Significance
Restriction factors are a component of the primate innate immune defense against viral pathogens. The recently discovered restriction factor SAMHD1 prevents lentiviruses such as human immunodeficiency virus and the related simian immunodeficiency virus from replicating in myeloid cells and resting T cells. Many lentiviruses encode accessory genes to antagonize SAMHD1 to replicate more efficiently, resulting in the rapid evolution of SAMHD1 to escape viral antagonism, characteristic of a molecular arms race between host and virus. HIV-1, surprisingly, does not encode a SAMHD1 antagonist. By examining viral adaptation to SAMHD1 polymorphism occurring in naturally infected primates, we show that SAMHD1 antagonism must be a factor contributing to the ability of lentiviruses to adapt to their primate hosts.
Keywords: Vpr, SIVagm, evolution
Abstract
Restriction factors are effectors of the innate immune response to viral pathogens that inhibit viral replication by operating as molecular barriers to steps of the viral life cycle. The restriction factor SAMHD1 blocks lentiviral reverse transcription in myeloid cells and resting CD4+ T cells. Many lineages of lentiviruses, including HIV-2 and other simian immunodeficiency viruses, encode accessory genes that serve to counteract host SAMHD1 restriction by causing degradation of the antiviral factor. The viral accessory protein Vpr is responsible for SAMHD1 degradation in some lineages of lentiviruses, whereas in others the related protein Vpx assumes this task. However, HIV-1 has no SAMHD1 degradation capability, leading to questions about the selective advantage of this activity. We use an evolutionary approach to examine the importance of SAMHD1 antagonism for viral fitness by studying adaptation to host SAMHD1 in natural simian immunodeficiency virus infections of African Green Monkeys. We identified multiple SAMHD1 haplotypes in African Green Monkeys and find that the vpr gene from different strains of Simian Immunodeficiency Virus has adapted to the polymorphisms of the African Green Monkey population in which it is found. Such evidence of viral adaptation to host restriction indicates that SAMHD1 antagonism is actively maintained in natural infections and that this function must be advantageous to viral fitness, despite its absence in HIV-1.
Simian immunodeficiency viruses (SIV) naturally infect over 40 species of African primates and have given rise to HIV-1 and HIV-2 in humans (1, 2). These primate lentiviruses have evolved to counteract host-specific, intracellular immune defenses called restriction factors, which can potently obstruct viral replication (3, 4). Viral accessory proteins are largely responsible for the circumvention of host restriction, and a defining feature of restriction factors is their engagement in a molecular “arms race” to continually escape recognition by these rapidly adapting viral antagonist proteins (5, 6).
SAMHD1 is deoxynucleoside triphosphate triphosphohydrolase that restricts lentiviral replication in myeloid and quiescent CD4+ T cells by suppressing cellular dNTP pools below the level required for reverse transcription and possibly by other mechanisms (7–12). The viral accessory proteins Vpr and Vpx relieve SAMHD1 inhibition by bridging the restriction factor to an ubiquitin ligase complex, targeting it for proteasomal degradation (13, 14). The viral genes vpr and vpx are paralogous, but only two major lineages of lentivirus encode vpx, whereas all encode vpr (15). In lineages that encode both genes, the Vpx protein is used for SAMHD1 antagonism, whereas in lentiviral lineages that do not encode vpx, the Vpr protein sometimes functions to degrade SAMHD1 (16). However, a subset of lentiviruses does not encode any SAMHD1 antagonist, including pandemic HIV-1 (16, 17). Neither HIV-1 nor its precursor, the SIV from chimpanzees (SIVcpz), encode a vpx gene, and their respective Vpr proteins do not degrade SAMHD1 due to a deletion that occurred during the generation of SIVcpz (18).
SAMHD1 exhibits classic features of a gene entrenched in virus–host genetic conflict, including episodes of rapid evolution and species specificity of the SAMHD1–Vpx/Vpr interaction (16, 17). Additionally, extant Vpx/Vpr proteins bind SAMHD1 at strikingly different interfaces, which also hints at the strength of selective pressure to recover antagonism after host switching or emergence of SAMHD1 escape variants (19). A lack of SAMHD1 degradation by HIV-1 is therefore perplexing, and additionally, in vivo studies show that the vpx gene, and thus presumably SAMHD1 antagonism, is critical for SIV dissemination and progression to AIDS in macaque models of infection (20–22). Therefore, we sought to examine the importance of SAMHD1 antagonism for viral fitness by an independent method of studying viral adaptation to host SAMHD1 in natural infections of extant primates.
SIV infections of African Green Monkeys (AGMs) provide a unique opportunity to study the evolutionary forces governing virus–host interactions. AGMs comprise at least four related species of the genus Chlorocebus. These primates inhabit most of Sub-Saharan Africa, although the individual species, commonly known as the sabaeus, vervet, grivet, and tantalus monkeys, are mostly geographically distinct (23). Although they share a most recent common ancestor less than 3 million years ago, each population is infected with a distinct subtype of SIVagm, named SIVagm.Sab, SIVagm.Ver, SIVagm.Gri, and SIVagm.Tan (24–26). Here we ask whether SIVagm subtypes adapt to variation in SAMHD1 in natural and experimental infections of AGMs. We find that SAMHD1 is polymorphic in AGMs, and variable sites alter sensitivity to degradation by the SIVagm SAMHD1 antagonist Vpr. Furthermore, we show that the specificity of Vpr for SAMHD1 involves both the N- and C-terminal regions of SAMHD1, and this specificity has evolved independently from adaptations of other Vpr proteins to their host SAMHD1. Evidence of viral adaptation to host restriction in AGMs indicates that SAMHD1 antagonism is indeed a component of viral fitness in the context of natural infections.
Results
SAMHD1 Is Polymorphic in AGM Species.
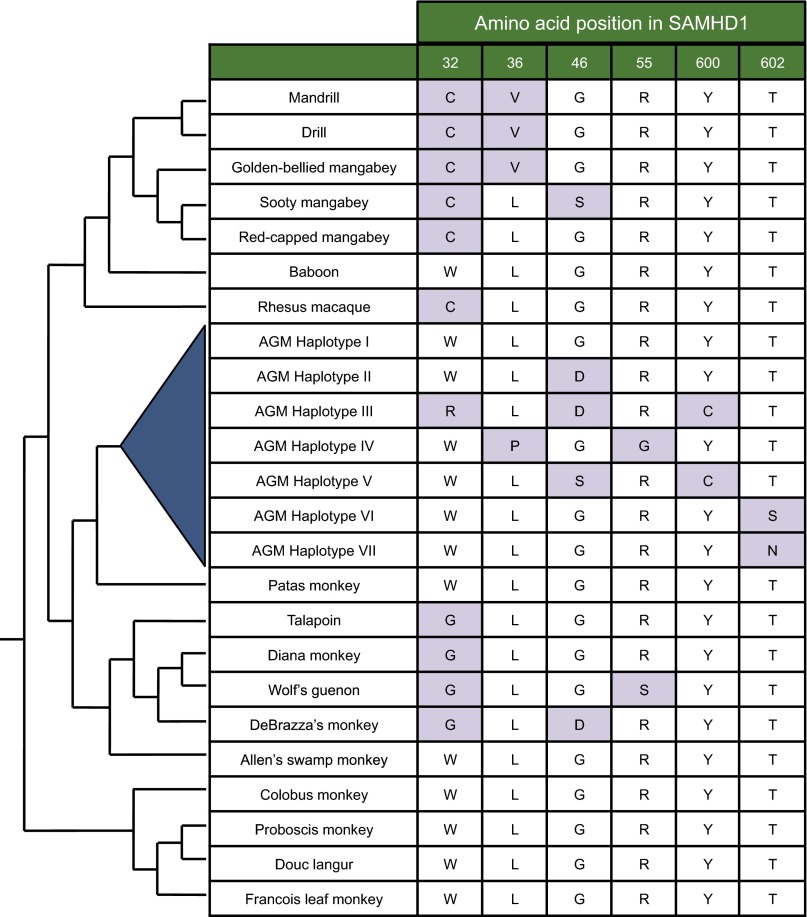
A key feature of the virus–host molecular arms race is the selection for amino acid–altering mutations in host antiviral proteins that disrupt binding by the viral antagonist. We sequenced SAMHD1 from four different AGM species to search for polymorphism that could affect the interaction between SAMHD1 and SIVagm Vpr (Fig. 1). Each of the species was represented by 9–11 samples, and two populations of sabaeus monkeys were included, one set taken from sabaeus monkeys in the original geographic range in West Africa and the other from a population introduced to the Caribbean islands (27). Sequence analysis identified seven distinct haplotypes of AGM SAMHD1 that yield variation at six amino acid positions (Fig. 1). The seven haplotypes are unique sequences that encompass all amino acid variation identified in AGM SAMHD1. Six of the 100 sequenced genes contain a silent SNP, but were counted as the haplotype of their amino acid sequence. Four of the variable amino acid sites are located at the N terminus of the protein and the other two are at the C terminus, but the middle of the gene is essentially devoid of both silent and amino acid–altering SNPs. The locations of variation are consistent with previous studies of positive selection in primate SAMHD1, where sites exhibiting strong signals of positive selection were identified mostly in the N and C termini of the protein (16, 17). The ancestral haplotype of AGM SAMHD1 (the version that contains the ancestral version of each amino acid at each variable site) is maintained in AGMs and was named haplotype I, whereas the other haplotypes vary from this ancestral haplotype by one to three amino acid changes. Sites 46 and 602 appear to have changed multiple times, as three different amino acids were observed at these sites. Variation occurs both at sites that are highly variable in other Old World monkey SAMHD1 as well as at sites that appear fixed in related primates (Fig. 1). For example, SAMHD1 of other Old World monkeys also vary at all N-terminal sites, with extensive variation between primates occurring at sites 32 and 46. However, the C terminus–altering polymorphisms have only been identified in AGM SAMHD1 and appear invariant in other Old World primates (Fig. 1).
Fig. 1.
AGM SAMHD1 is polymorphic. The seven identified SAMHD1 haplotypes (blue triangle) differ at six amino acid positions across the gene. No other nonsynonymous changes were found in any of the individuals outside of these sites, and only six of the 100 sequenced genes contain a synonymous SNP. Ancestral amino acids (white boxes) were inferred by maximum likelihood sequence reconstruction using Ancestral Sequence Reconstruction programs in Datamonkey (32, 33). Purple boxes denote derived amino acid changes. For comparison, the amino acid present in Old World monkey SAMHD1 sequences is shown for each location of AGM SAMHD1 variation. The sources of these sequences were previously reported (16, 17). The relationships of species according to Perelman et al. are shown by cladogram (24).
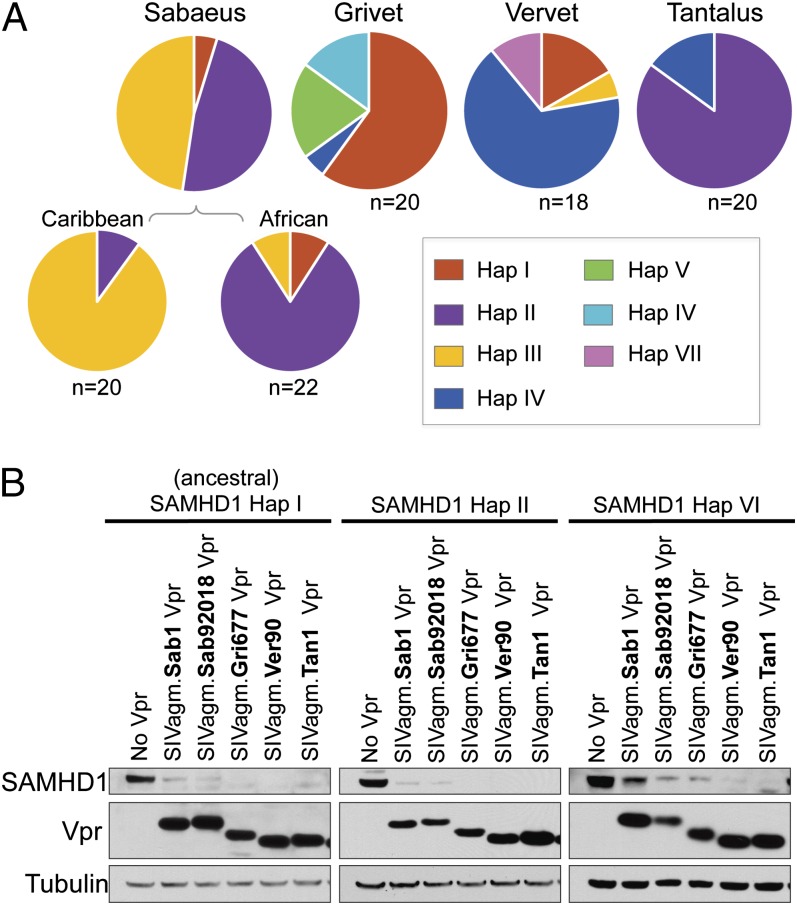
The distribution of SAMHD1 haplotypes among AGM species indicates that all populations harbor multiple versions of SAMHD1; thus, in no population has a haplotype drifted to fixation (Fig. 2A). However, there is a predominant haplotype in each species, which differ between populations. Several haplotypes were identified in at least three of the AGM species, including the reconstructed ancestral sequence, haplotype I, as well as haplotypes II and IV. The remaining haplotypes appear to have a more limited distribution. Haplotype III is found almost exclusively in sabaeus monkeys, although it was identified in one vervet sample. Haplotypes V, VI, and VII are unique to either grivet or vervet populations. Therefore, because each species exhibits a distinct composition of SAMHD1 alleles, we were able to test whether the autologous virus has adapted to antagonize the major and minor SAMHD1 variants in its host population.
Fig. 2.
All SIVagm Vprs have the ability to antagonize a subset of AGM SAMHD1 haplotypes. (A) Distribution of seven SAMHD1 haplotypes among AGM species. The grivet and tantalus pie charts represent 10 animals (20 SAMHD1 genes sequenced), whereas the vervet pie chart represents nine animals (18 SAMHD1 genes). Sabaeus samples originate from 10 Caribbean (20 genes) and 11 African (22 genes) animals. (B) Western blot analysis of HA-tagged AGM SAMHD1 expression in 293T cells with and without cotransfection of FLAG-tagged SIVagm Vprs. The Vprs of two SIVagm.Sab molecular clones are shown and appear to have slightly different degradation abilities. Tubulin was probed as a loading control.
AGM SAMHD1 Polymorphisms Have Functional Consequences for Degradation by Vpr.
To determine whether SIVagm Vprs demonstrate specific activity toward AGM SAMHD1, we cloned the seven SAMHD1 haplotypes into a mammalian expression vector and assayed them for sensitivity to Vpr-mediated degradation by viral proteins from each SIVagm subtype. Plasmids containing epitope-tagged SAMHD1 and Vpr were cotransfected into 293T cells and analyzed by immunoblotting. Levels of SAMHD1 in cellular lysates were compared with SAMHD1 expression in the absence of Vpr, and lower levels of SAMHD1 indicate that the tested Vpr is able to recognize and degrade the SAMHD1 variant.
Western blot analyses show that some variants of AGM SAMHD1 are degraded by Vpr of all SIVagm subtypes. For instance, Vpr from all SIVagm subtypes have the ability to degrade ancestral SAMHD1 (Fig. 2B). Additionally, Vpr from all SIVagm subtypes degraded the SAMHD1 variants encoded by haplotype II and haplotype VI, although one SIVagm.Sab Vpr appears slightly less efficient at degradation of the haplotype VI variant than the other SIVagm Vprs. Thus, the antagonism of ancestral SAMHD1 and two highly similar sequences is maintained by all SIVagm subtypes tested.
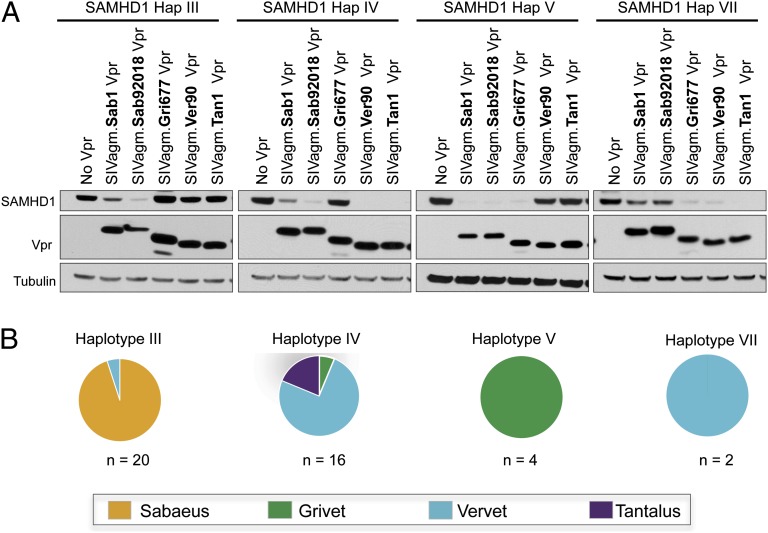
Although three AGM SAMHD1 variants are sensitive to degradation by Vpr from all SIVagm subtypes, we identified viral lineage-specific differences in Vpr antagonism of the other four SAMHD1 variants, those encoded by haplotypes III, IV, V, and VII (Fig. 3A). The variants encoded by these haplotypes are each degraded by a different subset of SIVagm Vprs. Additionally, each viral Vpr is able to degrade some, but not all, of these SAMHD1 variants. For example, haplotype III variant is degraded only by Vpr from viruses infecting sabaeus monkeys and is resistant to Vpr from all other SIVagm subtypes (Fig. 3, Left). This haplotype is found in both Caribbean and African sabaeus monkeys, suggesting SIVagm.Sab Vpr has adapted to antagonize this version of SAMHD1 due to its presence in the host population. Moreover, haplotype IV is completely degraded by SIVagm.Ver and SIVagm.Tan Vprs with Vpr from the two SIVagm.Sab viruses exhibiting full and partial degradation activities. This haplotype, haplotype IV, is found mostly in vervet and tantalus monkeys and is degraded by viruses naturally infecting these populations (Fig. 3). The haplotype V variant is found only in the grivet population, and SAMHD1 is degraded by Vpr proteins from viruses infecting grivet and sabaeus monkeys but is resistant to the other Vprs (Fig. 3). Finally, haplotype VII, which is found only in vervets, has partial resistance to Vpr of the virus infecting sabaeus population (Fig. 3). We also tested two additional SIVagm.Sab Vpr proteins and one additional SIVagm.Ver Vpr to determine if the pattern of degradation of SAMHD1 variants is consistent for a given SIVagm lineage. These vpr genes clustered with their respective SIVagm lineage on a phylogenetic tree, but were distinct from one another (Fig. S1). We found that the pattern of degradation for SAHMD1 variants was similar for each of the four SIVagm.Sab Vpr proteins. In addition, both SIVagm.Ver Vpr proteins exhibited identical phenotypes in degradation of AGM SAMHD1 haplotypes (Fig. S1). Thus, each lineage of SIVagm has evolved to recognize distinct SAMHD1 haplotypes.
Fig. 3.
Vpr has adapted to autologous SAMHD1. (A) Western blot analysis of 293T cells cotransfected with HA–AGM SAMHD1 and FLAG–Vpr from virus infecting each population. Probing for tubulin serves as a loading control. (B) Pie charts showing in which species each haplotype is most commonly found.
In summary, four AGM SAMHD1 variants demonstrate resistance to some but not all SIVagm Vprs, and this specificity is determined by only one to three amino acid changes in the SAMHD1 sequence. Two important points emerged from this analysis. First, SAMHD1 variants are always sensitive to Vpr from the population where the haplotype is most frequent (Fig. 3B). Second, every virus is able to antagonize the predominant haplotype in its population (Figs. 2A and 3A). In fact, despite the complex patterns of resistance and sensitivity observed for this interaction, each SIVagm Vpr is able to degrade all SAMHD1 variants found in the host population with two exceptions. These two exceptions are a single haplotype III sequence found in one heterozygous vervet and a single haplotype IV sequence found in one heterozygous grivet. The respective autologous SIVagm Vprs were not able to degrade the SAMHD1 proteins encoded by these haplotypes (Fig. 3). It is possible that the low frequency of these alleles may not pose a significant selective pressure on autologous virus, or that they represent rare instances of resistant hosts in a system experiencing ongoing genetic conflict between SAMHD1 and Vpr. Interestingly, we did not find evidence that SIVagm.Ver Vpr adapted to improve degradation of resistant SAMHD1 up to 1 y after experimental infection of AGMs expressing the SAMHD1 variant encoded by haplotype III (Fig. S2). This suggests that Vpr adaptation to SAMHD1 may require more than a single infection cycle to develop or possibly that SIVagm.Ver Vpr is inherently ill-equipped to acquire the ability to tolerate particular mutations in the haplotype III SAMHD1 variant. From a population-level perspective, however, SIVagm Vpr is always active against the majority of SAMHD1 variants in its host population, despite mutations that render SAMHD1 resistant to the related viruses. Therefore, SIVagm has adapted to antagonize host SAMHD1 in evolutionarily shallow time, indicating that this function provides a selective advantage to the virus in infections of extant primates.
SIVagm Vprs Are Sensitive to Independent Escape Mutations in AGM SAMHD1.
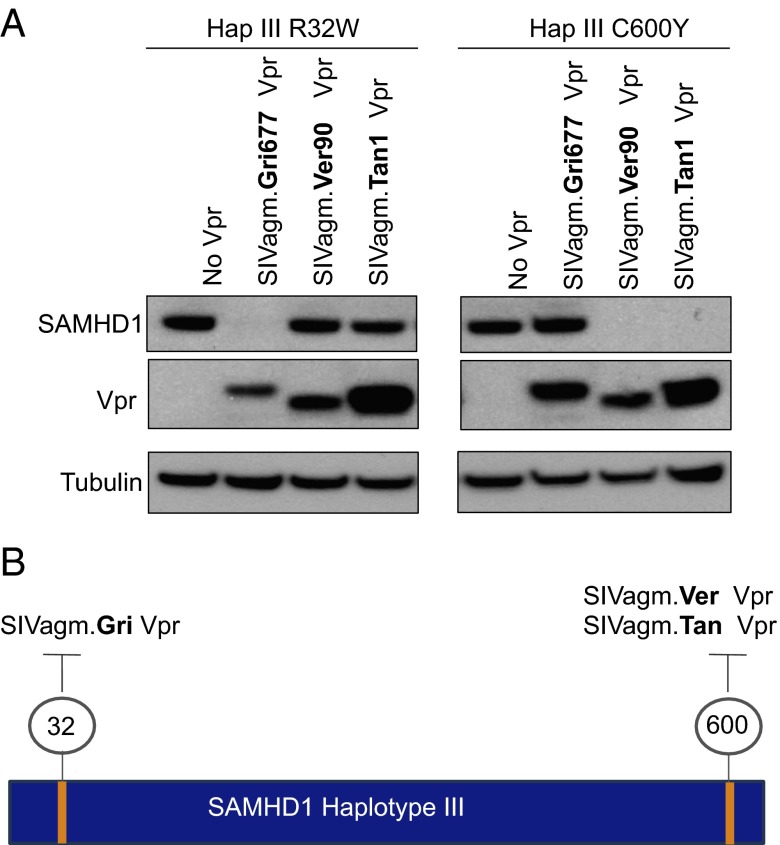
Vpx and Vpr proteins bind SAMHD1 at strikingly different interfaces, but most appear to target either the N or the C terminus of the protein (19). However, the region of SAMHD1 targeted by individual SIVagm Vpr proteins is not known, and variation at both the N and the C terminus appear to affect the outcome of degradation (Figs. 1 and 3A). We sought to map the individual amino acid changes responsible for resistance of a SAMHD1 variant to better understand how they affect interaction with Vpr. We chose the haplotype III variant because it contains mutations at both N and C termini and is resistant degradation by multiple SIVagm Vpr proteins. We hypothesized that either the derived change at the N terminus (site 32) or the derived change at the C terminus (site 600) was responsible for the resistance of the haplotype III SAMHD1 variant. As the third derived change in this haplotype is also present in haplotype II, which is sensitive to all SIVagm Vprs, it should not influence degradation, and we thus excluded it from our analysis. To determine which mutation is responsible for SAMHD1 haplotype III resistance, the residues were separately mutated to the ancestral amino acid. We first asked whether Vprs that cannot degrade the haplotype III SAMHD1 variant had activity against haplotype III R32W (Fig. 4A). Surprisingly, this change allowed for SAMHD1 to be completely degraded by SIVagm.Gri Vpr but had no effect on the activity of two other Vprs. Thus, change at site 32 has the ability to affect Vpr antagonism for one SIVagm subtype, but other features of haplotype III must affect resistance to SIVagm.Ver and SIVagm.Tan Vpr. Indeed the cysteine at site 600 prevents SIVagm.Ver and SIVagm.Tan Vprs from degrading the haplotype III variant. When this site is changed to the ancestral tyrosine, SIVagm.Ver and SIVagm.Tan Vprs are now capable of degrading SAMHD1 haplotype III (Fig. 4A). Thus, although haplotype III is resistant to all but SIVagm.Sab Vpr, separate changes at the N and C termini are independently responsible for resistance to Vpr of different viral lineages (Fig. 4B). Further, the mutations altering specificity occur at disparate regions of SAMHD1, indicating that Vpr from different subtypes SIVagm may bind SAMHD1 differently. Thus, although SIVagm vpr genes are related, there is remarkable diversity in their adaptation to SAMHD1, suggesting that these viruses have experienced selective pressure to counteract SAMHD1 restriction on an evolutionarily recent timescale.
Fig. 4.
Multiple amino acids independently alter sensitivity to SIVagm Vprs. (A) Western blot analysis of HA–SAMHD1 haplotype III point mutants R32W and C600Y. To assay for a gain of degradation ability, only SIVagm Vprs that lacked activity against wild-type SAMHD1 haplotype III were included. Probing for tubulin serves as a loading control. (B) Schematic showing how amino acids in a single SAMHD1 variant independently affect interaction with SIVagm Vprs.
Discussion
We show that SIVagm has adapted to polymorphism in the host restriction factor SAMHD1. Adaptation has occurred in the context of natural lentiviral infections and in an evolutionarily short time frame of less than 3 million years. SIVagm Vpr adaptation to these SAMHD1 variants occurred even in the presence of mutations that render the restriction factor resistant to Vpr-mediated degradation by closely related viruses. Additionally, we found that separate residues in SAMHD1 independently confer resistance to Vpr from different SIVagm lineages, implying that Vpr may use multiple target surfaces to bind SAMHD1 despite the relatedness of SIVagm vpr genes. Maintained Vpr function due to viral adaptation requires host selective pressure and indicates that SAMHD1 antagonism is a component of viral fitness in natural infections.
Polymorphism in restriction factors that are targeted by viral antagonists is a classic feature of the virus–host arms race, and within-species variation in restriction factors may be indicative of balancing selection, in which multiple alleles are maintained due to heterozygote advantage (28). AGM SAMHD1 haplotypes contain amino acid changes in regions of the gene known to be evolving under positive selection and contain almost no silent mutations (16, 17). The majority of nonsynonymous changes we identified in SAMHD1 affect the protein’s degradation by SIVagm Vpr, suggesting that amino acid variation occurs precisely at interfaces targeted by the viral antagonist. Such variation may have provided a historical selective advantage to hosts expressing resistant SAMHD1. The existence of minor alleles (1 of ∼20 gene sequences per species sampled) that are resistant to autologous virus may indicate that this conflict is ongoing.
The recognition of SAMHD1 by Vpx and Vpr proteins from different SIV lineages has recently been shown to be evolutionarily dynamic. Some Vpx/Vpr proteins target the N terminus of SAMHD1, whereas others target the C terminus (19). These seemingly extreme binding switches can be rationalized by the head-to-tail nature of the catalytically active SAMHD1 tetramer (29), as the far ends of the protein may be adjacent in physical space. Thus, antagonism could be reestablished after host escape by a Vpx/Vpr shift to target SAMHD1 at a new, but proximate, interface (19). The previous study suggested SAMHDl antagonists depend on only one terminus of SAMHD1 for degradation, and that C-terminal binding versus N-terminal binding correlates with the two major viral lineages encoding vpx genes. However, within the evolutionarily short amount of time encompassing SIVagm strain divergence, SIVagm Vprs seem to have acquired distinct binding interfaces on SAMHD1. For example, SIVagm.Ver and SIVagm.Tan Vpr are highly sensitive to a single C-terminal mutation in SAMHD1, but tolerate extensive N-terminal variation. SIVagm.Gri Vpr is sensitive to N-terminal variation but not changes at the C terminus, and further, SIVagm.Sab Vpr is broad-acting and tolerates both N- and C-terminal variation (Fig. 3A). The speed at which such complexity in the SAMHD1–Vpr interaction developed suggests that genetic conflict is ongoing and further supports the hypothesis that SAMHD1 antagonism is actively maintained and therefore advantageous to the virus.
The successful emergence of HIV-1 and its precursor SIVcpz is puzzling considering the evidence that SAMHD1 antagonism is a valuable component of viral fitness. We offer a possible explanation by suggesting that SAMHD1 antagonism is advantageous, but the cell-type–specific nature of SAMHD1 restriction may mean selective pressure on the virus is limited to particular stages of infection—namely, early stages that rely on productive infection of myeloid cells. The lack of restriction in cycling CD4+ T cells, the primary target cells, creates a unique scenario that may provide a window of opportunity for the virus to acquire compensatory mutations in cases where SAMHD1 is not effectively antagonized. We observed that SIVagm.Ver could replicate in AGMs, albeit at modest levels, even when at least the majority of Vpr clones lacked ability to cause the degradation of host SAMHD1 up to 1 y postinfection (Fig. S2) However, we did not test minor variants or the possibility that this function was acquired by another viral protein. Additionally, if SIVagm.Ver Vpr requires a large number of changes to acquire activity against the particular SAMHD1 variant tested, a failure to witness adaptation could be a consequence of the relatively short time frame of the study. However, because the samples used for tracking Vpr evolution in vivo were taken from animals infected by IV inoculation (30), we speculate that it may have bypassed SAMHD1 restriction. In contrast, restriction by the antiviral protein APOBEC3G (apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G) is constant, and antagonism evolved in the same experimental animals tested here (31). Thus, despite our demonstration that SAMHD1 antagonism must be an important component of viral fitness, the successful emergence of SIVcpz is not implausible. Initial transmission to chimpanzees via blood-borne exposure from hunting may similarly bypass the necessity of myeloid cell infection, and subsequent low-level replication could allow time for compensatory mutations to arise. Nonetheless, we show that in naturally infected populations, there is adaptation of SIVagm to actively maintain SAMHD1 antagonism, which underscores the importance of this viral function.
Methods and Materials
Amplification and Sequencing of AGM SAMHD1.
RNA and genomic DNA samples were derived from AGM peripheral blood mononuclear cells or cell lines, and sample origins and extractions were described in ref. 31. Additional samples of AGM fibroblasts were obtained from the Systems Bio Sample Repository at the University of California, Los Angeles. RNA and genomic DNA were extracted using RNeasy mini kit (QIAGEN) and the DNeasy blood and tissue mini kit (QIAGEN). SAMHD1 was amplified from RNA using the One-Step SuperScript III RT-PCR system. For each sample, bulk PCR products were sequenced from RT-PCR amplifications using Old World monkey SAMHD1 specific primers. In the case of heterozygotes, cDNA was TA cloned using the pGEM T-Easy vector system (Promega) and individual clones were sequenced. The first and last exon of each haplotype were also amplified and sequenced from genomic DNA to ensure there were no polymorphisms at the ends of the gene.
Expression Plasmids.
Each distinct AGM SAMHD1 haplotype was cloned from cDNA and ligated into the pLPCX vector containing a C-terminal HA tag as described in ref. 16. SIVagm.Gri677, SIVagm.Sab1, SIVagm.Ver9648, and SIVagm.Tan1 were amplified and cloned from proviral plasmids and ligated into the pcDNA3.1 vector with an N-terminal 3xFLAG epitope tag as described in ref. 16. SIVagm.Tan1 Vpr contains a stop codon at position 34, which was changed to the tryptophan conserved in all other sequenced SIVagm Vprs by site-directed mutagenesis (QuikChange II, Agilent Technologies). SIVagm.Sab92018 Vpr was amplified and cloned from viral RNA provided by Christian Apetrei (University of Pittsburgh). The SIVagm.Ver90 Vpr was cloned from cDNA amplified from virus isolated from a vervet monkey in ref. 30. SIVagm.Sab D30 and SIVagm.SabD42 vpr genes were synthesized (Integrated DNA Techologies). Point mutations in SAMHD1 and Vpr were created using site-directed mutagenesis (QuikChange II, Agilent Technologies).
Degradation Assays.
The 293T cells were plated in a 12-well dish at 1.6 × 105 cells per mL and transfected the following day using TransIT-LT1 (Mirus Bio). Cells were cotransfected with 200 ng pLPCX–HA–SAMHD1 expression plasmid and between 30 and 200 ng of pCDNA3.1 3xFLAG–Vpr expression plasmid. Different amounts of Vpr plasmid were transfected to normalize for similar expression, but appropriate empty vector was added to maintain constant total DNA transfected. Cells were harvested 48 h post-transfection for Western blot analysis. Cells were lysed in radioimmunoprecipitation assay buffer (RIPA) for 15 min on ice and spun at 16,000 × g for 10 min to remove cell debris. We heated 20 µg of protein in sample buffer for 5 min and loaded them onto NuPAGE Novex 4–12% Bis⋅Tris gradient gels (Invitrogen). Epitope-tagged proteins were detected using HA-specific antibody (Babco) and anti-FLAG M2 antibody (Sigma-Aldrich). Anti-tubulin (Sigma-Aldrich) antibody was used to ensure equal loading. Horseradish peroxidase-conjugated secondary antibody (SantaCruz Biotech) was used to detect primary antibodies.
Amplification of Viral Vpr Genes.
Viral RNA isolation from plasma of the experimentally infected sabaeus monkeys was described in ref. 31. Vpr sequences were amplified using the One-Step SuperScript III RT-PCR system and the following primers: GCTATAAGGGGAGAGAGATTCGTCTT (forward) and CAAAGCTGACAGTGATAGCAACACTT (reverse). Vpr cDNA was TA cloned using the pGEM T-Easy vector system (Promega), and ∼10 individual clones were sequenced for each time point.
Supplementary Material
Acknowledgments
We thank the following investigators for providing samples used in this study: M. Muller-Trutwin (AGM PBMC), C. Apetrei (AGM PBMC and SIVagm.Sab92018), and N. Freimer and A. Jasinska (AGM DNA). We thank M. Patel, L. Etienne, P. Mitchell, and O. Fregoso for comments on the manuscript. We thank the Fred Hutchinson Cancer Research Center Genomics for sequencing efforts. We thank the National Institutes of Health (NIH) Nonhuman Primate Research Resource for providing the V038 and AG23 AGM cells and SIVagm.Gri+ plasma. African sabaeus samples used in this study were collected as a part of the Systems Biology Sample Repository funded by NIH Grants R01RR016300 and R0100010980, with the particular sabaeus samples used in this study through the Department of Parks and Management Ministry of Forestry and the Environment and Medical Research Council Unit, The Gambia. This work was supported by NIH R01 AI030927 and R01 GM110570 (to M.E.), and C.J.S was supported by a National Science Foundation Graduate Research Fellowship and an NIH Training Grant in Viral Pathogenesis (T32AI083203).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession numbers KF741041–KF741096).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316839110/-/DCSupplemental.
References
- 1.Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: Scientific and public health implications. Science. 2000;287(5453):607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- 2.VandeWoude S, Apetrei C. Going wild: Lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev. 2006;19(4):728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emerman M, Malik HS. Paleovirology—Modern consequences of ancient viruses. PLoS Biol. 2010;8(2):e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duggal NK, Emerman M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 2012;12(10):687–695. doi: 10.1038/nri3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daugherty MD, Malik HS. Rules of engagement: Molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 6.Johnson WE. Rapid adversarial co-evolution of viruses and cellular restriction factors. Curr Top Microbiol Immunol. 2013;371:123–151. doi: 10.1007/978-3-642-37765-5_5. [DOI] [PubMed] [Google Scholar]
- 7.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 8.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldauf HM, et al. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med. 2012;18(11):1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cribier A, Descours B, Valadão AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3(4):1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 12.White TE, et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13(4):441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharova N, et al. Primate lentiviral Vpx commandeers DDB1 to counteract a macrophage restriction. PLoS Pathog. 2008;4(5):e1000057. doi: 10.1371/journal.ppat.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schüle S, et al. Restriction of HIV-1 replication in monocytes is abolished by Vpx of SIVsmmPBj. PLoS ONE. 2009;4(9):e7098. doi: 10.1371/journal.pone.0007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tristem M, Purvis A, Quicke DL. Complex evolutionary history of primate lentiviral vpr genes. Virology. 1998;240(2):232–237. doi: 10.1006/viro.1997.8929. [DOI] [PubMed] [Google Scholar]
- 16.Lim ES, et al. The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe. 2012;11(2):194–204. doi: 10.1016/j.chom.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laguette N, et al. Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe. 2012;11(2):205–217. doi: 10.1016/j.chom.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Etienne L, Hahn BH, Sharp PM, Matsen FA, Emerman M. Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe. 2013;14(1):85–92. doi: 10.1016/j.chom.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fregoso OI, et al. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog. 2013;9(7):e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirsch VM, et al. Vpx is required for dissemination and pathogenesis of SIV(SM) PBj: Evidence of macrophage-dependent viral amplification. Nat Med. 1998;4(12):1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belshan M, et al. Vpx is critical for SIVmne infection of pigtail macaques. Retrovirology. 2012;9:32. doi: 10.1186/1742-4690-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbs JS, et al. Progression to AIDS in the absence of a gene for vpr or vpx. J Virol. 1995;69(4):2378–2383. doi: 10.1128/jvi.69.4.2378-2383.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolfhiem J. Primates of the World: Distribution, Abundance, and Conservation. Seattle: University of Washington Press; 1998. [Google Scholar]
- 24.Perelman P, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7(3):e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allan JS, et al. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65(6):2816–2828. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wertheim JO, Worobey M. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog. 2007;3(7):e95. doi: 10.1371/journal.ppat.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VanderKuyl AC, Dekker JT, Goudsmit J. St Kitts green monkeys originate from West Africa: Genetic evidence from feces. Am J Primatol. 1996;40(4):361–364. doi: 10.1002/(SICI)1098-2345(1996)40:4<361::AID-AJP5>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 28.Newman RM, et al. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci USA. 2006;103(50):19134–19139. doi: 10.1073/pnas.0605838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan J, et al. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem. 2013;288(15):10406–10417. doi: 10.1074/jbc.M112.443796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein S, et al. Comparison of simian immunodeficiency virus SIVagmVer replication and CD4+ T-cell dynamics in vervet and sabaeus African green monkeys. J Virol. 2006;80(10):4868–4877. doi: 10.1128/JVI.80.10.4868-4877.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Compton AA, Hirsch VM, Emerman M. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe. 2012;11(1):91–98. doi: 10.1016/j.chom.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delport W, Poon AF, Frost SD, Kosakovsky Pond SL. Datamonkey 2010: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26(19):2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pupko T, Pe’er I, Shamir R, Graur D. A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol Biol Evol. 2000;17(6):890–896. doi: 10.1093/oxfordjournals.molbev.a026369. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.