Abstract
Jatropha curcas is a promising renewable feedstock for biodiesel and bio-jet fuel production. To study gene expression in Jatropha in different tissues throughout development and under stress conditions, we examined a total of 11 typical candidate reference genes using real-time quantitative polymerase chain reaction (RT-qPCR) analysis, which is widely used for validating transcript levels in gene expression studies. The expression stability of these candidate reference genes was assessed across a total of 20 samples, including various tissues at vegetative and reproductive stages and under desiccation and cold stress treatments. The results obtained using software qBasePLUS showed that the top-ranked reference genes differed across the sample subsets. The combination of actin, GAPDH, and EF1α would be appropriate as a reference panel for normalizing gene expression data across samples at different developmental stages; the combination of actin, GAPDH, and TUB5 should be used as a reference panel for normalizing gene expression data across samples under various abiotic stress treatments. With regard to different developmental stages, we recommend the use of actin and TUB8 for normalization at the vegetative stage and GAPDH and EF1α for normalization at the reproductive stage. For abiotic stress treatments, we recommend the use of TUB5 and TUB8 for normalization under desiccation stress and GAPDH and actin for normalization under cold stress. These results are valuable for future research on gene expression during development or under abiotic stress in Jatropha. To our knowledge, this is the first report on the stability of reference genes in Jatropha.
Keywords: physic nut, reference gene, RT-qPCR, developmental stage, abiotic stress, biofuels
1. Introduction
Physic nut (Jatropha curcas, hereafter referred to as Jatropha) is a perennial, oily seed-bearing plant belonging to the family Euphorbiaceae that most likely originated in Central America and is widely distributed in the tropics and subtropics [1,2]. Jatropha has gained attention in tropical and subtropical countries and has spread beyond its center of origin because of its simple propagation, rapid growth, hardiness, drought tolerance, adaptation to wide agro-climatic conditions, high oil content, and multiple uses [3–6].
Recently, Jatropha has received much attention as a potential renewable feedstock for producing biodiesel and bio-jet fuel that may replace petroleum in many developed and developing countries [7–9]. However, the full potential of Jatropha has not been realized due to several technological and economic factors, one of which is the lack of high-yielding varieties with a high oil content [10–13]. Intensive research efforts have been aimed at improving Jatropha growth and yield performance and enhancing beneficial traits, such as a reduced plant height, more natural branching, early maturity, more female flowers, high seed yield with a high oil content, resistance to pests and diseases, drought tolerance/resistance, and an improved composition of lipids suitable for bio-diesel production [14]. Transgenic approaches have the potential to significantly improve the seed productivity of Jatropha [15,16] but will require studies of Jatropha functional genomics. Because the genome of Jatropha has been sequenced [17,18] and genetic transformation technology has been established [19–22], the identification of a large number of agronomically important genes is now in progress.
Quantifying gene expression levels is important for unraveling the function of agronomically important genes. Real-time quantitative polymerase chain reaction (RT-qPCR) is a widely used technique for assessing gene expression and allows the rapid and reliable quantification of transcripts expressed at low levels [23]. However, for the accurate quantification of mRNA transcripts using RT-qPCR, the identification of stable reference genes is crucial for normalizing the levels of target mRNA [24]. Indeed, the use of unvalidated or unstable reference genes may have significant impacts on the results obtained by RT-qPCR and could lead to erroneous conclusions [25–27]. Although housekeeping genes or endogenous control genes that are thought to be unregulated are typically selected as the reference genes for RT-qPCR analysis, several studies have revealed significant variation in the expression levels of these reference genes under different experimental conditions. As no universal reference gene has been identified in plants or animals to date [26], it is therefore pivotal to identify the best potential reference genes for the experimental system under study [28,29].
In recent years, researchers have evaluated the expression stability of reference genes in many species, such as Arabidopsis [30–33] and rice [34,35]. A good reference gene is one that is essential for cell function and shows relatively constant expression in different tissues throughout different developmental stages or under different experimental conditions [36,37]. In the last decade, relevant tools for selecting reference genes for RT-qPCR analysis have become available, and several research groups have developed software packages, such as geNorm [38], NormFinder [39] and qBase [40], to identify the most stably expressed genes across a set of samples. These tools are freely available on the web, allowing the identification of the best reference gene for specific experiments. In addition, these programs allow the calculation of a normalization factor over multiple reference genes, which further improve the robustness of normalization. qBasePLUS[41] and GenEx [42] are currently considered to be the most powerful, flexible, and user-friendly softwares for RT-qPCR data analyses.
Although there have been many reports describing gene expression analysis under various experimental conditions using different reference genes (18S or actin) by semi-quantitative RT-PCR [43,44] or RT-qPCR [45–48] in Jatropha, a comparison of the different reference genes in Jatropha has not yet been reported. Accordingly, we evaluated the expression stability in Jatropha of 11 typical candidate reference genes by RT-qPCR among tissues at different developmental stages and under different abiotic stress treatments. Our results reveal that different reference genes should be selected according to the sample type and that a combination of the most stable reference genes provides a more accurate and reliable method of normalization in RT-qPCR analyses.
2. Results and Discussion
2.1. PCR Amplification Specificity and PCR Efficiency
The melting curves for the amplified products of all 11 candidate reference genes showed a single peak, corresponding to a specific melting temperature (Figure S1a), and agarose gel electrophoresis showed a single band at the correct molecular weight for each product (Figure S1b), indicating good specificity of all the primer pairs used in RT-qPCR. A significant linear relationship (R2 > 0.99) between the fractional cycle number and the log of the initial copy number was demonstrated for the 11 calibration curves (Table 1). The PCR efficiency of each candidate reference gene, which ranged from 90% for UBQ-LIKE to 109% for UBQ10, is shown in Table 1, and these efficiencies were used in the calculations for the subsequent expression data.
Table 1.
Slopes, amplification efficiency (E), correlation coefficient (R2), annealing temperature (Ta) and melting temperature (Tm) [30] of the selected reference genes.
| Gene symbol | Slope | E (%) | R2 | Ta (°C) | Tm (°C) |
|---|---|---|---|---|---|
| 18S | −3.21 | 105% | 0.999 | 58 | 85.57 |
| Actin | −3.299 | 101% | 0.995 | 58 | 83.90 |
| EF1α | −3.158 | 107% | 0.999 | 59 | 82.45 |
| GAPDH | −3.173 | 107% | 0.998 | 58 | 85.72 |
| TUA3 | −3.353 | 99% | 0.999 | 58 | 86.46 |
| TUB5 | −3.462 | 94% | 0.999 | 58 | 84.05 |
| TUB8 | −3.285 | 102% | 0.998 | 58 | 82.58 |
| UBQ10 | −3.129 | 109% | 0.998 | 59 | 79.79 |
| UBQ-LIKE | −3.595 | 90% | 0.996 | 59 | 84.38 |
| UEC | −3.149 | 107% | 0.999 | 58 | 85.79 |
| UEP | −3.23 | 104% | 0.999 | 58 | 86.24 |
2.2. Transcript Accumulation of Candidate Reference Genes
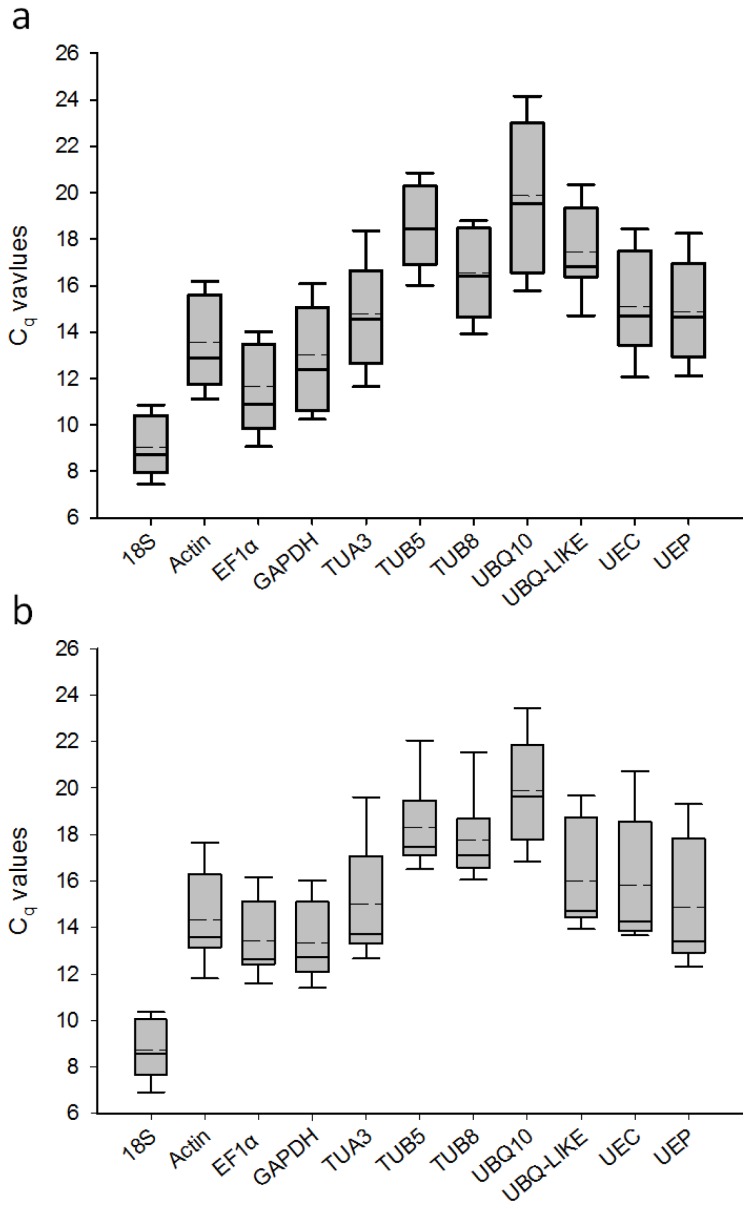
The Cq values, which indicate the cycle at which the fluorescence signal is significantly different from background, of the 11 candidate reference genes were obtained by RT-qPCR using two sample subsets. Figure 1a shows the transcript accumulation across 11 tissue samples at two different developmental stages, and Figure 1b shows the transcript accumulation across 9 tissue samples under two different abiotic stress treatments. In both sample subsets, the 18S rRNA gene always had the lowest Cq values, even though the cDNA templates were 50-fold diluted relative to any other genes. In contrast, UBQ10 consistently showed the highest Cq values. The other three genes with abundant expression levels were EF1α, GAPDH, and actin, with mean Cq values < 15 in all the samples. The average Cq values of each candidate reference gene across all the samples and their relative expression variation across all the samples are shown in Supplementary Material (Tables S1 and S2).
Figure 1.
Cq values of the 11 candidate reference genes in the RT-qPCR analysis (a) Eleven samples at two developmental stages; (b) Nine samples exposed to desiccation and cold stress treatments. The Cq values are shown as the first and third quartile. The vertical lines indicate the range of values. The median values are indicated by solid lines, and the mean values are indicated by broken lines.
2.3. Ranking of Candidate Reference Genes and Determination of Optimal Reference Genes
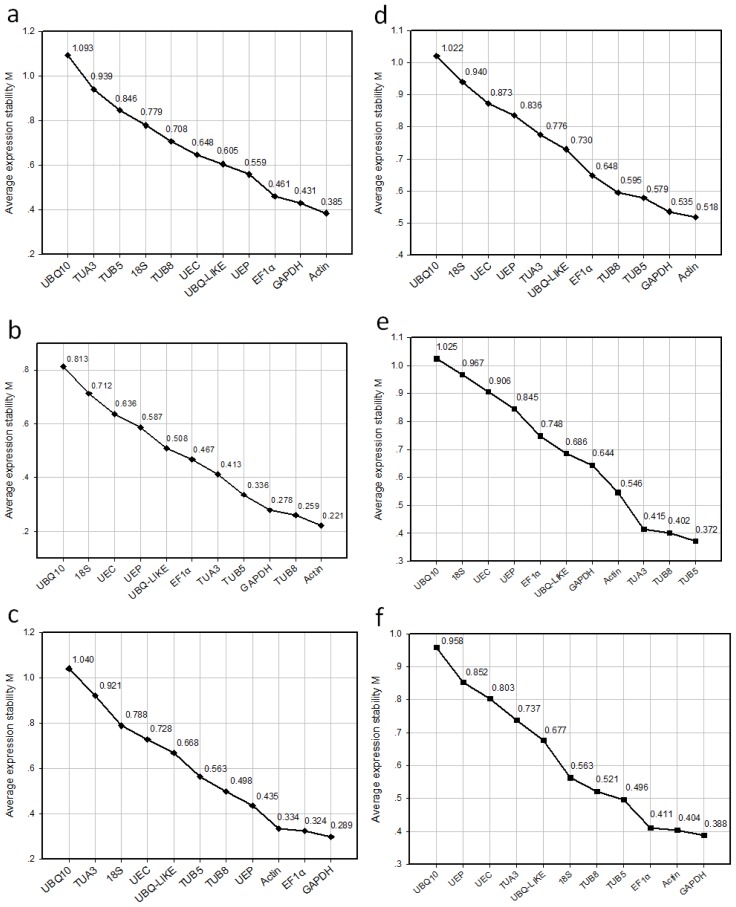
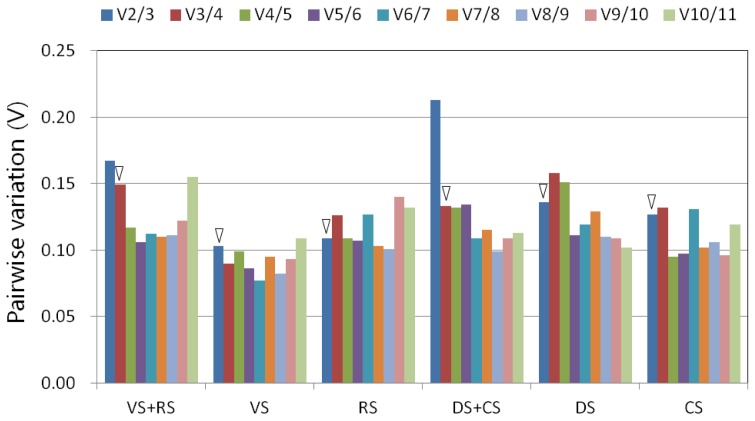
To minimize the bias generated by the assumptions underlying the different evaluation methods, the expression data in this study were analyzed using two different programs, qBasePLUS[40] and NormFinder [39], to rank the candidate reference genes. Based on calculations using the qBasePLUS software, the M values of the candidate reference genes in all six datasets tested were below the default limit of 1.5 (Figure 2). Among the 11 candidate reference genes, the most stable reference genes varied within the sample datasets. In the samples obtained at two different developmental stages, the three most stable transcripts were actin, GAPDH and EF1α (Figure 2a). The V value (V2/3 = 0.167) indicated that the third reference gene, EF1α, was required in the developmental series, and the optimal normalization factor can be calculated as the geometric mean of the reference targets actin, GAPDH, and EF1α (V3/4 = 0.149) (Figure 3). However, the inclusion of only the two most stably expressed genes as references was sufficient in cases in which the datasets contained only one developmental stage, as the addition of a third reference gene did not have a significant effect on the result (Figure 3). At the vegetative stage, the two most stably expressed genes were actin and TUB8 (Figure 2b), and the pairwise variation V2/3 was 0.103 (Figure 3), which is below the default cut-off value of 0.15. This result suggests that the combination of actin and TUB8 is optimal for transcript accumulation analysis at this developmental stage. At the reproductive stage, GAPDH and EF1α were the two most stably expressed genes (Figure 2c), with V2/3 = 0.109 (Figure 3), indicating that the addition of a third reference gene was also not necessary. Therefore, GAPDH and EF1α are sufficient for use as reference genes at the reproductive stage.
Figure 2.
Expression stability values (M) and ranking of the candidate reference genes based on qBasePLUS calculations. (a) Two different developmental stages; (b) Vegetative stage; (c) Reproductive stage; (d) Desiccation and cold stress treatments; (e) Desiccation stress treatment; (f) Cold stress treatment. The M value and ranking were calculated through a pairwise comparison and stepwise exclusion of the least stable gene. Low M values correspond to a high expression stability.
Figure 3.
Pairwise variation (V) analysis by qBasePLUS to determine the optimal number of reference genes needed for accurate normalization in all the tested samples. V values less than 0.15 indicate that no additional genes are needed to calculate a reliable normalization factor. The triangles indicate V values lower than 0.15. CS, cold stress; DS, desiccation stress; RS, reproductive stage; VS, vegetative stage.
In the dataset including the samples from the two abiotic stress treatments, the two most stably expressed genes were actin and GAPDH (Figure 2d), and the pairwise variation V2/3 value was 0.213 (Figure 3). When a third reference gene (TUB5) was added, the pairwise variation (V3/4 = 0.133) dropped significantly below the default cut-off value of 0.15 (Figure 3). Thus, the normalization factor should preferably contain at least three of the best reference genes (actin, GAPDH, and TUB5). However, the results were different when the samples obtained from the desiccation and cold stress treatments were analyzed separately. The use of two of the most stably expressed reference genes as internal control genes was sufficient for the samples obtained from each of the stress treatments. For the desiccation stress treatment, the combination of TUB5 and TUB8 (Figure 2e) was appropriate for gene expression normalization, whereas GAPDH and actin (Figure 2f) were the best combination for the cold stress treatment (Figure 3). An exclusion analysis was performed to test the possibility that TUB5 and TUB8 are co-regulated, whereby each gene was excluded in separate analyses. The ranking of the genes was the same, with very similar M values, indicating a lack of co-regulation between TUB5 and TUB8 (data not shown).
In addition to the analysis by qBasePLUS, we also evaluated the RT-qPCR data obtained in this study using the NormFinder software [39] and found a similar but not identical ranking of the reference genes in the different sample subsets (Table 2). Actin, EF1α, and GAPDH were found to be the three most stable reference genes in the dataset including samples from two different developmental stages (Table 2), which is consistent with the result determined by qBasePLUS (Figure 2a). In contrast, the ranking of EF1α and GAPDH differed. In the subset containing only samples from the vegetative or reproductive stage, the two most stably expressed genes determined by NormFinder were actin and GAPDH or EF1α and UEP, respectively (Table 2), whereas actin and TUB8 or GAPDH and EF1α were identified by qBasePLUS (Figure 2b,c).
Table 2.
Candidate reference genes ranked according to their expression stability (M value) as determined by NormFinder.
| Ranking | VS + RS | VS | RS | DS + CS | DS | CS |
|---|---|---|---|---|---|---|
|
| ||||||
| Gene (M value) | Gene (M value) | Gene (M value) | Gene (M value) | Gene (M value) | Gene (M value) | |
| 1 | Actin (0.112) | Actin (0.112) | EF1α (0.020) | Actin (0.139) | Actin (0.116) | Actin (0.133) |
| 2 | EF1α (0.194) | GAPDH (0.185) | UEP (0.110) | GAPDH (0.256) | UBQ-LIKE (0.268) | GAPDH (0.136) |
| 3 | GAPDH (0.230) | TUB8 (0.187) | GAPDH (0.200) | UBQ-LIKE (0.349) | GAPDH (0.309) | EF1α (0.279) |
| 4 | UEP (0.287) | UBQ-LIKE (0.229) | Actin (0.201) | TUB5 (0.419) | TUB5 (0.432) | UBQ-LIKE (0.387) |
| 5 | UBQ-LIKE (0.406) | EF1α (0.289) | TUB8 (0.429) | EF1α (0.434) | TUB8 (0.476) | TUB5 (0.436) |
| 6 | TUB8 (0.424) | TUA3 (0.364) | UEC (0.459) | TUA3 (0.467) | EF1α (0.489) | TUA3 (0.470) |
| 7 | UEC (0.486) | UEP (0.394) | UBQ-LIKE (0.509) | TUB8 (0.497) | TUA3 (0.549) | TUB8 (0.475) |
| 8 | 18S (0.646) | TUB5 (0.416) | TUB5 (0.597) | UEP (0.593) | UEP (0.586) | UEC (0.592) |
| 9 | TUB5 (0.678) | UEC (0.459) | 18S (0.654) | UEC (0.664) | UBQ10 (0.662) | UEP (0.631) |
| 10 | TUA3 (0.886) | 18S (0.692) | TUA3 (0.988) | 18S (0.763) | UEC (0.686) | 18S (0.634) |
| 11 | UBQ10 (1.185) | UBQ10 (0.807) | UBQ10 (1.030) | UBQ10 (0.848) | 18S (0.757) | UBQ10 (0.897) |
In the dataset including samples from the two abiotic stress treatments, NormFinder identified actin, GAPDH, and UBQ-like as the three most stable reference genes (Table 2), with actin and GAPDH also being identified by qBasePLUS (Figure 2d). In the subset containing only samples exposed to desiccation stress, actin and UBQ-like were identified by NormFinder as the two most stably expressed genes (Table 2), whereas TUB5 and TUB8 were identified by qBasePLUS (Figure 2e). However, GAPDH and actin were identified by both programs as the best combination of reference genes for the subset containing only samples treated with cold stress (Table 2, Figure 2f). Some discrepancies between the rankings obtained from qBasePLUS and NormFinder are expected because the two evaluation approaches are based on distinct statistical algorithms [49].
2.4. Validation of the Selected Reference Genes in Leaf Samples Treated with Desiccation or Cold Stress
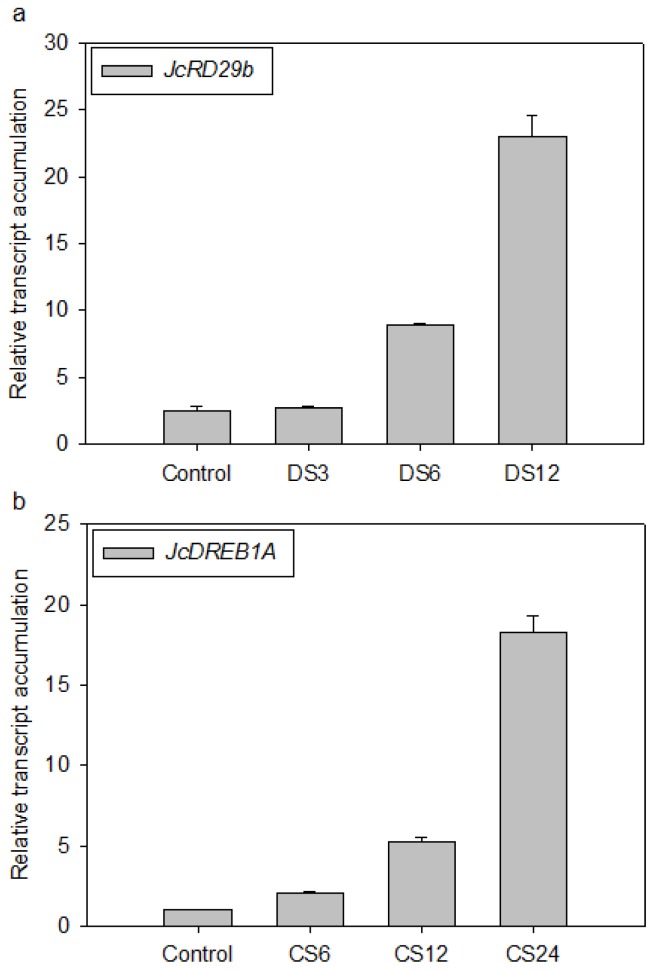
To validate the selected reference genes, the relative expression levels of two stress-responsive genes of Jatropha, JcRD29b and JcDREB1A, were evaluated in leaf samples from plants treated with desiccation or cold stress. The JcRD29b and JcDREB1A Arabidopsis homologs RD29b and DREB1A were reported to be induced by desiccation [47] and cold stress [50], respectively. Using the reference genes TUB5 and TUB8 (Figure 2e and Figure 3), our RT-qPCR analysis revealed that JcRD29b expression was induced at 6 h after the start of desiccation, with its mRNA level significantly increasing after 12 h (Figure 4a). For JcDREB1A, the reference genes GAPDH and actin (Figure 2f and Figure 3) were used to normalize the RT-qPCR data, and we found that the expression of JcDREB1A was induced by low-temperature treatment (4 °C) within 6 h, with the strongest expression after 12 h (Figure 4b).
Figure 4.
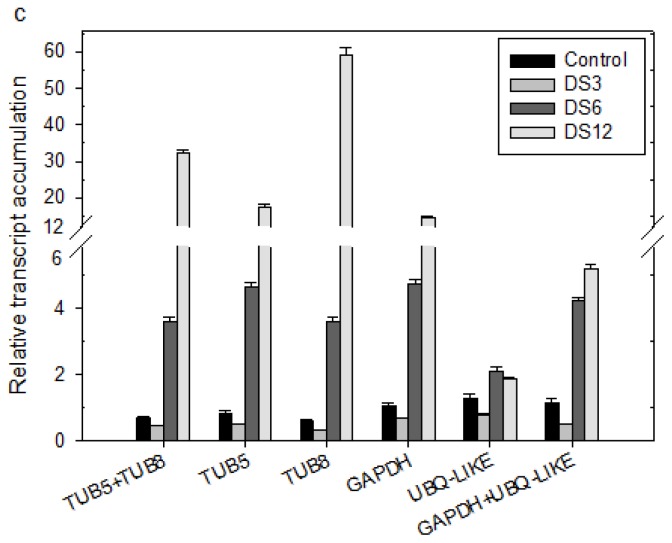
Normalized transcript accumulation of two stress-responsive genes of Jatropha, as determined by RT-qPCR analyses. (a) Expression profile of JcRD29b relative to TUB5 and TUB8 in samples exposed to desiccation stress (DS) for 0 (control), 3, 6 and 12 h; (b) Expression profile of JcDREB1A relative to GAPDH and actin in samples exposed to cold stress (CS) at 4 °C for 0 (control), 6, 12 and 24 h; (c) Comparison of JcRD29b expression profiles relative to the two most stable reference genes (TUB5 and TUB8) and the two unstable reference genes (GAPDH and UBQ-LIKE) in samples exposed to desiccation stress (DS) for 0 (control), 3, 6 and 12 h.
To further validate the suitability of the reference genes identified in this study, we used the two most stable reference genes, TUB5 and TUB8, and the reference genes ranked 5th and 6th by qBasePLUS, GAPDH and UBQ-LIKE (Figure 2e), to quantify the expression profile of JcRD29b in Jatropha seedlings exposed to desiccation stress. Although normalization using TUB5, TUB8, or GAPDH produced similar expression patterns for each gene, normalization using UBQ-LIKE resulted in a strong bias, significantly decreasing the desiccation-induced expression of JcRD29b (Figure 4c). This result demonstrates that UBQ-LIKE is an unstable reference gene for normalization under conditions of experimental desiccation stress, yet NormFinder defined UBQ-LIKE as the second most stable reference gene in the subset containing samples treated with desiccation stress (Table 2). So we conclude that qBasePLUS is preferred for the calculation of normalization factors at least in samples exposed to desiccation stress.
3. Experimental Section
3.1. Plant Materials and Stress Treatments
At the vegetative stage, tissues (roots, hypocotyls, cotyledons, young leaves, and mature leaves) were collected from Jatropha seedlings that were grown in a growth chamber for three weeks after germination (14 h light/day, 20–25 °C). At the reproductive stage, tissues (roots, stems, young leaves, mature leaves, flowers shoots, and seeds at 20 days after pollination [DAP]) were collected from three-year-old adult Jatropha plants that were grown in a field at the Xishuangbanna Tropical Botanical Garden (XTBG; 21°54′N, 101°46′E; 580 m elevation) of the Chinese Academy of Sciences located in Mengla County, Yunnan Province, southwest China. All the collected samples were immediately frozen in liquid nitrogen and stored at −80 °C.
The Jatropha seedlings that were treated with different abiotic stresses were grown under the same conditions as described above for two weeks after germination (at the three-leaf stage). For the desiccation stress treatment, the seedlings were carefully removed from the soil, and their roots were flushed with tap water. The seedlings were then placed on filter paper and left on the lab bench for the period of time indicated; the roots, hypocotyls, and young leaves were collected at 3, 6 and 12 h after the initiation of the desiccation treatment. For the cold stress treatment, the plants were subjected to 4 °C for 6, 12 and 24 h. Control tissues (roots, hypocotyls, and young leaves) were collected from plants that were untreated. After collection, all the samples were immediately frozen in liquid nitrogen and stored at −80 °C.
3.2. Candidate Reference Genes from Jatropha
Eleven typical candidate reference genes were selected for analysis in this study (Table 3). The nucleotide sequences of these genes were obtained from the Jatropha genomic database [51] or the National Center for Biotechnology Information (NCBI) nucleotide database [52]. The primer pairs used for RT-qPCR were designed using the software Primer Premier 5 and are listed in Table 3.
Table 3.
Primer sequences, PCR amplicon length of the selected reference genes and stress responsive genes in Jatropha.
| Gene ID | Gene symbol | Primer sequences | Amplicon length |
|---|---|---|---|
| Jcr4S06558.10 a | Actin | F: 5′-CTCCTCTCAACCCCAAAGCCAA-3′ R: 5′-CACCAGAATCCAGCACGATACCA-3′ |
147 bp |
| Jcr4U29393.10 a | GAPDH | F: 5′-TGAAGGACTGGAGAGGTGGAAGAGC-3′ R: 5′-ATCAACAGTTGGAACACGGAAAGCC-3′ |
140 bp |
| Jcr4S00045.200 a | UBQ10 | F: 5′-AAAGCAGTTGGAGGATGGAAGGAC-3′ R: 5′-GCGAAGCCTGAGAACAAGGTGAAG-3′ |
82 bp |
| Jcr4S10519.50 a | UBQ-LIKE | F: 5′-GGTGAGAGTGAAGTGTAATGATGACGAC-3′ R: 5′-CCTCAGAGTTATATGGTCCTTGTAAATGG-3′ |
136 bp |
| Jcr4508473.50 a | UEP | F: 5′-AATCCCTCCAGACCAGCAGCGACT-3′ R: 5′-GCTCTTGTAGAACTGAAGCACGGC-3′ |
220 bp |
| Jcr4S00542.10 a | EF1α | F: 5′-AAGATGATTCCCACCAAGCCCA-3′ R: 5′-CACAGCAAAACGACCCAGAGGA-3′ |
72 bp |
| GW880075.1 b | TUA3 | F: 5′-TTCAATCAGCGAAAATGAGAGAGTG-3′ R: 5′-TCACTGAAAAAGGTGTTGAAGGCA-3′ |
178 bp |
| GW877086.1 b | TUB8 | F: 5′-GCAGGGAATAACTGGGCTAAAGGT-3′ R: 5′-CTCCACCCAACGAATGACAAACTT-3′ |
136 bp |
| EZ114400.1 b | UEC | F: 5′-GTCCCTGATTTTGAGATGGCGTC-3′ R: 5′-CAATATGTCAAGACAAATGCTCCCG-3′ |
284 bp |
| GW878948.1 b | TUB5 | F: 5′-TATGTTCCCAGGGCGGTTCTAATG-3′ R: 5′-GGACTGCCCAAAGACAAAGTTATCG-3′ |
111 bp |
| AY823528.1 b | 18S | F: 5′-CTCAACCATAAACGATGCCGACC-3′ R: 5′-TTCAGCCTTGCGACCATACTCCC-3′ |
117 bp |
| Jcr4S01474.40 a | JcRD29b | F: 5′-AATCTCCGCAAAGAATGTTGTAGC-3′ R: 5′-CTCCCTGTCTCAGCAACTTTCTCATA-3′ |
180 bp |
| Jcr4S27135.10 a | JcDREB1A | F: 5′-CGGATGGACTTTTAGGGGATGAAT-3′ R: 5′-CACTGAGGTGGAGGCAACAACA-3′ |
160 bp |
accession number in Jatropha genome database [52];
GenBank accession number; F, forward primer; R. reverse primer.
3.3. Desiccation Stress- and Cold Stress-Responsive Genes from Jatropha
The Jatropha homologs of the two Arabidopsis stress-responsive genes RD29b [53] and DREB1A [50], JcRD29b and JcDREB1A, were chosen for the RT-qPCR analysis in this study. Arabidopsis RD29b is responsive to desiccation and was induced at 3 h after the start of desiccation; its mRNA level reached a maximum at 10 h [53]. The expression of Arabidopsis DREB1A, which encodes a dehydration response element-binding protein, was induced within 1 h of exposure to low temperature (4 °C), and the DREB1A mRNA level peaked after 2 h [50]. The nucleotide sequences of JcRD29b and JcDREB1A were obtained from the Jatropha genomic database [52]; the primer pairs used in RT-qPCR were designed with the software Primer Premier 5 and are listed in Table 3.
3.4. RNA Isolation and Purification and cDNA Synthesis
Two biological replicate samples were collected at each developmental stage and under each abiotic stress treatment. Total RNA was extracted using the silica particle-phenol/chloroform extraction method, as described by Ding et al. [54]. All RNA samples were analyzed spectrophotometrically, and their absorbance ratios at 260/280 and 260/230 nm were greater than 1.8. The RNA samples were also analyzed by agarose gel electrophoresis, showing well-defined bands corresponding to each rRNA and no evidence of nucleic acid degradation. The total RNA samples were purified using the RNAclean Kit (TIANGEN Biotech, Beijing, China), treated with DNase I to remove DNA, and immediately reverse transcribed using the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Bio. Inc., Dalian, China) according to the manufacturer’s protocol. Either a gene-specific primer mix or oligo-dT primers were used in the reverse transcription reactions. In a preliminary test with three different genes (actin, GAPDH and TUB8), consistent RT-qPCR expression patterns were observed in different tissues (roots, cotyledons, stems, young leaves, and mature leaves), regardless of whether the cDNAs were synthesized using the gene-specific primer mix or oligo-dT primers (Figure S2). Because more transcript accumulation was detected with cDNAs obtained using the gene-specific primer mix (Figure S2), the cDNAs used for subsequent experiments in this study were reverse transcribed with the gene-specific primer mix.
3.5. Primer Design and RT-qPCR Analysis
The RT-qPCR primers (Table 3) were designed using the software Primer Premier 5. To verify the sequences of the reference genes, all the amplicons of the 11 candidate genes were analyzed on 1.5% agarose gels and sequenced (Figure S1b and Table S3). Each PCR reaction contained 10 μL SYBR® Premix Ex Taq™ II (Tli RNaseH Plus, Takara Bio. Inc., Dalian, China), 200 nM each primer, and 2 μL diluted cDNA (18S cDNA was diluted 50-fold relative to all other genes) in a total volume of 20 μL. No-template controls were also included for each primer pair. The RT-qPCR reactions were performed using the Roche LightCycler 480 Real-time PCR Detection System (Roche Diagnostics, Mannheim, Germany). We chose the annealing temperature with the highest amplification and best product specificity, as determined by a melting curve analysis. The reactions were performed in a 96-well reaction plate under the following conditions: 5 s at 95 °C for the initial denaturation, followed by 40 cycles of 5 s at 95 °C, 20 s at the optimal temperature for each primer pair (Table 1), and 20 s at 72 °C for PCR amplification, and 1 cycle of 15 s at 95 °C, 20 s at 65 °C and 0.06 °C/s heating up to 97 °C for the melting curve. The machine measures the DNA concentration in each cycle when the temperature is at 72 °C. Three technical replicates were analyzed for each biological replicate. The amplification efficiency of each set of primers was tested prior to the expression studies and was calculated as E = −1 + 10(−1/slope)[55], where the slope is derived from a standard curve generated using four 10-fold serial dilutions of cDNA obtained from young leaves.
3.6. Data Analysis
We used a Roche LightCycler 480 Real-time PCR Detection System to collect the fluorescence data. The cycle threshold, Cq (the cycle at which the fluorescence signal is significantly different from the background), was determined for each reaction. The average Cq and standard deviation of Cq for each sample were estimated based on three replicates; outliers were removed from the average Cq calculation. Any replicates showing non-specific products in the dissociation curve analysis were also removed. At least two of the three technical replicates were included in the average Cq calculations.
Two algorithms—qBasePLUS and NormFinder—were used to determine the best reference genes for the normalization of the gene expression data. The software qBasePLUS V2.4 [41], combines proven geNorm and qBase technology, and is thus considered valid software for analyzing reference genes [40]. This method is based on the principle that the expression ratio of two ideal reference genes is identical in all samples, regardless of the cell type or treatment. The pairwise variation of a particular gene from all the other reference genes is calculated and defined as the M value, and the gene with the lowest M value is considered to have the most stable expression. The pairwise variations between two subsequent normalization factors (V value, Vn/n+1) are also calculated, indicating the effect of including one additional gene for normalization. The recommended cut-off of 0.15 for the V value was used in our study [38]. NormFinder uses an analysis of variance (ANOVA)-based model to estimate intra- and inter-group variations and combines these estimates to provide a direct measure of the variation in expression for each gene [39]. The candidates are ranked according to their expression stability.
4. Conclusions
This study represents the first attempt to select a set of commonly used candidate reference genes in Jatropha for the normalization of gene expression data using RT-qPCR. Our results indicate that more than one reference gene is preferred and that different reference genes should be selected according to the different sample type. According to the results calculated using the qBasePLUS software, the best combinations of reference genes for the normalization of gene expression profiles in Jatropha are as follows: actin + GAPDH + EF1α for all developmental stages; actin + TUB8 for the vegetative stage; GAPDH + EF1α for the reproductive stage; actin + GAPDH + TUB5 for desiccation and cold stress treatments; TUB5 + TUB8 for desiccation stress treatment; and GAPDH + actin for cold stress treatment. The results obtained in this study provide information that will ensure more accurate RT-qPCR-based gene expression quantification in Jatropha.
Supplementary Information
Acknowledgments
The authors thank Yaling Song for critically reading the manuscript. This work was funded by the Top Science and Technology Talents Scheme of Yunnan Province (2009CI123), the Natural Science Foundation of Yunnan Province (2011FA034), and the CAS 135 program (XTBG-T02) (to Z.-F. Xu) and the West Light Foundation of the Chinese Academy of Sciences (to Q.-T. Fu).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Carels N. Jatropha curcas: A review. Adv. Bot. Res. 2009;50:39–86. [Google Scholar]
- 2.Fairless D. Biofuel: The little shrub that could—Maybe. Nature. 2007;449:652–655. doi: 10.1038/449652a. [DOI] [PubMed] [Google Scholar]
- 3.Francis G., Edinger R., Becker K. A concept for simultaneous wasteland reclamation, fuel production, and socio-economic development in degraded areas in India: Need, potential and perspectives of Jatropha plantations. Nat. Resour. Forum. 2005;29:12–24. [Google Scholar]
- 4.Jones N., Miller J.H. Jatropha curcas: A Multipurpose Species for Problematic Sites. World Bank; Washington, DC, USA: 1992. pp. 1–12. [Google Scholar]
- 5.Kumar A., Sharma S. An evaluation of multipurpose oil seed crop for industrial uses (Jatropha curcas L.): A review. Ind. Crops Prod. 2008;28:1–10. [Google Scholar]
- 6.Makkar H.P., Becker K. Jatropha curcas, a promising crop for the generation of biodiesel and value–added coproducts. Eur. J. Lipid Sci. Technol. 2009;111:773–787. [Google Scholar]
- 7.Abdulla R., Chan E.S., Ravindra P. Biodiesel production from Jatropha curcas: A critical review. Crit. Rev. Biotechnol. 2011;31:53–64. doi: 10.3109/07388551.2010.487185. [DOI] [PubMed] [Google Scholar]
- 8.Bonnet S., Gheewala S. Potential of Jatropha as an Energy Crop. In: Carels N., Sujatha M., Bahadur B., editors. Jatropha, Challenges for a New Energy Crop. Vol. 1. Springer; New York, NY, USA: 2012. pp. 571–582. [Google Scholar]
- 9.Li L., Coppola E., Rine J., Miller J.L., Walker D. Catalytic hydrothermal conversion of triglycerides to non-ester biofuels. Energy Fuels. 2010;24:1305–1315. [Google Scholar]
- 10.Chikara J., Prakash A., Mastan S.G., Ghosh A. Genetic Improvement in Jatropha curcas through Selection and Breeding. In: Bahadur B., Sujatha M., Carels N., editors. Jatropha, Challenges for a New Energy Crop. Vol. 2. Springer; New York, NY, USA: 2013. pp. 119–133. [Google Scholar]
- 11.Divakara B.N., Upadhyaya H.D., Wani S.P., Gowda C.L.L. Biology and genetic improvement of Jatropha curcas L.: A review. Appl. Energy. 2010;87:732–742. [Google Scholar]
- 12.Pan B.-Z., Xu Z.-F. Benzyladenine treatment significantly increases the seed yield of the biofuel plant Jatropha curcas. J. Plant Growth Regul. 2011;30:166–174. [Google Scholar]
- 13.Sanderson K. Wonder weed plans fail to flourish. Nature. 2009;461:328–329. doi: 10.1038/461328a. [DOI] [PubMed] [Google Scholar]
- 14.Carels N. Towards the Domestication of Jatropha: The Integration of Sciences. In: Bahadur B., Sujatha M., Carels N., editors. Jatropha, Challenges for a New Energy Crop. Vol. 2. Springer; New York, NY, USA: 2013. pp. 263–299. [Google Scholar]
- 15.Gressel J. Transgenics are imperative for biofuel crops. Plant Sci. 2008;174:246–263. [Google Scholar]
- 16.Sujatha M., Reddy T.P., Mahasi M.J. Role of biotechnological interventions in the improvement of castor (Ricinus communis L.) and Jatropha curcas L. Biotechnol. Adv. 2008;26:424–435. doi: 10.1016/j.biotechadv.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Hirakawa H., Tsuchimoto S., Sakai H., Nakayama S., Fujishiro T., Kishida Y., Kohara M., Watanabe A., Yamada M., Aizu T., et al. Upgraded genomic information of Jatropha curcas L. Plant Biotechnol. 2012;29:123–130. [Google Scholar]
- 18.Sato S., Hirakawa H., Isobe S., Fukai E., Watanabe A., Kato M., Kawashima K., Minami C., Muraki A., Nakazaki N., et al. Sequence analysis of the genome of an oil-bearing tree, Jatropha curcas L. DNA Res. 2011;18:65–76. doi: 10.1093/dnares/dsq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kajikawa M., Morikawa K., Inoue M., Widyastuti U., Suharsono S., Yokota A., Akashi K. Establishment of bispyribac selection protocols for Agrobacterium tumefaciens- and Agrobacterium rhizogenes-mediated transformation of the oil seed plant Jatropha curcas L. Plant Biotechnol. 2012;29:145–153. [Google Scholar]
- 20.Kumar N., Reddy M., Sujatha M. Genetic transformation of Jatropha curcas: Current status and future prospects. In: Bahadur B., Sujatha M., Carels N., editors. Jatropha, Challenges for a New Energy Crop. Vol. 2. Springer; New York, NY, USA: 2013. pp. 535–546. [Google Scholar]
- 21.Li M.R., Li H.Q., Jiang H.W., Pan X.P., Wu G.J. Establishment of an Agrobacteriuim-mediated cotyledon disc transformation method for Jatropha curcas. Plant Cell Tiss. Organ Cult. 2008;92:173–181. [Google Scholar]
- 22.Pan J.L., Fu Q.T., Xu Z.F. Agrobacterium tumefaciens-mediated transformation of biofuel plant Jatropha curcas using kanamycin selection. Afr. J. Biotechnol. 2010;9:6477–6481. doi: 10.1007/s11816-015-0377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bustin S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 24.Phillips M.A., D’Auria J.C., Luck K., Gershenzon J. Evaluation of candidate reference genes for real-time quantitative PCR of plant samples using purified cDNA as template. Plant Mol. Biol. Report. 2009;27:407–416. doi: 10.1007/s11105-008-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guénin S., Mauriat M., Pelloux J., Van Wuytswinkel O., Bellini C., Gutierrez L. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 2009;60:487–493. doi: 10.1093/jxb/ern305. [DOI] [PubMed] [Google Scholar]
- 26.Gutierrez L., Mauriat M., Guénin S., Pelloux J., Lefebvre J.F., Louvet R., Rusterucci C., Moritz T., Guerineau F., Bellini C. The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription–polymerase chain reaction (RT–PCR) analysis in plants. Plant Biotechnol. J. 2008;6:609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- 27.Huggett J., Dheda K., Bustin S., Zumla A. Real-time RT-PCR normalisation; Strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 28.Bustin S.A. Why the need for qPCR publication guidelines?—The case for MIQE. Methods. 2010;50:217–226. doi: 10.1016/j.ymeth.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 30.Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekkers B.J.W., Willems L., Bassel G.W., van Bolderen-Veldkamp R.P., Ligterink W., Hilhorst H.W.M., Bentsink L. Identification of reference genes for RT–qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol. 2012;53:28–37. doi: 10.1093/pcp/pcr113. [DOI] [PubMed] [Google Scholar]
- 32.Hong S.M., Bahn S.C., Lyu A., Jung H.S., Ahn J.H. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 2010;51:1694–1706. doi: 10.1093/pcp/pcq128. [DOI] [PubMed] [Google Scholar]
- 33.Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227:1343–1349. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- 34.Jain M., Nijhawan A., Tyagi A.K., Khurana J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 35.Li Q.-F., Sun S.M., Yuan D.-Y., Yu H.-X., Gu M.-H., Liu Q.-Q. Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol. Biol. Report. 2010;28:49–57. [Google Scholar]
- 36.Qi J., Yu S., Zhang F., Shen X., Zhao X., Yu Y., Zhang D. Reference gene selection for real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage (Brassica rapa L. ssp. pekinensis) Plant Mol. Biol. Report. 2010;28:597–604. [Google Scholar]
- 37.Schmittgen T.D., Zakrajsek B.A. Effect of experimental treatment on housekeeping gene expression: Validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods. 2000;46:69–81. doi: 10.1016/s0165-022x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 38.Vandesompele J., de Preter K., Pattyn F., Poppe B., van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 40.Hellemans J., Mortier G., de Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biogazelle. [(accessed on 18 May 2012)]. http://www.biogazelle.com/qbaseplus.
- 42.Multid Analyses. [(accessed on 18 May 2012)]. http://www.multid.se/
- 43.Liu B., Wang W., Gao J., Chen F., Wang S., Xu Y., Tang L., Jia Y. Molecular cloning and characterization of a jasmonate biosynthetic pathway gene for allene oxide cyclase from Jatropha curcas. Acta Physiol. Plant. 2010;32:531–539. [Google Scholar]
- 44.Zhang F.-L., Niu B., Wang Y.-C., Chen F., Wang S.-H., Xu Y., Jiang L.-D., Gao S., Wu J., Tang L. A novel betaine aldehyde dehydrogenase gene from Jatropha curcas, encoding an enzyme implicated in adaptation to environmental stress. Plant Sci. 2008;174:510–518. [Google Scholar]
- 45.Li J., Li M.-R., Wu P.-Z., Tian C.-E., Jiang H.-W., Wu G.-J. Molecular cloning and expression analysis of a gene encoding a putative β-ketoacyl-acyl carrier protein (ACP) synthase III (KAS III) from Jatropha curcas. Tree Physiol. 2008;28:921–927. doi: 10.1093/treephys/28.6.921. [DOI] [PubMed] [Google Scholar]
- 46.Omar S.A., Fu Q.-T., Chen M.-S., Wang G.-J., Song S.-Q., Elsheery N.I., Xu Z.F. Identification and expression analysis of two small heat shock protein cDNAs from developing seeds of biodiesel feedstock plant Jatropha curcas. Plant Sci. 2011;181:632–637. doi: 10.1016/j.plantsci.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Xu R., Wang R., Liu A. Expression profiles of genes involved in fatty acid and triacylglycerol synthesis in developing seeds of Jatropha (Jatropha curcas L.) Biomass Bioenergy. 2011;35:1683–1692. [Google Scholar]
- 48.Yang J., Yang M.F., Wang D., Chen F., Shen S.H. JcDof1, a Dof transcription factor gene, is associated with the light-mediated circadian clock in Jatropha curcas. Physiol. Plant. 2010;139:324–334. doi: 10.1111/j.1399-3054.2010.01363.x. [DOI] [PubMed] [Google Scholar]
- 49.Exposito-Rodriguez M., Borges A., Borges-Perez A., Perez J. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol. 2008;8:131. doi: 10.1186/1471-2229-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jatropha Genome Database (Contents Release 4.5) [(accessed on 20 January 2011)]. http://www.kazusa.or.jp/jatropha.
- 52.National Center for Biotechnology Information. [(accessed on 8 February 2011)]. http://www.ncbi.nlm.nih.gov/
- 53.Yamaguchi-Shinozaki K., Shinozaki K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 1993;236:331–340. doi: 10.1007/BF00277130. [DOI] [PubMed] [Google Scholar]
- 54.Ding L.W., Sun Q.Y., Wang Z.Y., Sun Y.B., Xu Z.F. Using silica particles to isolate total RNA from plant tissues recalcitrant to extraction in guanidine thiocyanate. Anal. Biochem. 2008;374:426–428. doi: 10.1016/j.ab.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 55.Ramakers C., Ruijter J.M., Deprez R.H.L., Moorman A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.