Abstract
Isolates of the Salmonella enterica serotype Typhimurium definitive phage type (DT104) were found to contain the same prophage (designated phage ST104). The complete sequence of the DNA genome of prophage ST104 was determined. The entire DNA sequence consisted of 41,391 bp, including 64 open reading frames, and exhibited high similarity to P22 and to phage type conversion phage ST64T.
Recently, Salmonella enterica serotype Typhimurium multidrug-resistant strain definitive phage type 104 (DT104) has emerged and spread over many countries (4, 9, 14, 15). The organism has a core pattern of resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline. Previously, we used fluorescent amplified-fragment length polymorphism fingerprinting (FAFLP) analysis for molecular epidemiological investigation of serotype Typhimurium (13). Among 120 isolates from cattle, there were 17 FAFLP profiles that formed four distinct clusters (A to D). The isolates belonging to cluster A, in which all of the isolates of DT104 were included, have become increasingly common since 1992 in the northernmost island of Japan. The sequence of a polymorphic marker that is common to the strains of FAFLP cluster A has homology with the segment of the eae gene of phage P22. In this study, we isolated prophage ST104, which is common to isolates of DT104, and determined the whole sequence of this phage. The genomic architecture is similar to that of P22, and a number of regions are very similar to those of P22.
Isolation of prophage common to serotype Typhimurium DT104.
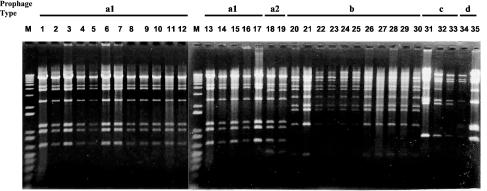
Forty-two serotype Typhimurium strains, including the 12 isolates of DT104 used in this study, were described previously (13). To investigate whether lysogenic prophages are present in serotype Typhimurium, we cultured the strains in the presence of mitomycin C at a concentration of 0.5 μg/ml as previously described (17). Thirty-four out of 42 strains released phages that produced plaques on lawns of serotype Typhimurium strain LT2. To characterize the isolated phages, the restriction patterns of their DNAs were compared. Endonuclease digestion with EcoRI revealed five DNA types, designated a1, a2, b, c, and d (Fig. 1). All of the phages isolated from strains that belong to FAFLP cluster A (13), including 12 isolates of DT104, show the same restriction pattern, type a1 (Fig. 1). We have named this phage ST104. The restriction patterns of the prophages from strains NET25 and NET26, both of which belong to FAFLP cluster B, are similar to that of ST104; however, an additional EcoRI site was observed (Fig. 1). Therefore, the DNA type of these phages was designated a2. The restriction patterns of the phages isolated from strains belonging to FAFLP cluster B or C are different from those of ST104. No phage was detected in the two strains that belong to FAFLP cluster D. Schicklmaier et al. (10) suggested that prophage restriction patterns in natural isolates of serotype Typhimurium could serve as markers for the epidemiologic classification of pathogenic strains. Our results suggest that the presence of prophage ST104 could be a marker of DT104 or its related strains.
FIG. 1.
Electrophoresis of endonuclease digests of phage DNA. Phage DNA was digested with EcoRI and subsequently separated at 10 mV cm−1 in a 0.8% agarose gel prepared with 89 mM Tris-2.5 M EDTA-89 mM boric acid. Lanes (FAFLP profiles are in parentheses): M, 1-kb ladder; 1, U1 (A2); 2, U2 (A2); 3, U3 (A2); 4, U4 (A2); 5, U5 (A2); 6, U6 (A1); 7, U7 (A2); 8, U8 (A2); 9, U9 (A1); 10, U17 (A2); 11, U18 (A1); 12, U20 (A1); 13, U1 (A1); 14, NET57 (A1); 15, NET2 (A2); 16, NET8 (A3); 17, H6(A4); 18, NET25 (B1); 19, NET26 (B1); 20, KT1 (B2); 21, KT2 (B2); 22, N78 (B2); 23, NET55 (B2); 24, N59 (B5); 25, NET37 (B5); 26, N34 (B5); 27, N79 (B5); 28, N81 (B5); 29, NET48 (B5); 30, NET52 (B6); 31, NET31 (B2); 32, NET40 (C1); 33, KT3 (C1); 34, 478 (B4); 35, N48 (C5).
Sequence of the ST104 genome.
The entire nucleotide sequence of ST104 was 41,391 bp in size. This sequence was homologous to the genomic sequences of both bacteriophages P22 (41,724 bp) and ST64T (40,679 bp). The overall homology between ST104 and P22 was 70.4%, and that between ST104 and ST64T was 74.5%. The average GC content of ST104 was calculated to be 47.3%, which is a level similar to those of phages P22 (47.1%) and ST64T (47.5%).
Schmieger and Schicklmaier (11) reported that all of the DT104 strains examined harbor a prophage, PDT17, that is related to P22 and is a generalized transducing phage, like P22. Our data show that the nucleotide sequence of prophage ST104 has high similarity to that of P22 and that all of the DT104 strains examined harbored ST104, suggesting that ST104 is the same prophage as PDT17.
Analysis and comparison of ORFs.
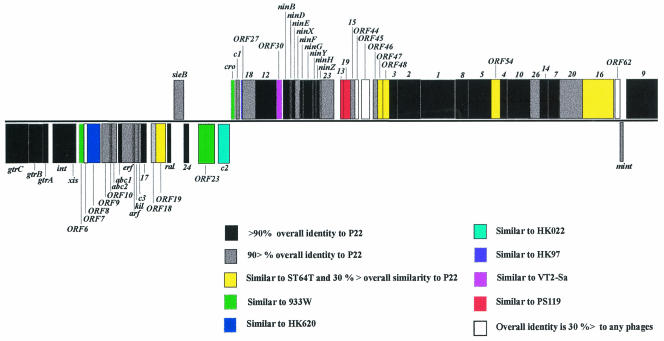
A search for open reading frames (ORFs) with WebGeneMark.hmm (5) revealed 64 ORFs larger than 144 nucleotides. A complete list of ST104 ORFs is given in Table 1. We labeled these ORFs consecutively from ORF1 to ORF64. Searches for homologous protein sequences were conducted with the BLAST software (2, 3) against the GenBank database. The characteristics of these ORFs and their corresponding predicted proteins are described in Table 1. Genes with functional assignments were named, and the names of equivalent P22 genes were used when possible. ST104 has similarities to both P22 and ST64T, not only at the sequence level but also at the gene organization level (Table 1; Fig. 2). Most of the ORFs of prophage ST104 are similar to particular ORFs present in these two bacteriophages. Beginning with a counterpart to P22 gtrC (92% identity) and ending with the gene 9 protein of P22 (98% identity), 27 of the translated products of these genes showed more than 90% amino acid sequence identity to P22 (Table 1; Fig. 2). These proteins included the functional serotype conversion cassette, integrase, excisionase, Abc1, superinfection exclusion (gp17), antitermination (gp24), Ral, helicase, NinB, NinD, NinE, NinF, NinG, NinY, NinH, packaging (gp3 and gp2), head (with the exception of gp26, gp20, and gp16), and tail proteins. Three corresponding ORFs, namely, ORF19, ORF47, and ORF48, are present in ST64T but absent in P22.
TABLE 1.
Characteristics of ST104 ORFs and deduced proteins
| ORF | Gene | Directiona | Start position | Stop position | Product length (amino acids) | Function of deduced protein | Accession no. and sequence similarity | % Identityb |
|---|---|---|---|---|---|---|---|---|
| ORF1 | gtrc | − | 17 | 1474 | 485 | O-antigen conversion | AAF74999; glucosyl transferase, phage P22 (S. enterica) | 92 (485) |
| AAL15475; glucosyl transferase; phage ST64T (S. enterica) | 93 (485) | |||||||
| ORF2 | gtrB | − | 1464 | 2396 | 310 | O-antigen conversion | AAF75000; bactoprenol glucosyltransferase, phage P22 (S. enterica) | 99 (310) |
| AAL15476; bactoprenol glucosyltransferase, phage ST64T (S. enterica) | 100 (310) | |||||||
| ORF3 | gtrA | − | 2393 | 2755 | 120 | O-antigen conversion | AAF75001; translocase, phage P22 (S. enterica) | 100 (120) |
| AAL15477; translocase, phage ST64T (S. enterica) | 100 (120) | |||||||
| ORF4 | int | − | 3104 | 4267 | 387 | Integrase | AAF75002; integrase, phage P22 (S. enterica) | 97 (387) |
| AAL15478; integrase, phage ST64T (S. enterica) | 98 (387) | |||||||
| ORF5 | xis | − | 4144 | 4494 | 116 | Excisionase | AAF75003; excisionase, phage P22 (S. enterica) | 97 (116) |
| AAL15479; excisionase, phage ST64T (S. enterica) | 97 (116) | |||||||
| ORF6 | − | 4706 | 4990 | 94 | AAD25409; hypothetical protein, phage 933W (E. coli) | 97 (94) | ||
| ORF7 | − | 4983 | 5267 | 94 | BAA84361; hypothetical protein, prophagte VT2-Sa (E. coli) | 28 (73) | ||
| ORF8 | − | 5267 | 6058 | 263 | AAK28855; hypothetical protein, phage HK620 (E. coli) | 88 (130) | ||
| AAD25483; hypothetical protein, phage 933W (E. coli) | 38 (316) | |||||||
| AAF75008; Ead, phage P22 (S. enterica) | 47 (122) | |||||||
| AAF75006; Eaa, phage P22 (S. enterica) | 39 (78) | |||||||
| AAL15482; Eaa2, phage ST64T (S. enterica) | 37 (79) | |||||||
| ORF9 | − | 6128 | 6637 | 169 | AAK28856; hypothetical protein, phage HK620 (E. coli) | 49 (169) | ||
| AAF75010; Eae, phage P22 (S. enterica) | 62 (86) | |||||||
| ORF10 | − | 6634 | 6804 | 56 | AAK28857; hypothetical protein, phage HK620 (E. coli) | 75 (56) | ||
| AAN52173; ORF56; phage ST64 (S. enterica) | 89 (56) | |||||||
| AAF75011; ORF56, phage P22 (S. enterica) | 60 (55) | |||||||
| ORF11 | abc2 | − | 6815 | 7108 | 97 | Anti-RecBCD protein | AAF7512; anti-Rec protein Abc2, phage P22 (S. enterica) | 71 (97) |
| AAL15488; anti-Rec protein Abc2, phage ST64T (S. enterica) | 72 (97) | |||||||
| ORF12 | abc1 | − | 7155 | 7439 | 94 | Anti-RecBCD protein | AAF75013; anti-Rec protein Abc1, phage P22 (S. enterica) | 97 (94) |
| AAL15489; anti-Rec protein Abc1, phage ST64T (S. enterica) | 97 (94) | |||||||
| ORF13 | erf | − | 7439 | 8146 | 235 | Recombination protein | AAF75014; recombination protein Erf, phage P22 (S. enterica) | 75 (71) |
| AAL15490; ORF235, phage ST64T (S. enterica) | 95 (235) | |||||||
| ORF14 | arf | − | 8143 | 8286 | 47 | Recombination protein | AF217253; recombination protein Arf, phage P22 (S. enterica) | 87 (47) |
| ORF15 | kil | − | 8276 | 8464 | 62 | Unknown | AAF75016; Kil, phage P22 (S. enterica) | 78 (62) |
| ORF16 | c3 | − | 8445 | 8603 | 52 | Regulatory protein | AAF75017; regulatory protein C3, phage P22 (S. enterica) | 75 (52) |
| AAL15492; regulatory protein C3, phage ST64T (S. enterica) | 93 (45) | |||||||
| ORF17 | 17 | − | 8688 | 9002 | 104 | Superinfection exclusion protein | AAF75018; superinfection exclusion protein, phage P22 (S. enterica) | 91 (103) |
| AAL15493; superinfection exclusion protein, phage ST64T (S. enterica) | 99 (104) | |||||||
| ORF18 | − | 9278 | 9565 | 95 | AAF75020; ORF78, phage P22 (S. enterica) | 46 (48) | ||
| ORF19 | − | 9599 | 10243 | 214 | AAL15494; ORF232, phage ST64T (S. enterica) | 76 (213) | ||
| ORF20 | ral | − | 10327 | 10521 | 64 | Antirestriction protein | AAF75021; antirestriction protein Ral, phage P22 (S. enterica) | 96 (63) |
| AAL15495; antirestriction protein Ral, phage ST64T (S. enterica) | 98 (64) | |||||||
| ORF21 | sieB | + | 10735 | 11322 | 195 | Superinfection exclusion protein | AAF75022; superinfection protein, phage P22 (S. enterica) | 88 (192) |
| ORF22 | 24 | − | 11335 | 11637 | 100 | Antitermination protein | AAF75023; antitermination protein, phage P22 (S. enterica) | 98 (100) |
| AAL15496; antitermination protein, phage ST64T (S. enterica) | 96 (75) | |||||||
| ORF23 | − | 12244 | 13323 | 359 | CAB39294; hypothetical protein, phage 933W (E. coli) | 34 (357) | ||
| ORF24 | c2 | − | 13488 | 14177 | 229 | Regulatory protein | CAA34222; regularoty protein CI, phage HK022 (E. coli) | 41 (234) |
| ORF25 | cro | + | 14288 | 14503 | 71 | Antirepressor | AAD25431; regulatory protein Cro, phage 933W (E. coli) | 32 (61) |
| ORF26 | c1 | + | 14614 | 14895 | 93 | Transcriptional activator | AAL15499; regulatory protein C1, phage ST64T (S. enterica) | 100 (93) |
| AAF75026; transcriptional protein, phage P22 (S. enterica) | 49 (93) | |||||||
| ORF27 | + | 14930 | 15091 | 53 | AAf31131; gp53, phage HK97 (E. coli) | 100 (53) | ||
| ORF28 | 18 | + | 15078 | 15899 | 273 | DNA replication protein | AAF31132; gp54, prophage HK97 (E. coli) | 98 (273) |
| AAF75028; DNA replication protein, phage P22 (S. enterica) | 51 (259) | |||||||
| ORF29 | 12 | + | 15896 | 17272 | 458 | Helicase | AAF75029; helicase, phage P22 (S. enterica) | 98 (458) |
| ORF30 | + | 17269 | 17538 | 89 | BAA84311; ORF28, phage VT2-Sa (E. coli) | 95 (89) | ||
| ORF31 | ninB | + | 17612 | 18049 | 145 | Unknown | AAF75030; NinB, phage P22 (S. enterica) | 97 (145) |
| AAL15506; NinB, phage ST64T (S. enterica) | 46 (102) | |||||||
| ORF32 | ninD | + | 18046 | 18219 | 57 | Unknown | AAF75031; NinD, phage P22 (S. enterica) | 96 (57) |
| AAL15507; NinD, phage ST64T (S. enterica) | 96 (57) | |||||||
| ORF33 | ninE | + | 18186 | 18362 | 58 | Unknown | AAF75032; NinE, phage P22 (S. enterica) | 98 (58) |
| AAL15508; NinE, phage ST64T (S. enterica) | 98 (58) | |||||||
| ORF34 | ninX | + | 18359 | 18706 | 115 | Unknown | AAF75033; NinX, phage P22 (S. enterica) | 57 (125) |
| AAL15509; NinX, phage ST64T (S. enterica) | 100 (115) | |||||||
| ORF35 | ninF | + | 18699 | 18875 | 58 | Unknown | AAF75034; NinF, phage P22 (S. enterica) | 96 (58) |
| AAL15510; NinF, phage ST64T (S. enterica) | 85 (57) | |||||||
| ORF36 | ninG | + | 18868 | 19479 | 203 | Unknown | AAF75035; NinG, phage P22 (S. enterica) | 94 (203) |
| ORF37 | ninY | + | 19476 | 19700 | 74 | Unknown | AAF75036; NinY, phage P22 (S. enterica) | 97 (74) |
| ORF38 | ninH | + | 19697 | 19900 | 67 | Unknown | AAF75037; NinH, phage P22 (S. enterica) | 98 (67) |
| AAL15514; NinH, phage ST64T (S. enterica) | 98 (67) | |||||||
| ORF39 | ninZ | + | 19881 | 20060 | 59 | Unknown | CAA55166; NinZ, phage P22 (S. enterica) | 89 (59) |
| AAL15515; NinZ, phage ST64T (S. enterica) | 93 (59) | |||||||
| ORF40 | 23 | + | 20057 | 20830 | 257 | Antitermination protein | AAG55478; antitermination protein, phage CP-933W (E. coli) | 43 (272) |
| AAA96595; antitermination protein, phage lambda (E. coli) | 42 (256) | |||||||
| AAF75038; antitermination protein, phage P22 (S. enterica) | 39 (256) | |||||||
| ORF41 | 13 | + | 21261 | 21464 | 67 | Holin | CAA09709; holin, phage PS119 (S. enterica) | 100 (67) |
| ORF42 | 19 | + | 21436 | 21939 | 167 | Lysozyme | CAA09710; lysozyme, phage PS119 (S. enterica) | 100 (167) |
| AAF75040; lysozyme, phage P22 (S. enterica) | 35 (131) | |||||||
| ORF43 | 15 | + | 21936 | 22403 | 155 | Endopeptidase | CAA09711; endopeptidase, phage PS119 (S. enterica) | 100 (155) |
| AAF75041; endopeptidase, phage P22 (S. enterica) | 55 (156) | |||||||
| AAL15519; endopeptidase, phage ST64T (S. enterica) | 39 (151) | |||||||
| ORF44 | + | 22156 | 22362 | 68 | BAB34244; putative lipoprotein precursor (E. coli) | 95 (68) | ||
| ORF45 | + | 22616 | 23146 | 176 | CAA33655; KilA, phage P1 (E. coli) | 31 (104) | ||
| ORF46 | + | 23369 | 23611 | 80 | AAL15520; ORF118, phage ST64T (S. enterica) | 100 (80) | ||
| AAF7506; ORF80, phage P22 (S. enterica) | 42 (80) | |||||||
| AAK28888; HkbM, prophage HK620 (E. coli) | 43 (80) | |||||||
| ORF47 | + | 23615 | 24004 | 129 | AAL15521; ORF129, phage ST64T (S. enterica) | 100 (117) | ||
| ORF48 | + | 24004 | 24408 | 134 | AAL15522; ORF134, phage ST64T (S. enterica) | 100 (134) | ||
| ORF49 | 3 | + | 24412 | 24900 | 162 | Terminase (small subunit) | AAF75043; terminase small subunit, phage P22 (S. enterica) | 96 (162) |
| AAL11524; terminase small subunit, phage ST64T (S. enterica) | 100 (162) | |||||||
| ORF50 | 2 | + | 24878 | 26377 | 499 | Terminase (large subunit) | AAF75044; terminase large subunit, phage P22 (S. enterica) | 99 (441) |
| AAL15523; terminase large subunit, phage ST64T (S. enterica) | 99 (499) | |||||||
| ORF51 | 1 | + | 26377 | 28554 | 725 | Portal protein | AAF75045; portal protein, phage P22 (S. enterica) | 98 (705) |
| AAL15525; portal protein, phage ST64T (S. enterica) | 99 (725) | |||||||
| ORF52 | 8 | + | 28568 | 29479 | 303 | Scaffolding protein | AAF75046; scaffolding protein, phage P22 (S. enterica) | 99 (303) |
| AAL15526; scaffolding protein, phage ST64T (S. enterica) | 99 (303) | |||||||
| ORF53 | 5 | + | 29479 | 30771 | 430 | Coat protein | AAF75047; coat protein, phage P22 (S. enterica) | 99 (430) |
| AAL15527; coat protein, phage ST64T (S. enterica) | 100 (430) | |||||||
| ORF54 | + | 30812 | 31372 | 186 | AAL15528; ORF186, phage ST64T (S. enterica) | 95 (186) | ||
| AAF75048; ORF69, phage P22 (S. enterica) | 36 (44) | |||||||
| ORF55 | 4 | + | 31356 | 31856 | 166 | DNA stabilization | AAF75049; DNA-stabilizing protein, phage P22 (S. enterica) | 99 (166) |
| AAL15529; DNA-stabilizing protein, phage ST64T (S. enterica) | 96 (166) | |||||||
| ORF56 | 10 | + | 31816 | 33234 | 472 | Packaged DNA stabilization | AAF75050; packaged DNA stabilization protein, phage P22 (S. enterica) | 95 (472) |
| AAL15530; packaged DNA stabilization protein, phage ST64T (S. enterica) | 98 (472) | |||||||
| ORF57 | 26 | + | 33238 | 33939 | 233 | Packaged DNA stabilization | AAF75051; packaged DNA stabilization protein, phage P22 (S. enterica) | 80 (233) |
| AAL15531; packaged DNA stabilization protein, phage ST64T (S. enterica) | 54 (233) | |||||||
| ORF58 | 14 | + | 33939 | 34394 | 151 | Unknown | AAF75052; gp14, phage P22 (S. enterica) | 92 (151) |
| AAL15532; gp14, phage ST64T (S. enterica) | 98 (151) | |||||||
| ORF59 | 7 | + | 34397 | 35086 | 229 | DNA transfer protein | AAF75053; DNA transfer protein, phage P22 (S. enterica) | 96 (229) |
| AAL15533; DNA transfer protein, phage ST64T (S. enterica) | 61 (230) | |||||||
| ORF60 | 20 | + | 35097 | 36533 | 478 | DNA transfer protein | AAF75054; DNA transfer protein, phage P22 (S. enterica) | 78 (495) |
| AAL15534; DNA transfer protein, phage ST64T (S. enterica) | 58 (449) | |||||||
| ORF61 | 16 | + | 36533 | 38509 | 658 | DNA transfer protein | AAL15535; DNA transfer protein, phage ST64T (S. enterica) | 100 (658) |
| AAF75055; DNA transfer protein, phage P22 (S. enterica) | 31 (618) | |||||||
| ORF62 | + | 38642 | 38941 | 99 | ZP00032952; hypothetical protein (Burkholderia fungorum) | 42 (87) | ||
| ORF63 | mnt | − | 38962 | 39210 | 82 | Regulatory protein | AAL15536; regulatory protein, phage ST64T (S. enterica) | 100 (82) |
| AAF75057; regulatory protein, phage P22 (S. enterica) | 60 (81) | |||||||
| ORF64 | 9 | + | 39346 | 41349 | 667 | Tailspike protein | AAF75060; tailspike protein, phage P22 (S. enterica) | 98 (667) |
| AAL15537; tailspike protein, phage ST64T (S. enterica) | 99 (667) |
A plus sigin indicates rightward orientation of the gene, and a minus sign indicates leftward orientation of the gene.
Each value in parentheses is the number of amino acids from which the sequence identity is calculated. The GenBank database was used for homology searches.
FIG. 2.
Schematic illustration of the ST104 genome. The predicted ORFs are illustrated. ORFs with a rightward orientation are above the black line; ORFs with a leftward orientation are below the line. The map was opened adjacent to a stem-loop structure located between gene 9 and gtrC as reported in the sequence of P22 (16).
P22 belongs to the formal tailed-phage family Podoviridae because its virions have very short tails (1). This phage also belongs to the informal category known as lambdoid phages. Originally, lambdoid phages were classified as such because they formed recombinant hybrids with phage lambda DNA. Lambdoid phages are often found to have mosaic structures (8). Sequencing of the complete ST104 genome revealed that the ST104 genome is a genetic mosaic composed of gene modules (Table 1; Fig. 2). The morphopoietic gene products (gp3 through gp9) and some other segments are almost identical to those of P22, whereas lysis genes 13, 19, and 15 resemble those of phage PS119. The immunity C (immC) region of P22 consists of the repressor gene (c2), antirepressor gene (cro), and transcriptional activator (c1). The deduced product of the c2 gene of ST104 exhibits no homology to that of P22 but 41% identity to the cI gene product of Escherichia coli lambdoid phage HK022, which is the most similar protein in the database. The deduced product of the cro gene shows a low-level similarity (32% identity) to that of E. coli phage 933W. The gene product of c1 is identical to that of ST64T. The product of replication protein gene 18 shows high similarity (98% identity) to gp54 of E. coli phage HK97. Indeed, ST104 is composed of genome segments typical of at least eight different members of the lambdoid phage family.
The genomic architecture of ST104 is similar not only to that of P22 but also to that of ST64T, which is a temperate phage induced by mitomycin C from DT64. The complete genome of ST64T was only found in DT64. ST64T is a generalized transducing phage, is heteroimmune to P22, and mediates phage type conversion (7). ST64T sequence analysis has confirmed that this bacteriophage has an immunity region different from that of P22 (6). It is likely that the phage type is primarily determined by the carriage of a template phage or the presence or absence of potential receptors on the bacterial cell surface, and it was proposed that integration of ST64T into the chromosome results in phage type conversion by changing immunity to the panel of the typing phage (7). As stated above, the products of c2 and cro, which consists of the immC region of ST104, are different from those of P22 and ST64T. In contrast to phage lambda and most other known lambdoid phages, P22 carries, in addition to the immC region, a second region, immI, that expresses an antirepressor, Ant, and two repressors, Arc and Mnt, that regulate the expression gene ant (12). ST104 has the mint gene; however, the ORFs corresponding to arc and ant are not present in ST104. Although we did not examine if ST104 mediates phage type conversion, ST104 might be resident as a prophage within DT104, and the expression of phage immunity proteins then influences the phage typing results. Further studies are needed to elucidate the function of ST104.
Nucleotide sequence accession numbers.
The DNA sequence data presented here have been submitted to the DDBJ database and appear under accession number AB102868.
Acknowledgments
This research was supported by a grant-in-aid from the Zoonosis Control Project of the Ministry of Agriculture, Forestry and Fisheries of Japan.
REFERENCES
- 1.Ackermann, H. W. 1998. Tailed bacteriophages: the order caudovirales. Adv. Virus. Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glynn, M. K., C. Bopp, W. Dewitt, P. Dabney, M. Mokhtar, and F. J. Angulo. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N. Engl. J. Med. 338:1333-1338. [DOI] [PubMed] [Google Scholar]
- 5.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mmolawa, P. T., H. Schmieger, C. P. Tucker, and M. W. Heuzenroeder. 2003. Genomic structure of the Salmonella enterica serovar Typhimurium DT64 bacteriophage ST64T: evidence for modular genetic architecture. J. Bacteriol. 185:3473-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mmolawa, P. T., R. Willmore, C. J. Thomas, and M. W. Heuzenroeder. 2002. Temperate phages in Salmonella enterica serovar Typhimurium: implications for epidemiology. Int. J. Med. Microbiol. 291:633-644. [DOI] [PubMed] [Google Scholar]
- 8.Oberto, J., S. B. Sloan, and R. A. Weisberg. 1994. A segment of the phage HK022 chromosome is a mosaic of other lambdoid chromosomes. Nucleic Acids Res. 22:354-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sameshima, T., M. Akiba, H. Izumiya, J. Terajima, K. Tamura, H. Watanabe, and M. Nakazawa. 2000. Salmonella typhimurium DT104 from livestock in Japan. Jpn. J. Infect. Dis. 53:15-16. [PubMed] [Google Scholar]
- 10.Schicklmaier, P., T. Wieland, and H. Schmieger. 1999. Molecular characterization and module composition of P22-related Salmonella phage genomes. J. Biotechnol. 73:185-194. [DOI] [PubMed] [Google Scholar]
- 11.Schmieger, H., and P. Schicklmaier. 1999. Transduction of multiple drug resistance of Salmonella enterica serovar Typhimurium DT104. FEMS Microbiol. Lett. 170:251-256. [DOI] [PubMed] [Google Scholar]
- 12.Susskind, M. M. 1980. A new gene of bacteriophage P22 which regulates synthesis of antirepressor. J. Mol. Biol. 138:685-713. [DOI] [PubMed] [Google Scholar]
- 13.Tamada, Y., Y. Nakaoka, K. Nishimori, A. Doi, T. Kumaki, N. Uemura, K. Tanaka, S. I. Makino, T. Sameshima, M. Akiba, M. Nakazawa, and I. Uchida. 2001. Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Threlfall, E. J., J. A. Frost, L. R. Ward, and B. Rowe. 1994. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet. Rec. 134:577. [DOI] [PubMed] [Google Scholar]
- 15.Threlfall, E. J., J. A. Frost, L. R. Ward, and B. Rowe. 1996. Increasing spectrum of resistance in multiresistant Salmonella typhimurium. Lancet 347:1053-1054. [DOI] [PubMed] [Google Scholar]
- 16.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 182:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yee, A. J., S. De Grandis, and C. L. Gyles. 1993. Mitomycin-induced synthesis of a Shiga-like toxin from enteropathogenic Escherichia coli H.I.8. Infect. Immun. 61:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]