20 min and heating at 95°C for 15 min through 35 cycles of amplification, with each cycle at 94°C for 15 s, 48°C for 20 s, and 72°C for 30 s. Tissue samples with more than 150 copies of β-actin mRNA were used for hOGG1 mRNA analyzing (Additional file 1: Figure S1, Additional file 2: Figure S2 and Additional file 3: Figure S3).
Statistical analysis
All statistical analyses were carried out using the statistical program SPSS version 13.0. Descriptive statistical values included mean ± SD values for continuous data and percentages for categorical data. Differences in protein and mRNA levels as well as in 8-OHdG concentrations among subgroups were evaluated using the student t-test, and t-test was used for categorical variables such as immunostaining results. In all cases, a p value of less than 0.05 was considered as statistically significant. The Cox proportional hazards model was used to assess the association between overall survival and clinicopathological characteristics including the 8-OHdG level. The Kaplan–Meier estimate was calculated by the log-rank test for overall survival and progression-free survival, values stratified by 8-OHdG alteration.
Results
8-OHdG levels increased in HG-SOC compared to tumor-adjacent normal tissue
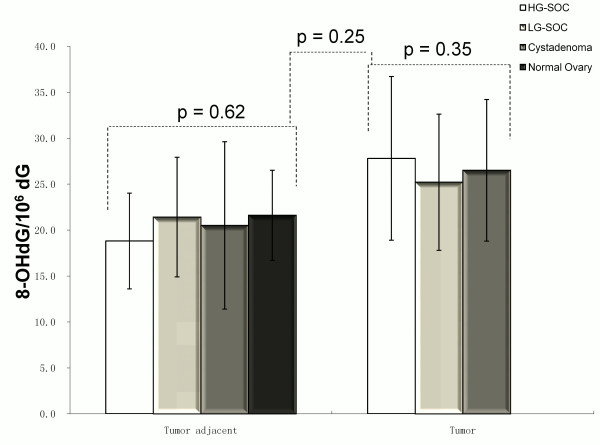
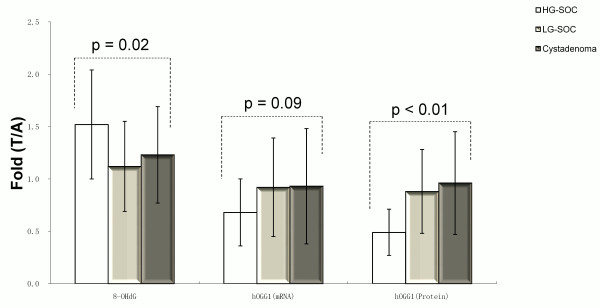
8-OHdG levels in tissue DNA were not statistically different in HG-SOC (27.8 ± 8.98/106dG), LG-SOC (25.2 ± 7.4/106dG), and benign serous cystadenoma (26.5 ± 7.7/106dG; Figure 1, p = 0.35). There was no difference in a parallel investigation of tumor-adjacent normal tissue in HG-SOC (18.8 ± 5.2/106dG), LG-SOC (21.4 ± 6.5/106dG), benign serous cystadenoma (20.5 ± 9.1/106dG) and non-tumor ovary (21.6 ± 4.9/106dG; Figure 1, p = 0.62). Higher fold of 8-OHdG expression in the tumor compared with that in the normal tissue adjacent to carcinoma was found in HG-SOC, but not in LG-SOC or serous cystadenoma (Figure 2, p = 0.02).
Figure 1.
8-OHdG DNA levels in tumors and matched adjacent normal tissues of HG-SOC, LG-SOC, serous cystadenoma, and normal ovaries (mean ± SEM).
Figure 2.
8-OHdG, hOGG1 mRNA and protein expression alteration in tumors and matched adjacent normal tissues of HG-SOC, LG-SOC, serous cystadenoma, and normal ovaries (mean ± SEM) (T, tumor tissue; A, adjacent normal tissue).
hOGG1 gene expression was lower in HG-SOC compared to tumor-adjacent normal tissue
hOGG1 mRNA expression
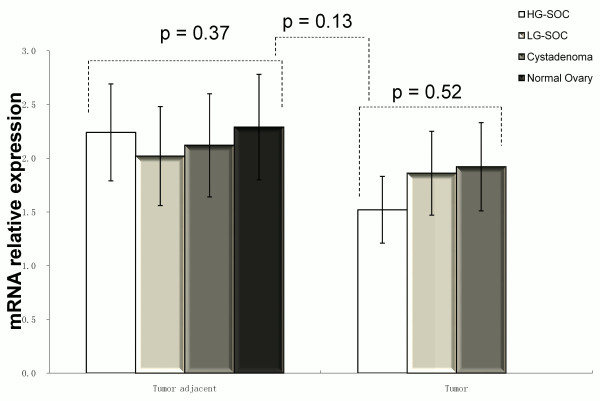
There was no difference of hOGG1 mRNA levels among HG-SOC, LG-SOC, and serous cystadenoma using RT-PCR analysis (p = 0.52, Figure 3). No difference was noted among the tumor-adjacent normal tissues from HG-SOC, LG-SOC, serous cystadenoma, and normal control (p = 0.37, Figure 3). hOGG1 mRNA level in tumors decreased compared with tumor-adjacent normal tissues in HG-SOC, but there was no statistical significance (p = 0.09, Figure 2).
Figure 3.
hOGG1 mRNA levels in tumors and matched adjacent normal tissues of HG-SOC, LG-SOC, serous cystadenoma, and normal ovaries (mean ± SEM).
hOGG1 protein immunoblot
There was no difference of hOGG1 protein expression in HG-SOC, LG-SOC, and serous cystadenoma (p = 0.31, Figure 4), as well as among tumor-adjacent normal tissues, including HG-SOC, LG-SOC, serous cystadenoma, and normal control (p = 0.60, Figure 4). hOGG1 protein expression level in tumors decreased compared with tumor-adjacent normal tissues and the degree was more remarkable in HG-SOC than in LG-SOC and serous cystadenoma (p < 0.01, Figure 2).
Figure 4.
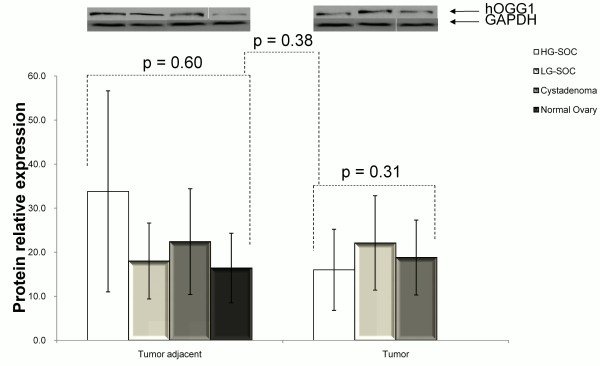
hOGG1 protein expression in tumors and matched adjacent normal tissues of HG-SOC, LG-SOC, serous cystadenoma, and normal ovaries (mean ± SEM). hOGG1 and GAPDH are apparent at 39 kDA and 37 kDa, respectively.
P53 positive-staining is associated with lower hOGG1 expression in HG-SOC
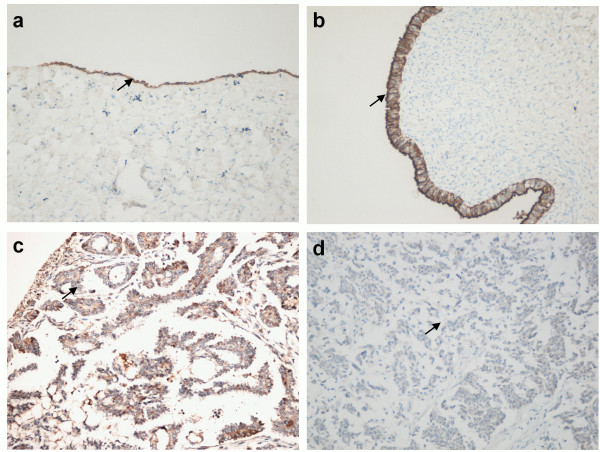
Negative IHC-P staining of hOGG1 was more frequently in HG-SOC than in LG-SOC and serous cystadenoma; however, no statistical significance (p = 0.19, Figure 5) was noted.
Figure 5.
IHC-P analysis of hOGG1 protein expression in serous ovarian cancer: (A) Normal ovarian tissue treated with anti-hOGG1 antibody, biotinylated anti-rabbit IgG secondary antibody, and avidin-biotin-peroxidase complex, and counterstained with p-dimethylaminobenzaldehyde reagent (magnification 100 x); (B) serous cystadenoma with positive staining (magnification 200 x); (C) LG-SOC tissue with moderate positive staining (magnification 200 x); and (D) HG-SOC with negative staining (magnification 200 x). Black arrow pointed at the immunostained epithelial cells.
Compared with that of LG-SOC, the positive-staining rate of p53 in HG-SOC tissues was significantly higher (75.0% vs. 20.8%, p < 0.01). P53 positive-staining was associated with hOGG1 lower expression in HG-SOC (p = 0.04) but not in LG-SOC (p = 0.55, Table 2).
Table 2.
p53 correlated with hOGG1 expression in ovarian cancer
| Subgroup | p53 immunostaining | hOGG1(+) | hOGG1(−) | p value |
|---|---|---|---|---|
| LG-SOC |
(+) |
4(16.7 %) |
1(4.2 %) |
0.55 |
| (−) |
15(62.5 %) |
4(16.7 %) |
||
| HG-SOC |
(+) |
10(20.8 %) |
26(54.2 %) |
0.04 |
| (−) | 8(16.7 %) | 4(8.3 %) |
Increasing 8-OHdG level is associated with poor prognosis in serous ovarian cancer
The Cox proportional hazards analysis revealed that increasing 8-OHdG level independently associated with overall survival in serous ovarian cancer (p < 0.01, Table 3). We further found that increasing 8-OHdG level (p = 0.03 and p = 0.001, Figure 6), but not hOGG1 expression alteration (Additional file 4: Figure S4), in tumor was associated with shorter overall and progression free survivals.
Table 3.
8-OHdG alteration associated with overall survival of serous ovarian cancer patients (N = 72)
|
Characteristic |
Univariate analysis |
p value |
Multivariate analysis |
p value |
||
|---|---|---|---|---|---|---|
| HR | 95 % CI | HR | 95 % CI | |||
| FIGO stage |
|
|
|
|||
| I |
1.00 |
Reference |
|
1.00 |
Reference |
|
| II |
1.42 |
0.68-6.25 |
|
1.21 |
0.43-8.02 |
|
| III |
6.78 |
1.66-18.41 |
|
2.25 |
1.24-6.08 |
|
| IV |
9.43 |
3.61-27.87 |
<0.01 |
3.28 |
1.47-9.54 |
<0.01 |
| Grade |
4.12 |
1.35-12.69 |
<0.01 |
2.57 |
1.62-3.45 |
<0.01 |
| Suboptimal CRS |
2.16 |
1.25-3.85 |
<0.01 |
1.65 |
1.19-2.55 |
0.02 |
| Ascites |
1.47 |
1.26-3.04 |
0.01 |
1.23 |
1.08-2.36 |
0.04 |
| Nadir CA-125 level |
1.02 |
1.01-1.04 |
<0.01 |
1.01 |
1.00-1.03 |
<0.01 |
| p53 positive |
1.30 |
1.09-4.39 |
0.02 |
1.13 |
0.96-2.41 |
0.10 |
| 8-OHdG (T/A) |
1.35 |
1.13-3.42 |
<0.01 |
1.18 |
1.01-2.07 |
<0.01 |
| hOGG1 protein (T/A) |
0.84 |
0.78-1.06 |
0.54 |
0.89 |
0. 88–1.02 |
0.65 |
| Age | 1.01 | 1.00-1.01 | 0.04 | 1.00 | 0.98-1.01 | 0.21 |
HR, Hazard ratio; CI, Confidence interval; CRS, Cytoreductive surgery. T/A, Tumor/Adjacent non-tumor tissue.
Figure 6.
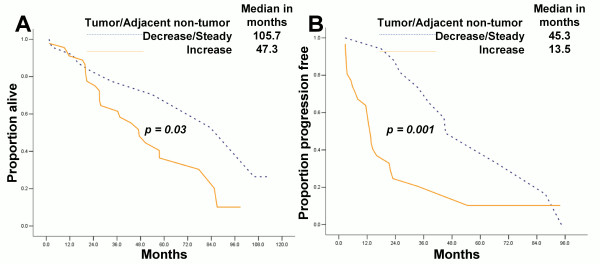
Increasing 8-OHdG expression in tumor DNA is associated with poor prognosis of serous ovarian cancer patients (N = 72). (A) 8-OHdG level associated with overall survival; (B) 8-OHdG level associated with progression-free survival.
Discussion
EOC is a serious health problem among the female population worldwide. Type II subtype of this tumor is responsible for 90% of deaths associated with this disease, and it has different tumorigenesis with type I patients. HG-SOC, the major subtype of type II ovarian cancer, is associated with p53 mutation and decreased ability of DNA damage repair [25,26]. We observed in HG-SOC, but not in LG-SOC and serous cystadenoma, that the average 8-OHdG/106dG level was significantly higher in tumor DNA, and protein expression of hOGG1 was remarkably decreased in the tumors, compared with those in matched non-tumor tissues. The status of p53 immunostaining was also associated with hOGG1 expression in HG-SOC.
Base excision repair, as one of the major DNA damage repair systems, includes several enzymes with cooperative and compensatory enzymatic activities and further associated with the susceptibility of tumor developments. It’s difficult for intuitively estimating the weight of one enzyme dysfunction as an alternative to the overall repair ability. Our results show that no difference existed between hOGG1 mRNA and protein expressions among HG-SOC, LG-SOC, and serous cystadenoma, as well as between tumor-adjacent tissues and normal ovaries. Previous studies led to conflicting conclusions on the relationship between hOGG1 expression and cancer risk. Li et al. [27] found that normal breast tissues from cancer patients had a significantly higher level of oxidative DNA damage. The elevated level of 8-oxo-dG in cancer patients was not related to deficiency of the hOGG1 expression. Other studies reported that hOGG1 gene expression was markedly suppressed in up to 38% of head and neck squamous cell carcinomas [28,29]. In the present research, we used paired specimens from tumor and adjacent non-tumor tissue to eliminate individual differences. We found that 8-OHdG level and hOGG1 protein expression were associated with the grade of SOC.
Serous carcinoma was reported to be correlated with p53 mutation [30]. Diminished base excision repair was also reported in p53 mutant and p53-null cells [31]. These cells exhibited very low activity of DNA β-polymerase, an enzyme required for repair. Immunohistochemical positive status of p53 is commonly thought to correlate closely with mutation status and lost the activity as the tumor suppresses the gene. We found that p53 positive immunostaining is related to decreasing protein level of hOGG1 in HG-SOC. Does p53 dysfunction induce a decrease in hOGG1 expression, or vice versa? Hence, further studies are needed to test the causality relationship between p53 mutation and hOGG1 expression on the carcinogenesis of HG-SOC.
This study also has several drawbacks. Firstly, tumor tissues and adjacent normal tissues were mostly sampled from surgical biopsies. 8-OHdG level and hOGG1 expression might have been influenced by neo-adjuvant chemotherapy in several cases, though our experiment had controls. Secondly, in spite of having tissue slide sections for quality control, non-tumor cells such as lymphocytes and fibroblasts may interfere in tumor cells during 8-OHdG and hOGG1 assays, which also affected the accuracy of our results. Thirdly, increase in oxidative damage products and decrease in damage repair ability linked with advancement in age was reported to be associated with age-related diseases [32,33]. Meanwhile, HG-SOC patients were relatively older than LG-SOC cases [34]. Is this a satellite phenomenon that requires serious consideration?
The poly (ADP-ribose) polymerases (PARPs) are a series of enzymes in DNA repair pathways, especially the BER for DNA single-strand breaks repair. Inhibition of PARPs can lead to cell death and genomic instability in DNA repair-defective tumors (like those with BRAC1/2 mutations), what is so called "synthetic lethality". Hooten et al. [35] identified an interaction between two DNA repair proteins, OGG1 and poly (ADP-ribose) polymerase 1 (PARP-1). They provided evidence that cells are more sensitive to PARP inhibitors as a single agent or in combination with a DNA-damaging agent, H2O2, in the absence of OGG1. These data imply that an identified DNA glycosylase dysfunction or a concurrent hOGG1-targeted therapy may possibly help in clinical trials involving patients with BRCA1/2-mutated or not ovarian carcinoma treated with PARP inhibitors [36-39].
In conclusion, we believe that defects in hOGG1 expression and increase in 8-OHdG levels may have significant influences in the development of HG-SOC. 8-OHdG and hOGG1 levels may be used as molecular markers to distinguish the two types of SOC. Moreover, specific genetic or regulation abnormalities in different EOC subtypes may help in the development of specific drugs.
Abbreviations
EOC: Epithelial ovarian cancer; HG-SOC: High-grade serous ovarian carcinoma; LG-SOC: Low-grade serous ovarian carcinoma; hOGG1: Human 8-oxoguanine (8-OHdG) DNA glycosylase.
Competing interests
All authors declare no conflicts of interest.
Authors’ contributions
XX, XXC and KJLparticipated in drafting the full manuscript and writing of this manuscript. FD and JN partly participated in clinical study design, coordination and data analysis. YW, WWG, YQZ and CML participated in collecting data, creating figures and tables. WWG contributed by writing specific sections of this manuscript. YW, YQZ and CML provided advice and participated in revising the manuscript. XXC participated in substantial contribution to conception and revising it critically for important intellectual content. All the authors in this manuscript have read and approved the final version.
Supplementary Material
Electrophoresis of RT-PCR products of hOGG1.
Amplification plot of hOGG1 mRNA level assay.
Melt curve of hOGG1 mRNA level assay.
hOGG1 protein expression is not associated with prognosis of serous ovarian cancer patients (N = 72). (A) hOGG1 protein level is not associated with overall survival; (B) hOGG1 protein level is not associated with progression-free survival.
Contributor Information
Xia Xu, Email: chenjsch@gmail.com.
Yan Wang, Email: wangyan_9999@sina.com.
Wenwen Guo, Email: hguowen@gmail.com.
Yiqing Zhou, Email: 1332019072@qq.com.
Chunmei Lv, Email: ChunmeiLv@163.com.
Xiaoxiang Chen, Email: cxxxxcyd@Gmail.com.
Kaijiang Liu, Email: Liukaijiang@renji.com.
Acknowledgements
We are grateful to Enpaper’s helpful editing of this manuscript. This work was supported by the Natural Science Foundation of Jiangsu (BK20131439), the Jiangsu Province Institute of Cancer Research Foundation (grant number: ZK201203), The transformation foundation of science and technology achievements in the Xinjiang Uygur Autonomous Region (20120021) and The Key Projects in the National Science & Technology Pillar Program during the Eleventh Five-Year Plan Period (2008BA157B00). Dr. Xiaoxiang Chen is a visiting scientist at MD Anderson Cancer Center, supported by the Jiangsu Health International Exchange Program in 2011.
References
- Jemal A, Siegel R, Xu J. et al. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Yang L, Parkin DM, Ferlay J. et al. Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer Epidemiol Biomarkers Prev. 2005;14:243–250. [PubMed] [Google Scholar]
- Fehrmann RS, Li XY, van der Zee AG. et al. Profiling studies in ovarian cancer: a review. Oncologist. 2007;12:960–966. doi: 10.1634/theoncologist.12-8-960. [DOI] [PubMed] [Google Scholar]
- Wang F, Ye Y, Xu X. et al. CA-125-indicated asymptomatic relapse confers survival benefit to ovarian cancer patients who underwent secondary cytoreduction surgery. J Ovarian Res. 2013;6:14. doi: 10.1186/1757-2215-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Wang Y, Wang F. et al. Nadir CA-125 level as prognosis indicator of high-grade serous ovarian cancer. J Ovarian Res. 2013;6:31. doi: 10.1186/1757-2215-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Ie M, Kurman RJ. Ovarian tumorigenesis: a proposed model based on morphological and molecular genetic analysis. Am J Pathol. 2004;164:1511–1518. doi: 10.1016/S0002-9440(10)63708-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang J, Zhang Z. et al. Cancer stem cells, epithelial-mesenchymal transition, and drug resistance in high-grade ovarian serous carcinoma. Hum Pathol. 2013;44:2373–2384. doi: 10.1016/j.humpath.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluggage WG. My approach to and thoughts on the typing of ovarian carcinomas. J Clin Pathol. 2008;61:152–163. doi: 10.1136/jcp.2007.049478. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Pharoah PD, Stratton JF, Mackay J. Screening for breast and ovarian cancer: the relevance of family history. Br Med Bull. 1998;54:823–838. doi: 10.1093/oxfordjournals.bmb.a011732. [DOI] [PubMed] [Google Scholar]
- Altieri F, Grillo C, Maceroni M. et al. DNA damage and repair: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:891–937. doi: 10.1089/ars.2007.1830. [DOI] [PubMed] [Google Scholar]
- Wilson DM 3rd, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst) 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Tuhkanen H, Anttila M, Kosma VM. et al. Genetic alterations in the peritumoral stromal cells of malignant and borderline epithelial ovarian tumors as indicated by allelic imbalance on chromosome 3p. Int J Cancer. 2004;109:247–252. doi: 10.1002/ijc.11733. [DOI] [PubMed] [Google Scholar]
- Manderson EN, Presneau N, Provencher D. et al. Comparative analysis of loss of heterozygosity of specific chromosome 3, 13, 17, and X loci and TP53 mutations in human epithelial ovarian cancer. Mol Carcinog. 2002;34:78–90. doi: 10.1002/mc.10051. [DOI] [PubMed] [Google Scholar]
- Arcand SL, Provencher D, Mes-Masson AM. et al. OGG1 Cys326 variant, allelic imbalance of chromosome band 3p253 and TP53 mutations in ovarian cancer. Int J Oncol. 2005;27:1315–1320. [PubMed] [Google Scholar]
- Sokhansanj BA, Wilson DM 3rd. Estimating the effect of human base excision repair protein variants on the repair of oxidative DNA base damage. Cancer Epidemiol Biomarkers Prev. 2006;15:1000–1008. doi: 10.1158/1055-9965.EPI-05-0817. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu X, Wang J. et al. Functional polymorphisms of the hOGG1 gene confer risk to type 2 epithelial ovarian cancer in Chinese. Int J Gynecol Cancer. 2011;21:1407–1413. doi: 10.1097/IGC.0b013e31823122c6. [DOI] [PubMed] [Google Scholar]
- Chen X, Wang J, Guo W. et al. Two functional variations in 5'-UTR of hoGG1 gene associated with the risk of breast cancer in Chinese. Breast Cancer Res Treat. 2011;127:795–803. doi: 10.1007/s10549-010-1284-2. [DOI] [PubMed] [Google Scholar]
- Xie H, Xia K, Rong H. et al. Genetic polymorphism in hOGG1 is associated with triple-negative breast cancer risk in Chinese Han women. Breast. 2013;29:707–712. doi: 10.1016/j.breast.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Karihtala P, Soini Y, Vaskivuo L. et al. DNA adduct 8-hydroxydeoxyguanosine, a novel putative marker of prognostic significance in ovarian carcinoma. Int J Gynecol Cancer. 2009;19:1047–1051. doi: 10.1111/IGC.0b013e3181ad0f0d. [DOI] [PubMed] [Google Scholar]
- Pylvas M, Puistola U, Kauppila S. et al. Oxidative stress-induced antioxidant enzyme expression is an early phenomenon in ovarian carcinogenesis. Eur J Cancer. 2010;46:1661–1667. doi: 10.1016/j.ejca.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Ahmed AA, Etemadmoghadam D, Temple J. et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Chen H, Tao J. et al. Association of base excision repair gene polymorphisms with ESRD risk in a Chinese population. Oxid Med Cell Longev. 2012;2012:928421. doi: 10.1155/2012/928421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karihtala P, Kauppila S, Puistola U. et al. Absence of the DNA repair enzyme human 8-oxoguanine glycosylase is associated with an aggressive breast cancer phenotype. Br J Cancer. 2012;106:344–347. doi: 10.1038/bjc.2011.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Shih IM. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer–shifting the paradigm. Hum Pathol. 2011;42:918–931. doi: 10.1016/j.humpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurman RJ, Shih IM. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang W, Zhu J. et al. Oxidative DNA damage and 8-hydroxy-2-deoxyguanosine DNA glycosylase/apurinic lyase in human breast cancer. Mol Carcinog. 2001;31:214–223. doi: 10.1002/mc.1056. [DOI] [PubMed] [Google Scholar]
- Fan CY, Liu KL, Huang HY. et al. Frequent allelic imbalance and loss of protein expression of the DNA repair gene hOGG1 in head and neck squamous cell carcinoma. Lab Invest. 2001;81:1429–1438. doi: 10.1038/labinvest.3780356. [DOI] [PubMed] [Google Scholar]
- Xie B, Freudenheim JL, Cummings SS. et al. Accurate genotyping from paraffin-embedded normal tissue adjacent to breast cancer. Carcinogenesis. 2006;27:307–310. doi: 10.1093/carcin/bgi215. [DOI] [PubMed] [Google Scholar]
- Chan WY, Cheung KK, Schorge JO. et al. Bcl-2 and p53 protein expression, apoptosis, and p53 mutation in human epithelial ovarian cancers. Am J Pathol. 2000;156:409–417. doi: 10.1016/S0002-9440(10)64744-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YR, Fishel ML, Amundson S. et al. Implication of p53 in base excision DNA repair: in vivo evidence. Oncogene. 2002;21:731–737. doi: 10.1038/sj.onc.1205129. [DOI] [PubMed] [Google Scholar]
- Blasiak J, Synowiec E, Salminen A. et al. Genetic variability in DNA repair proteins in age-related macular degeneration. Int J Mol Sci. 2012;13:13378–13397. doi: 10.3390/ijms131013378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabelof DC, Raffoul JJ, Ge Y. et al. Age-related loss of the DNA repair response following exposure to oxidative stress. J Gerontol A Biol Sci Med Sci. 2006;61:427–434. doi: 10.1093/gerona/61.5.427. [DOI] [PubMed] [Google Scholar]
- Vang R, Shih Ie M, Kurman RJ. Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol. 2009;16:267–282. doi: 10.1097/PAP.0b013e3181b4fffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren Hooten N, Kompaniez K, Barnes J. et al. Poly(ADP-ribose) polymerase 1 (PARP-1) binds to 8-oxoguanine-DNA glycosylase (OGG1) J Biol Chem. 2011;286:44679–44690. doi: 10.1074/jbc.M111.255869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin SN, Herzog TJ, Coleman RL. Investigational agents in development for the treatment of ovarian cancer. Invest New Drugs. 2013;31:213–229. doi: 10.1007/s10637-012-9837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audeh MW, Carmichael J, Penson RT. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- Fong PC, Yap TA, Boss DS. et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- Gelmon KA, Tischkowitz M, Mackay H. et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electrophoresis of RT-PCR products of hOGG1.
Amplification plot of hOGG1 mRNA level assay.
Melt curve of hOGG1 mRNA level assay.
hOGG1 protein expression is not associated with prognosis of serous ovarian cancer patients (N = 72). (A) hOGG1 protein level is not associated with overall survival; (B) hOGG1 protein level is not associated with progression-free survival.