Abstract
Penicillium marneffei is an emerging opportunistic dimorphic fungal pathogen that is endemic in Southeast Asia. A typing method based on the analysis of size polymorphisms in microsatellite loci was investigated. Three loci available from the GenBank database were identified to harbor microsatellites. PCR primers flanking the microsatellite repeats were designed with one primer in the set fluorescently labeled. PCR products were then sized by automated capillary electrophoresis. As expected for a haploid fungus, a single band was observed for each microsatellite locus for all isolates. Polymorphic microsatellite marker (PMM) analysis detected a total of 22 different allelic types for 35 isolates of P. marneffei with a high discriminatory power (D = 0.956). Microsatellites I, II, and III detected 14, 10, and 7 alleles, respectively. The reproducibility of length polymorphisms was confirmed by using different DNA preparations from the same isolate or by repeated runs from the same DNA preparation. PMM profiles for eight isolates passaged in vitro for 7 to 8 weeks were identical to the original culture, demonstrating short-term stability and reproducibility. PCR products were not observed for other dimorphic fungi or human DNA. Comparison of allelic frequencies in isolates obtained from China and Thailand identified distinct allele combinations, suggesting the potential geographic isolation of populations. Due to the high discriminatory power, reproducibility, and potential for high throughput, PMM analysis may provide a good typing method for epidemiologic and surveillance investigations of P. marneffei.
The fungus Penicillium marneffei is a dimorphic, opportunistic pathogen for humans. The fungus is believed to be haploid, with at least six large chromosomes with a genome size estimated from 17.8 to 26.2 Mb (51). P. marneffei was first isolated by Capponi et al. (9) from hepatic abscess tissue samples obtained from an infected bamboo rat, Rhizomys sinensis, from Vietnam. The first naturally acquired clinical case of human penicilliosis by P. marneffei was described in 1973 in a patient with Hodgkin's disease after travel to Southeast Asia (15). P. marneffei is endemic in Southeast Asia, especially in Southern China, Thailand, Vietnam, Laos, Malaysia, and Indonesia. P. marneffei can cause widespread disseminated infections in persons with defects in cell-mediated immunity such as are associated with AIDS (12, 22). The emergence of this pathogen in Southeast Asia closely parallels the increase in the incidence of human immunodeficiency virus infections in this region (12). For instance, in Thailand, P. marneffei is currently the fourth most common opportunistic infection in human immunodeficiency virus-positive persons and has been classified as an AIDS-defining pathogen (26, 42, 43). In comparison, nearly 10% of the persons with AIDS living in Hong Kong have been infected by P. marneffei (50). Although most cases of penicilliosis, P. marneffei has been reported in persons with impaired cell-mediated immunity, infections have also been reported in immunocompetent individuals, especially in persons with a history of travel to Southeast Asia (16). Until highly active antiretroviral therapy becomes available to treat patients with AIDS, it is likely that there will be a continuing increase in the incidence of P. marneffei infections in many countries in regions where this organism is endemic (10).
Although progress has been made on understanding the P. marneffei genome (51) and control of the dimorphic transition (2), little is known regarding either its environmental sources or routes of transmission. In nature, P. marneffei has been isolated from four species of bamboo rats, as well from soil and feces samples obtained from bamboo rat burrows (1). When case-control studies were performed to identify risk factors for P. marneffei infections, neither exposure to, nor consumption of, bamboo rats was identified as a significant risk factor for infection (11). Cases were found associated with recent occupational exposure to soil especially during the rainy season. Routes of transmission from environmental sources to humans remain obscure. Acquisition of P. marneffei may be by three potential routes: through skin abrasions, via the digestive tract or, most commonly, by inhalation of conidia followed by dissemination (23).
Molecular strain typing of clinical and environmental isolates of P. marneffei may provide answers to important public health questions, including the sources and routes of transmission, the existence of pathogenic or drug-resistant strains, the genetic relatedness of isolates, and the persistence of strains after relapse. Epidemiologic investigations could be facilitated by better methods of typing; however, fast, reliable, and discriminatory methods are lacking. Biotyping methods have been used to type 32 strains of P. marneffei into 17 biotypes (49). By testing the susceptibility to five antifungal agents, eight antibiotypes were observed for 20 isolates obtained from 11 patients. In contrast, the cellular fatty acid methyl profiles of these isolates were similar (24). Although these methods are considered generally easy to perform for laboratories without access to sophisticated equipment, phenotypic methods are prone to problems such as low discriminatory power and, more importantly, phenotypic variability. Genotypic methods include restriction endonuclease analysis with HaeIII (47). A total of 46 isolates were differentiated into two types based on HaeIII digestion patterns. Discriminatory power was increased by using a combination of six RAPD [random(ly) amplified polymorphic DNA] primers to type 20 isolates obtained from patients in Taiwan (24). Eight different RAPD patterns were observed. However, RAPD profiles have been shown to be unstable in Aspergillus fumigatus or too sensitive to reaction conditions (4, 27). Macrorestriction digestion profiles of NotI restriction fragments resolved by pulsed-field gel electrophoresis for 69 P. marneffei isolates obtained from AIDS patients in Thailand detected 54 patterns (46). Although it provided good discriminatory power, NotI restriction fragment length polymorphism analysis was reported to lack concordance with HaeIII analysis. The method requires specialized equipment not available in many laboratories and tedious sample preparation (46).
Methods that are easy and rapid to perform, reproducible, and highly discriminatory and that facilitate interlaboratory comparisons are the hallmarks for a good typing procedure (41). Microsatellites are short 2- to 10-bp multiple tandem repeats found dispersed throughout all eukaryotic genomes (44). Microsatellite markers have been found to be useful for molecular typing since length polymorphisms are often detected between isolates within the same species. The molecular mechanism for producing differences in allele sizes is primarily due to polymerase slippage errors (40). Microsatellites have successfully been used to type other pathogenic fungi, including Aspergillus fumigatus (3, 27), Coccidioides immitis (19), Saccharomyces cerevisiae (21), and especially Candida albicans (6, 13, 14, 18),
The goals of this investigation were to identify and evaluate the use of polymorphic microsatellite marker (PMM) analysis as a method for molecular typing of isolates of P. marneffei. Instead of screening a genomic library, P. marneffei microsatellites were identified by examining sequences deposited in the GenBank database. Three PMM loci were chosen from P. marneffei sequences deposited in the GenBank databases for typing since they were shown to provide a high degree of reproducibility and discriminatory power.
MATERIALS AND METHODS
Isolates.
The 35 isolates of P. marneffei and their sources used in this investigation are listed in Table 1. Ten isolates (B-6317 to B-6326) were provided by the West China Hospital of Sichuan University, Chengdu, China. Ten isolates (IFM 47279 to 47282 and IFM 47284 to 47289) were provided by Kazuko Nishimura, Research Center for Pathogenic Fungi and Microbial Toxicoses, Chiba, Japan. Five isolates (NCPF 4090, 4147, 4157, 4158, and 4176) were obtained from the National Collection of Pathogenic Fungi held at the PHLS Mycology Reference Laboratory, Bristol, United Kingdom. The remaining isolates of P. marneffei were obtained from the culture collection maintained by the Mycotic Diseases Branch at the Centers for Disease Control and Prevention, as were isolates of Candida albicans, Candida tropicalis, Blastomyces dermatitidis, Histoplasma capsulatum, Sporothrix schenckii, Trichophyton rubrum, Aspergillus fumigatus, and A. flavus. Species identification of P. marneffei isolates was based on the ability of the culture to convert from yeast to mold and on the production of a red pigment and by morphological identification of colonies and conidiophores (12, 38, 49). Isolates were stored in frozen glycerol phosphate buffer (27) or 8% (vol/vol) dimethyl sulfoxide (Sigma, St. Louis, Mo.). Cultures were maintained either as yeast by incubating slants at 37°C or at 25°C for mycelial phase cells on Bacto brain heart infusion agar (Becton Dickinson, Sparks, Md.).
TABLE 1.
P. marneffei isolates, isolate sources, and genotypes determined by PMM analysis
| Isolatea | Source | Geographic location | PMM type |
|---|---|---|---|
| ATCC 64101* | Human | China | 1 |
| ATCC 64102* | Rat | China | 2 |
| NCPF 4090 | Human | Hong Kong | 3 |
| NCPF 4147 | Human | Not known | 4 |
| NCPF 4157 | Human | Thailand | 4 |
| NCPF 4158 | Human | Thailand | 5 |
| NCPF 4176 | Human | Not known | 6 |
| B-6317 | Human | China | 7 |
| B-6318 | Human | China | 8 |
| B-6319 | Human | China | 7 |
| B-6320 | Human | China | 7 |
| B-6321 | Human | China | 7 |
| B-6322 | Human | China | 9 |
| B-6323 | Human | China | 10 |
| B-6324* | Human | China | 11 |
| B-6325 | Human | China | 12 |
| B-6326 | Human | China | 12 |
| ATCC 201564 | Soil | Thailand | 13 |
| ATCC 24100 | Human | Not known | 14 |
| ATCC 58950 | Human | Not known | 15 |
| ATCC 200050 | Human | Thailand | 13 |
| ATCC 200051 | Human | Thailand | 16 |
| ATCC 201013* | Human | Thailand | 13 |
| ATCC 18224 | Rat | Vietnam | 17 |
| IFM 47279* | Human | Thailand | 19 |
| IFM 47280 | Human | Thailand | 13 |
| IFM 47281 | Human | Thailand | 13 |
| IFM 47282 | Human | Thailand | 18 |
| IFM 47284* | Human | Thailand | 20 |
| IFM 47285 | Human | Thailand | 13 |
| IFM 47286* | Human | Thailand | 21 |
| IFM 47287 | Human | Thailand | 22 |
| IFM 47288* | Human | Thailand | 16 |
| IFM 47289* | Human | Thailand | 13 |
| ATCC 56573 | Human | Not known | 6 |
Isolates used to evaluate PMM stability are indicated by an asterisk.
Isolation of genomic DNA.
The surface of potato dextrose agar (Difco, Detroit, Mich.) plates was inoculated with P. marneffei, followed by incubation at 25°C for 1 to 3 weeks to allow conidial production. Conidia were harvested by gently rubbing the surface of the plates with sterile 0.9% (wt/vol) sodium chloride by using a disposable “hockey stick.” Conidia were washed twice in a 50-ml conical screw-cap centrifuge tube at 1,000 × g in a Beckman Model TJ-6 clinical centrifuge. After washing, conidial suspensions were counted in a hemocytometer and used to inoculate 100 ml of Sabouraud glucose broth (Difco) to a final concentration of 107 conidia/ml. Cells were grown at 25°C until germlings were visible. Spheroplasts were produced, and DNA purified as described by Vanittanakom et al. (47). DNA was purified from nonsporulating isolates by pulverizing cultures in liquid nitrogen as described by Lasker (27) and then eluting lysates through Genomic G-100 columns (Qiagen, Inc., Valencia, Calif) by using the buffers and procedures supplied by the manufacturer.
Identification of microsatellites.
All available P. marneffei DNA sequences deposited in the GenBank database were screened by using the FindPatterns program available in the Wisconsin Package (version 10.2; Accelrys; GCG, San Diego, Calif.). Selection of sequences was based solely on microsatellite length; sequences were predicted to have six or more dinucleotide repeat units. Three loci that met these criteria were evaluated. Oligonucleotide primers pairs were then designed to be complementary to sequences closely flanking the region containing microsatellite repeats. One primer in the set was 5′ labeled with 6-carboxyfluorescein (FAM) or with 4,7,2′,4′,5′,7′-hexachloro-6-carboxy-fluorescein (HEX) (27). Primer sequences for microsatellites I, II, and III and their origins are shown in Table 2.
TABLE 2.
Characteristics of three polymorphic microsatellite markers of P. marneffei
| Microsatellite | GenBank accession no. | Microsatellite sequencea | Primer sequences (5′ to 3′) | Range of PCR (bp) | No. of alleles |
|---|---|---|---|---|---|
| I | AL684924 | (GA)7AC(GA)22 | AGTAGTTCATCGGCCGAA | 172-266 | 14 |
| GTACCCTTCAAGGTGGTA | |||||
| II | AL686035 | (CA)7TA(CA)7 | AGCTGGAACTGCACCTCTGA | 136-157 | 8 |
| CTTTTGGAGCTGGGTCTGCA | |||||
| III | AL684847 | (CA)6TA(CA)6 | CTATGCCGGGAAGGCAGTGA | 206-226 | 7 |
| ACCCTACGCACTAGACGGAA |
Based on P. marneffei strain PM1.
PMM analysis.
PCRs were performed in a volume of 20 μl containing 20 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM concentrations of each of four deoxynucleoside triphosphates (Roche Diagnostic Corp.), 0.1 μM (each) forward and reverse primers, 1.0 U of TaqDNA Polymerase (Roche), 5% (vol/vol) dimethyl sulfoxide (Sigma), and 30 mg of genomic DNA template. PCR amplification was conducted by using a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Inc., Foster City, Calif.). The amplification program consisted of an initial denaturation at 96°C for 4 min, followed by 30 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s, followed by a final extension step of 72°C for 30 min. After PCR, a 0.5- to 2-μl aliquot of each sample was added to 30 μl of formamide (Applied Biosystems) containing 0.7 μl of GeneScan 500 6-carboxytetramethylrhodamine size standards (Applied Biosystems). Amplicons were denatured at 95°C for 8 min and then rapidly chilled on ice. Denatured samples were resolved by capillary electrophoresis with a 47-cm-long, 0.75-μm-inside-diameter capillary filled with POP-4 polymer (Applied Biosystems) in an ABI Prism 310 genetic analyzer (Applied Biosystems). Molecular sizes of amplified alleles were automatically analyzed and calculated by using GeneScan Analysis software (version 2.1; Applied Biosystems). Due to length polymorphisms, isolates were assigned to a new microsatellite genotype based on one or more differences in band size (Table 1).
Discriminatory power and reproducibility.
The mathematical probability that two unrelated isolates randomly chosen from a test population can be demonstrated to belong to a separate group is known as discriminatory power. The discriminatory power (D) for PMM analysis was calculated in the manner as described by Hunter (25). Reproducibility was examined by analyzing the same DNA preparations repeatedly and by repeated analysis of a second DNA preparation from the same isolate. The short-term stability of the microsatellite markers was examined by PMM analysis of DNA preparations purified from eight isolates serially passaged twice a week in vitro for 7 to 8 weeks at 25°C.
RESULTS
From a total of 1,771,694 bp consisting of 2,334 entries for P. marneffei gene sequences or random sequence tags deposited in the GenBank database and screened for microsatellites, three sequences were selected based on the overall length of the predicted dinucleotide motifs. The three chosen sequences were designated microsatellites I, II, and III. Their gene accession numbers are provided in Table 2.
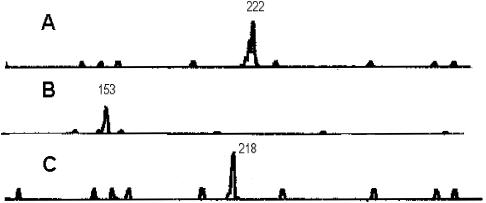
A representative PCR profile for PMM I, II, and III, for P. marneffei strain ATCC 64101 is shown in Fig. 1. As expected for a haploid microorganism, a single PCR amplification product was observed for all 35 isolates at each locus, suggesting the cultures were not composed of a mixture of P. marneffei strains. The size of the PCR products was automatically determined by the GeneScan software. Less-intense stutter or shadow bands, defined as PCR products a few base pairs shorter than the main peak, were frequently observed, as shown in Fig. 1A. Only the major peak was scored for allele size. The ability to type all isolates in a test population, defined as typeability, by PMM analysis was 100%.
FIG. 1.
PCR analysis of PCR profiles for P. marneffei ATCC 64102. Analyses for PMM I (A) and PMM II (B) were performed with a FAM-labeled primer; analysis for PMM III (C) was performed with a primer labeled with HEX. The number above the peak refers to the size of the PCR amplification product.
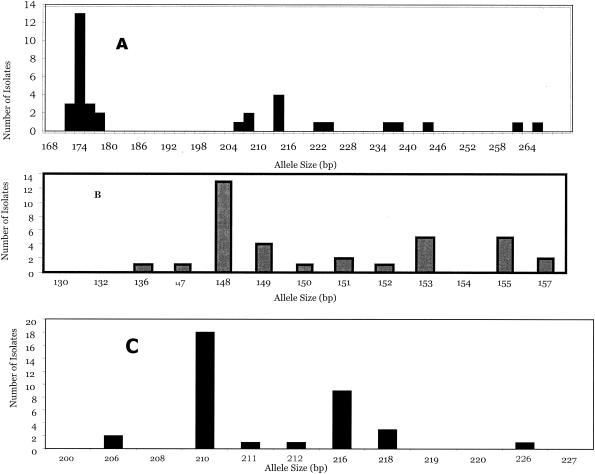
The highest level of discriminatory power was observed for PMM I (D = 0.845). PMM I analysis detected a total of 14 different-size alleles, with PCR products ranging in size from 172 to 266 bp. The distribution of allele sizes for PMM I is shown in Fig. 2A. The most frequent allele was at 174 bp (frequency, 37%) and was only found among isolates from Thailand. The second most frequent allele at 214 bp included only isolates obtained from China (Fig. 3A). Eight isolates were represented by a single type. The number of different alleles for PMM II was 10. The size of the PCR products ranged from 136 to 157 bp with a moderate degree of discrimination (D = 0.822). The most common alleles were at 148, 153, and 155 bp consisting of 13, 5, and 5 isolates, respectively (Fig. 2B). The lowest degree of discriminatory power was for PMM III (D = 0.676). A total of seven different alleles were identified, ranging in size from 206 to 226 bp (Fig. 2). The most common alleles were at 210 and 216 bp with 15 and 8 isolates, respectively (Fig. 2). Interestingly, all isolates harboring the allele at 210 bp were obtained from Thailand, whereas 7 of 8 isolates harboring the allele at 216 bp were obtained from China (Fig. 3). Differences in band size by one PMM were used to assign alleles to a different PMM type.
FIG. 2.
Allele size distributions for PMM I (A), PMM II (B), and PMM III (C) for 35 P. marneffei isolates.
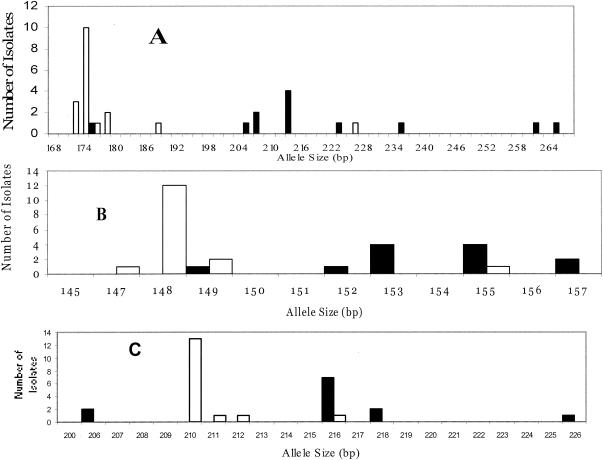
FIG. 3.
Allele size distributions for PMM I (A), PMM II (B), and PMM III (C) for 12 P. marneffei clinical isolates obtained from China (▪) and 16 clinical isolates obtained from Thailand (□).
By combining all three PMMs, a high degree of discriminatory power was observed (D = 0.956). A total of 22 different PMM types were observed with 16 types represented by single isolates. The most frequent PMM types, 13 and 7, consisted of seven and four isolates each (Table 1). PMM types 1, 3, 7, 8, 9, 10, 11, and 12 were observed only for isolates obtained from China. The PMM types for the Chinese isolates were clearly different from the PMM types observed for isolates obtained from Thailand and included PMM types 4, 5, 13, 16, 18, 19, 20, 21, and 22 (Table 1). The distribution of alleles between these two populations is shown in Fig. 3. PMM type 17 was an environmental isolate from Vietnam, and PMM types 4, 6, 14, and 15 were obtained from clinical isolates derived from individuals in the United States or the United Kingdom who had previously lived in or traveled to Southeast Asia (Table 1). The PMM type observed from an environmental soil isolate from Thailand ATCC 201564 was consistent with the PMM type observed for Thailand clinical isolates. Likewise, alleles observed from the single environmental isolate obtained from a bamboo rat in China (ATCC 64102) were consistent with alleles of 153 and 218 bp for PMMs II and III, respectively, observed only in clinical isolates from China.
To determine the molecular origin of length polymorphisms in PMMs I, II, and III, three different-size PCR products from each locus were directly sequenced. Alignment of sequences was aided by using the sequences obtained from the loci submitted to the GenBank database for P. marneffei strain PM1 (Table 2). The variation observed in the length of the PCR products was correlated with the number of dinucleotide repeat units.
No PCR products were observed for PMM I, II, or III for dimorphic fungi such as Candida albicans, Candida tropicalis, Blastomyces dermatitidis, Histoplasma capsulatum, or Sporothrix schenckii. Also, no PCR products were observed for Trichophyton rubrum, Aspergillus fumigatus, Aspergillus flavus, or human placenta.
Reproducibility of PMM analysis was evaluated by analysis of different DNA preparations from the same isolate and by repeated analysis of the same DNA preparation. In both cases, the reproducibility was high. Each analysis included a standard sample panel of isolates with known allele sizes to identify run-to-run variation. When yeast cells were transferred to fresh broth at 37°C, cultures were found to die off by the third to sixth passages. Therefore, stability of the PMM profiles was examined by transferring eight different isolates in broth cultures at 25°C for 7 to 8 weeks. The isolates examined were ATCC 64101, ATCC 64102, ATCC 201013, IFM 47279, IFM 47284, IFM 47286, IFM 47288, IFM 47289, and B-6324. For all eight isolates, the PMM profiles obtained from the original cultures were indistinguishable from the profiles obtained by cultures passaged in vitro.
DISCUSSION
Due to the increasing public health significance of human infections by P. marneffei, the goals of the present study were to develop and evaluate PMM analysis as a viable typing method. This is the first investigation to use PMM analysis for typing P. marneffei clinical and environmental isolates. By using a combination of three PMMs, a high degree of discriminatory power was observed (D = 0.956), and it was possible to distinguish between closely related strains (Table 1). A high degree of discriminatory power is an important performance factor for any typing system (7, 41). The high degree of discrimination achieved for P. marneffei is in good agreement with reports for typing other fungi by PMM analysis. For the pathogenic yeast C. albicans, the discriminatory power based on PMM analysis was reported to range from 0.87 with three markers to type 60 strains (14) to 0.97 for typing 114 strains (37). Likewise, a high degree of discrimination (D = 0.989) was observed by using four PMM for typing A. fumigatus isolates (3). A higher degree of discriminatory power may be achieved in the future as more P. marneffei DNA sequences become available for screening.
The ability to assign an identical type to the same isolate after repeated assays defines reproducibility and is another important criterion for evaluating a typing system (41). The reproducibility of PMM was evaluated by using different DNA preparations of the same isolate and by repeated analysis of the same DNA preparation. In both cases reproducibility was high. This may be due to both stringent PCR conditions and sample preparation. Amplification of target sequences was conducted at high annealing temperatures (58°C), thereby reducing the chance for mismatch hybridization, which has been commonly observed for RAPD analysis (33). Furthermore, PMM primers may be specific for P. marneffei since no PCR signal was observed for related pathogenic fungi such as A. fumigatus and A. flavus (2, 51). The lack of PCR products from human DNA template suggests that PMM analysis may be modified to directly type P. marneffei DNA obtained from tissue samples. The size of the PCR products is automatically calculated and can be accurately measured due to the direct incorporation of size markers in each sample. A high degree of reproducibility by PMM analysis is consistent with other investigations of pathogenic fungi (3, 30, 37) or Leishmania (8).
In addition to a high degree of discrimination, reproducibility, and 100% typeability, PMM analysis has several other advantages for typing P. marneffei isolates. Because the procedure is PCR based, PMM analysis requires relatively small amounts (∼30 mg per reaction) of template DNA compared to other methods. Large numbers of isolates may be typed since sample preparation is simplified and does not require tedious and labor-intensive sample preparation of highly purified DNA such as by pulsed-field gel electrophoresis or Southern blotting. Analysis may be simplified by multiplex PCR assays by using PMMs labeled with two or more different fluorescent dyes, resulting in higher throughput. Using a standard panel of test strains and PMMs, interlaboratory comparisons and databases may then be feasible. Finally, PMM analysis may be specific and should allow the detection of mixed cultures, which would appear as a PCR profile harboring two or more bands from a DNA sample.
Microsatellites are dispersed and abundant in all eukaryotes. Although the exact mechanism is not known, the generation of microsatellite length variation is thought to be the result of polymerase slippage during DNA replication (29). The mutation rate for microsatellite loci ranges from 10−2 to 10−6 (17, 34). Microsatellite mutation rates were found to be dependent on repeat type, species, and primarily on the length of the microsatellite repeat (36). Generally, the longer the repeat the faster the rate (48). In the present study DNA sequence analysis was used to confirm that length polymorphisms observed in PMMs I, II, and III were consistent with the number of microsatellite repeat units. PMM primers were designed to anneal as closely as possible to the microsatellite repeat units in order to reduce the possibility of detecting insertion and/or deletions within regions flanking the microsatellite repeats. The mutation rate for PMM in P. marneffei is not known. Despite the higher mutation rate for microsatellites compared to spontaneous mutations, the ability to detect microevolutionary events and relative short-term stability of markers is an important characteristic in evaluating the requirements of a typing method (41, 45). In this investigation, the short-term stability of PMM profiles was analyzed by monitoring eight isolates passaged in vitro for 7 to 8 weeks. The identical PMM profiles observed between the nonpassaged and comparable passaged samples demonstrated the short-term stability by PMM analysis. This result suggests the speed of the PMM molecular clock is sufficient and appropriate for molecular strain typing of outbreak and surveillance isolates. Although PMM analysis has been shown to discriminate between members of a species, the use of PMM loci for phylogenetic analysis remains uncertain due to constraints on allele size (20, 28, 32). Size constraints on allele size are believed to reduce the ability to detect the true genetic distance.
One disadvantage of PMM analysis is the need for specialized reagents, software, and expensive equipment which may not be available in many laboratories. This problem can be partially circumvented by resolution of PCR products with a high-resolution agarose gel system, as previously reported for A. fumigatus (5, 35) or denaturing polyacrylamide gel electrophoresis, as reported for S. cerevisiae (21) or C. albicans (6, 7). Even when high-stringency conditions for PCR amplification are used, potential PCR artifacts have been observed. The addition of an extra adenosine residue to the 3′ end of the amplified product is a commonly observed artifact. To aid in the uniform addition of the extra A residue to the ends of all PCR amplification products, the final extension was lengthened to 30 min at 72°C as described previously (3, 4, 27). Alternatively, PCR amplification with a DNA polymerase with 3′-to-5′ proofreading exonuclease activity such as Pfu, Tma, or Pwo should result in the majority of PCR fragments without an additional 3′-A overhang. Due to the high sensitivity of the fluorescent detection system for capillary electrophoresis, PCR stutter products are commonly observed (Fig. 1A). These stutter fragments are generated during PCR amplification, resulting in PCR products that differ by multiples of the microsatellite repeat length (39). Stutter products were greatly reduced by the incorporation of 5% (vol/vol) dimethyl sulfoxide into the PCRs and was also observed to enhance the yield of PCR products as reported previously (3, 27). Stutter bands, although present, did not interfere with analysis since allele size is denoted by the highest observed sized band for analysis (3, 39).
This investigation distinguished 22 different PMM types among 32 clinical isolates, 2 isolates from bamboo rats, and 1 soil isolate (Table 1). In contrast, Vanittanakom et al. (47) detected only two HaeIII types among 46 isolates of P. marneffei. More recently, pulsed-field gel electrophoresis of NotI restriction endonuclease fragments was shown to be reproducible and highly discriminatory between isolates despite laborious sample preparation (46). The results of typing by PMM analysis in the present study of isolates ATCC 64102, 24100, 18224, and 64101 was consistent with the typing results obtained by NotI analysis of the same isolates by Vanittanakom et al. (47); both methods showed these isolates to be unrelated. Only a moderate degree of discrimination of P. marneffei isolates has been achieved by RAPD analysis (24). Eight RAPD types were identified for 20 isolates of P. marneffei obtained from 11 patients in Taiwan. Even though RAPD analysis has been considered to be easy to perform, the interpretation of RAPD profiles should proceed with caution since an instability of RAPD profiles was observed in A. fumigatus (4) due to an extreme sensitivity to reaction conditions (33).
Nine different PMM types were observed among 15 clinical isolates of P. marneffei obtained from Thailand; these included isolates from Chiang Rai and Lam-Pang provinces. In comparison, eight different PMM types were detected among 12 clinical isolates obtained from China. Interestingly, all of the PMM types differed between these two populations (Table 1), indicating the potential for geographic isolation. The apparent allopatric nature of these two populations may be due, at least in part, to several factors. One factor might be the small sample size: the clinical isolates may not be fully representative of these geographic regions. Second, only a very small number (n = 3) of nonclinical isolates were included in this analysis. Third, our isolates were also not matched temporally. It is also possible that several alleles may be artificially overrepresented due to analysis of the isolates obtained from different body sites from the same patient. Lastly, alleles may be distorted by homoplasy (31). In this case, identical genotypes may not be derived from a common ancestor but may be the result of molecular events such as convergence, parallelisms, or reversions. However, the observed differences in PMM types and allele frequencies by geographic region is concordant with analyses of isolates obtained from the environment or from rats. For instance, PMM analysis of isolate ATCC 64102 from a rat from China showed that it harbors two alleles predominant in Chinese clinical isolates at 153 and 218 bp for PMMs II and III, respectively (Fig. 3). Likewise, a soil isolate from Thailand (ATCC 201564) had a PMM profile identical to those observed in six clinical isolates obtained from Thailand (Table 1). However, despite our findings of identical PMM profiles among clinical and environmental isolates, it is still speculative to associate an environmental source with direct transmission to patients. The PMM type observed for ATCC 18224 from Vietnam differed from the PMM types observed in isolates obtained from China and Thailand.
Taken as a whole, the results of PMM analysis displayed a high degree of discriminatory power, reproducibility, ease of use and interpretation, typeability, and a high-throughput potential. Molecular typing of P. marneffei by PMM analysis should be useful for characterizing individuals in populations and to help answer important epidemiologic questions regarding the transmission of infection, the prevalence of pathogenic strains, and the analysis of ecological niches. At present, it remains to be determined whether a single method or a combination of methods is more appropriate for typing. With the addition of new markers, PMM analysis should assist the further characterization of the reproductive mode and transmission of P. marneffei isolates.
Acknowledgments
This research was supported by an appointment of Y.R. to the International Emerging Infectious Disease Fellowship Program administered by the Association of Public Health Laboratories and funded by the Centers for Disease Control and Prevention.
REFERENCES
- 1.Ajello, L., A. A. Padhye, S. Sukroongreung, C. H. Nilakul, and S. Tantimavanich. 1995. Occurrence of Penicillium marneffei in Thailand. Mycopathologia 131:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Andrianopoulos, A. 2002. Control of morphogenesis in the human fungal pathogen Penicillium marneffei. Int. J. Med. Microbiol. 292:331-347. [DOI] [PubMed] [Google Scholar]
- 3.Bart-Delabesse, E., J.-F. Humbert, E. Delabesse, and S. Bretagne. 1998. Microsatellite markers for typing Aspergillus fumigatus isolates. J. Clin. Microbiol. 36:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bart-Delabesse, E., J. Sarfati, J.-P. Debeaupuis, W. van Leeuwen, A. van Belkum, S. Bretagne, and J.-P. Latgé. 2001. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analysis for fingerprinting Aspergillus fumigatus isolates. J. Clin. Microbiol. 39:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertout, S., F. Renaud, R. Barton, F. Symoens, J. Burnod, M.-A. Piens, B. Lebeau, M.-A. Viviani, F. Chapuis, J.-M. Grillot, M. Mallíe, et al. 2001. Genetic polymorphisms of Aspergillus fumigatus in clinical samples from patients with invasive aspergillosis: investigation using multiple typing methods. J. Clin. Microbiol. 39:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botterel, F., C. Desterke, C. Costa, and S. Bretagne. 2001. Analysis of microsatellite markers of Candida albicans used for rapid typing. J. Clin. Microbiol. 39:4076-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bretagne, S., J.-M. Costa, C. Besmond, R. Carsique, and R. Calderone. 1997. Microsatellite polymorphism in the promoter sequence of the elongation factor 3 gene of Candida albicans as the basis for a typing system. J. Clin. Microbiol. 35:1777-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulle, B., L. Millon, J.-M. Bart, M. Gállego, F. Gambarelli, M. Portús, L. Schnur, C. L. Jaffe, S. Fernandez-Barredo, J. M. Alunda, and R. Piarroux. 2002. Practical approach for typing strains of Leishmania infantum by microsatellite analysis. J. Clin. Microbiol. 40:3391-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capponi, M., P. Sureau, and R. C. Mathews. 1956. Penicillose de Rhizomys sinensis. Bull. Soc. Pathol. Exot. Fil. 49:418-421. [PubMed] [Google Scholar]
- 10.Chariyalertsak, S., T. Sirisanthana, K. Supparatpinyo, and K. E. Nelson. 1996. Seasonal variation of disseminated Penicillium marneffei infections in Northern Thailand: a clue to the reservoir? J. Infect. Dis. 173:1490-1493. [DOI] [PubMed] [Google Scholar]
- 11.Chariyalertsak, S., T. Sirisanthana, K. Supparatpinyo, J. Praparattanapan, and K. E. Nelson. 1997. Case-control study of risk factors for Penicillium marneffei infection in human immunodeficiency virus-infected patients in northern Thailand. Clin. Infect. Dis. 24:1080-1086. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, C. R., and M. R. McGinnis. 1997. Pathology of Penicillium marneffei. Arch. Pathol. Lab. Med. 121:798-804. [PubMed] [Google Scholar]
- 13.Dalle, F., L. Dumont, N. Franco, D. Mesmacque, D. Caillot, P. Bonnin, C. Moiroux, O. Vagner, B. Cuisenier, S. Lizard, and A. Bonnin. 2003. Genotyping of Candida albicans oral strains from healthy individuals by polymorphic microsatellite locus analysis. J. Clin. Microbiol. 41:2203-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalle, F., N. Franco, J. Lopez, O. Vagner, D. Caillot, P. Chavanet, B. Cuisenier, S. Oho, S. Lizard, and A. Bonnin. 2000. Comparative genotyping of Candida albicans bloodstream and non-bloodstream isolates at a polymorphic microsatellite locus. J. Clin. Microbiol. 38:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiSalvo, D. A., A. M. Fickling, and L. Ajello. 1973. Infection caused by Penicillium marneffei: description of the first natural infection in man. Am. J. Clin. Pathol. 60:259-263. [DOI] [PubMed] [Google Scholar]
- 16.Drouhet, E. 1993. Penicilliosis due to Penicillium marneffei: a new emerging systemic mycosis in AIDS patients traveling or living in Southeast Asia: review of 44 cases reported in HIV-infected patients during the last 5 years compared to 44 cases of non-AIDS patients reported over 20 years. J. Mycol. Med. 4:195-224. [Google Scholar]
- 17.Ellegren, H. 2000. Microsatellite mutations in the germline: implications for evolutionary inference. Trends Genet. 16:551-558. [DOI] [PubMed] [Google Scholar]
- 18.Field, D., L. Eggert, D. Metzgar, R. Rose, and C. Wills. 1996. Use of polymorphic short and clustered coding-region microsatellites to distinguish strains of Candida albicans. FEMS. Immunol. Med. Microbiol. 15:73-79. [DOI] [PubMed] [Google Scholar]
- 19.Fisher, M. C., G. Koenig, T. J. White, and J. W. Taylor. 2000. A test for concordance between the multilocus genealogies of genes and microsatellites in the pathogenic fungus Coccidioides immitis. Mol. Biol. Evol. 17:1164-1174. [DOI] [PubMed] [Google Scholar]
- 20.Garza, J. C., M. Slatkin, and N. B. Freimer. 1995. Microsatellite allele frequencies in humans and chimpanzees, with implications for constraints on allele size. Mol. Biol. Evol. 12:594-603. [DOI] [PubMed] [Google Scholar]
- 21.Hennequin, C., A. Thierry, G. F. Richard, G. Lecointre, H. V. Nguyen, C. Gaillardin, and B. Dujon. 2001. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 39:551-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hien, T. V., P. P. Loc, N. T. T. Hoa, N. M. Duong, V. M. Quang, M. M. McNeil, N. T. Dung, and D. A. Ashford. 2001. First cases of disseminated penicilliosis marneffei infection among patients with acquired immunodeficiency syndrome in Vietnam. Clin. Infect. Dis. 32:78-80. [DOI] [PubMed] [Google Scholar]
- 23.Hilmarsdottir, I., J. L. Meynard, O. Rogeaux, G. Guermonprez, A. Datry, C. Katlama, G. Brucker, A. Coutellier, M. Danis, and M. Gentilini. 1993. Disseminated Penicillium marneffei infection associated with human immunodeficiency virus: a report of two cases and a review of 35 published cases. J. Acquir. Immune Defic. Syndr. 6:466-471. [PubMed] [Google Scholar]
- 24.Hsueh, P.-R., L.-J. Teng, C.-C. Hung, J.-H. Hsu, P.-C. Yang, S.-W. Ho, and K.-T. Luh. 2000. Molecular evidence for strain dissemination of Penicillium marneffei: an emerging pathogen in Taiwan. J. Infect. Dis. 181:1706-1712. [DOI] [PubMed] [Google Scholar]
- 25.Hunter, P. R. 1990. Reproducibility indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imwidthya, P. 1994. Update of penicilliosis marneffei in Thailand. Mycopathologia 127:135-137. [DOI] [PubMed] [Google Scholar]
- 27.Lasker, B. A. 2002. Evaluation of performance of four genotypic methods for studying the genetic epidemiology of Aspergillus fumigatus isolates. J. Clin. Microbiol. 40:2886-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehmann, T., A. Hawley, and F. H. Collins. 1996. An evaluation of evolutionary constraints on microsatellite loci using null alleles. Genetics 144:1155-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levinson, G., and G. A. Gutman. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4:203-224. [DOI] [PubMed] [Google Scholar]
- 30.Metzgar, D., A. V. Belkum, D. Fields, R. Haubrich, and C. Wills. 1998. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J. Clin. Microbiol. 36:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzgar, D. D. Field, R. Haubrich, and C. Wills. 1998. Sequence analysis of a compound coding-region microsatellite in Candida albicans resolves homoplasies and provides a high-resolution tool for genotyping. FEMS. Immunol. Med. Microbiol. 20:103-109. [DOI] [PubMed] [Google Scholar]
- 32.Ortí, G., D. E. Pearse, and J. C. Avise. Phylogenetic assessment of length variation at a microsatellite locus. Proc. Natl. Acad. Sci. USA 94:10745-10749. [DOI] [PMC free article] [PubMed]
- 33.Penner, G. A., A. Bush, R. Wise, W. Kim, L. Domier, K. Kasha, A. Laroche, G. Scoles, S. J. Molnar, and G. Fedak. 1993. Reproducibility of random amplified polymorphic (RAPD) analysis among laboratories. PCR Methods Appl. 2:112-116. [DOI] [PubMed] [Google Scholar]
- 34.Richard, G.-F., C. Hennequin, A. Thierry, and B. Dujon. 1999. Trinucleotide repeats and other microsatellites in yeasts. Res. Microbiol. 150:589-602. [DOI] [PubMed] [Google Scholar]
- 35.Rosehart, K., M. H. Richards, and M. J. Bidochka. 2002. Microsatellite analysis of environmental and clinical isolates of the opportunistic fungal pathogen Aspergillus fumigatus. J. Med. Microbiol. 51:1128-1134. [DOI] [PubMed] [Google Scholar]
- 36.Rubinsztein, D. C., W. Amos, J. Leggo, S. Goodburn, S. Jain, S. H. Li, R. L. Margolis, C. A. Ross, and M. A. Fergson-Smith. 1995. Microsatellite evolution-evidence for directionality and variation in rate between species. Nat. Genet. 10:337-343. [DOI] [PubMed] [Google Scholar]
- 37.Sampaio, P., L. GusmÂo, C. Alves, C. Pina-Vaz, A. Amorim, amd C. Pais. 2003. Highly polymorphic microsatellite for identification of Candida albicans strains. J. Clin. Microbiol. 41:552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segretain, G. 1959. Penicillium marneffei n. sp. agent d'une mycose du systeme reticuloendothelial. Mycopathol. Mycol. Appl. 11:327-353. [DOI] [PubMed] [Google Scholar]
- 39.Shinde, D., Y. Lai, F. Sun, and N. Arnheim. 2003. TaqDNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis:(CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res. 31:974-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strand, M., T. Prolla, R. Liskay, and T. Petes. 1994. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365:274-276. [DOI] [PubMed] [Google Scholar]
- 41.Stuelens, M. J., et al. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 42.Supparatpinyo, K., S. Chiewchanvit, P. Hirunsri, C. Uthammachai, K. E. Nelson, and T. Sirisanthana. 1992. Penicillium marneffei infection in patients with human immunodeficiency virus. Clin. Infect. Dis. 14:871-874. [DOI] [PubMed] [Google Scholar]
- 43.Supparatpinyo, K., J. Perriens, K. E. Nelson, and T. Srisanthana. 1998. A controlled trial of itraconazole to prevent relapse of Penicillium marneffei infection in patients infected with human immunodeficiency virus. N. Engl. J. Med. 339:1739-1743. [DOI] [PubMed] [Google Scholar]
- 44.Tautz, D. 1998. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 17:6463-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor, J. W., D. M. Geiser, A. Burt, and V. Koufopanou. 1999. The evolutionary biology and population genetics underlying strain typing. Clin. Microbiol. Rev. 12:126-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trewatcharegon, S., S. Sirisinha, A. Romsai, B. Eampokalap, R. Teanpaisan, and S. C. Chaiyaroj. 2001. Molecular typing of Penicillium marneffei isolates from Thailand by NotI macrorestriction and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:4544-4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanittanakom, N., C. R. Cooper, Jr., S. Chariyaletsak, S. Youngchim, K. E. Nelson, and T. Sirisanthana. 1996. Restriction endonuclease analysis of Penicillium marneffei. J. Clin. Microbiol. 34:1834-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber, J. L., and C. Wong. 1993. Mutation of human short tandem repeats. Hum. Mol. Genet. 2:1123-1128. [DOI] [PubMed] [Google Scholar]
- 49.Wong, S. S. Y., T. Y. C. Ho, A. H. Y. Ngan, P. C. Y. Woo, T.-L. Que, and K.-Y. Yuen. 2001. Biotyping of Penicillium marneffei reveals concentration-dependent growth inhibition by galactose. J. Clin. Microbiol. 39:1416-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong, K. H., and S. S. Kee. 1998. Comparing the first and second hundred AIDS cases in Hong Kong. Singapore Med. J. 39:236-240. [PubMed] [Google Scholar]
- 51.Yuen, K.-Y., G. Pascal, S. S. Y. Wong, P. Glaser, P. C. Y. Woo, F. Kunst, P. Glaser, P. C. Y. Woo, F. Kunst, J. J. Cai, E. Y. L. Cheung, C. Médigue, and A. Danchin. 2003. Exploring the Penicillium marneffei genome. Arch. Microbiol. 179:339-353. [DOI] [PubMed] [Google Scholar]