Significance
Adaptation to local environmental conditions is common, but the genetic mechanisms of adaptation are poorly known. We produced recombinant inbred lines (RILs) of the model plant Arabidopsis thaliana by crossing populations that inhabit drastically different climates in Sweden and Italy, grew the RILs at the parental sites for 3 y, and genetically mapped quantitative trait loci (QTL) for fitness. The results demonstrate that surprisingly few QTL explain much of the adaptive divergence between the two plant populations. Moreover, we find strong evidence for tradeoffs (i.e., adaptation to one environment reduces performance elsewhere). The results shed light on processes governing the evolution of biological diversity and the potential for adaptive evolution in response to environmental change.
Keywords: divergent selection, genetic drift, inbreeding, QTL mapping, RIL population
Abstract
Organisms inhabiting different environments are often locally adapted, and yet despite a considerable body of theory, the genetic basis of local adaptation is poorly understood. Unanswered questions include the number and effect sizes of adaptive loci, whether locally favored loci reduce fitness elsewhere (i.e., fitness tradeoffs), and whether a lack of genetic variation limits adaptation. To address these questions, we mapped quantitative trait loci (QTL) for total fitness in 398 recombinant inbred lines derived from a cross between locally adapted populations of the highly selfing plant Arabidopsis thaliana from Sweden and Italy and grown for 3 consecutive years at the parental sites (>40,000 plants monitored). We show that local adaptation is controlled by relatively few genomic regions of small to modest effect. A third of the 15 fitness QTL we detected showed evidence of tradeoffs, which contrasts with the minimal evidence for fitness tradeoffs found in previous studies. This difference may reflect the power of our multiyear study to distinguish conditionally neutral QTL from those that reflect fitness tradeoffs. In Sweden, but not in Italy, the local genotype underlying fitness QTL was often maladaptive, suggesting that adaptation there is constrained by a lack of adaptive genetic variation, attributable perhaps to genetic bottlenecks during postglacial colonization of Scandinavia or to recent changes in selection regime caused by climate change. Our results suggest that adaptation to markedly different environments can be achieved through changes in relatively few genomic regions, that fitness tradeoffs are common, and that lack of genetic variation can limit adaptation.
Organisms inhabiting different environments are often locally adapted, as demonstrated by experiments showing that local populations grown in their native sites outperform other populations (1–3). Despite a large body of theory, the genetic basis of local adaptation is poorly understood. Key unanswered questions include the number and effect sizes of adaptive loci, whether locally favored loci reduce fitness elsewhere (i.e., fitness tradeoffs), and whether the adaptive potential of populations is limited by insufficient genetic variation (4–11). The Fisherian view that adaptation is a function of many genes of small effect (12) has been challenged on theoretical grounds (4, 6), and recent empirical work indicates that genes with both large and small phenotypic effects contribute to differentiation in putatively adaptive traits (e.g., refs. 8 and 13). However, there are few direct estimates of the number of loci underlying fitness variation in native environments (14).
Because natural environments are variable in both time and space, temporally replicated experiments are particularly important for distinguishing whether adaptive alleles are conditionally neutral [i.e., favored locally but neutral elsewhere (7, 15)] or are favored in one environment but disfavored in the other, reflecting an adaptive tradeoff (2, 7, 16). Evidence for conditional neutrality is far more common than that for tradeoffs (15), but this disparity may be attributable to the more rigorous statistical criterion for detecting tradeoffs (11), a hurdle alleviated in part by long-term field experiments.
Although adaptation is ubiquitous, genetic drift may, under some conditions, counter or slow the rate of adaptation (7, 13, 17). The efficacy of selection is inversely proportional to the effective-population size (18), and small populations may therefore often lose beneficial mutations and fix deleterious mutations attributable to drift. Particularly low effective-population sizes are expected in highly inbred species such as those with a high rate of self-fertilization (19, 20). Moreover, reductions in effective-population size can occur during range expansions and at range margins because of repeated bottlenecks and reduced gene flow among populations (21, 22). Thus, even natural populations that are locally adapted may not possess the optimal phenotype, because they may lack the genetic variation necessary to achieve the highest possible fitness.
Using recombinant inbred lines (RILs), we examined the genetic basis of adaptive differentiation between two populations of the highly self-fertilizing, model plant Arabidopsis thaliana, one from Sweden and one from Italy. A recent study using reciprocal transplant experiments carried out over 5 y demonstrated that these populations are locally adapted (23). This system combines a number of features that makes it uniquely suited for studies of the genetic basis of adaptive divergence. First, the two ecotypes grow under markedly different environmental conditions (23), having been collected from the northern (Sweden) and southern (Italy) limits of the native range (24). This provides an expectation that factors such as winter temperature and day length during growth and flowering contribute to differences in selection regime and adaptive differentiation. Second, the two populations are adapted to their local environments. This is important because only then can we attribute fitness differences to natural selection and evaluate whether high fitness of local alleles at quantitative trait loci (QTL) in the native environment is associated with reduced fitness in a different environment (tradeoff) or whether different QTL are important for adaptation in different environments (conditional neutrality). Third, a large RIL population derived from a cross between the parental ecotypes is available for genetic-mapping studies. Finally, the relative short lifespan of A. thaliana makes it possible to assess temporal variation in the relationship between genotype and fitness.
A number of studies have used the genetic resources available for A. thaliana to investigate the genetic basis of adaptation. For example, Weinig et al. (25) planted RILs derived from A. thaliana accessions with European ancestry (Columbia and Landsberg erecta) and identified QTL associated with survival and fecundity at two field sites in North America. Similarly, a large-effect QTL associated with seed dormancy and overall fitness was identified in a field study in which a new set of RILs were planted in two novel environments (26). Genome-wide association (GWA) mapping using a wide diversity of A. thaliana accessions has revealed a number of putative cases of adaptive differentiation (27–29), and recent studies that incorporate GWA mapping have identified correlations between environmental factors and genetic polymorphisms, some of which were also associated with fitness in field experiments (30, 31). Nevertheless, no study of the genetic basis of adaptation in A. thaliana has been conducted on locally adapted populations grown in their native environments; thus, it is not currently possible to determine the adaptive significance of allelic variation or the prevalence of genetically based tradeoffs at the loci underlying local adaptation.
To examine the genetic basis of adaptive differentiation, in 3 consecutive years, we planted seedlings of the two parents and of 398 genotyped RILs in experimental gardens at the parental sites and mapped QTL for survival, fruit production of survivors, and overall fitness. In total, we grew 43,964 plants, and counted 565,070 fruits. We asked the following. (i) How many QTL are involved in adaptation and what are their effect sizes? (ii) Do locally favored QTL reduce fitness elsewhere [i.e., are there fitness tradeoffs]? (iii) Are some local alleles maladaptive, which would suggest that adaptive evolution is constrained by lack of genetic variation? (iv) Do QTL affect fitness because of effects on survival, fecundity, or both?
Results
Fitness of Parents and RILs.
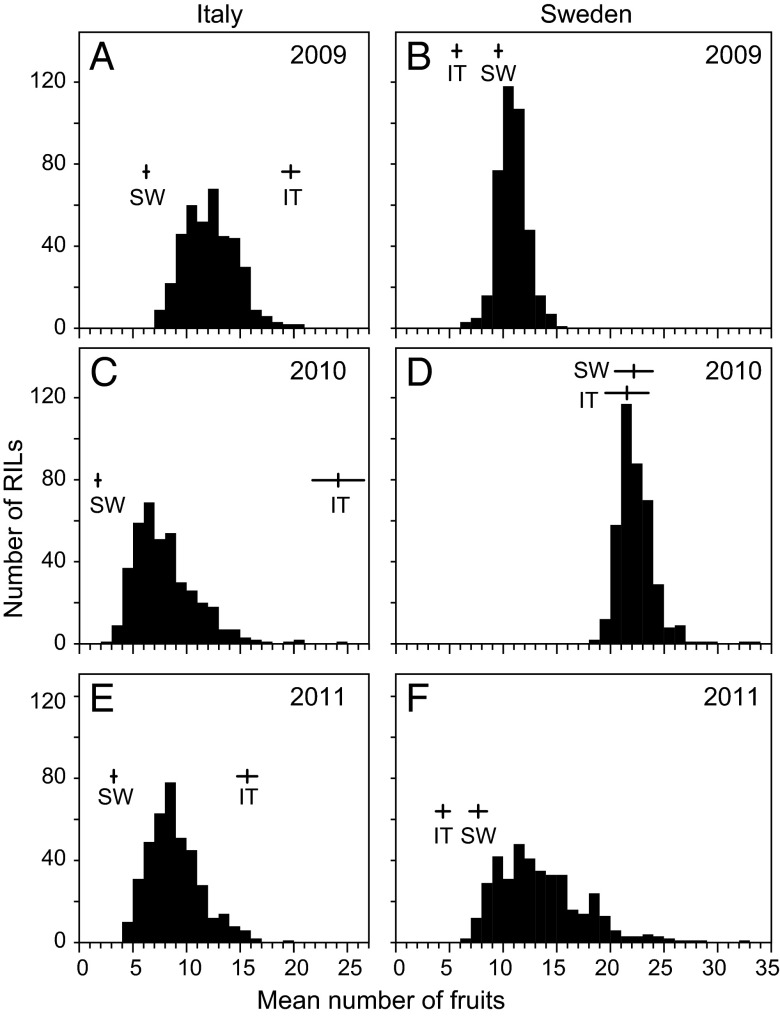
The local parent had significantly higher fitness (fruit production per planted seedling) at both sites in all years (contrasts in ANOVA, P < 0.05; Fig. 1 and Table S1), except for Sweden in 2010, when winter temperatures at ground level were exceptionally warm (23) and when native voles nonselectively destroyed about half of the plants. In Italy, RIL mean fitness was intermediate to that of the parental lines, whereas in Sweden, considerable transgressive variation was observed (Fig. 1). Fitness varied significantly among RILs, and in the 3 y of study, genotype (RIL) accounted for 18.1%, 9.9%, and 14.7% of the variance in fitness among individual plants in Italy and for 9.5%, 1.4%, and 5.8% of that in Sweden, respectively.
Fig. 1.
Distribution of RIL means for fitness (mean number of fruits per seedling planted) in field experiments established in Italy (A, C, and E) and Sweden (B, D, and F) in 2009, 2010, and 2011. Means for parental lines and associated 95% confidence intervals are indicated.
One potential limitation of QTL mapping studies in natural populations is that the within-population variation in the trait of interest may not be captured in a single cross between two lines. To address this, we grew 18 replicates from each of eight independent lines per population at each site in 2012/2013. The local population produced on average 9.2 times more fruits compared with the nonlocal population at the Italy site (contrast Italy vs. Sweden population in ANOVA model: t = 10.8, P < 0.0001) and 1.9 times more fruits at the Sweden site (t = 2.3, P = 0.03). Among-line variation in fitness within populations was not statistically significant (P > 0.05) at either of the two sites, and the range of line mean fruit production did not overlap between the Italy and the Swedish populations (back-transformed least square mean number of fruits: range, 36.5–60.9 vs. 3.8–6.1 fruits per planted seedling in Italy; 4.6–9.3 vs. 11.2–14.4 fruits per planted seedling in Sweden). This suggests that although the RIL population is derived from a cross between only two genotypes, it is likely to segregate for the genes underlying the major fitness differences between the two populations.
Number and Effect of QTL for Fitness.
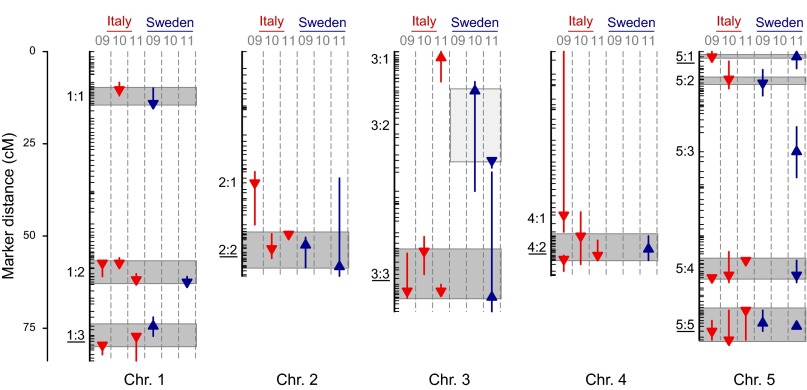
In the 3 y of study, we identified nine, eight, and eight fitness QTL in Italy, together explaining on average 61% (range 55–66%) of the variance in mean fitness among RILs (Fig. 2 and Tables S2 and S3). In Sweden five, one, and nine fitness QTL were identified, together explaining on average 28% (range 4–49%) of the variance in mean fitness among RILs (Fig. 2 and Tables S2 and S3). A total of 13 distinct QTL were found in Italy and 12 in Sweden. For both sites and all years taken together, 15 distinct QTL for fitness were identified (with nonoverlapping 95% Bayesian credible intervals; Methods), of which 3 were observed only in Italy, 2 only in Sweden, and 10 were shared across sites (Fig. 2 and Table S2).
Fig. 2.
Fitness QTL detected in field experiments in Italy and Sweden in 2009, 2010, and 2011. Arrows indicate QTL position and the effect of the Swedish genotype (upward, fitness increased; downward, fitness decreased). The vertical line gives the 95% Bayesian credible interval. Of the 15 distinct QTL identified, 11 were detected in more than one site × year combination [indicated with light gray (QTL observed at just one site) and dark gray boxes (QTL shared across sites)]. Boxes specify the range of point estimates for distinct QTL detected more than once. The six fitness tradeoff QTL, where the local genotype was favored at each site, are underlined. LOD profiles are given in Fig. S2A.
The local genotype was favored for 92% of the fitness QTL in Italy (n = 13) but for only 58% of those in Sweden (n = 12; Fig. 2 and Table S2). For those QTL where the local allele was favored, the mean effect sizes observed at the two sites were very similar, 1.2 fruits per plant (range 0.7–1.7) in Italy and 1.3 fruits per plant (range 0.1–3.7) in Sweden (Table S2). However, the relative effect size (i.e., the ratio of the absolute effect to the difference in mean fruit production of the two ecotypes) differed substantially between sites, with a mean of 0.07 in Italy (range 0.04–0.12) and 0.46 in Sweden (range 0.02–1.14). The mean effect size (fruits per plant) for those QTL in Sweden where the Swedish allele decreased fitness (1.0) was only slightly smaller than that of QTL where the Swedish allele was favored (1.3; Table S2). By comparison, the effect size of the single QTL in Italy where the local allele decreased fitness was the lowest of all QTL detected at this site (0.66 fruits per plant; Table S2).
Epistatic Interactions.
Epistatic interactions were observed only in Sweden: two QTL pairs in 2009 and one pair in 2011 (Table S2). In these cases, RILs with the Swedish genotype at one QTL and the Italian genotype at the second QTL outperformed RILs with local or nonlocal genotypes only (Fig. S1).
Fitness Tradeoffs.
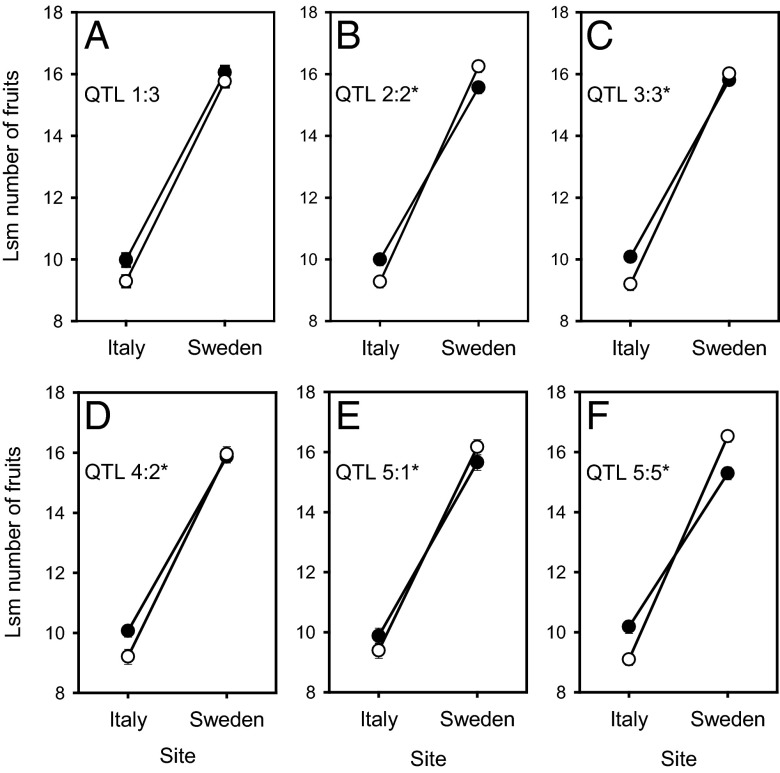
Mapping results for individual year × site combinations identified tradeoffs between sites for 6 fitness QTL (Fig. 2 and Table S2), 5 of which were supported by an analysis of the effects of the 15 distinct fitness QTL across all 3 y (Fig. 3 and Table S4). The mapping results indicated conditional neutrality for three of the five fitness QTL detected at only one of the sites (2:1, 4:1, 5:3) and a negative effect of the local allele at QTL 3:1 in Italy (Fig. 2). Finally, for four shared fitness QTL (1:1, 1:2, 5:2, and 5:4), the Italian allele was favored at both sites (Fig. 2).
Fig. 3.
Least-square mean number of fruits (±2 SE) of the Italian (closed symbol) and Swedish genotype (open symbol) at the six distinct fitness QTL (A–E) for which mapping results indicated a tradeoff between the Italian and Swedish sites across the 2009, 2010, and 2011 experiments. Least-square means were derived from an ANOVA including genotype at each of the 15 distinct QTL, site, year, and interactions as independent variables (Table S4). The five fitness QTL for which the ANOVA supported a tradeoff are indicated with an asterisk.
QTL for Survival and Fecundity.
Fitness QTL influenced both the proportion of plants surviving to reproduce and the fecundity of reproducing plants. Eight fitness QTL in Italy and 6 in Sweden colocalized with QTL for survival in at least 1 y, whereas 12 of 13 distinct fitness QTL in Italy and 9 of 12 fitness QTL in Sweden colocalized with QTL for fecundity (Figs. S2 and S3 and Tables S2, S3, S5, and S6). In all cases where Swedish alleles showed evidence of local maladaptation, the Swedish genotype was disfavored because of effects on fecundity (Table S2), whereas the local genotype was favored at all survival QTL (Table S5). In each of the 14 cases (QTL × site × year combinations) where credible intervals of fecundity and survival QTL overlapped, the direction of the effect on the two fitness components was the same (Fig. S3) (i.e., there was no evidence of conflicting selection through survival and fecundity).
Discussion
We find that surprisingly few QTL can explain much of the adaptive divergence between two natural populations of the model plant A. thaliana inhabiting markedly different climates, that fitness tradeoffs are common, and that lack of genetic variation can limit adaptation. A total of 15 distinct fitness QTL were detected, of which 10 were shared across sites, and taken together, these explained a considerable proportion of the total genetic variance in fitness at each of the two sites.
One potential weakness of the QTL mapping approach is the inability to detect small effect QTL and the subsequent overestimation of the effects of the detected QTL [the Beavis effect (32)]. We have several reasons to believe that these effects are not severe in our study. First, the Beavis effect is expected to diminish as the number of lines increases, with only modest effects for mapping populations as large as those used here (33). Second, using a RIL mapping population provides increased precision in estimating the phenotype over an F2 population such as used in Beavis’ simulation. Finally, multiple QTL modeling implemented in the software R/qtl increases the ability to detect small effect QTL (34). We detected QTL that explained as little as 1.5% of the variance in RIL means. Still, the number of fitness QTL we identified must be considered a minimum estimate because many small effect QTL were certainly not detected.
In addition to the statistical limitations of identifying QTL, the design of our experiment, with individuals planted at the seedling stage, should have reduced the effects of genes affecting seed dormancy and the timing of germination. As a result, it is likely that experiments initiated at the seed stage would identify additional QTL for fitness (cf., refs. 26 and 35).
Both the results of QTL mapping conducted separately by year and site, and the analysis of QTL effects across sites and years by ANOVA, provide evidence of fitness tradeoffs for 5 of the 15 fitness QTL identified. That one-third of all fitness QTL were involved in tradeoffs in our study contrasts with the minimal evidence of fitness tradeoffs in earlier studies of the genetic basis of local adaptation in plants (15, 36). Our mapping results indicated conditional neutrality for three of the five fitness QTL detected at only one of the sites (2:1, 4:1, 5:3; Fig. 2), but this designation should be considered tentative because some of our putative tradeoff QTL were only identified after 2 or more years of study. Had our study been conducted in a single year, we would have identified two (2009), none (2010), or four (2011) fitness tradeoffs, illustrating that the classification of fitness QTL depends strongly on the duration of field experiments. This underscores the value of temporally replicated studies (11) such as those presented here. Credible intervals around some of the fitness QTL were wide (Fig. 2), and further work is required to determine whether tradeoffs at the level of QTL are also detectable at the level of individual genes.
Small effective-population size in highly selfing species such as A. thaliana should increase the likelihood that deleterious mutations of moderate to large effect are fixed by drift (37, 38). Consistent with this expectation, we find many cases in Sweden where the local allele was deleterious, and the mean effect size (fruits per plant) for these QTL was only slightly smaller than that of QTL where the Swedish allele was favored. In contrast, only a single QTL was observed in Italy for which the local allele was deleterious and its effect size was the smallest of all QTL observed at this site. Both of our study populations are highly selfing in the field and laboratory, and although we have not conducted quantitative studies of the mating system, we have no reason to suspect that the populations differ in their degree of self-fertilization. Thus, we do not believe that the greater frequency and effect sizes of deleterious alleles in Sweden are attributable to differences between populations in the degree of self-pollination.
Instead a number of other factors, potentially acting in concert, could explain the higher incidence of deleterious alleles in Sweden. First, selection against the nonlocal ecotype is much weaker in Sweden (selection coefficient; mean, 0.29) than in Italy (mean, 0.80; Table S1), and selection on fitness QTL was less consistent across years in Sweden (Fig. 2 and Table S2). This weaker selection in Sweden provides a greater opportunity for the fixation of maladaptive alleles through random genetic drift. Second, reduced genetic variation in the northern part of the range, consistent with population bottlenecks during postglacial expansion (39), may have limited the adaptive potential of Swedish populations relative to Italian populations. Finally, recent climatic warming in Sweden, as reflected in marked shifts in tree lines to higher elevation (40), may have increased the relative fitness of some southern alleles. Climatic warming should reduce the fitness difference between the southern and northern genotype at the northern site but possibly increase the difference at the southern site.
For those QTL where the local allele was favored, the mean effect sizes observed at the two sites were very similar when estimated as the absolute number of fruits per plant (1.2 in Italy vs. 1.3 in Sweden). In contrast, the relative effect size, estimated as the mean fruit number relative to the difference in mean fruit production between the two parental lines, was much larger in Sweden than in Italy. Thus, in Italy both the absolute and relative effect sizes of fitness QTL were rather small, whereas in Sweden, the relative effect sizes of locally adaptive QTL were much higher.
In Sweden, many RILs outperformed the Swedish parental line (Fig. 1). Both fixation of deleterious mutations in the Swedish parental line and epistatic interactions apparently contribute to this transgressive variation. All three epistatic interactions observed in Sweden were negative (i.e., RILs with the Swedish genotype at one QTL and the Italian genotype at the second QTL outperformed RILs with local or nonlocal genotypes only). These negative epistatic interactions should contribute to the right-skewed distribution of the transgressive variation.
A comparison of map positions indicated that fitness QTL influenced fitness because of effects on both survival to reproduction and fecundity of reproducing plants. Most fitness QTL colocalized with QTL for fecundity, and about half colocalized with QTL for survival in at least 1 y. In Sweden, all fitness QTL that showed evidence of local maladaptation were disfavored because of effects on fecundity, whereas the local genotype was favored at all survival QTL. This may reflect historically more consistent selection on genes affecting survival at the Swedish site, where the local genotype had higher survival than the nonlocal genotype in all 3 y but higher fecundity in only 1 of 3 y (Table S1). In no case was a tradeoff between effects on fecundity and survival at a given site recorded.
Field observations suggest that differences in flowering time and cold tolerance contribute to adaptive differentiation of the studied populations (23), and preliminary comparisons of QTL for these traits and for fitness in the field support this hypothesis. In the vernalization treatment of a recent QTL study of flowering using F2 plants produced from our parental populations, three flowering time QTL were identified (VF1, VF3, and VF6; ref. 41) that mapped to the same regions as fitness QTL showing tradeoffs between the Italy and Sweden sites (QTL 1:3, 3:3, and 5:5; Fig. 2), and at all three loci, the Swedish genotype was associated with later flowering (41). Flowering time QTL VF1 maps to the same region as the candidate gene MADS AFFECTING FLOWERING 1 (MAF1), whereas VF3 does not have an obvious candidate gene (41). Flowering time QTL VF6 maps to a region with several flowering time candidate genes, including VIN3, VIP4, ELF5, and MAF2-5 (41), of which at least MAF2 exhibits geographically widespread functional variation (42). FRIGIDA (FRI) and FLOWERING LOCUS C (FLC), two genes that have received considerable attention for their effects on flowering (43, 44), and that show geographic variation across Europe consistent with the action of selection (45, 46), did not contribute to fitness tradeoffs observed in our study. FRI does not influence flowering time differences between our parental populations (41); both a large effect QTL for flowering time identified in the F2 study (VF4) (41) and fitness QTL 5:2 map to the same region as FLC does. However, the Italian genotype at QTL 5:2 was favored at both sites (Fig. 2), indicating that although variation at FLC may have influenced fitness in this experiment, it was not involved in a fitness tradeoff.
Fitness QTL 4:2 maps to the same region as the transcription factor CBF2, a major regulatory hub of a freezing tolerance gene network in A. thaliana (47, 48). The fitness QTL at this location also shows a tradeoff between Sweden and Italy and maps to a region with QTL for both survival and fecundity at both sites (Table S2). This is consistent with the hypothesis that freezing tolerance is associated with a cost that is expressed in warmer climates but may also reflect relaxed selection for freezing tolerance at southern latitudes (49). Taken together, these results support the hypothesis that flowering time and freezing tolerance contribute to the adaptive differentiation of these populations. More generally, they exemplify how QTL mapping of fitness can guide attempts to identify genes underlying fitness tradeoffs in natural populations.
Using a unique population of RILs in a multiyear field experiment, we have demonstrated that a limited number of QTL are responsible for much of the adaptive differentiation between the ecotypes examined and that tradeoffs, as well as epistatic interactions and genetic drift, contribute to differentiation among natural populations of A. thaliana.
Methods
Source Populations.
To maximize environmental differences contributing to geographic adaptation, we chose populations near the northern and southern limits of the native geographic range; one in north-central Sweden (Rödåsen; 62°48′N, 18°12′E) and one in central Italy (Castelnuovo; 42°07′N, 12°29′E). Phylogeographic analysis indicates that the genetic makeup of the study populations is typical of that found within each geographic region (39). Both populations grow on steep, rocky slopes and are winter annuals; seeds germinate in the autumn and overwinter as rosettes. Plants flower during March to April in Italy and May to June in Sweden (23). Seeds were collected in the source populations in June 2002 in Sweden and in April 2003 in Italy.
RIL Construction.
We produced RILs derived from a cross between an individual from the Swedish locality (♂) with an individual from Italy (♀). The F1 hybrid was selfed (S1), and 544 independent lines were generated by autonomous selfing and single seed descent for nine generations (S9). Seeds of the RILs and associated genotype information will be deposited at the Arabidopsis Biological Resource Center.
RIL Genotyping and Construction of the Linkage Map.
We resequenced the two parental lines using the Illumina GAII platform. Paired-end reads from each sample were aligned to the A. thaliana reference sequence (the Arabidopsis Information Resource version 9 of the genome annotation of the Columbia reference), using the short-read alignment program Bowtie (50). We identified 141,437 single-nucleotide differences between the Italy and Sweden parent. Of these, we selected 384 SNPs evenly spaced across all five nuclear chromosomes of the Columbia physical map. These were genotyped using the Illumina Golden Gate Assay on a set of 544 RILs. SNPs that were redundant had >5% missing data and/or appeared to be genotyping errors (as determined in R/qtl; ref. 51) were removed. The remaining 348 SNPs were mapped to five linkage groups using the maximum likelihood algorithm with the Kosambi mapping function in JoinMap 4 (52) (see Table S7 for list of markers used).
For the 348 SNP markers in the linkage map, we compared the centimorgan position of each marker with the physical map position in Columbia (Table S7). For each chromosome, there is a region of low recombination that corresponds to the centromere. In addition, a heterochromatic region on chromosome 4 that was sequenced and published with the Columbia genome in 2000 (53) is apparent as an inversion.
Overall, the cross shows some segregation distortion, in particular on chromosome 4 where Italy alleles are overrepresented across the entire chromosome (range 53–63%; Table S7). We analyzed the effect of segregation distortion by mapping the location of a single QTL with a set of 2,000 simulated populations, which either mimicked the maximal level of segregation distortion observed in our data or had equal proportions of alleles at all loci. Neither the mean QTL position nor the error associated with the estimate differed between the two simulations. The results of the simulations suggest that in mapping experiments like ours with many RILs, estimates of QTL positions are robust to this level of segregation distortion, because there are still hundreds of observations of the rarer alleles.
Field Experiment.
To identify QTL for fitness and for components of fitness (survival, fecundity), in 3 consecutive years (2009 to 2011), we planted seedlings of 398 randomly selected RILs and the two parents in experimental gardens established in natural vegetation at the sites of the source populations. Across the 3 y, 404 RILs were included in the experiment; 390 RILs were represented in all site × year combinations.
Seeds were planted in Petri dishes on agar, cold-stratified in the dark at 4 °C for 1 wk, and then moved to a growth room (22 °C/16 °C; 16-h day at photosynthetically active radiation of 150 μE⋅m−2⋅s−1; 8-h dark) where the seeds germinated. Nine days after germination, seedlings were transplanted to randomized positions in plug trays comprised of 299 cells (cell size: 20 mm × 20 mm × 40 mm) filled with local soil in Italy and with an equal mixture of local sand, gravel, and unfertilized peat in Sweden. In 2009, 20 seedlings of each RIL and 184 seedlings of each parent were transplanted. To reduce edge effects in this year, we excluded plants in the outer three rows of the array, giving a final sample size of 12–20 (median 17) plants per site × RIL combination and about 150 plants per site × parental line combination (23). In 2010 and 2011, we established three rows of “border” plants (all RILs contributed equally) that were not considered in subsequent analyses. In these years, we transplanted 18 seedlings of each RIL and 180 seedlings of each parent to positions inside the border.
During transplantation, plug trays were kept in a greenhouse at about 18 °C/12 °C and 16-h day/8-h night. Within 6 d, when the seedlings were at approximately the same stage of development as naturally germinating plants in the source populations, the trays were transported to the field sites where they were sunk into the ground (on September 16, 2009; September 10, 2010; and September 8, 2011 in Sweden and on November 7, 2009; October 30, 2010; and November 7, 2011 in Italy). Plants were misted once directly after transplanting and plots were weeded several times during the experiment.
We scored survival to reproduction and number of fruits per reproducing plant and quantified total fitness as the number of fruits produced per seedling planted. Within a week of the transplant to the field, we recorded the survival of transplanted seedlings. Seedling mortality during this first week was attributed to transplant shock, and these seedlings were excluded from subsequent analyses. At fruit maturation, we recorded survival and the number of fruits produced by reproducing plants. Fruit production is positively correlated with total seed production in both parental lines (23).
QTL Analysis.
We analyzed QTLs for fitness and its components: proportion of seedlings surviving to reproduce and mean fecundity of reproducing plants. For each RIL × site × year combination, we estimated fitness as the least-square mean number of fruits per seedling planted from mixed-model ANOVA that included RIL (random effect) and two variables controlling for position effects (tray and row; both treated as fixed effects). For each RIL, we also calculated the proportion of plants surviving to reproduction and mean fecundity (mean number of fruits per surviving plant). QTL mapping was conducted for each of the six site × year combinations in R/qtl (54) using the following settings: (i) Haley–Knott regression using genotype probabilities of the genetic markers and pseudomarkers in gaps >2 cM; (ii) 10,000 permutations to calculate thresholds for incorporating additive QTL and epistatic interactions at experiment-wise α = 0.05; (iii) automated stepwise model selection scanning for additive QTL and interactions (34); and (iv) refining the QTL positions and fitting the model with ANOVA to calculate the effect size and percentage variance explained for each QTL. Haley–Knott regression was used because our marker density was high (average marker spacing, ∼1 cM), and the proportion of missing data were low (∼1%). Because the automated stepwise procedure is sensitive to departures from normality, we first transformed the data by quantile normalization (51). We then fitted this model with the nonnormalized data to generate genotypic effect sizes on the raw scale.
To identify fitness QTL that were shared across years within sites, and across sites over all years, we considered 95% Bayesian credible intervals and point estimates of QTL. Within sites, QTL were considered distinct had they been identified in the analysis of data from a single site × year combination. QTL detected in different years and with nonoverlapping credible intervals were also considered distinct unless their intervals overlapped that of a QTL in a third site × year combination. By these criteria, we identified 13 fitness QTL in Italy and 12 fitness QTL in Sweden (Fig. 2 and Table S2). Across sites, QTL with overlapping confidence intervals were considered shared with two exceptions: the 95% Bayesian credible intervals of the QTL at 58.3 cM on chromosome 2 and at 1.4 cM on chromosome 5 in Sweden 2011 each overlapped with the credible intervals of two distinct QTL in Italy. In these cases, the QTL with the most similar point estimates were considered the same. These criteria yielded a total of 15 distinct QTL, of which 10 were shared between sites.
Using 95% Bayesian credible intervals to identify shared QTL might be problematic if by invoking this criterion, widely spaced QTL are considered the same. However, we find that the point estimates for shared QTL were often very similar. Within sites, the maximum distance between point estimates for QTL considered to be the same based on overlapping 95% Bayesian intervals ranged from 2.4 to 10.8 cM (median, 4.5 cM; n = 7) in Italy and from 0.9 to 18.8 cM (median, 5.8 cM; n = 3) in Sweden. Point estimates were identical in 2 of 3 y for QTL 1:2 and 3:3 in Italy. Between sites, the closest point estimates for QTL considered to be the same were always within 3.8 cM (median, 1.2 cM; range, 0–3.8 cM). QTL 5:1 was observed just once at each site, but its point estimate was identical at the two sites. In 1 of the 3 y that QTL 5:4 was observed in Italy, the point estimate was identical to that of the single year it was observed in Sweden. Given these results, we suggest that we have strong evidence for shared QTL among years and across sites.
Using 1.5-logarithm of the odds (LOD) intervals rather than 95% Bayesian credible intervals to identify distinct QTL yielded identical results, except that one rather than two distinct QTL early on chromosome 5 was identified (Table S2), resulting in a total of 14 distinct QTL of which 9 were shared between sites.
Analysis of QTL Effects with ANOVA.
To further explore differences between sites and years in QTL effects, for each of the 15 distinct QTL identified by 95% Bayesian credible intervals, we first identified marker loci closest to mean map positions (weighted by LOD score) and then estimated the effects of QTL genotype (Italy or Sweden), site, year, the three pairwise QTL × QTL interactions detected, and the interactions of QTL with site and year on least-square mean fitness. Significant QTL × site and/or QTL × site × year interactions were detected for all 15 QTL except 2 (QTL 1:3 and 5:4; Table S4), and contrasts were used to compare QTL effects on fitness for each site × year combination. Variance inflation factors were all less than five, suggesting that collinearity of independent variables was not strong (55).
Map Positions of QTL for Fitness Components and Overall Fitness.
To assess whether QTL affected fitness because of effects on survival, fecundity, or both, we determined whether 95% credible intervals of fitness QTL overlapped those of QTL for survival and fecundity, respectively.
Supplementary Material
Acknowledgments
We thank H. D. Bradshaw, Jr., C. Jones, and T. Mitchell-Olds for comments on the manuscript, and F. Spada, E. Carli, S. Lundemo, and a team of highly skilled assistants for help with the field experiment. This study was financially supported by the Swedish Research Council (J.Å.), US National Science Foundation Awards DEB 1022202 (to D.W.S.) and DEB 1022196 (to J.K.M.), and the Wenner-Gren Foundation (J.Å.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316773110/-/DCSupplemental.
References
- 1. Clausen J, Keck DD, Hiesey WM (1940) Experimental Studies on the Nature of Species. I. Effect of Varied Environment on Western North American Plants (Carnegie Institution of Washington, Washington), Publications No. 520.
- 2.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7(12):1225–1241. [Google Scholar]
- 3.Hereford J. A quantitative survey of local adaptation and fitness trade-offs. Am Nat. 2009;173(5):579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- 4.Orr HA, Coyne JA. The genetics of adaptation: A reassessment. Am Nat. 1992;140(5):725–742. doi: 10.1086/285437. [DOI] [PubMed] [Google Scholar]
- 5.Barton NH, Keightley PD. Understanding quantitative genetic variation. Nat Rev Genet. 2002;3(1):11–21. doi: 10.1038/nrg700. [DOI] [PubMed] [Google Scholar]
- 6.Orr HA. The genetic theory of adaptation: A brief history. Nat Rev Genet. 2005;6(2):119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 7.Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8(11):845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- 8.Jones FC, et al. Broad Institute Genome Sequencing Platform & Whole Genome Assembly Team The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484(7392):55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson-Manning CF, Wagner MR, Mitchell-Olds T. Adaptive evolution: Evaluating empirical support for theoretical predictions. Nat Rev Genet. 2012;13(12):867–877. doi: 10.1038/nrg3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockman MV. The QTN program and the alleles that matter for evolution: All that’s gold does not glitter. Evolution. 2012;66(1):1–17. doi: 10.1111/j.1558-5646.2011.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson JT, Lee C-R, Rushworth CA, Colautti RI, Mitchell-Olds T. Genetic trade-offs and conditional neutrality contribute to local adaptation. Mol Ecol. 2013;22(3):699–708. doi: 10.1111/j.1365-294X.2012.05522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Oxford University Press; 1930. [Google Scholar]
- 13.Hall MC, Lowry DB, Willis JH. Is local adaptation in Mimulus guttatus caused by trade-offs at individual loci? Mol Ecol. 2010;19(13):2739–2753. doi: 10.1111/j.1365-294X.2010.04680.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett RDH, Hoekstra HE. Molecular spandrels: Tests of adaptation at the genetic level. Nat Rev Genet. 2011;12(11):767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- 15.Anderson JT, Willis JH, Mitchell-Olds T. Evolutionary genetics of plant adaptation. Trends Genet. 2011;27(7):258–266. doi: 10.1016/j.tig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fry JD. The evolution of host specialization: Are trade-offs overrated? Am Nat. 1996;148:S84–S107. [Google Scholar]
- 17.Alleaume-Benharira M, Pen IR, Ronce O. Geographical patterns of adaptation within a species’ range: Interactions between drift and gene flow. J Evol Biol. 2006;19(1):203–215. doi: 10.1111/j.1420-9101.2005.00976.x. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M. On the probability of fixation of mutant genes in a population. Genetics. 1962;47(6):713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nordborg M. Linkage disequilibrium, gene trees and selfing: An ancestral recombination graph with partial self-fertilization. Genetics. 2000;154(2):923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glémin S, Bazin E, Charlesworth D. Impact of mating systems on patterns of sequence polymorphism in flowering plants. Proc Biol Sci. 2006;273(1604):3011–3019. doi: 10.1098/rspb.2006.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pujol B, Zhou SR, Sanchez Vilas J, Pannell JR. Reduced inbreeding depression after species range expansion. Proc Natl Acad Sci USA. 2009;106(36):15379–15383. doi: 10.1073/pnas.0902257106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barringer BC, Kulka EA, Galloway LF. Reduced inbreeding depression in peripheral relative to central populations of a monocarpic herb. J Evol Biol. 2012;25(6):1200–1208. doi: 10.1111/j.1420-9101.2012.02510.x. [DOI] [PubMed] [Google Scholar]
- 23.Ågren J, Schemske DW. Reciprocal transplants demonstrate strong adaptive differentiation of the model organism Arabidopsis thaliana in its native range. New Phytol. 2012;194(4):1112–1122. doi: 10.1111/j.1469-8137.2012.04112.x. [DOI] [PubMed] [Google Scholar]
- 24.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 25.Weinig C, et al. Heterogeneous selection at specific loci in natural environments in Arabidopsis thaliana. Genetics. 2003;165(1):321–329. doi: 10.1093/genetics/165.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, et al. The earliest stages of adaptation in an experimental plant population: Strong selection on QTLS for seed dormancy. Mol Ecol. 2010;19(7):1335–1351. doi: 10.1111/j.1365-294X.2010.04557.x. [DOI] [PubMed] [Google Scholar]
- 27.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet. 2010;11(12):867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- 29.Long Q, et al. Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet. 2013;45(8):884–890. doi: 10.1038/ng.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fournier-Level A, et al. A map of local adaptation in Arabidopsis thaliana. Science. 2011;334(6052):86–89. doi: 10.1126/science.1209271. [DOI] [PubMed] [Google Scholar]
- 31.Hancock AM, et al. Adaptation to climate across the Arabidopsis thaliana genome. Science. 2011;334(6052):83–86. doi: 10.1126/science.1209244. [DOI] [PubMed] [Google Scholar]
- 32.Beavis WD. In: Molecular Dissection of Complex Traits. Paterson AH, editor. New York: CRC; 1998. pp. 145–162. [Google Scholar]
- 33.Xu S. Theoretical basis of the Beavis effect. Genetics. 2003;165(4):2259–2268. doi: 10.1093/genetics/165.4.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manichaikul A, Moon JY, Sen S, Yandell BS, Broman KW. A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics. 2009;181(3):1077–1086. doi: 10.1534/genetics.108.094565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kronholm I, Picó FX, Alonso-Blanco C, Goudet J, de Meaux J. Genetic basis of adaptation in Arabidopsis thaliana: Local adaptation at the seed dormancy QTL DOG1. Evolution. 2012;66(7):2287–2302. doi: 10.1111/j.1558-5646.2012.01590.x. [DOI] [PubMed] [Google Scholar]
- 36.Anderson JT, Lee C-R, Mitchell-Olds T. Strong selection genome-wide enhances fitness trade-offs across environments and episodes of selection. Evolution. 2013 doi: 10.1111/evo.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bustamante CD, et al. The cost of inbreeding in Arabidopsis. Nature. 2002;416(6880):531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- 38.Flowers JM, Hanzawa Y, Hall MC, Moore RC, Purugganan MD. Population genomics of the Arabidopsis thaliana flowering time gene network. Mol Biol Evol. 2009;26(11):2475–2486. doi: 10.1093/molbev/msp161. [DOI] [PubMed] [Google Scholar]
- 39.Beck JB, Schmuths H, Schaal BA. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol Ecol. 2008;17(3):902–915. doi: 10.1111/j.1365-294X.2007.03615.x. [DOI] [PubMed] [Google Scholar]
- 40.Kullman L. 20th century climate warming and tree-limit rise in the southern Scandes of Sweden. Ambio. 2001;30(2):72–80. doi: 10.1579/0044-7447-30.2.72. [DOI] [PubMed] [Google Scholar]
- 41.Grillo MA, Li C, Hammond M, Wang L, Schemske DW. Genetic architecture of flowering time differentiation between locally adapted populations of Arabidopsis thaliana. New Phytol. 2013;197(4):1321–1331. doi: 10.1111/nph.12109. [DOI] [PubMed] [Google Scholar]
- 42.Rosloski SM, Jali SS, Balasubramanian S, Weigel D, Grbic V. Natural diversity in flowering responses of Arabidopsis thaliana caused by variation in a tandem gene array. Genetics. 2010;186(1):263–276. doi: 10.1534/genetics.110.116392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stinchcombe JR, et al. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA. 2004;101(13):4712–4717. doi: 10.1073/pnas.0306401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61(6):1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 45.Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD. Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA. 2004;101(44):15670–15675. doi: 10.1073/pnas.0406232101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shindo C, et al. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005;138(2):1163–1173. doi: 10.1104/pp.105.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54(5):767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- 48.Thomashow MF. Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiol. 2010;154(2):571–577. doi: 10.1104/pp.110.161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhen Y, Ungerer MC. Relaxed selection on the CBF/DREB1 regulatory genes and reduced freezing tolerance in the southern range of Arabidopsis thaliana. Mol Biol Evol. 2008;25(12):2547–2555. doi: 10.1093/molbev/msn196. [DOI] [PubMed] [Google Scholar]
- 50.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. New York: Springer; 2009. [Google Scholar]
- 52.Van Ooijen JW. Joinmap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Wageningen, The Netherlands: Kyazma; 2006. [Google Scholar]
- 53.McCombie WR, et al. The complete sequence of a heterochromatic island from a higher eukaryote. The Cold Spring Harbor Laboratory, Washington University Genome Sequencing Center, and PE Biosystems Arabidopsis Sequencing Consortium. Cell. 2000;100(3):377–386. [PubMed] [Google Scholar]
- 54.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 55.Quinn GP, Keough MJ. Experimental Design and Data Analysis for Biologists. Cambridge, UK: Cambridge Univ Press; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.