Abstract
pbp2b gene alterations were analyzed in 102 clinical isolates of Streptococcus pneumoniae (30 penicillin susceptible, 23 intermediate, and 49 resistant) from Korea. On the basis of PBP2B amino acid sequences, penicillin-nonsusceptible isolates of S. pneumoniae belonged to six groups, and 76% of the isolates in groups I to IV showed the same divergent block of amino acid alterations. Thirteen isolates (group II) also possessed a divergent block that was identical to that of Streptococcus oralis. The pbp2b genes of most Korean isolates showed novel mosaic mutations due to horizontal gene transfer. The Thr252→Ala substitution, previously thought to be associated only with penicillin-nonsusceptible strains, was also found in three penicillin-susceptible strains. On the basis of their pbp2b nucleotide sequences, all penicillin-nonsusceptible isolates can be detected by multiplex PCR, which can be used as a novel method for detection of antibiotic-resistant pneumococcal strains in clinical specimens.
The increasing prevalence of antimicrobial resistance in Streptococcus pneumoniae has become a serious concern in medical practice since the 1990s. Korea was reported to be one of the countries with the highest prevalence of penicillin resistance among pneumococcal isolates (22). The rapid emergence of pneumococcal resistance in Korea was partly due to the spread of resistant clones that had been introduced from other countries (22, 23).
The classic mechanism of penicillin resistance in pneumococci is alteration of penicillin-binding proteins (PBPs), especially PBP1A, -2X, and -2B (3, 20). The pbp2b transpeptidase-encoding region (TER) sequences of penicillin-nonsusceptible S. pneumoniae isolates were highly divergent from those of penicillin-susceptible S. pneumoniae (PSSP) isolates (7, 20). In addition, considerable variations in the genetic structure of the pbp2b genes of penicillin-resistant strains were also reported, which are due to horizontal gene transfer by secondary transformation (2, 5, 7). Previous reports documented that the pbp2b genes of penicillin-nonsusceptible strains could be classified into two major classes (A and B) based on their nucleotide and amino acid sequences (7, 24). Class A genes showed more extensive variations than class B genes (7). However, some pneumococcal strains did not belong to these classes and cannot be detected by class-specific PCR assays (24, 25). Genetic analysis of pbp2b sequences in penicillin-resistant strains were reported from Japan, Spain, Iceland, Canada, France, the United Kingdom, Papua New Guinea, South Africa, and the United States (7, 10, 11, 17, 19, 20, 21, 25). In Korea, the pbp2b gene sequences of multidrug-resistant S. pneumoniae isolates were reported (23). This study was performed to characterize the genetic structure of the pbp2b gene of penicillin-nonsusceptible strains of S. pneumoniae from Korea, as well as to explore a potential new method for detection of penicillin-resistant pneumococci based on common mutation patterns of the pbp2b gene.
Bacterial isolates.
One hundred two isolates of S. pneumoniae that were obtained from Korean patients between 1998 and 2002 were analyzed in this study. MICs of penicillin were determined by the NCCLS agar dilution method (15, 16). Of 102 isolates, 30 were penicillin susceptible (MIC, ≤0.06 μg/ml), 23 were penicillin intermediate (MIC, 0.125 to 1.0 μg/ml), and 49 were penicillin resistant (MIC, ≥2.0 μg/ml).
Amplification and sequencing of the pbp2b gene.
The 1.5-kb DNA fragments encoding the PBP2B TER were amplified from the chromosomal DNAs of S. pneumoniae isolates by PCR. Template DNA for PCR was prepared by the simple boiling-lysis method. Briefly, colonies were suspended in lysis buffer (100 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, 1% Triton X-100) and incubated at 70°C for 10 min. The mixture was then centrifuged for a moment, and the aqueous phase was used as a template for PCR. The primers used for amplification, which were designed on the basis of the published sequences of strain R6, were as follows: forward (1-25), 5′-GAT CCT CTA AAT GAT TCT CAG GTG G-3′; reverse (1504-1480), 5′-CAA TTA GCT TAG CAA TAG GTG TTG G-3′. PCR was carried out in a final volume of 100 μl containing 10 mM Tris-HCl (pH 8.8), 50 mM KCl, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphate, 1 μM each oligonucleotide primer, 2.5 U of Taq polymerase, and 1 to 10 ng of DNA template. The PCR conditions used were 30 cycles of 95°C for 1 min, 55°C for 2 min, and 72°C for 2 min. PCR products were purified with a Core-One purification kit (CoreBio System, Seoul, Korea) for sequencing. DNA sequences were determined with an ABI prism Rhodomine terminator cycle sequencing kit (PE Biosystems, Foster City, Calif.) and an ABI 377 automated sequencer (PE Biosystems). Amino acid sequences were deduced by the MegAlign program in DNASTAR (DNASTAR, Madison, Wis.). Nucleotide and deduced amino acid sequences were also analyzed with the MegAlign program in DNASTAR. The amino acid sequences were aligned and compared with the corresponding sequences of PSSP laboratory strain R6 (GenBank accession no. AJ243051).
Amino acid alterations of pbp2b genes.
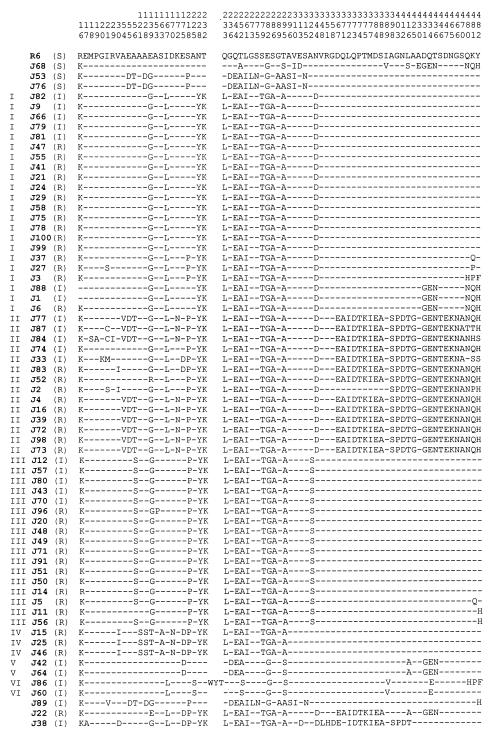
Most of the isolates of PSSP had nucleotide and amino acid sequences highly homologous to those of R6, with more than 99.4% nucleotide and amino acid similarity. However, the PBP2B sequences of three PSSP isolates (J68, J53, and J76) contained several amino acid substitutions, as in penicillin-nonsusceptible isolates. These isolates were confirmed as susceptible on the basis of repeated MIC tests. The amino acid sequences of J68, which were highly diverged from those of R6, were very similar to those of two penicillin-intermediate S. pneumoniae (PISP) isolates, J42 and J64 (94.5 and 97.4% nucleotide and amino acid similarity, respectively) (Fig. 1). Two other PSSP isolates, J53 and J76, showed pbp2b sequences nearly identical to that of a penicillin-intermediate isolate, J89 (99.9 and 99.6% nucleotide and amino acid similarity, respectively). However, these sequence changes observed in PSSP isolates were not related to penicillin resistance.
FIG. 1.
Deduced amino acid sequences encoded by the pbp2b genes of Korean PSSP and PNSP isolates. Only those amino acids that differ from the sequences of R6 are shown. Dashes indicate residues identical to those of R6. The numbers of sites represented above are according to Dowson et al. (7). The roman numeral to the left of each isolate number is the group that the isolate belongs to. The letter in parentheses to the right of each isolate number is the isolate's susceptibility to penicillin (S, susceptible, I, intermediate; R, resistant). The sequences of 27 PSSP isolates that differ from the R6 sequence at one or two sites are not shown. Amino acid abbreviations: A, alanine; R, arginine; N, asparagine; D, aspartic acid; C, cysteine; E, glutamic acid; Q, glutamine; G, glycine; H, histidine; I, isoleucine; L, leucine; K, lysine; M, methionine; F, phenylalanine; P, proline; S, serine; T, threonine; W, tryptophan; Y, tyrosine; V, valine.
As a whole, the pbp2b sequences of penicillin-nonsusceptible pneumococcal isolates from Korea showed a maximum of 16.6 and 11.1% dissimilarity with respect to that of R6 on the nucleotide and amino acid levels, respectively. Of the 467 amino acid sequences analyzed in this study, 71 positions of penicillin-nonsusceptible pneumococcal isolates differed from those of R6 (Fig. 1). Of these 71 substitutions, 41 amino acid alterations were noted exclusively in penicillin-nonsusceptible isolates. In particular, the amino acid alterations Thr252→Ala, Glu282→Gly, and Thr295→Ala were noted in all of the PRSP and most of the PISP isolates, while Thr252→Ser or Thr295→Ser was also noted in some PISP strains. Previous studies reported that Thr252→Ala was a common and specific mutation only in penicillin-nonsusceptible strains (6, 18, 20). However, our data documented the Thr252→Ala mutation in three penicillin-susceptible isolates, J68, J53, and J76, and replacement of Thr at position 252 or 295 with Ser instead of Ala in two penicillin-intermediate isolates, J89 and J86. Replacement of Glu with Gly at position 282 was also noted in all penicillin-susceptible strains (Fig. 1). An insertion of nine nucleotides was detected in J86, which was also reported in some Japanese strains (25).
Grouping of penicillin-nonsusceptible isolates on the basis of amino acid sequences.
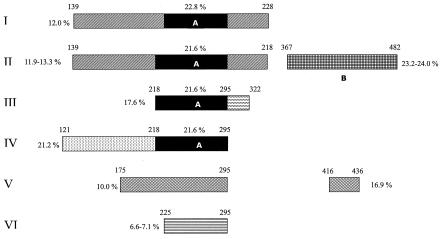
Of 72 penicillin-nonsusceptible pneumococcal isolates, 69 belonged to six groups based on the amino acid sequence encoded by the pbp2b gene. Groups I (15 PRSP, 7 PISP), II (9 PRSP, 4 PISP), and III (12 PRSP, 5 PISP) were major groups, while groups IV (3 PRSP), V (2 PISP), and VI (2 PISP) included fewer isolates (Fig. 1). Three isolates (J22, J38, and J89) with distinct sequence variations did not belong to any group. Compared with the PBP2B sequence of R6, all group I isolates showed the same 13 amino acid alterations, i.e., Glu139→Gly, Ile167→Leu, Asn228→Tyr, Thr232→Lys, Gln233→Leu, Gln244→Glu, Thr252→Ala, Leu261→Ile, Ser279→Thr, Glu282→Gly, Ser286→Ala, Thr296→Ala, and Asn344→Asp. However, these amino acid substitutions were not specific for group I isolates. Two penicillin-susceptible isolates (J53 and J76) also showed some of the alterations found in group I isolates (Glu139→Gly, Gln244→Glu, Thr252→Ala, Leu261→Ile, Glu282→Gly, and Thr295→Ala). The most remarkable finding in the pbp2b sequences of Korean pneumococcal isolates was a divergent block (block A, positions 228 to 295) common to all of the isolates in groups I to IV (Fig. 2). Although this divergent block A was not found in group V and VI isolates, it was present in 76% (55 of 72) of the penicillin-nonsusceptible isolates from Korea.
FIG. 2.
Schematic representation of the divergent blocks of PBP2B in groups of penicillin-nonsusceptible pneumococcal isolates from Korea. The numbers above the ends of the blocks are amino acid positions (7). Percentages indicate nucleotide dissimilarities of corresponding divergent blocks from R6. Block A is a divergent block common to groups I to IV, and block B is a sequence similar to that of penicillin-resistant S. oralis 3626.
Group II isolates showed another blocked mutation (block B, positions 367 to 482), except for strain J2 (Fig. 1 and 2). The sequences of block B were closely related to those of penicillin-resistant Streptococcus oralis 3626 (MIC, 4 μg/ml) (Fig. 3) (8). Part of the block B sequence (position 398 to the 3′ end) was also identical to those of an Icelandic isolate, IC221 (19), and a French isolate, SP22861 (10). Previous reports have already documented that alterations in the pbp genes of S. pneumoniae are caused by interspecies recombinational event from S. oralis or S. mitis to S. pneumoniae (12, 13, 14). Our data also confirmed that there would be an interspecies recombinational process between S. oralis and S. pneumoniae on the basis of the pbp2b sequences of group II isolates.
FIG. 3.
Alignment of amino acid sequences (positions 367 to 482) corresponding to block B of the pbp2b gene. One isolate each of groups I and II to VI, 14 isolates of group II, and S. oralis 3626 are represented. Sequences identical to those of R6 are represented by dots.
Nine isolates that belonged to group II (J77, J87, J84, J4, J16, J39, J72, J98, and J73) had the short mosaic sequence V-D-T at positions 54 to 56 (Fig. 1). The nucleotide and amino acid sequences of group III isolates corresponded to those of class B isolates, as reported by Dowson et al. (7). Group IV isolates showed another distinct block (121 to 175) in addition to block A (218 to 295) (Fig. 2). There were characteristic alterations in the amino acid sequences of group V isolates (Thr295→Ser and Leu416→Ala) and group VI isolates (Ala225→Ser, Thr252→Ser, Thr295→Ser, and Ile389→Val).
The PBP2B amino acid sequences of group V isolates were identical to those of three Icelandic and French isolates, IC100, IC141, and SP1053. The whole pbp2b gene sequences of isolates of groups I, II, IV, and VI did not match any published sequences according to the BLAST search (1). It suggested that novel mosaic mutations of pbp2b due to horizontal gene exchange are related to reduced susceptibility to penicillin in Korean isolates (4, 7, 8, 9, 19, 20).
Multiplex PCR to detect penicillin-nonsusceptible isolates.
Rapid and sensitive detection of penicillin-nonsusceptible pneumococcal isolates is very critical in the treatment of various pneumococcal diseases, especially pneumococcal meningitis. Direct detection of penicillin-nonsusceptible pneumococcal isolates from clinical specimens without conventional culture methods could make targeted antimicrobial treatment against antibiotic-resistant pneumococci possible from the start of therapy.
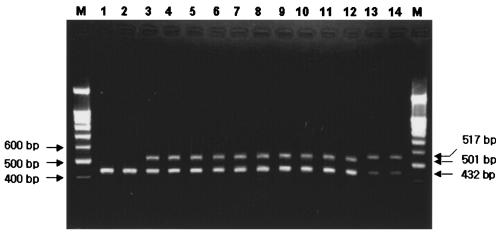
On the basis of the pbp2b sequences determined in this study and retrieved from a public database (GenBank), we developed a multiplex PCR method to detect penicillin-nonsusceptible isolates. Two forward primers (149-F1, 5′-TTA TGC AGT TGC TTT GAA T-3′; 155-F2, 5′-AAC CCW AAR ACA GGT GCT-3′; nucleotides 447 to 465 and 463 to 480 in R6, respectively) and one reverse primer (322-R, 5′-GTT ATC AAA CTG CCC AAA GGC-3′; nucleotides 963 to 984 in R6) were used in the multiplex PCR. Primer 149-F1 is specific for group II isolates, while primer 155-F2 corresponds to all other penicillin-nonsusceptible isolates. Another primer set amplifying the pneumolysin (ply) gene (432 bp) and known to be specific for S. pneumoniae (Ply-F, 5′-GCT CCG ATG ACT TAT AGT AT-3′; Ply-R, 5′-ACA CTC GAA ATA TAG ACC AA-3′) was simultaneously applied. The multiplex PCR conditions used were 30 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s. A multiplex PCR with five primers (149-F1, 155-F2, 322-R, Ply-F, and Ply-R) generated two amplicons (432 and 517 bp for group II and 432 and 501 bp for the other penicillin-nonsusceptible isolates) in penicillin-nonsusceptible isolates and only one amplicon (432 bp) from penicillin-susceptible ones (Fig. 4). The multiplex PCR used in this study not only confirmed the presence of S. pneumoniae but also differentiated all penicillin-nonsusceptible pneumococcal isolates from penicillin-susceptible ones.
FIG. 4.
Multiplex PCR amplification of penicillin-nonsusceptible pneumococcal isolates. Lanes 1 and 2 are penicillin-susceptible strains. Lanes 3 to 14 are penicillin-nonsusceptible strains. Lanes: M, 100-bp ladder marker; 1, R6 (reference strain); 2, J23 (penicillin-susceptible strain); 3, J3 (group I); 4, J82 (group I); 5, J43 (group III); 6, J80 (group III); 7, J25 (group IV); 8, J46 (group IV); 9, J42 (group V); 10, J64 (group V); 11, J60 (group VI); 12, J86 (group VI); 13, J72 (group II); 14, J73 (group II).
In conclusion, penicillin-nonsusceptible pneumococcal isolates from Korea showed relatively homogeneous pbp2b gene nucleotide and amino acid sequence patterns. Sequence variations of pbp2b in Korean isolates were due to novel mosaic mutations due to horizontal gene transfer. The successful detection of all penicillin-nonsusceptible strains by multiplex PCR demonstrated in this study can be developed into a novel diagnostic method for use in clinical practice.
Nucleotide sequence accession numbers.
The pbp2b gene sequences determined in this study have been deposited in the GenBank database and assigned accession no. AY515392 to AY515397.
Acknowledgments
This study was partly supported by a grant from the Korea Health 21 R&D Project, Ministry of Health and Welfare, Seoul, Republic of Korea (01-PJ10-PG6-01GM03-0002), and by a Samsung Biomedical Research Institute grant (C-A3-214-1).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffey, T. J., S. Berron, M. Daniels, M. E. Garcia-Leoni, E. Cercenado, E. Bouza, A. Fenoll, and B. G. Spratt. 1996. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988-1994): novel major clones of serotypes 14, 19F, and 15F. Microbiology 142:2747-2757. [DOI] [PubMed] [Google Scholar]
- 3.Coffey, T. J., M. Daniels, L. K. McDougal, C. G. Dowson, F. C. Tenover, and B. G. Spratt. 1995. Genetic analysis of clinical isolates of Streptococcus pneumoniae with high-level resistance to expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 39:1306-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffey, T. J., C. G. Dowson, M. Daniels, J. Zhou, C. Martin, B. G. Spratt, and J. M. Musser. 1991. Horizontal transfer of multiple penicillin-binding protein genes and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol. Microbiol. 5:2255-2260. [DOI] [PubMed] [Google Scholar]
- 5.Coffey, T. J., C. G. Dowson, M. Daniels, and B. G. Spratt. 1993. Horizontal spread of an altered penicillin-binding protein 2B gene between Streptococcus pneumoniae and Streptococcus oralis. FEMS Microbiol. Lett. 110:335-340. [DOI] [PubMed] [Google Scholar]
- 6.Dowson, C. G., T. J. Coffey, C. Kell, and R. A. Whiley. 1993. Evolution of penicillin resistance in Streptococcus pneumoniae: the role of Streptococcus mitis in the formation of a low affinity PBP2B in S. pneumoniae. Mol. Microbiol. 3:635-643. [DOI] [PubMed] [Google Scholar]
- 7.Dowson, C. G., A. Hutchison, J. A. Branningan, R. C. George, D. Hansman, J. Linares, A. Tomasz, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowson, C. G., A. Hutchison, J. A. N. Woodford, A. P. Johnson, R. C. George, and B. G. Spratt. 1990. Penicillin-resistant viridans streptococci have obtained altered penicillin-binding protein genes from penicillin-resistant strains of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 87:5858-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., and B. G. Spratt. 1999. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol. Biol. E vol. 16:1687-1695. [DOI] [PubMed] [Google Scholar]
- 10.Ferroni, A., and P. Berche. 2001. Alterations to penicillin-binding proteins 1A, 2B, and 2X amongst penicillin-resistant clinical isolates of Streptococcus pneumoniae serotype 23F from the nasopharyngeal flora of children. J. Med. Microbiol. 50:828-832. [DOI] [PubMed] [Google Scholar]
- 11.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hakenbeck, R. 1998. Mosaic genes and their role in penicillin-resistant Streptococcus pneumoniae. Electrophoresis 19:597-601. [DOI] [PubMed] [Google Scholar]
- 13.Hakenbeck, R., K. Kaminski, A. Köning, M. van der Linden, J. Paik, P. Reichmann, and D. Zahner. 1999. Penicillin-binding proteins in β-lactam resistant Streptococcus pneumoniae. Microb. Drug Resist. 5:91-99. [DOI] [PubMed] [Google Scholar]
- 14.Hakenbeck, R., A. Köning, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, fifth edition. Approved standard M7-A5. National Committee for Clinical Laboratory Standard, Villanova, Pa.
- 16.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing. Eleventh informational supplement M100-S11. National Committee for Clinical Laboratory Standard, Villanova, Pa.
- 17.Nichol, K. A., G. G. Zhanel, and D. J. Hoban. 2002. Penicillin-binding protein 1A, 2B, and 2X alterations in Canadian isolates of penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:3261-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reichmann, P., A. König, J. Liñares, F. Alcaide, F. C. Tenover, L. McDougal, S. Swidsinski, and R. Hakenbeck. 1997. A global gene pool for high-level cephalosporin resistance in commensal Streptococcus species and Streptococcus pneumoniae. J. Infect. Dis. 176:1001-1012. [DOI] [PubMed] [Google Scholar]
- 19.Sá-Leão, R., S. E. Vihelmsson, H. de Lencastre, K. G. Kristinsson, and A. Tomasz. 2002. Diversity of penicillin-nonsusceptible Streptococcus pneumoniae circulating in Iceland after the introduction of penicillin-resistant clone Spain6B-2. J. Infect. Dis. 186:966-975. [DOI] [PubMed] [Google Scholar]
- 20.Smith, A. M., and K. P. Klugman. 1995. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 39:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, A. M., K. P. Klugman, T. J. Coffey, and B. G. Spratt. 1993. Genetic diversity of penicillin-binding protein 2B and 2X genes from Streptococcus pneumoniae in South Africa. Antimicrob. Agents Chemother. 37:1938-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song, J. H., N. Y. Lee, S. Ichiyanma, R. Yoshida, Y. Hirakata, W. Fu, A. Chongthaleong, N. Aswapokee, C. H. Chiu, M. K. Lalitha, K. Thomas, J. Perera, T. T. Yee, F. Jamel, U. C. Warsa, B. X. Vinh, M. R. Jacobs, P. C. Appelbaum, and C. H. Pai. 1999. Spread of drug-resistant Streptococcus pneumoniae in Asian countries; Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Clin. Infect. Dis. 28:1206-1211. [DOI] [PubMed] [Google Scholar]
- 23.Song, J. H., J. W. Yang, J. H. Jin, S. W. Kim, C. K. Kim, H. Lee, K. R. Peck, S. M. Kim, N. W. Lee, M. R. Jacobs, P. C. Appelbaum, and The Asian Network for Surveillance of Resistant Pathogens (ANSORP) Study Group. 2000. Molecular characterization of multidrug-resistant Streptococcus pneumoniae isolates in Korea. J. Clin. Microbiol. 38:1641-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ubukata, K., Y. Asahi, A. Yamane, and M. Konno. 1996. Combinational detection of autolysin and penicillin-binding protein 2B gene of Streptococcus pneumoniae by PCR. J. Clin. Microbiol. 34:592-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamane, A., H. Nakano, Y. Asahi, K. Ubukata, and M. Konno. 1996. Directly repeated insertion of 9-nucleotide sequence detected in penicillin-binding protein 2B gene of penicillin-resistant Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:1257-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]