Abstract
A carrageenan-degrading marine Cellulophaga lytica strain N5-2 was isolated from the sediment of carrageenan production base. A κ-carrageenase (EC 3.2.1.83) with high activity was purified to electrophoretic homogeneity from the culture supernatant by a procedure of ammonium sulfate precipitation, dialyzing and gel filtration on SephadexG-200 and SephadexG-75. The purified enzyme was verified as a single protein on SDS-PAGE, and whose molecular weight was 40.8 kDa. The κ-carrageenase yielded a high activity of 1170 U/mg protein. For κ-carrageenase activity, the optimum temperature and pH were 35 °C and pH 7.0, respectively. The enzyme was stable at 40 °C for at least 2.5 h. The enzyme against κ-carrageenan gave a Km value of 1.647 mg/mL and a Vmax value of 8.7 μmol/min/mg when the reaction was carried out at 35 °C and pH 7.0. The degradation products of the κ-carrageenase were analyzed by thin layer chromatography (TLC), high performance liquid chromatography (HPLC), electrospray ionization time-of-flight mass spectroscopy (ESI-TOF-MS) and 13C-NMR spectroscopy, and the results indicated that the enzyme was specific of the β-1,4 linkage and hydrolyzed κ-carrageenan into κ-neocarraoctaose-sulfate and κ-neocarrahexaose-sulfate first, and then broke κ-neocarraoctaose-sulfate into κ-neocarrabiose-sulfate and κ-neocarrahexaose-sulfate.
Keywords: κ-carrageenase, Cellulophaga lytica strain N5-2, characterization, degradation product
1. Introduction
Carrageenans are linear sulfated galactans extracted from many species of red seaweeds and share a common backbone of d-galactose with alternating α-1,3 and β-1,4 linkages. They are classified according to the number and the position of sulfate ester groups. The main industrially exploited carrageenans are κ- and ι-carrageenans owing to their gelling properties. κ-carrageenan is conventionally described as the repetition of the disaccharide motif 4-sulfate-O-β-d-galactopyranosyl-(1,4)-3,6-anhydro-α-d-galactose. In most reports, κ-carrageenase, which hydrolyzes β-1,4 linkages in κ-carrageenan, is a novel family-16 glycoside hydrolases member [1,2]. κ-carrageenase as a tool enzyme for carrageenan degradation has a wide range of applications in the formation of algal protoplast [3], preparation of carrageenan oligosaccharides [4], structure and function study of carrageenan [5,6], etc. Furthermore, compared with carrageenan, carrageenan oligosaccharide and its derivants have more biological activities including anti-tumor [7], anti-oxidation [8], anti-cogulant, immunoregulation and anti-angiogenesis [9]. Among the previous reports, κ-carrageenases have been isolated from Pseudoalteromonas [10], Cytophaga [11], Alteromonas carrageenovora and Pseudomonas carrageenovora [12,13]. In the present study, a κ-carrageenase was purified from Cellulophaga lytica strain N5-2, and the hydrolyzed products of the enzyme were analyzed.
2. Results and Discussion
2.1. Identification of Marine Cellulophaga lytica Strain N5-2
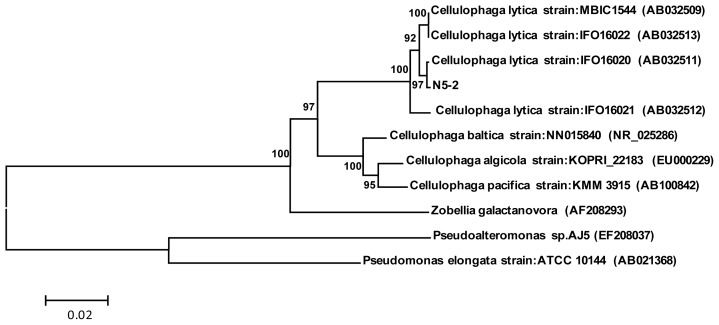
Marine Cellulophaga lytica strain N5-2 produced yellow pigmentation and displayed gram negative, the cell size was 0.6 μm × (2.5–3.7) μm; colonies were yellow with irregular edge and granular surface protuberance. The 16SrRNA sequence of this strain demonstrated 99% similarity with the Cellulophaga lytica strain IFO: 16020 (AB032511). Therefore, the phylogenetic tree suggested that the strain may be a new strain in the Cellulophaga lytica (Figure 1), called Cellulophaga lytica strain: N5-2. The nucleotide sequence of 16S rRNA of the strain was submitted to GenBank nucleotide sequence database. The sequence is available in the GenBank nucleotide sequence database with the accession number of GU129978.
Figure 1.
Neighbor-joining phylogenetic tree based on 16SrRNA gene sequencing showed the relationships between the strain of N5-2 and other related genera. Neighbor-joining phylogenetic tree based on 16Sr RNA gene sequencing showed the relationships between the strain N5-2 and other related genera. Numbers at nodes are levels of bootstrap support (%). Scale bar represents two nucleotides substitution per 100 nucleotides.
2.2. Purification of κ-Carrageenase
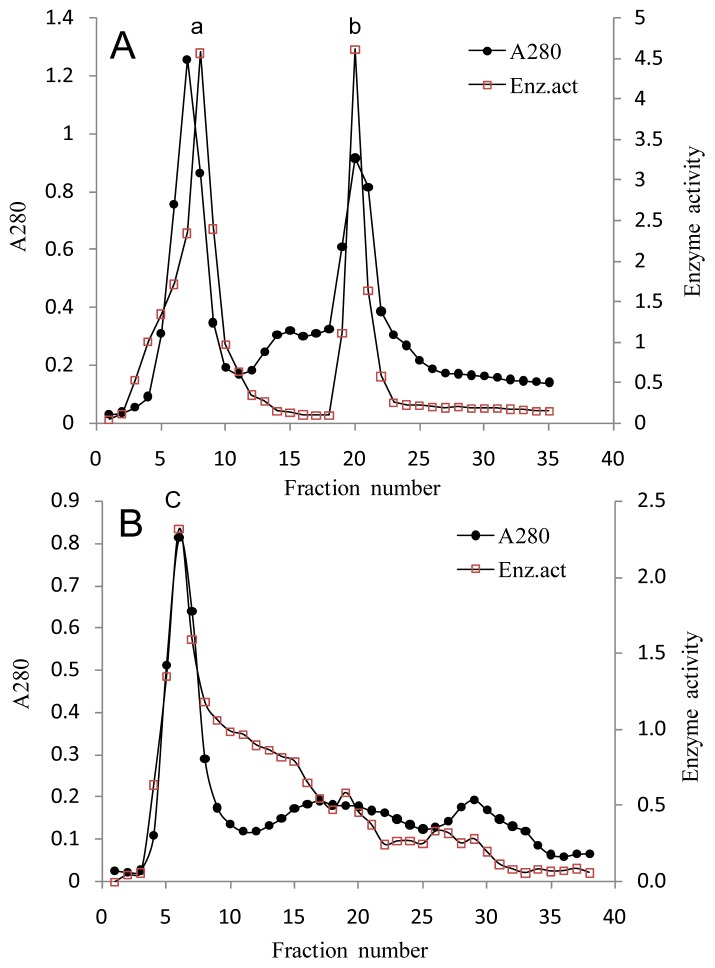
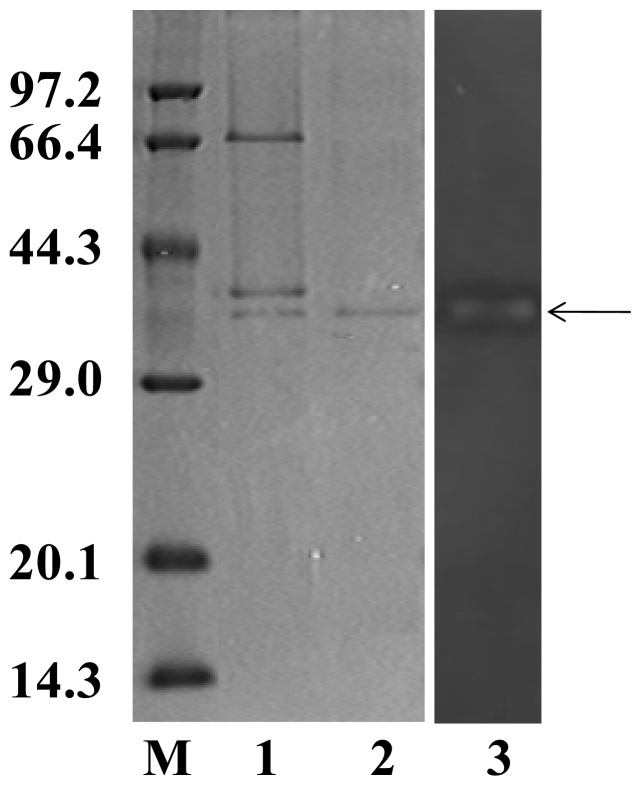
As shown in Table 1, after precipitated with 40% and 80% (NH4)2SO4, about 80% foreign protein was removed and the specific activity was increased about 4 folds with a significant loss of total activity (69.5% recovery). Sephadex G-200 column chromatography separated 2 peaks with κ-carrageenase activity (Figure 2A), by this step the specific activity was increased 23 folds. The further purification of κ-Carrageenase was obtained by Sephadex G-75 column chromatography, from which a single activity peak (Figure 2B) was achieved. The result of SDS-PAGE showed that this activity peak contained a single protein band (Figure 3 line 2) with molecular mass of 40.8 kDa. A clear zone on zymography further stained the band could degrade κ-carrageenan (Figure 3 line 3). The final purified enzyme yielded significantly high activity of 1170 U/mg protein and a fold of 40 (Table 1), at the same time much of the total activity was lost with 17.3% recovery only.
Table 1.
Purification of the κ-carrageenase from Cellulophaga lytica strain N5-2.
| Step | Volume (mL) | Total protein a (mg) | Total activity b (units) | Specific activity (units/mg) | Folds | Recovery (%) |
|---|---|---|---|---|---|---|
| Cell-free medium | 1000 | 56.5 | 1648 | 29 | 1 | 100 |
| 40% and 80% (NH4)2SO4 precipitate | 80 | 11 | 1145 | 104 | 4 | 69.5 |
| SephadexG-200 gel-filtration | 5 | 0.86 | 580 | 675 | 23 | 35.2 |
| SephadexG-75 gel-filtration | 2 | 0.235 | 285 | 1170 | 40 | 17.3 |
Protein contents were determined by Bradford method;
Carrageenase activity was determined by the reducing power method of 3,5-dinitrosalicylic acid (DNS).
Figure 2.
Gel filtration chromatography of κ-carrageenase. (A) Sephadex G-200 gel filtration chromatography; and (B) Sephadex G-75 gel filtration chromatography. The sample was eluted at a flow rate of 0.1 mL/min with Tris-HCl buffer at pH 7.0. The eluates were monitored for protein (A280) and κ-carrageenase activity.
Figure 3.
SDS-PAGE pattern and zymography of purified κ-carrageenase. SDS-PAGR was conducted in a 5%/15% discontinuous polyacrylamide gel at 25 mA at room temperature and stained with Coomassie brilliant blue R-250. The peaks from Sephadex G-200 contained three protein bands (1); Only one protein band, with an apparent molecular mass of 40.8 kDa, was detected in the activity peak from Sephadex G-75 (2); The sample of the activity peak from Sephadex G-75 was applied to zymography experiment, stained with alcian blue. A degradated band was detected at the same position, just as pointed by the arrow (3). Line M was protein molecular mass marker.
2.3. Properties of the κ-Carragcenase
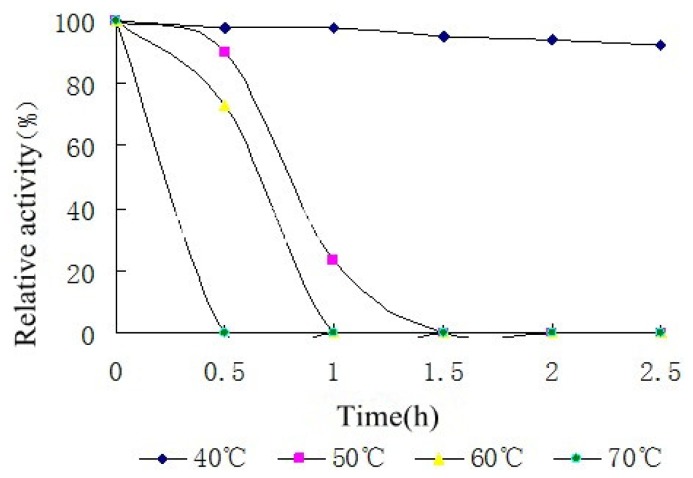
κ-carrageenase was stable in a broad range of temperature (20 °C to 60 °C) and 35 °C was optimum temperature. Furthermore, temperature stability result showed that the κ-carrageenase had a good thermal stability at 40 °C, with the prolongation of time the enzyme activity remained stable at least for 2.5 h. When the temperature was above 40 °C, the enzyme activity decreased significantly as time prolonging (Figure 4).
Figure 4.
Temperature stability of the κ-carrageenase from Cellulophaga lytica strain N5-2. The enzyme solution was incubated at each temperature (20–70 °C) for 0.5 to 2.5 h and then the residual enzyme activity was measured. The activity of untreated enzyme was regarded as 100% and relative activity was determined.
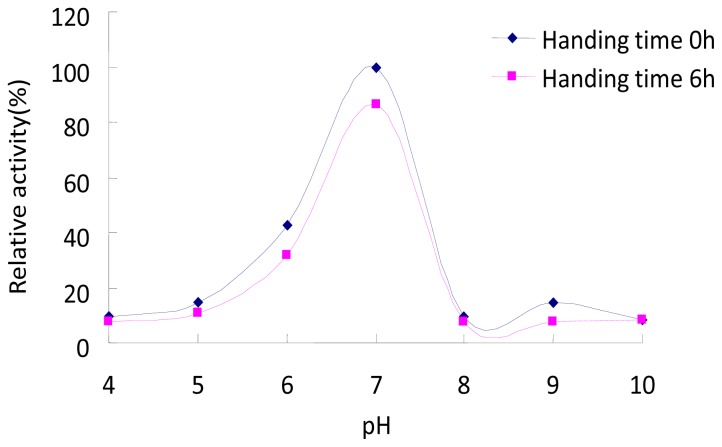
The optimum pH of the κ-carrageenase was 7.0. The enzyme was inactivated or denaturated at extreme values of pH, and handling time had a weak effect on the enzyme activity (Figure 5).
Figure 5.
pH stability of the κ-carrageenase from Cellulophaga lytica strain N5-2. The different pH buffer was: Na2HPO4-Citric acid buffer (pH 4.0–7.0), Tris-HCl buffer (pH 8.0 and 9.0) and glycine-NaOH buffer (pH 10.0). Pre-incubating the enzyme solution at each pH (4.0–10.0) at 35 °C for 6 h and then the enzyme activity was determined in the same pH buffer. The activity of untreated enzyme was regarded as 100% and relative activity was determined.
As calculated from Lineweaver plots, the aparent Km and Vmax values of the κ-carrageenase were 1.647 mg/mL and 8.7 μmol/min/mg respectively.
2.4. Analysis of Degradation Products
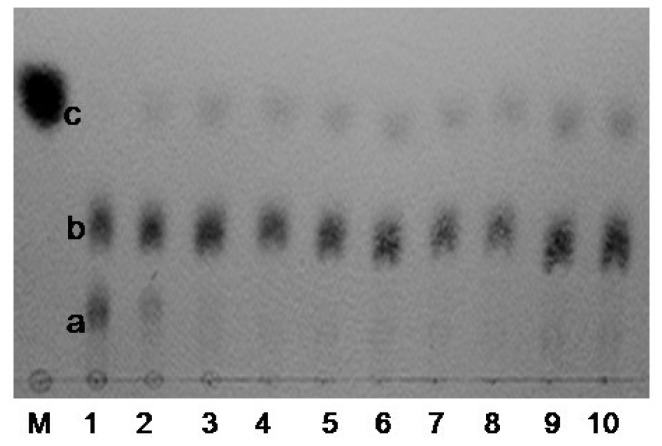
Analysis of the time course of the oligosaccharide release showed that at first κ-carrageenan was hydrolyzed into two high-molecular-mass components a and b (Figure 6), with the prolongation of hydrolysis time, a smaller molecular-mass component c emerged and component-a decreased gradually. The hydrolyzed product contained three components a, b and c were separated with HPLC, three dominant peaks appeared on HPLC mass spectrum (Figure 7A–D).
Figure 6.
Thin layer chromatography (TLC) of the κ-carrageenase degradation products. Enzymatic hydrolysis of κ-carrageenan was conducted under standard condition with 0.5% κ-carrageenan as substrate and reaction for 5 min (1); 30 min (2); 1 h (3); 2 h (4); 3 h (5); 6 h (6); 9 h (7); 12 h (8); 24 h (9) and 48 h (10), respectively. M, control (galactose). Combined with HPLC and mass spectrometry results, a, b and c were ê-neocarraoctaose, κ-neocarrabiose and κ-neocarrabiose respectively.
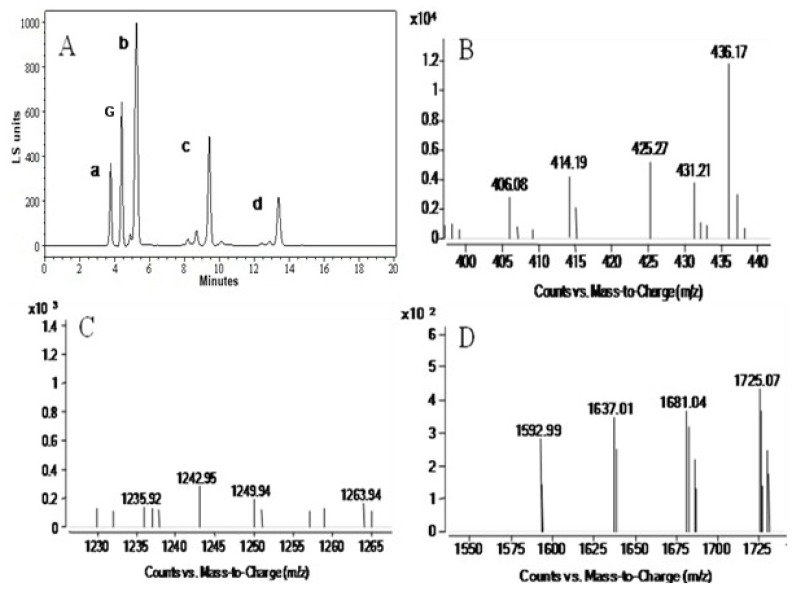
Figure 7.
HPLC (A) and MSI-TOF-MS (B–D) analysis of the κ-carrageenase degradation products. (A) HPLC spectrum of the κ-carrageenase degradation products. κ-carrageenan was hydrolyzed with the enzyme under standard condition for 48 h, and the product was separated with HPLC. Three components were detected and separated (b, c and d), a is solvent peak, G is control (galactose) peak. The components obtained from HPLC were analyzed with MSI-TOF-MS to determine the molecular mass distribution; (B–D) MSI-TOF-MS analysis of the κ-carrageenase degradation products b, c and d (in HPLC) respectively.
In order to identify the molecular mass of the hydrolyzed products, the three components obtained from HPLC was analyzed with MSI-TOF-MS, and the result demonstrated that the molecular mass of three components were 425.27, 1242.95 and 1681.04 Da respectively (Figure 7B–D). It is well known that the base unit of the κ-carrageenan was a dimer (426 Da) composed of one β-d-galactopyranose residue (G-unit) and one 3, 6-anhydro-α-d-galactopyranose residue (A-unit). So the 425.27 Da component could be κ-neocarrabiose-sulfate, the 1242.95 Da component could be κ-neocarrahexaose-sulfate, and the molecular mass of acetylated κ-neocarraoctaose was 1681.04 Da.
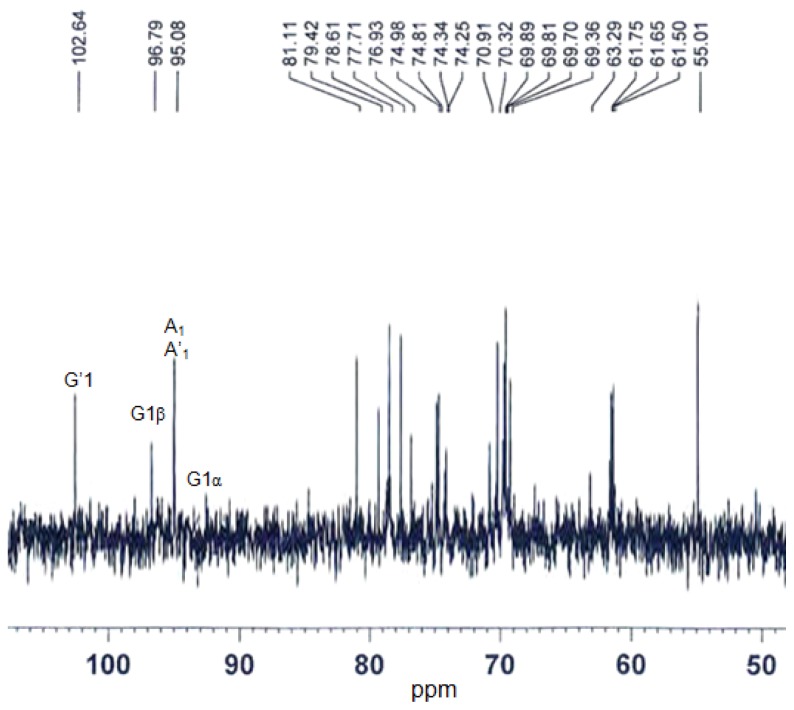
The structure information of the products was further obtained by 13C-NMR. A typical pattern for oligosaccharide was observed according to the NMR spectrum (Figure 8). Only one signal was observed at around 102.64 ppm, which is the characteristic signal of the G-unit at non-reducing end of the oligosaccharides. The resonance at 95.08 ppm was assigned to be the carbon of the A-unit. The signal at 92.5 and 96.79 ppm were assigned to be the carbon of the G-unit at the reducing end for α and β configuration respectively. According to above results, the reducing end of the oligosaccharides was G-unit. This indicated that the κ-carrageenase from N5-2 is specific of the β-1,4 linkage. So the hydrolyzed products of the κ-carrageenase were κ-neocarrabiose-sulfate, κ-neocarrahexaose-sulfate and κ-neocarraoctaose-sulfate respectively.
Figure 8.
13C-NMR spectrum of the κ-carrageenase degradation products. The degradation products were dissolved in 2H2O and processed at 30 °C. Spectrum was recorded on an AVANCE 500 MHz apparatus (Bruker, Karlsruhe, Germany).
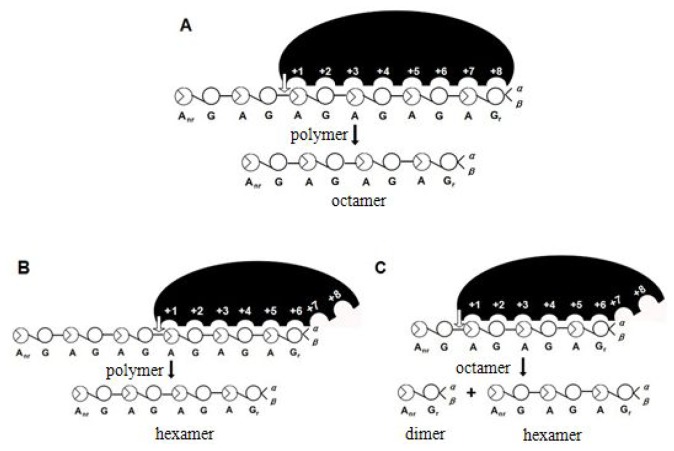
It is surprising that neocarratetraose-sulfate was not detected suggesting that the recognition units of the enzyme were κ-neocarraoctaose and κ-neocarrahexaose, and then κ-neocarraoctaose was broken into κ-neocarrabiose and κ-neocarrahexaose (Figure 9).
Figure 9.
Degradation mode of κ-carrageenase. The recognition units of the enzyme are κ-neocarraoctaose (A) and κ-neocarrahexaose (B); and then κ-neocarraoctaose is broken into κ-neocarrabiose and κ-neocarrahexaose (C). The end-products of the κ-carrageenase are κ-neocarrabiose and κ-neocarrahexaose.
3. Materials and Methods
3.1. Microorganism
Cellulophaga lytica strain N5-2 was isolated from the sediment of carrageenan production base in Hainan (Hainan, China), and cultured according to our previous method [14].
3.2. Chemicals
κ-carrageenan was purchased from the Changhang Colloid Technological Co., Ltd. (Changzhou, Jiangsu, China). SephadexG-200 and SephadexG-75 are of Pharmacia products (Pharmacia Company, Stockholm, Sweden). All other reagents were of analytical grade.
3.3. κ-Carrageenase Assay
κ-carrageenase activity was determined by measuring the increase in the concentration of reducing sugar as described by Kidby [15]. The assay system consisted of 1.0 mL substrate [0.5% κ-carrageenan in Na2HPO4-citric acid buffer (pH 7.0)] and 1.0 mL enzyme, and boiled inactivated enzyme with the same treatment was used as control. One unit of κ-carrageenase activity was defined as the amount of enzyme needed to release 1 μmol reducing sugars (d-galactose equivalent) per min.
3.4. Purification of κ-Carrageenase
Strain N5-2 was propagated in a medium containing: κ-carrageenan (2 g), FeSO4·7H2O (0.02 g), polypepton (3 g) in 1000 mL layout seawater with shaking for 20 h at 35 °C. The following operations were carried out at 4 °C. The culture medium was centrifuged (10,000 g, 20 min) and the cell-free supernatant was fractionated at 40% and 80% ammonium sulfate saturation. The precipitated protein with 40% ammonium sulfate saturation was discarded, and the precipitated protein with 80% ammonium sulfate saturation was resuspended in distilled water and dialyzed in dialysis bag (MWCO: 8000–14,000 Da) against the distilled water, and freeze-dried successively. Protein contents were determined by Bradford method [16]. The obtained enzyme powder was dissolved in 5 mL Tris-HCl buffer (pH 7.0) and the end concentration was 4%, then the enzyme solution was applied to Sephadex G-200 (Pharmacia company, Stockholm, Sweden) column chromatography (100 × 1.6 cm) equilibrated with the same buffer, and eluted at a flow rate of 0.1 mL/min. The eluates were monitored continuously at 280 nm for protein and fractions were assay for activity against κ-carrageenan. All the fractions of the first peak from Sephadex G-200 column chromatography containing κ-carrageenase activity were gathered, concentrated and applied to another column of Sephadex G-75, furthermore equilibrated with the same eluent. Fractions were collected and monitored for the presence of κ-carrageenase. The purity of the fractions was assessed by SDS-PAGE. Pure fractions with activity were stored at −20 °C.
3.5. Zymography Experiment
15% Separating gel was prepared by adding 0.2% κ-carrageenan, the content of other reagents was as same as the normal SDS-PAGE. The zymography electrophoresis was carried out as normal discontinuous SDS-PAGE. After electrophoresis, above all, the separating gel was renaturated in renaturation solution (0.05 mol/L Tris–HCl, 0.2 mol/L NaCl, 0.01 mol/L CaCl2, 1% TritonX-100, pH 7.0) at 35 °C for 5 h, then stained with 0.1% alcian blue (in 0.1 M HCl, pH 1.0).
3.6. Dependence and Stability of the Enzyme Activity on Temperature and pH
The optimum temperature for κ-carrageenase activity was determined under the standard assay condition by varying the incubation temperature from 20 to 70 °C. The thermal stability of the κ-carrageenase was determined by incubating the enzyme solution at each temperature (20–70 °C) for 0.5 to 2.5 h and then measuring the residual enzyme activity. The activity of untreated enzyme was regarded as 100% and relative activity was determined.
The effect of pH on κ-carrageenase activity was assayed by replacing Na2HPO4-citric acid buffer (pH 7.0) with: Na2HPO4-citric acid buffer (pH 4.0–7.0), Tris-HCl buffer (pH 8.0 and 9.0) and glycine-NaOH buffer (pH 10.0) at 35 °C. The pH stability of the κ-carrageenase was determined by pre-incubating the enzyme solution at each pH (4.0–10.0) at 35 °C for 6 h and then the enzyme activity was determined in the same pH buffer. The activity of untreated enzyme was regarded as 100% and relative activity was determined.
3.7. Kinetic Studies
The initial reaction rate of κ-carrageenan degradation were assayed at several κ-carrageenan concentrations (0.3125, 0.625, 1.25, 2.5 and 5.0 mg/mL) by DNS method and the reaction system contained 0.5 mg/L pure enzyme protein. The Michaelis constant (Km) and the reaction rate at infinite substrate concentration (Vmax) were determined according to the Lineweaver-Burk plotting method [17].
3.8. Analysis of Degradation Products
Enzymatic hydrolysis of κ-carrageenan was conducted under standard condition with 0.5% κ-carrageenan as substrate and reaction for 5 min, 30 min, 1 h, 2 h, 3 h, 6 h, 9 h, 12 h, 24 h and 48 h, respectively. The products of different reaction time was analyzed by TLC (thin-layer chromatograthy) method with the developing agent composed of butanol–acetic-acid–water (2:1:1, v/v/v) [3], and the saccharide was visualized with a diphenylamine-aniline-phosphate reagent.
κ-carrageenan was hydrolyzed with the enzyme under standard condition for 48 h, and the product was separated with HPLC (Agilent 1100, Agilent Technologies, Santa Clara, CA, USA), and the separation condition was moving phase 2.5 mM ammonium acetate:methyl alcohol (3:2). The components obtained from HPLC were analyzed with ESI-TOF-MS (Agilent G1969A, Agilent Technologies, Santa Clara, CA, USA) to determine the molecular mass distribution. Structure of the product was identified by 13C-NMR spectroscopy (Bruker AVANCE 500 MHz, Bruker, Karlsruhe, Germany), 10 mg sample was dissolved in 0.5 mL H2O and assay was carried out at 30 °C.
4. Conclusions
In this paper, a marine bacterium with high κ-carrageenase activity was isolated and was identified as marine Cellulophaga lytica by 16SrRNA, named strain N5-2. After purification, the specific activity of the enzyme was 1170 U/mg. The molecular mass of the enzyme was 40.8 kDa, and it possessed thermal, pH stabilities and satisfactory kinetic property. This κ-carrageenase from N5-2 was specific of the β-1,4 linkage, and the recognition units of the enzyme were κ-neocarraoctaose and κ-neocarrahexaose, and then κ-neocarraoctaose was broken into κ-neocarrabiose and κ-neocarrahexaose. So the hydrolyzed products of the enzyme were κ-neocarrabiose-sulfate, κ-neocarrahexaose-sulfate and κ-neocarraoctaose-sulfate respectively.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81070973 and 81373429).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Barbeyron T., Gerard A., Potin P. The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases. Mol. Biol. Evol. 1998;15:528–537. doi: 10.1093/oxfordjournals.molbev.a025952. [DOI] [PubMed] [Google Scholar]
- 2.Strohmeier M., Hrmova M., Fischer M., Harvey A.J., Fincher G.B., Pleiss J. Molecular modeling of family GH16 glycoside hydrolases: Potential roles for xyloglucan transglucosylases/hydrolases in cell wall modification in the poaceae. Protein Sci. 2004;13:3200–3213. doi: 10.1110/ps.04828404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zablackis E., Vreeland V., Kloareg B. Isolation of protoplasts from kappaphycus alvarezii var. tambalang (Rhodophyta) and secretion of carrageenan fragments by cultured cells. J. Exp. Bot. 1993;44:1515–1522. [Google Scholar]
- 4.Marion G., Patrick B. Composition and distribution of carrabiose moieties in hybrid κ-/ι-carrageenans using carrageenases. Biomacromolecules. 2008;9:408–415. doi: 10.1021/bm701109r. [DOI] [PubMed] [Google Scholar]
- 5.Lemonnier-Le P.C., Chatelet C., Kloareg B. Carrageenan oligosaccharides enhance stress-induced microspore embryogenesis in Brassica oleracea var. italica. Plant Sci. 2001;160:1211–1220. doi: 10.1016/s0168-9452(01)00372-7. [DOI] [PubMed] [Google Scholar]
- 6.Nyval P., Maud L., Richard D. Enzymatic degradation of κ-carrageenan in aqueous solution. Biomacromolecules. 2009;10:1757–1767. doi: 10.1021/bm9001766. [DOI] [PubMed] [Google Scholar]
- 7.Yuan H.M., Song L.M., Li X.G. Immunomoduladtion and antitumor activity of κ-carrageenan oligosaccharides. Cancer Lett. 2006;243:228–234. doi: 10.1016/j.canlet.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 8.Yuan H.M., Zhang W.W., Li X.G. Preparation and in vitro antioxidant activity of κ-carrageenan oligosaccharides and their over sulfated, acetylated and phosphorylated derivatives. Carbohydr. Res. 2005;340:685–692. doi: 10.1016/j.carres.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 9.Keisuke I., Michal K.W., Takayuki T., Siro S., Hiroyuki O. Novel heparin sulfatemimetic compounds as antitumor agents. Chem. Biol. 2004;11:367–377. doi: 10.1016/j.chembiol.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 10.Gauthier G., Gauthier M., Christen R. Phylogenetic analysis of the genera Alteromonas, Shewanella, and Moritella using genes coding for small-subunit rRNA sequences and division of the genus Alteromonas into two genera, Alteromonas (emended) and Pseudoalteromonas gen. nov., and proposal of twelve new species combinations. Int. J. Syst. Bacteriol. 1995;45:755–761. doi: 10.1099/00207713-45-4-755. [DOI] [PubMed] [Google Scholar]
- 11.Golam S., Seiken M., Hiroshi O. Purification of a κ-carrageenase from marine Cytophaga species. Microbiol. Immunol. 1987;31:869–877. doi: 10.1111/j.1348-0421.1987.tb03148.x. [DOI] [PubMed] [Google Scholar]
- 12.Khambhty Y., Mody K., Jha B. Statistical optimization of medium components for κ-carrageenase production by Pseudomonas elongata. Enzyme Microbial Technol. 2007;40:813–822. [Google Scholar]
- 13.Michel G., Chantalat L., Duee E., Barbeyron T., Henrissat B., Kloareg B., Dideberg O. The κ-carrageenase of P carrageenovora features a tunnel-shaped active site: A novel insight in the evolution of Clan-B glycoside hydrolases. Structure. 2001;9:513–525. doi: 10.1016/s0969-2126(01)00612-8. [DOI] [PubMed] [Google Scholar]
- 14.Dun J.X., Yao Z.A., Wu H.G., Wang F.F., Zhu N.N., Du Y.G. Screening and characterization of a κ-carrageenase producing strain and the initial analysis of products. Food Sci. Technol. 2010;35:11–16. (in Chinese) [Google Scholar]
- 15.Kidby D.K., Davidson D.J. A convenient ferricyanide estimation of reducing sugars in the nanomole range. Anal. Biochem. 1973;55:321–325. doi: 10.1016/0003-2697(73)90323-0. [DOI] [PubMed] [Google Scholar]
- 16.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Lineweaver H., Bruk D. The determination of enzyme dissociation constants. Chem. Soc. Rev. 1934;56:658–666. [Google Scholar]