Significance
In mammals, the sense of odors relies on the peculiar expression pattern of olfactory receptors (ORs). Each single neuron chooses one, and only one, from all ∼1,400 OR genes that are present in a mouse genome. In neurobiology, a long-standing mystery is how such singularity can be achieved. We show theoretically that a simple kinetic scheme of OR activation followed by feedback can be solely responsible for the observed singularity, as long as the two timescales—slow activation by epigenetic modification and fast feedback by transcriptional regulation—are well separated. Our work provides the theoretical underpinning behind the choice of ORs, and demonstrates how the nervous system utilizes the kinetics of epigenetic changes to direct neurogenesis.
Keywords: neurogenesis, stochastic gene expression, histone modification, kinetics modeling, negative feedback
Abstract
Mammals sense odors through the gene family of olfactory receptors (ORs). Despite the enormous number of OR genes (∼1,400 in mouse), each olfactory sensory neuron expresses one, and only one, of them. In neurobiology, it remains a long-standing mystery how this singularity can be achieved despite intrinsic stochasticity of gene expression. Recent experiments showed an epigenetic mechanism for maintaining singular OR expression: Once any ORs are activated, their expression inhibits further OR activation by down-regulating a histone demethylase Lsd1 (also known as Aof2 or Kdm1a), an enzyme required for the removal of the repressive histone marker H3K9me3 on OR genes. However, it remains unclear at a quantitative level how singularity can be initiated in the first place. In particular, does a simple activation/feedback scheme suffice to generate singularity? Here we show theoretically that rare events of histone demethylation can indeed produce robust singularity by separating two timescales: slow OR activation by stepwise H3K9me3 demethylation, and fast feedback to turn off Lsd1. Given a typical 1-h response of transcriptional feedback, to achieve the observed extent of singularity (only 2% of neurons express more than one ORs), we predict that OR activation must be as slow as 5–10 d—a timescale compatible with experiments. Our model further suggests H3K9me3-to-H3K9me2 demethylation as an additional rate-limiting step responsible for OR singularity. Our conclusions may be generally applicable to other systems where monoallelic expression is desired, and provide guidelines for the design of a synthetic system of singular expression.
In mammals, the ability to sense odors relies on the singular expression of the olfactory receptor (OR) gene family. Despite the enormous family size (∼1,400 genes in the mouse genome), each sensory neuron only expresses one single allele of OR (1–4). It has long been thought that this singularity stems from a negative feedback in which the expression of one OR specifically silences all other ORs (5–7). However, this hypothesis has led to the question of how the one, and only one, active OR can escape its own silencing effect. In other words, it was unclear how such silencing can be biologically feasible.
A series of recent studies provoked a different mechanism for OR singularity in light of epigenetic regulation. Contrary to previous belief, Magklara et al. (8) found that silencing of OR genes precedes OR expression: The histone marker H3K9me2 on OR genes become methylated into H3K9me3 as early as in the stage of neuronal progenitors, which do not yet express any ORs. H3K9me2 is a marker for localization of inactive genes into facultative heterochromatin; its further methylation into H3K9me3, however, marks OR genes for constitutive heterochromatin—nuclear compartments that normally contain pericentromeric and telomeric regions. With this marker, all OR genes are deeply repressed in nuclear aggregates before activation (9). The existence of negative feedback was also confirmed recently: Once a choice of ORs is made, the histone demethylase Lsd1 (also known as Aof2 or Kdm1a) will be transcriptionally repressed by OR expression (10, 11). Lsd1 down-regulation will prevent further activation and freeze the system after the choice because this enzyme is essential for OR activation.
Taken together, these studies provided a clear mechanism for maintaining a choice of OR genes through inhibition of histone demethylation; however, it remains unclear how a singular choice of OR can be initiated in the first place. Although these studies hypothesized that a slow activation may help singularity (8–10), it is not obvious at a quantitative level whether this scheme is feasible. The critical assessment is whether a slow kinetics can really generate singularity and, if so, what the requirements are for the kinetic parameters. In this paper, we theoretically prove that rare events of histone modifications indeed produce robust singularity on a timescale compatible with experiments. We find that the key to singular expression is a combination of slow activation and fast feedback. In particular, it is the ratio between the two timescales that determines the extent of singularity in a wide range of models. Clowney et al. (9) suggested the inaccessibility of aggregated heterochromatin as the reason for slow activation. As an alternative, our model predicts that a rate-limiting step of H3K9me3-to-H3K9me2 demethylation, catalyzed by an unidentified enzyme, can also be responsible for OR singularity; this is in contrast to the previous proposal that Lsd1 (H3K9me2 demethylation) is a rate-limiting enzyme (10).
Materials and Methods
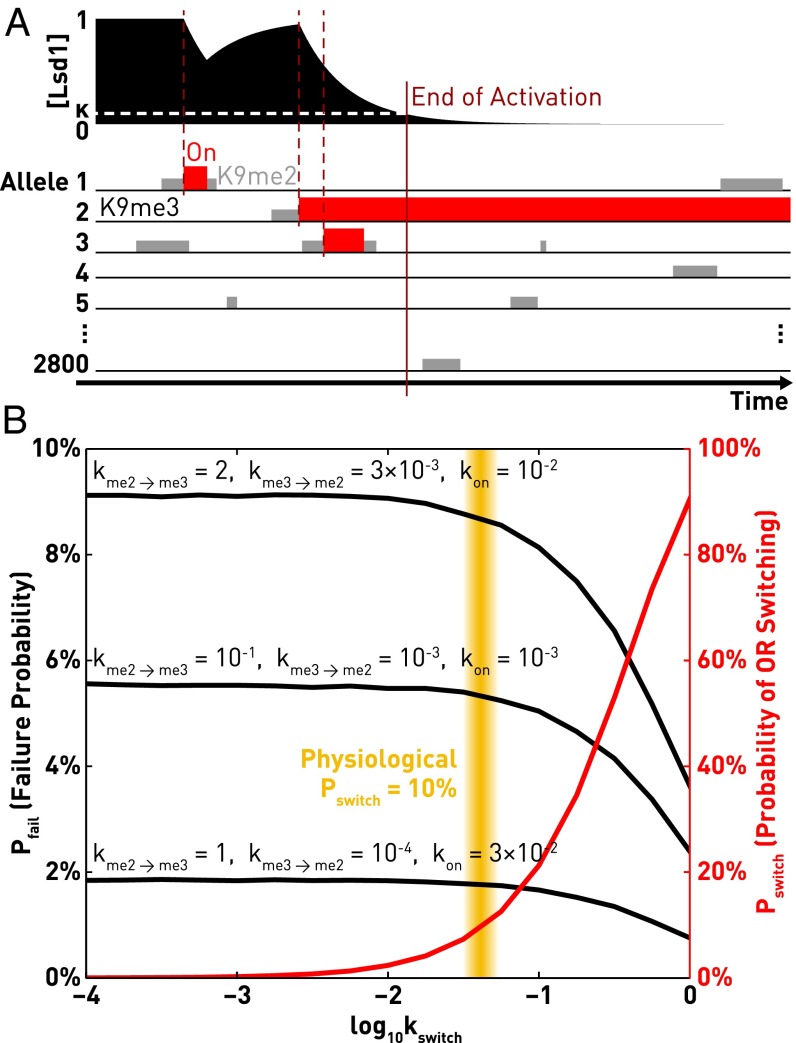
We model each allele of OR with three epigenetic states (Fig. 1A): an “off” state that contains H3K9me3 (constitutive heterochromatin), an “intermediate” state that contains H3K9me2 (facultative heterochromatin), and an “on” state that actively expresses the OR. Neuronal commitment begins with extensive methylation of H3K9me2 into H3K9me3 (intermediate→off, with a rate kme2→me3) (8); therefore, we initialize each allele of OR in the off state. The activation of an OR—namely, the complete demethylation of its repressive H3K9me3 marker—involves two consecutive steps: H3K9me3-to-H3K9me2 demethylation by an unidentified demethylase (off→intermediate, with a rate kme3→me2), and then H3K9me2 demethylation by Lsd1 (intermediate→on, with a rate kon) (10, 12, 13). Notice that here we focus on the activator role of Lsd1; its second role as a repressor is discussed in the last subsection of Results. Once fully activated, OR expression will transcriptionally down-regulate Lsd1 via a recently identified cascade (OR activates the adenylate cyclase Adcy3, which in turn represses Lsd1) (10, 11), whose response time is denoted Δt. If the feedback is instant, namely, Δt = 0, singularity will be perfect; however, this is not physiological because Lsd1 depletion takes time (thus Δt > 0). Although here Δt is assumed to be deterministic, our model still works if the feedback is stochastic, in which case Δt will denote the mean response time (Fig. S1).
Fig. 1.
A kinetic model for the choice of olfactory receptors. (A) Each allele of OR is modeled by three epigenetic states: an off state that contains H3K9me3, an intermediate state that contains H3K9me2, and an on state that supports active expression. Between the states off and intermediate, the rates of conversion are kme2→me3 (for intermediate→off) and kme3→me2 (for off→intermediate). Only the intermediate state is poised for further activation by Lsd1, whose rate is kon. Once fully activated, OR expression will transcriptionally down-regulate Lsd1 within a fixed response time Δt (red dashed arrow). (B) In each simulation, all n = 2,800 alleles of ORs (including both the paternal and the maternal ones) are initialized in the off state (black line), and then allowed to transition into the intermediate state (gray box) and finally into the on state (red box). The earliest activation down-regulates Lsd1 after time Δt, thus terminating any further activation (dark red vertical line, end of activation). Singular OR expression will be achieved if no other alleles become activated during the response time Δt. (Left) Simulation parameters are kme2→me3 = 1, kme3→me2 = 0.01, kon = 0.01, Δt = 1. (Right) The simulation has the same parameters except for a higher kme3→me2 = 0.1. (C) The waiting time of each allele follows a probability distribution that contains a linear rise on a short timescale and an exponential fall on a long timescale. In this example, the parameters are kme2→me3 = kme3→me2 = kon = 1.
The choice of ORs is simulated by initializing all n = 2,800 alleles (including both the paternal and the maternal ones) in the off state (H3K9me3) and then waiting for the earliest activation to occur at any allele, which would turn off Lsd1 after the response time Δt of the feedback loop. Two representative simulations are shown in Fig. 1B. Starting from the off state (K9me3), each allele occasionally loses one methyl group and transitions into the intermediate state (K9me2). In some cases, a methyl group is added back, bringing the allele back into the off state, whereas in other cases Lsd1 takes over and further demethylates the allele for active expression. Singularity will be achieved if no further activation occurs in time Δt after the first activation, as shown in the simulation on Fig. 1B, Left. After Δt, Lsd1 becomes down-regulated and kon falls to zero, thus prohibiting other ORs from being activated. However, if any other ORs are activated before the feedback takes effect, as shown in Fig. 1B, Right, singularity will be broken because Lsd1 regulation only ensures the maintenance of previous OR choices (10).
Results
Our Kinetic Model Generates Robust Singular OR Expression by a Combination of Slow Activation and Fast Feedback.
In our three-state model, the waiting time for each allele to reach the on state can be solved analytically by analogy to the Michaelis–Menten kinetics (14). Although the system relies on three parameters, kme2→me3, kme3→me2, and kon, the distribution of waiting times depends on only two factors: The geometric mean of kme3→me2 (the rate of off→intermediate) and kon (the rate of intermediate→on)—denoted kbottleneck ≡ √kme3→me2kon—describes how slow the two demethylation reactions are, whereas the sum of all three rates—denoted ktotal ≡ kme2→me3+kme3→me2+kon—describes how fast the system transitions between epigenetic states. For each allele, the mean waiting time before activation is ktotal/k2bottleneck, and its probability density function contains a linear rise from zero on a short timescale (t ∼1/ktotal) and an exponential fall on a long timescale (t ∼ ktotal/k2bottleneck; Fig. 1C).
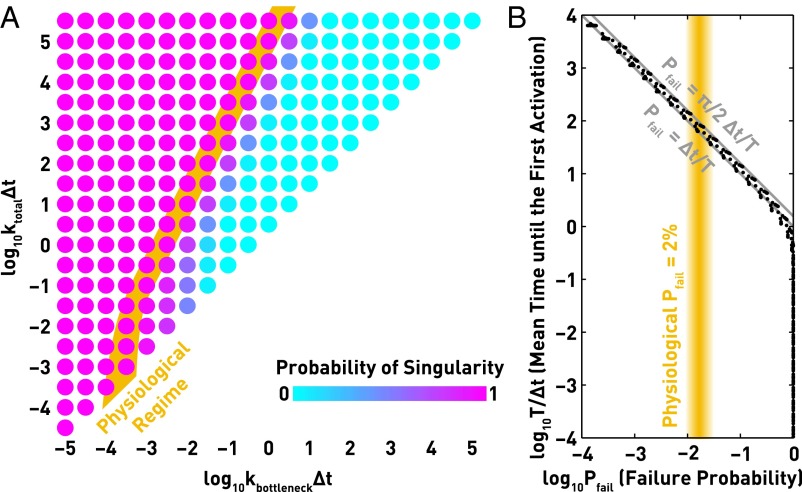
With all n = 2,800 alleles competing for the earliest activation, our model exhibits singular OR expression for a wide range of values for the two dimensionless parameters kbottleneckΔt and ktotalΔt (Fig. 2A). In particular, two aspects can contribute to singularity: a kinetic bottleneck in activation (small kbottleneckΔt), and rapid switching between epigenetic states (large ktotalΔt); in terms of the original parameters, this is equivalent to having two slow demethylation reactions (small kme3→me2 and kon) or fast methylation back into the off state (large kme2→me3). It is intriguing to notice that either condition will severely delay the first activation of any ORs by decreasing the opportunity of two consecutive demethylation events into the on state. For example, in the simulation in Fig. 1B, Left (corresponding to ktotalΔt = 1.02 and kbottleneckΔt = 0.01), each allele spends most of its time in the off state and only transiently switches to the intermediate state, where it may be further demethylated into the on state. These observations suggest that the rarity of complete demethylation may be responsible for singular OR expression.
Fig. 2.
Our kinetic model generates singular OR expression by slow activation and fast feedback. (A) Singularity is achieved with a wide range of parameter values. The system is characterized by two dimensionless parameters: kbottleneckΔt ≡ Δt √kme3→me2kon describes how slow the two demethylation reactions are, whereas ktotalΔt ≡ (kme2→me3+kme3→me2+kon)Δt describes how fast the system transitions between epigenetic states. Singular OR expression can be established by a small kbottleneckΔt or a large ktotalΔt, which is equivalent to having a kinetic bottleneck in the two demethylation reactions (small kme3→me2 and kon) or rapid methylation back into the H3K9me3 state (large kme2→me3). Either condition should severely delay the first activation of any OR by limiting the opportunity of complete H3K9me3 demethylation. (B) The extent of singularity, quantified by the failure probability Pfail (the probability of expressing more than one OR), is determined by the ratio of two timescales: the mean time before the first OR activation T and the response time of the negative feedback Δt. We find that Pfail = c ⋅ Δt/T is a general conclusion for all kinetic models with an irreversible step of activation. Our model corresponds to c = 1 or π/2, and simulated data (black dots) fit well with the two theoretical predictions (gray lines). In both panels, orange regions correspond to the physiological failure probability Pfail ∼ 2%. Each data point is the mean of 106 simulations.
Consistent with this heuristic, we find that the extent of singularity is determined by the ratio of two timescales—the rate of OR activation and the response time of the negative feedback, a general conclusion that holds for a wide range of kinetic models. For each choice of parameters, we recorded T, defined as the mean time before the earliest activation (namely, the smallest among all n first-passage times to reach an on state), and the failure probability Pfail, defined as the probability of choosing more than one allele of ORs (namely, the percentage of neurons that are not singular and thus defective). Slower activation with respect to the feedback response (larger T/Δt) almost always leads to better singularity (smaller Pfail; black dots in Fig. 2B). This relationship is actually a general property for all kinetic models with an irreversible step of activation that can be turned off by negative feedback. We show theoretically that when the number of alleles n is sufficiently large, the failure probability Pfail = c ⋅ Δt/T, where c is a constant, holds regardless of the details of the kinetic model if Δt << T (Fig. S1A). The constant c is independent of n or any kinetic parameters; instead, it only depends on the categorization of the model based on the shape of its waiting time distribution near zero. Our model corresponds to c = 1 or π/2, depending on whether the linear rise or the exponential fall of the waiting time distribution (Fig. 1C) dominates under our choice of n = 2,800. Simulated data (black dots in Fig. 2B) fit well with the two theoretical predictions (gray lines). We also derived a full analytical solution in which Δt is not necessarily much smaller than T (Fig. S1B). In conclusion, slow activation and fast feedback can generate robust singularity regardless of the details of the activation process.
The Timescales Required for Singularity Are Compatible with Experimental Observations.
Our kinetic model suggests an intrinsic tradeoff between time and singularity. Singular OR expression (a very small failure probability Pfail) is achieved by sacrificing the rate of neuronal maturation (a long time before the first activation T). Although the exact extent of singularity has not been investigated for all ∼1,400 ORs, RNA FISH in the septal organ—a small fraction of the olfactory epithelium where most neurons choose from a pool of only nine ORs—showed a 1–2% probability of expressing more than one OR in newborn mice or in sensory-deprived adults (in contrast, normal adults would have a lower failure probability of ∼0.2% due to apoptosis of neurons with more than one OR) (15).
To achieve a failure probability of the physiological value 2%, the first activation must be ∼100× slower than the response of the negative feedback (T/Δt ∼ 100, orange regions in Fig. 2). Although the kinetics of this feedback has not been measured, the extremely high transcription levels of ORs and Adcy3 (8), the utilization of unfolded protein response (11), and the intrinsic instability of Lsd1 (rapid precipitation and proteasomal degradation when not bound by cofactors) (16, 17) may help to accelerate the response. Given a typical timescale Δt ∼ 1–2 h for transcriptional regulations (18), the activation of the first OR—namely, the maturation of olfactory sensory neurons—should be ∼5–10 d, which is consistent with observations after olfactory bulbectomy experiments, where the regeneration of sensory neurons occurred 5–10 d after the operation (19).
H3K9me3-to-H3K9me2 Demethylation May Be the Rate-Limiting Step Responsible for the Slow OR Activation.
In addition to predicting a slow kinetics of OR activation, our model also provides information on the rate-limiting step that leads to this kinetic bottleneck. In the analytical solution, the two rates of demethylation, kme3→me2 (for off→intermediate) and kon (for intermediate→on) (Fig. 1), play equal roles because they jointly contribute to singularity via the kinetic bottleneck kbottleneck ≡ √kme3→me2kon. Therefore, singular OR expression can in principle be generated in multiple ways: a slow rate of K9me3-to-K9me2 demethylation by an unidentified enzyme, a slow rate of K9me2 demethylation by Lsd1, or both. However, changes in kme3→me2 and kon will leave different impacts on the relative abundance of epigenetic states: If Lsd1 (kon for intermediate→on) is the only rate-limiting step, we would expect accumulation of alleles in the intermediate state (K9me2), all waiting for further demethylation by Lsd1. In contrast, OR genes were found mostly with K9me3 instead of K9me2 in globose basal cells, neuronal progenitors, and mature neurons in the olfactory epithelium (8). Furthermore, Lsd1 is expressed at a relatively high level as revealed by RNA FISH and immunostaining (10, 20), which is not compatible with Lsd1 being a rate-limiting enzyme. Therefore, a more likely explanation will be a rate-limiting step of K9me3-to-K9me2 demethylation (kme3→me2 for off→intermediate), probably mediated by an enzyme of very low abundance. It is also possible that Lsd1 and this H3K9me3 demethylase work in a protein complex, as in the case of regulating androgen receptor-dependent genes (21); if so, the low abundance of the complex will account for the bottleneck. The identification of this H3K9me3 demethylase will provide key insights into the cause of singular OR expression.
Experimentally Observed Switching Between ORs May Enhance Singularity, but the Improvement Is Marginal Given the Physiological Frequency of Switching.
So far our model has assumed an irreversible OR activation by Lsd1; however, lineage-tracing experiments showed that ∼10% of sensory neurons once abandoned their initial OR choice and switched to a different one (22). Lyons et al. (10) further suggested that the other function of Lsd1—demethylation of the active histone marker H3K4me2—may be responsible for turning off an already activated OR. This observation poses the question of whether the reversibility of OR activation contributes to singularity. The ability of Lsd1 to turn off ORs can help to cease the transcription of pseudogenes—nonfunctional OR genes that cannot induce the negative feedback; however, its influence on the singularity of intact OR genes has not been investigated.
We include OR switching in our model by adding an Lsd1-mediated reaction from the active state back into the intermediated state, with a rate kswitch (on→intermediate). To model explicitly the negative feedback, we assume that its response time Δt is primarily determined by the rate of Lsd1 down-regulation, because other components of the feedback, ORs and Adcy3, are extremely highly transcribed (8) and will thus respond much more rapidly. Our conclusions still hold if OR or Adcy3 accumulation is instead the primary determinant of Δt (Fig. S2). In addition, we assumed a decay rate α for Lsd1 when any ORs are active, a steady-state Lsd1 concentration of one when ORs are absent, and a concentration threshold κ below which Lsd1 can no longer induce any further activation.
We find that reversible OR activation can improve singularity by turning off the competing allele that may have otherwise induced nonsingular expression, but the extent of enhancement is marginal under the physiological condition. Once the first allele of OR is activated, the down-regulation of Lsd1 from one to κ takes Δt = log(1/κ)/α. To induce nonsingularity, the second, competing allele must not only be activated during this time window, but also remain activated until the end of activation (Fig. 3A). Therefore, OR switching may protect against nonsingular expression by turning the second allele off. However, any significant reduction in the failure probability must be accompanied by a very high frequency of OR switching Pswitch (Fig. 3B). For example, a mere twofold reduction in Pfail (which requires kswitch ∼ 1/Δt) requires a switching probability Pswitch (defined as the percentage of neurons that at least once abandoned an initial OR choice) of almost 90%. Analytical calculation shows that, when OR switching is infrequent, the fold reduction in Pfail is equal to Pswitch/2 in most situations (Fig. S3A). Given the physiological switching probability of ∼10% (22), which corresponds to kswitch ∼ 0.1/Δt, the failure probability is reduced by a mere 5% (orange region in Fig. 3B). Therefore, singularity can only be affected marginally. If 98.0% of all neurons are singular without switching, the physiological OR switching will only improve it to 98.1%. Note that there also exists an edge case where kon is significantly larger than both 1/T and kme2→me3 (namely, an allele that is turned off will quickly revive). In this case, OR switching actually harms singularity (Fig. S1B). In conclusion, the kinetic constraint that we previously derived still holds in the presence of OR switching.
Fig. 3.
Switching between ORs may facilitate singular OR expression, but the enhancement is marginal under the physiological condition. (A) A schematic of OR switching, modeled by the ability of ORs to be turned off by Lsd1 (off→intermediate) with a rate kswitch. We assume that the response of the negative feedback is primarily determined by Lsd1 depletion, although our conclusion holds if the primary determinant is instead OR/Adcy3 accumulation (Fig. S2). Therefore, we model Lsd1 by a decay rate α when ORs are active, a steady-state concentration of one when ORs are absent, and a threshold κ (white dashed line) below which no further activation can occur. Switching between ORs (such as the turning off of alleles 1 and 3) can enhance singular OR expression by deactivating the competing allele (in this case, allele 3) before the end of activation. (B) Any significant enhancement in singularity requires a large kswitch and thus a high probability of OR switching. Each representative trace is produced by fixing the values of kme2→me3, kme3→me2, kon and then tuning the turning-off rate kswitch. The probability of OR switching Pswitch is defined as the percentage of simulations where at least one OR was once activated and turned back off. The values of Pswitch are almost identical between the three choices of parameters; therefore only one trace is shown [red curve, which fits well with the analytical prediction Pswitch = 1 − exp(−kswitchΔt)]. The orange region corresponds to the physiological switching probability Pswitch ∼ 10%. Each data point is the mean of 106 simulations, with α = 1 and κ = 0.1 (corresponding to Δt ∼ 2.30).
Although OR switching has limited effects under the physiological condition, it can provide extra protection against large disruptions in Δt. When the feedback response is artificially prolonged such that Δt >> 1/kswitch, rather than producing neurons with more than one OR (losing singularity according to Pfail ∝ Δt), the system will get stuck in the high-Lsd1 phase and keep switching between ORs, because the mean lifetime of the active state (1/kswitch) is too short to fully down-regulate Lsd1; this is indeed the observation after Adcy3 knockout or after constitutive Lsd1 expression (10). Therefore, the ability of Lsd1 to turn off ORs provides a quality control against Δt disruptions, preventing loss of singularity beyond a baseline failure probability 1/(kswitch T). Such a better-safe-than-sorry strategy may provide an evolutionary advantage.
Discussion
The singular expression of ORs is a fascinating phenomenon in neurobiology and has attracted numerous modeling efforts. Various models have attempted to explain how these neurons overcome the competition of multiple ORs to generate robust singularity, but none was completely satisfactory. One model suggested that the expression of each OR might specifically silence all ORs but itself (6, 7). Just as in a bistable switch, the mutual repression between ORs can in principle generate multistability in which each stable state only supports one active allele. However, this model was challenged by the extreme difficulty for each OR to distinguish between all ∼2,800 alleles and avoid silencing itself. In another model, a short DNA sequence—the H element—was hypothesized to act as a global OR activator (23); if this is the case, intrinsically the system is only compatible with a singular OR choice, because there is one, and only one, copy of the H element in the genome. However, later experiments cast doubt on this model because the deletion of the H element does not affect the expression of most ORs (24, 25).
A recent epigenetic study by Magklara et al. (8) provided crucial evidence for a completely different model of OR singularity; in particular, they showed that OR silencing is not induced by OR expression but rather precedes it. This observation manifests that OR expression must involve an activation phase that releases some ORs from their initially silent state, which may be followed by a maintenance phase that preserves the choice of activation. At first glance, this mechanism seems less ideal than previous models in the sense that if an initial choice activated more than one OR, the maintenance phase will not reject it. The singularity of OR expression must completely rely on the kinetics of the activation process; therefore, it is unclear whether this is a feasible mechanism and what kind of kinetics is required.
In this paper, we show theoretically that such an activation-maintenance scheme can indeed generate robust singularity. In contrast to the recent series of studies (8–10), which emphasized the dichotomy between activation and maintenance during OR expression, we suggest that these two phases should not be viewed separately. A slow or fast kinetics of either phase cannot guarantee singularity; instead, it is the ratio between the two timescales that determines the extent of singular expression. This conclusion holds regardless of the details of the kinetic model or in the presence of OR switching (10, 22), and does not rely on ad hoc assumptions of enzymatic cooperativity (26, 27). Robust singularity can be achieved if OR activation—namely, the complete demethylation of H3K9me3—occurs as a rare, discrete event, as a result of a combination of slow activation and fast feedback. Now that the feedback loop (OR, Adcy3, Lsd1) underlying the maintenance phase has been identified (10, 11), the only missing piece in OR singularity is the activation step responsible for the required kinetic bottleneck. It is even possible that both the H3K9me3 and H3K9me2 demethylases are down-regulated by the feedback, which may enhance the sensitivity and robustness of the feedback.
In general, a slow epigenetic response coupled by a fast transcriptional response in our model is also applicable to other systems where monoallelic expression is desired, such as V(D)J recombination in the immune system (28–31) and the expression of protocadherins (32–34). Furthermore, understanding the kinetic constraints of singular expression may provide a guiding principle for the design of a synthetic circuit that assigns one unique barcode to each single cell in vivo.
Supplementary Material
Acknowledgments
The authors would like to thank Yuntao Steve Mao, Alec R. Chapman, and Jun Yong for helpful discussions, and Attila Szabo and Yang Shi for critical comments. This work is supported by National Institutes of Health (5RO1EB010244) to X.S.X.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321511111/-/DCSupplemental.
References
- 1.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci USA. 2004;101(7):2156–2161. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5(2):124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 3.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78(5):823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 4.Buck L, Axel R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell. 1991;65(1):175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts P. Odorant receptor gene choice in olfactory sensory neurons: The one receptor-one neuron hypothesis revisited. Curr Opin Neurobiol. 2004;14(1):31–36. doi: 10.1016/j.conb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 6.Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci USA. 2004;101(4):1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serizawa S, et al. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302(5653):2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 8.Magklara A, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145(4):555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clowney EJ, et al. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151(4):724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyons DB, et al. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154(2):325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155(2):321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13(5):297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 13.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 14.English BP, et al. Ever-fluctuating single enzyme molecules: Michaelis–Menten equation revisited. Nat Chem Biol. 2006;2(2):87–94. doi: 10.1038/nchembio759. [DOI] [PubMed] [Google Scholar]
- 15.Tian H, Ma M. Activity plays a role in eliminating olfactory sensory neurons expressing multiple odorant receptors in the mouse septal organ. Mol Cell Neurosci. 2008;38(4):484–488. doi: 10.1016/j.mcn.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zibetti C, et al. 2010. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci 30(7):2521–2532.
- 17.Forneris F, Binda C, Battaglioli E, Mattevi A. LSD1: Oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem Sci. 2008;33(4):181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Yosef N, Regev A. Impulse control: Temporal dynamics in gene transcription. Cell. 2011;144(6):886–896. doi: 10.1016/j.cell.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gokoffski KK, et al. 2010. Feedback regulation of neurogenesis in the mammalian olfactory epithelium: New insights from genetics and systems biology. The Neurobiology of Olfaction. Frontiers in Neuroscience, ed Menini A (CRC, Boca Raton, FL), Chap 10. [Google Scholar]
- 20.Krolewski RC, Packard A, Schwob JE. Global expression profiling of globose basal cells and neurogenic progression within the olfactory epithelium. J Comp Neurol. 2013;521(4):833–859. doi: 10.1002/cne.23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wissmann M, et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat Cell Biol. 2007;9(3):347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- 22.Shykind BM, et al. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117(6):801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126(2):403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 24.Fuss SH, Omura M, Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130(2):373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Khan M, Vaes E, Mombaerts P. Regulation of the probability of mouse odorant receptor gene choice. Cell. 2011;147(4):907–921. doi: 10.1016/j.cell.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 26.Alsing AK, Sneppen K. Differentiation of developing olfactory neurons analysed in terms of coupled epigenetic landscapes. Nucleic Acids Res. 2013;41(9):4755–4764. doi: 10.1093/nar/gkt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolterman BE, Iossifov I, Koulakov AA. 2012. A race model for singular olfactory receptor expression. ArXiv:1201:2933.
- 28.Farago M, et al. Clonal allelic predetermination of immunoglobulin-κ rearrangement. Nature. 2012;490(7421):561–565. doi: 10.1038/nature11496. [DOI] [PubMed] [Google Scholar]
- 29.Cedar H, Bergman Y. Epigenetics of haematopoietic cell development. Nat Rev Immunol. 2011;11(7):478–488. doi: 10.1038/nri2991. [DOI] [PubMed] [Google Scholar]
- 30.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21(2):133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlimgen RJ, Reddy KL, Singh H, Krangel MS. Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nat Immunol. 2008;9(7):802–809. doi: 10.1038/ni.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esumi S, et al. Monoallelic yet combinatorial expression of variable exons of the protocadherin-alpha gene cluster in single neurons. Nat Genet. 2005;37(2):171–176. doi: 10.1038/ng1500. [DOI] [PubMed] [Google Scholar]
- 33.Monahan K, et al. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-α gene expression. Proc Natl Acad Sci USA. 2012;109(23):9125–9130. doi: 10.1073/pnas.1205074109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magklara A, Lomvardas S. Stochastic gene expression in mammals: Lessons from olfaction. Trends Cell Biol. 2013;23(9):449–456. doi: 10.1016/j.tcb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.