Abstract
Severe acute respiratory syndrome (SARS) is a novel infectious disease caused by the SARS-associated coronavirus (SARS-CoV). There are four major structural proteins in the SARS-CoV, including the nucleocapsid, spike, membrane, and small envelope proteins. In this study, two sets of truncated fragments of spike protein were generated, the first were approximately 210-bp nonoverlapping fragments and the second were overlapping segments of 750 to 900 bp. From these 23 fragments, we identified a fragment of 259 amino acids (amino acids 441 to 700) that is a major immunodominant epitope. This fragment was highly expressed, and the purified fragment C could detect all 33 SARS patient serum samples tested, collected from 7 to 60 days after the onset of fever, but had no reactivity with all 66 healthy human serum samples tested. Thus, fragment C of spike protein was identified as an immunodominant antigen and could be used for serological detection of SARS-CoV infection.
Severe acute respiratory syndrome (SARS) was first reported in the Guangdong province of China in late 2002. The disease is characterized by fever, nonproductive cough, and dyspnea (15, 23, 27). The SARS-associated coronavirus (SARS-CoV), a novel CoV (order Nidovirales, family Coronaviridae, genus Coronavirus), has been determined to be the cause of SARS by laboratory investigations utilizing electron microscopy, virus-discovery microarrays containing conserved nucleotide sequences characteristic of many virus families, randomly primed reverse transcription (RT)-PCR, serological tests, and a monkey model (2, 4, 12, 18, 20, 24).
The Coronaviridae family comprises enveloped, positive-stranded RNA viruses that cause respiratory and enteric diseases in humans and animals. There are three groups of CoVs: groups 1 and 2 contain mammalian viruses and group 3 contains only avian viruses. Their genome, about 30,000 nucleotides, is the largest found in RNA viruses and encodes 23 putative proteins, including four major structural proteins: nucleocapsid (N), spike (S), membrane (M), and small envelope (E) (3, 7, 14). S is a large membrane glycoprotein and forms 180- to 190-kDa peplomers that bind to receptors on CoV-susceptible cells and induce cell fusion. Phylogenetic analysis of the genome sequence of the SARS-CoV indicated that the newly found virus is not closely related to any of the previously characterized CoVs and forms a distinct group within the genus Coronavirus (14, 17).
As the SARS epidemic spreads, rapid viral diagnosis will become increasingly critical, both for the control of the epidemic and for the management of patients. Although the real time PCR-based diagnostic test for SARS is reported to perform well for early identification of infections (sensitivity of 79% and specificity of 98%) (22), specific antibody or antigen detection tests will be technologically simpler and less expensive; hence, they will be urgently needed in hospitals of the epidemic region.
The S, M, and N mature proteins all contribute to generating the host immune response in transmissible gastroenteritis CoV (TGEV), infectious bronchitis virus (IBV), pig respiratory CoV, and mouse hepatitis virus. However, the S protein, a projection on the viral surface, is the major neutralizing antigen of the known CoVs (1, 6, 10, 11, 19). Because of the low level of similarity (20 to 27% pairwise amino acid identity) between the predicted amino acid sequence of the S protein of SARS-CoV and other CoVs, comparison of primary amino acid sequences does not provide insight into the antigenic properties of the SARS-CoV S protein. The specific objectives of this study were, thus, to analyze the natural immune response of SARS patients to S protein and to identify the immunodominant epitopes or domains within S protein which might serve as candidate antigens for the detection of SARS-CoV infection.
MATERIALS AND METHODS
Viruses and cells.
SARS-CoV (SIN2774, GenBank accession number AY283798) was provided by the Singapore General Hospital. Spodoptera frugiperda (SF9) cells were maintained at 27°C in SFM-900 II medium. Infection of the cells with recombinant viruses and plaque titration of virus stocks were performed according to standard protocols (Invitrogen, Carlsbad, Calif.).
Sera.
The source and nature of human serum samples used in this study are listed in Table 1. Sera of IBV-infected chicken and TGEV-infected swine were developed in this study according to the methods described previously (28).
TABLE 1.
Nature and source of sera used in the immunoblot assays
| Serum group | No. of samples | Origin of serum samples (n) |
|---|---|---|
| Confirmed SARS patient serum (collected 7-60 days after onset of fever) | 6 | National Environmental Agency, Singapore (1) |
| Center for Disease Control, Guangzhou, China (5) | ||
| 27 | Singapore General Hospital (12) | |
| Tan Tock Seng Hospital, Singapore (15) | ||
| Healthy human serum | 66 | Singapore General Hospital (10) |
| Tan Tock Seng Hospital, Singapore (24) | ||
| Voluntary blood donors in Temasek Life Science Laboratory (32) |
cDNA cloning.
SARS-CoV RNA was extracted from the tissue culture supernatant of SARS-CoV-infected Vero cells by using Trizol (Invitrogen). cDNA was synthesized from the SARS-CoV RNA by using Superscript II RNase H− reverse transcriptase (Invitrogen) and a primer specific for the s gene of SARS-CoV representing nucleotide positions 3741 to 3768 (downstream primer 5′-TTATGTGTAATGTAATTTGACACCCTTG-3′). The RT reaction was carried out for 1 h at 40°C in the presence of 1 mM deoxynucleoside triphosphate mix and 10 mM dithiothreitol in the 1× reaction buffer. The second strand of DNA was synthesized by PCR amplification with primers corresponding to different domains of the s gene. In this study, two sets of s gene fragments were amplified through the RT-PCR approach. Eighteen nonoverlapping linear fragments (s1 to s18) covering the whole s gene were designed for expression as glutathione S-transferase (GST) fusion proteins (s1 to s17 were 210 bp in length [each] and s18 was 195 bp in length); five overlapping fragments representing the whole s gene (sa, sb, sc, sd, and se) were cloned into pFastBacHta vector (Invitrogen) for expression in insect cells as His6-tagged peptides (Fig. 1 and 2 and Table 2). Correct clones were confirmed by sequencing. After immunologic screening, the antigenic sc fragment was further cloned into the pQE30 vector and highly expressed in Escherichia coli M15 (Qiagen, Hilden, Germany) (Table 2).
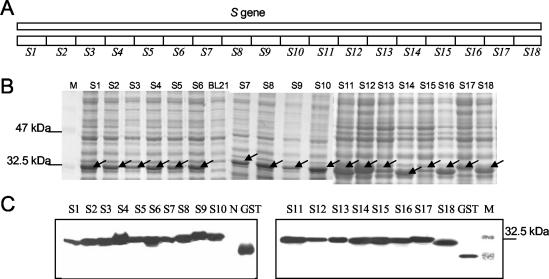
FIG. 1.
SDS-PAGE and Western blot analysis of 18 nonoverlapping fragments covering the whole S protein expressed as GST fusion proteins. (A) Schematic diagram of 18 nonoverlapping fragments within the S gene. (B) SDS-PAGE of the 18 induced GST fusion fragments. GST fusion proteins around 32 kDa were expressed in all of the induced cells. M, protein marker; BL21, control BL21 cellular extract; S1 to S18, total cellular extracts harvested 4 h post-IPTG induction of the BL21 cells transformed with recombinant pGEX constructs bearing s1 to s18, respectively. (C) Western blot analysis of the expressed GST fusion fragments with monoclonal mouse anti-GST antibody. M, protein marker; GST, control GST protein; N, control BL21 cellular extract; S1 to S18, samples as described for panel A. The arrows indicate the expressed fusion proteins.
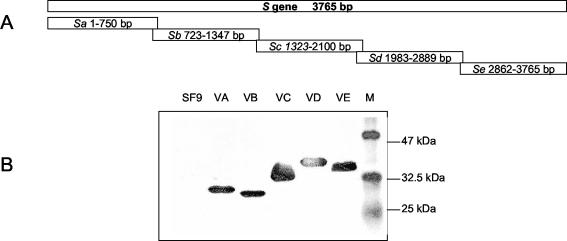
FIG. 2.
Expression of five overlapping domains within the S protein in SF9 cells. (A) Schematic diagram of five overlapping fragments within the S protein. (B) Western blot analysis of the expressed His6-tagged fragments. SF9, control SF9 cells; VA, fragment A; VB, fragment B; VC, fragment C; VD, fragment D; VE, fragment E; M, protein marker.
TABLE 2.
cDNA cloning primers
| Fragment | Primers used for cDNA amplification and cloning (sequence) | Cloning vector and cloning sites |
|---|---|---|
| Sa | HTAs+1 (CGCGGATCCGATGTTTATTTTCTTATTATTTCTT) | pFastBac Hta, BamHI, KpnI |
| S-5 (CGGGGTACCTGCAGCTGACGTGCCCCAAATGTC) | ||
| Sb | HTAs+2 (CGCGGATCCGATTTGGGGCACGTCAGCTGCAGCC) | pFastBac Hta, BamHI, KpnI |
| S-4 (CGGGGTACCCCTAAGCTTGCCATGTCTAAGATA) | ||
| Sc | HTAs+3 (CGCGGATCCGTATCTTAGACATGGCAAGCTTAGG) | pFastBac Hta, BamHI, KpnI |
| S-3 (CGGGGTACCAAAGTTAGTAGGTATAGCAAT) | ||
| Sd | HTAs+4 (CGCGGATCCGCATACAGTTTCTTTATTACGTAGT) | pFastBac Hta, BamHI, KpnI |
| S-2 (CGGGGTACCAAGGATATCATTTAGCACACTTGA) | ||
| Se | HTA+5 (CGCGGATCCGTCAAGTGTGCTAAATGATATCCTT) | pFastBac Hta, BamHI, KpnI |
| S-1 (CGGGGTACCTTATGTGTAATGTAATTTGACACC) | ||
| Sc | S+3 (CGCGGATCCTATCTTAGACATGGCAAGCTTAGG) | pQE30, BamHI, KpnI |
| S-3 (CGGGGTACCAAAGTTAGTAGGTATAGCAAT) |
Protein expression and purification in E. coli.
For expression and purification of GST fusion proteins, 18 gene fragments covering the whole s gene (s1 to s17, 210 bp; s18, 195 bp) (Fig. 1) were cloned into the pGEX4T-3 vector and transformed into E. coli BL21; after induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h, the GST fusion proteins were expressed and purified according to the standard protocol (Amersham Bioscience, Piscataway, N.J.).
For His6-tagged protein expressed from the pQE30 vector, the recombinant proteins were induced in E. coli M15 with 1 mM isopropyl-β-d-thiogalactopyranoside, after which the clarified cell extracts were mixed with Ni2+-charged resin (Ni2+-nitrilotriacetic acid) in binding buffer (100 mM NaCl, 10 mM Tris-Cl, and 8 M urea [pH 8.0]). Batch purification of His6-tagged proteins under denatured conditions was performed according to the handbook along with the resin (Qiagen).
Protein expression in baculovirus expression system.
Construction of recombinant Autographa calfornica multiple nucleopolyhedroviruses expressing His6-tagged proteins from the polyhedrin promoter was performed according to the protocol of the Bac-to-Bac system (Invitrogen), a fast approach for generating a recombinant baculovirus by site-specific transposition with Tn7 to insert foreign genes into bacmid DNA (Autographa californica multiple nucleopolyhedrovirus expression vector) propagated in E. coli. For protein expression, 106 cells were infected at a multiplicity of infection of 5 and harvested at 72 h postinfection. The cells were then washed one time with cold 1× phosphate-buffered saline and stored as a frozen pellet at −70°C.
Electrophoresis of proteins and Western blotting.
Electrophoresis of proteins and Western blot assays were performed according to standard protocol (25).
RESULTS
Antigenic analysis of 18 fragments expressed as GST fusion proteins.
To search for potential small antigenic epitopes that might serve as a basis for the localization of the antigenic domains within the s gene, 18 fragments (s1 to s18) encompassing the whole S protein were cloned into the pGEX-4T-3 vector, expressed, and fused with GST. The locations of these fragments are shown in Fig. 1A. A set of GST fusion proteins of about 32 kDa were successfully expressed in bacteria as demonstrated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1B) and Western blot analysis with monoclonal anti-GST antibody (Fig. 1C).
After purification, these fusion proteins were subjected to Western blot analysis with 10 randomly selected SARS-infected patient serum samples. Fragment S9 reacted with 6 of the 10 serum samples while other fragments either had no reaction with all the sera (s1, s2, s3, s4, s5, s15, s16, and s18) or only reacted with three samples or less (s6, s7, s8, s10, s11, s12, s14, s14, and s17). None of the truncated fragments were able to detect antibodies against SARS in serum sample no. 2, 5, and 7 (Table 3).
TABLE 3.
Reactivity of 18 GST fusion fragments against 10 convalescent SARS-positive serum samples
| Serum no. | Result for fragment:
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S15 | S16 | S17 | S18 | |
| 1 | − | − | − | − | − | + | + | + | + | + | + | − | − | + | − | − | + | − |
| 2 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 3 | − | − | − | − | − | − | + | + | + | + | + | + | + | + | − | − | − | − |
| 4 | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| 5 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 6 | − | − | − | − | − | + | + | − | + | − | − | − | − | − | − | − | − | − |
| 7 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| 8 | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| 9 | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − |
| 10 | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| Total no. of reactive sera | 2 | 3 | 3 | 6 | 2 | 2 | 1 | 1 | 2 | 1 | ||||||||
Since none of the identified antigenic fragments could detect all 10 serum samples individually or in combination, we proceeded to study the antigenicities of larger fragments within the S protein by expressing them in insect cells.
Expression of five overlapping polypeptides covering the whole S protein with a baculovirus expression system.
Five overlapping fragments (named Sa, Sb, Sc, Sd, and Se) representing the whole S gene were amplified from the SARS-CoV genome by RT-PCR as detailed in Materials and Methods. They were subsequently cloned into the pFastBacHTa vectors under the control of the polyhedral promoter and integrated into the bacmid genome by transposition after these recombinant vectors were transformed into the DH10Bac competent cell individually. The recombinant baculoviruses were obtained after transfection of these bacmids into insect SF9 cells. For protein expression, SF9 cells were infected with the five recombinant baculoviruses (named VA, VB, VC, VD, and VE) with a multiplicity of infection of 5 individually. The cells were harvested at 72 h postinfection. Total cell lysate from each infection was submitted to Western blot analysis with the anti-His5 monoclonal antibody, since each of the expressed recombinant proteins will be tagged with a His6 epitope at the N terminus. Five proteins representing the five overlapping domains (A, B, C, D, and E) of S protein at the expected sizes were successfully expressed and detected in the immunoblot assay (Fig. 2).
Identification of antigenic domains within S protein.
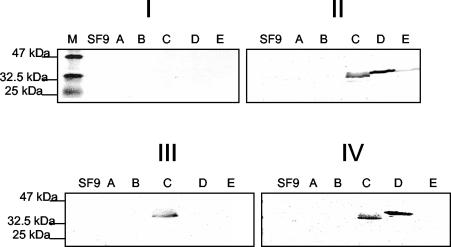
Immunoblot analysis was used to identify the antigenic domains within S protein. We first tested the reactivity of 10 randomly selected healthy human serum samples with the total cellular extracts described above, and we found no observed reactivity between five domains with the serum, as shown in Fig. 3I. Then the randomly selected 10 SARS-infected patient serum samples described above were utilized to screen the antigenic domains within S protein. None of the 10 serum samples reacted with domains A and B (Table 4 and Fig. 3II, III, and IV, lanes A and B); All of the 10 serum samples reacted with domain C (Table 4 and Fig. 3II, III, and IV, lane C) while 6 of the 10 samples reacted with domain D (Table 4 and Fig. 3IV, lane D) and only one sample reacted with domain E (Table 4 and Fig. 3II, III, and IV, lane E).
FIG. 3.
Representative immunoblot assays with sera from the 10 randomly selected SARS patients. Total cellular extracts of fragments A to E were subjected to SDS-12% PAGE and immunoblot analysis. For all of the immunoblot assays, sera were used at 1:100 and peroxidase-conjugated goat anti-total Ig polyclonal sera were used at 1:1,000. M, protein marker; SF9, cellular extracts from null-infected SF9 cells; A to E, cellular extracts from SF9 cells infected with VA, VB, VC, VD, and VE, respectively. (I) Immunoblot with one healthy human serum sample as a control. (II) Immunoblot with serum sample no. 1 that reacted with domains C, D, and E. (III) Immunoblot with serum sample no. 5 that recognized only the C domain of S protein. (IV) Immunoblot with serum sample no. 8 that recognized both the C and D domains.
TABLE 4.
Reactivities of 10 SARS patient serum samples with fragments of S protein expressed from insect cells
| Serum fragment | Normal serum result | Result for sample no.:
|
No. of reactive sera | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |||
| A | − | − | − | − | − | − | − | − | − | − | − | 0 |
| B | − | − | − | − | − | − | − | − | − | − | − | 0 |
| C | − | + | + | + | + | + | + | + | + | + | + | 10 |
| D | − | + | − | + | + | − | + | − | + | − | + | 6 |
| E | − | + | − | − | − | − | − | − | − | − | − | 1 |
Since only domain C could be identified by all 10 SARS-positive sera in the immunoblot screening, domain C was identified as a major immunodominant domain within S protein.
Expression, purification, and immunoblot analysis of the C fragment in a bacterial system.
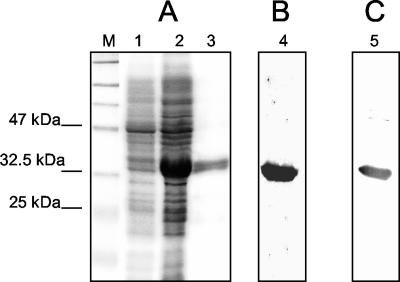
To further characterize humoral immune response to the C domain, we proceeded to express it in a bacterial system, since it could produce the antigen much more efficiently. The Sc fragment encoding the C domain was cloned into the pQE30 expression vector and transformed into E. coli strain M15. The N terminal His6-tagged C domain was highly expressed and purified according to the standard protocol from Qiagen (Fig. 4A). The protein purification method was efficient, as about 1 mg of purified protein was obtained from approximately 100 ml of bacterial culture.
FIG. 4.
Expression, purification, and immunoblot analysis of the His6-tagged C domain from E. coli strain M15. (A) SDS-PAGE of samples eluted from affinity chromatography purification. M, protein marker; lane 1, total cellular extract before IPTG induction; lane 2, total cellular extract 4 h post-IPTG induction; lane 3, eluted protein sample from affinity chromatography purification. (B) Immunoblot of fragment C with the monoclonal anti-His5 antibody. Lane 4, purified protein sample. (C) Immunoblot of purified fragment C with a pooled serum sample from the 10 positive serum samples. Lane 5, purified protein sample as described for lane 3.
As expected, Western blot analysis demonstrated that the purified protein reacted strongly with the anti-His5 monoclonal antibody (Fig. 4B). Furthermore, the pooled serum sample from the 10 SARS-positive serum samples described above was shown to react strongly with the purified protein (Fig. 4C).
To further study humoral immune response to the C domain of S protein, another 56 healthy human serum samples and 23 SARS-infected patient serum samples were tested against the bacterially expressed C domain. The purified 32-kDa antigenic domain C was recognized by all of the confirmed SARS-infected patient serum samples (n = 33), and no antigenic reactivity could be detected when healthy human serum samples (n = 66) were tested (data not shown).
From phylogenic analysis, the SARS-CoV S protein appears to be related to the avian, dog, cat, pig, and rat CoV S proteins but does not show a closer relationship to human CoV S protein (16, 23). Western blot analysis was performed to test whether the purified antigen could react with sera from chickens experimentally infected with IBV and swine experimentally infected with TGEV. No antigenic cross-reactivity could be detected (data not shown). Thus, the C domain appeared to be a very specific antigen for the SARS-CoV.
DISCUSSION
The pathophysiology of the CoV diseases of humans and animals has been studied extensively; there is still a lack of information on the recent outbreak, particularly regarding the development of humoral immune response. In general, each CoV causes disease in only one animal species. In immunocompetent hosts, infection elicits neutralizing antibodies and cell-mediated immune responses that kill infected cells (8, 9). Sequence analysis reveals that the SARS-CoV has all the characteristics of a CoV but is sufficiently different from all previously known CoVs to represent a new CoV group (18, 24). The SARS-CoV genome contains five major open reading frames that encode the replicase protein and four major structural proteins: S, E, M, and N. Among these structural proteins of CoVs, S protein is the key protein that allows the virus to dock with cells and initiate infection (5). Previous reports show that the S protein has been determined to be the major neutralizing antigen for other known CoVs (1, 6). S protein also was reported to be a factor of virulence in CoVs. Evidence comes mainly from comparative studies of different naturally occurring mouse hepatitis virus strains in which the alterations in virus virulence were most closely associated with differences in the S gene (13, 21, 26). Besides that, the unique projection of S protein from E protein is the earliest recognized antigenic epitope by the host immune system compared to other structural proteins (1, 6, 10, 26).
To identify the immunogenic sites and putative locations of the major immunodominant epitope of S protein, we first cloned the 18 truncated nonoverlapping fragments of S protein and expressed them in an E. coli system. Immunoblot assays, with 10 SARS-infected patient serum samples and 10 healthy human serum samples randomly chosen from the panel of screening sera (33 SARS-infected patient serum samples and 66 healthy human serum samples), to screen the reactivity of these fragments showed that none of the 18 fragments, either individually or in combination, could recognize the 10 SARS-infected patient serum samples. These small nonoverlapping fragments lack sensitivity because of a lack of antigenicity. They were unable to detect the SARS antibodies, as the immunogenic sites are composed of discontinuous epitopes.
In parallel, to identify potential candidate antigens for use as subunit vaccines against the SARS-CoV, five larger overlapping fragments of S protein were cloned, expressed in the baculovirus system, and reacted with the same randomly selected SARS-infected patient serum samples and healthy patient serum samples. Domain C was the only fragment reacted with all the 10 SARS infected patient serum samples. Thus, domain C contains major immunogenic sites of the SARS-CoV (Table 4 and Fig. 4). In this study, domains A and B, both originating from the N-terminal region of the S protein, showed no reactivity with the SARS patient serum samples. This correlated with the findings of the lack of antigenicity of the truncated fragments S1 to S5 (A and B domains). The surface probability test with the S protein sequences showed that domains A and B have a low surface probability, indicating that these two regions might be buried in the three dimensional structure of S protein. This also applies to domain E, which correlated with the truncated fragments S15 to S18. In contrast, domains C and D have a high surface probability, which may contribute to their antigenicity.
To further characterize domain C, which contains the immunogenic sites of the SARS-CoV, this fragment C was subcloned into an E. coli expression system to increase the yield of expression. Purified fragment C was used to develop Western blots, and the fragment elicited a strong immune response with all 33 SARS-infected patient serum samples, including the 10 randomly selected in the previous studies and 23 additional SARS-infected patient serum samples, but showed no reactivity towards all 66 healthy human serum samples.
The 33 SARS-infected patient serum samples used in this study were collected from patients 7 to 60 days after the onset of fever. We have used the C fragment of the S protein to test the immunoglobulin G (IgG) and IgM levels in the SARS-infected serum samples. This result with the C fragment was compared to that obtained with the truncated N protein (unpublished data) developed by our lab. The results showed that the C fragment of the S protein was able to detect IgM SARS antibodies in two infected patient serum samples collected 9 and 11 days after the onset of fever, and these two samples were not detectable by the truncated N protein for IgM SARS antibodies. This suggests that the truncated S protein may be a potential antigen for diagnostics used in the early detection of SARS infection. This correlated with the findings that the S glycoprotein was the earliest recognized antigen by the host immune system, whereas the N protein induced antibody responses during the later stage of infection (1). It has also been reported that SARS patients first developed serum IgM specific for the SARS-CoV, although the IgM level begins to decline 3 weeks after the onset of fever (16).
However, the early detection of SARS infection by assay also depends on the nature of the assay. Following SARS infection, no humoral response to the SARS-CoV is detectable for several days. This is known as the lag period. SARS-infected patients tested negative for IgM and IgG at week 1 after the onset of symptoms (16). A more sensitive detection method, such as an enzyme-linked immunosorbent assay might need to be developed to obtain a higher detection rate in the early stage of infection. Thus, to verify the sensitivity of the C fragment of S protein in the early detection of SARS infection, more sera of SARS-CoV infection at different times during the course of infection are needed.
Further characterization was done with various sera against other animal CoVs, and no cross-reaction was detected between the C fragment of S protein and other related CoV-infected animal serum samples. Thus, our results suggest that fragment C may be a good candidate to develop a specific and effective antibody detection test for SARS-CoV infection. Today, although the SARS outbreak has now ceased, the volume of tests for SARS is likely to remain a significant portion of the diagnostic laboratory's workload, especially in the coming winter months in the northern hemisphere. It is important to have the reliable assay for the SARS-CoV ready.
In conclusion, fragment C of the S protein has been identified as a major immunodominant antigen for the SARS-CoV. This fragment C could serve as a useful tool to monitor the antibody response from suspected SARS patients, individually or in combination with other antigenic proteins from the SARS-CoV, such as the N protein.
REFERENCES
- 1.Callebaut, P., L. Enjuanes, and M. Pensaert. 1996. An adenovirus recombinant expressing the spike glycoprotein of porcine respiratory coronavirus is immunogenic in swine. J. Gen. Virol. 77:309-313. [DOI] [PubMed] [Google Scholar]
- 2.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 3.Enjuanes, L., et al. 2000. Coronaviridae, p. 845-849. In M. H. V. van Regenmortal, C. M. Fauqet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Academic Press, New York, N.Y.
- 4.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez, N., C. Carrillo, J. Salinas, F. Parra, M. V. Borca, and J. M. Escribano. 1998. Expression of immunogenic glycoprotein S polypeptides from transmissible gastroenteritis coronavirus in transgenic plants. Virology 249:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes, K. V. 2001. Coronaviridae, p. 1187-1203. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, New York, N.Y.
- 8.Holmes, K. V. 2003. SARS coronavirus: a new challenge for prevention and therapy. J. Clin. Investig. 111:1605-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmes, K. V., and L. Enjuanes. 2003. The SARS coronavirus: a postgenomic era. Science 300:1377-1378. [DOI] [PubMed] [Google Scholar]
- 10.Homberger, F. R. 1994. Nucleotide sequence comparison of the membrane protein genes of three enterotropic strains of mouse hepatitis virus. Virus Res. 31:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackwood, M. W., and D. A. Hilt. 1995. Production and immunogenicity of multiple antigenic peptide (MAP) constructs derived from the S1 glycoprotein of infectious bronchitis virus (IBV). Adv. Exp. Med. Biol. 380:213-219. [DOI] [PubMed] [Google Scholar]
- 12.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 13.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1186. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, New York, N.Y.
- 15.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 16.Li, G., X. Chen, and A. Xu. 2003. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 349:508-509. [DOI] [PubMed] [Google Scholar]
- 17.Lin, X. Q., K. L. O'Reilly, J. Storz, C. W. Purdy, and R. W. Loan. 2000. Antibody responses to respiratory coronavirus infections of cattle during shipping fever pathogenesis. Arch. Virol. 145:2335-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 19.Ndifuna, A., A. K. Waters, M. Zhou, and E. W. Collisson. 1998. Recombinant nucleocapsid protein is potentially an inexpensive, effective serodiagnostic reagent for infectious bronchitis virus. J. Virol. Methods 70:37-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon, L. L., O. K. Wong, K. H. Chan, W. Luk, K. Y. Yuen, J. S. Peiris, and Y. Guan. 2003. Rapid diagnosis of a coronavirus associated with severe acute respiratory syndrome (SARS). Clin. Chem. 49:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 24.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Sanchez, C. M., A. Izeta, J. M. Sanchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Duran, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1977-1985. [DOI] [PubMed] [Google Scholar]
- 28.Yu, L., W. Liu, W. M. Schnitzlein, D. N. Tripathy, and J. Kwang. 2001. Study of protection by recombinant fowl poxvirus expressing C-terminal nucleocapsid protein of infectious bronchitis virus against challenge. Avian Dis. 45:340-348. [PubMed] [Google Scholar]