Significance
In a few milliliters of blood from a cancer patient, one can isolate a few circulating tumor cells (CTCs). Originating from the primary tumor, CTCs seed metastases, which account for the majority of cancer-related deaths. We demonstrate the analyses of the whole genome of single CTCs, which are highly needed for personalized treatment. We discovered that copy number variations (CNVs), one of the major genomic variations, are specific to cancer types, reproducible from cell to cell, and even from patient to patient. We hypothesize that CNVs at certain genomic loci are selected for and lead to metastasis. Our work shows the prospect of noninvasive CTC-based cancer diagnostics.
Keywords: cancer diagnostics, personalized therapy
Abstract
Circulating tumor cells (CTCs) enter peripheral blood from primary tumors and seed metastases. The genome sequencing of CTCs could offer noninvasive prognosis or even diagnosis, but has been hampered by low single-cell genome coverage of scarce CTCs. Here, we report the use of the recently developed multiple annealing and looping-based amplification cycles for whole-genome amplification of single CTCs from lung cancer patients. We observed characteristic cancer-associated single-nucleotide variations and insertions/deletions in exomes of CTCs. These mutations provided information needed for individualized therapy, such as drug resistance and phenotypic transition, but were heterogeneous from cell to cell. In contrast, every CTC from an individual patient, regardless of the cancer subtypes, exhibited reproducible copy number variation (CNV) patterns, similar to those of the metastatic tumor of the same patient. Interestingly, different patients with the same lung cancer adenocarcinoma (ADC) shared similar CNV patterns in their CTCs. Even more interestingly, patients of small-cell lung cancer have CNV patterns distinctly different from those of ADC patients. Our finding suggests that CNVs at certain genomic loci are selected for the metastasis of cancer. The reproducibility of cancer-specific CNVs offers potential for CTC-based cancer diagnostics.
As a genomic disease, cancer involves a series of changes in the genome, starting from primary tumors, via circulating tumor cells (CTCs), to metastases that cause the majority of mortalities (1–3). These genomic alterations include copy number variations (CNVs), single-nucleotide variations (SNVs), and insertions/deletions (INDELs). Regardless of the concentrated efforts in the past decades, the key driving genomic alterations responsible for metastases are still elusive (1).
For noninvasive prognosis and diagnosis of cancer, it is desirable to monitor genomic alterations through the circulatory system. Genetic analyses of cell-free DNA fragments in peripheral blood have been reported (4–6) and recently extended to the whole-genome scale (7–9). However, it may be advantageous to analyze CTCs, as they represent intact functional cancer cells circulating in peripheral blood (10). Although previous studies have shown that CTC counting was able to predict progression and overall survival of cancer patients (11, 12), genomic analyses of CTCs could provide more pertinent information for personalized therapy (13). However, it is difficult to probe the genomic changes in DNA obtainable from the small number of captured CTCs. To meet this challenge, a single-cell whole-genome amplification (WGA) method, multiple annealing and looping-based amplification cycles (MALBAC) (14), has been developed to improve the amplification uniformity across the entire genome over previous methods (15, 16), allowing precise determination of CNVs and detection of SNVs with a low false-positive rate in a single cell. Here, we present genomic analyses of CTCs from 11 patients (SI Appendix, Table S1) with lung cancer, the leading cause of worldwide cancer-related deaths. CTCs were captured with the CellSearch platform using antibodies enrichment after fixation, further isolated with 94% specificity (Materials and Methods), and then subjected to WGA using MALBAC before next-generation sequencing.
Results
Single-Cell Exome Sequencing Reveals SNV/INDEL Profiles in Individual CTCs and Provides Information Needed for Personalized Therapy.
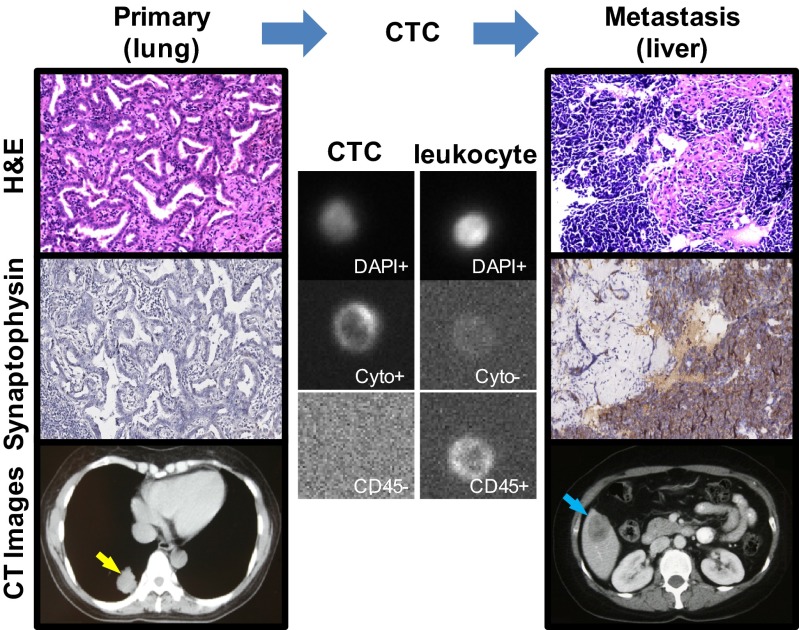
To detect SNVs/INDELs, we performed exome sequencing of 24 individual CTCs from four lung adenocarcinoma (ADC) patients (patients 1–4) and compared them with the exomes of their primary and/or metastatic tumors. Unlike the other three ADC patients, patient 1 had undergone a phenotypic transition from lung ADC to small-cell lung cancer (SCLC) in the liver, which was evidenced by H&E and immunohistochemical staining (Fig. 1).
Fig. 1.
Primary and metastatic tumors and CTC from patient 1 who experienced a phenotypic transition from ADC to SCLC. The H&E staining and immunohistochemical staining for synaptophysin (SYN) show a typical ADC in the lung (Left) and a typical SCLC in the liver (Right) (image magnification: 200x). The CT images show the preoperative primary tumor in the lower lobe of the right lung (yellow arrow) and the metastatic posttreatment tumor in the right lobe of the liver (blue arrow). In the Center, a circulating tumor cell is identified by positive staining for DAPI and cytokeratin (Cyto), and negative staining for CD45. As a control, a leukocyte is also shown (DAPI+, Cyto−, CD45+).
Bulk exome sequencing identified 54 nonsynonymous SNVs and INDELs, mutations that cause amino acid changes in proteins, in the primary and metastatic tumors of patient 1 (Fig. 2A). Single-cell sequencing of eight individual CTCs from patient 1 showed a total of 44 nonsynonymous SNVs and INDELs (Fig. 2A), each of which was called if a SNV or INDEL in a CTC was also detected in two other CTCs or in primary/metastatic tumors to eliminate false calls due to amplification errors (Materials and Methods). CTCs showed a large similarity with the metastatic but not the primary tumor in SNVs/INDELs. This difference was partially due to the low abundance of a given SNV/INDEL in the primary tumor. A Venn diagram depicts the overlap of nonsynonymous SNVs and INDELs across primary tumor, CTCs, and the metastatic tumor in patient 1 (Fig. 2B). Similar results were seen in the other ADC patients: sequencing of patient 2’s (patient 3’s) six (five) CTCs identified 106 (145) out of 146 (170) nonsynonymous SNVs/INDELs in the metastatic tumor. Although a few key SNVs/INDELs were enriched in CTCs, other point mutations (Fig. 2A and SI Appendix, Fig. S1) are heterogeneous from cell to cell, as previously reported for solid tumors (16, 17).
Fig. 2.
Detection of somatic mutations (SNVs and INDELs) in CTCs and primary/metastatic tumors of patient 1. (A) Nonsynonymous heterozygous (hetero.) and homozygous (homo.) mutations in the lung primary (Pri.) tumor, eight CTCs, and the liver metastatic (Meta.) tumor. The blank region represents no sequence coverage. The mutated genes are listed in the right column. (B) Venn diagram of the nonsynonymous SNVs and INDELs among the lung primary tumor, CTCs, and the liver metastatic tumor of patient 1. (C) Venn diagram of the nonsynonymous SNVs and INDELs that are reported in the COSMIC database.
We next focused on those SNVs/INDELs reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) (18), which may play critical roles in cancer. In patient 1, all COSMIC mutations that appeared in the primary and/or metastatic tumors have been detected in CTCs, as shown in the Venn diagram (Fig. 2C). Among these mutations, one INDEL in the epidermal growth factor receptor (EGFR) gene (p.Lys746_Ala750del), which is a target for tyrosine kinase inhibitors (TKIs) (19), was identified in the primary and metastatic tumors as well as in CTCs. This illustrates the utility of CTC sequencing for identifying therapeutic target for personalized treatment.
The other three COSMIC mutations in the phosphatidylinositol 3-kinase catalytic subunit α (PIK3CA) (p.Glu545Lys), tumor protein 53 (TP53) (p.Thr155Ile), and retinoblastoma (RB1) (p.Arg320*) genes were only shared between the liver metastatic tumor and CTCs. The fact that these three mutations were not detected in the primary tumor was due to their low abundance. Indeed, the use of PCR amplification together with deep sequencing revealed these mutations in the primary tumor (SI Appendix, Fig. S2). Despite its low abundance in the primary tumor, the PIK3CA mutation was detected in seven of eight CTCs from patient 1. The PIK3CA mutation has been implicated in drug resistance of erlotinib (20). Consistently, patient 1 underwent rapid disease progression in the liver metastasis after 1 mo of EGFR TKI treatment with erlotinib.
Concurrent mutations in RB1 and TP53 were commonly found in SCLC (21) and have been reported to be able to efficiently transform other cells to SCLC (22). We observed RB1 and TP53 mutations in most of the CTCs in this lung ADC patient. Subsequent needle biopsy of the liver confirmed this transition (Fig. 1). A standard SCLC treatment with etoposide plus cisplatin for six cycles led to a dramatic clinical response. This demonstrated again that CTC sequencing might provide an a priori indication of phenotypic transition and guide the selection of therapeutic regimens.
CNV Patterns of Individual CTCs in Each Patient Are Highly Reproducible.
Capitalizing on MALBAC’s ability to precisely determine a single cell’s CNVs (14), another major form of genetic variations in cancers (23–26), we now examine whether CNVs also exhibit heterogeneity from cell to cell. We performed whole-genome sequencing (∼0.1× sequencing depth) of CTCs from patient 1. Fig. 3A shows the CNV patterns (segmented with a hidden Markov model) across the genome for the eight CTCs of patient 1, along with the bulk sequencing of her primary and metastatic tumors. As a control experiment, the CNV patterns in the single leukocyte were consistent with that of the blood bulk DNA, confirming the uniformity of single-cell WGA with MALBAC and excluding the possibility that the amplification procedure produced artifacts in CNVs. The CNV patterns in each CTC were distinctly different from the normal single leukocyte as shown in SI Appendix, Fig. S3. Surprisingly, we found that all CTCs of patient 1 exhibited reproducible gain and loss CNV patterns (an average of 83% of the gain and loss regions was shared between any two CTCs).
Fig. 3.
CNVs in CTCs from six patients with ADC and one patient with a mixture of ADC and SCLC. (A) All eight CTCs in patient 1 with reproducible CNV patterns. The copy numbers were segmented (blue and red lines) with HMM. (B) CNV patterns of CTCs from six ADC patients (patients 1–6) and a patient with a mixture of ADC and SCLC (patient 7). Patient 1 experienced a phenotypic transition from ADC in the lung to SCLC in the liver. Patient 7 was diagnosed as a mixture of ADC and SCLC in the lung. In each patient, sequencing data from all CTCs were combined for CNV analyses. (C) Clustering analyses of CTCs based on the CNVs. CTCs from patients 1 and 7 were separated from CTCs from other five ADC patients according to the analyses.
Such reproducible global CNV patterns were hidden in bulk-sequencing analyses of tumors, and only made visible by the high accuracy afforded by MALBAC. The gain and loss regions accounted for ∼33% and ∼8% of the entire genome of CTCs from patient 1, respectively. The CNV patterns of CTCs in patient 1 resembled more closely those of the metastatic tumor than those of the primary tumor, raising the possibility that our captured CTCs came from the metastatic tumor. However, we observed that the EGFR mutation was homozygous in bulk sequencing of the liver metastatic tumor, but was 50% heterogeneous in the eight CTCs. A mixture of wild-type and homozygous mutant genotypes led to an appearance of heterozygous EGFR mutations in the primary tumor. The EGFR mutation frequency in CTCs is close to that in the primary tumor, suggesting that a large proportion of CTCs originated from the primary tumor and were an intermediary for metastasis. Furthermore, both primary and metastatic tumors had more than 70% of tumor cell content (SI Appendix, Table S1), which excluded the possibility of low tumor content in complicating our observation.
Our finding suggests that during the metastatic process gain and loss of copy numbers at certain chromosome regions are selected for cancer cells to enter or survive in the circulatory system, becoming CTCs. The reproducible CNV patterns might come from the possibility that CTCs originated from one subclone in the primary tumor or due to the CTC selection criterion. This is unlikely given the heterogeneity of SNVs in single CTCs.
We examined the reproducibility of the CNV patterns among five other patients (patients 2–6) with ADC and one patient (patient 7) with a mixture of ADC and SCLC in the lung. Again, individual CTCs from the same patient showed reproducible CNV patterns (SI Appendix, Figs. S4–S9). These commonly occurring CNVs were discernible in bulk sequencing of the matched metastases (SI Appendix, Figs. S4 and S5). The mean CNVs (average over all CTCs in each patient) of patients 2–6 were plotted and segmented in Fig. 3B.
CTC’s CNV Patterns of Different Patients of the Same ADC Are Similar.
Patients 2–6 with ADC exhibited almost identical global CNV patterns; an average of 78% of the gain and loss regions was shared between any two of these patients. Given the different clinical characteristics of these patients, such as different sexes and ages, the observation of five ADC patients with almost identical global CNV patterns is striking, providing not only the basis for potential diagnosis of ADC via CTCs, but also clues for metastasis.
We list the common copy number gain (in >16 CTCs) and loss (in >7 CTCs) regions, together with some important cancer-related genes, of the five ADC patients’ CTCs in SI Appendix, Table S2. Most of these regions were consistent with a previous statistical analysis of CNVs on 528 snap-frozen lung adenocarcinoma resection specimens (25). The statistical significance of the CNVs in 19 CTCs from these five patients is illustrated in SI Appendix, Fig. S10. Although CNVs spanned a large portion of the chromosome arm, a few genes in the common CNV regions have crucial roles in cancer. For example, the gain region in chromosome 8q contains the c-Myc gene, which is associated with cell proliferation and differentiation. Likewise, all five ADC patients showed significant gain in chromosome 5p, which contains the telomerase reverse transcriptase (TERT) gene that prevents the chromosome ends from degradation. We confirmed the amplification of the c-Myc and TERT genes in a CTC but not in a normal leukocyte with digital PCR (SI Appendix, Fig. S11). Four particular chromosomal regions, 3q29, 17q22, 17q25.3, and 20p13, have significant gain in all 19 CTCs of ADC patients 2–6 we sequenced. None of the genes in these regions are listed in the Cancer Gene Census (27). The functional roles of those genes in metastasis of adenocarcinoma warrant further investigation.
CTC’s CNV Patterns of Patients with Different Cancer Subtypes Are Dissimilar.
Patients 1 and 7 are different from patients 2–6 with ADC in that patient 1 underwent an ADC-to-SCLC transition, whereas patient 7 has a mixture of ADC and SCLC in the lung. Interestingly, the CNV patterns of patients 1 and 7 were dissimilar to patients 2–6 with ADC. Such dissimilarity is further proven by hierarchical clustering analyses of their CNV patterns (Fig. 3C), confirming the distinction among patients 1 and 7 and the other five ADC patients 2–6. In particular, a significant response following standard SCLC treatment in patient 1 was observed, indicating the potential for a therapeutic stratification of ADC patients based on their CTCs’ CNV patterns.
Fig. 4A shows the CNV patterns of CTCs from patients 8–11 with SCLC without phenotypic transitions, yielding further evidence for different cancer subtypes exhibiting distinct CNV patterns. The SCLC patients showed global CNV patterns different from ADC patients 2–6. An average of 42% of the gain and loss regions was shared between any two patients. Interpatient heterogeneity is generally associated with aggressive cancer subtypes, such as is the case for SCLC, which is prone to metastasis and has poor prognosis (21). Nevertheless, similarity still existed among all ADC and SCLC patients. For example, a common copy number gain spanning chromosome 6p, the human leukocyte antigen (HLA) region, was seen and has been associated with the tumor progression (28). Regardless of the heterogeneity among SCLC patients, it is important to note that the CNV patterns of individual CTCs from the same patient were still reproducible (SI Appendix, Figs. S12–S15). The fact that CNV patterns of ADC and SCLC were different implied these patterns were cancer subtype-specific, which is of diagnostic significance.
Fig. 4.
CNVs and SNVs/INDELs of SCLC. (A) Four SCLC patients (patients 8–11) with heterogeneities in their CNV patterns. In each patient, sequencing data from all CTCs were combined for CNV analyses. (B) Fraction of mutation frequency of 152 SNVs/INDELs across CTCs before (blue), and during the first-line (red) and second-line (green) chemotherapy (chemo.) in patient 8. (C) CTCs from patient 8 with constant CNVs at different therapeutic stages. Four CTCs from each stage were shown in this plot (see SI Appendix, Figs. S12 and S17, for CNVs of all CTCs from this patient).
The SNVs/INDELs in CTCs Change During Treatment, Whereas the CNV Patterns Remain Constant.
Important to predict disease progression during drug treatment is the ability to monitor the genomic changes of CTCs over time, given that repeat biopsy is not desirable. We performed sequential CTC isolation and sequencing on one SCLC patient (patient 8) at three time points: before chemotherapy, after partial response to first-line chemotherapy with etoposide plus platinum, and after disease progression to second-line chemotherapy with topotecan. Tumor responses were evaluated according to the RECIST1.1 criteria. Mutation frequencies of SNVs/INDELs across CTCs clearly varied with time (Fig. 4B and SI Appendix, Fig. S16). For the 23 genes with significantly increased mutation frequencies in response to chemotherapy, we performed a gene ontology (GO) analysis using GeneCodis 3.0 (29), which revealed that 6 genes (ALPK2, KIF16B, TP53, MYH7, TTLL2, PAK2) were enriched in the GO category of “ATP binding” (GO: 0005524) and perhaps responsible for the disease progression in this patient. Interestingly, the CTCs’ CNV patterns, at a whole-genome scale, do not change at different therapeutic stages (Fig. 4C), indicating that the reproducible CNV patterns observed were not affected by drug treatment. This further supports that CNVs at certain chromosomal loci are not only selected for at the onset of metastasis but also remain constant throughout.
Discussion
Monitoring the emergence and alteration of SNVs/INDELs is essential in the process of targeted therapy. Consistent with previous work (30), our present work showed that the genomic profiles in the metastatic tumors are distinct from those of the primary tumor. Genomic analyses of multiple metastatic sites could provide important information related to treatment (31). However, it is difficult in clinical practice for most patients to undertake repeat biopsies at multiple tumor regions. Although the SNVs/INDELs in CTCs are heterogeneous from cell to cell (32), genomic analyses of a few CTCs can provide the overall SNV/INDEL profiles that are present in the metastatic tumor tissues during the treatment. Noticeably, some important tumor-related genes, including those involved in drug resistance and phenotypic transitions, were frequently mutated in CTCs. Such enrichment may represent a selective advantage of CTCs to escape targeted therapy.
Given the above observations of highly reproducible CNVs in the CTCs of individual (and even different) patients, we hypothesize that copy number changes are the key events of metastasis: in the evolution of cancer, gain and loss in copy numbers of certain chromosome regions are selected for metastases. The CNVs at a certain combination of gene loci (such as c-Myc, TERT, HLA) could alter the gene expression of different pathways, conferring a selective advantage for metastasis. With regard to the underlying selection mechanism, one possibility is that cancer cells in the primary tumor with certain CNVs affecting a large scale of genes could invade the surrounding tissues and intravasate more efficiently. It is also possible that this selection happens in the circulatory system where CTCs survive the immune surveillance. For example, the common gain region in chromosome 6p could elevate the expression levels of HLA proteins and inhibit natural killer (NK) cells (33). This gain could be a critical requirement to become a CTC, given the abundance of NK cells in blood compared with their scarcity in the primary tumor.
A broad survey of CNV patterns in CTCs of different cancers is underway to examine whether the same degree of reproducibility also occurs in other types of cancers. Although the underlying molecular mechanism is yet to be illustrated, the observation of reproducible genomic alterations is highly instructive for the understanding of metastasis or even genesis of cancer. The reproducible CNV patterns that are characteristic of different cancers might allow noninvasive cancer diagnostics and classification through sequencing of CTCs.
Materials and Methods
This study was approved by the institutional ethics committee at Peking University Cancer Hospital and Institute and the Committee on the Use of Human Subjects in Research at Harvard University. Written informed consent was obtained from all patients. A total of 16 patients were enrolled. Among them, 11 patients were chosen for sequencing study. A summary of patient information is listed in SI Appendix, Table S1. The patient recruitment and clinical information are described in SI Appendix.
CTC Capture and Isolation.
Circulating tumor cells from 7.5 mL of blood sample were captured with the CellSearch Epithelial Cell Kit (Veridex, LLC) using magnetic bead conjugated to anti-epithelial cell adhesion molecule (EpCAM) antibodies. The captured CTCs were stained with 4′,6-diamidino-2-phenylindole (DAPI), anti–cytokeratin-phycoerythrin and anti–CD45-allophycocyanin antibodies to distinguish cancer cells from carryover leukocytes. We then isolated individual CTC (DAPI+, anti-cytokeratin+, anti-CD45−) and leukocyte (DAPI+, anti-cytokeratin−, anti-CD45+) under fluorescence microscope by separating individual cells manually through micropipetting. An additional FITC channel was added to ensure that fluorescence signal in the anti–cytokeratin-phycoerythrin channel was not due to other fluorophores. Approximately 30% of CTCs (DAPI+, anti-cytokeratin+, anti-CD45−) originally captured with CellSearch were further excluded as potential false positives by this procedure. Each selected CTC was washed multiple times in droplets of UV-exposed water to minimize DNA contamination. A total number of 72 CTCs were sequenced. Four CTCs were later determined to be normal leukocytes based on their CNV and SNV/INDEL profiles and were excluded from further analyses, which gave a specificity of 94%.
Whole-Genome Amplification.
The DNA in a single CTC was amplified following the steps in ref. 14. Quantitative PCR (qPCR) was performed in eight randomly selected loci to check for the genomic integrity of the amplification product. DNAs with seven out of eight loci amplified with reasonable Ct number in qPCR can be used for further sequencing study. Seventy percent of CTCs have passed this filter.
Exome Library Preparation and Sequencing.
The coding exons plus UTRs were captured with SureSelect All Exon V4 (Agilent Technologies) according to ref. 34 with a few modifications. A total of 150 ng to 1 µg of DNA extracted from tumor tissues or amplified from a CTC by MALBAC was sheared into fragments around 175 bp using the Covaris system (Covaris). The sheared DNA was purified with Agencourt AMPure XP SPRI beads (Beckman Coulter). The DNA was blunted with 5′-phosphorylated ends using the NEB Quick Blunting Kit and ligated to truncated PE P7 adaptors and barcoded P5 adaptors using NEBNext Quick Ligation Module. After cleanup with Agencourt AMPure XP SPRI beads and nick fill-in with Bst polymerase Large Fragment (New England Biolabs), the DNA fragments with adaptors were enriched by PCR. A total amount of 500 ng of DNA pooled from four barcoded libraries was used for hybridization and posthybridization amplification following the manufacturer’s protocol (SureSelectXT Target Enrichment System for Illumina Paired-End Sequencing Library, version 1.3.1, February 2012, pp. 37–60). The posthybridization amplification product was quality checked and sequenced with Illumina HiSEq. 2000/2500 2 × 100-bp paired-end (PE) reads. The coverage information of exome sequencing is shown in SI Appendix, Table S3.
Whole-Genome Library Preparation and Sequencing.
Libraries for whole-genome sequencing were prepared from the adaptor-ligated DNA before the pooling step in exome library preparation. Enrichment PCR was performed on an aliquot of adaptor-ligated DNA to complete the adaptor for Illumina PE sequencing. The PCR product was quality checked and sequenced with Illumina HiSEq. 2000/2500 2 × 100-bp PE reads or MiSEq. 300 2 × 150-bp PE reads at ∼0.1× sequencing depth. The cost for this low sequence depth is affordable for clinical study.
Exome Sequencing Data Analysis for SNVs/INDELs.
Sequencing reads were aligned to the University of California, Santa Cruz, human reference genome (hg19) using the Burrows–Wheeler Aligner (35). The aligned reads were sorted and merged with Samtools 0.1.18 (36). The INDEL realignment was done with the Genome Analysis Toolkit (GATK 2.1–8) (37), and mate pair fix and duplicate removal were done with Picard-tools 1.76 (http://Picard.Sourceforge.net). The base quality was recalibrated and population variations were detected by GATK using dbSNP 135 (www.ncbi.nlm.nih.gov/projects/SNP/). The functional effect of variants was annotated with SNPEFF 3.0 (38). Variations that presented in dbSNP 135 and the National Heart, Lung, and Blood Institute Exome Sequencing Project but not in COSMIC v61 (18) were filtered out. SNVs/INDELs were called for variations that presented in tumor tissue specimens or more than two CTCs but not in the matched blood genomic DNA or single leukocyte. A detailed list of the SNVs/INDELs for all four patients is shown in SI Appendix, Table S4, some of which were validated by Sanger sequencing.
Copy Number Determination from Whole-Genome Sequencing Data.
The copy number variant regions were identified according to the procedure in ref. 14. See SI Appendix for the procedure.
Significance Analysis of Gain and Loss Regions in CTCs of ADC Patients.
Significance analysis of gain and loss regions in all 19 CTCs of ADC patients (patients 2–6) followed GISTIC algorithm (23, 39), which is originally intended for CNV data from multiple distinct individuals. The sequence data are separated into two sets to calculate P values for gain and loss regions, respectively. All of the bins that have CNV < 2 were reassigned as 2 for P values calculation in gain regions and those that have CNV > 2 were reassigned as 2 for P values calculation in loss regions. A value of 0.8 was set for CNV = 0. Then we replaced the copy numbers with an amplitude (a = log2CNV − log22). In each dataset, we obtained a G score for every bin in the chromosome considering both amplitude and its frequency across all 19 CTCs [G = a × freq.]. A null distribution for G score was determined by permuting the data within each CTC. Compared with the null distribution, we obtained a P value for each bin in the chromosome. After false-discovery rate P value adjustment (40), a q value for each bin was assigned. A significant level of 10−4.76 for gains and 10−4.18 for losses are given according to the q values of gains and losses in eight normal leukocytes; no gain or loss regions have been observed in the normal leukocytes based on those significant levels.
Validation of SNVs and CNVs.
Digital PCR, Sanger sequencing, and deep sequencing were used to validate SNVs and CNVs. The detailed validation procedure was described in SI Appendix.
Clustering Analysis.
Clustering analysis based on the whole-genome CNVs was applied to distinguish CTCs from different patients using algorithms implemented in R package (www.R-project.org). First, we normalized sequence reads of each CTC with sequence reads from all normal leukocytes to remove whole-genome amplification bias. We then determined the copy number sequence (a1, a2, a3,…; b1, b2, b3,…;…) at a bin size of 500,000 along the genome by comparing the normalized reads with reads from diploid regions found by hidden Markov model (HMM). The Euclidean distance between pairs of copy number sequence of CTCs was calculated by the following:
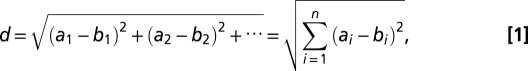 |
where a and b represent different CTCs. i is the index for the bins.
Based on the above Euclidean distances, Ward’s linkage criterion (41, 42) was applied to create clusters of CTCs (Fig. 3C).
Supplementary Material
Acknowledgments
We thank all of the patients for their participation in this study. We thank Amy Ly at Massachusetts General Hospital for assistance; Dengfeng Cao at the Department of Pathology at Peking University Cancer Hospital/Institute for help in the histological diagnosis; Fuchou Tang, Yanyi Huang, and Hao Ge at Biodynamic Optical Imaging Center (BIOPIC), Rui Xing at Beijing Cancer Hospital, and Hong Wu at University of California, Los Angeles, for useful discussions; Yun Zhang and Wenping Ma at BIOPIC for assistance with sequencing; Yun Zhu and Yingying Pu for assistance with TA clone; and Ruoyan Li, Jing Sun, and Yang Xu at BIOPIC for assistance with data processing. This work was supported by Peking University (PKU) 985 Special Funding for Collaborative Research with PKU Hospitals (to J.W., F.B., and X.S.X.), the National Natural Science Fund for Distinguished Young Scholars of China (no. 81025012) (to J.W.), and the 1000 Young-Elite Scholar Program (to F.B.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The raw sequence data have been deposited with the National Center for Biotechnology Information Sequence Read Archive (SRA), www.ncbi.nlm.nih.gov (study accession no. SRP029757).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320659110/-/DCSupplemental.
References
- 1.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sethi N, Kang Y. Unravelling the complexity of metastasis—molecular understanding and targeted therapies. Nat Rev Cancer. 2011;11(10):735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465(7297):473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 4.Vasioukhin V, et al. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br J Haematol. 1994;86(4):774–779. doi: 10.1111/j.1365-2141.1994.tb04828.x. [DOI] [PubMed] [Google Scholar]
- 5.Bai H, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27(16):2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 6.Forshew T, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4(136):136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 7.Dawson SJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;368(13):1199–1209. doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 8.Leary RJ, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4(162):162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 10.Maheswaran S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristofanilli M, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 12.Krebs MG, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol. 2011;29(12):1556–1563. doi: 10.1200/JCO.2010.28.7045. [DOI] [PubMed] [Google Scholar]
- 13.Magbanua MJ, et al. Isolation and genomic analysis of circulating tumor cells from castration resistant metastatic prostate cancer. BMC Cancer. 2012;12:78. doi: 10.1186/1471-2407-12-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forbes SA, et al. COSMIC: Mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 20.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3(75):75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peifer M, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44(10):1104–1110. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sutherland KD, et al. Cell of origin of small cell lung cancer: Inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell. 2011;19(6):754–764. doi: 10.1016/j.ccr.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 23.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 24.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 25.Weir BA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450(7171):893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang YC, Amon A. Gene copy-number alterations: A cost-benefit analysis. Cell. 2013;152(3):394–405. doi: 10.1016/j.cell.2012.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos GC, Zielenska M, Prasad M, Squire JA. Chromosome 6p amplification and cancer progression. J Clin Pathol. 2007;60(1):1–7. doi: 10.1136/jcp.2005.034389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012;40(Web Server issue):W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vermaat JS, et al. Primary colorectal cancers and their subsequent hepatic metastases are genetically different: Implications for selection of patients for targeted treatment. Clin Cancer Res. 2012;18(3):688–699. doi: 10.1158/1078-0432.CCR-11-1965. [DOI] [PubMed] [Google Scholar]
- 31.Kang Y, Pantel K. Tumor cell dissemination: Emerging biological insights from animal models and cancer patients. Cancer Cell. 2013;23(5):573–581. doi: 10.1016/j.ccr.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasch C, et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem. 2013;59(1):252–260. doi: 10.1373/clinchem.2012.188557. [DOI] [PubMed] [Google Scholar]
- 33.Smyth MJ, Hayakawa Y, Takeda K, Yagita H. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat Rev Cancer. 2002;2(11):850–861. doi: 10.1038/nrc928. [DOI] [PubMed] [Google Scholar]
- 34.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22(5):939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104(50):20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Storey JD. A direct approach to false discovery rates. J R Stat Soc B. 2002;64(3):479–498. [Google Scholar]
- 41.Ward JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58(301):236–244. [Google Scholar]
- 42.McQuitty LL. Similarity analysis by reciprocal pairs for discrete and continuous data. Educ Psychol Meas. 1966;26:825–831. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.