Abstract
Autophagy regulates cell death both positively and negatively, but the molecular basis for this paradox remains inadequately characterized. We demonstrate here that transient cell-to-cell variations in autophagy can either promote cell death or survival depending on the stimulus and cell type. By separating cells with high and low basal autophagy by flow cytometry, we demonstrate that autophagy determines which cells live or die in response to death receptor activation. We have determined that selective autophagic degradation of the phosphatase Fap-1 promotes Fas apoptosis in Type I cells. Conversely, autophagy inhibits apoptosis in Type II cells or upon treatment with TRAIL in either Type I or II cells. These data illustrate that differences in autophagy in a cell population determine cell fate in a stimulus- and cell type-specific manner. This example of selective autophagy of an apoptosis regulator may represent a general mechanism for context-specific regulation of cell fate by autophagy.
INTRODUCTION
Macroautophagy (hereafter autophagy) is a catabolic process that facilitates cell survival in response to stress by providing nutrients, biosynthetic monomers and by mitigating cellular damage1, 2. Several studies have suggested that autophagy is capable of regulating apoptosis but, surprisingly, autophagy can both promote or inhibit cell death in different cellular contexts3, 4. The molecular underpinnings of this duality remain poorly defined despite the fact that they have important implications in human disease5–7. Despite many links between specific proteins of the autophagy and apoptosis pathways, surprisingly little is known about how the overall process of autophagy determines whether cells live or die in response to cell death stimuli8–11. Apoptosis is known to control autophagy (both positively and negatively) through molecular mechanisms that have been described12–14 and many autophagy regulators also control the apoptotic apparatus15–18. However, mechanisms responsible for regulation of apoptosis by the overall process of autophagy are less clear19–21. Except in the case of salivary gland cell death in Drosophila22 and the autophagic degradation of catalase23, precise mechanisms responsible for direct promotion of cell death by autophagy are unknown.
In populations of cells treated with apoptotic stimuli some cells will escape death for reasons that have only recently been addressed but which have important clinical consequences, particularly in cancer therapy. Non-genetic heterogeneity, stochastic state differences and variation in levels of apoptotic proteins between cells have recently received attention as determinants of cell fate that govern which cells live and which die in a population24–26 but underlying cellular processes that alter or regulate these activities have not been identified. We hypothesized that basal variability in autophagy could determine cell fate by altering levels of critical apoptosis regulators. Here, we reveal high steady-state variability in basal autophagy in a cell population, which acts as a non-genetic determinant of cell fate through the selective autophagic degradation of a key apoptosis regulatory protein. This provides an example of how variation in autophagy can regulate cell fate and identifies a specific mechanism by which autophagy can promote apoptosis in a cell type and stimulus-specific manner.
RESULTS
Quantitative cell-to-cell differences in basal autophagy in a homogeneous cell population
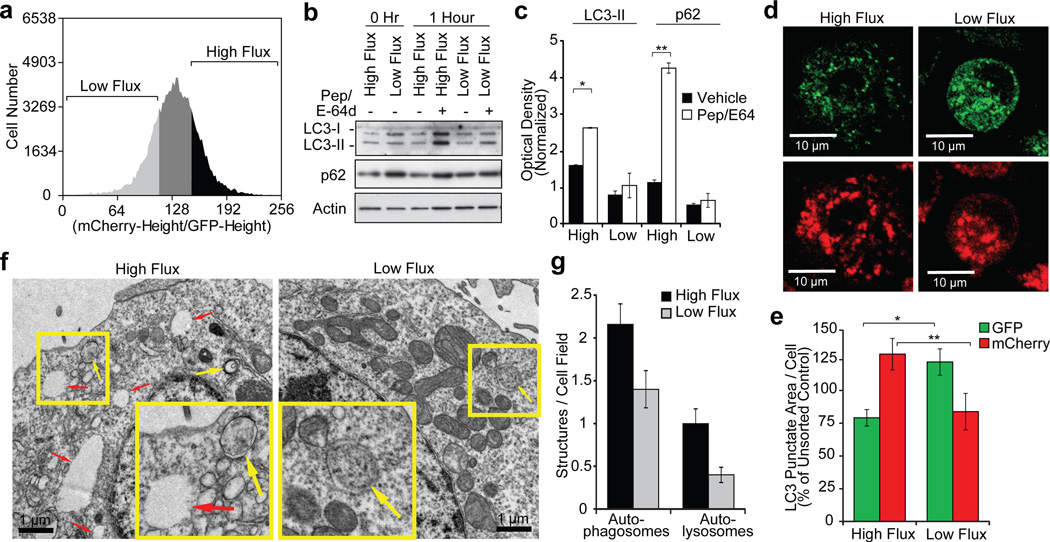
Differences in basal autophagy have been associated with certain oncogenes but the role of role of basal autophagy in cancer cell death has not been examined27, 28. Stochastic variability in critical apoptotic proteins has been identified as a determinant of cell fate24, 26. Therefore, variability in a cellular process capable of altering the levels of apoptotic proteins would also be predicted to determine cell fate. We sought to quantitate stochastic differences in basal autophagy in a cell population and determine the role of these differences in basal autophagy on cell death in response to specific apoptotic stimuli. To accomplish this, we used flow cytometry to sort cells based on their relative levels of autophagic flux using mCherry-EGFP-LC3 as a reporter29 (Supplementary Fig. 1a). This reporter for autophagic flux takes advantage of the higher sensitivity of EGFP fluorescence to the acidic environment of the autolysosome relative to mCherry30: cells with higher flux are less green due to autophagosome fusion with lysosomes, thereby increasing the mCherry/EGFP ratio (Fig. 1a, Supplementary Figs. 1a, b). This method to measure flux has been extensively validated and accurately quantitates autophagic flux induction by multiple stimuli and chemical and genetic inhibition of autophagy (Fig. 1, Supplementary Figs. 1, 2). To examine differences between high and low autophagic flux cells under basal conditions, BJAB B-cell lymphoma cells were maintained near log phase in growth medium, harvested and flow sorted into low and high flux populations (Fig. 1a). Immunoblots for LC3 and p62 autophagy proteins confirmed the relative levels of flux present in the sorted cells (Fig. 1b,c) and quantitation of autophagosomes and lysosomes by saponin extraction31 followed by flow cytometry revealed substantial differences in the lysosome and autolysosome number between the high and low flux subpopulations (Supplementary Fig. 1d–f). Furthermore, fluorescent and electron microscopy further substantiated that the cells differ in their levels of autophagic flux (Figs. 1d–g, Supplementary Figs. 2a, b), and that these differences in flux were independent of cell size, cell cycle or spontaneous apoptosis (sub-G1 events) (Supplementary Figs. 2c–f). Importantly, we found that differences in basal flux were transient, persisting for at least 4 hours after sorting but returning back to steady state by 24 hours (Figs. 2a–c); this rapid reversion of the sorted cells indicates that genetic heterogeneity in the starting population was not responsible for differences in flux. Together, these data show that substantial differences in autophagic flux exist within cell populations, even under optimal growth conditions, likely due to transient stochastic fluctuations in the level or activity of key autophagy regulators.
Figure 1. Log-phase proliferating cells in optimal growth media exhibit significant steady-state differences in autophagic flux.
a, b, BJAB lymphoma cells stably expressing mCherry-GFP-LC3 were serially cultured at log phase followed by FACS sorting for cells with high and low autophagic flux using the ratio of mCherry/GFP (a). The high and low 20% were sorted (a), replated and treated with lysosomal protease inhibitors pepstatin and E-64d for 1 hour; lysates were then immunoblotted for the indicated proteins (b). c, Densitometry of LC3-II and p62 westerns (normalized to actin and hour 0, mean ± s.e.m., n=3 blots from 2 independent experiments, *p=0.051, **p=0.0091). d, e, HeLa Cherry-GFP-LC3 cells were sorted as in (a), cytospun onto slides, fixed and visualized by confocal microscopy (d); autophagic LC3 puncta were assessed by quantitative microscopy (e) (punctate area per cell, mean±s.e.m., n=50 fields, *p=0.010, **p=0.053). f, Electron micrographs of HeLa mCherry-GFP-LC3 cells sorted for autophagic flux as in (a). Yellow arrows denote autophagosomes; red arrows indicate autolysosomes. g, Quantitation of autophagosomes and autolysomes from electron micrographs (mean ± s.e.m., n=50 fields).
Figure 2. Differences in basal autophagic flux are transient but determine apoptotic response in a stimulus-specific manner.
a–c, BJAB lymphoma cells stably expressing mCherry-GFP-LC3 were serially cultured at log phase followed by FACS sorting for cells with high and low autophagic flux using the ratio of mCherry/GFP. Cells were then re-plated in growth medium and autophagic flux was again measured by flow cytometry at the indicated timepoints (median ratio of mCherry/GFP fluorescence ± s.e.m., n=3 wells). Lysates from cells harvested at the indicated timepoints were immunoblotted with the indicated antibodies (b). Representative flow cytometry histograms for cells at 0 hours and 24 hours after sorting (c). d, BJAB mCherry-GFP-LC3 cells were sorted for autophagic flux as in (a) (top and bottom 20%). Following treatment with Fas ligand (1.5 ng/mL) or TRAIL (4 ng/mL), cell viability was determined by MTS assay at 24 hours (% of untreated control, mean ± s.e.m., n=3 wells, *p=2.3×10−4, **p=0.0036). e, Long term growth of BJAB mCherry-GFP-LC3 cells sorted for autophagic flux followed by treatment with Fas ligand (4 ng/mL) or TRAIL (15 ng/mL) for 24 hours. Cells were then re-plated and allowed to recover for 5 (Fas Ligand) or 6 (TRAIL) days then assayed for viability (% of no ligand control, mean ± s.e.m., n=3 wells, *p=0.012). f, BJAB mCherry-GFP-LC3 cells were sorted for autophagic flux as above and apoptosis was measured at 1 hour by flow cytometry using AnnexinV and DAPI (mean ± s.e.m., n=3 wells, *p=0.0046). g, cells sorted as in (a), were re-plated and, starting at the indicated times following sorting, treated with Fas ligand (4 ng/mL) for 24 hours; cell viability was then determined by MTS (% of untreated control, mean ± s.e.m., n=3 wells).
Differences in basal autophagic flux dictate the apoptotic response
To establish whether cell-to-cell differences in basal autophagic flux control cell fate, we treated flow-sorted high and low autophagic flux populations with the death receptor agonists Fas ligand and TRAIL. BJAB cells with higher basal autophagic flux were more sensitive to Fas ligand-induced apoptosis than low flux cells (Figs. 2d, e, Supplementary Fig. 3a). Surprisingly, this was not found with TRAIL treatment– high and low autophagy cells (sorted at the same time from the same population) tended towards the opposite in their sensitivity to TRAIL (Figs. 2d, e, Supplementary Fig. 3b). Caspase-3/7 activity, AnnexinV staining and inhibition by the caspase inhibitor zVAD, all indicated that the cell death we observed was apoptotic (not necrotic or autophagic) but independent of Bid (Fig. 2f, Supplementary Figs. 3c–f, 4a, b). Similar to the transience of high and low autophagic flux observed in sorted cells (Figs. 2a–c), the differential response to Fas-induced apoptosis between sorted high and low flux populations persisted for 4–6 hours and returned to steady state by 24 hours (Fig. 2g). These data indicate that transient differences in the level of basal autophagy between cells in a population determine the response to death receptor agonists. Furthermore, autophagy modulated apoptosis in opposing directions depending on the stimulus; high autophagy cells were more sensitive to Fas ligand-induced apoptosis but less sensitive to TRAIL, while low autophagy cells showed the opposite. While the effect of autophagy on TRAIL-induced apoptosis was generally cytoprotective, the differences were less pronounced and consistent than those with Fas ligand.
Autophagy facilitates apoptosis in a stimulus and cell type-specific manner
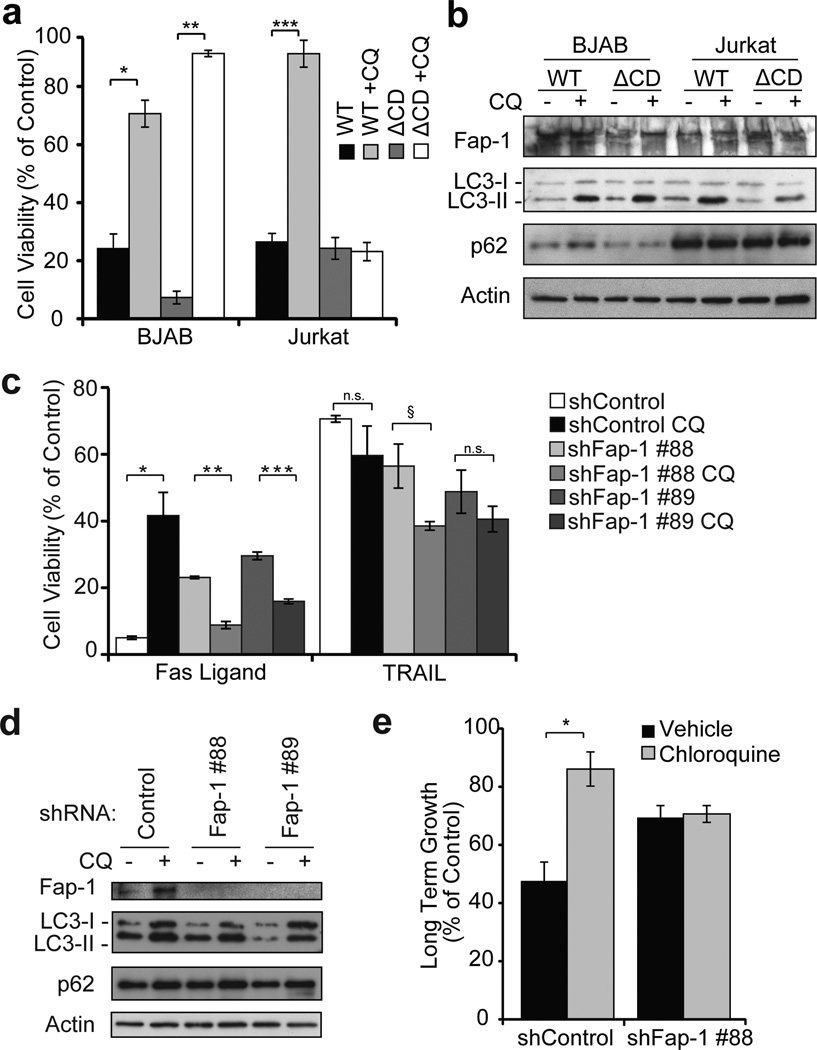
Consistent with our results in sorted low autophagy cells, pharmacologic and genetic inhibition of autophagy in BJAB cells resulted in increased viability upon treatment with Fas ligand but not TRAIL or other cytotoxic drugs. To assess whether the effect of autophagy on Fas apoptosis was cell type-specific, we tested the effect of autophagy inhibition with chloroquine on Fas apoptosis in Jurkat A3 cells and found that, contrary to what we observed in BJAB cells, autophagy inhibition instead caused a modest decrease in cell viability in Jurkat cells treated with Fas ligand (Figs. 3a–c). Inhibition of autophagy by shRNA knockdown of Atg5 or Atg7 also resulted in significant inhibition of cell death induced by Fas ligand in the BJAB cells but had no substantial effect on TRAIL-induced killing (Figs. 3d–f). Knockdown of Atg5 or Atg7 led to no difference in cell death with either Fas ligand or TRAIL in Jurkat cells (Figs. 3d–f). Furthermore, combined genetic and pharmacologic inhibition via Atg5 knockdown and chloroquine treatment had no additive effect on Fas ligand-induced death in BJAB cells (Supplementary Fig. 4e). Autophagy inhibition via transient overexpression of autophagy dominant-negative Atg4b-C74A or Rab7-Q79L mutants also reduced BJAB cell death in response to Fas ligand (Supplementary Fig. 4d). Conversely, treatment of BJAB cells with autophagy inducers (EBSS and trehalose) led to more cell death in response to Fas ligand treatment despite some reduced cell viability in the absence Fas ligand (Supplementary Figs. 4f, g). Together these data show that autophagy is necessary for efficient cell killing by Fas ligand (but not TRAIL) in BJAB cells, but autophagy is dispensable for Fas ligand-induced apoptosis in Jurkat cells and is dispensable for TRAIL-induced apoptosis in both cell types.
Figure 3. Autophagy inhibition suppresses Fas ligand induced cell killing in a cell type-specific manner.
a, BJAB and Jurkat cells were treated with vehicle or chloroquine (BJAB 20 μM, Jurkat 10 μM) for 16 hours followed by Fas ligand (1.5 ng/mL). Cell viability was determined by MTS 24 hours later (% of control (no ligand), mean ± s.e.m., n=3 wells, *p=0.0058). b, Immunoblots of cells in (a, c) probed for the indicated antibodies. c, BJAB and Jurkat cells were treated with chloroquine (BJAB 20 μM, Jurkat 10 μM) for 16 hours followed by treatment with Fas ligand (4 ng/mL) for 24 hours (same technical replicate as (a)). Cells were then re-plated at low density in growth media and allowed to recover for 5 days, then assayed for viability (% of no ligand control, mean ± s.e.m., n=3 wells, *p=0.0024). d, BJAB and Jurkat cells were transduced with the indicated lentiviral shRNA constructs, followed by 3 days of puromycin selection. Selected cells were plated and treated with Fas ligand (1.5 ng/mL) or TRAIL (4 ng/mL) for 24 hours and viability was assessed by MTS (% of control (no ligand), mean ± s.e.m., n=3, *p=1.7×10−4, **p=0.024). e, Immunoblots for Atg5, Atg7 and p62 confirm protein depletion and autophagy inhibition in (d, f). f, BJAB and Jurkat cells expressing control or Atg5 shRNA were treated with Fas ligand (15 ng/mL) or TRAIL (12.5 ng/mL) for 24 hours (same technical replicate as (d)). Cells were then re-plated at low density and allowed to recover for 5–6 days then assayed for viability (% of no ligand control, mean ± s.e.m., n=3 wells, *p=0.0089). Atg7 knockdown data were not included for long term viability due to growth suppressive effect of Atg7 knockdown in the absence of drug treatment.
Autophagy modulates Fas-induced death via the phosphatase Fap-1
The tyrosine phosphatase Fap-1 (Fas-associated phosphatase 1 or PTPN13, PTP-L1) specifically modulates Fas but not TRAIL signaling through dephosphorylation of Fas, which reduces its cell surface expression and activity32–35. Fap-1 has been implicated in several cancers including liver cancer in which autophagy has been implicated as a tumor suppressor and Fas plays a major role in liver cell apoptosis36–39. Fas-induced apoptosis can occur in two ways – with (Type II) or without mitochondrial involvement (Type I)40–42. Reduced Fap-1 expression in Type II cells is responsible (at least in part) for phenotypic differences between Fas Type I and Type II cells39. Flow-sorted low autophagic flux BJAB cells (Type I) displayed higher Fap-1 levels than high flux cells that correlated with higher cell viability (Figs. 4a, b) and increased Fas receptor at the cell surface (Supplementary Fig. 5a). This was not seen in Jurkat cells (Type II), which express nearly undetectable levels of Fap-143 (Figs. 4a, b). Type I SKW6.4 cells and Type II CEM-CCRF (CEM) cells displayed a similar pattern of cell viability with autophagy inhibition and Fas ligand treatment as the BJAB and Jurkat cells (Figs. 4c, d) and Fap-1 protein levels were substantially higher in the Type I cells than in the Type II cells (Fig. 4c). When autophagy was blocked by chloroquine or shRNA knockdown of Atg5, Atg7 or Vps34, Fap-1 levels increased in the Type I cells, correlating with the reduced level of Fas ligand-induced apoptosis (Figs. 4e, f). No Fap-1 was detected with or without autophagy inhibition in the Type I Jurkat cells (Fig. 4c, f, Supplementary Fig. 5b). Conversely, autophagy acted in a cytoprotective manner in Jurkat cells with autophagy gene knockdown leading to decreased viability in response to Fas ligand. Together, these data led us to hypothesize that autophagy was modulating Fas apoptosis by selectively degrading Fap-1.
Figure 4. Autophagy facilitates Fas apoptosis in Type I cells and correlates with Fap-1 expression.
a–b, BJAB and Jurkat cells stably expressing mCherry-GFP-LC3 were flow sorted for high and low autophagic flux, treated with Fas ligand (1.5 ng/mL) for 24 hours and viability was assessed by MTS (a) (% of no ligand control, mean ± s.e.m., n=3 wells, *p=0.0046). b, Immunoblots following sorting of the samples in (a). c–d, The indicated cell lines were treated with chloroquine (BJAB, CEM 20 μM; SKW6.4, 25 μM; Jurkat, 10 μM) for 16 hours, followed by Fas ligand (BJAB, 12.5 ng/mL; SKW6.4, 50 ng/mL; Jurkat, 0.4 ng/mL; CEM, 40 ng/mL). Lysates from cells harvested after chloroquine treatment were immunoblotted with the indicated antibodies (c). Cell viability was assessed by MTS 24 hours following Fas ligand treatment (d) (% of control (no ligand), mean ± s.e.m., n=3 wells, *p=2.7×10−4, **p=1.6×10−4). e–f, BJAB and Jurkat cells were transduced with control, Atg5, Atg7 or Vps34 shRNA lentiviruses, followed by 3 days of puromycin selection. Cells were then treated with Fas ligand (1.25 ng/mL) or TRAIL (1.25 ng/mL) for 24 hours and viability was assessed by MTS (e) (% of control (no ligand), mean ± s.e.m., n=3 wells, *p=1.4×10−6, **p=2.3×10−5, ***p=3.9×10−5, §p=9.2×10−6, §§p=3.4×10−6, §§§p=1.7×10−5). Immunoblots demonstrate Atg5, Atg7 and Vps34 knockdown, autophagy inhibition and altered Fap-1 levels (f).
To determine the role for Fap-1 in autophagy modulation of autophagy we sought to determine whether removal of Fap-1 could make BJAB cells respond to Fas ligand like Type II cells and if exogenous expression of Fap-1 could make the Jurkat cells respond to Fas ligand like Type I cells. As predicted, overexpression of wild-type (WT) Fap-1 caused Jurkat cells to behave more like Type I cells in response to autophagy inhibition and Fas ligand, while expression of catalytically inactive (ΔCD) Fap-1 had no effect; WT or ΔCD Fap-1 had no effect on the BJAB cells (Figs. 5a, b). Conversely, Fap-1 shRNA knockdown abrogated the increase in cell death caused by chloroquine in response to Fas ligand in Type I BJAB cells (Figs. 5c–e, Supplementary Figs. 5c, d) but did not have a substantial effect on TRAIL-induced viability (Figs. 5c). These results indicate that Fap-1 is necessary and sufficient for autophagy to modulate Fas-induced apoptosis.
Figure 5. Autophagy facilitates apoptosis via selective degradation of Fap-1.
a–b, BJAB and Jurkat cells expressing the indicated Fap-1 wild-type (WT) and catalytically-inactive (ΔCD) constructs were treated with chloroquine (BJAB, 20 μM; Jurkat, 10 μM) for 16 hours followed by Fas ligand (BJAB, 1.5 ng/mL; Jurkat 15 ng/mL) for 24 hours. Cell viability was determined by MTS (a) (% of control (no ligand), mean ± s.e.m., n=3 wells, *p=2.7×10−4, **p=2.4×10−5, ***p=0.0024). Cell lysates were blotted and probed with the indicated antibodies (b). c–e, BJAB cells expressing control or Fap-1 shRNAs were treated with 20 µM chloroquine for 16 hours followed by Fas ligand (12.5 ng/mL) or TRAIL (25 ng/mL) for 24 hours; cell viability was then determined by MTS (c) (% of control (no ligand), mean ± s.e.m., n=3 wells, *p=4.8×10−6, **p=8.1×10−5, ***p=8.0×10−4, §p=0.013, n.s. p>0.05). Immunoblots of lysates harvested following chloroquine treatment (d). Long term growth was assessed in cells from (c) by re-plating at low density and allowing to recover for 5 days, followed by viability assay (% of untreated control, mean ± s.e.m., n=3 wells, *p=0.043) (e).
Fap-1 is targeted for autophagic degradation via the selective autophagy adaptor p62
The adaptor protein p62/SQSTM1 and its family members regulate selective autophagy of specific cargoes5, 44–48. Furthermore, p62 associates with the death inducing signaling complex (DISC) and modulates death receptor signaling18, 44. To determine if Fap-1 autophagic degradation occurs via p62, we depleted p62 protein, which resulted in increased Fap-1 and concomitant inhibition of Fas ligand-induced apoptosis (Figs. 6a, b). We were also able to co-immunoprecipitate endogenous Fap-1 and p62 in a chloroquine and Fas ligand-dependent manner (Fig. 6c, Supplementary Fig. 5e). Therefore, p62 associates directly with Fap-1 enabling it to recruit Fap-1 for autophagic degradation in response to Fas receptor activation. Together, these data show that p62-mediated recruitment of Fap-1 to autophagosomes regulates the apoptotic response to Fas activation.
Figure 6. p62 is required for Fap-1 degradation by autophagy and p62 and Fap-1 interact directly.
a–b, Cell viability was determined by MTS assay following Fas ligand (4 ng/mL) treatment in BJAB cells expressing control or p62 lentiviral shRNAs (a) (% of control (no ligand), mean ± s.e.m., n=3 wells, *p=3.2×10−5, **p=1.8×10−6). Immunoblots confirm p62 depletion and Fap-1 levels (b). c, Co-immunoprecipitation of endogenous p62 and Fap-1 in BJAB cells treated with 20 µM chloroquine for 16 hours, followed by treatment with Fas ligand (50 ng/mL) for 2 hours at 4 °C. d, Model of the mechanism for autophagy promotion of Fas apoptosis in Type I cells.
DISCUSSION
Our results demonstrate that transient and significant differences in autophagic flux are present at baseline in a population of actively proliferating cells. This cell-to-cell variability in autophagy leads to substantial differences in apoptosis induced by Fas ligand and TRAIL such that high and low autophagy cells from the same population display opposing sensitivities to these two death ligands. These differences arise even under optimized cell culture conditions with clonally derived cells; we predict that such differences may be magnified in a more heterogeneous population of cells exposed to greater fluctuations in nutrients and environmental stresses that affect autophagy, such as those in a tumor.
Autophagy can thus determine which cells live and which cells die in a population but these effects are both stimulus- and cell type-specific. Our work addresses an area that has been confusing – does autophagy protect or kill cells?49 Our data show that even in the same population of cells, it can do both and it is the context (i.e. specific stimulus and cell type) that determines whether autophagy is pro- or anti-apoptotic. We don’t know specifically or generally how autophagy is able to generally protect against apoptosis in our system although it has been reported that autophagy can degrade activated Caspase-8 to protect against TRAIL-induced death in other contexts19. However, our work does reveal a defined mechanism by which autophagy promotes apoptosis in mammalian cells. For Fas ligand, autophagy-dependent differences in apoptotic response depend on p62-mediated selective autophagic degradation of Fap-1 (Fig. 6d). This model explains both the cell type specific differences whereby only Type I cells require autophagy for efficient apoptosis and the stimulus specific effect whereby autophagy promotes Fas-induced death but not TRAIL-induced death. Our work here establishes a molecular mechanism for an autophagy-dependent switch from apoptosis inhibition to promotion and provides an example whereby stochastic variability in autophagy can determine cell fate. These findings also underscore the importance of autophagy in cancer therapy, specifically in the case of cancers where Fap-1 is implicated50 and more generally by demonstrating that autophagy can switch from a death promoter to a death inhibitor in different contexts. Current attempts to modulate autophagy in a clinical setting51 are focused on using autophagy inhibition combined with other anti-cancer drugs and are the focus of over three dozen clinical trials. Such trials should consider the implications of context-specific promotion and inhibition of apoptosis by autophagy.
Supplementary Material
Acknowledgements
We thank all members of the Thorburn lab for thoughtful discussions and critical comments. We are grateful to K. Helm, L. Acosta and C. Childs of the University of Colorado Cancer Center Flow Cytometry Core for their guidance and assistance. We also thank D. Dill and the University of Colorado Anschutz Medical Campus (UCAMC) Electron Microscopy Core for E.M. images and technical assistance, and R. Moldovan at the UCAMC Advanced Light Microscopy Core for assistance with confocal imaging. This work was supported by National Institutes of Health grants R01 CA111421 and CA150925 (A.T.), HL68628 (D.W.H.R.), and Shared Resources supported by P30 CA46934. J.M.G. was previously supported by 5T32CA82086-10 (UCAMC Department of Pediatrics) and is now an American Cancer Society Postdoctoral Fellow.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions J.M.G. and A.T. designed the study; J.M.G. and L.S. performed experiments; A.B. and D.W.H.R. provided Fap-1 reagents and expertise; J.M.G. designed experiments and analyzed data; J.M.G. and A.T. wrote the manuscript which was commented on by all authors.
The authors declare no competing financial interests.
REFERENCES
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maycotte P, Thorburn A. Autophagy and cancer therapy. Cancer Biol Ther. 2011;11:127–137. doi: 10.4161/cbt.11.2.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White EJ, et al. Autophagy regulation in cancer development and therapy. Am J Cancer Res. 2011;1:362–372. [PMC free article] [PubMed] [Google Scholar]
- 5.Dikic I, Johansen T, Kirkin V. Selective autophagy in cancer development and therapy. Cancer Res. 2010;70:3431–3434. doi: 10.1158/0008-5472.CAN-09-4027. [DOI] [PubMed] [Google Scholar]
- 6.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 8.Gump JM, Thorburn A. Autophagy and apoptosis: what is the connection? Trends Cell Biol. 2011;21:387–392. doi: 10.1016/j.tcb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- 10.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 11.Yonekawa T, Thorburn A. Autophagy and cell death. Essays Biochem. 2013;55:105–117. doi: 10.1042/bse0550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norman JM, Cohen GM, Bampton ET. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010;6:1042–1056. doi: 10.4161/auto.6.8.13337. [DOI] [PubMed] [Google Scholar]
- 13.Luo S, et al. Bim Inhibits Autophagy by Recruiting Beclin 1 to Microtubules. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirawan E, et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. doi: 10.1038/cddis.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radoshevich L, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinstein AD, Eisenstein M, Ber Y, Bialik S, Kimchi A. The autophagy protein Atg12 associates with antiapoptotic Bcl-2 family members to promote mitochondrial apoptosis. Mol Cell. 2011;44:698–709. doi: 10.1016/j.molcel.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Yousefi S, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 18.Jin Z, et al. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 19.Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891–900. doi: 10.4161/auto.6.7.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young MM, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fullgrabe J, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McPhee CK, Logan MA, Freeman MR, Baehrecke EH. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta PB, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 25.Kreso A, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaudet S, Spencer SL, Chen WW, Sorger PK. Exploring the contextual sensitivity of factors that determine cell-to-cell variability in receptor-mediated apoptosis. PLoS Comput Biol. 2012;8:e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo JY, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaminskyy VO, Piskunova T, Zborovskaya IB, Tchevkina EM, Zhivotovsky B. Suppression of basal autophagy reduces lung cancer cell proliferation and enhances caspase-dependent and -independent apoptosis by stimulating ROS formation. Autophagy. 2012;8:1032–1044. doi: 10.4161/auto.20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hundeshagen P, Hamacher-Brady A, Eils R, Brady NR. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 31.Eng KE, Panas MD, Hedestam GB, McInerney GM. A novel quantitative flow cytometry-based assay for autophagy. Autophagy. 2010;6 doi: 10.4161/auto.6.5.12112. [DOI] [PubMed] [Google Scholar]
- 32.Sato T, Irie S, Kitada S, Reed JC. FAP-1: a protein tyrosine phosphatase that associates with Fas. Science. 1995;268:411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, et al. Negative regulation of Fas-mediated apoptosis by FAP-1 in human cancer cells. Int J Cancer. 2000;87:473–479. doi: 10.1002/1097-0215(20000815)87:4<473::aid-ijc3>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 34.Ivanov VN, et al. FAP-1 association with Fas (Apo-1) inhibits Fas expression on the cell surface. Mol Cell Biol. 2003;23:3623–3635. doi: 10.1128/MCB.23.10.3623-3635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov VN, Ronai Z, Hei TK. Opposite roles of FAP-1 and dynamin in the regulation of Fas (CD95) translocation to the cell surface and susceptibility to Fas ligand-mediated apoptosis. J Biol Chem. 2006;281:1840–1852. doi: 10.1074/jbc.M509866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takamura A, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ying J, et al. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20:1173–1175. doi: 10.1038/sj.leu.2404193. [DOI] [PubMed] [Google Scholar]
- 38.Yeh SH, et al. Genetic characterization of fas-associated phosphatase-1 as a putative tumor suppressor gene on chromosome 4q21.3 in hepatocellular carcinoma. Clin Cancer Res. 2006;12:1097–1108. doi: 10.1158/1078-0432.CCR-05-1383. [DOI] [PubMed] [Google Scholar]
- 39.Schickel R, Park SM, Murmann AE, Peter ME. miR-200c regulates induction of apoptosis through CD95 by targeting FAP-1. Mol Cell. 2010;38:908–915. doi: 10.1016/j.molcel.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol. 2003;15:185–193. doi: 10.1016/s1044-5323(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 41.Algeciras-Schimnich A, et al. Molecular ordering of the initial signaling events of CD95. Mol Cell Biol. 2002;22:207–220. doi: 10.1128/MCB.22.1.207-220.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz I, Walczak H, Krammer PH, Peter ME. Differences between CD95 type I and II cells detected with the CD95 ligand. Cell Death Differ. 1999;6:821–822. doi: 10.1038/sj.cdd.4400569. [DOI] [PubMed] [Google Scholar]
- 43.Meinhold-Heerlein I, et al. Expression and potential role of Fas-associated phosphatase-1 in ovarian cancer. Am J Pathol. 2001;158:1335–1344. doi: 10.1016/S0002-9440(10)64084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 46.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective Autophagy of the Adaptor Protein Bcl10 Modulates T Cell Receptor Activation of NF-kappaB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ichimura Y, Komatsu M. Selective degradation of p62 by autophagy. Semin Immunopathol. 2010;32:431–436. doi: 10.1007/s00281-010-0220-1. [DOI] [PubMed] [Google Scholar]
- 48.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 49.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–2688. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu JH, et al. Protein tyrosine phosphatase PTPN13 negatively regulates Her2/ErbB2 malignant signaling. Oncogene. 2008;27:2525–2531. doi: 10.1038/sj.onc.1210922. [DOI] [PubMed] [Google Scholar]
- 51.Amaravadi RK, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.